ABSTRACT
Tiankeng acts as a refugium for biodiversity amid a changing global climate, and a previous study has shown that some ancient (Alsophila spinulosa) and unique plants (cool-adapted plants) are present in Tiankeng. However, there are few reports on Ascomycota from the Tiankeng karst region. In this research, the species diversity of Cordyceps-like fungi in Monkey-Ear Tiankeng was investigated. Seven species in the genera Akanthomyces, Beauveria, Cordyceps, and Samsoniella were identified based on internal transcribed spacer sequences and morphological characteristics. Eight new species in the genera Akanthomyces, Cordyceps, and Samsoniella were established and described according to a multilocus phylogenetic analysis and morphological characteristics. Our results revealed that Cordyceps-like fungi were abundant in Monkey-Ear Tiankeng, providing new insights into the diversity of Ascomycota in this special eco-environment.
IMPORTANCE Karst Tiankeng has a special eco-environment and acts as a refugium for biodiversity. However, there are few reports on Ascomycota from the Tiankeng karst region. In this research, seven known species and eight new species in the genera Akanthomyces, Beauveria, Cordyceps, and Samsoniella were reported. The results showed that Cordyceps-like fungi are abundant in Monkey-Ear Tiankeng. Interestingly, the month of the sampling was November, which is not an active period of growth and reproduction for Cordyceps-like fungi. These results revealed that unconventional time sampling should not be ignored, especially for a special eco-environment, and provided new insights into the diversity of Ascomycota in this special eco-environment.
KEYWORDS: Tiankeng, ascomycete, Cordyceps-like fungi, phylogenetic analysis, morphological
INTRODUCTION
Tiankeng is a kind of negative karst terrain, which was first named by Zhu in 2001 (1), and it has developed from a carbonate rock stratum, is connected with an underground river at the bottom, and is surrounded by steep rock walls and an aquifer vadose zone with a continuous sedimentary thickness (2). The unique geological landform of Tiankeng creates a microclimate that is different from those of its surrounding areas and acts as a haven of biodiversity in the context of changing climates (3–5).
Research on the Tiankeng karst region has been conducted for many years and has focused on geology (including the morphology, formation, and evolution mechanism of Tiankeng) (6–10), animals (11, 12), plants (13–15), soil microbiology (16–18), the value of Tiankeng's tourism resources (19), and the settlement of organic pollutants in Tiankeng (20, 21). A high floristic diversity, abundant species, a remarkably high uniqueness, a water-bearing capability, and endemic species have been found in the Tiankeng karst region (4). In particular, there is a large amount of water and nutrients present at the bottom of the Tiankeng karst region, which strongly affects the composition and structure of the vegetation (22, 23).
Jiang et al. (24) noted that the soil fungus Trichoderma sp. was able to degrade feathers efficiently. Lan et al. (25) reported that the fungus Exophiala sp., which was isolated from the soil of the Tiankeng karst region, was able to improve the drought resistance and growth of Zenia insignis Chun and Caesalpinia sappan Linn. Long et al. (18) reported a new species, Tiankengomelania guangxiense, which was shown to be able to promote the growth of the medicinal orchid Dendrobium officinale Kimura et Migo and was isolated from the rhizosphere soils of a virgin forest in the Baidong Tiankeng. However, there are few reports regarding the Cordyceps-like fungi isolated from the Tiankeng karst region.
During a survey of entomopathogenic fungi from Southwest China, some Cordyceps-like fungi were found in Monkey-Ear Tiankeng. Eight new species distributed across the three genera Akanthomyces, Cordyceps, and Samsoniella were established based on a multilocus phylogeny as well as their morphological and ecological characteristics.
RESULTS
Phylogenetic analyses.
