ABSTRACT
Necrotizing fasciitis is a severe infectious disease that results in significant mortality. Streptococcus pyogenes (group A Streptococcus, GAS) is one of the most common bacterial pathogens of monomicrobial necrotizing fasciitis. The early diagnosis of necrotizing fasciitis is crucial; however, the typical cutaneous manifestations are not always presented in patients with GAS necrotizing fasciitis, which would lead to miss- or delayed diagnosis. GAS with spontaneous inactivating mutations in the CovR/CovS two-component regulatory system is significantly associated with destructive diseases such as necrotizing fasciitis and toxic shock syndrome; however, no specific marker has been used to identify these invasive clinical isolates. This study evaluated the sensitivity and specificity of using CovR/CovS-controlled phenotypes to identify CovR/CovS-inactivated isolates. Results showed that the increase of hyaluronic acid capsule production and streptolysin O expression were not consistently presented in CovS-inactivated clinical isolates. The repression of SpeB is the phenotype with 100% sensitivity of identifying in CovS-inactivated isolates among 61 clinical isolates. Nonetheless, this phenotype failed to distinguish RopB-inactivated isolates from CovS-inactivated isolates and cannot be utilized to identify CovR-inactivated mutant and RocA (Regulator of Cov)-inactivated isolates. In this study, we identified and verified that PepO, the endopeptidase which regulates SpeB expression through degrading SpeB-inducing quorum-sensing peptide, was a bacterial marker to identify isolates with defects in the CovR/CovS pathway. These results also inform the potential strategy of developing rapid detection methods to identify invasive GAS variants during infection.
IMPORTANCE Necrotizing fasciitis is rapidly progressive and life-threatening; if the initial diagnosis is delayed, deep soft tissue infection can progress to massive tissue destruction and toxic shock syndrome. Group A Streptococcus (GAS) with inactivated mutations in the CovR/CovS two-component regulatory system are related to necrotizing fasciitis and toxic shock syndrome; however, no bacterial marker is available to identify these invasive clinical isolates. Inactivation of CovR/CovS resulted in the increased expression of endopeptidase PepO. Our study showed that the upregulation of PepO mediates a decrease in SpeB-inducing peptide (SIP) in the covR mutant, indicating that CovR/CovS modulates SIP-dependent quorum-sensing activity through PepO. Importantly, the sensitivity and specificity of utilizing PepO to identify clinical isolates with defects in the CovR/CovS pathway, including its upstream RocA regulator, were 100%. Our results suggest that identification of invasive GAS by PepO may be a strategy for preventing severe manifestation or poor prognosis after GAS infection.
KEYWORDS: CovR/CovS, PepO, RocA, SpeB, SIP, group A Streptococcus
INTRODUCTION
Necrotizing fasciitis is a bacterial infectious disease with mortality exceeding 70%, characterized by rapidly progressive soft tissue necrosis, sepsis, and toxic shock syndrome (1). Among Gram-positive bacteria, Streptococcus pyogenes (group A Streptococcus, GAS) is the leading pathogen causing monomicrobial necrotizing fasciitis (1). Cutaneous manifestations are not always initially present in patients with GAS necrotizing fasciitis (1, 2). Therefore, the infection is often misdiagnosed or the correct diagnosis is delayed (1, 3), which could result in severe acute complications in patients. Developing predictive markers for identifying high-risk patients is crucial for preventing and diagnosing necrotizing fasciitis. Currently, early diagnosis of necrotizing fasciitis is dependent on the observation of manifestations in the clinic and the detection of surrogate markers (e.g., The Laboratory Risk Indicator for Necrotizing Fasciitis [LRINEC] scoring system) in the laboratory (1, 4). Nonetheless, whether the LRINEC scoring system is useful for the early recognition of GAS necrotizing fasciitis is still under debate (5–8). Also, no bacterial biomarker is available for the early diagnosis of GAS necrotizing fasciitis.
GAS isolates are classified by the widely used single-locus sequence emm-typing system. More than 250 different emm types have been identified, with emm1- and emm3-GAS frequently associated with severe clinical manifestations (9–11). Furthermore, spontaneous inactivating mutations in genes encoding the CovR/CovS (control of virulence) two-component regulatory system are significantly related to necrotizing fasciitis and toxic shock syndrome (12–14). CovR is phosphorylated by CovS and the phosphorylated response CovR protein mediates the transcriptional repression of virulence factors including streptolysin S (SLS), streptolysin O (SLO), streptokinase, DNase Sda1, hyaluronic acid capsule, M protein, and C5a peptidase (15–19). The spontaneous CovR/CovS-inactivated mutations occur during GAS infection; the derepression of virulence factors expression in both covS and covR mutants contributes to increased resistance to immune clearance, enhances invasion activity, and may increase the risk of systemic infection (14, 18, 20, 21). Intriguingly, the phenotypes of the covS-deletion and covR-deletion mutants are not identical. The expression of SpeB cysteine protease, DNase Spd3, and protein G-related α2 M-binding protein (Grab) is upregulated in the covR-deletion mutant but downregulated in the covS-deletion mutant (14, 20–22). In this study, therefore, we referred to the covS-deletion, CovS-truncation, and CovS kinase-inactivated isolates as the CovS-inactivated isolates (expressing nonphosphorylated CovR) and to the covR-deletion and CovR-truncation isolates as the CovR-inactivated isolates (have no CovR protein). The phosphorylation of CovR is also modulated by RocA (regulator of Cov) in a CovS-dependent manner (23–26). Previous studies have demonstrated that rocA mutants show decreased levels of CovR phosphorylation and increased bacterial virulence (26–28). Notably, emm3 isolates have a truncated RocA; this truncation is responsible for increased capsule expression and may contribute to the association of emm3 GAS with severe manifestations (29, 30).
The emm typing and identification of rocA truncation or CovR/CovS spontaneous mutations by Sanger sequencing are time- and labor-consuming. The increased hyaluronic acid capsule expression in the covR/covS mutants would result in an encapsulated or mucoid colony morphology; however, we found that 69% (20/29) of CovS-inactivated clinical isolates did not have a distinguishable encapsulated colony morphology, indicating that colony morphology is not a reliable characteristic of CovS-inactivated isolates. The early diagnosis of necrotizing fasciitis is critically important. As an alternative indicator of infection severity (e.g., the LRINEC scoring system), this study suggested that the bacterial marker PepO endopeptidase could be utilized to monitor the appearance of CovR/CovS- and RocA-inactivated mutants during GAS infection. Also, this finding reveals a novel strategy, identification of bacterial markers of CovR/CovS-inactivated mutants during infection, for the prevention of invasive GAS infection.
RESULTS
Encapsulated colony morphology is not a reliable phenotype for the identification of CovS-inactivated clinical isolates.
