Current psychiatric research, evaluation, and treatment rely on a diagnostic system based almost exclusively on subjective data, such as self-reported experiences and questionnaires, rather than on more objective markers. Recent advances in computational power have enabled the increasing use of data-driven computational approaches aiming to identify objective neural markers for diagnosing psychopathology (i.e., classifying subjects as either patients or healthy control subjects), predicting illness onset and development, and predicting treatment response. The pursuit of objective markers is mostly in its infancy, highlighting the need for additional efforts, better collaborative endeavors across labs, and large datasets, which would further advance the clinical applications of data-driven computational approaches in psychiatry.
In the current issue of Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, Fitzgerald et al. (1) examined whether whole-brain hippocampal resting-state functional connectivity (rsFC), assessed shortly after exposure to trauma with functional magnetic resonance imaging (fMRI), can forecast future onset of posttraumatic stress disorder (PTSD) symptoms (1). Ninety-eight patients with traumatic injury were recruited during or immediately after hospitalization in the emergency department. Patients completed an rsFC scan within the first month after the injury and a clinical evaluation of PTSD 6 months later, based on the Clinician-Administered PTSD Scale for DSM-5 (CAPS-5). Whole-brain rsFC with bilateral hippocampi seeds were extracted, and a multivariate pattern analysis (MVPA) approach was used to examine the utility of whole-brain distributed hippocampal rsFC patterns in predicting individual PTSD symptoms. The results indicated that acute hippocampal rsFC significantly predicted CAPS-5 scores 6 months later, confirming previous findings indicating the key role the hippocampus plays in PTSD, possibly owing to its involvement in memory functions (consolidation and retrieval) and fear-related learning processes (extinction recall). This study is particularly interesting in that it shows that functional integration of the hippocampal rsFC across the whole brain, measured acutely after exposure to trauma, can function as a significant biomarker for the future development of PTSD symptoms.
The key strengths of the current study are its longitudinal design, the type of data used (both resting-state fMRI and clinical data), and the data analyses methods used. First, using a longitudinal design, this study incorporates neuroimaging data collected in the acute aftermath of trauma exposure to predict the risk of developing PTSD 6 months later. Predicting PTSD symptomology based on hippocampus whole-brain connectivity patterns immediately after trauma exposure extends our current knowledge on the role of the hippocampus in the onset of PTSD and of its course. Second, using fMRI-based rsFC data has several advantages over traditional task-based fMRI research. Unlike task-based fMRI research, which is characterized by limited test–retest reliability and failure to replicate findings—two features crucial to trusting emerging scientific results (2)—rsFC methods have been increasingly shown to yield greater reliability and reproducibility, especially when large-scale connectivity patterns between distributed brain networks are examined. In addition, low performance demands from participants during scans yields high compliance, which in turn minimizes behavioral confounds that are normally found in longer task-based fMRI scanning. These advantages, as well as the standardization and relatively low cost of rsFC, make rsFC especially suitable for clinical applications. Third, by using an MVPA method, Fitzgerald et al. (1) addressed well-documented shortcomings of group-level univariate analysis, which most rsFC studies in PTSD have been using to identify biomarkers. As univariate methods permit inference only at the group level and assume that the covariance across neighboring voxels is not informative, the application of univariate methods to clinical population is significantly limited.
Data-driven computational approaches and MVPA have gained traction in recent years. They enable better access to whole-brain spatial distribution patterns of neural activation and to FC patterns across multiple voxels when predicting experimental variables (3), providing highly valuable information at the individual level. Several machine learning approaches have been used for multivariate pattern analysis in rsFC studies, offering the opportunity to search for or identify the most useful disorder-specific neural patterns across the entire brain, free of the constraints of traditional univariate schemes. Specifically, the MVPA method helps clarify how rsFC of multiple voxels might collectively correspond to a given diagnosis, symptom severity, or cognitive event. To date, there are several MVPA methods (Figure 1A), including methods based on correlation (4), regression (5), support vector machines (6), support vector regression (7), and deep learning neural networks (8). Fitzgerald et al. (1) used a kernel ridge regression MVPA approach to explore whether the functional integration of the hippocampi across the whole brain can forecast future PTSD symptom severity.