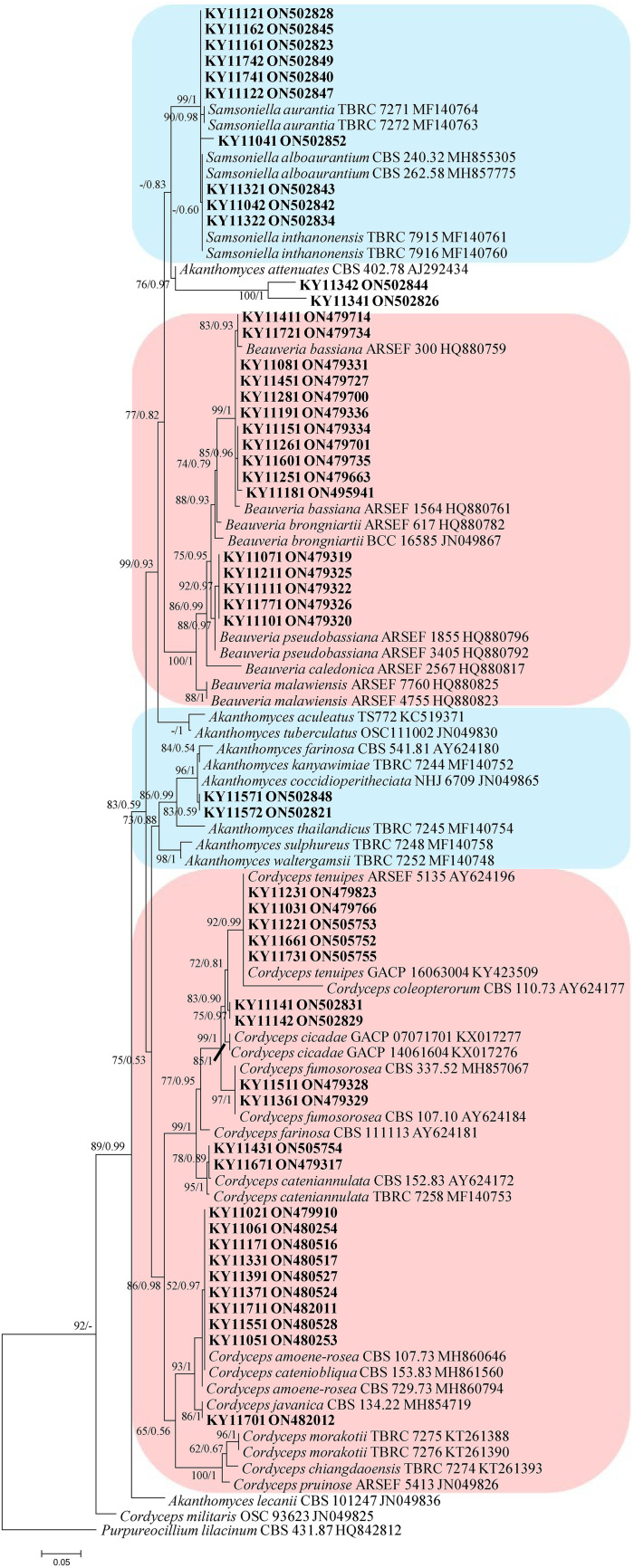
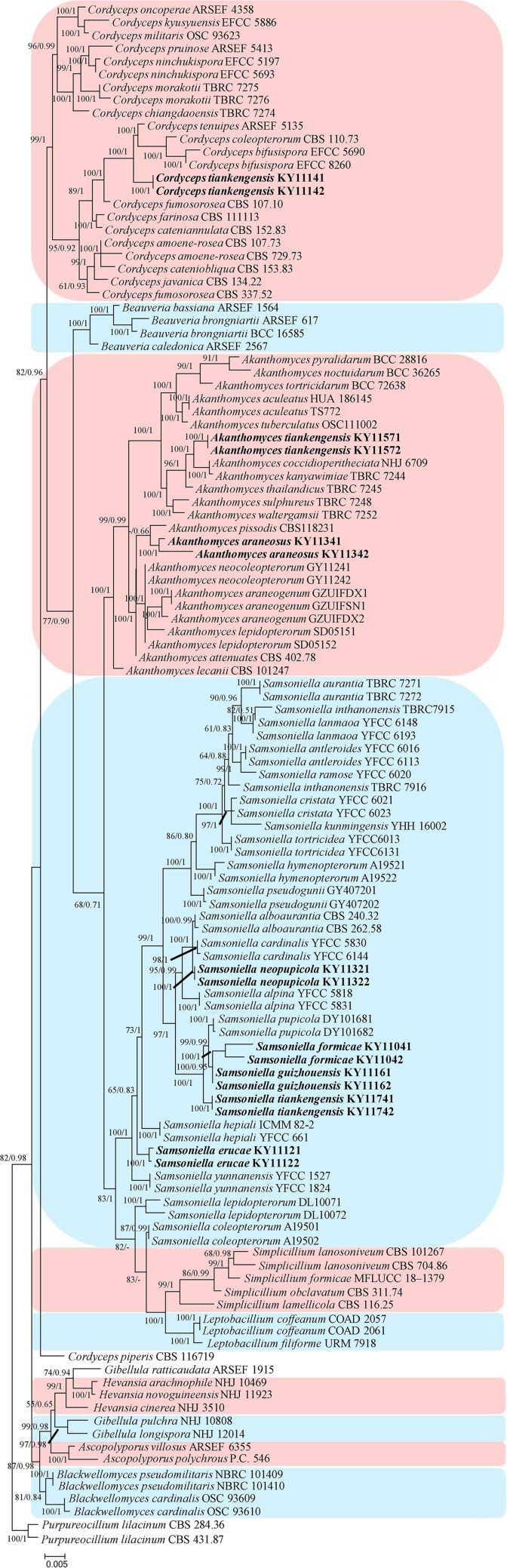
In the phylogenetic tree of analysis 1 (establishing the genus placement of the new strains) and analysis 2 (determining the establishment of the new species) (Fig. 1 and 2, respectively), Purpureocillium lilacinum (Thom) Luangsa-ard, Houbraken, and Hywel-Jones & Samson (CBS 431.87) were used as the outgroup in analysis 1, whereas P. lilacinum (CBS 284.36 and CBS 431.87) was used as the outgroup in analysis 2. The concatenated sequences of analyses 1 and 2 included 32 and 74 taxa, respectively, and consisted of 516 (internal transcribed spacer [ITS], 516) and 2,480 (ITS, 540; large subunit rRNA [LSU], 586; RNA polymerase II largest subunit 2 [RPB2], 671; and translation elongation factor 1 alpha [TEF], 683) characters with gaps, respectively.