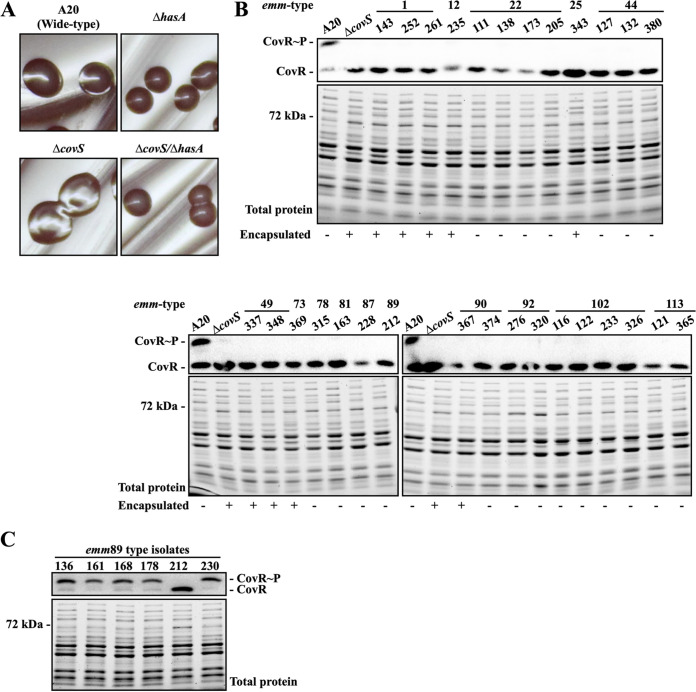
CovR/CovS regulates hyaluronic acid capsule synthesis by inhibiting transcription of the hasABC operon (16, 19); therefore, an encapsulated (or mucoid) colony morphology is the typical phenotype of covR and covS mutants (Fig. 1A). Our previous study identified, by Phostag Western blotting (Fig. 1B), 29 CovS-inactivated clinical isolates (31). In this study, we found that only nine isolates (31%) exhibited a distinguishable encapsulated colony morphology (Fig. 1B and data not shown). hasA and hasB, but not hasC, are required for the hyaluronic acid capsule synthesis (32). Therefore, the hasA and hasB sequences of the 20 CovS-inactivated isolates with a normal colony morphology were analyzed, and the results showed that SPY163 (emm81), SPY228 (emm87), SPY173 (emm22), and SPY121 (emm113) had frameshift mutations in hasA or hasB (Table 1). In addition, the emm89-type isolate (SPY212) had no hasABC operon (data not shown). Due to the lack of a hasABC operon, the colony morphology of the CovS-inactivated SPY212 was not distinguishable from those of other emm89 isolates (Fig. 1C and Fig. S1). The remaining 15 CovS-inactivated isolates possessed no deletion or frameshift mutations in hasA and hasB. The emm22, emm102, and emm113 isolates had missense mutations in hasA and hasB (Table 1); however, whether these amino acid replacements are involved in inactivating HasA and HasB activity is unknown. These results indicated that in isolates with or without the functional hasABC operon, an encapsulated colony morphology is not a reliable phenotype for identifying CovS-inactivated isolates.
FIG 1.
Colony morphology and expression of phosphorylated CovR in the selected clinical isolates and in the wild-type A20 strain and its covS isogenic mutants. (A) Colony morphology of the wild-type A20 strain, the covS mutant, and their hasA mutants. (B) Expression of phosphorylated CovR and the total protein profile upon SDS-PAGE of selected clinical isolates. (C) Phosphorylation level of CovR in the emm89 isolates. The phosphorylated CovR protein was detected by Phostag Western blotting. Encapsulated colonies are denoted by a plus (+) and unencapsulated colonies are denoted by a minus (−) as shown below the SDS-PAGE gel and Western blot images.
TABLE 1.
Nucleotide sequence of hasA and hasB in the phosphorylated CovR-negative and non-encapsulated GAS isolatesa
| Isolate no. | emm type | hasA | hasB |
|---|---|---|---|
| SPY111 | 22 | S109N, D402V | |
| SPY138 | 22 | S109N | |
| SPY173 | 22 | S109N; c394t = stop at aa 132 | |
| SPY205 | 22 | S109N | |
| SPY127 | 44 | ||
| SPY132 | 44 | ||
| SPY380 | 44 | ||
| SPY315 | 78 | ||
| SPY163 | 81 | g224t = stop at aa 82 | |
| SPY228 | 87 | 1 bp insert = stop at aa 47 | |
| SPY212 | 89 | Absent | Absent |
| SPY374 | 90 | ||
| SPY276 | 92 | ||
| SPY320 | 92 | ||
| SPY116 | 102 | I41V | I235V, R305Q, N400G |
| SPY122 | 102 | I41V | I235V, R305Q, N400G |
| SPY233 | 102 | I41V | I235V, R305Q, N400G |
| SPY326 | 102 | I41V | I235V, R305Q, N400G |
| SPY121 | 113 | Y52S; 1 bp delete = stop at aa 105 | |
| SPY365 | 113 | Y232D | Y52S |
aa, amino acid. Reference sequences (NCBI accession no.): emm22: NZ_CP035438.1, NZ_CAAIOC010000006.1, NZ_CAAJES010000006.1, NZ_CAAIJZ010000003.1; emm44: NZ_CAAIYK010000002.1, NZ_CAAIXB010000002.1; emm78: CP035437.1, NZ_CAAJDX010000002.1; emm81: CP027771.1, NZ_CAAJCN010000008.1; emm87: NZ_CAAIZD010000010.1, NZ_CAAIUZ010000009.1; emm90: CP035444.1, NZ_CAAJFV010000002.1; emm102: NZ_CAAIOC010000006.1, NZ_CAAHKE010000006.1, NZ_CAAJES010000006.1; emm113: NZ_CAAIYN010000002.1, NZ_CAAIXY010000002.1, NZ_CAAIXF010000003.1, CAAHKQ010000017.1.
SpeB expression level is the negative predictive marker of CovS-inactivated isolates.
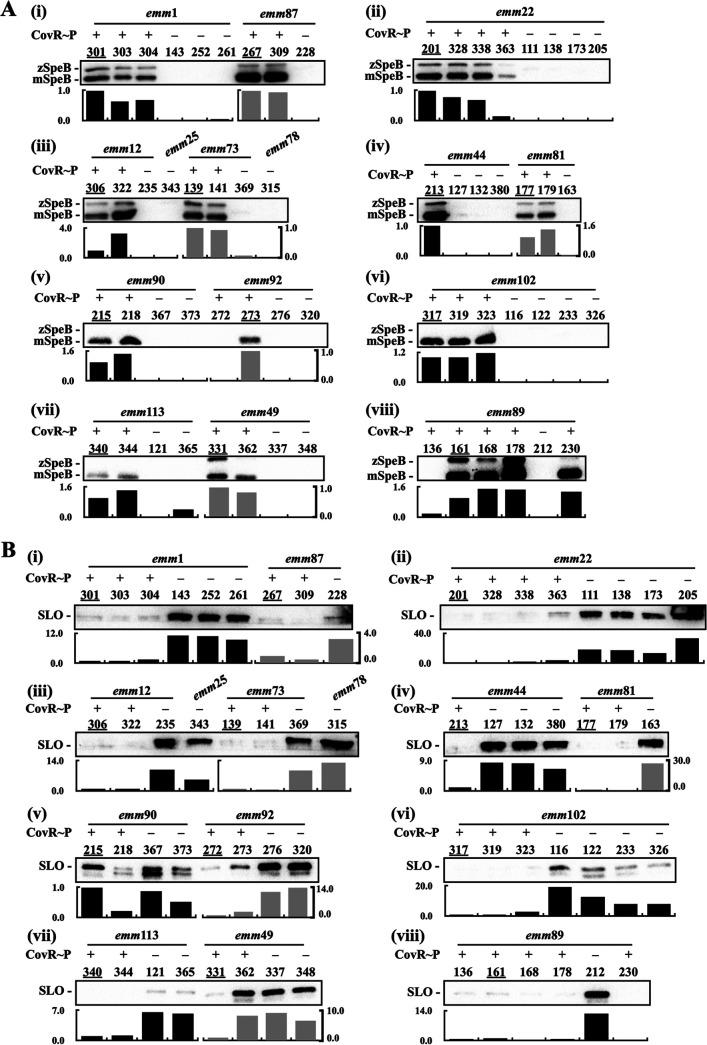
CovR is phosphorylated by CovS, and the expression of the cysteine protease SpeB is inhibited by the phosphorylated CovR (17, 33). The expression of SpeB in the covR-deletion mutant is increased compared to that in the wild-type strain; however, in the covS mutant, in which CovR cannot be phosphorylated, SpeB expression is repressed (14, 20–22). Therefore, the repression of SpeB is the phenotype which has been utilized for screening CovS-inactivated mutants in mouse infection models (18, 34, 35). To evaluate whether the SpeB expression could be utilized as the predictive marker for the CovS-inactivated isolates, the 61 CovR/CovS-activated and CovS-inactivated clinical isolates were included, and the supernatant from these isolates was collected for detecting SpeB cysteine protease by Western blot hybridization. The expression of SpeB, including the zymogen and mature forms, was detected in all isolates which expressed phosphorylated CovR except for SPY272 (emm92) and SPY136 (emm89) (Fig. 2A, subpanels v and viii). All CovS-inactivated isolates showed no SpeB expression (Fig. 2A). Based on these results, the positive and negative predictive values for detecting CovS-inactivated isolates by SpeB are 0.94 and 1.0, respectively. The sensitivity and specificity are 100% and 93.75%, respectively (Table 2 and Table S1 in the supplemental material).
