ABSTRACT
To study the effect of different tolerable levels of constitutive mcr-1 expression on Escherichia coli, and to provide direct evidence for moderate resistance mediated by mcr-1, construction of E. coli strains carrying mcr-1 on the chromosome with promoters of different strengths was conducted using λ-red recombination. Our results demonstrated that over-high expression of mcr-1 cannot be tolerated, and seven constructs with more than 200-fold mcr-1 transcriptional expression differences were obtained. The colistin MICs of the seven strains increased with the increase of MCR-1 levels, and the highest MIC was 8 μg/mL. Lower expression of mcr-1 didn’t demonstrate many effects on bacteria, while higher tolerable expression of mcr-1 tended to show fitness costs in growth rate, competitive ability, and cell structures, but no obvious change of virulence was observed in mice. Bacteria demonstrated colistin MICs of 4–8 μg/mL at mcr-1 expression levels similar to clinical isolates, which were the mcr-1 expression levels with relatively lower fitness costs.
IMPORTANCE The effects of relatively lower tolerable levels of mcr-1 were not evaluated thoroughly, and direct evidence for moderate resistance mediated by mcr-1 was lacking. In the present study, we made constructs carrying mcr-1 on the E. coli K12 chromosome under the control of serial constitutive promoters of different strengths and studied the effects of different tolerable levels of mcr-1 expression in vitro and in vivo. The results demonstrated that generally, except QH0007 (the construct with the highest mcr-1 expression that showed some extent of cell death), the fitness costs of tolerable mcr-1 expression on bacteria were not apparent or low. Bacteria demonstrated colistin MICs of 4–8 μg/mL at mcr-1 expression levels similar to clinical isolates, which corresponded to the lower levels of mcr-1 expression that can lead to colistin resistance, indicating the cleverness of bacteria to balance the benefit and cost of MCR-1-mediated colistin resistance.
KEYWORDS: tolerable mcr-1 expression, colistin resistance, fitness cost
INTRODUCTION
Polymyxins are a group of nonribosomally synthesized, cationic, cyclic peptide antibiotics discovered in 1947 (1). The mechanism by which polymyxins (polymyxin B and colistin) kill Gram-negative pathogens relies on disruption of membrane permeability through polar and hydrophobic interactions (2). Of these interactions, an electrostatic interaction is present between positively charged residues of polymyxins and the negatively charged lipid A moieties of lipopolysaccharides (LPS) anchored on the outer leaflet of the bacterial membrane. Potential nephrotoxicity and neurotoxicity of polymyxins hamper their wide introduction into regular clinical therapies. However, with the emergence of carbapenem-resistant bacteria (especially in Pseudomonas aeruginosa, Acinetobacter baumannii, Klebsiella pneumoniae and Escherichia coli), polymyxins have become the last line of defense against drug-resistant Gram-negative bacterial infections worldwide.
With increased use in clinics, animal husbandry, and agriculture, the therapeutic effect of colistin on Gram-negative bacteria has been relatively reduced, and colistin resistance emerged. One of the most worrisome is the mobile colistin resistance gene mcr-1 (usually plasmid borne), first discovered in animal-derived E. coli in 2015 (3). The inner membrane protein MCR-1 can catalyze the transfer of phosphoethanolamine (PEA) to lipid A, resulting in colistin resistance (4). Colistin MICs displayed by mcr-1-positive E. coli are moderate (usually 2–8 μg/mL) compared to the level of colistin resistance (usually 8–256 μg/mL) mediated by, for example, increased expression of pmrA/pmrB, inferring that the expression of mcr-1 (and hence phosphoethanolamine modification of lipid A) is tightly controlled. This, in part, is supported by the report that mcr-1 was usually carried by plasmids of a relatively low copy number (5).
Several groups have studied the in vitro and in vivo fitness costs of mcr-1-mediated colistin resistance, but with inconsistent results (6–10). These may at least partly relate to the complexity of the plasmid structures, as the plasmid backbone was reported to have strong effects on fitness burden (11). As certain genes in the mcr-1-carrying plasmid may compensate for the fitness cost in colistin-resistant strains, the fitness cost of a plasmid may be different according to plasmid type and bacterial host (10). The fitness of high expression levels of mcr-1 was studied by induction of an overexpressing plasmid carrying mcr-1, and decreased growth rate, cell viability, competitive ability, and significant degradation in cell membrane and cytoplasmic structures were found (12). However, the effects of relatively lower tolerable levels of mcr-1 were not evaluated thoroughly, and direct evidence for moderate resistance mediated by mcr-1 was lacking.
Here, we made constructs carrying mcr-1 on the E. coli K12 chromosome under the control of serial constitutive promoters of different strengths and studied the effects of different tolerable levels of mcr-1 expression in vitro and in vivo in aspects of cell growth rate, antibiotic susceptibility, competitive ability, cell membrane and cytoplasmic structures, and virulence. The results will provide more information for understanding how bacteria manage to benefit from mcr-1 expression with lower costs, and why usually moderate resistance was mediated.
RESULTS
Strain construction and mcr-1 transcriptional expression levels.
Isogenic strains were constructed with mcr-1 transcriptionally fused to each of the seven constitutive promoters of different strengths on the chromosome. The strains were named QH0001 (J23213), QH0002 (J23112), QH0003 (J23113), QH0004 (J23117), QH0005 (J23115), QH0006 (J23105) and QH0007 (J23110), respectively, with the corresponding promoters listed in the parentheses. The transcriptional expression levels of mcr-1 varied over a 200-fold range as measured by qPCR, with the strongest promoter expressing mcr-1 about 186-fold the level of the control genes (Table 1). The three clinical mcr-1-carrying isolates, i.e., E. coli 08–85, E. coli 13–66, E. coli 13–43, demonstrated mcr-1 transcriptional expression of 17- to 34-fold of the control genes, corresponding to levels between QH0004 and QH0005 in the serial constructed strains (Table 1).
TABLE 1.