FIG 1.
Phylogenetic relationships among the new strains and their allies based on an internal transcribed spacer (ITS) sequence. Statistical support values (≥0.5/50%) are shown at the nodes for maximum likelihood (ML) bootstrap support/Bayesian inference (BI) posterior probabilities.
FIG 2.
Phylogenetic relationships among the new strains and their allies based on a multilocus data set (ITS, large subunit rRNA [LSU], RNA polymerase II largest subunit 2 [RPB2], and translation elongation factor 1 alpha [TEF]). Statistical support values (≥0.5/50%) are shown at the nodes for ML bootstrap support/BI posterior probabilities.
Analysis 1: The final value of the highest scoring tree was –3,525.3844, which was obtained from a maximum likelihood (ML) analysis of ITS sequence. The parameters of the general time-reversible (GTR) model used to analyze the data set were estimated using the following frequencies: A = 0.2280, C = 0.3330, G = 0.2552, and T = 0.1838 with the substitution rates AC = 1.9959, AG = 2.6768, AT = 1.0000, CG = 1.9959, CT = 6.7579 and GT = 1.0000. The gamma distribution shape parameter was α = 0.4764. The selected model for the Bayesian inference (BI) analysis was GTR+F+G4 (ITS). The phylogenetic trees (Fig. 1) constructed using the ML and BI analyses were largely congruent and strongly supported in most branches. The new strains were clustered into to the following genera: Akanthomyces Lebert, Cordyceps Fr., Beauveria Vuill., and Samsoniella Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard species, respectively. Strains KY11411, KY11721, KY11081, KY11451, KY11281, KY11191, KY11151, KY11261, KY11601, KY11251, and KY11181 were clustered into the subclade of Beauveria bassiana (Bals.-Criv.) Vuill. with a high bootstrap value (99/1). Strains KY11071, KY11211, KY11111, KY11771, and KY11101 were clustered into the subclade of B. pseudobassiana S.A. Rehner & Humber with a high bootstrap value (92/0.97). Strains KY11231, KY11031, KY11221, KY11661, and KY11731 were clustered into the subclade of Cordyceps tenuipes (Peck) Kepler, B. Shrestha & Spatafora with a high bootstrap value (92/0.99). Strains KY11511 and KY11361 were clustered into the subclade of C. fumosorosea (Wize) Kepler, B. Shrestha & Spatafora with a high bootstrap value (99/1). Strains KY11431 and KY11671 were clustered into the subclade of C. cateniannulata (Z.Q. Liang) Kepler, B. Shrestha & Spatafora with a high bootstrap value (95/1). Strains KY11021, KY11061, KY11171, KY11331, KY11391, KY11371, KY11711, KY11551, and KY11051 were clustered with C. amoene-rosea (Henn.) Kepler, B. Shrestha & Spatafora and C. cateniobliqua (Z.Q. Liang) Kepler, B. Shrestha & Spatafora. Strain KY11701 was clustered into the subclade of C. javanica (Bally) Kepler, B. Shrestha & Spatafora with a high bootstrap value (86/1). Strains KY11121, KY11122, KY11161, KY11162, KY11741, KY11742, KY11041, KY11042, KY11321 ,and KY11322 were clustered into the clade of the genus Samsoniella Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard. Strains KY11341, KY11342, KY11571, and KY11572 were all clustered with the Akanthomyces species. Strains KY11141 and KY11142 were clustered into the clade of the genus Cordyceps.
Comparing their typical morphological characteristics, strains KY11411, KY11721, KY11081, KY11451, KY11281, KY11191, KY11151, KY11261, KY11601, KY11251, and KY11181 were identified as Beauveria bassiana. Strains KY11071, KY11211, KY11111, KY11771, and KY11101 were identified as B. pseudobassiana. Strains KY11231, KY11031, KY11221, KY11661, and KY11731 were identified as Cordyceps tenuipes. Strains KY11511 and KY11361 were identified as C. fumosorosea. Strains KY11431 and KY11671 were identified as C. cateniannulata. Strains KY11021, KY11061, KY11171, KY11331, KY11391, KY11371, KY11711, KY11551, and KY11051 were identified as C. cateniobliqua. Strain KY11701 was identified as C. javanica. Multilocus phylogenetic analysis was required for the further identification of the strains KY11121, KY11122, KY11161, KY11162, KY11741, KY11742, KY11041, KY11042, KY11321, KY11322, KY11341, KY11342, KY11571, KY11572, KY11141, and KY11142.