FIG 2.
Expression of SpeB and SLO in the CovR/CovS-activated and CovS-inactivated isolates. Group A Streptococcus (GAS) clinical isolates were grown to an optical density at 600 nm (OD600) of 1.0 and the bacterial culture supernatants were collected for analysis. Expression of SpeB (A) and SLO (B) in the CovR/CovS-activated and CovS-inactivated isolates was analyzed by Western blot hybridization. Lower panels show relative band density (relative to the isolates labeled with an underline on each blot). CovR~P denotes phosphorylated CovR; CovR denotes non-phosphorylated CovR. zSpeB, SpeB zymogen form; mSpeB, SpeB mature form.
TABLE 2.
Positive predictive value, negative predictive value, sensitivity, and specificity of SpeB, SLO, and PepO as markers for identifying CovS-inactivated isolatesa
| Marker and no. of isolates | TP | FP | TN | FN | PPV | NPV | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|
| SpeB | CovR~P (−)/SpeB (−) | CovR~P (+)/SpeB (−) | CovR~P (+)/SpeB (+) | CovR~P (−)/SpeB (+) | TP/(TP+FP) | TN/(TN+FN) | TP/(TP+FN) | TN/(FP+TN) |
| Isolates (n) | 29 | 2 | 30 | 0 | 0.94 | 1.0 | 100 | 93.75 |
| SLO | CovR~P (−)/SLO (++) | CovR~P (+)/SLO (++) | CovR~P (+)/SLO (−/+) | CovR~P (−)/SLO (−/+) | ||||
| Isolates (n) | 21 | 2 | 30 | 8 | 0.91 | 0.79 | 72.41 | 93.25 |
| PepO | CovR~P (−)/PepO (++) | CovR~P (+)/PepO (++)b | CovR~P (+)/PepO (−/+) | CovR~P (−)/PepO (−/+) | ||||
| Isolates (n) | 29 | 1 | 31 | 0 | 0.97 | 1.0 | 100 | 96.88 |
TP, true positive; FP, false positive; TN, true negative; FN, false negative; PPV, positive predictive value; NPV, negative predictive value.
RocA-inactivated isolate (SPY362).
The expression of streptolysin O is inhibited by both phosphorylated and nonphosphorylated CovR (21); therefore, the expression of SLO is upregulated in CovR- and CovS-inactivated mutants. The anti-SLO antibody is commercially available and the SLO expression level was utilized in this study for identifying CovS-inactivated isolates. Higher levels of SLO expression were generally observed in the CovS-inactivated isolates compared to the CovS-activated isolates (except SPY215, see Fig. 2B panel v); however, a strong, clear SLO signal was not revealed in some of the CovS-inactivated isolates, including SPY228 (emm87, Fig. 2B subpanel i), SPY373 (emm90, subpanel v), SPY223, SPY326 (emm102, subpanel vi), SPY121, and SPY365 (emm113, subpanel vii). Therefore, the positive and negative predictive values for detecting CovS-inactivated isolates by SLO are 0.91 and 0.79, respectively. The specificity is 93.25%; however, the sensitivity is only 72.41% (Table 2 and Table S1). Notably, it was difficult to evaluate SLO expression levels in some GAS isolates because SLO expression levels were not equally similar among different emm type isolates [e.g., the emm90, emm92, and emm102 isolates; Fig. 2B subpanels v and vi).
Identification of an overexpressed 72-kDa protein in the covR and covS mutants.
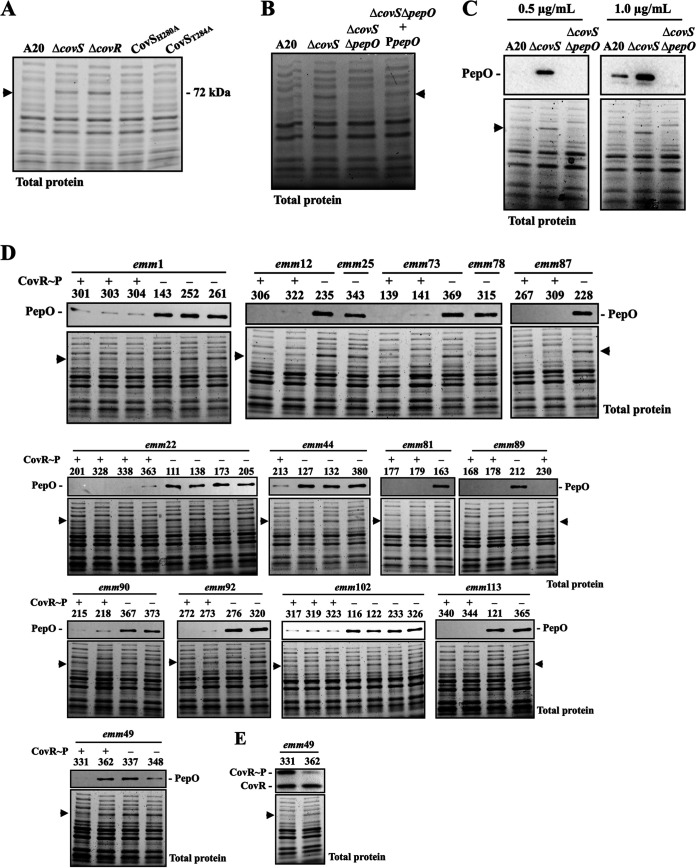
In this study, we identified an unknown protein (about 70 to 72 kDa) which can be clearly visualized on 10% SDS-PAGE of total proteins from all CovS-inactivated isolates (Fig. 1B) and the covS mutant, covR mutant, and CovS kinase-inactivated mutant (CovSH280A) (Fig. 3A); however, this unknown protein was not detected in total proteins from the wild-type A20 strain or its CovS phosphatase-inactivated mutant (CovST284A) (Fig. 3A). These findings suggest that this unknown protein could be a potential marker for identifying both CovR- and CovS-inactivated isolates. Protein identification by mass spectrometry analysis showed that the three most abundant proteins were endopeptidase PepO (72 kDa), proline tRNA ligase (69 kDa), and oligoendopeptidase F (69 kDa). To confirm the identity of the unknown protein, a ΔcovS/ΔpepO mutant was constructed and the total protein of the wild-type strain, covR mutant, covS mutant, and ΔcovS/ΔpepO mutant were extracted and analyzed by SDS-PAGE. The results showed that the 70- to 72-kDa protein signal was not observed in the wild-type strain nor in the ΔcovS/ΔpepO mutant (Fig. 3B). Furthermore, the expression of this 70- to 72-kDa protein was restored in the pepO trans-complementary strain (Fig. 3B), indicating that this previously unidentified protein is PepO.
FIG 3.