Strain characteristics as a function of mcr-1 transcription levela
| Strainsb | Description | Promoter sequence | mRNA levelc Mean ± SD |
Relative growth rated Mean ± SD | Colistin MICse |
Protein levelf Mean ± SD |
NPN uptake [%]g Mean ± SD |
|---|---|---|---|---|---|---|---|
| K12 (DA5438) | MG1655 | 1.00 ± 0.01 | 0.125 | 0.04 ± 0.02 | 0 ± 0.72 | ||
| QH0001 | J23213-mcr-1 | 5′-CTG ATG GCT AGC TCA GTC CTA GGG ATA GTG CTA GC-3′ | 0.7 ± 0.22 | 0.98 ± 0.01 | 0.25 | 0.34 ± 0.06 | −0.35 ± 0.34 |
| QH0002 | J23112-mcr-1 | 5′-CTG ATA GCT AGC TCA GTC CTA GGG ATT ATG CTA GC-3′ | 1.4 ± 0.78 | 0.98 ± 0.01 | 0.5 | 0.48 ± 0.18 | 0.23 ± 0.41 |
| QH0003 | J23113-mcr-1 | 5′-CTG ATG GCT AGC TCA GTC CTA GGG ATT ATG CTA GC-3′ | 2.5 ± 0.69 | 0.98 ± 0.02 | 1 | 0.51 ± 0.42 | 0.13 ± 0.54 |
| QH0004 | J23117-mcr-1 | 5′-TTG ACA GCT AGC TCA GTC CTA GGG ATT GTG CTA GC-3′ | 10.0 ± 2.37 | 0.98 ± 0.01 | 4 | 0.83 ± 0.45 | 0.12 ± 0.63 |
| QH0005 | J23115-mcr-1 | 5′-TTT ATA GCT AGC TCA GCC CTT GGT ACA ATG CTA GC-3′ | 102.6 ± 66.05 | 0.97 ± 0.01 | 8 | 1.29 ± 0.27 | 0.86 ± 0.35 |
| QH0006 | J23105-mcr-1 | 5′-TTT ACG GCT AGC TCA GTC CTA GGT ACT ATG CTA GC-3′ | 177.8 ± 96.50 | 0.98 ± 0.01 | 8 | 3.14 ± 0.57 | 0.90 ± 0.14 |
| QH0007 | J23110-mcr-1 | 5′-TTT ACG GCT AGC TCA GTC CTA GGT ACA ATG CTA GC-3′ | 185.7 ± 34.28 | 0.88 ± 0.02 | 8 | 7.96 ± 2.64 | 16.69 ± 0.36 |
| E. coli 08-85 | mcr-1 carrying | NA | 33.96 ± 11.74 | ND | 4 | ND | ND |
| E. coli 13-66 | mcr-1 carrying | NA | 24.76 ± 8.53 | ND | 8 | ND | ND |
| E. coli 13-43 | mcr-1 carrying | NA | 16.95 ± 3.63 | ND | 8 | ND | ND |
NA, not applicable; ND, not determined.
QH0001 to QH0007 are constructs isogenic to MG1655 with the mcr-1 gene being placed downstream of J23-series promoters in the galK locus. E. coli 08–85, E. coli 13–66, and E. coli 13–43 are clinical mcr-1-carrying strains.
mRNA levels of mcr-1 relative to control genes hcaT and cysG, with standard deviations.
Exponential growth rates relative to E. coli K12 with standard deviations; the experiment was conducted three times on different days, and representative results from one experiment are shown.
MICs of colistin against E. coli 08–85, E. coli 13–66, and E. coli 13–43 were determined previously (13).
MCR-1 protein levels quantified by western blot band intensity analysis with a densitometer (n = 3).
NPN uptake rates with 0.1% Triton X-100-treated cells set as the 100% NPN uptake control.
MCR-1 detection by Western blotting.
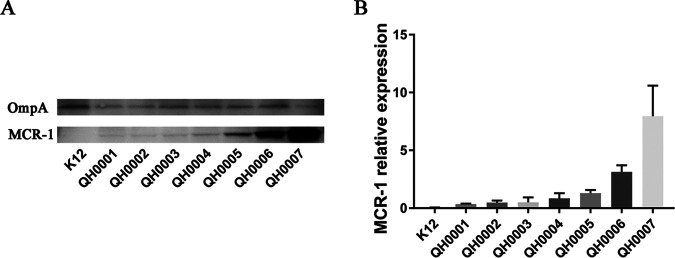
The MCR-1 protein level in each strain was detected by Western blotting with OmpA as the internal control. As shown in Fig. 1, while the control protein OmpA showed similar levels in each strain, the MCR-1 levels differed. The parental strain E. coli K12 (without mcr-1) showed no band in MCR-1 detection, and the mcr-1-carrying strains (QH0001–QH0007) demonstrated gradually increased MCR-1 protein levels with the increase of promoter strength. The band intensity results analyzed with a densitometer showed that with the increase of the promoter activity, mcr-1 expression levels increased from 0.34 in QH0001 to 7.96 in QH0007 (Table 1).
FIG 1.
Western blot detection of MCR-1 protein levels in the mcr-1 constructs. (A) Western blot image for MCR-1 protein detection; (B) MCR-1 band intensity quantification using a densitometer. The experiment was repeated three times; the representative image is shown in panel A, and the MCR-1 quantification results from the three experiments are shown in panel B.
Susceptibility of the strains to different antibiotics.
The broth microdilution method was used to determine the MICs of different antibiotics against the strains (Table 2). The MICs of colistin and polymyxin B increased as the expression of mcr-1 increased, the highest MICs of the two antibiotics were 8 μg/mL in the serial strains. However, the MICs of aztreonam, ceftazidime, gentamicin, and rifampin showed no apparent change as the expression of MCR-1 increased.
TABLE 2.
MICs of compounds against strains expressing different levels of mcr-1
| Compounds | ATCC 25922 | MIC (μg/mL) against different E. colia |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| K12 | QH0001 | QH0002 | QH0003 | QH0004 | QH0005 | QH0006 | QH0007 | ||
| Colistin | 0.25 | 0.125 | 0.25 | 0.5 | 1 | 4 | 8 | 8 | 8 |
| Polymyxin B | 0.5 | 0.25 | 0.25 | 0.5 | 1 | 2 | 4 | 8 | 4 |
| Aztreonam | 0.06 | 0.125 | 0.125 | 0.25 | 0.125 | 0.125 | 0.25 | 0.125 | 0.125 |
| Ceftazidime | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 | 0.5 | 0.5 | 0.25 | 0.25 |
| Gentamycin | 0.25 | 0.25 | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 | 0.25 | 0.5 |
| Rifampin | 8 | 8 | 16 | 8 | 8 | 8 | 8 | 8 | 8 |
MICs were determined in CAMH broth by microdilution methods according to CLSI guidelines.
