ABSTRACT
The increasing awareness of emerging tickborne pathogens (TBPs) has inspired much research. In the present study, the coinfections of TBPs both in ticks and their wild hedgehog hosts in Jiangsu province, Eastern China were determined by metagenome next-generation sequencing and nested PCR. As a result, Rickettsia japonica (81.1%), novel Rickettsia sp. SFGR-1 (5.1%), Anaplasma bovis (12%), A. platys (6.3%), novel Ehrlichia spp. Ehr-1 (16%) and Ehr-2 (0.6%), E. ewingii-like strain (0.6%), Coxiella burnetii (10.9%), and a novel Coxiella-like endosymbiont (CLE) strain (61.1%) were detected in Haemaphysalis flava ticks. A. bovis (43.8%), Ehrlichia sp. Ehr-1 (83.3%), and C. burnetii (80%) were detected in Erinaceus amurensis hedgehogs. Coinfection rates with various TBPs were 71.5% and 83.3% in ticks and hedgehogs, respectively, both with double-pathogen/endosymbiont coinfection rates over 50%. We found the following. (i) Er. amurensis hedgehogs seem to contribute to the natural cycles of R. japonica, A. bovis, Ehrlichia sp., and C. burnetii and may be reservoirs of them except for R. japonica, and A. bovis is proved to infect hedgehogs for the first time. (ii) H. flava is proved to harbor various TBPs as a reservoir host, including CLE identified for the first time, which could inhibit coinfection of C. burnetii while promoting that of Rickettsia spp. in H. flava. (iii) Four novel TBP species were identified. This study provides useful epidemiological information crucial for assessing the potential infection risks to humans, thus benefiting the development of strategies to prevent and control tick-borne diseases.
IMPORTANCE In the present study, we found the following. (i) Er. amurensis hedgehogs seem to contribute to the natural cycles of R. japonica, A. bovis, Ehrlichia sp., and C. burnetii and may be reservoirs of them except for R. japonica, and A. bovis is proved to infect hedgehogs for the first time. (ii) H. flava is proved to harbor various tickborne pathogens (TBPs) as a reservoir host, including Coxiella-like endosymbiont (CLE) identified for the first time, which could inhibit coinfection of C. burnetii while promoting that of Rickettsia spp. in H. flava. (iii) Four novel TBP species were identified. This study provides useful epidemiological information on TBPs harbored and transmitted by ticks and their hosts, for assessing the potential infection risks to humans, thus benefiting the developing strategies for tick-borne diseases prevention and control.
KEYWORDS: Haemaphysalis flava, coinfection, tick, hedgehog, Rickettsia, mNGS, tick-borne pathogen
INTRODUCTION
Ticks transmit various pathogens, such as bacteria, viruses, protozoa, and helminths, causing serious diseases or even death in both animals and humans (1 to 3). In light of the ongoing geographical expansion of ticks caused by climatic change, as well as their increased ability to harbor new pathogens, public health concerns have been raised for both humans and animals (4, 5). During 1950 to 2018, China reported 103 tick-borne agents, including 29 species (subspecies) of pathogens, with most discovered in the last decades (5), indicating the serious public health threat that the tickborne diseases have imposed in China.
Hosts of ticks include domestic animals, wild animals, and humans; wild hedgehogs tend to be highly infested by ticks and highly at risk for tick-borne diseases (6, 7). In urban or suburban areas, hedgehogs are thought to play a significant role in the dissemination and transmission of various tick-borne pathogens (TBPs) such as Borrelia spielmanii, B. bavariensis, B. afzelii, Rickettsia helvetica, and Anaplasma phagocytophilum as well as their enzootic cycles (8). Various pathogens have different geographical distributions, and the role of hedgehogs and their parasitic ticks in tick-borne diseases is still not fully understood (6). Hedgehogs' ecological habits dictate their close relationship with humans. Also, nowadays, with an expanding range of human activities, there is a much greater chance of exposure of humans to hedgehogs and their harboring ticks. Consequently, studies focusing on this topic should be undertaken (8).
The present study tested for the existence and coexistence of TBPs within Haemaphysalis flava ticks and their hedgehog hosts collected in Jiangsu province, Eastern China, by combining metagenomic next-generation sequencing (mNGS) and nested PCR. We hoped the findings could provide a deeper epidemiological understanding of the pathogens harbored by ticks and their hedgehog hosts as well as valuable data for evaluating the potential risk of tick-borne diseases from contact with wild hedgehogs or their contaminated environment, thus benefiting the prevention and control of the potential tick-borne diseases.
RESULTS
Taxonomic classification.
According to the partial mitochondrial large subunit rRNA gene sequencing and alignment (GenBank: ON016529), all the obtained hedgehogs were classified as Erinaceus amurensis. Six (3.3%) of the 181 adult ticks feeding on hedgehogs were identified to be Haemaphysalis longicornis, and the other 175 (96.7%) were identified as H. flava (GenBank: ON016525 to ON016528). Only H. flava was analyzed in this study due to the small sample sizes of H. longicornis. Female H. flava accounted for 74.9% (131/175) and male ones accounted for 25.1% (44/175).
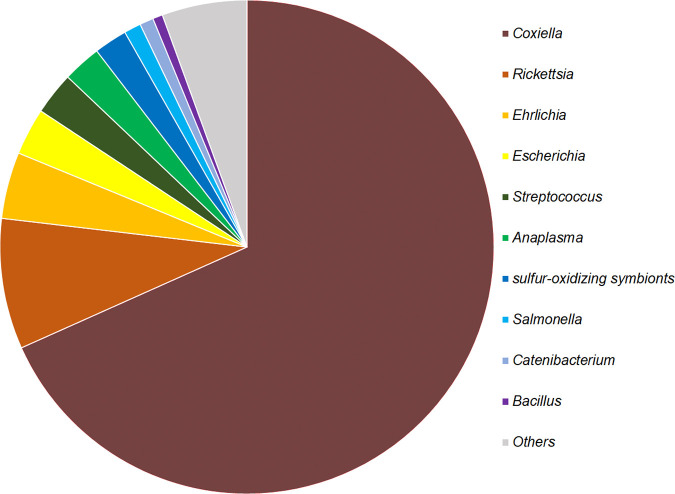
In mNGS of the pooled DNA samples, sequencing yielded a total of 90 million reads and 13.5 billon bases with GC bases ratio of 48.32% and high quality of Q20 = 95.28%, which rose to 97.20% after treatment with Trimmomatic. The relative abundance was measured at various taxonomic levels (Fig. S2). Briefly, 11.2% of the obtained open reading frames (ORFs) belonged to microorganisms, including bacteria (10.98%), archaea (0.04%), viruses (0.05%), and fungi (0.13%). Taxonomic profiles at the genus level confirmed that genera Rickettsia, Anaplasma, Ehrlichia, and Coxiella existed in the pooled sample, which all predominated the top 6 in relative abundance (Fig. 1 and Fig. S2). Genus Coxiella was most abundant in the pool.
FIG 1.
Relative abundances of potential top 10 pathogens at the genus level in the pooled H. flava sample analyzed by metagenomic next-generation sequencing.
Prevalence of TBPs in ticks and hedgehogs.