Identification and verification of the 70- to 72-kDa protein marker in the covR/covS mutants and the CovR/CovS-activated and -inactivated isolates. (A) Total protein profiles of the wild-type A20 strain, covS mutant (ΔcovS), covR mutant (ΔcovR), CovS kinase-inactivated mutant (CovSH280A), and CovS phosphatase-inactivated mutant (CovST284A). (B) Total protein profiles of A20, covS mutant (ΔcovS), covR mutant (ΔcovR), covS/pepO double mutant (ΔcovS/ΔpepO), and pepO trans-complementary strain (ΔcovS/ΔpepO+PpepO). (C) Western blot of PepO expression in A20, covS mutant (ΔcovS), and covS/pepO double mutant (ΔcovS/ΔpepO) detected by the anti-PDTTYYEEGNEKAEELR antibody. (D) Comparison of PepO expression in CovR/CovS-activated and CovS-inactivated clinical isolates. The PepO protein was detected by the anti-PDTTYYEEGNEKAEELR antibody in a total protein extract. (E) Phosphorylation levels of CovR in the emm49 SPY331 and SPY362 isolates. CovR~P denotes phosphorylated CovR; CovR denotes non-phosphorylated CovR. Arrows indicate when a signal from only the CovR/CovS-inactivated mutants was identified. The lower sections of the image in panels C, D, and E show total protein as the loading control.
Western blot hybridization analysis was performed to confirm the identity of the 70- to 72-kDa protein as PepO. In brief, we produced a polyclonal antibody against the synthetic predicted antigenic PepO peptide (PDTTYYEEGNEKAEELR) to detect the protein. As a negative control, we observed no signal in the 70- to 72-kDa range for the ΔcovS/ΔpepO mutant (Fig. 3C). Furthermore, PepO expression was upregulated in the covR and covS mutants compared to that in the wild-type A20 strain from the log to the stationary growth phases (Fig. S2). These results confirm that the overexpressed 70- to 72-kDa protein in the CovR/CovS-inactivated mutants is PepO. Additionally, Western blotting with the anti-PepO antibody (0.1 μg/mL) was used to investigate whether PepO could be used as a marker to identify CovS-inactivated clinical isolates. As shown in Fig. 3D, with only one exception, SPY362 (emm49), the CovS-inactivated isolates showed a stronger PepO signal than the CovR/CovS-activated isolates. SPY362 showed a weak phosphorylated CovR signal but expressed PepO at a level comparable to that of the CovS-inactivated emm49 isolates (SPY337 and SPY348; Fig. 3D and E). Genome sequencing analysis (using the MiSeq system) of SPY362 showed single-nucleotide polymorphisms (SNPs) in CovR/CovS, HasA, HasC, and SpeB compared to the emm49 reference NZ131 strain (NCBI accession no. CP000829.1; Table S2). The amino acid sequence of CovR/CovS in SPY362 was identical to that of the emm1-type A20 strain (NCBI accession no. CP003901.1) except for a common SNP (I332V) found in CovS, suggesting that CovR/CovS should be functional in SPY362. Nonetheless, the deletion of a single unit of the GAAGGA variable-number tandem-repeat (VNTR) in the rocA promoter region was identified by Sanger sequencing. Based on these results, the positive and negative predictive values for identifying CovR/CovS-inactivated isolates by PepO are 0.97 and 1.0, respectively. The sensitivity and specificity are 100% and 96.88%, respectively (Table 2 and Table S1).
PepO could be utilized to identify RocA-inactivated isolates and emm3-type isolates.
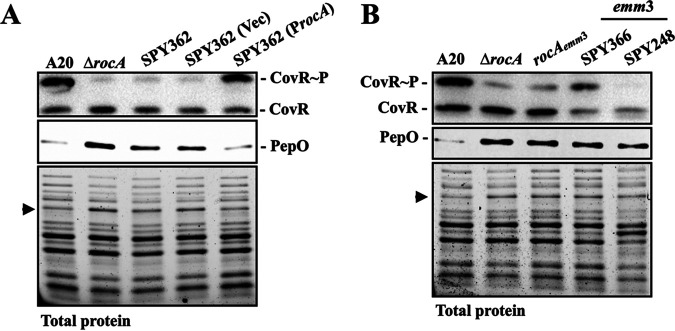
RocA is an accessory protein that modulates CovR phosphorylation through CovS (23–26, 36). Zhu et al. (28) showed that deletion of a single unit of the GAAGGA in the rocA promoter region results in the downregulation of CovR phosphorylation. To determine whether the decrease in CovR phosphorylation in the SPY362 isolate was mediated by RocA, the wild-type rocA gene from the emm1-type A20 strain was trans-complemented into SPY362, and the expression of PepO and phosphorylated CovR in the complementary strain was analyzed. As the experimental control, the rocA isogenic mutant showed a decrease in phosphorylated CovR expression compared to the wild-type A20 strain (Fig. 4A). Complementation of the rocA gene from the emm1-type A20 strain into SPY362 upregulated the expression of phosphorylated CovR in comparison with SPY362 and its vector-control strain (Fig. 4A). Expression of PepO was increased in the A20 rocA isogenic mutant compared with that in the wild-type A20 strain; furthermore, the expression of PepO was decreased in the rocA trans-complementary strain to a level similar to that in A20 (Fig. 4A). These results indicate that the upregulation of PepO expression in SPY362 is mediated by RocA. Also, these results indicated that the repression of SpeB cannot be utilized to detect a RocA-inactivated isolate (SPY362, Fig. 2A panel vii). After including the RocA-inactivated isolate, the positive predictive value for detecting CovS-inactivated clinical isolates by PepO increased from 0.97 to 1.0 and the specificity increased from 96.88% to 100%.
FIG 4.
Expression levels of PepO and phosphorylated CovR in the wild-type strains, the rocA mutants, the rocA trans-complementary strain, and the emm3-type clinical isolates. (A) The expression level of PepO and phosphorylated CovR in the emm1 wild-type A20 strain, its rocA mutant (ΔrocA), SPY362 (emm49), and its vector-control (Vec), and the rocA trans-complementary strain (ProcA). (B) Expression levels of PepO and phosphorylated CovR in the emm1 wild-type A20 strain, its rocA mutant (ΔrocA), the RocA-truncated mutant (rocAemm3), and the emm3-type isolates. PepO and phosphorylated CovR were detected by Western blotting and Phostag Western blotting, respectively, from a total protein extract. CovR~P denotes phosphorylated CovR; CovR denotes non-phosphorylated CovR. Lower panels of images show total protein as the internal loading control. An arrow indicates the PepO signal on SDS-PAGE.
Studies have shown that emm3-type isolates have a truncated RocA and express lower levels of phosphorylated CovR than emm1-type isolates (29, 30). Therefore, we further verified whether PepO could be used to identify emm3 isolates. First, we analyzed whether the expression of PepO was upregulated in the RocA-truncated mutant. To achieve this, the rocA gene of the emm1 A20 strain was replaced by the rocA gene from the emm3 isolate (SPY248), producing a RocA-truncated mutant (rocAemm3, Fig. 4B) (26). Next, we used Western blot hybridization to determine whether PepO could be a marker for identifying the RocA-truncated mutant and emm3 isolates. Phostag Western blot analysis showed the presence of phosphorylated CovR in the A20 rocA isogenic mutants, the RocA-truncated mutant, and SPY366 (emm3), but not in SPY248 (emm3) (Fig. 4B). The expression of PepO was significantly upregulated in the rocA mutants and emm3 isolates compared to that in the wild-type A20 strain (Fig. 4B). Also, the PepO signal in rocA mutants and emm3 isolates was also visualized by SDS-PAGE (Fig. 4B). These results indicate that PepO can also be used to identify emm3 isolates.
PepO mediates the decrease of SIP-inducing speB expression in the covR mutant.
Brouwer et al. (37) showed that the speB expression is negatively regulated by PepO. The expression of PepO was upregulated in the CovR/CovS- and RocA-inactivated isolates (Fig. 3 and 4); however, the repression of speB is found in the covS mutant but not in the covR mutant (14, 20–22, 27). Therefore, the present study further elucidated the role of PepO in regulating SpeB expression in the covR mutant. We constructed a ΔcovR/ΔpepO mutant and compared speB expression in the covR mutant (ΔcovR) to that in the ΔcovR/ΔpepO mutant. The results showed that the deletion of pepO in the covR mutant upregulated speB expression (Fig. 5A). In addition, the trans-complementation of pepO in the ΔcovR/ΔpepO mutant repressed speB expression (Fig. 5A). These results indicated that PepO acts as the repressor of speB transcription in the covR mutant.