Figure 1.
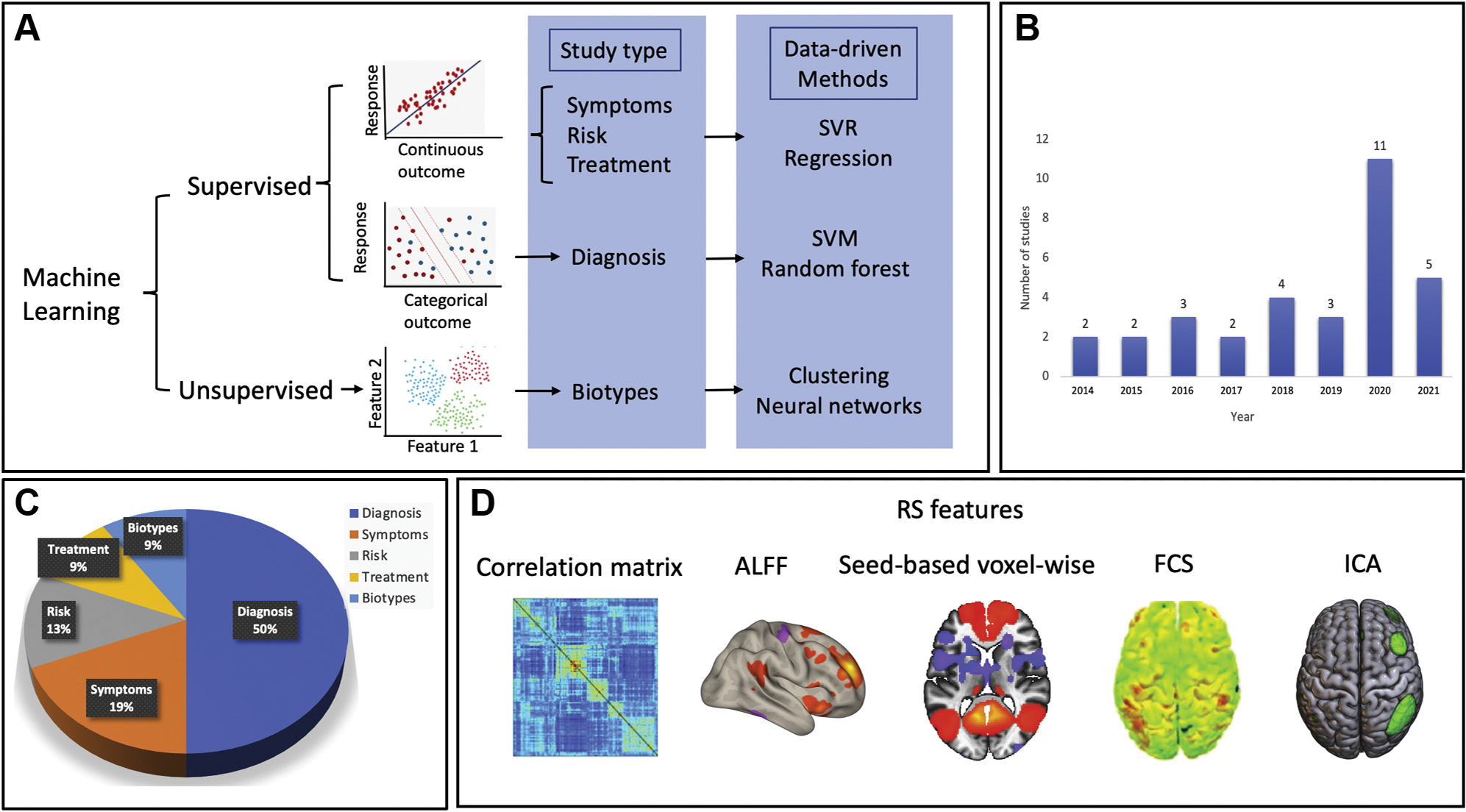
Snapshot of resting-state functional magnetic resonance imaging (RS fMRI) studies using data-driven approaches/multivariate predictive models. We searched PubMed for original neuroimaging research articles using RS fMRI in posttraumatic stress disorder published between January 2000 and October 2021. The search terms included “posttraumatic stress disorder,” “machine learning” or “data-driven,” and “resting-state fMRI” or “functional connectivity.” Nonhuman and nonclinical studies were excluded, as well as those that did not use multivariate pattern analysis. The initial search yielded 87 studies, of which 32 studies were selected based on a full-text review. (A) (Top) Categories of machine learning methods. (B) Growth of machine learning studies using RS fMRI in posttraumatic stress disorder since 2015. (C) Use of machine learning models. “Diagnosis” refers to patient versus control subject classification, “symptoms” refers to the prediction of continuous symptom scores, “risk” refers to classification of groups that are at high risk, “treatment” refers to predictions of individual differences in disease progression and response to an intervention, and “biotypes” refers to the identification of subgroups of patients based on brain patterns. (D) Frequently used RS functional connectivity measures in these studies including region of interest to region of interest–based correlation matrix, amplitude of low-frequency fluctuation (ALFF), seed-based region of interest–to-voxel whole-brain measures, voxel-to-voxel whole-brain–wise functional connectivity strength (FCS), and independent component analysis (ICA). SVM, support vector machine; SVR, support vector regression.