Determination of exponential growth rates.
In order to determine whether the expression of mcr-1 has fitness costs on bacterial growth, Bioscreen was used to determine the growth rates of strains expressing different levels of mcr-1. As shown in Table 1, QH0001, QH0002, QH0003, QH0004, QH0005 and QH0006 showed no apparent effects on growth rates in comparison to the parental strain, with relative growth rates being 0.97 to 0.98. QH0007 (the one with the strongest promoter in the current study), however, demonstrated significantly lowered growth rate, with a fitness cost (difference of the relative growth rate of QH0007 in comparison to the parental strain) of about 12%, indicating the adverse effect of over-high expression of mcr-1 on cell growth.
In vitro competition assay.
In vitro competition assay results (Fig. 2) of the constructs versus parental strain (E. coli K12) demonstrated that QH0004 had a relative fitness of 1.041 to 1.068 at 24 h, 48 h and 72 h, indicating no apparent fitness costs. However, QH0005 (relative fitness of 0.923–0.998), QH0006 (relative fitness of 0.884–0.958), and QH0007 (relative fitness of 0.109–0.645) demonstrated gradually increased fitness costs as the expression level of mcr-1 increased.
FIG 2.
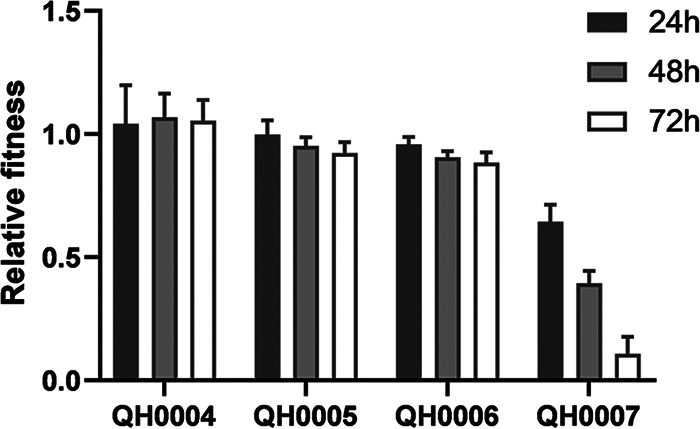
In vitro competition assay results of the mcr-1 constructs versus the parental strain (n = 9). The experiments were performed three times on different days with three independent samples each time.
E. coli morphology observation with SEM and TEM.
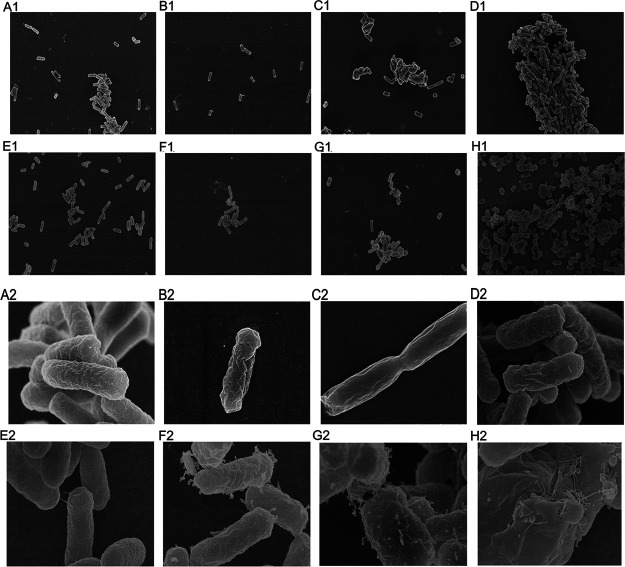
MCR-1 catalyzes the modification of lipid A in the bacterial outer membrane, hence, bacteria expressing MCR-1 may show defects in cellular morphology. We first visualized the morphology of the entire E. coli cells by scanning electron microscopy (SEM). Control cells demonstrated smooth, homogenous cell surfaces and rod-shaped morphology types (Fig. 3A1 and 3A2). QH0001 (Fig. 3B1 and 3B2), QH0002 (Fig. 3C1 and 3C1), QH0003 (Fig. 3D1 and 3D2), and QH0004 (Fig. 3E1 and 3E2) with relatively lower MCR-1 expression showed no significant changes in surface characteristics. QH0005 (Fig. 3F1 and 3F2) had some abnormal secretions on the surface of the cells; QH0006 (Fig. 3G1 and 3G2) demonstrated increased abnormal secretions, a small portion of cell membrane broke, and leakage of intracellular substance was found. The cells of QH0007 (the strain with the highest MCR-1 expression; Fig. 3H1 and 3H2) lost their normal morphology, the abnormal secretions increased, and broken cells were found.
FIG 3.
SEM micrographs of E. coli expressing different levels of mcr-1. A1–H1: K12, QH0001, QH0002, QH0003, QH0004, QH0005, QH0006, and QH0007 at 4,000-fold enlargement; A2–H2: K12, QH0001, QH0002, QH0003, QH0004, QH0005, QH0006, and QH0007 at 40,000-fold enlargement.