FIG 5.
Expression of SpeB in the covR mutant and its ΔpepO and SIP-inactivated (SIP*) mutants. (A) Expression of speB in the ΔcovR mutant, ΔcovR/ΔpepO mutant, the vector-control (Vec) strain, and the pepO trans-complementary (PpepO) strain. (B and C) Transcription of (B) speB and (C) expression of SpeB in the SIP* and SIP*/ΔcovR mutants in the treatment with the synthetic SIP peptide and scramble peptide (SCRA). (D and E) The expression of (D) speB and (E) SpeB in the SIP*/ΔcovR and SIP*/ΔcovR/ΔpepO mutants in the treatment with synthetic SIP and SCRA peptides. GAS strains were grown to OD600 = 0.8 in tryptic soy broth supplemented with yeast (TSBY broth), and bacterial pellets were collected and incubated in acidic TSBY broth (pH 6.0) supplemented with different concentrations of SIP and SCRA for 1 h. Culture supernatant was used for detecting SpeB. zSpeB, zymogen form of SpeB; mSpeB, mature form of SpeB. Bacterial RNA was extracted for real-time quantitative PCR analysis. The expression of speB was normalized to that of gyrA. *, P < 0.05.
speB expression is activated by the quorum-sensing RopB regulator with binding to the SpeB-inducing peptide (38–40). PepO is an endopeptidase which disrupts the RopB-independent quorum-sensing pathway (Rgg2/3) by degrading the quorum-sensing short hydrophobic peptide (41). This study, therefore, elucidated whether the upregulation of PepO in the covR mutant would degrade SIP to downregulate SIP-inducing SpeB expression. To exclude the signal from the endogenous SIP, SIP-inactivated mutants (SIP*) of the wild-type strain and the covR mutant were utilized for analyses. The results showed that supplementation with SIP but not scrambled peptide (SCRA) activated speB transcription in the SIP* mutant and SIP*/ΔcovR mutant; noticeably, the 0.1- and 0.5-μM SIP treatments induced a 12- and 52-fold increases in speB transcription in the SIP* mutant, while 4- and 17-fold increases in the SIP*/ΔcovR mutant were observed (Fig. 5B). In line with Fig. 5B, a Western blot analysis showed that the SIP* mutant expressed a higher level of SpeB than the SIP*/ΔcovR mutant under 0.5-μM SIP treatments (Fig. 5C), indicating that the SIP*/ΔcovR mutant is less susceptible to SIP stimuli than the SIP* mutant.
To demonstrate that PepO could modulate the intracellular concentration of SIP in the covR mutant, a pepO mutant of the SIP*/ΔcovR mutant was constructed and SpeB expression was analyzed in the SIP*/ΔcovR and SIP*/ΔcovR/ΔpepO mutants under treatment with different concentrations of SIP. Supplementation with SIP but not SCRA during incubation activated SpeB expression at both the transcriptional and translational levels in the SIP*/ΔcovR and SIP*/ΔcovR/ΔpepO mutants (Fig. 5D and E). Under treatments with the same concentrations of SIP, quantitative PCR (qPCR) and Western blot analyses showed that the SIP*/ΔcovR/ΔpepO mutant expressed a higher level of SpeB than the SIP*/ΔcovR mutant (Fig. 5D and E), suggesting that PepO regulates speB expression by degrading SIP in the covR mutant.
DISCUSSION
An encapsulated, or mucoid, colony morphology is one of the most identifiable phenotypes of CovR/CovS-inactivated mutants (16). However, in the present study, 69% (20/29) of CovS-inactivated isolates did not exhibit an encapsulated colony morphology. We did not determine whether the transcription of hasA was upregulated in these CovS-inactivated isolates; however, our results suggest that the presence of encapsulated colony morphology is not a reliable characteristic for identifying CovS-inactivated clinical isolates. Importantly, the unexpectedly low rate of encapsulated phenotype expression in CovS-inactivated GAS isolates may result in underestimation of their prevalence.
An anti-SLO antibody is commercially available, and increased SLO expression is observed in both CovR- and CovS-inactivated mutants (14, 20, 21, 42). Nonetheless, the negative predictive value and the sensitivity of identifying CovS-inactivated isolates by SLO were only 0.79 and 72.41%, respectively, indicating that SLO is not a sensitive marker for identifying these invasive isolates. The cysteine protease SpeB is the most abundantly secreted protein in GAS culture supernatant (43, 44). Although the repression of SpeB is not a phenotype for identifying CovR-inactivated mutants, this phenotype is highly sensitive to identifying CovS-inactivated isolates. Nonetheless, SpeB expression is regulated by multiple regulators, and mutations in the SpeB-positive regulator RopB could also result in repressed SpeB expression (45). Further, SpeB expression is restricted during the specific growth phase (45–47), and quantification of secreted proteins is difficult because of the lack of reliable internal control proteins in bacterial culture supernatants. These factors increased the difficulty of using SpeB as a bacterial maker for identifying CovS-inactivated isolates.
Phosphorylation of CovR is modulated by RocA in a CovS-dependent manner (24–26). There is no phosphorylation of CovR in the CovS-inactivated mutants; however, in the rocA mutant, CovR phosphorylation is decreased during the exponential growth phase but increases to a level similar to that in the wild-type strain during the stationary phase (26, 27). Similarly, in the rocA mutant, SpeB expression is repressed during the exponential growth phase but upregulated during the stationary phase compared with that in the wild-type strain (27). Therefore, although rocA-inactivating mutations result in the inactivation of CovR phosphorylation during the exponential phase of growth, rocA mutants cannot be identified by the detection of phosphorylated CovR or SpeB. The present study showed that PepO expression was upregulated in the rocA mutant and emm3 isolates (with native truncated RocA), indicating that PepO could also be utilized as a marker for identifying RocA-inactivated isolates, including RocA-truncated emm3-type isolates.
Do et al. (39) showed that SpeB expression is activated by the pH-sensitive RopB-SIP quorum-sensing pathway. The present study suggested that PepO endopeptidase could degrade SIP to downregulate SpeB expression. In the covS mutant, the transcription of ropB is repressed by the nonphosphorylated CovR (22). Consequently, the decrease in intracellular SIP and the low level of RopB could result in the downregulation of SpeB in the covS mutant. In the covR mutant, the expression of ropB is derepressed (33, 48). Therefore, during the stationary phase of growth, the increase levels of RopB and SIP could compensate for the effect of PepO degradation to activate SpeB expression. These results suggest that the increased PepO expression could contribute to the repression of speB in the covS mutant and reveal that CovR/CovS could modulate RopB-SIP via controlling PepO expression.