Analysis 2: The final value of the highest scoring tree was −28,566.0385, which was obtained from the ML analysis of the data set (ITS+LSU+RPB2+TEF). The parameters of the GTR model used to analyze the data set were estimated based on the following frequencies: A = 0.2351, C = 0.2835, G = 0.2710, and T = 0.2104 with the substitution rates AC = 1.0000, AG = 2.2438, AT = 1.0000, CG = 1.0000, CT = 5.0707, and GT = 1.0000. The gamma distribution shape parameter was α = 0.5592. The selected models for the BI analysis were GTR+F+I+G4 (ITS, TEF), GTR+F+G4 (LSU), and SYM+G4 (RPB2). The phylogenetic trees (Fig. 2) constructed using the ML and BI analyses were largely congruent and strongly supported in most branches. Most of the genera were clustered into their independent clades. Strains KY11141 and KY11142 were clustered with Cordyceps tenuipes (Peck) Kepler, B. Shrestha & Spatafora, C. coleopterorum (Samson & H.C. Evans) Kepler, B. Shrestha & Spatafora, and C. bifusispora O.E. Erikss. in a subclade. Strains KY11571 and KY11572 were clustered with Akanthomyces coccidioperitheciata (Kobayasi & Shimizu) Spatafora, Kepler & B. Shrestha, A. kanyawimiae Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard, and A. thailandicus Mongkols., Spatafora & Luangsa-ard in a subclade. Strains KY11341 and KY11342 were clustered with A. pissodis (Kope & I. Leal) W.H. Chen, Y.F. Han & Z.Q. Liang in a subclade. Strains KY11321 and KY11322 were clustered with Samsoniella alboaurantia (G. Sm.) Mongkols., Noisrip., Thanakitp., Spatafora & Luangsa-ard and S. cardinalis H. Yu, Y.B. Wang, Y. Wang, Q. Fan & Zhu L. Yang in a subclade. Strains KY11041, KY11042, KY11161, and KY11162 were clustered with S. pupicola W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang in a subclade. Strains KY11741, KY11742, KY11121, and KY11122 were clustered into two independent clades.
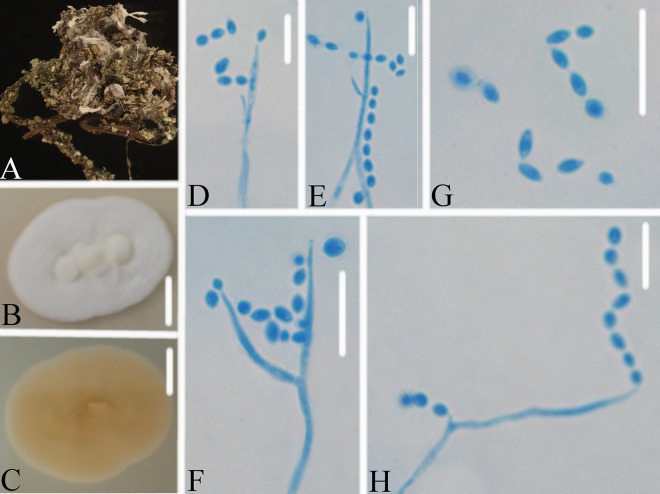
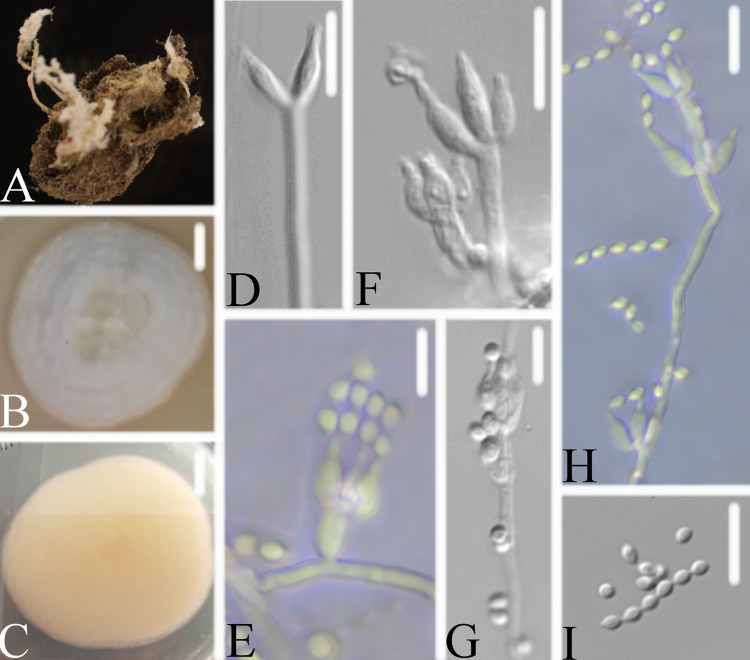
Eight new species distributed in the three genera Akanthomyces, Cordyceps and Samsoniella, were established based on a multi-locus phylogeny, their morphological and ecological characteristics. The morphological and ecological characteristics of the new species were as follows. Akanthomyces araneosus W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov (MycoBank: 844985) (Fig. 3).
