ABSTRACT
High-resolution and efficient typing for the bacterial pathogen is essential for tracking the sources, detecting or diagnosing variants, and conducting a risk assessment. However, a systematic in-field investigation of Salmonella along the food chain has not been documented. This study assessed 12 typing methods, such as antimicrobial-resistance (AMR) gene profile typing, Core Genome Multilocus Sequence Typing (cgMLST), and CRISPR multi-virulence locus sequence typing (CRISPR-MVLST), to evaluate their effectiveness for use in routine monitoring of foodborne Salmonella transmission along the poultry production chain. During 2015-16, a total of 1,064 samples were collected from poultry production chain, starting from breeding farms and slaughterhouses to the markets of Zhejiang province in China. A total of 61 consecutive unique Salmonella isolates recovered from these samples were selected for genome sequencing and further comparative typing analysis. Traditional typing methods, including serotyping, AMR phenotype-based typing, as well as modern genotyping approaches, were evaluated and compared by their discrimination index (DI). The results showed that the serotyping method identified nine serovars. The gold standard cgMLST method indicated only 18 different types (DI = 0.8541), while the CRISPR-MVLST method detected 30 types (DI = 0.9628), with a higher DI than all examined medium-resolution WGS-based genotyping methods. We demonstrate that the CRISPR-MVLST might be used as a tool with high discriminatory power, comparable ease of use, ability of tracking the source of Salmonella strains along the food chain and indication of genetic features especially virulence genes. The available methods with different purposes and laboratory expertise were also illustrated to assist in rational implementation.
IMPORTANCE In public health field, high-resolution and efficient typing of the bacterial pathogen is essential, considering source-tracking and risk assessment are fundamental issues. Currently, there are no recommendations for applying molecular characterization methods for Salmonella along the food chain, and a systematic in-field investigation comparing subtyping methods in the context of routine surveillance was partially addressed. Using 1,064 samples along a poultry production chain with a considerable level of Salmonella contamination, we collected representative isolates for genome sequencing and comparative analysis by using 12 typing techniques, particularly with whole-genome sequence (WGS) based methods and a recently invented CRISPR multi-virulence locus sequence typing (CRISPR-MVLST) method. CRISPR-MVLST is identified as a tool with higher discriminatory power compared with medium-resolution WGS-based typing methods, comparable ease of use and proven ability of tracking Salmonella isolates. Besides, we also offer recommendations for rational choice of subtyping methods to assist in better implementation schemes.
KEYWORDS: Salmonella, typing method, CRISPR-MVLST, CoreSNP, cgMLST, antimicrobial resistance, poultry production chain
INTRODUCTION
Foodborne diseases, caused by Salmonella and many other pathogens, are critical and sustaining threats to global public health (1, 2). Improved control of foodborne bacterial transmission requires various aspects of investment, such as investigation of epidemiological prevalence, detection of contaminated point, and bacterial typing. The capabilities for quick, reliable, and convenient differentiation of typing approaches are invaluable for diagnosis, treatment and epidemiological surveillance of bacterial infections (3–7).
The conventional typing methods, i.e., bacteriophage typing, serotyping, and biochemical typing, have played important roles in understanding the nature of diversity among clinically relevant bacterial agents (8, 9). Alongside this, antibiogram typing or antimicrobial resistance profiling has been used for epidemiological source prediction or typing purposes (8–11). These phenotyping assays aim to elucidate regional- and national-scale outbreaks due to specific bacterial strains. Though they are also useful for particular purposes, they have several practical limitations that render them unsuitable for comprehensive studies of bacterial population structure or dynamic variants. Nowadays, advanced molecular typing or genotyping have been widely adopted. While having different levels of resolution and time-output efficiency, they require a range of varying expertise for practical implementation.
There are various molecular typing or genotyping methods used in the veterinary public health and food safety field (12). These include: (i) Pulsed-field Gel Electrophoresis (PFGE), (ii) Multi Locus VNTR Analysis (MLVA), (iii) Restriction Fragment Length Polymorphism (RFLP), (iv) Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR), (v) Multilocus Sequence Typing (MLST) and (vi) Whole-Genome Single-Nucleotide Polymorphism (SNP). Currently, there are no recommendations for applying molecular characterization methods for Salmonella, although the food industry regularly uses banding pattern-based and sequence-based subtyping methods for incident investigations (13). Two decades ago, US CDC introduced PFGE for routine use in surveillance and set up a PulseNet International network. Its disadvantages, however, were labor-intensity, low robustness, poor comparability of results among different laboratories, and limited resolution in source tracking of disease outbreaks associated with foodborne bacteria, including Salmonella. Recently, the whole-genome sequence (WGS) approach started to take place. Although it offers apparent advantages, expensive WGS infrastructure and use of downstream bioinformatic toolkits remain key bottlenecks for academic and surveillance staff.
In general, phenotypic methods are not promising for tracking sources, as in most cases, they are very time- and labor-intense, and usually require well-trained technicians. Nevertheless, serotyping and AMR profiling are still routinely used in food safety, particularly for typical foodborne pathogens, i.e., Salmonella. Given the increasing need to detect emerging clones or hazards, identifying the distinct types and pinpointing the source of Salmonella isolates is critical for improving surveillance and implementing control measures for such risks along the production chain (14, 15). Although a range of typing methods have been presented, to our knowledge, no studies have systematically examined or compared different typing methods in the field for their practical application potential.
CRISPR multi-virulence locus sequence typing (CRISPR-MVLST) was proved to have good discriminatory power, but only a few studies reported other traits of this method. Previous research showed that bacteria from distant geographic locations had extremely different spacer arrangements because of the existence of unique phage or plasmid pools in those different geographic locations (16). It is suggested that spacer arrangements may be a good indicator of bacterial adaptation to diversified microenvironments (17). Nevertheless, CRISPRs may evolve much faster than virulence genes (18). Besides, the loss or duplication of a single spacer and its associated direct repeat could frequently cause small allelic differences in CRISPR arrays between different Salmonella isolates (19, 20). While CRISPR-MVLST sequence type (CST) was reported to be associated with AMR in Salmonella Typhimurium (21), as far as we know, few studies have reported its relationship with the AMR gene and compared it with other genetic features.
For proof of concept, we used the newly produced data in the poultry production chain with a focus on Salmonella. We aimed to provide a reference for the practical application of 12 methods in the context of routine epidemiological surveillance, including serotyping, MLST, MIC profile typing, AMR profile typing, CRISPR typing (CT), CRISPR-MVLST, Virulence Factor (VF) gene profile typing, plasmid profile typing, AMR gene profile typing, core genome multilocus sequence typing (cgMLST), whole-genome MLST (wgMLST), and CoreSNP typing. Our results showed that CRISPR-MVLST would be the best choice for traceability and ease of use for Salmonella isolates along the food chain. Further, we proved close correlations between CRISPR-MVLST results and AMR genes or VF genes.
RESULTS
Prevalence and distribution of Salmonella.
Among the 1,064 samples collected along the poultry production chain, a total of 253 Salmonella positive samples were detected, representing an overall prevalence rate of 23.78% (Table 1). The prevalence rates of Salmonella in breeding farms, slaughterhouses, and markets were 10.27% (15/146), 20.19% (152/753), and 52.12% (86/165), respectively. Our results showed a high level of contamination along the poultry production chain, especially in the market, which could be a potential risk for consumers. No isolate was found in the samples collected from breeding farms, possibly because positive samples were few, and most of the contaminations were caused by Salmonella Gallinarum biovar Pullorum, which are highly avian-adapted and grew too slow to be isolated in selective enrichment medium used during the sampling periods (22). Due to dominance of S. Pullorum in China (23, 24) and its host restriction, any contamination in breeding farms is of lesser priority within the scope of the foodborne pathogen. Therefore, this study focused on the contamination in slaughterhouses and markets. We eliminated the copy isolates within the same sample origin (copy isolates here are defined as clones with the same colony morphology during isolation, serovar and ST from the same sample) and selected a representative unique collection of 61 Salmonella isolates, including 40 isolates from slaughterhouses and 21 isolates from markets, to further evaluate subtyping methods.
TABLE 1.
Samples and prevalence rate
| Sampling place | Sampling size | Source | Positive samples | Prevalence |
|---|---|---|---|---|
| Huzhou | 40 | Breeding farm A | 3 | 7.50% |
| Hangzhou | 42 | Breeding farm B | 6 | 14.29% |
| Yiwu | 64 | Breeding farm C | 6 | 9.37% |
| Yiwu | 46 | Slaughterhouse A | 13 | 28.26% |
| Yiwu | 448 | Slaughterhouse B | 92 | 20.54% |
| Huzhou | 259 | Slaughterhouse C | 47 | 18.15% |
| Hangzhou | 42 | Supermarket | 30 | 71.43% |
| Hangzhou | 22 | Market A | 5 | 22.73% |
| Hangzhou | 47 | Market B | 9 | 19.15% |
| Hangzhou | 54 | Market C | 42 | 77.78% |
| Total | 1,064 | 253 | 23.78% |
Phenotyping analysis: AMR phenotype-based typing and serotyping.
