Introduction
Precision medicine with targeted therapies based on genetic alterations has greatly advanced the treatment of many solid tumors. Although tissue genotyping is presently considered the gold standard for molecular analysis in solid tumors, it is invasive and only provides a snapshot of the molecular aberration in the tumor. When combined with the need for on-going disease monitoring, recurring tissue biopsies create undue patient burden and limit our ability to manage disease progression.
Liquid biopsy is rapidly emerging to address the unmet clinical need for noninvasive detection of actionable mutations in cancers. Liquid biopsy based on cell-free circulating tumor DNA (ctDNA), which can serve as surrogates for the entire tumor genome, can be used to monitor response to treatments and assess the emergence of drug resistance leading to targeted therapies such as actionable epidermal growth factor receptor (EGFR) mutations in nonsmall cell lung carcinoma (NSCLC) patients.1
Pharmaceutical companies have developed several effective therapies, such as gefitinib (Tarceva), erlotinib (Iressa), and osimertinib (AZD9291), that use small-molecule tyrosine kinase inhibitors (TKIs) to target distinct tyrosine kinase domains of the EGFR gene. TKIs have demonstrated impressive response rates and survival benefits2 and have been approved by the Food and Drug Administration (FDA) for the treatment of advanced NSCLC. Thus, knowing the EGFR mutation status is critical to guide precision therapy and prognostic assessment of patient health.
Our group has performed two prospective blinded clinical studies to detect actionable EGFR mutations in NSCLC patients with the novel electric field-induced release and measurement (EFIRM) electrochemical liquid biopsy technology.3,4 Using plasma and saliva from NSCLC patients, we had previously found that two front-line actionable EGFR mutations (L858R and exon19 del) were detectable in saliva of both blinded cohorts of 44 and 67 NSCLC patients with 96% and 100% sensitivity/concordance, respectively, compared with biopsy-based genotyping. These studies confirmed the existence of ctDNA in the saliva of NSCLC patients.
EFIRM is an electrochemical platform that uses electric fields to enhance molecular hybridization reactions with immobilized DNA probes and followed by enzymatic signal amplification to detect ctDNA.5 This electrochemical liquid biopsy technology has been developed to specifically capture and monitor actionable oncogene mutations in biofluids.3,5,6 Besides advanced lung cancer patients, a recent study reported that EFIRM is capable of detecting ctDNA in patients with early-stage NSCLC.7
Methods
Sample preparation
Two groups of saliva samples were procured from four healthy individuals. The first group was collected with Spectrum SDNA-1000 collector (Spectrum Solutions Inc., Draper, UT) following the direction of the manual. The control saliva sample was collected from each donor without adding the preservative solution. Three synthetic oligos harboring one of three EGFR mutations (p.L858R, g.Exon19 Del and p.T790M) were spiked into each saliva sample with the same concentration of 100 pM. To test the linearity of each assay with/without preservative, the mixture of all three mutated oligos was diluted with each saliva sample to final 75%, 50%, 25%, and 10% of the original 100 pM concentration.
Mutation detection with EFIRM assays
All the saliva samples with spike-in oligos or oligo mixtures were blinded and randomized by a biostatistician from the University of California Los Angeles Medicine Statistics Core. Three EFIRM assays targeting EGFR p.L858R, g.Exon19 Del, and p.T790M mutations were performed as described previously8 and each sample was measured in duplicate on two plates.
Results
In this study, we tested three EFIRM assays with the saliva collected using Spectrum SDNA-1000 collector and matched to saliva without any preservative. The three assays targeted two of the most common baseline sensitizing EGFR mutations: p.L858R and g.Exon19 Del, and one common acquired resistant mutation p.T790M, respectively. Oligos harboring one of the three mutations were spiked into saliva samples collected from four healthy donors with/without Spectrum preservative solution. Each group containing four individual saliva samples was blinded and tested with three EFIRM assays for each mutation detection.