FIG 3.
Akanthomyces araneosus. (A) Infected spider (Araneae). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–I) Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–I).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138'' N, 107°0'48.42'' E). On a dead spider (Araneae), 16 November 2020, Wanhao Chen, GZAC KY1134 (holotype), ex-type living cultures, KY11341.
Description: Spider host was completely covered by white mycelium. Conidiophores were mononematous and arose from the lateral hyphae. Colonies on potato dextrose agar (PDA) were 2.4 to 2.6 cm in diameter after 14 d at 25°C, white, and comprised of a basal felt and a floccose hyphal overgrowth with the reverse yellowish. Prostrate hyphae were smooth, septate, hyaline, and 1.0 to 2.5 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of two. Phialides were 16.9 to 18.1 × 1.3 to 1.9 μm with a cylindrical basal portion and tapered into a short, distinct neck. Conidia were hyaline, fusiform, one-celled, and 3.1 to 5.0 × 1.0 to 1.8 μm. The sexual state was not observed.
Host: Spider (Araneae).
Locality: Kaiyang County (27°5'12.138'' N, 107°0'48.42'' E), Guiyang, Guizhou Province, China.
Etymology: Referring to the mycelium covering the spider like a spider web.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138'' N, 107°0'48.42'' E). On a dead spider (Araneae), 16 November 2020, Wanhao Chen, KY11342.
Remarks: Akanthomyces araneosus was easily identified as Akanthomyces, according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1 and 2), and has a close relationship with A. pissodis. When comparing the typical characteristics, A. araneosus was easily distinguished from A. pissodis by its fusiform, smaller conidia (3.1 to 5.0 × 1.0 to 1.8 μm) and its spider host.
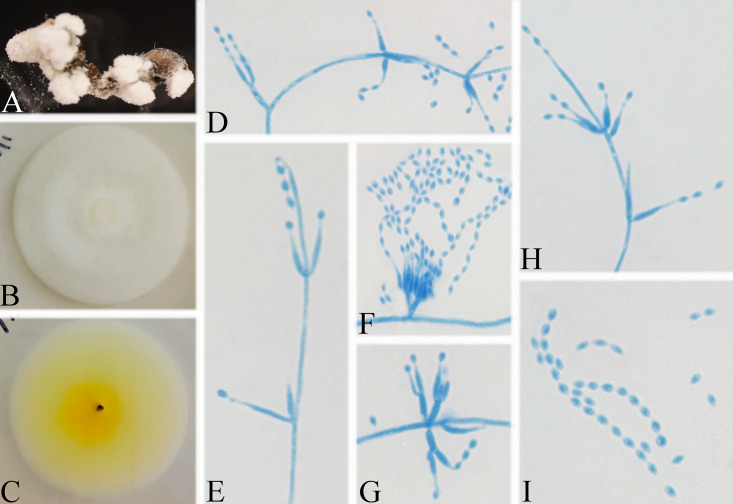
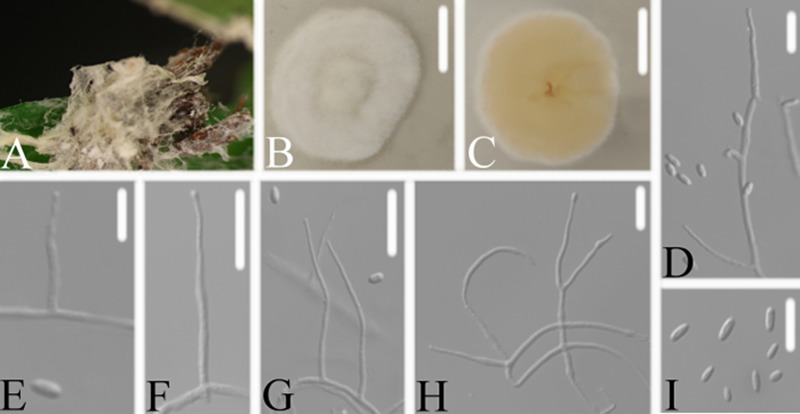
Akanthomyces tiankengensis W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844986) (Fig. 4).
FIG 4.
Akanthomyces tiankengensis. (A) Infected spider (Araneae). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–H) Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–H).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138'' N, 107°0'48.42'' E). On a dead spider (Araneae), 16 November 2020, Wanhao Chen, GZAC KY1157 (holotype), ex-type living cultures, KY11571.