Although the mutation rate is not high, it is generally accepted that CovR/CovS-inactivated mutants are selected by immune stress during infection, and these mutants are capable of invading deep tissues, resisting phagocytic attack, and causing severe disease manifestations (12–14, 34). This study demonstrated that PepO, the endopeptidase which regulates SpeB expression by degrading the quorum-sensing peptide SIP, could be used as a marker for the identification of these invasive GAS isolates. PepO is a cytosolic protein and flow cytometry analysis showed that the anti-PepO antibody cannot detect PepO with whole-cell GAS (Fig. S3). The cytosolic protein is not preferred as the detection target; however, the increased production of the hyaluronic acid capsule in the covR and covS mutants would mask the surface antigens, which may limit the use of surface antigens as targets for identifying these mutants. In conclusion, monitoring the emergence of CovR/CovS-inactivated mutants from patients with wound, deep tissue, or blood GAS infection using PepO could be a potential strategy for early detection of high-risk patients to prevent severe manifestations or poor prognoses.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
GAS A20 is the emm1-type strain and has been described previously (49). Strain AP3 is a mouse-passaged isolate of A20 which has a frameshift deletion in the covS gene (33) and was designed as the covS mutant in this study. The rocA mutant, RocA-truncated mutant, hasA mutants, and CovR/CovS-inactivated clinical isolates have been described previously (26, 31, 33). GAS strains were cultured on Trypticase soy agar containing 5% sheep blood or in tryptic soy broth (Becton, Dickinson and Co., Sparks, MD, USA) supplemented with 0.5% yeast extract (TSBY). E. coli DH5α was purchased from Yeastern (Yeastern Biotech Co., Ltd.; Taipei, Taiwan) and was cultured in lysogeny broth (LB) at 37°C with vigorous aeration. SpeB-inducing peptide (MWLLLLFL; purity: 94.469%) and scrambled control peptide (LLFLWLLM; purity: 92.822%) (38) were purchased from Leadgene Biomedical, Inc. (Tainan, Taiwan). These synthetic peptides were suspended in 100% dimethyl sulfoxide (DMSO) to prepare a 10-mM stock solution and stored at −20°C until use. Working solutions were prepared by diluting the stock solution with 25% DMSO. In SIP- and SCRA-supplemented cultures, bacteria were grown to an optical density at 600 nm (OD600) of 0.8. Bacterial pellets were collected via centrifugation and resuspended in acidic TSBY broth (pH 6.0) containing different concentrations of SIP and SCRA for 1 h at 37°C. When appropriate, the antibiotics chloramphenicol (25 and 3 μg/mL for E. coli and GAS, respectively) and spectinomycin (100 μg/mL) were used for selection.
Bacterial colony morphology.
GAS strains were cultured on blood agar plates (Becton, Dickinson and Company; Sparks, MD, USA) at 37°C with 5% CO2 supplementation for 24 h. The images were acquired using a Gel Doc XR+ system (Bio-Rad Laboratories, Inc; Hercules, CA, USA).
DNA and RNA manipulations.
Bacterial genomic DNA extraction, RNA extraction, and reverse transcription were performed as previously described (50). For sequencing of hasA and hasB, the hasA and hasB genes were amplified by PCR with the primers PhasA-F and hasB-R-1, and the PCR products were sequenced with the primers hasA-seq-F, hasA-seqR, hasB-seq-F, and hasB-R-1 (Table 3). Real-time PCR was performed in a 20-μL reaction mixture containing 1 μL of cDNA, 0.8 μL of primers (10 μM), and 10 μL of SensiFAST SYBR Lo-ROX pre-mixture (Bioline Ltd; London, United Kingdom) according to the manufacturer’s instructions. Biological replicate experiments were performed using three independent RNA preparations in duplicates. The expression level of pepO was normalized to gyrA and analyzed using the ΔΔCT method (Roche LightCycler 96 System, Roche Molecular Systems, Inc.; Pleasanton, CA, USA). All values of the control and experimental groups were divided by the mean of the control samples before statistical analysis (51). Primers used for real-time PCR analysis (Table 3) were designed using Primer3 (v0.4.0, http://frodo.wi.mit.edu) according to the MGAS5005 sequence (NCBI accession no. CP000017.2).
TABLE 3.
Primers used in this studya
| Primer | Use | Sequence (5′–3′)b | Reference |
|---|---|---|---|
| PhasA-F | PCR | tcagatgaagttgtactccctgaa | This study |
| hasB-R-1 | PCR/sequencing | gctcaatcataccaccaact | This study |
| hasA-seq-F | Sequencing | atcgaggtccctgtctttca | This study |
| hasA-seq-R | Sequencing | cgttttgaagtgataaaagaactcc | This study |
| hasB-seq-F | Sequencing | ggaatggggaacacgtaaaa | This study |
| pepO-F | qPCR | tgcctttaaagagcgcacag | This study |
| pepO-R | qPCR | ctaccccacctagatcagcg | This study |
| Orf-1-F | Construction | gttaaaaggaggcgcctactTAgtggttattgttactatttttg | 38 |
| Orf-1-R | Construction | caaaaatagtaacaataaccacTAagtaggcgcctccttttaac | 38 |
| SIP-F-1 | Construction | gcgggatccggtcaatagccagatgcgata | 38 |
| SIP-R-1 | Construction | gcgggatcctcgtgatagggtccacaaca | 38 |
| gyrA-F-3 | qPCR | cgtcgtttgactggtttgg | 33 |
| gyrA-F-3 | qPCR | ggcgtgggttagcgtattta | 33 |
| PepO-BamHI-F | Construction | gcg ggatcc cgagacatcaatgtcgaaaca | This study |
| PepO-BamHI-R | Construction | gcg ggatcc gaatctcgtgcaatggttgat | This study |
| PepO-SacII-F | Construction | tcc ccgcgg taattaggtagttgataaacc | This study |
| PepO-SacII-R | Construction | tcc ccgcgg cattcgtatctccttatatca | This study |
| promoter-pepO-F | Construction | gcg ggatcc aacgccatttaggtgaccag | This study |
| promoter-pepO-R | Construction | gcg ggatcc tgcttttgcacgttttgaag | This study |
qPCR, quantitative PCR.
The underline indicates the restriction enzyme site.
Construction of the SIP-inactivated mutants, pepO mutants, pepO trans-complementary strain, and rocA trans-complementary strain.
To inactivate SIP translation, the start codon of the SIP open reading frame was replaced with a stop codon (TAG) in the chromosome, according to a previous report (38). The 454-bp PCR product was amplified using the primers SIP-F-1 and SIP-R-1 and ligated into pCN143 through the BamHI site (designated as pCN215). To construct the pepO mutants, the pepO gene with its upstream (538 bp) and downstream (556 bp) regions was amplified using the primers PepO-BamHI-F and PepO-BamHI-R (Table 3). PCR amplicons were digested with BamHI and ligated into the temperature-sensitive vector pCN143 (33). The pepO gene was removed via inverted PCR using the primers PepO-SacII-F and PepO-SacII-R (Table 3) and replaced with the chloramphenicol cassette from Vector 78 (52) to generate pCN210. These plasmids were transformed into GAS strains via electroporation, and the transformants were selected as described previously (33). The deletion of pepO and the replacement of TAG in the SIP open in the transformants was confirmed via Sanger sequencing. To complement the pepO expression, the pepO gene with its native promoter (2,553 bp) was amplified using the primers promoter-pepO-F and promoter-pepO-R from the wild-type A20 strain, ligated into the low-copy number vector pTRKL2, and transformed into the pepO mutant. The rocA trans-complementary strain was constructed as described previously (26).
Anti-PepO antibody.
The antigenic region of PepO was predicted, and the peptide PDTTYYEEGNEKAEELR and the rabbit anti-PDTTYYEEGNEKAEELR polyclonal antibody were purchased from Leadgene Biomedical, Inc. (Tainan, Taiwan).
Western blot and Phostag Western blot hybridization.
Bacteria were cultured in TSBY broth to a turbidity of OD600 = 1.0. The bacterial culture supernatants were collected, filtrated by a 0.22-μm filter, and used to analyze SpeB and SLO expression. The bacterial cells were disrupted using a bead beater (Mini-Beadbeater, BioSpec Products Inc; Bartlesville, OK, USA). The bacterial cell lysate was centrifuged and total protein in the supernatant was collected for analysis. Total protein was mixed with 6× protein loading dye, boiled for 5 min, and subjected to 10% SDS-PAGE. For Phostag Western blot analysis, the bacterial proteins were mixed with 6× protein loading dye (without boiling) and loaded into a 10% SDS-PAGE containing 10 μM Phostag (Wako Pure Chemical Industries Ltd; Richmond, VA, USA) and 0.5 μM MnCl2. The separated proteins were transferred onto polyvinylidene fluoride membranes (Millipore; Billerica, MA, USA). The membranes were blocked with 5% skim milk in PBST buffer (phosphate-buffered saline containing 0.2% vol/vol Tween 20) at 37°C for 1 h. CovR protein was detected using anti-CovR serum (33) and PepO was detected by the anti-PDTTYYEEGNEKAEELR antibody. SpeB and SLO proteins were detected by anti-SpeB (Toxin Technology, Inc; Sarasota, IL, USA) and anti-SLO antibodies (GeneTex; Irvine, CA, USA), respectively. After hybridization, the membrane was washed with PBST buffer and hybridized with a secondary antibody, peroxidase-conjugated goat anti-rabbit IgG (Cell Signaling Technology, Inc; Danvers, MA, USA) at room temperature (25°C to 28°C) for 1 h. The blot was developed using Pierce ECL Western blotting substrate (Thermo Fisher Scientific Inc; Rockford, IL, USA) and the signals were detected using a Gel Doc XR+ system (Bio-Rad Laboratories, Inc; Hercules, CA, USA).