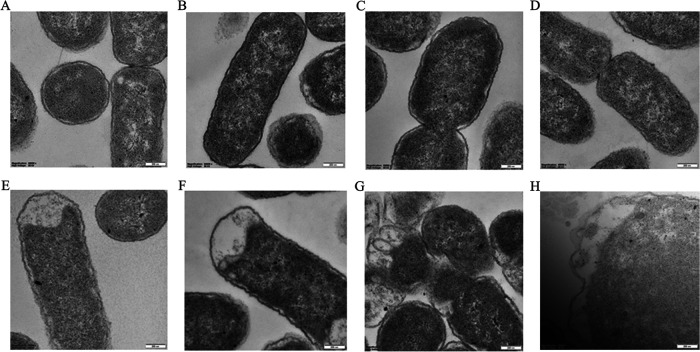
Transmission electron microscopy (TEM) images are shown in Fig. 4. The ultrastructural characteristics of the E. coli K12 cells showed homogeneous cytoplasm and intact cell membrane (Fig. 4A). There were no significant changes in cell membrane and internal structure in QH0001 (Fig. 4B) and QH0002 (Fig. 4C), the strains with relatively weaker promoters. Burr-like folds appeared in the cell membrane structure of QH0003 (Fig. 4D), indicating occurrence of damages. The vacuolar-like structure appeared in the cells of QH0004 (Fig. 4E), and the number of burr-like folds increased. The cell morphology was slightly deformed in QH0005 (Fig. 4F), a vacuolar-like structure appeared, and the cell membrane fold deformation intensified. Cell deformation was more obvious in QH0006 (Fig. 4G), the proportion of cell vacuolar structure increased, and the cell membrane structure became significantly unstable. Cell membrane structures were incomplete in some of the cells of QH0007 (Fig. 4H), and the intracellular structure was blurred.
FIG 4.
TEM micrographs of E. coli expressing different levels of mcr-1. A–H: K12, QH0001, QH0002, QH0003, QH0004, QH0005, QH0006, and QH0007 at 80,000-fold enlargement.
Membrane integrity assays.
As over-high expression of mcr-1 posted a significant effect on the cell membrane morphology of E. coli, we further tested the membrane integrity by measuring the 1-N-phenyl-naphthylamine (NPN) uptake rate. As shown in Fig. 5 and Table 1, QH0001, QH0002, QH0003, QH0004, QH0005, and QH0006 demonstrated almost the same levels of NPN uptake as the parental strain; there was no significant difference between these groups and the K12 group (P > 0.05 as demonstrated by one-way ANOVA), indicating relatively good integrity of the cell membranes. However, a significant increase in NPN uptake was found in QH0007 (16.69% when 0.1% Triton X-100 treated cells was set as the 100% NPN uptake control; P < 0.0001 in comparison to E. coli K12 as demonstrated by one-way ANOVA), suggesting damage on the cell membranes.
FIG 5.
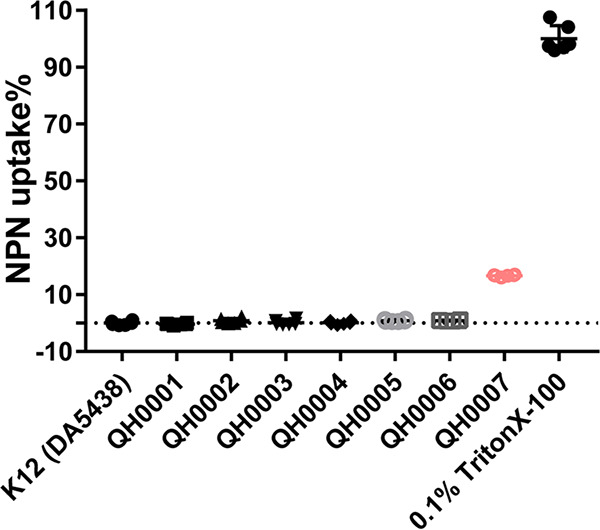
NPN uptake percentages of different strains. Three biological replicates and two technical replicates were used for each strain (six samples for each strain). E. coli K12 (DA5438) treated by 0.1% Triton X-100 was used as the positive control (NPN 100% uptake).
Virulence of different strains to mice.
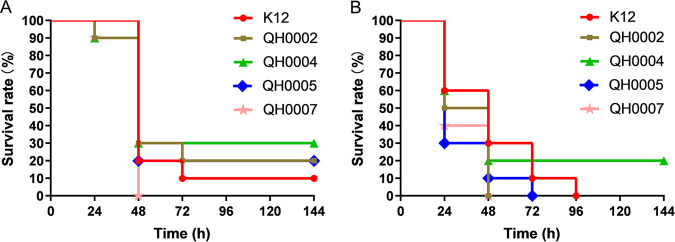
To assess the effect of different levels of MCR-1 on bacterial virulence, the mouse systemic infection model by intraperitoneal injection was used with strains E. coli K12 (DA5438), QH0002, QH0004, QH0005, and QH0007 as the challenge organisms. In the presence of 5% mucin, the infection doses were 3.0 × 106, 3.7 × 106, 3.3 × 106, 3.3 × 106, and 1.9 × 106 CFU/mouse for E. coli K12 (DA5438), QH0002, QH0004, QH0005 and QH0007, respectively. In the absence of 5% mucin, the corresponding infection doses were 1.6 × 109, 1.8 × 109, 1.8 × 109, 1.9 × 109, and 1.0 × 109 CFU/mouse, respectively. As shown in Fig. 6, no apparent virulence change can be found for the strains with the increase of the mcr-1 expression in the presence of 5% mucin (Fig. 6A) as well as in the absence of 5% mucin (Fig. 6B).
FIG 6.
Virulence of strains expressing different levels of mcr-1. (A) Mouse systemic infection by intraperitoneal injection in the presence of 5% mucin. Infection doses: E. coli K12, 3.0 × 106 CFU/mouse; QH0002, 3.7 × 106 CFU/mouse; QH0004, 3.3 × 106 CFU/mouse; QH0005, 3.3 × 106 CFU/mouse; and QH0007, 1.9 × 106 CFU/mouse; n = 10. (B) Mouse systemic infection by intraperitoneal injection in the absence of 5% mucin. Infection doses: E. coli K12, 1.6 × 109 CFU/mouse; QH0002, 1.8 × 109 CFU/mouse; QH0004, 1.8 × 109 CFU/mouse; QH0005, 1.9 × 109 CFU/mouse; and QH0007, 1.0 × 109 CFU/mouse; n = 10.
DISCUSSION
Decreased growth rate, cell viability, competitive ability, and significant degradation in cell membrane and cytoplasmic structures were found in E. coli by induction of an overexpressing plasmid carrying mcr-1 (12). However, the effects of lower tolerable levels of mcr-1, including mcr-1 levels found in clinical isolates, were not evaluated thoroughly, and direct evidence for moderate resistance mediated by mcr-1 was still lacking. In the present study, we conducted construction of isogenic strains with mcr-1 under the control of constitutive promoters of different strengths at the galK locus in the chromosome of E. coli K12. Chromosomal integration avoids complications arising from issues such as plasmid segregation or plasmid maintenance-associated metabolic burden, constant antibiotic selection pressure required for maintenance of plasmids and bacterial artificial chromosomes (BACs) in the cell (14–15).