Description: Spider host was completely covered by white mycelium. Conidiophores were mononematous and arose from the lateral hyphae. Colonies on PDA were 2.8 to 3.6 cm in diameter after 14 d at 25°C, white, and comprised of a basal felt and a floccose hyphal overgrowth with the reverse yellowish. Prostrate hyphae were smooth, septate, hyaline, and 1.2 to 2.9 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of two. Phialides were 13.9 to 17.1 × 1.1 to 1.6 μm with a cylindrical basal portion and tapered into a short, distinct neck. Conidia were hyaline, fusiform, one-celled, and 2.3 to 3.0 × 1.5 to 2.3 μm. The sexual state was not observed.
Host: Spider (Araneae).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its location in Tiankeng.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On a dead spider (Araneae), 16 November 2020, Wanhao Chen, KY11572.
Remarks: Akanthomyces tiankengensis was easily identified as Akanthomyces, according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1 and 2), and has a close relationship with A. coccidioperitheciata, A. kanyawimiae, and A. thailandicus. When comparing the typical characteristics, A. tiankengensis was easily distinguished from A. coccidioperitheciata by the absence of a teleomorph and the presence of a spider host. It was distinguished from A. kanyawimiae by its longer phialides (13.9 to 17.1 × 1.1 to 1.6 μm) and fusiform conidia. It was distinguished from A. thailandicus by its smaller conidia (2.3 to 3.0 × 1.5 to 2.3 μm) and cylindrical phialides.
Cordyceps tiankengensis W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844987) (Fig. 5).
FIG 5.
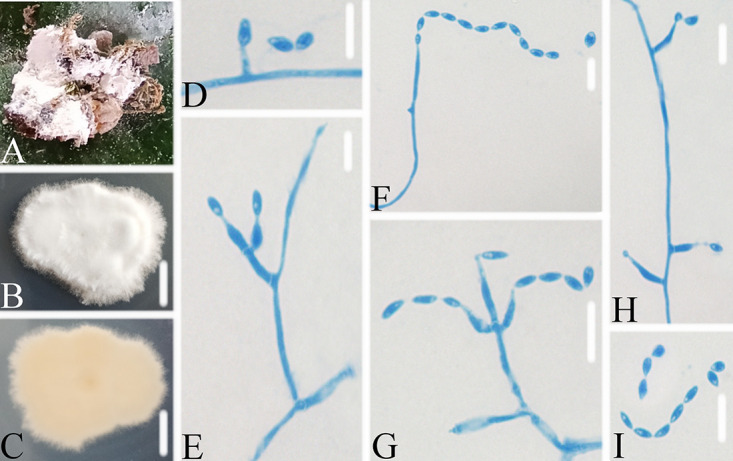
Cordyceps tiankengensis. (A) Infected pupa (Lepidoptera). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–I). Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–I).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, GZAC KY1114 (holotype), ex-type living cultures, KY11141.
Description: Insect pupa was completely covered by white mycelium. Conidiophores were mononematous and arose from the lateral hyphae. Colonies on PDA were 2.2 to 3.2 cm in diameter after 14 d at 25°C, white, and comprised of a basal felt and a floccose hyphal overgrowth with the reverse yellowish. Prostrate hyphae were smooth, septate, hyaline, and 1.3 to 1.9 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of two. Phialides were 7.1 to 12.2 × 1.9 to 2.5 μm with a cylindrical basal portion and tapered into a short, distinct neck. Conidia were in chains, hyaline, fusiform, one-celled, and 4.1 to 5.1 × 1.8 to 2.7 μm. The sexual state was not observed.
Host: Pupa (Lepidoptera).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its location in Tiankeng.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, KY11142.
Remarks: Cordyceps tiankengensis was easily identified as Cordyceps, according to the phylogenetic analysis of combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1 and 2), and has a close relationship with C. tenuipes and C. coleopterorum. When comparing the typical characteristics, C. tiankengensis was easily distinguished from C. tenuipes by its longer phialides (7.1 to 12.2 × 1.9 to 2.5 μm), and it was easily distinguished from C. coleopterorum by its smaller conidia (4.1 to 5.1 × 1.8 to 2.7 μm).
Samsoniella formicae W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844988) (Fig. 6).
FIG 6.
Samsoniella formicae. (A) Infected ant (Formicidae). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–I) Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–I).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E). On an ant (Formicidae), 16 November 2020, Wanhao Chen, GZAC KY1104 (holotype), ex-type living cultures, KY11041.
Description: Ant host was completely covered by white mycelium. Conidiophores were mononematous and arose from the lateral hyphae. Colonies on PDA were 5.0 to 5.1 cm in diameter after 14 d at 25°C, white, and comprised of a basal felt and a floccose hyphal overgrowth with the reverse yellowish or pale orange. Prostrate hyphae were smooth, septate, hyaline, and 1.3 to 1.9 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of two. Phialides were 8.3 to 13.5 × 1.2 to 1.8 μm with a cylindrical basal portion and tapered into a short, distinct neck. Conidia were in chains, hyaline, fusiform, one-celled, and 2.5 to 3.2 × 1.6 to 2.2 μm. The sexual state was not observed.