Phenotypic AMR of the 61 isolates was evaluated using the MIC of 15 antimicrobials, and the results are summarized in Fig. S1 and Table S1. When the isolates categorized as intermediate were also considered as resistant, all the studied isolates (n = 61) were resistant to more than 3 antimicrobial classes and were classified as multi-drug resistance (MDR) (Fig. S2). Additionally, we found the resistance rate varied dramatically among 15 examined antimicrobials (Fig. S1). Therefore, it would be appropriate to convey 2 AMR phenotype-based typing methods here. All isolates were categorized using the MIC value profile (MIC data matrix of 15 antimicrobials), with 51 types identified using the AMR profile (AMR data matrix of 15 antimicrobials).
The 61 studied isolates comprised 9 serovars (Table S1). Serotyping by the slide agglutination showed identical results to the in silico prediction method. One exception resulted from a misjudgement during the slide agglutination test and was revealed by the in silico prediction results.
Non-WGS-based genotyping analysis: MLST, CT, and CRISPR-MVLST.
The 61 studied Salmonella isolates comprised 11 STs (Table S1), including S. Indiana ST17 (34.43%; 21/61), S. Kentucky ST314 (21.31%; 13/61), S. Kentucky ST198 (6.56%; 4/61), S. Cerro ST367 (13.11%; 8/61), S. Typhimurium ST19 (8.20%; 5/61), S. Typhimurium ST34 (3.28%; 2/61), S. Apeyeme ST1546 (4.92%; 3/61), S. Albany ST292 (3.28%; 2/61), S. Anatum ST64 (1.64%; 1/61), S. Montevideo ST4 (1.64%; 1/61), and S. Rissen ST469 (1.64%; 1/61). The MLST results obtained from the Sanger sequences were consistent with those from WGS. Among all 61 isolates, MLST discriminated 11 types while serotyping discriminated 9 types. The outcomes of these 2 methods are shown with a similar color scheme and appear well-matched (Fig. 1).
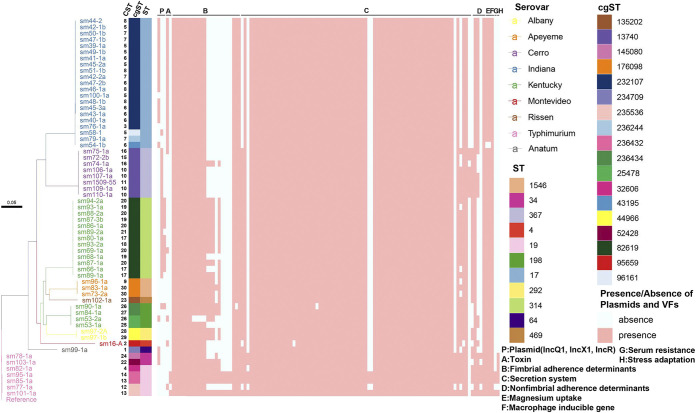
FIG 1.
Results of Salmonella typing and virulence gene detection of 61 isolates. A phylogenetic tree was built to show the genetic relationship of 61 isolates and the visualization of core genome SNP typing. Different serovars were labeled with different colors, as shown in the right part near the heatmap. A parallel matching heatmap was aligned to the phylogenetic tree. The left side of the heatmap shows the CST, cgST, and ST results. Different colors are used to distinguish different types, as noted on the right side of the heatmap. The right part shows a detailed matrix of plasmids (IncQ1, IncX1, IncR), and VF genes. The presence of a plasmid or VF gene in an isolate is marked as pink, otherwise, it is marked as light blue.
The typing and cluster analysis of isolates was undertaken by determining CRISPR- pattern-based on the spacer sequences of CRISPR1 (C1) and CRISPR2 (C2) loci. The phylogenetic tree and heatmap were generated only for serovars with a high number of isolates, including Indiana, Typhimurium, Cerro, and Kentucky, in which 2, 6, 2, and 2 CRISPR-patterns were identified, respectively (Fig. 2). The spacer sequences contained with different CRISPR-patterns are given in detail in Table S2. The CRISPR-MVLST sequence types (CSTs) were classified by adding the loci information of 2 VF genes sseL and fimH to 2 basic CRISPR loci as previous studies reported (19, 25–27). Our findings showed the presence of 30 different CSTs among these 61 Salmonella isolates (Table S3). In serovar Indiana, 17 types of C1 spacer and 17 types of C2 spacer were detected, with 1 ST (ST17) and 5 CSTs (Fig. 2a). In serovar Kentucky, 60 types of C1 spacer and 55 types of C2 spacer were detected, with 2 STs (ST314 and ST198, each ST showed a fully distinguished CRISPR-pattern) and 8 CSTs (Fig. 2b). In serovar Typhimurium, 31 types of C1 spacer and 32 types of C2 spacer were detected, with 2 STs (ST19 and ST34) and 6 CSTs (Fig. 2c). In serovar Cerro, 23 types of C1 spacer and 21 types of C2 spacer were detected, with 1 ST (ST367) and 4 CSTs (Fig. 2d).
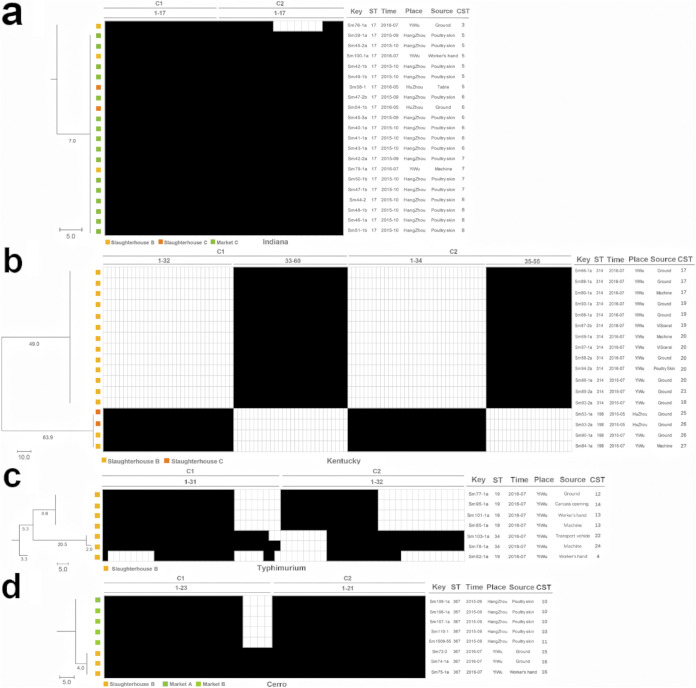
FIG 2.
CRISPR-pattern of the studied Salmonella serovars, including Indiana (a), Kentucky (b), Typhimurium (c), and Cerro (d). CRISPR cluster analysis diagram of 4 serovars: (a) Indiana, (b) Typhimurium, (c) Cerro, (d) Kentucky. These phylogenies were made to cluster isolates based on their CRISPR spacer profiles (CRISPR-patterns). Key variables and isolate information are marked around the trees and heatmap of the CRISPR-pattern matrixes: CRISPR 1 is abbreviated as C1 and CRISPR 2 as C2. The tree scale bar represents a standard distance estimated by a neighbor-joining method. Time of sample collection (time), location of sample isolation (place), CRISPR-MVLST sequence type (CST), MLST sequence type (ST). The yellow and orange color represents the isolate from slaughterhouses A and B, respectively. The dark green, light green and standard green color represent the isolate from market A, B and C, respectively. In the CRISPR-pattern heatmap, each square represents a spacer sequence and those marked in black indicates the presence of the spacer in that isolate, while remaining blank indicates the absence of the spacer. Due to the excessive number of spacers, they are marked and labeled with numbers on the diagram for convenience. Take (b) as an example, 1-32 means spacers of C1 from the 1st to the 32nd; 1-55 means all the spacers of C2 from the first to the end. A line is drawn to segregate the necessary section and facilitate locating the spacer.
WGS-based genotyping analysis: cgMLST, wgMLST, CoreSNP, AMR gene, VF gene, and plasmid profile typing.
The whole-genome sequences of these Salmonella isolates (n = 61) were analyzed to predict the plasmid replicons, AMR genes, VF genes, and calculate cgST, CoreSNP type, and wgMLST type (summarized in Table S1, wgMLST profile matrix in Table S4).
The results of plasmid replicon prediction showed existence of 18 different plasmids. All isolates were divided into 18 types using plasmid profiles (data matrix of plasmid presence/absence table).
The results of AMR gene detection showed the presence of 59 different AMR genes and 2 patterns of AMR chromosomal mutations at varying levels (Fig. S3 and Fig. S4). These isolates were divided into 32 types using the AMR gene profile (data matrix of AMR gene presence/absence).
The results of VF gene detection showed the presence of 156 genes (117~139 gene per isolate, Fig. 1, and Table S1). The isolates were divided into 36 types using VF gene profile (data matrix of VF gene presence/absence).
Comparative analysis of various typing methods.
The comparison of discriminatory power among the 6 non-WGS genotyping or phenotyping methods in this study, including serotyping, MLST, CRISPR-MVLST, CRISPR, AMR profile, and MIC profile, were carried out based on the discriminatory index (DI). The results showed that, except for AMR phenotype-based methods, the CRISPR-MVLST method provided higher discrimination power of the 61 studied isolates (DI = 0.9628), followed by the CT (DI = 0.8377), MLST (DI = 0.8158), and serotyping method (DI = 0.7820) (Table 2 and 3).
TABLE 2.