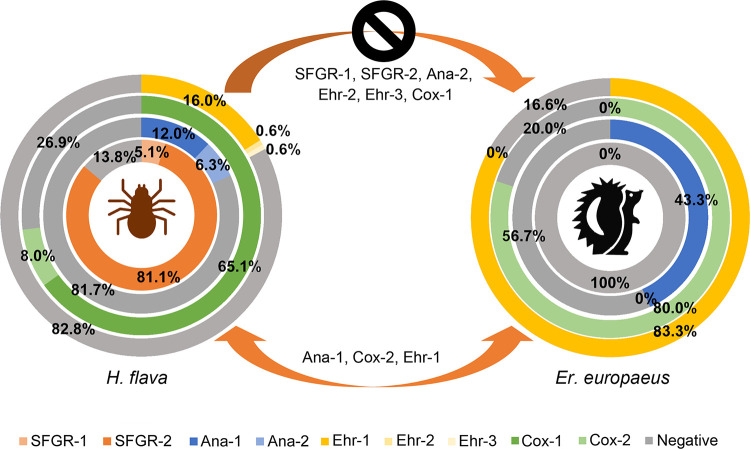
Nested PCR was performed on each sample of H. flava to confirm Rickettsia, Anaplasma, Ehrlichia, and Coxiella. Representative results of agarose gel electrophoresis analysis of the amplified products of nested PCR toward partial rrs genes of Anaplasma spp. and Ehrlichia spp are shown in Fig. S3. As a result (Fig. 2), in H. flava ticks from hedgehogs, 86.3% (151/175) were positive for Rickettsia, 15.4% (27/175) positive for Anaplasma, 17.1% (30/175) positive for Ehrlichia, and 71.4% (125/175) positive for Coxiella. Overall, in the 175 H. flava ticks, only 6 were negative for any of the pathogens of the 4 genera of pathogens.
FIG 2.
The observed positive rates and potential mutual communications of various pathogens in the H. flava ticks (n = 175) and Er. amurensis hedgehogs (n = 30). The identified Rickettsia spp. SFGR-1 and SFGR-2, Anaplasma spp. Ana-1 and Ana-2, Ehrlichia spp. Ehr-1, Ehr-2, and Ehr-3, and Coxiella spp. Cox-1 and Cox-2 in this study were indicated. The black forbidden symbol indicates the pathogens in H. flava didn’t spread to or establish infection in hedgehogs.
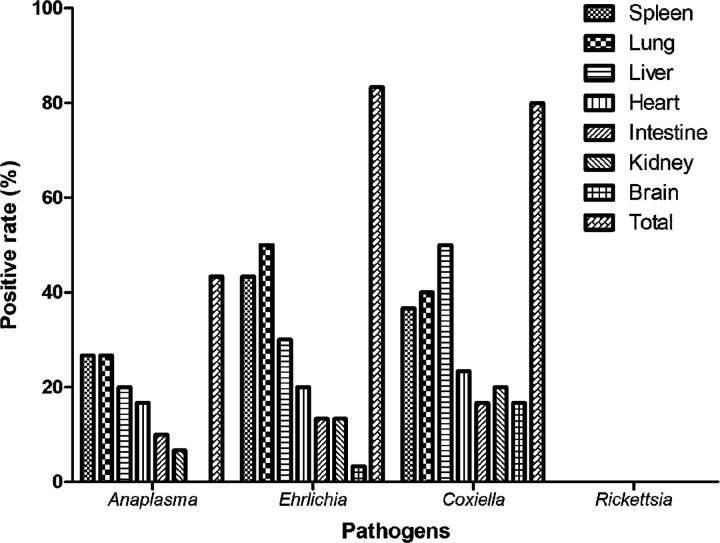
In hedgehogs, different organs performed different positive rates for the TBPs (Fig. 3). In total, spleens, lungs, and livers represented the top 3 organs harboring Anaplasma, Ehrlichia, and Coxiella with the highest positive rates. The total positive rates of Anaplasma, Ehrlichia, and Coxiella in the hedgehogs were 43.8% (13/30), 83.3% (25/30), and 80% (24/30), respectively. To our surprise, the rrs gene of Rickettsia was not detected in any organs of the hedgehogs. Another set of primers targeting the gltA gene of spotted fever group Rickettsia (SFGR) was further used for nested PCR amplification of the samples from these hedgehogs, and the same result was observed, confirming the absence of Rickettsia in the hedgehogs. Overall, in the 30 hedgehogs, only 2 were negative for all of the 4 kinds of pathogens.
FIG 3.
Positive rates of the tick-borne pathogens in different organs of the hedgehogs (n = 30).
Coinfection in individual ticks and hedgehogs.
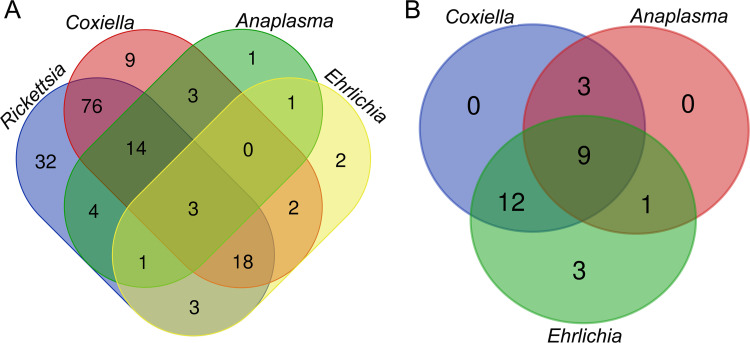
As shown in Table 1 and Fig. 4, 74.9% (131/175) of the H. flava ticks were coinfected with more than one pathogen identified in this study. The dual coinfection with Coxiella spp. and Rickettsia spp. was identified as predominantAs shown in (43.4%), and the triple coinfection with Coxiella spp., Ehrlichia spp., and Rickettsia spp. was also frequent. The quadra coinfection with all 4 species of pathogens was observed, though with a low rate of 1.7%.
TABLE 1.
Infection and coinfection with tick-borne pathogens at genus level in individual H. flava ticks and Er. amurensis hedgehogs
| Pathogens (genus) | No. of positive individuals |
|
|---|---|---|
| Ticks (n = 175) | Hedgehogs (n = 30) | |
| Quadruple | 3 (1.7%) | |
| Anaplasma, Coxiella, Ehrlichia, Rickettsia | 3 (1.7%) | |
| Triple | 33 (18.9%) | 9 (30%) |
| Anaplasma, Coxiella, Rickettsia | 14 (8%) | |
| Coxiella, Ehrlichia, Rickettsia | 18 (10.3%) | |
| Anaplasma, Ehrlichia, Rickettsia | 1 (0.6%) | |
| Anaplasma, Ehrlichia, Coxiella | 9 (30%) | |
| Double | 89 (50.9%) | 14 (53.3%) |
| Coxiella, Rickettsia | 76 (43.4%) | |
| Anaplasma, Rickettsia | 4 (2.3%) | |
| Ehrlichia, Rickettsia | 3 (1.7%) | |
| Anaplasma, Coxiella | 3 (1.7%) | 3 (10%) |
| Coxiella, Ehrlichia | 2 (1.1%) | 12 (40%) |
| Anaplasma, Ehrlichia | 1 (0.6%) | 1 (3.3%) |
| Single | 44 (25.1%) | 3 (10%) |
| Rickettsia | 32 (18.3%) | |
| Coxiella | 9 (5.1%) | |
| Anaplasma | 1 (0.6%) | |
| Ehrlichia | 2 (1.1%) | 3 (10%) |
| None | 6 (3.4%) | 2 (6.7%) |
FIG 4.
Coexistences of various pathogens in H. flava ticks (A) and their Er. amurensis hedgehog hosts (B) at genus level. Pathogens and numbers of individuals harboring various pathogens are indicated.
In hedgehogs, the dual pathogen coinfections with Coxiella spp. and Ehrlichia spp. and triple pathogen coinfections with Anaplasma spp., Coxiella spp., and Ehrlichia spp. took the lead with rates of 40% and 30%, respectively. The quadra coinfection was not observed due to the absence of Rickettsia spp. in hedgehogs.
Additionally, one species of each of the three genera, Anaplasma, Ehrlichia, and Coxiella, was found to infect both ticks and hedgehogs (Fig. 2).
Phylogenetic analysis.