A total of seven strains were constructed, with mcr-1 transcriptional expression levels changing over 200 folds as shown by qPCR determination, and the mcr-1 expression levels were also confirmed by Western blotting (with a range of about 23 folds). The three mcr-1-carrying clinical E. coli with colistin MICs of 4 to 8 μg/mL demonstrated mcr-1 transcriptional expression levels 17- to 34-fold of the control genes by qPCR, corresponding to mcr-1 levels between those of QH0004 and QH0005 (Table 1). The effects of different tolerable levels of mcr-1 on bacteria were then studied by growth rate determination, MIC determination, electron microscopy observation, membrane integrity assay, in vitro competition assay, and virulence to mice.
MIC results demonstrated that as mcr-1 expression increased, the MICs of colistin and polymyxin B increased, and the highest MIC that could be obtained was 8 μg/mL, consistent with clinical observations (3). The lower levels of mcr-1 expression that can confer colistin resistance in the serial strains were those of QH0004 and QH0005 (with colistin MICs of 4–8 μg/mL), which were similar to the mcr-1 expression levels in the three clinical isolates (Table 1). When a bacterium acquires resistance to an antibiotic, it usually pays a certain cost, i.e., the fitness cost (6, 16). In this study, mcr-1 expressions in QH0001 to QH0006 had no apparent or lower fitness costs on growth rate, membrane integrity, and susceptibility to other antibiotics. However, in QH0007 with the highest mcr-1 expression, a fitness cost of 12% was seen in relative growth rate, and dramatic effects on membranes (NPN uptake assay and morphology observation with SEM and TEM) were found. Consistent with the dramatic effects of mcr-1 expression on cells in QH0007, construction of strains with mcr-1 expression levels higher than that of QH0007 was not successful in our study (data not shown), indicating that the effects higher than that of QH0007 mcr-1 expression in cells may be too heavy to be tolerated. In vitro competition and electron microscopy results demonstrated the gradual effects of mcr-1 expression on strains: as the mcr-1 expression levels increased, the competition strength decreased, and cell morphology and cell structure changed.
It has been reported that colistin resistance may affect bacterial virulence in K. pneumoniae through affecting the amount of polysaccharides (17). Yang et al. (12) also reported highly attenuated virulence of clinical E. coli when transformed with mcr-1 plasmids in a Galleria mellonella infection model. However, studies from Tietgen et al. (8). stated no substantial effect of mcr-1 expression on virulence in Galleria mellonella larvae both in E. coli and K. pneumoniae. The inconsistency may lie in the variation of the strains and mcr-1-carrying plasmids used (10). To study the effects of pure tolerable mcr-1 expression on virulence, we used four of the isogenic strains together with E. coli K12 in the mouse systemic infection model by intraperitoneal injection in the presence or absence of 5% mucin. The results demonstrated no apparent virulence changes of the strains, even with the construction of the highest mcr-1 expression (Fig. 6).
In Gram-negative bacteria, LPS is synthesized in the cytoplasm via the Raetz pathway (18) and introduced into the inner leaflet of the cytoplasmic membrane (CM), which is then flipped to the outer leaflet of the CM by MsbA and transported to the outer membrane (OM) via the LptABCDEFG machinery (19). In the mcr-1-carrying isolate (normal mcr-1 expression level), MCR-1 modifies lipid A with phosphoethanolamine (pEtN) moiety as it passes through the CM on its way to the OM, resulting in most of LPS in the CM modified and colistin resistance. However, a relatively large quantity of unmodified LPS is present in the OM and serves as a target for colistin (20). Under normal expression levels of mcr-1 (between those of QH0004 and QH0005), there is a sufficient quantity of unmodified LPS in the OM for maintenance of cell integrity, and hence no apparent fitness costs were seen. As the expression level of mcr-1 increases (from QH0004 to QH0007), the quantity of unmodified LPS in the OM decreases and damage to cell membrane appears (Fig. 3 and 4). When the quantity of unmodified LPS drops to a certain point (lower than that in QH0007), cell membrane damages can be too severe to be tolerated, consistent with our observation that construction of strains with mcr-1 levels higher than that of QH0007 was not successful. In clinical isolates, bacteria manage to keep the expression of mcr-1 at the levels (between the levels of QH0004 and QH0005) that can manifest colistin resistance by LPS modification in the CM, while keeping a large quantity of unmodified LPS in the OM for normal function.
In conclusion, our results demonstrated the effects of tolerable pure mcr-1 expression on bacteria. Generally, except QH0007 (the one with the highest mcr-1 expression that showed some extent of cell death), the fitness costs of tolerable mcr-1 expressions on bacteria are not apparent, or low, supporting its wide spread in clinic. Bacteria can demonstrate colistin MICs of 4–8 μg/mL at mcr-1 expression levels similar to clinical isolates, which correspond to the lower levels of mcr-1 expression that can lead to colistin resistance, indicating the cleverness of bacteria to balance the benefit and cost of MCR-1-mediated colistin resistance.
MATERIALS AND METHODS
Media.
LB broth and LB agar were used as liquid and solid media for bacterial growth. Chloramphenicol was used at 30 μg/mL where indicated, and counter-selection of sacB was done on LB agar supplemented with 5% sucrose. BBL TM Mueller-Hinton (MH) II agar (Becton, Dickinson & Company, France) was used for MIC determinations.
Strains.