Comparison of three examined phenotyping methods in this studya
| Assessment indicator | Serotyping | AMR profile | MIC profile |
|---|---|---|---|
| Repeatability | Good | Moderate | Moderate |
| Reproducibility | Moderateb | Moderate | Poor |
| Discriminatory power (DI)c | 0.7820 | 0.9940 | 1 |
| Discriminated types | 9 | 51 | 61 |
| Scheme standardized or notd | Yes | No | No |
| Ease of interpretation of data generatede | Good | Excellent | Excellent |
| Ease of use | Poor to moderatef | Good to moderateg | Poor to moderateh |
| Throughput | No | Yes | Yes |
| Cost | Moderate to highi | Moderate to highj | High |
| Time required (days)k | 2~17# (usually > 5 days for expt) (13) | 3 | 3 |
Ranking: 1. DI in order (good to poor): MIC profile = wgMLST > CoreSNP > AMR profile > CRISPR-MVLST > VF gene profile > AMR gene profile > plasmid profile > cgMLST > CRISPR > MLST > serotyping. 2. Ease of use (good to poor): CRISPR > CRISPR-MVLST > MLST > cgMLST = wgMLST > plasmid profile = AMR gene profile = VF gene profile = CoreSNP > AMR profile = MIC profile = serotyping. 3. Cost (low to high): CRISPR < CRISPR-MVLST < MLST = VF gene profile = AMR gene profile = plasmid profile < wgMLST = cgMLST = coreSNP < AMR profile < serotyping < MIC profile. 4. Time required (short to long): MLST < CRISPR < CRISPR-MVLST < AMR profile = MIC profile < plasmid profile = VF gene profile = AMR gene profile < wgMLST < CoreSNP = cgMLST < serotyping.
We summarized three phenotyping methods in a similar manner of a previous study (28).
Using the Discriminatory index (DI) for a description of discriminatory power.
If there is 1~2 universally acknowledged standard for this typing or not.
Intended as unequivocal interpretation.
Using in silico prediction will be easier and faster.
Using disc agar diffusion test will be easier.
Become easier if the lab is doing antimicrobial resistance-related research.
Using in silico prediction will be cheaper.
Using disc agar diffusion test will be more affordable.
The approximate number of days to get typing results is estimated by excluding the interval of time to obtain a single pure colony suitable to be handled by the method.
TABLE 3.
Comparison of nine examined genotyping methods in this studya
| Assessment indicator |
non-WGS-based typing |
WGS-based typing |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Low-resolutionb |
High-resolution |
Medium-resolution |
High-resolution |
||||||
| 7-gene legacy MLST | CRISPR | CRISPR-MVLST | cgMLST | Plasmid profile | AMR gene profile | VF gene profile | CoreSNP | wgMLST | |
| Repeatability | Excellent | Excellent | Excellent | Excellent | Excellent | Excellent | Excellent | Excellent | Excellent |
| Reproducibility | Excellent | Excellent | Excellentc | Excellent | Excellent | Excellent | Excellent | Excellent | Excellent |
| Discriminatory power (DI)d | 0.8158 | 0.8377 | 0.9628 | 0.8541 | 0.8852 | 0.9361 | 0.9497 | 0.9967 | 1 |
| Discriminatory types | 11 | 15 | 30 | 18 | 18 | 32 | 36 | 56 | 61 |
| Scheme standardized or note | Yes | Yes | Yes | Yes | No | No | No | Not yet | Not yet |
| Ease of interpretation of data generatedf | Excellent | Excellent | Excellent | Good | Excellent | Excellent | Excellent | Moderate | Moderate |
| Ease of use | Good to moderateg | Moderate | Moderate | Moderate | Good | Good | Good | Poor | Moderate |
| High throughput | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Cost | Moderate | Low | Low | Moderate | Moderate | Moderate | Moderate | Moderate | Moderate |
| Time required (days)h | 1~2 | 1~2 | 1~2 | 2~7 | 2~7 | 2~7 | 2~7 | 2~7 | 2~7 |
Rankings 1. DI in order (good to poor): MIC profile = wgMLST > CoreSNP > AMR profile > CRISPR-MVLST > VF gene profile > AMR gene profile > plasmid profile > cgMLST > CRISPR > MLST > serotyping. 2. Easy of use (good to poor): CRISPR > CRISPR-MVLST > MLST > cgMLST = wgMLST > plasmid profile = AMR gene profile = VF gene profile = CoreSNP > AMR profile = MIC profile = serotyping. 3. Cost (low to high): CRISPR < CRISPR-MVLST < MLST = VF gene profile = AMR gene profile = plasmid profile < wgMLST = cgMLST = coreSNP < AMR profile < serotyping < MIC profile. 4. Time required (short to long): MLST < CRISPR < CRISPR-MVLST < AMR profile = MIC profile < plasmid profile = VF gene profile = AMR gene profile < wgMLST < CoreSNP = cgMLST < serotyping.
For reading ease, Low/Medium/High resolution classification is set up for genotyping methods according to their DI results in this study.
We summarized nine methods in a similar manner of a previous study (28).
dUsing the Discriminatory index (DI) for a description of discriminatory power.
If there is 1~2 universally acknowledged standard for this typing or not.
Intended as unequivocal interpretation.
Using in silico prediction will be easier.
The approximate number of days to get typing results is estimated by excluding the interval of time to obtain a single pure colony suitable to be handled by the method.
On the other hand, for 6 WGS-based genotyping methods, cgMLST discriminated 18 different types among these 61 isolates and thus presented a DI of 0.8541, which was higher than that of 7-gene legacy MLST. However, our results showed that cgMLST could not distinguish isolates with close relationships, i.e., S. Cerro and S. Kentucky, while CRISPR-MVLST could clearly distinguish these isolates. The cgMLST showed a lower DI than most of the other tested methods, including the non-WGS-based method CRISPR-MVLST. VF gene profile typing (DI = 0.9497), AMR gene profile typing (DI = 0.9361) and plasmid profile typing (DI = 0.8852) gave a moderate level of discrimination power. Additionally, our results showed that WGS-based high-resolution methods presented remarkable discrimination power (DI > 0.9960, [Table 3]).
Thus, based on these findings, the typing methods can be arranged in decreasing order of their DIs as, MIC profile ≈ wgMLST > CoreSNP > AMR profile > CRISPR-MVLST > VF gene profile > AMR gene profile > plasmid profile > cgMLST > CRISPR > 7-gene legacy MLST > Serotyping.
A summary table of molecular typing methods, including MLST, was reported previously (28). We summarized 12 tested methods in a similar manner (Table 2 and 3) to provide a comprehensive evaluation, along with a ranking heatmap of 12 methods (Fig. 3).
FIG 3.
A visualized ranking of 12 examined typing methods in this study. The x axis shows the ranking position for 12 methods. The y axis shows all assessment indicators for these methods. Blocks in red represent methods using spacer sequences of CRISPR loci. Blocks in orange represent methods using a relatively global loci/site at a whole-genome level. Blocks in green represent the 7-gene legacy MLST method. Blocks in deep blue represent methods based on phenotypic antimicrobial resistance (AMR) data. Blocks in light blue represent methods based on AMR or VF loci. Blocks in light blue represent methods using plasmid profiles. Blocks in purple represent methods based on the White–Kauffman serotyping scheme.
CRISPR-MVLST tracking Salmonella isolates in the poultry production chain.
After demonstrating advantageous traits of the CRISPR-MVLST method, we tried to find out if the method could be applied to Salmonella source-tracking. The available results not only accurately revealed major lineages (STs) of 61 studied Salmonella isolates, but also clearly suggested cross-contamination points in 4 different serovars.
In serovar Indiana (Fig. 2a), all isolates recovered from markets came from the market C in Hangzhou. Isolate Sm100-1a (from the hand of a worker in slaughterhouse B in Yiwu), isolates Sm45-2a and Sm39-1a (from the skin of chilled poultry carcasses in market C) clustered together with consistent CRISPR-patterns and CSTs. Similar results were obtained in CST 6 and 7, including Sm54-1b (from slaughterhouse C in Huzhou) and Sm47-2b from market C. In addition, there was only one locus difference (in C2) between Sm76-1a (CST 3) and Sm39-1a (CST 5). This supported the close genetic relationship among isolates from market C and those from slaughterhouse B.
In serovar Kentucky (Fig. 2b), isolates Sm69-1, Sm87-1a, Sm88-2a, Sm94-2a, and Sm86-1a (recovered from equipment in the lairage area, viscera room, and skin before washing in one slaughterhouse) showed the same CST, suggesting that there may have been a strong viscera contamination before leaching. S. Kentucky probably had also contaminated the ground of the lairage, the equipment of the plucking room, and the ground of the carcass washing room (Sm68-1: CST19, Sm80-1a: CST17, Sm93-2a: CST18), suggesting that they could survive for a long time on the ground or equipment. The CRISPR-patterns of Sm90-1a (from the storage room) and Sm84-1 (from the viscera room) from slaughterhouse B (Yiwu) were identical to the other 2 isolates (Sm53-1a and Sm53-2a) from the lairage area of slaughterhouse C in Huzhou (Fig. 2b). The CSTs of Sm90-1a and Sm53-2a were also identical. It is likely that slaughterhouses B and C might have been contaminated with clones of the same origin in the breeding farms.