Host: Ant (Formicidae).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its ant host.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On an ant (Formicidae), 16 November 2020, Wanhao Chen, KY11042.
Remarks: Samsoniella formicae was easily identified as Samsoniella, according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1 and 2), and has a close relationship with S. pupicola and S. guzhouensis. When comparing the typical characteristics, S. formicae was easily distinguished from S. pupicola by its longer phialides (8.3 to 13.5 × 1.2 to 1.8 μm) and its ant host. It was distinguished from S. guzhouensis by its longer phialides (8.3 to 13.5 × 1.2 to 1.8 μm) and its bigger conidia (2.5 to 3.2 × 1.6 to 2.2 μm).
Samsoniella erucae W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844989) (Fig. 7).
FIG 7.
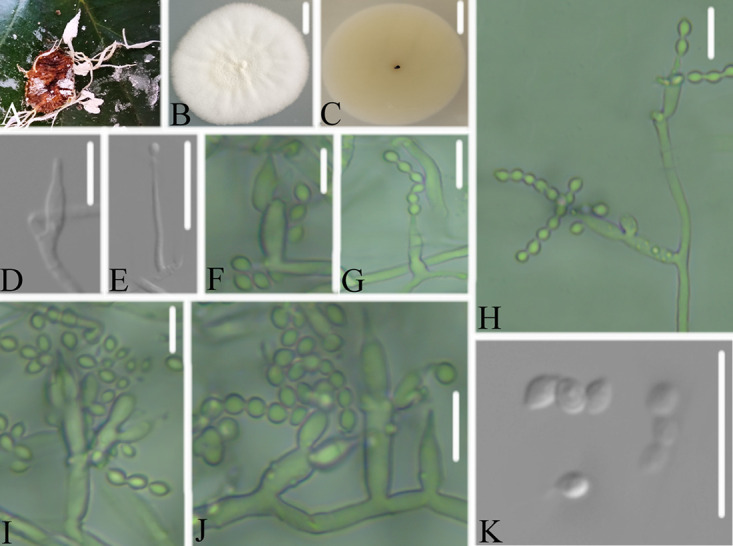
Samsoniella erucae. (A) Infected caterpillar (Lepidoptera). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–K). Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–K).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E). On a caterpillar (Lepidoptera), 16 November 2020, Wanhao Chen, GZAC KY1112 (holotype), ex-type living cultures, KY11121.
Description: Synnemata arose from different parts of the insect host. Conidiophores were synnematous and arose from the lateral hyphae of the synnemata. Colonies on PDA were 4.6 to 4.8 cm in diameter after 14 d at 25°C, white, comprised of a basal felt hyphal overgrowth, and powdery in the middle during mass sporulation with the reverse light yellowish. Prostrate hyphae were smooth, septate, hyaline, and 1.4 to 1.9 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of three. Phialides were 6.8 to 13.7 × 1.1 to 1.5 μm with a cylindrical or ellipsoidal basal portion and tapered into a short, distinct neck. Conidia were in chains, hyaline, fusiform to ellipsoidal, one-celled, and 2.3 to 2.9 × 1.1 to 1.5 μm. The sexual state was not observed.
Host: Caterpillar (Lepidoptera).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its caterpillar host in the order Lepidoptera.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On a caterpillar (Lepidoptera), 16 November 2020, Wanhao Chen, KY11122.
Remarks: Samsoniella erucae was easily identified as Samsoniella according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1). When comparing the typical characteristics, S. erucae was morphologically close to S. coleopterorum by its fusiform to ellipsoidal conidia, and it was distinguished from S. coleopterorum by its longer phialides (6.8 to 13.7 × 1.1 to 1.5 μm) and its bigger conidia (2.3 to 2.9 × 1.1 to 1.5 μm). S. erucae clustered into an independent subclade (Fig. 2) and was distinguished from other species.
Samsoniella guizhouensis W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844990) (Fig. 8).
FIG 8.
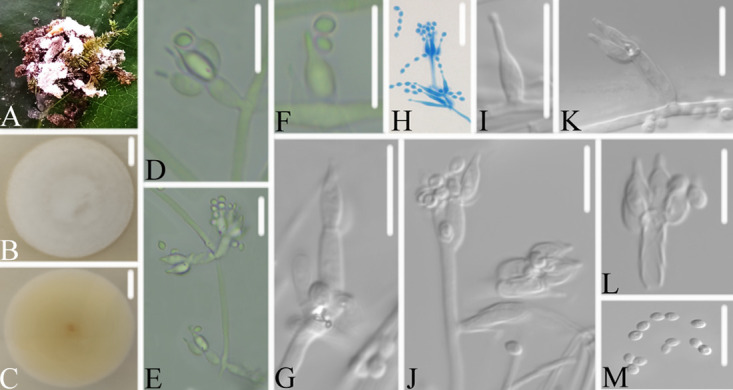
Samsoniella guizhouensis. (A) Infected pupa (Lepidoptera). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–M). Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–M).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, GZAC KY1116 (holotype), ex-type living cultures, KY11161.