E. coli 08–85, E. coli 13–43, and E. coli 13–68 were used as the mcr-1-carrying clinical isolates from the Chinese Academy of Medical Sciences Collection Center of Pathogen Microorganisms, CAMS-CCPM-AP. The E. coli K12 (DA5438) strain was obtained from Dan I Andersson of Uppsala University. A set of isogenic strains carrying constitutive promoters of different strengths upstream of a cat–sacB cassette at the galK locus in E. coli K12 (21) was used for construction of strains with mcr-1 expressing at different tolerable levels; the promoter sequences were listed in Table 1. The coding sequence for mcr-1 was amplified from a mcr-1-carrying clinical isolate (E. coli 08–85), and transcriptionally fused by recombineering each of the promoters by replacing the cat–sacB cassette using methods listed in literature (22). PCR and DNA sequencing (Invitrogen) were used to confirm the genetic constructions. The resulting strains were named QH0001, QH0002, QH0003, QH0004, QH0005, QH0006, and QH0007. The primers used were as follows: mcr-1 amplification primers, mcr1-galK-FP: 5′-AGC AGT ACT GTT ACT AGA GAA AGA GGA GAA ATA CTA GAT GAT GCA GCA TAC TTC TGT GTG-3′, mcr1-galK-RP: 5′-AAC GCA AAA AGC CCC GAG CGG TTA AAC TCA GGG CTT TAT TTT TAT CAG CGG ATG AAT GCG GTG CGG TC-3′; Insertion check primers, sYFP-FP: 5′-CAT GGA TCA GCT AAT TTC CG-3′, galk-RP: 5′-TTG TAT TCG CTG CCA ACC AG-3′.
Laboratory animals.
CD-1 (ICR) mice (18–21 g, half male, half female) were purchased from Vital River Laboratories (Beijing, China). All animals were housed under controlled humidity (30–70%) and temperature (22 ± 3°C), and a 12 h light-dark cycle. Animals had free access to food and water during the study. All the animal studies complied with the animal husbandry guidelines, and all animal experiments were performed according to national standards for laboratory animals in China (GB/T 35892-2018) (23), with approval from the Laboratory Animal Welfare and Ethics Committee in Institute of Medicinal Biotechnology, Peking Union Medical College.
RNA extraction and qPCR.
RNA extractions were made in three biological replicates. RNA was isolated with cultures of OD600 at 0.3–0.4 using an RNAPrep Pure Cell/Bacteria kit (TIANGEN, cat no. DP430) and quantified using a NanoDrop ND-1000 spectrophotometer. FastQuant RT kits (with gDNase) (TIANGEN, cat no. KR106) were used to synthesize cDNA. Quantitation of the mRNA levels for the mcr-1 was performed on the 7500 Fast real-time PCR system (Applied Biosystems) with the PowerUp SYBR green master mix (Applied Biosystems) according to the manufacturer’s instructions. Primers for RT-PCR were synthesized by Sangon Biotech (Shanghai). PCR amplification was carried out in a total volume of 20 μL, containing 8 μL 1:10, 1:100, 1:1000, and 1:10000 diluted cDNA solution, 10 μL of 2 × PowerUp SYBR green master mix, and 1 μL of each primer. Control housekeeping genes were cysG and hcaT. Primers used were as follows: qPCR-mcr1-FP: 5′-GCT CCA AAA TGC CCT ACA GA-3′; qPCR-mcr1-RP: 5′-CTT GGT AGC ACA CCC AAA CC-3′; qPCR-cysG-FP: 5′-TTG TCG GCG GTG GTG ATG TC-3′; qPCR-cysG –RP: 5′-ATG CGG TGA ACT GTG GAA TAA ACG-3′; qPCR-hcaT-FP: 5′-GCT GCT CGG CTT TCT CAT CC-3′; qPCR-hcaT-RP: 5′-CCA ACC ACG CTG ACC AAC C-3′.
Western blot analysis.
A total of 4–5 single colonies were inoculated in 3 mL LB medium and incubated at 37°C overnight. Cell pellets from 2 mL of overnight cultures were collected by centrifugation at 8,000 rpm for 10 min, washed three times with physiological saline, and resuspended in 200 μL of radioimmunoprecipitation (RIPA) lysate (Beyotime, cat no. P0013C). The cell suspensions were sonicated for 1.5 min (work 3 s, stop 6 s, with frequency of 28 KHz), centrifuged at 14,000 rpm, 4°C for 10 min, and the supernatants were collected. Equal amounts of proteins (100 μg/lane) from each bacterial lysate were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Hybond-P; GE Healthcare). Blots were probed with the Polymyxin resistance protein MCR-1 primary antibody (polyclonal mouse anti-E. coli polymyxin resistance protein MCR-1 antibody; LS Bio), followed by horseradish enzyme labeled goat antimouse IgG (ZB-2305; ZSGB-Bio). Protein bands were visualized using an enhanced chemiluminescence (ECL) detection method (Bio-Rad), and band intensities were analyzed with a densitometer (LAS-4000; GE Healthcare). The experiment was repeated 3 times. OmpA, measured quantitatively using an OmpA antibody (polyclonal rabbit anti-Salmonella typhi OmpA antibody; LS Bio), followed by horseradish enzyme labeled goat anti-rabbit IgG (ZB-2301; ZSGB-Bio), was used as the control.
MIC determination.
The MICs of the antibacterial agents for all constructed E. coli isolates were determined by the broth microdilution method according to CLSI guidelines (24). The final inoculum in each well was about 5 × 105 CFU/mL. The microtiter plates were incubated at 37°C for about 18 h, and the results were recorded by naked eyes. MICs were defined as the lowest concentrations of antibiotics that can inhibit the visible growth of the tested organisms.
Measurement of growth rates.
The exponential growth rates of E. coli K12 and the mcr-1-carrying constructs were measured in MH broth at 37°C by taking optical density at 600 nm (OD600) every 4 min in a Bioscreen C reader (Oy Growth Curves Ab Ltd.) (25). Four independent cultures per strain were grown overnight until saturation. The cultures were diluted 1,000-fold and aliquoted into a Bioscreen C plate in duplicate (0.3 mL/well). The growth rates were estimated from the OD600 interval between 0.01 and 0.1, where the growth was observed to be exponential. Relative growth rates of the strains were calculated by comparing the growth rates with that of E. coli K12.
Competition experiments in vitro.