In serovar Typhimurium (Fig. 2c), it was found that ST19 and ST34 were not fully distinguished; Sm82-1a (ST19) shared all its spacers with Sm78-1a (ST34) but only 80% of its spacers with other ST19 isolates. Besides, there were differences in three (75%) loci of CRISPR-MVLST profiles between the 2 ST34 isolates. It was worth noting that the isolate recovered from the hands of workers at the end of the slaughter chain had high genetic congruence to those from the ground of plucking room, the machine of viscera room and carcass opening before washing (Fig. 2c, and Table S1 and S3). For example, the CST of Sm101-1a (from the worker in the packaging room) was the same as that of Sm85-1a (from the machine in the viscera room). This suggested the presence of a certain degree of cross-spatial contamination in slaughterhouse B.
In serovar Cerro (Fig. 2d), a high similarity of CRISPR sequences (sharing 91% spacers, 40/44) was observed between 3 isolates (from slaughterhouse B in Yiwu) and 5 isolates (from market A or market B in Hangzhou), suggesting that slaughterhouse B could be a potential source of S. Cerro in market A and B. Additionally, 4 out of these 5 isolates (Sm106-1a, Sm107-1a, Sm110-1, and Sm1509-55), which were all recovered from the skin of chilled poultry in market B, presented the identical CRISPR-pattern, and the former 3 were all identified as CST 10, suggesting the existence of frequent cross-contamination in market B. In the scalding room in slaughterhouse B, 2 isolates sharing one CST (CST 16) were isolated from the worker’s hand (Sm75-1a) and the ground (Sm74-1a).
Correlation analysis between CRISPR-MVLST results and genetic features.
CRISPR-MVLST was further examined to investigated if it could deliver indications of genetic features including AMR determinants (ARD), VF genes, and plasmid replicons from certain perspective. We also compared it with some standard methods as controls, including serotyping, MLST and cgMLST. Firstly, to find genetic features that were locally associated with certain CST in the poultry production chain, such as ARDs or major plasmid replicons, additional heatmaps were projected for visualization (Fig. 1 and Fig. S3).
Secondly, to locate significant correlation between CRISPR-MVLST results and genetic features, we tried to number the CST in the order of the total number of C1 and C2’s spacers (from small to big) and calculated the Spearman’s rank correlation coefficient between the numerical number of CST (nCST) and ARD/VF gene/plasmid. Some close correlation (correlation coefficient >0.6 or <−0.6, P < 0.01) was detected.
The most abundant AMR genes were aph(6)-Id (67.21%) encoding resistance to aminoglycosides (Fig. S4), tet(A) (72.13%) encoding resistance to tetracyclines, floR (63.93%) encoding resistance to phenicols, and sul1 (60.66%) encoding resistance to sulfonamides, while no significant feature was found to be associated with certain CST. Nevertheless, the isolates with the same ST or cgST carried similar AMR gene patterns. nCST was close negative correlation with aadA5, dfrA17, oqxA, and oqxB (correlation coefficient was −0.64, −0.68, −0.69, and −0.69, respectively, P < 0.01).
For VF genes, pefABCD gene cluster, rck, gogB, spvC, spvR, and sodCI were only detected in CST13 and CST14 of S. Typhimurium ST19 (Fig. 1). cdtB encoding typhoid toxin-producing was closely related to S. Indiana ST17 while spvB was closely related to S. Typhimurium ST19. The lpf gene cluster, encoding the long polar fimbriae, mediating attachment to the Peyer's patches, was associated with S. Kentucky ST314 and ST198, S. Typhimurium ST19, S. Apeyeme ST1546, and S. Albany ST292. A high correlation between sspH1 and S. Cerro ST367 was detected. sspH2 was detected in all serovars except S. Indiana ST17. Moreover, sseI/srfH was highly correlated with ST19 and ST34, while ratB (non-fimbrial adhesin) was detected in S. Indiana ST17, S. Kentucky ST198, S. Cerro ST367, S. Typhimurium (ST19, ST34), and S. Montevideo ST4. nCST was close positive correlation with avrA, lpfA, lpfB, lpfC, lpfE, and sseK1 (correlation coefficient was 0.74, 0.67, 0.67, 0.67, 0.64, and 0.63, respectively, P < 0.01), and was close negative correlation with cdtB, hsiC1/vipB, and ratB (the correlation coefficient was −0.64, −0.73, and −0.62, respectively, P < 0.01).
Of the 18 different plasmids, among which IncX1 (37.70%; 23/61) and IncQ1 (34.43%; 21/61) were the most prevalent in these studied isolates. Col156 was predicted to exist in CST6 only. Moreover, the distribution of 18 plasmids among serovars showed that S. Indiana ST17 harbored more diversified plasmids (n = 9), followed by S. Typhimurium ST19 (n = 6) and S. Kentucky ST198 (n = 5) (Fig. S3). No close correlation was detected between nCST and plasmid replicons.
The results of chromosomal AMR mutation detection showed 2 mutation patterns in the quinolone-resistance-determining region (QRDR), including the pattern with a single mutation in the gyrA gene (S83F) (39.3%) and a pattern with double mutations in the gyrA gene (S83F and D87G) (6.6%) (Fig. S4). No close correlation was detected between nCST and chromosomal AMR mutations.
These results indicated close correlations between CRISPR-MVLST results (or total No. of C1&C2’s spacers) and ARG or VF genes but not between CRISPR-MVLST results (or total No. of C1&C2’s spacers) and plasmids or AMR mutations in Salmonella isolates. And some VF genes were found to be associated with certain CSTs.
DISCUSSION
The poultry production chain is considered the main vehicle for Salmonella human infections. Here, we report a high prevalence of Salmonella in different stages of production, including breeding farm, slaughterhouse, and market. Additionally, we demonstrate that the contamination rate increased from upstream (the stunning point at the slaughterhouse) to downstream (storage/sales of finished products at the markets) of the poultry value chain, reflecting the issue of cross-contamination in the slaughterhouse, transport, and market as in previous studies (29–31). Moreover, the study also shows that majority of the Salmonella isolates were highly MDR, and harbored various ARDs and VF genes, which is considered a significant concern to public health.
Currently, different typing methods have been used to track the movement of Salmonella along with the food chain and to correlate the disease outbreaks with the probable source. Serotyping was considered the classical yet innovative typing method for Salmonella, which could identify the major groups that caused human salmonellosis (32, 33). Thereafter, several typing methods based on the analysis of amplified, restricted, or sequenced DNA profiles of Salmonella isolates have been developed to provide more accurate typing results (13, 34–36).
The choice of the appropriate method is influenced by various factors, including (i) the discriminatory ability to distinguish between non-clonal isolates, (ii) the ability to generate interpretable data, (iii) the reproducibility of results among different personnel and laboratories, (iv) the time required to return typing results, (v) the need for a standardized scheme, and (vi) the technical complexity, including bioinformatics skills and the resources in terms of equipment, personnel and cost (34). A full evaluation would determine the most appropriate method for source-tracking or variant detecting along the food chain.
In the last 2 decades, PFGE has been used as the gold standard typing method by the PulseNet network before being emergence of WGS. Despite its advantages, PFGE has some inherent limits: time-consuming and low discrimination power for all unrelated isolates. MLVA is used as a typing method that can compensate for the low discriminatory power of PFGE in some Salmonella serovars but probably will be replaced by WGS (13). Moreover, MLST based on the sequence analysis of seven housekeeping genes, or its recent version based on the core genome sequences (cgMLST), has been used to provide appropriate sequence types of Salmonella isolates and has succeeded in discriminating some AMR-related Salmonella clones like S. Kentucky ST198, S. Indiana ST17, the monophasic variant of S. Typhimurium ST34 (37–39). Furthermore, CRISPR typing and its updated version CRISPR-MVLST have been recently used to provide high discrimination ability of Salmonella isolates, especially CRISPR-MVLST (19, 26, 40).
Although WGS is currently used as a gold standard method for typing foodborne pathogens, it seems that the traditional phenotypic serotyping and Sanger sequencing approaches are more suitable for initinal monitoring, especially in developing countries. Our findings showed that among the 61 isolates of our study, serotyping identified 9 serovars, 7-gene legacy MLST identified 11 STs, and CRISPR-MVLST identified 30 subtypes, while cgMLST identified 18 cgSTs, VF gene profile identified 36 subtypes, and CoreSNP identified 56 subtypes. Altogether, CRISPR-MVLST (DI = 0.9628) provided a high discrimination power compared with most of other methods, except CoreSNP, wgMLST or 2 AMR phenotype-based methods. CRISPR-MVLST has shown a high discriminatory ability (DI = 0.980) in typing S. Dublin recovered from humans and animals (41). CT identified 76 types with a discriminatory power of 97.6% among 180 clinical Salmonella strains isolated during 2017–2018 (42). Several previous studies have proven the efficiency of CRISPR-MVLST in typing different Salmonella serovars, including Typhimurium, Newport, and Enteritidis, suggesting the use of this method to complement and validate results obtained by PFGE (43–46). A study has demonstrated that CRISPR-MVLST could separate the common PFGE patterns of S. Heidelberg, providing significantly greater discriminatory power, and proposed using CRISPR-MVLST as an alternative to PFGE (26). Based on the performance shown in this study, CRISPR-MVLST appeared as an ideal typing method with high discriminatory power, proven ability to track isolate and general applicability (intended as standardized, reproducible, and low requirements in technicality level and equipment) for foodborne pathogen surveillance (Table 3). For 2 AMR phenotype-based methods, though with considerable DIs, mediocre repeatability, poor general applicability, and high cost imply less efficiency during surveillance, which may not be acceptable for most sentinel laboratories. More importantly, the AMR gene and plasmid are frequently linked with mobile elements, therefore, bacterial strain might rapidly change due to high frequency of horizontal gene transfer.