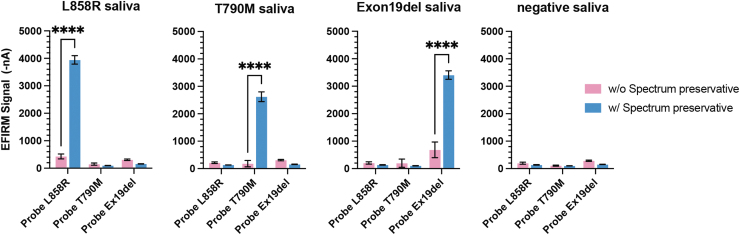
Compared with the whole saliva without any preservative, the EFIRM signals for different mutations have an average 5.0- to 14.4-fold increase (p < 0.0001) in saliva with Spectrum preservative (Fig. 1). Intriguingly, the intra-assay coefficient of variability (CV) of all three different assays with Spectrum-preserved saliva is 5.0%, which is significantly decreased from 26.8% with nonpreserved saliva samples.
FIG. 1.
Comparison of EGFR mutation detection with mutation-harboring oligos spiked-in saliva samples with or without Spectrum preservative solution. Saliva samples collected from four different healthy subjects with or without Spectrum preservative solution. Three EFIRM assays for L858R, T790M, and exon19 del detection were tested. The p-value was determined by two-way analysis of variance and the error bar represents the standard error in duplicates of four samples. ****Means p < 0.0001. EFIRM, electric field-induced release and measurement; EGFR, epidermal growth factor receptor.
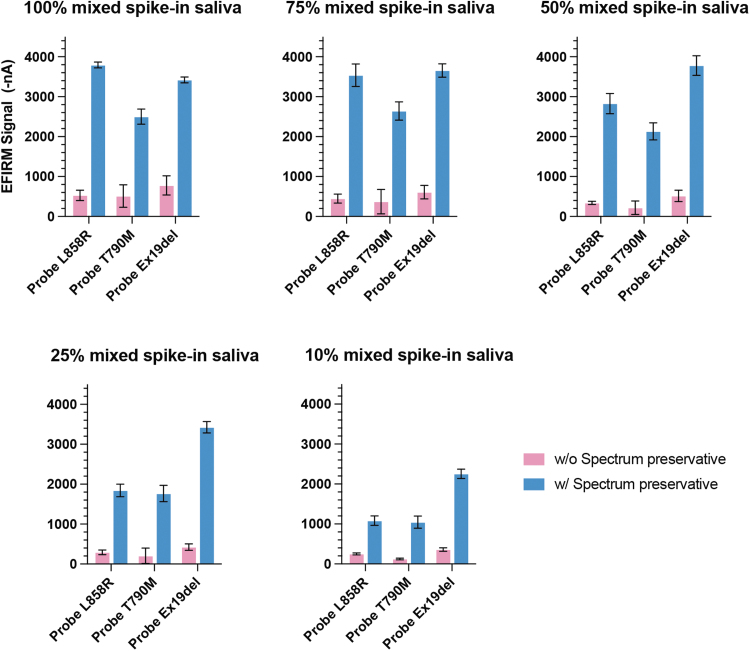
We further test the mixture of all three oligos spiked into saliva samples collected from four healthy individuals with or without Spectrum-preserved solution. The concentrations of spike-in oligo mixture were diluted to 75%, 50%, 25%, and 10% with preserved or nonpreserved saliva from each donor. As shown in Figure 2, each EFIRM assay with preserved saliva (blue bars) has much higher signals than those with nonpreserved saliva (purple bars) at different target concentrations. The average fold increases of the EFIRM signal with saliva from the Spectrum device are 6.8, 7.7, and 6.4 with L858R, T790M, and Exon19 del assay, respectively. The decrease of intra-assay CV from 35.5% to 7.5% with the Spectrum preservative solution was observed compared with neat saliva.
FIG. 2.
Comparison of EGFR mutation detection with different concentrations of mixed spike-in oligos in saliva samples with or without Spectrum preservative solution. Saliva samples were collected from four different healthy subjects with or without Spectrum preservative solution. The mixed spike-in oligos harboring EGFR L858R, T790M, and exon19 del, respectively, were diluted with each preserved or nonpreserved saliva sample.
Discussion
The Spectrum SDNA-1000 collector significantly boosted the EFIRM detection of EGFR ctDNA oligos in saliva. Considering the half-life of DNA in saliva at room temperature is ∼4.2 hours,9 the signal amplification likely is not primarily from the protection of DNA targets by Spectrum preservative solution. A possible reason for the enhancement in the sensitivity for DNA oligo hybridization might be within the proprietary Spectrum preservative, which was initially developed for viral RNA detection.