To assess the fitness cost of different mcr-1 expression levels, the four mcr-1-carrying strains with relatively strong promoters (QH0004, QH0005, QH0006, and QH0007, which showed high colistin MICs that can be selected out on plates containing 2 μg/mL colistin) were subjected to in vitro competition assay against E. coli K12. Strains were cultured in LB broth overnight, after that, strain cultures were adjusted to the 0.5 McFarland standard and 10-μL aliquots of each competitor was mixed with E. coli K12 at a 1:1 ratio. The mixtures were then inoculated in 10 mL LB broth, and incubated at 37°C with shaking, the mixed population was diluted 1,000-fold into fresh LB broth every 24 h until the competition experiment had lasted for 72 h. The total number of bacteria- and colistin-resistant cells were determined by spreading properly diluted samples of each competition mixture on nonselective (without colistin) and selective (with 2 μg/mL colistin) LB agar plates at 0, 24, 48, and 72 h. The formula RF = (log10 S1t− log10 S10)/(log10 S2t− log10 S20) (26) was used to calculate the relative fitness (RF), where S1 and S2 represent CFU densities of the constructed isolates and E. coli K12, respectively, and t is the measure time in hours. If there exists a fitness cost between the competitors, then RF<1; if not, RF>1.
Morphology observation with scanning electron microscopy (SEM) and transmission electron microscopy (TEM).
The morphology of E. coli strains with different levels of mcr-1 expression was observed by TEM and SEM. E. coli strains were grown in cation-adjusted Mueller-Hinton (CAMH) broth overnight at 37°C; 2 mL of the stationary-phase cell cultures were collected and fixed with 2.5% glutaraldehyde for at least 24 h at 4°C. For SEM (27), the samples were then centrifuged to remove glutaraldehyde and resuspended in phosphate buffer. The bacterial suspensions were spotted on a polished silicon wafer and dried overnight in a biosafety cabinet. After drying, the samples were coated with chromium and subjected to SEM imaging using Hitachi-SEM-SU8010. For TEM (12), the fixed organisms were washed and further fixed with 1% osmium tetroxide. The samples were washed, dehydrated in a graded series of ethanol, and embedded in Epon Araldite. Ultrathin sections containing the cells were then placed on copper grids, stained with uranyl acetate and lead citrate, observed, and photographed with a JEM-1400 plus.
Outer membrane integrity assays.
Overnight cell cultures were diluted 50-fold in fresh LB medium and incubated until midlog phase (OD600 of about 0.5). The cell cultures were then centrifuged, the pellets were washed 3 times with 5 mM HEPES buffer containing 20 mM glucose (pH 7.2) and resuspended in the same buffer. Then, 100-μL cell suspensions were then mixed with 100 μL of the 20 μM 1-N-phenyl-naphthylamine (NPN) solution (prepared in the same buffer as the cell suspensions) in each well of the 96-well black clear-bottom plates. Fluorescence was read immediately (within 3 min) in an EnSpire 2300 (excitation wavelength 355 nm, emission wavelength 420 nm). Three biological replicates and two technical replicates were used for each strain. E. coli K12 (DA5438) treated by 0.1% Triton X-100 was used as the positive control. Percent NPN uptake was calculated for each strain as previously described (3, 28):
where Fobs is the observed fluorescence of a given strain, F0 is the initial fluorescence of NPN with E. coli K12 (DA5438) without treatment, and F100 is the fluorescence of NPN with E. coli K12 (DA5438) upon addition of 0.1% Triton X-100.
Murine systemic infection model.
E. coli strains K12, QH0002, QH0004, QH0005, and QH0007 were subjected to in vivo virulence comparison using the murine systemic infection model by intraperitoneal injection in the presence or absence of 5% mucin. For infections in the presence of 5% mucin, cell pellets from fresh overnight cell cultures were first washed and resuspended in saline (adjusted to 0.5 McFarland), then further diluted 10-fold in 5% mucin. Then, 0.5 mL of the bacterial suspensions in 5% mucin were injected intraperitoneally to each mouse randomly allocated to different groups with a final infection dose of 2–4 × 106 CFU/mouse. Animal deaths were recorded at 24, 48, 72, 96, 120, and 144 h postinfection, and the cumulative survival rates for each time point were calculated. For infections in the absence of 5% mucin, cell pellets from fresh overnight cell cultures were washed in saline, and then resuspended in saline and adjusted to 50 McFarland; 0.2 mL of the bacterial suspensions were then injected intraperitoneally to each mouse randomly allocated to different groups with a final infection dose of 1–2 × 109 CFU/mouse. Animal deaths were recorded at 24, 48, 72, 96, 120, and 144 h postinfection, and the cumulative survival rates for each time point were calculated.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (32141003), the CAMS Innovation Fund for Medical Sciences (CIFMS) (2021-I2M-1-030, 2021-1-12M-039, and 2022-I2M-2-002), the Fundamental Research Funds for the Central Universities (2021-PT350-001), and the National Science and Technology Infrastructure of China (NPRC-32).
We declare no conflicts of interest.
Contributor Information
Congran Li, Email: congranli@imb.pumc.edu.cn.
Xuefu You, Email: xuefuyou@imb.pumc.edu.cn.