We hypothesize that most laboratories have at least the ability to perform the basic serotyping by phenotypic method and the best subtyping methods for different levels of laboratory expertise are summarized here:
(i) Without any sequencing ability, only have phenotyping ability.
BEST OPTION: Serotyping or (for local surveillance only) AMR phenotype-based methods.
(ii) Sanger sequencing available but without WGS.
BEST OPTION: CRISPR–MVLST in general, and AMR phenotype-based methods can be used as supplements if available.
(iii) Sanger sequencing & WGS available but without good bioinformatics skill or calculation capability.
BEST OPTION: wgMLST (online website based) or CRISPR–MVLST.
(iv) Sanger sequencing & WGS available with good bioinformatics skill & calculation capability.
BEST OPTION: wgMLST or CoreSNP.
Our findings showed a high efficiency of CRISPR-MVLST in tracking the source of Salmonella along the poultry production chain. The subtype CST26 was identified in the samples from the waiting room of slaughterhouse C and the storage room of slaughterhouse B, suggesting that the isolates could have come from the same farm, where the animals were colonized with Salmonella before being transported to different slaughterhouses. Additionally, the findings demonstrated the presence of isolates such as CST13, CST19, and CST20 with the same CRISPR-pattern, at different processing steps in slaughterhouse B, indicating the persistence of Salmonella isolates after applying sanitization operations and a high frequency in contamination as well as cross-contamination of poultry carcass in this slaughterhouse. The slaughtering process is a critical step in the poultry production chain, and bacteria can disseminate from intestinal content during the evisceration process and then contaminate and/or cross-contaminate poultry carcasses, facilities, and workers’ hands along with the slaughtering process steps (15). The subtypes CST5 and CST7 were identified both in slaughterhouse B and poultry carcasses in market C, indicating that the poultry might have been contaminated in the slaughterhouse before being supplied to the market. The same reasoning is valid for the CST6 subtype isolates recovered from slaughterhouse C and poultry carcasses sold in the market C. These results also suggest the implementation of robust and efficient disinfection systems and personal hygiene to prevent and control the dissemination of Salmonella at those critical points of the poultry production chain. Interestingly, there was no obvious evidence supporting that a CST could indicate the existence of certain type of AMR gene or mutation. This seems not fitting well with the previous point of view based on AMR phenotype (21). A limitation of this study is that PCR specific for S. Pullorum wasn’t carried out during the sampling periods to prove the dominance of S. Pullorum in Salmonella contamination in the breeding farms.
Collectively, this study demonstrates that CRISPR-MVLST is an ideal choice, close to CoreSNP based on WGS, for typing Salmonella as it well-distinguished the isolates in the same serovar or ST or even cgST. The study also shows that its advantage in tracing back to the contaminating isolates in the slaughterhouse and market. CRISPR-MVLST meets the requirements of large-scale epidemiological investigation and tracing the major lineages of Salmonella in the poultry production chain. Further studies, for samplings from different food commodities, a diverse Salmonella serovars, and isolates from a large time scale, are needed, as well as samplings in other scenarios such as outbreak investigation.
MATERIALS AND METHODS
Ethical approval.
The experimental protocols regarding the animal handlings were approved by the Laboratory Animal Management Committee of Zhejiang University (Approval No. 2015016).
Sampling and isolation.
A total of 1,064 samples were collected from the poultry production chain, including breeding farms, slaughterhouses, and markets during 2015–2016 in Zhejiang province, China. The samples consisted of swabs (n = 146) from breeding farms; water samples (n = 153) and swabs of various sources (n = 600) from slaughterhouses; and carcass swabs (n = 165) from markets (Table S5). All samples were transported to the laboratory in ice packs on the same day. The swab samples were collected in 2 mL sterile phosphate-buffered saline (PBS) in Eppendorf (EP) tubes. The water samples (5 mL) from the disinfection tank and chilling tank in slaughterhouses were collected in sterile 7 mL Falcon tubes. For preliminary enrichment, Buffered Peptone Water (BPW, Haibo Biotechnology Co) was used in a 1:9 dilution (sample in PBS: BPW) and incubated at 37°C for 16–18 h in a rotatory incubator set at 180 rpm. For selective enrichment, Tetrathionate Broth Base (TTB, Land bridge Biotechnology Co), supplemented with iodine solution (Land bridge Biotechnology Co) and brilliant green solution (Land bridge Biotechnology Co) was used at a dilution of 1:10 (sample in BPW: TTB) and incubated at 42°C for 22–26 h in a rotatory incubator set at 180 rpm. For primary screening of Salmonella, bacterial DNA was extracted using the TIANamp bacteria DNA kit (Tiangen Biotech) according to the manufacturer’s instructions, and the DNA was subjected to PCR based identification as described previously (47). Pure Salmonella colonies were isolated from the positive samples by subculturing the selectively enriched samples on Xylose Lysine Deoxycholate (XLD, Land bridge Technology Co) agar with an incubation of 18–22 h at 37°C. Typical and pure colonies were picked up after subculturing on XLD agar and were transferred intoLB broth and incubated for 18–22 h at 37°C in a rotatory incubator set at 180 rpm. For confirmation of Salmonella isolates, genomic DNA was extracted using the TIANamp bacteria DNA kit from overnight cultures of the pure colonies and PCR was conducted using genus-specific primers as mentioned previously (47, 48).
Serotyping.
The PCR confirmed Salmonella isolates were serotyped by the slide agglutination method as previously described (24, 49).
Antimicrobial susceptibility.
The antimicrobial susceptibility of the confirmed isolates was performed by MIC assay using a panel of 15 antimicrobial agents, including penicillin (ampicillin: AMP, 0.25–128 μg/mL); β-lactamase inhibitor combinations (amoxicillin-clavulanic acid: AMC, 0.125/0.062–128/64 μg/mL); cephems (ceftiofur: CF, 0.125–128 μg/mL; cefoxitin: CX, 0.125–128 μg/mL); carbapenems (imipenem: IPM, 0.03–16 μg/mL), aminoglycosides (gentamicin: GEN, 0.031–64 μg/mL; kanamycin: KAN, 0.25–128 μg/mL; streptomycin: STR, 1–128 μg/mL); tetracyclines (tetracycline: TET, 0.062–128 μg/mL); (fluoro)quinolones (ciprofloxacin: CIP, 0.015–16 μg/mL; nalidixic acid: NAL, 0.5–128 μg/mL); sulfonamides (trimethoprim-sulfamethoxazole: TST, 0.25/4.75-32/608 μg/mL); polypeptides (colistin: COL, 0.031–64 μg/mL); macrolides (azithromycin: AZI, 0.25–128 μg/mL), and phenicol (chloramphenicol: CHL, 0.5–128 μg/mL), as elaborated in our previous studies (50–52).
Multilocus sequence typing.
Genomic DNA of isolates was extracted as mentioned above and quantified using the Qubit Broad Range assay kit (Invitrogen, Carlsbad), as per the manufacturer’s instructions. The 7-gene legacy MLST was conducted with the Sanger sequencing platform at Sunya Biotechnology Co., Ltd (Zhejiang, China) using the methods previously reported (53). Determination of STs was based on the sequence analysis of seven housekeeping genes using the Enterobase database (54). Among all confirmed isolates, 61 unique isolates were selected for further analysis after removing copy isolates in the same sample based on serovar, sequence type (ST) and colony morphology.
Genome sequencing and assembling.
Whole-genome sequencing was performed on an Illumina Nextseq platform using PE150 strategies. The raw reads were checked for sequence quality as described previously (55). The quality of sequencing reads was checked by FastQC toolkit v0.72, and low-quality sequences or joint sequences were removed using Trimmomatic v0.39 as described (56). De novo assembly was performed using SPAdes v3.12.0 on an in-house Galaxy platform (31).
CRISPR typing, CRISPR-MVLST, and data visualization.
To obtain more complete CRISPR sequences, the CRISPR1 and CRISPR2 sequences were collected using the Sanger sequencing platform of Sunya Biotechnology Co., Ltd (Zhejiang, China) according to the method described previously (19, 25–27). These sequences were consistent with 2 CRISPR loci’s sequences extracted from WGS. Analysis of CRISPR1 and CRISPR2 was carried out by CRISPRCasFinder (57) to locate and obtain the spacer information contained in the corresponding isolates. A local database of CRISPR1 and CRISPR2 spacers was established by R v3.6.1 to project the binary matrix of CRISPR-pattern (combination of both CRISPR loci’s spacers), which could be used for the phylogenetic tree, heatmap (produced by Microsoft Excel 2016), CT and CRISPR-MVLST. By GrapeTree v1.5.0, a neighbor-joining method (FastME V2) was used to calculate the tree (58). Simultaneously, a non-redundant database of CRISPR-pattern was built, and each CRISPR-pattern was numbered for CT.
Genome data (introduced here as a more efficient way, but Sanger sequencing data also works in the same manner) was imported into the in-house Galaxy platform as mentioned previously (59–63), and the location information of the corresponding fimH and sseL sequences of the isolates were detected by ABRicate v0.8 and Virulence Factor Database (VFDB) deployed to the platform (similarity >90%, coverage >60%) (64). Then, FASTA sequences were incorporated into Geneious v8.1.6 for visualization and extraction of these sequences. Compared with the previous literature (18, 26, 27), we numbered fimH and sseL sequences, assigned new numbers to the newly detected sseL and fimH sequences and build a non-redundant database.