We suspect that chaotropic anions at higher concentrations can lower the electrostatic repelling of the oligo backbone, which impacts the structure and thermostability of the oligos. The advantageous characteristics of chaotropic anions have been successfully applied in nucleic acid hybridization.10 The significant decrease of intra-assay CV indicated the Spectrum preservative might reduce the effects of saliva matrix on hybridization reactions.
The Spectrum preservative solution is designed for nucleic acid protection. The preservative used in this study was designed for nucleic acid protection/stabilization in saliva. The DNA has been stored in the preservative across a range of temperatures from −80°C to 40°C. The long-term stability has remained intact throughout. No significant changes in the quantity and integrity of DNA targets were observed after three frozen and thaw cycles when tested with Polymerase Chain Reaction (PCR) or Next Generation Sequencing (NGS) methods. The performance of this preservative solution with blood/serum samples remains to be verified in the future.
There are some limitations of this study. The concentration of spike-in oligo used in this analytical study is much higher than the ctDNA concentration under physiological conditions. The impact of the preservative on the detection limit of different EFIRM assays needs to be further investigated. Furthermore, the performance of the Spectrum preservative solution with clinical samples and the potential mechanism of the stimulation of the EFIRM signal remain to be investigated.
In summary, the Spectrum preservative solution dramatically and significantly increased the sensitivity of EFIRM detection of all three EGFR mutations with up to ∼14-folds signal boost (p < 0.0001). It also improved the reproducibility of the assay by decreasing the variability with saliva collected between individuals. These attributes are important foundational utilities for liquid biopsy for early cancer assessment and treatment monitoring.
Author Disclosure Statement
D.T.W.W. has equity in Liquid Diagnostics LLC and RNAmeTRIX. He is a consultant to Colgate-Palmolive and GSK. David Vigerust is CSO for Spectrum Solutions and holds financial interest. D.V. is the VP of product development for Spectrum Solutions.
Funding Information
This study was supported by Spectrum Solutions Research Grant 20212918 and the National Institutes of Health (PHS grants UH2 CA206126 and U01 CA233370 [D.T.W.W.]; R21 CA239052 [F.L.]).
References
- 1. Heitzer E, Ulz P, Geigl JB. Circulating tumor DNA as a liquid biopsy for cancer. Clin Chem 2015;61(1):112–123; doi: 10.1373/clinchem.2014.222679. [DOI] [PubMed] [Google Scholar]
- 2. Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12(8):735–742; doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 3. Wei F, Lin CC, Joon A, et al. Noninvasive saliva-based EGFR gene mutation detection in patients with lung cancer. Am J Respir Crit Care Med 2014;190(10):1117–1126; doi: 10.1164/rccm.201406-1003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pu D, Liang H, Wei F, et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: A pilot study. Thorac Cancer 2016;7(4):428–436; doi: 10.1111/1759-7714.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wei F, Yang J, Wong DT. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens Bioelectron 2013;44:115–121; doi: 10.1016/j.bios.2012.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Li F, Wei F, Huang WL, et al. Ultra-short circulating tumor DNA (usctDNA) in plasma and saliva of non-small cell lung cancer (NSCLC) patients. Cancers (Basel) 2020;12(8):2041; doi: 10.3390/cancers12082041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wei F, Strom CM, Cheng J, et al. Electric field-induced release and measurement liquid biopsy for noninvasive early lung cancer assessment. J Mol Diagn 2018;20(6):738–742; doi: 10.1016/j.jmoldx.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tu M, Cheng J, Chen YL, et al. Electric field-induced release and measurement (EFIRM): Characterization and technical validation of a novel liquid biopsy platform in plasma and saliva. J Mol Diagn 2020;22(8):1050–1062; doi: 10.1016/j.jmoldx.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yao W, Mei C, Nan X, et al. Evaluation and comparison of in vitro degradation kinetics of DNA in serum, urine and saliva: A qualitative study. Gene 2016;590(1):142–148; doi: 10.1016/j.gene.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 10. Tao SC, Li Y, Liu YH, et al. Room-temperature hybridization of target DNA with microarrays in concentrated solutions of guanidine thiocyanate. Biotechniques 2003;34(6):1260–1262; doi: 10.2144/03346mt01. [DOI] [PubMed] [Google Scholar]