Matthew J. Culyba, University of Pittsburgh
REFERENCES
- 1.Ainsworth GC, Brown AM, Brownlee G. 1947. Aerosporin, an antibiotic produced by Bacillus aerosporus Greer. Nature 159:263. doi: 10.1038/160263a0. [DOI] [PubMed] [Google Scholar]
- 2.Hancock RE. 1997. The bacterial outer membrane as a drug barrier. Trends Microbiol 5:37–42. doi: 10.1016/S0966-842X(97)81773-8. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 4.Wei P, Song G, Shi M, Zhou Y, Liu Y, Lei J, Chen P, Yin L. 2018. Substrate analog interaction with MCR-1 offers insight into the rising threat of the plasmid-mediated transferable colistin resistance. FASEB J 32:1085–1098. doi: 10.1096/fj.201700705R. [DOI] [PubMed] [Google Scholar]
- 5.Bontron S, Poirel L, Nordmann P. 2016. Real-time PCR for detection of plasmid-mediated polymyxin resistance (mcr-1) from cultured bacteria and stools. J Antimicrob Chemother 71:2318–2320. doi: 10.1093/jac/dkw139. [DOI] [PubMed] [Google Scholar]
- 6.Nang SC, Morris FC, McDonald MJ, Han ML, Wang J, Strugnell RA, Velkov T, Li J. 2018. Fitness cost of mcr-1-mediated polymyxin resistance in Klebsiella pneumoniae. J Antimicrob Chemother 73:1604–1610. doi: 10.1093/jac/dky061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Liao K, Gao H, Wang Q, Wang X, Li H, Wang R, Wang H. 2017. Decreased itfness and virulence in ST10 Escherichia coli harboring blaNDM-5 and mcr-1 against a ST4981 strain with blaNDM-5. Front Cell Infect Microbiol 7:242. doi: 10.3389/fcimb.2017.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tietgen M, Semmler T, Riedel-Christ S, Kempf VAJ, Molinaro A, Ewers C, Göttig S. 2018. Impact of the colistin resistance gene mcr-1 on bacterial fitness. Int J Antimicrob Agents 51:554–561. doi: 10.1016/j.ijantimicag.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Ma K, Feng Y, Zong Z. 2018. Fitness cost of a mcr-1-carrying IncHI2 plasmid. PLoS One 13:e0209706. doi: 10.1371/journal.pone.0209706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi Y, Lee JY, Lee H, Park M, Kang K, Lim SK, Shin D, Ko KS. 2020. Comparison of fitness cost and virulence in chromosome- and plasmid-mediated colistin-resistant Escherichia coli. Front Microbiol 11:798. doi: 10.3389/fmicb.2020.00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Humphrey B, Thomson NR, Thomas CM, Brooks K, Sanders M, Delsol AA, Roe JM, Bennett PM, Enne VI. 2012. Fitness of Escherichia coli strains carrying expressed and partially silent IncN and IncP1 plasmids. BMC Microbiol 12:53. doi: 10.1186/1471-2180-12-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Q, Li M, Spiller OB, Andrey DO, Hinchliffe P, Li H, MacLean C, Niumsup P, Powell L, Pritchard M, Papkou A, Shen Y, Portal E, Sands K, Spencer J, Tansawai U, Thomas D, Wang S, Wang Y, Shen J, Walsh T. 2017. Balancing mcr-1 expression and bacterial survival is a delicate equilibrium between essential cellular defence mechanisms. Nat Commun 8:2054. doi: 10.1038/s41467-017-02149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang Z, Pang J, Hu X, Nie T, Lu X, Li X, Wang X, Li C, Yang X, You X. 2021. Low prevalence of mcr-1 among clinical Enterobacteriaceae isolates and co-transfer of mcr-1 and blaNDM-1 from separate donors. Microb Drug Resist 27:476–484. doi: 10.1089/mdr.2020.0212. [DOI] [PubMed] [Google Scholar]
- 14.Guan L, Liu Q, Li C, Zhang Y. 2013. Development of a fur-dependent and tightly regulated expression system in Escherichia coli for toxic protein synthesis. BMC Biotechnol 13:25. doi: 10.1186/1472-6750-13-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham DS, Koepsel RR, Ataai MM, Domach MM. 2009. Factors affecting plasmid production in Escherichia coli from a resource allocation standpoint. Microb Cell Fact 8:27. doi: 10.1186/1475-2859-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson DI, Levin BR. 1999. The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493. doi: 10.1016/s1369-5274(99)00005-3. [DOI] [PubMed] [Google Scholar]
- 17.Choi MJ, Ko KS. 2015. Loss of hypermucoviscosity and increased fitness cost in colistin-resistant Klebsiella pneumoniae sequence type 23 strains. Antimicrob Agents Chemother 59:6763–6773. doi: 10.1128/AAC.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem 76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sabnis A, Hagart KL, Klöckner A, Becce M, Evans LE, Furniss RCD, Mavridou DA, Murphy R, Stevens MM, Davies JC, Larrouy-Maumus GJ, Clarke TB, Edwards AM. 2021. Colistin kills bacteria by targeting lipopolysaccharide in the cytoplasmic membrane. Elife 10:e65836. doi: 10.7554/eLife.65836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garoff L, Yadav K, Hughes D. 2018. Increased expression of Qnr is sufficient to confer clinical resistance to ciprofloxacin in Escherichia coli. J Antimicrob Chemother 73:348–352. doi: 10.1093/jac/dkx375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci USA 97:5978–5983. PMCID: PMC18544. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Standardization Administration of the People’s Republic of China. 2018. Laboratory animal -- Guideline for ethical review of animal welfare. GB/T 35892–2018, Beijing, China. https://www.chinesestandard.net/PDF.aspx/GBT35892-2018. [Google Scholar]
- 24.Clinical and Laboratory Standards Institute (CLSI). 2020. Performance standards for antimicrobial susceptibility testing. M100, 30th ed. CLSI. Malvern, PA. [Google Scholar]
- 25.Ahmad A, Zachariasen C, Christiansen LE, Græsbøll K, Toft N, Matthews L, Damborg P, Agersø Y, Olsen JE, Nielsen SS. 2015. Pharmacodynamic modelling of in vitro activity of tetracycline against a representative, naturally occurring population of porcine Escherichia coli. Acta Vet Scand 57:79. doi: 10.1186/s13028-015-0169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Machuca J, Briales A, Labrador G, Díaz-de-Alba P, López-Rojas R, Docobo-Pérez F, Martínez-Martínez L, Rodríguez-Baño J, Pachón ME, Pascual A, Rodríguez-Martínez JM. 2014. Interplay between plasmid-mediated and chromosomal-mediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J Antimicrob Chemother 69:3203–3215. doi: 10.1093/jac/dku308. [DOI] [PubMed] [Google Scholar]
- 27.Qu Y, Wang J, Zhou H, Ma Q, Zhang Z, Li D, Shen W, Zhou J. 2016. Concentration-dependent effects of carbon nanotubes on growth and biphenyl degradation of Dyella ginsengisoli LA-4. Environ Sci Pollut Res Int 23:2864–2872. doi: 10.1007/s11356-015-5532-1. [DOI] [PubMed] [Google Scholar]
- 28.MacNair CR, Stokes JM, Carfrae LA, Fiebig-Comyn AA, Coombes BK, Mulvey MR, Brown ED. 2018. Overcoming mcr-1 mediated colistin resistance with colistin in combination with other antibiotics. Nat Commun 9:458. doi: 10.1038/s41467-018-02875-z. [DOI] [PMC free article] [PubMed] [Google Scholar]