Numbers of the above 4 loci (CRISPR1, CRISPR2, fimH and sseL) of each isolate were combined to produce the CRISPR-MVLST profiles (18, 26, 27), and a local CRISPR-MVLST sequence type (CST) database was established using a matrix consisting of these profiles. Finally, the CST number of each isolate was assigned using this database.
Core genome multilocus sequence typing.
cgMLST analysis was performed using fastq data of all 61 isolates based on cgMLSTFinder software (65). An in-house Python3 script was used to convert the new cgMLST profile database matrix from Enterobase (http://enterobase.warwick.ac.uk/species/index/senterica, downloaded on 2021-04-23) to a usable format for cgMLSTFinder to produce updated cgMLST results.
Whole-genome MLST.
Cano-wgMLST_BacCompare Platform was used (66) for wgMLST analysis. This platform employed 2 main processes, namely, whole-genome scheme extraction (GSE) and discriminatory loci refinement (DLR), and the “feature importance” algorithm was used (67). In the GSE step, “contig annotation” and “pan-genome allele database (PGAdb) creation” were used to process all 61 fasta format files and generate locus starting with “SAL”. Finally, we obtained all strains’ wgMLST allele_profile matrix, and the number of types was calculated and assigned using this matrix following the similar method of CRISPR-MVLST.
WGS-based CoreSNP typing.
S. Typhimurium LT2 was used as a reference genome, and 115,469 core genome single nucleotide polymorphisms (Core SNPs) were identified by Snippy v4.4.4 as described in our previous publications (5, 50, 55). Core SNPs alignment was transformed into profile matrix, which was used for CoreSNP typing following the similar method of CRISPR-MVLST.
WGS-based plasmid profile typing.
The assembled genomes were analyzed for plasmid replicons based on the CGE PlasmidFinder database (similarity >95%, coverage >60%) (68) using ABRicate v0.8 (69) and in-house script as previously used (70). The produced 0/1 plasmid profile matrix (0 for absence, 1 for presence) was analyzed, following the similar method of CRISPR-MVLST.
WGS-based AMR gene profile typing.
The assembled genomes were analyzed for AMR gene based on the CGE ResFinder database (68) (similarity >90%, coverage >60%) using ABRicate v0.8 deployed to the in-house Galaxy platform (69) and in-house script as previously used (70). The resulted 0/1 profile matrix (0 for absence, 1 for presence) was analyzed following the similar method of CRISPR-MVLST.
WGS-based VF gene profile typing.
The assembled genomes were analyzed for potential virulence factor (VF) genes based on the Virulence Factor Database (VFDB) database (64) (similarity >90%, coverage >60%) using ABRicate v0.8 (69) and in-house script as previously used (70). The produced 0/1 profile matrix (0 for absence, 1 for presence) was analyzed following the similar method of CRISPR-MVLST.
AMR phenotype-based typing.
For AMR profile typing method and MIC profile typing method, the number of types was calculated and assigned using corresponding profile matrix following the similar method of CRISPR-MVLST.
The discriminatory power of the methods.
To evaluate the typing potential of the 12 studied methods, the individual Discriminatory Index (DI) was calculated based on the following formula (71):
where N is the total number of strains in the sample population, S is the total number of types described, and Mi is the number of strains belonging to the ith type.
Bioinformatics analysis for genomic epidemiology.
Serovar prediction for 61 studied isolates was carried out with 2 different methods, SISTR (72) and SeqSero2 (55, 73). These methods cross-validated the accuracy of prediction results. The Core SNPs alignment mentioned above was used for building a maximum-likelihood phylogenetic tree (1000 bootstraps) using IQ-TREE v1.6.12 (74) with the best model TVM+F+ASC+R3. The plasmid replicon, AMR gene, and VF gene were predicted or detected as mentioned above. The AMR mutations were detected by RGI v5.1.1 with CARD database v3.1.0 (similarity >90%, coverage >60%) as previously reported (70, 75). The plasmid, AMR determinant and VF gene with the phylogenetic tree were visualized by R-studio v1.1 with R v3.6.1 and R packages (ggplot2, ggtree, treeio, phytools, ape, maps, phangorn, Rcpp, vctrs, tidyverse, and gheatmap).
Data availability.
All the raw data have been deposited into CNGB Sequence Archive (CNSA) (76) of China National GenBank DataBase (CNGBdb) (77) with a project title ‘poultry production chain’ and accession number CNP0001590.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Program on Key Research Project of China (2019YFE0103900) as well as the European Union's Horizon 2020 Research and Innovation Program under Grant Agreement No. 861917 – SAFFI, the National Program on Key Research Project of China (2018YFD0701001), the National Natural Science Foundation of China (31872837 & 32150410374), Natural Science Foundation of Zhejiang Province (LR19C180001), Zhejiang Provincial Key R&D Program of China (2022C02024, 2021C02008, 2020C02032), and China Postdoctoral Science Foundation (2022M712785).
We appreciate the efforts of all the participants in China who contributed to the sampling and data collection. We also thank all those individuals involved in the characterization of Salmonella isolates and giving advice, especially associate chief technician Xuebin Xu of Shanghai Municipal CDC, Miss Qingqing Wu, Xiao Zhou and Tanveer Muhammad of Zhejiang University.
M.Y., X. Li, and W.F. designed the study. H.P., C.J., N.P., and C.K. wrote the initial draft. H.P., C.J., F.L., and J.M. did genomic data analysis, figures, and tables. H.P., N.P., X. Liu, X. Liao, and X. Li collected the samples. N.P., F.L., J.M., X. Liu, C.D., K.Z., X. Liao, J.G., did the microbiological investigations. M.Y. conceived the idea, finalized the manuscript, and managed the project. All the authors contributed to the article and approved the submitted version.
Footnotes
Supplemental material is available online only.
Contributor Information
Min Yue, Email: myue@zju.edu.cn.
Xianqin Yang, Agriculture and Agriculture-Food Canada.
REFERENCES
- 1.Yang C, Li Y, Jiang M, Wang L, Jiang Y, Hu L, Shi X, He L, Cai R, Wu S, Qiu Y, Lu L, Zuo L, Chen Q, Wu Y, Martinez-Urtaza J, Wan C, Yang R, Cui Y, Hu Q. 2022. Outbreak dynamics of foodborne pathogen Vibrio parahaemolyticus over a seventeen year period implies hidden reservoirs. Nat Microbiol 7:1221–1229. doi: 10.1038/s41564-022-01182-0. [DOI] [PubMed] [Google Scholar]
- 2.Paudyal N, Pan H, Liao X, Zhang X, Li X, Fang W, Yue M. 2018. A meta-analysis of major foodborne pathogens in Chinese food commodities between 2006 and 2016. Foodborne Pathog Dis 15:187–197. doi: 10.1089/fpd.2017.2417. [DOI] [PubMed] [Google Scholar]
- 3.Hu B, Hou P, Teng L, Miao S, Zhao L, Ji S, Li T, Kehrenberg C, Kang D, Yue M. 2022. Genomic investigation reveals a community typhoid outbreak caused by contaminated drinking water in China, 2016. Front Med (Lausanne) 9:753085. doi: 10.3389/fmed.2022.753085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elbediwi M, Pan H, Zhou X, Rankin SC, Schifferli DM, Yue M. 2021. Detection of mcr-9-harbouring ESBL-producing Salmonella Newport isolated from an outbreak in a large-animal teaching hospital in the USA. J Antimicrob Chemother 76:1107–1109. doi: 10.1093/jac/dkaa544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu H, Elbediwi M, Zhou X, Shuai H, Lou X, Wang H, Li Y, Yue M. 2020. Epidemiological and genomic characterization of Campylobacter jejuni isolates from a foodborne outbreak at Hangzhou, China. Int J Mol Sci 21. doi: 10.3390/ijms21083001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu Y, Zhou X, Jiang Z, Qi Y, Ed-Dra A, Yue M. 2021. Antimicrobial resistance profiles and genetic typing of Salmonella Serovars from chicken embryos in China. Antibiotics (Basel) 10:1156. doi: 10.3390/antibiotics10101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han X, Liu Y, Yin J, Yue M, Mu Y. 2021. Microfluidic devices for multiplexed detection of foodborne pathogens. Food Res Int 143:110246. doi: 10.1016/j.foodres.2021.110246. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Biswas S, Paudyal N, Pan H, Li X, Fang W, Yue M. 2019. Antibiotic resistance in Salmonella Typhimurium isolates recovered from the food chain through national antimicrobial resistance monitoring system between 1996 and 2016. Front Microbiol 10:985. doi: 10.3389/fmicb.2019.00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan H, Paudyal N, Li X, Fang W, Yue M. 2018. Multiple food-animal-borne route in transmission of antibiotic-resistant Salmonella Newport to humans. Front Microbiol 9:23. doi: 10.3389/fmicb.2018.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H, Jia C, Li H, Yin R, Chen J, Li Y, Yue M. 2022. Paving the way for precision diagnostics of antimicrobial-resistant bacteria. Front Mol Biosci 9:976705. doi: 10.3389/fmolb.2022.976705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan H, Zhou X, Chai W, Paudyal N, Li S, Zhou X, Zhou K, Wu Q, Wu B, Li G, Rajkovic A, Fang W, Rankin SC, Li Y, Xu X, Schifferli DM, Yue M. 2019. Diversified sources for human infections by Salmonella enterica serovar newport. Transbound Emerg Dis 66:1044–1048. doi: 10.1111/tbed.13099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue M, Bai L, Song H, Fang W. 2021. Impacts of microbial food safety in China and beyond. Foodborne Pathog Dis 18:508–509. doi: 10.1089/fpd.2021.29015.int. [DOI] [PubMed] [Google Scholar]