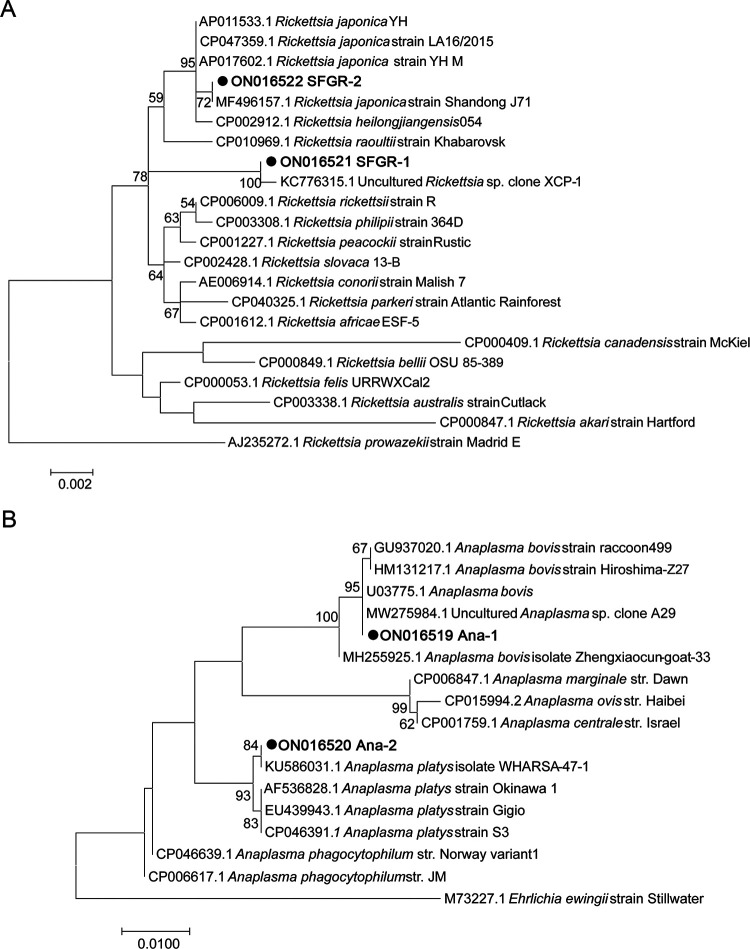
Two different partial rrs gene sequences of SFGR (SFGR-1 and SFGR-2 as indicated in Fig. 5A, GenBank: ON016521 and ON016522) were obtained from tick samples by nested PCR, indicating that two different SFGR strains existed in the samples. The sequence SFGR-1, amplified from 9 H. flava ticks, performed the highest homology (99.92% identity rate) with an uncultured Rickettsia sp. sequence (GenBank: KC776315.1) formerly identified in H. longicornis ticks in Beijing, China (9). They formed a separate branch in the phylogenetic tree (Fig. 5A), indicating they belonged to an unidentified novel species. The sequence SFGR-2, which was amplified from 142 ticks, performed the highest homology with the partial sequence from R. japonica strain Shandong J71 (99.71% identity rate) (GenBank: MF496157.1) identified in Shandong province, Northern China. Phylogenic analysis indicated that SFGR-2 was a strain of R. japonica, because it clustered with R. japonica and R. heilongjiangensis but was more closely related to R. japonica (Fig. 5A).
FIG 5.
Phylogenetic analysis based on the partial rrs gene sequence of Rickettsia (A) and Anaplasma (B) species using MEGA 7.0 software. Maximum likelihood method with 1,000 replicates was done to generate the tree. A bootstrap value of greater than 50% was indicated. Nucleotide substitutions per site are indicated by the scale bar. The reference species and GenBank accession number of the sequence are shown on each line.
For identification of Anaplasma spp., a longer rrs gene was amplified, and two different sequences were obtained (GenBank: ON016519 and ON016520). Ana-1, performing 100% identity rate with the partial rrs gene sequence of an uncultured Anaplasma sp. clone A29 (GenBank: MW275984.1) found in H. flava in China as well as an undefined A. bovis strain found in South Africa (GenBank: U03775.1), formed a cluster with various A. bovis strains in phylogenic analysis (Fig. 5B). This A. bovis sequence was amplified in 21 ticks and 13 hedgehogs. However, Ana-2, performing 100% identity rate with the partial rrs gene sequence of an isolate of A. platys (GenBank: KU586031.1), formed a cluster with various strains of this species as well. It was only detected in 11 ticks but none of the hedgehogs. Interestingly, some tick samples were found to contain both kinds of sequences judged by overlapping peaks in the sequencing map, indicating coinfection of various Anaplasma species existed in H. flava ticks (data not shown).
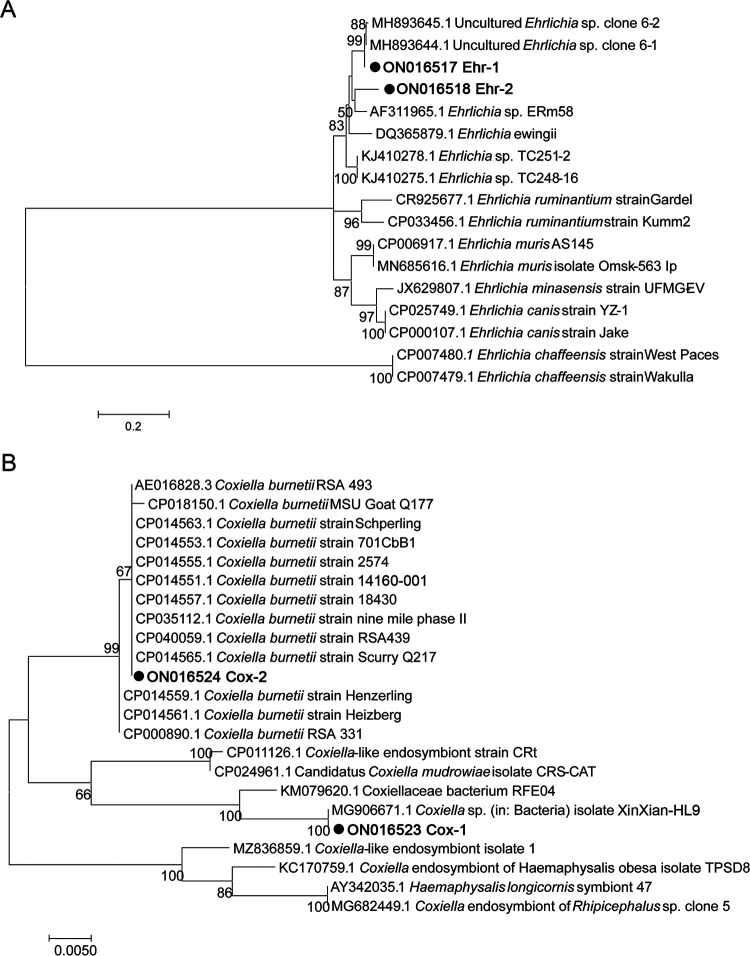
For Ehrlichia spp. identification, three different short sequences of the rrs gene were amplified from ticks (GenBank: ON016514 to ON016516), while only two partial gltA gene sequences were successfully amplified (GenBank: ON016517 and ON014518). Sequence of Ehr-1 performed 99.26% and 98.77% identity rates with partial sequences of gltA gene of two uncultured Ehrlichia species (GenBank: MH893644.1 and MH893645.1) hosted in Er. amurensis, respectively. They formed a cluster in the phylogenic tree (Fig. 6A). Similarly, Ehr-2 performed 91.11% identity rate with an undefined Ehrlichia sp. Erm58 (GenBank: AF311965.1) reported to belong to the E. canis group (10), which was identified in Rhipicephalus muhsamae ticks in African. Nevertheless, both Ehr-1 and Ehr-2 were believed to belong to novel Ehrlichia species. Though the gltA gene of the third Ehrlichia sp. was not amplified, the rrs sequence of this strain performed 99.59% identity rate with various strains or isolates of E. ewingii in the alignment analysis by BLAST (data not shown). In all, Ehr-1, Ehr-2, and the E. ewingii-like strain were identified in 28, 1, and 1 ticks, respectively, while only Ehr-1 existed in the hedgehogs with a high positive rate of 83.3% (25/30).