In recent years, the application of MVPA methods aided by data-driven computational approaches/machine learning has been acknowledged by multiple labs, resulting in a dramatic increase in the number of machine learning studies using rsFC in PTSD (Figure 1B). Machine learning research can be broadly grouped into two categories: supervised and unsupervised (Figure 1A). In supervised learning, a priori known categorical (e.g., diagnostic labels of groups) or continuous (e.g., PTSD symptom severity) outcome variables are used to train or “teach” a model to make accurate prediction of a new “unseen” dataset. In unsupervised learning, mathematical algorithms are used to reveal or uncover a presumably existing structure in the data, without a priori knowing the true labels of the data (Figure 1A). Initially, supervised machine learning efforts in PTSD primarily focused on the development of automatic tools for identifying a PTSD diagnosis. About 50% of the rsFC studies we reviewed (n = 16) focused on distinguishing PTSD patients from control subjects or one PTSD subtype from another (e.g., PTSD and major depressive disorder vs. PTSD alone; PTSD with vs. without dissociative symptoms) (Figure 1C) (6), yielding moderate to high accuracy rates. These studies suggest that the underlying neural circuitry of PTSD, as reflected in rsFC data, may begin to characterize the phenotype of PTSD. Only 6 studies (19%) used rsFC to predict symptom severity collected cross-sectionally at the same time point as the imaging data, with even fewer studies using baseline rsFC to predict a future PTSD diagnosis or treatment response (4 studies [13%] used rsFC to predict PTSD and 3 studies [9%] used rsFC to predict treatment response) (Figure 1C).
Key barriers still exist before such tools can be clinically useful, as PTSD is not only a highly heterogeneous disorder but also a highly comorbid disorder, especially with major depressive disorder (as seen in approximately 50% of patients with PTSD), complicating efforts to identify brain markers for diagnosis, risk prediction, and treatment efficacy (9). Since techniques allowing for complex multiple diagnoses in psychiatry are challenging, a shift toward the exploration of unique biotypes that go beyond the boundaries of traditional diagnoses using unsupervised machine learning (e.g., a PTSD–major depressive disorder biotype) may be useful. Yet to date, only 3 studies (9%) have examined subtypes of PTSD using unsupervised machine learning methods (Figure 1A).