Statistical analysis.
Statistical analyses were performed using Prism software version 5 (GraphPad Software, Inc; San Diego, CA, USA). Significant differences between multiple groups were determined using analysis of variance (ANOVA). Post-tests for ANOVA were performed using Tukey’s honestly significant difference test. Statistical significance was set at P < 0.05.
ACKNOWLEDGMENTS
This work was supported by parts of grants from the Chang Gung Memorial Hospital at Linkou, Taiwan (CMRPD1J0031-3) and the Ministry of Science and Technology, Taiwan (MOST 110-2628-B-182-012 and MOST 111-2628-B-182-005).
We acknowledge the assistance from the Core Instrument Center, Chang Gung University (Taiwan) for flow cytometry analysis.
The authors affirm no conflicts of interest relative to any source of funding, sponsorship, or financial benefit.
Footnotes
Supplemental material is available online only.
Contributor Information
Chuan Chiang-Ni, Email: entchuan@gap.cgu.edu.tw.
John M. Atack, Griffith University
REFERENCES
- 1.Stevens DL, Bryant AE. 2017. Necrotizing soft-tissue infections. N Engl J Med 377:2253–2265. doi: 10.1056/NEJMra1600673. [DOI] [PubMed] [Google Scholar]
- 2.Bisno AL, Stevens DL. 1996. Streptococcal infections of skin and soft tissues. N Engl J Med 334:240–245. doi: 10.1056/NEJM199601253340407. [DOI] [PubMed] [Google Scholar]
- 3.Olsen RJ, Musser JM. 2010. Molecular pathogenesis of necrotizing fasciitis. Annu Rev Pathol 5:1–31. doi: 10.1146/annurev-pathol-121808-102135. [DOI] [PubMed] [Google Scholar]
- 4.Wong CH, Khin LW, Heng KS, Tan KC, Low CO. 2004. The LRINEC (Laboratory Risk Indicator for Necrotizing Fasciitis) score: a tool for distinguishing necrotizing fasciitis from other soft tissue infections. Crit Care Med 32:1535–1541. doi: 10.1097/01.CCM.0000129486.35458.7D. [DOI] [PubMed] [Google Scholar]
- 5.Liao CI, Lee YK, Su YC, Chuang CH, Wong CH. 2012. Validation of the laboratory risk indicator for necrotizing fasciitis (LRINEC) score for early diagnosis of necrotizing fasciitis. Tzu Chi Medical J 24:73–76. doi: 10.1016/j.tcmj.2012.02.009. [DOI] [Google Scholar]
- 6.El-Menyar A, Asim M, Mudali IN, Mekkodathil A, Latifi R, Al-Thani H. 2017. The laboratory risk indicator for necrotizing fasciitis (LRINEC) scoring: the diagnostic and potential prognostic role. Scand J Trauma Resusc Emerg Med 25:28. doi: 10.1186/s13049-017-0359-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernando SM, Tran A, Cheng W, Rochwerg B, Kyeremanteng K, Seely AJE, Inaba K, Perry JJ. 2019. Necrotizing soft tissue infection: diagnostic accuracy of physical examination, imaging, and LRINEC score: A systematic review and meta-analysis. Ann Surg 269:58–65. doi: 10.1097/SLA.0000000000002774. [DOI] [PubMed] [Google Scholar]
- 8.Bruun T, Kittang BR, de Hoog BJ, Aardal S, Flaatten HK, Langeland N, Mylvaganam H, Vindenes HA, Skrede S. 2013. Necrotizing soft tissue infections caused by Streptococcus pyogenes and Streptococcus dysgalactiae subsp. equisimilis of groups C and G in western Norway. Clin Microbiol Infect 19:E545–E550. doi: 10.1111/1469-0691.12276. [DOI] [PubMed] [Google Scholar]
- 9.Gherardi G, Vitali LA, Creti R. 2018. Prevalent emm types among invasive GAS in Europe and North America since year 2000. Front Public Health 6:59. doi: 10.3389/fpubh.2018.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sekizuka T, Nai E, Yoshida T, Endo S, Hamajima E, Akiyama S, Ikuta Y, Obana N, Kawaguchi T, Hayashi K, Noda M, Sumita T, Kokaji M, Katori T, Hashino M, Oba K, Kuroda M. 2017. Streptococcal toxic shock syndrome caused by the dissemination of an invasive emm3/ST15 strain of Streptococcus pyogenes. BMC Infect Dis 17:774. doi: 10.1186/s12879-017-2870-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamayo E, Montes M, Garcia-Medina G, Garcia-Arenzana JM, Perez-Trallero E. 2010. Spread of a highly mucoid Streptococcus pyogenes emm3/ST15 clone. BMC Infect Dis 10:233. doi: 10.1186/1471-2334-10-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friaes A, Pato C, Melo-Cristino J, Ramirez M. 2015. Consequences of the variability of the CovRS and RopB regulators among Streptococcus pyogenes causing human infections. Sci Rep 5:12057. doi: 10.1038/srep12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikebe T, Ato M, Matsumura T, Hasegawa H, Sata T, Kobayashi K, Watanabe H. 2010. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog 6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. 2006. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog 2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Federle MJ, McIver KS, Scott JR. 1999. A response regulator that represses transcription of several virulence operons in the group A Streptococcus. J Bacteriol 181:3649–3657. doi: 10.1128/JB.181.12.3649-3657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levin JC, Wessels MR. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol 30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- 17.Miller AA, Engleberg NC, DiRita VJ. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol Microbiol 40:976–990. doi: 10.1046/j.1365-2958.2001.02441.x. [DOI] [PubMed] [Google Scholar]
- 18.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med 13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 19.Bernish B, van de Rijn I. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem 274:4786–4793. doi: 10.1074/jbc.274.8.4786. [DOI] [PubMed] [Google Scholar]
- 20.Tran-Winkler HJ, Love JF, Gryllos I, Wessels MR. 2011. Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus. PLoS Pathog 7:e1002361. doi: 10.1371/journal.ppat.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trevino J, Perez N, Ramirez-Pena E, Liu Z, Shelburne SA, Musser JM, Sumby P. 2009. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect Immun 77:3141–3149. doi: 10.1128/IAI.01560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiang-Ni C, Kao CY, Hsu CY, Chiu CH. 2019. Phosphorylation at the D53 but not the T65 residue of CovR determines the repression of rgg and speB transcription in emm1- and emm49-type group A streptococci. J Bacteriol 201:e00681-18. doi: 10.1128/JB.00681-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain I, Miller EW, Danger JL, Pflughoeft KJ, Sumby P. 2017. RocA is an accessory protein to the virulence-regulating CovRS two-component system in group A Streptococcus. Infect Immun 85:e00274-17. doi: 10.1128/IAI.00274-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynskey NN, Velarde JJ, Finn MB, Dove SL, Wessels MR. 2019. RocA binds CsrS to modulate CsrRS-mediated gene regulation in group A Streptococcus. mBio 10:e01495-19. doi: 10.1128/mBio.01495-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jain I, Danger JL, Burgess C, Uppal T, Sumby P. 2020. The group A Streptococcus accessory protein RocA: regulatory activity, interacting partners and influence on disease potential. Mol Microbiol 113:190–207. doi: 10.1111/mmi.14410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chiang-Ni C, Chiou HJ, Tseng HC, Hsu CY, Chiu CH. 2020. RocA regulates phosphatase activity of virulence sensor CovS of group A Streptococcus in growth phase- and pH-dependent manners. mSphere 5:e00361-20. doi: 10.1128/mSphere.00361-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feng W, Minor D, Liu M, Li J, Ishaq SL, Yeoman C, Lei B. 2017. Null mutations of group A Streptococcus orphan kinase RocA: selection in mouse infection and comparison with CovS mutations in alteration of in vitro and in vivo protease SpeB expression and virulence. Infect Immun 85:e00790-16. doi: 10.1128/IAI.00790-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu L, Olsen RJ, Horstmann N, Shelburne SA, Fan J, Hu Y, Musser JM. 2016. Intergenic variable-number tandem-repeat polymorphism upstream of rocA alters toxin production and enhances virulence in Streptococcus pyogenes. Infect Immun 84:2086–2093. doi: 10.1128/IAI.