FIG 6.
Phylogenetic analysis based on the partial gene sequences of gltA of Ehrlichia species (A) and rrs of Coxiella species (B) using MEGA 7.0 software. Maximum likelihood method with 1,000 replicates was done to generate the tree. A bootstrap value of greater than 50% is indicated. Nucleotide substitutions per site are indicated by the scale bar. The reference species and GenBank accession number of the sequence are shown on each line.
In phylogenic analysis of the obtained two partial sequences of the rrs gene (GenBank: ON016523 and ON016524) of Coxiella spp., Cox-1, performing high homologies of 99.44% and 99.31% with the sequences of two uncultured and undefined Coxiella spp. (GenBank: MG906671.1 and KC776319.1), respectively, and forming a separate branch together with “Candidatus C. mudrowiae isolate” (GenBank: CP011126.1) and Coxiella-like endosymbiont strain CRt (GenBank: CP024961.1), was difficult to be determined a novel C. burntii strain or an endosymbiont strain, while Cox-2 was believed to be a C. burntii strain by forming a cluster with various other C. burntii strains (Fig. 6B). Cox-1 was found in 114 ticks and none of the hedgehogs, while Cox-2 was found in 14 ticks and 24 hedgehogs. Also, 3 tick samples were found to contain both kinds of Coxiella spp., judged by overlapping peaks in the sequencing maps of Anaplasma species.
Mutual effects of various pathogens.
Whether one pathogen/endosymbiont infection could promote or inhibit the other’s coinfection in ticks or hedgehogs was statistically analyzed. In the H. flava ticks, the Coxiella sp. (Cox-1) was observed to have the ability to significantly inhibit the coinfection of C. burnetii (continuity adjusted χ2 = 10.8, P = 0.00102) and promote the coinfection of Rickettsia spp. (χ2 = 4.57, P = 0.0326). However, mutual effects between other pathogens were not observed either in the ticks or hedgehogs (P > 0.05).
DISCUSSION
In recent years, the increasing awareness of emerging TBPs has greatly inspired studies on ticks and tick-borne diseases in China (11). Epidemiological information on ticks and their hosts, and their harboring and transmitting pathogens, can be useful for the development of tick-borne diseases prevention and control (12). Despite the existence of a variety of TBPs, few studies have focused on coinfection with multiple pathogens (13), and this is the first study investigating the coinfection of TBPs both in wild hedgehogs and their harboring ticks in China, to our knowledge.
Though various species of ticks have been found to infest Erinaceidae in previous studies depending on the diversity of geographical environment they inhabit (14, 15), such as Hyalomma spp., Rhipicephalus spp., Haemaphysalis spp., Ixodes spp., et al., in this study in the Jiangsu province of Eastern China, the predominant ticks infesting Er. amurensis were H. flava (96.7%). This is consistent with other studies conducted in Jiangxi province, Eastern China, and Hubei province, Central China, in which all the 13 and 125 ticks collected from Erinaceidae were H. flava (11, 16). These studies, together with the present study, to some extent, reflect the parasite characteristics of Erinaceidae in China, while more studies are needed to verify the prediction, due to the small number of studies focusing on this area.
To get a general picture of the pathogen profile of ticks, mNGS has been conducted, which is useful to investigate the microbiomes and microbial diversity of ticks (17). Jiao et al. applied mNGS to investigate TBPs in cattle-attached ticks in Northern China, and proved that various pathogens including R. raoultii, “Candidatus R. tarasevichiae,” Anaplasma sp. Mongolia, Coxiella-like endosymbiont (CLE), and Babesia venatorum coinhabited Dermacentor nuttalli and Ixodes persulcatus (13). Another similar study in Japan confirmed the existence of Coxiella spp., Ehrlichia spp., and Rickettsia spp. in salivary glands of various ticks (including H. flava) by mNGS (18). Similarly, in the present study, the presence of 4 kinds of important genera, including Rickettsia, Anaplasma, Ehrlichia, and Coxiella, were detected by mNGS in the pooled sample of ticks, with Coxiella the most abundant. The mNGS procedure effectively cleared the fog in front of our targets, and reduced ineffective labor, so that we could evaluate the prevalence of specific genera of pathogens in the nested PCR. In general, coupling mNGS with PCR, 9 and 4 species of TBPs were identified in H. flava and Er. amurensis in the present study, respectively.
R. japonica is still prevalent in China and Japan in recent years and can cause Oriental spotted fever (19 to ,22). Interestingly, the H. flava ticks had a high positive rate (81.1%) of R. japonica, and their hedgehog hosts should also have a high R. japonica-positive rate. However, the reality was just the opposite, with neither the gltA nor the rrs gene of R. japonica detectable in the organs of these Er. amurensis hedgehogs. We supposed that R. japonica didn’t cause chronic infection and the pathogen transmitted by tick biting would be eliminated by the hedgehog's immune system quickly, based on the following three reasons. First, nowadays, the only existent Rickettsia sp. in hedgehogs reported was R. helvetica (8), which is also the only species that could establish chronic infection in SFGR (23). Second, C. burnetii, A. bovis, and Ehrlichia sp. could be detected in hedgehogs in the present study, and all of them have the ability to establish chronic infections. Third, in our previous study (24), when we did a preliminary experiment using R. heilongjiangensis, a species with a high degree of similarity with R. japonica at the genomic level, to infect healthy C3H/HeN mice, the bacterial load in mouse organs (including spleen, liver, and lung) dropped quickly since day 3 postinfection, until total elimination in days 7 to 10, and the elimination procedure of R. japonica might also happen in hedgehogs. Thus, very few organisms existed in the organs beyond the range of detection limit of nested PCR. In a recent study in Tunisia, similar results were observed in Atelerix algirus hedgehogs, who were infested with Rickettsia-positive ticks (positive rate 47.2%) and fleas (positive rate 79.2%), while the gltA and ompB genes of Rickettsia were not detected in the hedgehog organ samples by conventional or nested PCR (25).
Considering the high prevalence of R. japonica in the ticks, we hypothesize that Er. amurensis hedgehogs just play a role of “bridge” that transmits R. japonica from an infected H. flava tick to an uninfected one attached by the same host (26), but not as a reservoir host or infectious source. In this “bridge transmission pattern,” hedgehogs may acquire either a short-term systemic infection or a nonsystemic infection that is limited to the parts where the infected ticks are feeding to transmit R. japonica to uninfected H. flava ticks. Actually, the latter transmission pattern, with systemic infection in host unnecessary, has been reported in Borrelia burgdorferi transmission between ticks feeding on sheep (27). However, the hypothesis here needs further confirmation with animal model experiments considering the small number of hedgehogs used, and it is also possible that the pathogen is vertically transmitted in ticks, and the hedgehog blood meal simply allows for propagation of SFGR in ticks without being transmitted to mammals. The other sequence SFGR-1 performed highest homology with an uncultured Rickettsia sp. sequence (KC776315.1) identified in H. longicornis in Beijing (9), indicating it may be a novel Rickettsia sp. while further characterizations of its sequences and infectivity are needed.
Both A. platys and A. bovis were identified in H. flava, while only A. bovis was found in organs of the hedgehogs. A. bovis was predominant in ticks compared with A. platys, and this is not unexpected, because hedgehogs may play roles in the circulation of A. bovis. A. platys is a pathogen that infects platelet of dogs and could also cause human infections (28 to 30), while A. bovis causes anaplasmosis in animals including bovines, raccoons, cats, deer, etc. (31 to 34). To our knowledge, this is the first time A. bovis has been identified in hedgehogs.