- 13.Tang S, Orsi RH, Luo H, Ge C, Zhang G, Baker RC, Stevenson A, Wiedmann M. 2019. Assessment and comparison of molecular subtyping and characterization methods for Salmonella. Front Microbiol 10:1591. doi: 10.3389/fmicb.2019.01591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams MS, Ebel ED, Saini G, Nyirabahizi E. 2020. Changes in Salmonella contamination in meat and poultry since the introduction of the pathogen reduction and hazard analysis and critical control point rule. J Food Prot 83:1707–1717. doi: 10.4315/JFP-20-126. [DOI] [PubMed] [Google Scholar]
- 15.Boubendir S, Arsenault J, Quessy S, Thibodeau A, Fravalo P, ThEriault WP, Fournaise S, Gaucher ML. 2021. Salmonella contamination of broiler chicken carcasses at critical steps of the slaughter process and in the environment of two slaughter plants: prevalence, genetic profiles, and association with the final carcass status. J Food Prot 84:321–332. doi: 10.4315/JFP-20-250. [DOI] [PubMed] [Google Scholar]
- 16.Shariat N, Dudley E. 2021. CRISPR typing of Salmonella isolates. Methods Mol Biol 2182:39–44. doi: 10.1007/978-1-0716-0791-6_5. [DOI] [PubMed] [Google Scholar]
- 17.Kunin V, He S, Warnecke F, Peterson SB, Garcia Martin H, Haynes M, Ivanova N, Blackall LL, Breitbart M, Rohwer F, McMahon KD, Hugenholtz P. 2008. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res 18:293–297. doi: 10.1101/gr.6835308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu F, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Novel virulence gene and clustered regularly interspaced short palindromic repeat (CRISPR) multilocus sequence typing scheme for subtyping of the major serovars of Salmonella enterica subsp. enterica. Appl Environ Microbiol 77:1946–1956. doi: 10.1128/AEM.02625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabre L, Zhang J, Guigon G, Le Hello S, Guibert V, Accou-Demartin M, de Romans S, Lim C, Roux C, Passet V, Diancourt L, Guibourdenche M, Issenhuth-Jeanjean S, Achtman M, Brisse S, Sola C, Weill FX. 2012. CRISPR typing and subtyping for improved laboratory surveillance of Salmonella infections. PLoS One 7:e36995. doi: 10.1371/journal.pone.0036995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vosik D, Tewari D, Dettinger L, M'Ikanatha NM, Shariat NW. 2018. CRISPR typing and antibiotic resistance correlates with polyphyletic distribution in human solates of Salmonella Kentucky. Foodborne Pathog Dis 15:101–108. doi: 10.1089/fpd.2017.2298. [DOI] [PubMed] [Google Scholar]
- 21.DiMarzio M, Shariat N, Kariyawasam S, Barrangou R, Dudley EG. 2013. Antibiotic resistance in Salmonella enterica serovar Typhimurium associates with CRISPR sequence type. Antimicrob Agents Chemother 57:4282–4289. doi: 10.1128/AAC.00913-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou X, Kang X, Zhou K, Yue M. 2022. A global dataset for prevalence of Salmonella Gallinarum between 1945 and 2021. Sci Data 9:495. doi: 10.1038/s41597-022-01605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui L, Liu Q, Jiang Z, Song Y, Yi S, Qiu J, Hao G, Sun S. 2021. Characteristics of Salmonella from Chinese native chicken breeds fed on conventional or antibiotic-free diets. Front Vet Sci 8:607491. doi: 10.3389/fvets.2021.607491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Zhou X, Jiang Z, Qi Y, Ed-Dra A, Yue M. 2020. Epidemiological investigation and antimicrobial resistance profiles of Salmonella isolated from breeder chicken hatcheries in Henan. China Front Cell Infect Microbiol 10:497. doi: 10.3389/fcimb.2020.00497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li QC, Yin KQ, Xie XL, Zhao F, Xia J, Chen Y, Hu YC, Xu LJ, Chen X, Jiao XA. 2017. Detection and CRISPR subtyping of Salmonella spp. isolated from whole raw chickens in Yangzhou from China. Food Control 82:291–297. doi: 10.1016/j.foodcont.2017.07.008. [DOI] [Google Scholar]
- 26.Shariat N, Sandt CH, DiMarzio MJ, Barrangou R, Dudley EG. 2013. CRISPR-MVLST subtyping of Salmonella enterica subsp. enterica serovars Typhimurium and Heidelberg and application in identifying outbreak isolates. BMC Microbiol 13:254. doi: 10.1186/1471-2180-13-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Kariyawasam S, Jayarao BM, Barrangou R, Gerner-Smidt P, Ribot EM, Knabel SJ, Dudley EG. 2011. Subtyping Salmonella enterica serovar enteritidis isolates from different sources by using sequence typing based on virulence genes and clustered regularly interspaced short palindromic repeats (CRISPRs). Appl Environ Microbiol 77:4520–4526. doi: 10.1128/AEM.00468-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ranjbar R, Karami A, Farshad S, Giammanco GM, Mammina C. 2014. Typing methods used in the molecular epidemiology of microbial pathogens: a how-to guide. New Microbiol 37:1–15. [PubMed] [Google Scholar]
- 29.Rajan K, Shi Z, Ricke SC. 2017. Current aspects of Salmonella contamination in the US poultry production chain and the potential application of risk strategies in understanding emerging hazards. Crit Rev Microbiol 43:370–392. doi: 10.1080/1040841X.2016.1223600. [DOI] [PubMed] [Google Scholar]
- 30.Vinueza-Burgos C, Baquero M, Medina J, De Zutter L. 2019. Occurrence, genotypes and antimicrobial susceptibility of Salmonella collected from the broiler production chain within an integrated poultry company. Int J Food Microbiol 299:1–7. doi: 10.1016/j.ijfoodmicro.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Jiang J, Ed-Dra A, Li X, Peng X, Xia L, Guo Q, Yao G, Yue M. 2021. Prevalence and genomic investigation of Salmonella isolates recovered from animal food-chain in Xinjiang, China. Food Res Int 142:110198. doi: 10.1016/j.foodres.2021.110198. [DOI] [PubMed] [Google Scholar]
- 32.Alghoribi MF, Doumith M, Alrodayyan M, Al Zayer M, Koster WL, Muhanna A, Aljohani SM, Balkhy HH, Desin TS. 2019. S. Enteritidis and S. Typhimurium Harboring SPI-1 and SPI-2 are the predominant serotypes associated with human Salmonellosis in Saudi Arabia. Front Cell Infect Microbiol 9:187. doi: 10.3389/fcimb.2019.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langridge GC, Wain J, Nair S. 2012. Invasive Salmonellosis in Humans. EcoSal Plus 5. doi: 10.1128/ecosalplus.8.6.2.2. [DOI] [PubMed] [Google Scholar]
- 34.Foley SL, Lynne AM, Nayak R. 2009. Molecular typing methodologies for microbial source tracking and epidemiological investigations of Gram-negative bacterial foodborne pathogens. Infect Genet Evol 9:430–440. doi: 10.1016/j.meegid.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 35.Ranieri ML, Shi C, Moreno Switt AI, den Bakker HC, Wiedmann M. 2013. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. J Clin Microbiol 51:1786–1797. doi: 10.1128/JCM.03201-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferrari RG, Panzenhagen PHN, Conte-Junior CA. 2017. Phenotypic and genotypic eligible methods for Salmonella Typhimurium source tracking. Front Microbiol 8:2587. doi: 10.3389/fmicb.2017.02587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Le Hello S, Bekhit A, Granier SA, Barua H, Beutlich J, Zając M, Münch S, Sintchenko V, Bouchrif B, Fashae K, Pinsard J-L, Sontag L, Fabre L, Garnier M, Guibert V, Howard P, Hendriksen RS, Christensen JP, Biswas PK, Cloeckaert A, Rabsch W, Wasyl D, Doublet B, Weill F-X. 2013. The global establishment of a highly-fluoroquinolone resistant Salmonella enterica serotype Kentucky ST198 strain. Front Microbiol 4:395. doi: 10.3389/fmicb.2013.00395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elbediwi M, Beibei W, Pan H, Jiang Z, Biswas S, Li Y, Yue M. 2020. Genomic characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 Clone Isolated from pigs in China. Front Bioeng Biotechnol 8:663. doi: 10.3389/fbioe.2020.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong J, Zeng X, Zhang P, Zhang D, Wang C, Lin J. 2019. Characterization of the emerging multidrug-resistant Salmonella enterica serovar Indiana strains in China. Emerg Microbes Infect 8:29–39. doi: 10.1080/22221751.2018.1558961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, Wang Y, Gao Y, Li C, Ma B, Wang H. 2021. Antimicrobial resistance and CRISPR typing among Salmonella isolates from poultry farms in China. Front Microbiol 12:730046. doi: 10.3389/fmicb.2021.730046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilela FP, Dos Prazeres Rodrigues D, Costa RG, Casas MRT, Falcao JP, Campioni F. 2020. High similarity and high frequency of virulence genes among Salmonella Dublin strains isolated over a 33-year period in Brazil. Braz J Microbiol 51:497–509. doi: 10.1007/s42770-019-00156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang K, Zhang Y, Wang Z, Li Y, Xu H, Jiao X, Li Q. 2021. Characterization of CRISPR array in Salmonella enterica from asymptomatic people and patients. Int J Food Microbiol 355:109338. doi: 10.1016/j.ijfoodmicro.2021.109338. [DOI] [PubMed] [Google Scholar]