Lastly, to develop reliable, replicable, and clinically useful markers for diagnosis, illness course, and treatment response, large, heterogeneous datasets are highly needed. To date, most studies are single-site studies exploring small homogeneous samples, which tend to yield better performance compared with studies using larger samples owing to what is known as overfitting—when a machine learning model is too complex and, though working well in the training dataset, fails to generalize to an unseen one. Unfortunately, in PTSD, no study has explored the reproducibility of findings across multiple sites. The results of single-site studies are also more difficult to interpret and compare across studies owing to major differences in analytical approaches, scanners, acquisition parameters, and data processing pipelines. In addition, most studies estimate the model’s performance within a single cohort dataset, in which all samples are used in building the prediction model. Thus, no testing of the model’s performance is conducted using an independent unseen test dataset. Importantly, however, for machine learning models to be useful in real-world clinical settings, an evaluation of the model’s generalizability and robustness using unseen independent cohorts is needed, which can be achieved only when using samples that are large enough. Fortunately, several large-scale studies have been made possible by the recent establishment of several research consortia across healthy participants (the Human Connectome Project [HCP]), adolescents (the Adolescent Brain Cognitive Development [ABCD] Study), older participants (the Alzheimer’s Disease Neuroimaging Initiative [ADNI]), and more importantly and relevant in the present context, across psychiatric patient populations (Enhancing Neuro Imaging Genetics through Meta Analysis [ENIGMA]) (10). These consortia promote model development based on large samples, which are more likely to generalize across scanners and commonly encountered variations in study procedures, enabling tests on independent datasets with different characteristics. The increasing number of collaborative efforts will allow us to share predictive models that have been rigorously tested and make an important contribution to translational psychiatric research.
Acknowledgments and Disclosures
This work was supported by National Institute of Mental Health Grant Nos. R01 MH105355 and R01 MH072833 (to YN), Israel Science Foundation Grant No. 374/20 (to AL), and a Brain and Behavior Research Foundation Grant and National Institute of Mental Health Grant No. K01 MH122774 (to XZ).
Footnotes
The authors report no biomedical financial interests or potential conflicts of interest.
Contributor Information
Yuval Neria, Department of Psychiatry, Columbia University Irving Medical Center, New York, New York; New York State Psychiatric Institute, Columbia University Irving Medical Center, New York, New York.
Amit Lazarov, School of Psychological Sciences, Tel Aviv University, Tel Aviv, Israel..
Xi Zhu, Department of Psychiatry, Columbia University Irving Medical Center, New York, New York; New York State Psychiatric Institute, Columbia University Irving Medical Center, New York, New York.
References
- 1.Fitzgerald JM, Webb EK, Weis CN, Huggins AA, Bennett KP, Miskovich TA, et al. (2022): Hippocampal resting-state functional connectivity forecasts individual posttraumatic stress disorder symptoms: A data-driven approach. Biol Psychiatry Cogn Neurosci Neuroimaging 7:139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elliott ML, Knodt AR, Ireland D, Morris ML, Poulton R, Ramrakha S, et al. (2020): What is the test–retest reliability of common task-functional MRI measures? New empirical evidence and a meta-analysis. Psychol Sci 31:792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis T, LaRocque KF, Mumford JA, Norman KA, Wagner AD, Poldrack RA (2014): What do differences between multi-voxel and univariate analysis mean? How subject-, voxel-, and trial-level variance impact fMRI analysis. Neuroimage 97:271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P (2001): Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293:2425–2430. [DOI] [PubMed] [Google Scholar]
- 5.Zandvakili A, Barredo J, Swearingen HR, Aiken EM, Berlow YA, Greenberg BD, et al. (2020): Mapping PTSD symptoms to brain networks: A machine learning study. Transl Psychiatry 10:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zilcha-Mano S, Zhu X, Suarez-Jimenez B, Pickover A, Tal S, Such S, et al. (2020): Diagnostic and predictive neuroimaging biomarkers for posttraumatic stress disorder. Biol Psychiatry Cogn Neurosci Neuroimaging 5:688–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebois LAM, Li M, Baker JT, Wolff JD, Wang D, Lambros AM, et al. (2021): Large-scale functional brain network architecture changes associated with trauma-related dissociation. Am J Psychiatry 178:165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sheynin S, Wolf L, Ben-Zion Z, Sheynin J, Reznik S, Keynan JN, et al. (2021): Deep learning model of fMRI connectivity predicts PTSD symptom trajectories in recent trauma survivors. Neuroimage 238:118242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neria Y (2021): Functional neuroimaging in PTSD: From discovery of underlying mechanisms to addressing diagnostic heterogeneity. Am J Psychiatry 178:128–135. [DOI] [PubMed] [Google Scholar]
- 10.Thompson PM, Jahanshad N, Ching CRK, Salminen LE, Thomopoulos SI, Bright J, et al. (2020): ENIGMA and global neuroscience: A decade of large-scale studies of the brain in health and disease across more than 40 countries. Transl Psychiatry 10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]