Three kinds of Ehrlichia spp. were identified in ticks and only one kind (Ehr-1) was detected in organs of Er. amurensis. In phylogenetic analysis, Ehr-1 and Ehr-2 clustered together with uncultured or undefined Ehrlichia spp., indicating they were novel Ehrlichia species and genetic diversity existed in this area. Also, they are taxonomically related to E. ewingii by sharing the same clade in the phylogenetic tree. E. ewingii has been proved to cause human granulocytic ehrlichiosis (HGE) (35), and Ehr-1 in this study was proved to infect hedgehogs. So, whether the newly identified Ehrlichia spp. could infect humans still needs further confirmation. Moreover, Ehr-1 was predominant in both ticks and hedgehogs, indicating mutual communications existed between them, and hedgehogs might play some roles in the Ehrlichia circulation.
Interestingly, the phenomenon observed in Coxiella spp. is the opposite of that observed in Anaplasma spp. or Ehrlichia spp. Here, the predominant Cox-1 in H. flava ticks was not identified in Er. amurensis hedgehogs, while the predominant C. burnetii strain (Cox-2) detected in hedgehogs had a low positive rate in ticks. This indicates Cox-1 may not have the ability to infect hedgehogs and is most likely a symbiotic bacterium, considering it shared the same clade with Coxiella-like endosymbiont strain CRt (CP024961.1) in the phylogenic tree (Fig. 6B). Cox-1 has a positive rate of 61.1% in ticks, which is also consistent with the typically high infection frequencies of CLE in infected tick populations (36 to 39). These bacteria act as obligate nutrient providers for ticks by encoding pathways needed for essential amino acids, major B vitamins, and cofactors synthesis (40 to 44). The phenomenon that C. burnetii performed at a high positive rate of 80% in hedgehogs, while a low positive rate of 10.9% in parasitic ticks indicates the horizontal transmission of C. burnetii between the hedgehogs and their infesting ticks was inhibited. Coinfected TBPs can work synergistically, indifferently, or antagonistically within their host (45). The statistical analysis on possible interactions between these identified pathogens verified that the inhibition effect on C. burnetii infection in H. flava was provided by the CLE (Cox-1), with a very low P value. The presence of endosymbiont species has been reported to exclude R. rickettsii as a pathogen in ticks in rickettsial interactions (45 to 48), while the similar phenomenon in Coxiella spp. is the first time it was observed in the world, suggesting the possibility of biological intervention to reduce the prevalence of C. burnetii among H. flava hosts. Moreover, it is also the first time it was observed that the existence of CLE is able to facilitate the coinfection of Rickettsia spp., though with a relatively bigger P value (0.1 < P < 0.5) in the statistical analysis. Nowadays, there is still too little research on CLE, and the mechanisms behind the phenomenon need deep digging. CLE was previously detected in pools of ticks containing H. longicornis, H. flava, Rh. sanguineus, and Rh. turanicus in China, which actually could not confirm its presence in H. flava. So, according to our knowledge, this is the first identification of CLE in H. flava.
C. burnetii, the causative agent of the worldwide zoonosis, Q fever, is also a biological warfare agent (49). Ticks are the principal vector and reservoir of C. burnetii (49), and H. flava was reported to harbor C. burnetii previously (11). The high positive rate of C. burnetii in hedgehogs (80%) suggests Er. amurensis may be a potential reservoir host of this pathogen and contribute to its circulation. Moreover, the contact with Er. amurensis or its feces in this area may cause Q fever for humans. However, as the rrs gene is highly conserved, further investigations are needed to genotype the strain and confirm its virulence.
Coinfections of multiple pathogen/endosymbiont were observed to be in the majority in both ticks and their hosts in the present study, which may be a result of mutual infections between parasites and hosts during blood feeding. Similar observations on coinfections were reported in ticks collected from bovines in north China (13) and in ticks, fleas, and their hedgehog hosts in Tunisia (25). The results suggest the potential roles of hedgehogs and their parasites as simultaneous wild reservoirs and melting pots for TBPs, which may impose a threat to humans. Wild hedgehogs have become common in suburban and urban areas, searching for food and shelter. People are becoming more and more likely to be exposed to zoonotic pathogens carried by hedgehogs and their parasites. Therefore, future studies on the prevalence of coinfection with TBPs in humans living near their natural habitat are needed.
One of the limitations of our study is that we cannot conclude whether the presence of different TBPs can be entirely attributed to the ticks and their pathogenic contents, or they may be influenced by previous blood meal hosts, which is also the problem met by studies using engorged ticks (45). The coexistence of A. bovis and A. platys or Cox-1 and C. burnetii in the few ticks may be due to the host blood that the ticks fed on.
In conclusion, in the present study, we find the following. (i) Er. amurensis hedgehogs seem to contribute to the spread, transmission, and enzootic cycles of R. japonica, A. bovis, Ehrlichia sp., and C. burnetii, and they may be the reservoirs of A. bovis, Ehrlichia sp., and C. burnetii but not R. japonica, in which A. bovis is proved to infect hedgehogs for the first time in the world. (ii) H. flava is proved to harbor various TBPs as a reservoir host, including CLE identified for the first time, and the existence of CLE may inhibit coinfection of C. burnetii but facilitate that of SFGR in H. flava. (iii) Four novel TBP species were identified, including one Rickettsia sp., two Ehrlichia spp., and one CLE. Though some hypotheses in this study still need further investigations and confirmations, the prevalence and coinfection data of various important TBPs in Er. amurensis hedgehogs and their parasites H. flava provide useful information and data for evaluating the potential risk of human infections, thus benefiting the early warning, prevention, and control of potential diseases. Moreover, the more we learn, the more we want to know, including whether hedgehogs can maintain these pathogens in natural cycles independently, their contributions to the TBPs’ enzootic cycle, and the transmission mechanisms involving hedgehogs and ticks, all of which remain to be investigated.
MATERIALS AND METHODS
Hedgehogs and ticks.
Thirty hedgehogs were collected in the town of Tianquanhu (E 118°35′20″, N 32°47′20″), Huaian city, Jiangsu province, China during 2016 to 2018 (Fig. S1). All of them died from car accidents or some other unknown reasons and were donated by the local villagers. These hedgehogs died not long (within 48 h) before we got them. After the ticks were collected, the hedgehogs were dissected for collection of various organs including spleens, livers, lungs, intestines, brains, hearts, and kidneys.
All the 181 adult ticks collected were feeding on the hedgehogs. In a simple random sampling method, 4 to 10 ticks for each animal were collected and initially identified by morphological features, followed by molecular methods described previously (50).
Animal samples were used only with the approval of the Ethics Committee at Huadong Research Institute for Medicine and Biotechniques.
DNA purification.
Ticks that had been stored in ethanol were thoroughly washed twice with phosphate-buffered saline (PBS) before being homogenized individually in 1 mL of PBS. An appropriate amount of each homogenate or organ sample was used to extract their DNA using a QIAamp Fast DNA Tissue kit (Qiagen, Dusseldorf, Germany) in accordance with the manufacturer's instructions and stored at −20°C prior to analysis.
Metagenome next-generation sequencing and data analysis.
A pooled DNA sample was prepared by mixing 20 μL of individual DNA extractions from 10 randomly selected H. flava ticks for complete microbial genome sequencing using mNGS as described previously (13). Sequencing was conducted using the Illumina HiSeq platform by Sangon Biotech company (Shanghai, China). For quality control, the obtained raw data were trimmed and validated using Trimmomatic software to get clean reads (51). Raw data were filtered to remove reads with more than 10 “N” bases. Low-quality bases (quality value <20) on both ends as well as the adapters on the reads were removed. The clean reads were assembled to contigs using IDBA-UD software based on the De Bruijn graph mechanism (52). A Prodigal program was used to predict the open reading frames of the contigs (≥100 bp) and translate them into amino acid sequences (53). CD-HIT (V4.5.8) was used to establish a nonredundant gene catalog, and Bowtie2 was used to align the clean reads (54). The number of successfully aligned reads was calculated using Samtools (55), and based on the number and the length of the genes, the relative abundance of genes was calculated as previously described (13). The obtained amino acid sequences were aligned with the NCBI NR (NCBI nonredundant protein sequences) database using BLAST software to obtain functional annotations and homologous species information. Meanwhile, according to the NCBI microbial taxonomic information database, the annotation information of species classification of genes was obtained, and each taxonomic level was evaluated, including the kingdom, phylum, class, order, family, and genus.