- 43.Shariat N, DiMarzio MJ, Yin S, Dettinger L, Sandt CH, Lute JR, Barrangou R, Dudley EG. 2013. The combination of CRISPR-MVLST and PFGE provides increased discriminatory power for differentiating human clinical isolates of Salmonella enterica subsp. enterica serovar Enteritidis. Food Microbiol 34:164–173. doi: 10.1016/j.fm.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Almeida F, Medeiros MIC, Rodrigues DDP, Allard MW, Falcao JP. 2017. Molecular characterization of Salmonella Typhimurium isolated in Brazil by CRISPR-MVLST. J Microbiol Methods 133:55–61. doi: 10.1016/j.mimet.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 45.Very KJ, Kirchner MK, Shariat N, Cottrell W, Sandt CH, Dudley EG, Kariyawasam S, Jayarao BM. 2016. Prevalence and spatial distribution of Salmonella infections in the Pennsylvania raccoon (Procyon lotor). Zoonoses Public Health 63:223–233. doi: 10.1111/zph.12222. [DOI] [PubMed] [Google Scholar]
- 46.Shariat N, Kirchner MK, Sandt CH, Trees E, Barrangou R, Dudley EG. 2013. Subtyping of Salmonella enterica serovar Newport outbreak isolates by CRISPR-MVLST and determination of the relationship between CRISPR-MVLST and PFGE results. J Clin Microbiol 51:2328–2336. doi: 10.1128/JCM.00608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold T, Scholz HC, Marg H, Rosler U, Hensel A. 2004. Impact of invA-PCR and culture detection methods on occurrence and survival of salmonella in the flesh, internal organs and lymphoid tissues of experimentally infected pigs. J Vet Med B Infect Dis Vet Public Health 51:459–463. doi: 10.1111/j.1439-0450.2004.00808.x. [DOI] [PubMed] [Google Scholar]
- 48.Zhu C, Yue M, Rankin S, Weill FX, Frey J, Schifferli DM. 2015. One-step identification of five prominent chicken Salmonella serovars and biotypes. J Clin Microbiol 53:3881–3883. doi: 10.1128/JCM.01976-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grimont PAD, Weill F-X. 2007. Antigenic formulae of the Salmonella servovars, 9th Edition, pp 15–107. WHO Collaborating Centre for Reference and Research on Salmonella. Paris, France. [Google Scholar]
- 50.Paudyal N, Pan H, Elbediwi M, Zhou X, Peng X, Li X, Fang W, Yue M. 2019. Characterization of Salmonella Dublin isolated from bovine and human hosts. BMC Microbiol 19:226. doi: 10.1186/s12866-019-1598-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Z, Paudyal N, Xu Y, Deng T, Li F, Pan H, Peng X, He Q, Yue M. 2019. Antibiotic resistance profiles of Salmonella recovered from finishing pigs and slaughter facilities in Henan, China Front Microbiol 10. doi: 10.3389/fmicb.2019.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu B, Ed-Dra A, Pan H, Dong C, Jia C, Yue M. 2021. Genomic investigation of Salmonella isolates recovered from a pig slaughtering process in Hangzhou, China. Front Microbiol 12:704636. doi: 10.3389/fmicb.2021.704636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Achtman M, Wain J, Weill FX, Nair S, Zhou Z, Sangal V, Krauland MG, Hale JL, Harbottle H, Uesbeck A, Dougan G, Harrison LH, Brisse S, Group SEMS . 2012. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog 8:e1002776. doi: 10.1371/journal.ppat.1002776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Achtman M, Zhou Z, Alikhan NF, Tyne W, Parkhill J, Cormican M, Chiou CS, Torpdahl M, Litrup E, Prendergast DM, Moore JE, Strain S, Kornschober C, Meinersmann R, Uesbeck A, Weill FX, Coffey A, Andrews-Polymenis H, Curtiss Rd R, Fanning S. 2020. Genomic diversity of Salmonella enterica -the UoWUCC 10K genomes project. Wellcome Open Res 5:223. doi: 10.12688/wellcomeopenres.16291.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X, Chen Y, Pan H, Pang Z, Li F, Peng X, Ed-Dra A, Li Y, Yue M. 2020. Genomic characterization of Salmonella Uzaramo for human invasive infection. Microb Genom 6:1–8. doi: 10.1099/mgen.0.000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carrico JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng X, Ed-Dra A, Yue M. 2022. Whole genome sequencing for the risk assessment of probiotic lactic acid bacteria. Crit Rev Food Sci Nutr, in press. [DOI] [PubMed] [Google Scholar]
- 60.Teng L, Liao S, Zhou X, Jia C, Feng M, Pan H, Ma Z, Yue M. 2022. Prevalence and genomic investigation of multidrug-resistant Salmonella isolates from companion animals in Hangzhou, China. Antibiotics (Basel) 11:625. doi: 10.3390/antibiotics11050625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Kang X, Ed-Dra A, Zhou X, Jia C, Muller A, Liu Y, Kehrenberg C, Yue M. 2022. Genome-based assessment of antimicrobial resistance and virulence potential of isolates of non-Pullorum/Gallinarum Salmonella Serovars recovered from dead poultry in China. Microbiol Spectr 10:e0096522. doi: 10.1128/spectrum.00965-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Anwar TM, Pan H, Chai W, Ed-Dra A, Fang W, Li Y, Yue M. 2022. Genetic diversity, virulence factors, and antimicrobial resistance of Listeria monocytogenes from food, livestock, and clinical samples between 2002 and 2019 in China. Int J Food Microbiol 366:109572. doi: 10.1016/j.ijfoodmicro.2022.109572. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Ed-Dra A, Tang B, Kang X, Muller A, Kehrenberg C, Jia C, Pan H, Yang H, Yue M. 2022. Higher tolerance of predominant Salmonella serovars circulating in the antibiotic-free feed farms to environmental stresses. J Hazard Mater 438:129476. doi: 10.1016/j.jhazmat.2022.129476. [DOI] [PubMed] [Google Scholar]
- 64.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. doi: 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Clausen P, Aarestrup FM, Lund O. 2018. Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinformatics 19:307. doi: 10.1186/s12859-018-2336-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu YY, Lin JW, Chen CC. 2019. cano-wgMLST_BacCompare: a bacterial genome analysis platform for epidemiological investigation and comparative genomic analysis. Front Microbiol 10:1687. doi: 10.3389/fmicb.2019.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Geurts P, Ernst D, Wehenkel L. 2006. Extremely randomized trees. Mach Learn 63:3–42. doi: 10.1007/s10994-006-6226-1. [DOI] [Google Scholar]
- 68.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paudyal N, Pan H, Wu B, Zhou X, Zhou X, Chai W, Wu Q, Li S, Li F, Gu G, Wang H, Hu Q, Xu X, Li Y, Yue M. 2020. Persistent asymptomatic human infections by Salmonella enterica Serovar Newport in China. mSphere 5:e00163-20. doi: 10.1128/mSphere.00163-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshida CE, Kruczkiewicz P, Laing CR, Lingohr EJ, Gannon VP, Nash JH, Taboada EN. 2016. The Salmonella in Silico Typing Resource (SISTR): an open web-accessible tool for rapidly typing and subtyping draft Salmonella genome assemblies. PLoS One 11:e0147101. doi: 10.1371/journal.pone.0147101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, Lauer AC, Fields PI, Deng X. 2019. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol 85:e01746-19. doi: 10.1128/AEM.01746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo X, Chen F, Gao F, Li L, Liu K, You L, Hua C, Yang F, Liu W, Peng C, Wang L, Yang X, Zhou F, Tong J, Cai J, Li Z, Wan B, Zhang L, Yang T, Zhang M, Yang L, Yang Y, Zeng W, Wang B, Wei X, Xu X. 2020. CNSA: a data repository for archiving omics data. Database (Oxford) 2020:baaa055. doi: 10.1093/database/baaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen FZ, You LJ, Yang F, Wang LN, Guo XQ, Gao F, Hua C, Tan C, Fang L, Shan RQ, Zeng WJ, Wang B, Wang R, Xu X, Wei XF. 2020. CNGBdb: China National GeneBank DataBase. Yi Chuan 42:799–809. doi: 10.16288/j.yczz.20-080. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download spectrum.02479-22-s0001.pdf, PDF file, 0.7 MB (765.3KB, pdf)
Tables S1 to S5. Download spectrum.02479-22-s0002.xlsx, XLSX file, 3.3 MB (3.3MB, xlsx)
Data Availability Statement
All the raw data have been deposited into CNGB Sequence Archive (CNSA) (76) of China National GenBank DataBase (CNGBdb) (77) with a project title ‘poultry production chain’ and accession number CNP0001590.