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lynskey NN, Turner CE, Heng LS, Sriskandan S. 2015. A truncation in the regulator RocA underlies heightened capsule expression in serotype M3 group A streptococci. Infect Immun 83:1732–1733. doi: 10.1128/IAI.02892-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horstmann N, Sahasrabhojane P, Saldana M, Ajami NJ, Flores AR, Sumby P, Liu CG, Yao H, Su X, Thompson E, Shelburne SA. 2015. Characterization of the effect of the histidine kinase CovS on response regulator phosphorylation in group A Streptococcus. Infect Immun 83:1068–1077. doi: 10.1128/IAI.02659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiang-Ni C, Liu YS, Lin CY, Hsu CY, Shi YA, Chen YM, Lai CH, Chiu CH. 2021. Incidence and effects of acquisition of the phage-encoded ssa superantigen gene in invasive group A Streptococcus. Front Microbiol 12:685343. doi: 10.3389/fmicb.2021.685343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashbaugh CD, Alberti S, Wessels MR. 1998. Molecular analysis of the capsule gene region of group A Streptococcus: the hasAB genes are sufficient for capsule expression. J Bacteriol 180:4955–4959. doi: 10.1128/JB.180.18.4955-4959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiang-Ni C, Chu TP, Wu JJ, Chiu CH. 2016. Repression of Rgg but not upregulation of LacD.1 in emm1-type covS mutant mediates the SpeB repression in group A Streptococcus. Front Microbiol 7:1935. doi: 10.3389/fmicb.2016.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Liu G, Feng W, Zhou Y, Liu M, Wiley JA, Lei B. 2014. Neutrophils select hypervirulent CovRS mutants of M1T1 group A Streptococcus during subcutaneous infection of mice. Infect Immun 82:1579–1590. doi: 10.1128/IAI.01458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu G, Feng W, Li D, Liu M, Nelson DC, Lei B. 2015. The Mga regulon but not deoxyribonuclease Sda1 of invasive M1T1 group A Streptococcus contributes to in vivo selection of CovRS mutations and resistance to innate immune killing mechanisms. Infect Immun 83:4293–4303. doi: 10.1128/IAI.00857-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernard PE, Duarte A, Bogdanov M, Musser JM, Olsen RJ. 2020. Single amino acid replacements in RocA disrupt protein-protein interactions to alter the molecular pathogenesis of group A Streptococcus. Infect Immun 88:e00386-20. doi: 10.1128/IAI.00386-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brouwer S, Cork AJ, Ong CY, Barnett TC, West NP, McIver KS, Walker MJ. 2018. Endopeptidase PepO regulates the SpeB cysteine protease and is essential for the virulence of Invasive M1T1 Streptococcus pyogenes. J Bacteriol 200:e00654-17. doi: 10.1128/JB.00654-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Do H, Makthal N, VanderWal AR, Rettel M, Savitski MM, Peschek N, Papenfort K, Olsen RJ, Musser JM, Kumaraswami M. 2017. Leaderless secreted peptide signaling molecule alters global gene expression and increases virulence of a human bacterial pathogen. Proc Natl Acad Sci USA 114:E8498–E8507. doi: 10.1073/pnas.1705972114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Do H, Makthal N, VanderWal AR, Saavedra MO, Olsen RJ, Musser JM, Kumaraswami M. 2019. Environmental pH and peptide signaling control virulence of Streptococcus pyogenes via a quorum-sensing pathway. Nat Commun 10:2586. doi: 10.1038/s41467-019-10556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Makthal N, Do H, VanderWal AR, Olsen RJ, Musser JM, Kumaraswami M. 2018. Signaling by a conserved quorum sensing pathway contributes to growth ex vivo and oropharyngeal colonization of human pathogen group A Streptococcus. Infect Immun 86. doi: 10.1128/IAI.00169-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkening RV, Chang JC, Federle MJ. 2016. PepO, a CovRS-controlled endopeptidase, disrupts Streptococcus pyogenes quorum sensing. Mol Microbiol 99:71–87. doi: 10.1111/mmi.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiang-Ni C, Tseng HC, Hung CH, Chiu CH. 2017. Acidic stress enhances CovR/S-dependent gene repression through activation of the covR/S promoter in emm1-type group A Streptococcus. Int J Med Microbiol 307:329–339. doi: 10.1016/j.ijmm.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 43.Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Kotb M. 2004. Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol 51:123–134. doi: 10.1046/j.1365-2958.2003.03797.x. [DOI] [PubMed] [Google Scholar]
- 44.Persson H, Vindebro R, von Pawel-Rammingen U. 2013. The streptococcal cysteine protease SpeB is not a natural immunoglobulin-cleaving enzyme. Infect Immun 81:2236–2241. doi: 10.1128/IAI.00168-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neely MN, Lyon WR, Runft DL, Caparon M. 2003. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol 185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Unnikrishnan M, Cohen J, Sriskandan S. 1999. Growth-phase-dependent expression of virulence factors in an M1T1 clinical isolate of Streptococcus pyogenes. Infect Immun 67:5495–5499. doi: 10.1128/IAI.67.10.5495-5499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol 9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 48.Graham MR, Smoot LM, Migliaccio CA, Virtaneva K, Sturdevant DE, Porcella SF, Federle MJ, Adams GJ, Scott JR, Musser JM. 2002. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA 99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiang-Ni C, Zheng PX, Ho YR, Wu HM, Chuang WJ, Lin YS, Lin MT, Liu CC, Wu JJ. 2009. emm1/sequence type 28 strains of group A streptococci that express covR at early stationary phase are associated with increased growth and earlier SpeB secretion. J Clin Microbiol 47:3161–3169. doi: 10.1128/JCM.00202-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang CH, Chiang-Ni C, Kuo HT, Zheng PX, Tsou CC, Wang S, Tsai PJ, Chuang WJ, Lin YS, Liu CC, Wu JJ. 2013. Peroxide responsive regulator PerR of group A Streptococcus is required for the expression of phage-associated DNase Sda1 under oxidative stress. PLoS One 8:e81882. doi: 10.1371/journal.pone.0081882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Valcu M, Valcu CM. 2011. Data transformation practices in biomedical sciences. Nat Methods 8:104–105. doi: 10.1038/nmeth0211-104. [DOI] [PubMed] [Google Scholar]
- 52.Chiang-Ni C, Zheng PX, Tsai PJ, Chuang WJ, Lin YS, Liu CC, Wu JJ. 2012. Environmental pH changes, but not the LuxS signalling pathway, regulate SpeB expression in M1 group A streptococci. J Med Microbiol 61:16–22. doi: 10.1099/jmm.0.036012-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S2, Fig. S1-S3. Download spectrum.02033-22-s0001.pdf, PDF file, 2.4 MB (2.5MB, pdf)