PCR and sequencing.
The presence of TBPs identified by mNGS was further confirmed by genus-/group-specific nested PCR targeting rrs genes of Anaplasma spp., Ehrlichia spp., Coxiella spp., and spotted fever group Rickettsia (SFGR) in individual H. flava ticks and hedgehogs as described previously (11, 13). For SFGR, primers for the gltA gene were also used to detect samples from hedgehog organs. For Anaplasma spp. and Ehrlichia spp., first, a universal nested PCR primer set resulting in a short partial sequence of the rrs gene around 280 bp was used as described previously (13), and then by nested PCR, a longer partial rrs gene and gltA gene were amplified for Anaplasma- and Ehrlichia-positive samples, respectively.
Species of each hedgehog and tick was identified by PCR amplification of corresponding partial mitochondrial 16S rRNA gene fragment as described previously (50, 56), and the amplification procedure for ticks was also used as a quality control to ensure the successful extraction of DNA.
In ordinary PCR and the nested PCR, 1 μL of the extracted DNA sample was used as the template. Premix Taq kit (TaKaRa, Beijing, China) was used to carry out the PCR amplifications according to the manufacturer’s instructions. Negative and positive controls were conducted using ddH2O and DNA of corresponding pathogens, respectively. Amplified products were analyzed by agarose gel electrophoresis, and then sequenced by the Sangon Biotech company (Shanghai, China). The primers used are indicated in Table 2.
TABLE 2.
PCR primers used in this study
| Object species | Target genes | Primer names | Sequences (5′–3′) | Round |
|---|---|---|---|---|
| Spotted fever group Rickettsia | rrs | S1 | TGATCCTGGCTCAGAACGAAC | 1st |
| S2 | TAAGGAGGTAATCCAGCCGC | 1st | ||
| S3 | AACACATGCAAGTCGRACGG | 2nd | ||
| S4 | GGCTGCCTCTTGCGTTAGCT | 2nd | ||
| gltA | CS2d | ATGACCAATGAAAATAATAAT | 1st | |
| CSEndr | CTTATACTCTCTATGTACA | 1st | ||
| RpCS.877p | GGGGACCTGCTCACGGCGG | 2nd | ||
| RpCS.1258n | ATTGCAAAAAGTACAGTGAACA | 2nd | ||
| Anaplasma spp. and Ehrlichia spp. | rrs | Eh-out1 | TTGAGAGTTTGATCCTGGCTCAGAACG | 1st |
| Eh-out2 | CACCTCTACACTAGGAATTCCGCTATC | 1st | ||
| Eh-gs1 | GTAATAACTGTATAATCCCTG | 2nd | ||
| Eh-gs2 | GTACCGTCATTATCTTCCCTA | 2nd | ||
| Anaplasma spp. | rrs | An16s1 | GTCACTGACCCAACCTTAAATGGCTGC | 1st |
| An16s2 | ATCCTGGCTCAGAACGAACGCTGG | 1st | ||
| An16s3 | GCGCCCTTCCGTTAAGAAGGATCTA | 2nd | ||
| An16s4 | AGCTTAACACATGCAAGTCGAACGGA | 2nd | ||
| Ehrlichia spp. | gltA | e-gltAwf | TTCTCAGGAATACATGCCACC | 1st |
| e-gltAwr | ACCATTGAGCAGACCAGCCA | 1st | ||
| e-gltAnf | AATTGCAGGGATAGTGGCAA | 2nd | ||
| e-gltAnr | CTGTGGCCAAAACCCATCAA | 2nd | ||
| Coxiella spp. | rrs | Cox16SF1 | CGTAGGAATCTACCTTRTAGWGG | 1st |
| Cox16SR2 | GCCTACCCGCTTCTGGTACAATT | 1st | ||
| Cox16SF1 | CGTAGGAATCTACCTTRTAGWGG | 2nd | ||
| Cox16SR1 | ACTYYCCAACAGCTAGTTCTCA | 2nd | ||
| Tick | Large subunit ribosomal RNA gene | TickHF | GGTATTTTGACTATACAAAGGTATTG | 1st |
| TickHR | TTATTACGCTGTTATCCCTAGAGTATT | 1st |
Phylogenetic analysis.
SnapGene software (snapgene.com) was used to evaluate the quality of the sequencing data, and only data of high quality were used. The obtained nucleotide sequences, with primer sequences removed on both ends, were analyzed using the BLAST search engine online (https://blast.ncbi.nlm.nih.gov/Blast.cgi) in the GenBank database to obtain sequences with high homology. Corresponding sequences from whole genomes were preferentially selected when available.
Multiple sequence alignments among the obtained sequences and the selected sequences were performed using the ClustalW tool of MEGA 7.0 software, and phylogenetic analyses were performed for rrs of SFGR, Coxiella spp., and Anaplasma spp., and for gltA of Ehrlichia spp. using the maximum likelihood method with the number of bootstrap samples at 1,000 in MEGA 7.0 software.
Statistical analysis.
The mutual effect that the presence of one pathogen imposed on the coinfection of another pathogen was statistically analyzed. Determined by the sample volume (n) and theoretical frequencies (T), the appropriate test chosen from chi-square test, continuity adjusted chi-square test, and Fisher's exact test was used as described previously (57). A P of <0.05 indicated a significant correlation between the pathogens. The promotion or inhibition effects of individual pathogens on the other pathogens were judged by their actual frequencies.
Data availability.
The generated mNGS data have been deposited into the GenBank database under accession number PRJNA841387 and are accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA841387. The nucleotide sequences obtained in the present study were deposited into the GenBank database under accession numbers from ON016515 to ON016529.
ACKNOWLEDGMENTS
Conceptualization: Y.Q., L.A., and W.T. Data curation: Y.Q. and L.A. Formal analysis: C.Z. and Y.M. Funding acquisition: W.T. and Y.Q. Methodology: R.L., J.W., and N.L. Software: F.Y. Writing—original draft: Y.Q. Writing—review and editing: W.T. and Y.Q.
Projects from Jiangsu Social Development Project (BE2017620), National Natural Science Foundation of China (U1602223), and Medical Science and Technology Project (19SWAQ04).
Footnotes
Supplemental material is available online only.
Contributor Information
Weilong Tan, Email: mosquito_2008@126.com.
Biao He, Changchun Veterinary Research Institute.
REFERENCES
- 1.Dantas-Torres F, Chomel BB, Otranto D. 2012. Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol 28:437–446. doi: 10.1016/j.pt.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Song K, Ji Y, Sun S, Yue X, Wang C, Luo T, Moming A, Song Y, Zhang Y, Yang R. 2021. Bacterial microbiota in unfed ticks (Dermacentor nuttalli) from Xinjiang detected through 16S rDNA amplicon sequencing and culturomics. Zoonoses 1(1). doi: 10.15212/ZOONOSES-2021-0007. [DOI] [Google Scholar]
- 3.Bezerra-Santos MA, de Macedo LO, Nguyen VL, Manoj RR, Laidoudi Y, Latrofa MS, Beugnet F, Otranto D. 2022. Cercopithifilaria spp. in ticks of companion animals from Asia: new putative hosts and vectors. Ticks Tick Borne Dis 13:101957. doi: 10.1016/j.ttbdis.2022.101957. [DOI] [PubMed] [Google Scholar]
- 4.Sonenshine DE. 2018. Range expansion of tick disease vectors in North America: implications for spread of tick-borne disease. Int J Environ Res Public Health 15:478. doi: 10.3390/ijerph15030478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao GP, Wang YX, Fan ZW, Ji Y, Liu MJ, Zhang WH, Li XL, Zhou SX, Li H, Liang S, Liu W, Yang Y, Fang LQ. 2021. Mapping ticks and tick-borne pathogens in China. Nat Commun 12:1075. doi: 10.1038/s41467-021-21375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruszkowski JJ, Hetman M, Turlewicz-Podbielska H, Pomorska-Mól M. 2021. Hedgehogs as a potential source of zoonotic pathogens—a review and an update of knowledge. Animals 11:1754. doi: 10.3390/ani11061754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lesiczka PM, Hrazdilová K, Majerová K, Fonville M, Sprong H, Hönig V, Hofmannová L, Papežík P, Růžek D, Zurek L, Votýpka J, Modrý D. 2021. The role of peridomestic animals in the eco-epidemiology of Anaplasma phagocytophilum. Microb Ecol 82:602–612. doi: 10.1007/s00248-021-01704-z. [DOI] [PubMed] [Google Scholar]
- 8.Jahfari S, Ruyts SC, Frazer-Mendelewska E, Jaarsma R, Verheyen K, Sprong H. 2017. Melting pot of tick-borne zoonoses: the European hedgehog contributes to the maintenance of various tick-borne diseases in natural cycles urban and suburban areas. Parasit Vectors 10:134. doi: 10.1186/s13071-017-2065-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhuang L, Zhang Z, An X, Fan H, Ma M, Anderson BD, Jiang J, Liu W, Cao W, Tong Y. 2014. An efficient strategy of screening for pathogens in wild-caught ticks and mosquitoes by reusing small RNA deep sequencing data. PLoS One 9:e90831. doi: 10.1371/journal.pone.0090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parola P, Inokuma H, Camicas JL, Brouqui P, Raoult D. 2001. Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg Infect Dis 7:1014–1017. doi: 10.3201/eid0706.010616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang LZ, Lei SC, Yan ZJ, Xiao X, Liu JW, Gong XQ, Yu H, Yu XJ. 2021. Detection of multiple intracellular bacterial pathogens in Haemaphysalis flava ticks collected from hedgehogs in Central China. Pathogens 10:115. doi: 10.3390/pathogens10020115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chisu V, Loi F, Foxi C, Chessa G, Masu G, Rolesu S, Masala G. 2020. Coexistence of tick-borne pathogens in ticks collected from their hosts in Sardinia: an update. Acta Parasitol 65:999–1004. doi: 10.1007/s11686-020-00240-z. [DOI] [PubMed] [Google Scholar]
- 13.Jiao J, Lu Z, Yu Y, Ou Y, Fu M, Zhao Y, Wu N, Zhao M, Liu Y, Sun Y, Wen B, Zhou D, Yuan Q, Xiong X. 2021. Identification of tick-borne pathogens by metagenomic next-generation sequencing in Dermacentor nuttalli and Ixodes persulcatus in Inner Mongolia, China. Parasit Vectors 14:287. doi: 10.1186/s13071-021-04740-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orkun Ö, Çakmak A, Nalbantoğlu S, Karaer Z. 2019. Molecular detection of a novel Babesia sp. and pathogenic spotted fever group rickettsiae in ticks collected from hedgehogs in Turkey: Haemaphysalis erinacei, a novel candidate vector for the genus Babesia. Infect Genet Evol 69:190–198. doi: 10.1016/j.meegid.2019.01.028. [DOI] [PubMed] [Google Scholar]
- 15.Krawczyk AI, van Leeuwen AD, Jacobs-Reitsma W, Wijnands LM, Bouw E, Jahfari S, van Hoek AH, van der Giessen JW, Roelfsema JH, Kroes M, Kleve J, Dullemont Y, Sprong H, de Bruin A. 2015. Presence of zoonotic agents in engorged ticks and hedgehog faeces from Erinaceus europaeus in (sub) urban areas. Parasit Vectors 8:210. doi: 10.1186/s13071-015-0814-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng WQ, Xuan XN, Fu RL, Tao HY, Liu YQ, Liu XQ, Li DM, Ma HM, Chen HY. 2018. Tick-borne pathogens in Ixodid ticks from Poyang Lake Region, Southeastern China. Korean J Parasitol 56:589–596. doi: 10.3347/kjp.2018.56.6.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greay TL, Gofton AW, Paparini A, Ryan UM, Oskam CL, Irwin PJ. 2018. Recent insights into the tick microbiome gained through next-generation sequencing. Parasit Vectors 11:12. doi: 10.1186/s13071-017-2550-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qiu Y, Nakao R, Ohnuma A, Kawamori F, Sugimoto C. 2014. Microbial population analysis of the salivary glands of ticks; a possible strategy for the surveillance of bacterial pathogens. PLoS One 9:e103961. doi: 10.1371/journal.pone.0103961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Zhang PH, Du J, Yang ZD, Cui N, Xing B, Zhang XA, Liu W. 2019. Rickettsia japonica infections in humans, Xinyang, China, 2014–2017. Emerg Infect Dis 25:1719–1722. doi: 10.3201/eid2509.171421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Hu W, Wu T, Li HB, Hu W, Sun Y, Chen Z, Shi Y, Zong J, Latif A, Wang L, Yu L, Yu XJ, Liu BY, Liu Y. 2018. Japanese spotted fever in Eastern China, 2013. Emerg Infect Dis 24:2107–2109. doi: 10.3201/eid2411.170264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu Q, Yu J, Yu L, Zhang Y, Chen Y, Lin M, Fang X. 2018. Rickettsia japonica infections in humans, Zhejiang Province, China, 2015. Emerg Infect Dis 24:2077–2079. doi: 10.3201/eid2411.170044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujikawa T, Yoshikawa T, Kurosu T, Shimojima M, Saijo M, Yokota K. 2021. Co-infection with severe fever with thrombocytopenia syndrome virus and Rickettsia japonica after tick bite, Japan. Emerg Infect Dis 27:1247–1249. doi: 10.3201/eid2704.203610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scarpulla M, Barlozzari G, Salvato L, De Liberato C, Lorenzetti R, Macrì G. 2018. Rickettsia helvetica in human-parasitizing and free-living Ixodes ricinus from urban and wild green areas in the metropolitan city of Rome, Italy. Vector Borne Zoonotic Dis 18:404–407. doi: 10.1089/vbz.2017.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi Y, Xiong X, Duan C, Jiao J, Gong W, Wen B. 2013. Recombinant protein YbgF induces protective immunity against Rickettsia heilongjiangensis infection in C3H/HeN mice. Vaccine 31:5643–5650. doi: 10.1016/j.vaccine.2013.09.064. [DOI] [PubMed] [Google Scholar]
- 25.Balti G, Galon C, Derghal M, Souguir H, Guerbouj S, Rhim A, Chemkhi J, Guizani I, Bouattour A, Moutailler S, M’ghirbi Y. 2021. Atelerix algirus, the North African hedgehog: suitable wild host for infected ticks and fleas and reservoir of vector-borne pathogens in Tunisia. Pathogens 10:953. doi: 10.3390/pathogens10080953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voordouw MJ. 2015. Co-feeding transmission in Lyme disease pathogens. Parasitology 142:290–302. doi: 10.1017/S0031182014001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogden NH, Nuttall PA, Randolph SE. 1997. Natural Lyme disease cycles maintained via sheep by co-feeding ticks. Parasitology 115:591–599. doi: 10.1017/S0031182097001868. [DOI] [PubMed] [Google Scholar]
- 28.Maggi RG, Mascarelli PE, Havenga LN, Naidoo V, Breitschwerdt EB. 2013. Co-infection with Anaplasma platys, Bartonella henselae and Candidatus Mycoplasma haematoparvum in a veterinarian. Parasit Vectors 6:103. doi: 10.1186/1756-3305-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arraga-Alvarado CM, Qurollo BA, Parra OC, Berrueta MA, Hegarty BC, Breitschwerdt EB. 2014. Case report: molecular evidence of Anaplasma platys infection in two women from Venezuela. Am J Trop Med Hyg 91:1161–1165. doi: 10.4269/ajtmh.14-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wen B, Cao W, Pan H. 2003. Ehrlichiae and ehrlichial diseases in China. Ann N Y Acad Sci 990:45–53. doi: 10.1111/j.1749-6632.2003.tb07335.x. [DOI] [PubMed] [Google Scholar]
- 31.Im JH, Baek J, Durey A, Kwon HY, Chung M-H, Lee J-S. 2019. Current status of tick-borne diseases in South Korea. Vector Borne Zoonotic Dis 19:225–233. doi: 10.1089/vbz.2018.2298. [DOI] [PubMed] [Google Scholar]
- 32.Iqbal N, Mukhtar MU, Yang J, Sajid MS, Niu Q, Guan G, Liu Z, Yin H. 2019. First molecular evidence of Anaplasma bovis and Anaplasma phagocytophilum in bovine from Central Punjab, Pakistan. Pathogens 8:155. doi: 10.3390/pathogens8030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki H, Ichikawa Y, Sakata Y, Endo Y, Nishigaki K, Matsumoto K, Inokuma H. 2012. Molecular survey of Rickettsia, Ehrlichia, and Anaplasma infection of domestic cats in Japan. Ticks Tick Borne Dis 3:308–311. doi: 10.1016/j.ttbdis.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 34.Sashika M, Abe G, Matsumoto K, Inokuma H. 2011. Molecular survey of Anaplasma and Ehrlichia infections of feral raccoons (Procyon lotor) in Hokkaido, Japan. Vector Borne Zoonotic Dis 11:349–354. doi: 10.1089/vbz.2010.0052. [DOI] [PubMed] [Google Scholar]
- 35.Buller RS, Arens M, Hmiel SP, Paddock CD, Sumner JW, Rikhisa Y, Unver A, Gaudreault-Keener M, Manian FA, Liddell AM, Schmulewitz N, Storch GA. 1999. Ehrlichia ewingii, a newly recognized agent of human ehrlichiosis. N Engl J Med 341:148–155. doi: 10.1056/NEJM199907153410303. [DOI] [PubMed] [Google Scholar]
- 36.Almeida AP, Marcili A, Leite RC, Nieri-Bastos FA, Domingues LN, Martins JR, Labruna MB. 2012. Coxiella symbiont in the tick Ornithodoros rostratus (Acari: Argasidae). Ticks Tick Borne Dis 3:203–206. doi: 10.1016/j.ttbdis.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Clay K, Klyachko O, Grindle N, Civitello D, Oleske D, Fuqua C. 2008. Microbial communities and interactions in the lone star tick, Amblyomma americanum. Mol Ecol 17:4371–4381. doi: 10.1111/j.1365-294x.2008.03914.x. [DOI] [PubMed] [Google Scholar]
- 38.Duron O, Binetruy F, Noël V, Cremaschi J, McCoy KD, Arnathau C, Plantard O, Goolsby J, Pérez de León AA, Heylen DJA, Van Oosten AR, Gottlieb Y, Baneth G, Guglielmone AA, Estrada-Peña A, Opara MN, Zenner L, Vavre F, Chevillon C. 2017. Evolutionary changes in symbiont community structure in ticks. Mol Ecol 26:2905–2921. doi: 10.1111/mec.14094. [DOI] [PubMed] [Google Scholar]
- 39.Machado-Ferreira E, Vizzoni VF, Balsemão-Pires E, Moerbeck L, Gazeta GS, Piesman J, Voloch CM, Soares CA. 2016. Coxiella symbionts are widespread into hard ticks. Parasitol Res 115:4691–4699. doi: 10.1007/s00436-016-5230-z. [DOI] [PubMed] [Google Scholar]
- 40.Guizzo MG, Parizi LF, Nunes RD, Schama R, Albano RM, Tirloni L, Oldiges DP, Vieira RP, Oliveira WHC, Leite MS, Gonzales SA, Farber M, Martins O, Vaz IDS, Jr, Oliveira PL. 2017. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep 7:17554. doi: 10.1038/s41598-017-17309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li LH, Zhang Y, Zhu D. 2018. Effects of antibiotic treatment on the fecundity of Rhipicephalus haemaphysaloides ticks. Parasit Vectors 11:242. doi: 10.1186/s13071-018-2807-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang CM, Li NX, Zhang TT, Qiu ZX, Li Y, Li LW, Liu JZ. 2017. Endosymbiont CLS-HI plays a role in reproduction and development of Haemaphysalis longicornis. Exp Appl Acarol 73:429–438. doi: 10.1007/s10493-017-0194-y. [DOI] [PubMed] [Google Scholar]
- 43.Gottlieb Y, Lalzar I, Klasson L. 2015. Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-like endosymbionts in ticks. Genome Biol Evol 7:1779–1796. doi: 10.1093/gbe/evv108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith TA, Driscoll T, Gillespie JJ, Raghavan R. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the Lone Star tick. Genome Biol Evol 7:831–838. doi: 10.1093/gbe/evv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cutler SJ, Vayssier-Taussat M, Estrada-Peña A, Potkonjak A, Mihalca AD, Zeller H. 2021. Tick-borne diseases and co-infection: current considerations. Ticks Tick Borne Dis 12:101607. doi: 10.1016/j.ttbdis.2020.101607. [DOI] [PubMed] [Google Scholar]
- 46.Moutailler S, Valiente Moro C, Vaumourin E, Michelet L, Tran FH, Devillers E, Cosson JF, Gasqui P, Van VT, Mavingui P, Vourc'h G, Vayssier-Taussat M. 2016. Co-infection of ticks: the rule rather than the exception. PLoS Negl Trop Dis 10:e0004539. doi: 10.1371/journal.pntd.0004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macaluso KR, Sonenshine DE, Ceraul SM, Azad AF. 2002. Rickettsial infection in Dermacentor variabilis (Acari: Ixodidae) inhibits transovarial transmission of a second Rickettsia. J Med Entomol 39:809–813. doi: 10.1603/0022-2585-39.6.809. [DOI] [PubMed] [Google Scholar]
- 48.Baldridge GD, Burkhardt NY, Simser JA, Kurtti TJ, Munderloh UG. 2004. Sequence and expression analysis of the ompA gene of Rickettsia peacockii, an endosymbiont of the Rocky Mountain wood tick, Dermacentor andersoni. Appl Environ Microbiol 70:6628–6636. doi: 10.1128/AEM.70.11.6628-6636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kazar J. 2005. Coxiella burnetii infection. Ann N Y Acad Sci 1063:105–114. doi: 10.1196/annals.1355.018. [DOI] [PubMed] [Google Scholar]
- 50.Liu H, Li Q, Zhang X, Li Z, Wang Z, Song M, Wei F, Wang S, Liu Q. 2016. Characterization of rickettsiae in ticks in northeastern China. Parasit Vectors 9:498. doi: 10.1186/s13071-016-1764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng Y, Leung HC, Yiu S-M, Chin FY. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 53.Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langdon WB. 2015. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min 8:1. doi: 10.1186/s13040-014-0034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarri C, Stamatis C, Sarafidou T, Galara I, Godosopoulos V, Kolovos M, Liakou C, Tastsoglou S, Mamuris Z. 2014. A new set of 16S rRNA universal primers for identification of animal species. Food Control 43:35–41. doi: 10.1016/j.foodcont.2014.02.036. [DOI] [Google Scholar]
- 57.Hu L. 2009. Medical statistics. People's Military Doctor Press, Beijing, China. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.02138-22-s0001.pdf, PDF file, 0.4 MB (463.2KB, pdf)
Data Availability Statement
The generated mNGS data have been deposited into the GenBank database under accession number PRJNA841387 and are accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/PRJNA841387. The nucleotide sequences obtained in the present study were deposited into the GenBank database under accession numbers from ON016515 to ON016529.