Description: Insect pupa was covered completely covered by white mycelium. Conidiophores were mononematous and arose from the lateral hyphae. Colonies on PDA were 4.4 to 4.5 cm in diameter after 14 d at 25°C, white, and comprised of a basal felt and a floccose hyphal overgrowth with the reverse yellowish to pale brown or green. Prostrate hyphae were smooth, septate, hyaline, and 1.3 to 2.5 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of two. Phialides were 4.9 to 7.9 × 1.7 to 2.1 μm with an ellipsoidal basal portion and tapered into a short, distinct neck. Conidia were in chains, hyaline, fusiform, one-celled, and 2.2 to 2.5 × 1.5 to 1.9 μm. The sexual state was not observed.
Host: Pupa (Lepidoptera).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its location in Guizhou Province.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, KY11162.
Remarks: Samsoniella guizhouensis was easily identified as Samsoniella, according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1), and has a close relationship with S. pupicola and S. formicae. When comparing the typical characteristics, S. guizhouensis was easily distinguished from S. pupicola and S. formicae by its smaller conidia (2.2 to 2.5 × 1.5 to 1.9 μm) and its shorter phialides (4.9 to 7.9 × 1.7 to 2.1 μm).
Samsoniella neopupicola W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844991) (Fig. 9).
FIG 9.
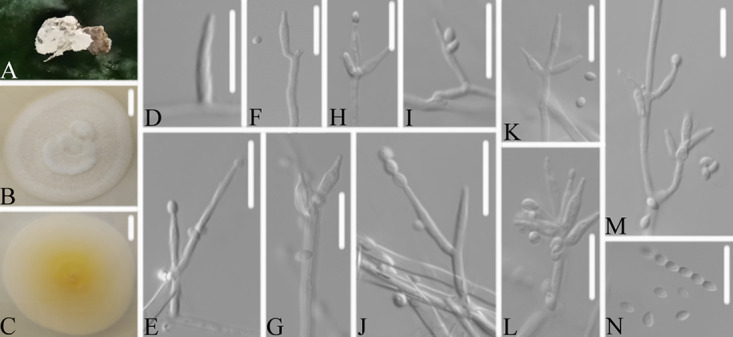
Samsoniella neopupicola. (A) Infected pupa (Lepidoptera). (B, C). PDA-containing culture viewed from above (B) and below (C). (D–N) Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–N).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, GZAC KY1132 (holotype), ex-type living cultures, KY11321.
Description: Insect host was completely covered by white mycelium. Conidiophores were mononematous and arose from the lateral hyphae. Colonies on PDA were 5.5 to 5.7 cm in diameter after 14 d at 25°C, white, and comprised of a basal felt and a floccose hyphal overgrowth with the reverse yellowish or pale orange. Prostrate hyphae were smooth, septate, hyaline, and 1.2 to 2.4 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of five. Phialides were 8.2 to 11.7 × 1.5 to 2.3 μm with a cylindrical basal portion and tapered into a short, distinct neck. Conidia were in chains, hyaline, fusiform, one-celled, and 2.5 to 3.0 × 1.6 to 2.3 μm. The sexual state was not observed.
Host: Pupa (Lepidoptera).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its pupa host in the order Lepidoptera.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, KY11322.
Remarks: Samsoniella neopupicola was easily identified as Samsoniella, according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1), and has a close relationship with S. alboaurantia and S. cardinalis. When comparing the typical characteristics, S. neopupicola was easily distinguished from S. alboaurantia by its longer phialides (8.2 to 11.7 × 1.5 to 2.3 μm) and its Lepidoptera pupa host. It was distinguished from S. cardinalis by its smaller phialides (8.2 to 11.7 × 1.5 to 2.3 μm).
Samsoniella tiankengensis W.H. Chen, Y.F. Han, J.D. Liang & Z.Q. Liang, sp. nov. (MycoBank: 844992) (Fig. 10).
FIG 10.
Samsoniella tiankengensis. (A) Infected pupa (Lepidoptera). (B, C) PDA-containing culture viewed from above (B) and below (C). (D–I) Phialides and conidia. Scale bars: 10 mm (B, C) and 10 μm (D–I).
Type: China, Guizhou, Guiyang, Kaiyang County, Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, GZAC KY1174 (holotype), ex-type living cultures, KY11741.
Description: Synnemata arose from different parts of the insect pupa. Conidiophores were synnematous and arose from the lateral hyphae of the synnemata. Colonies on PDA were 5.3 to 5.6 cm in diameter after 14 d at 25°C, white to light pink, and comprised of a basal felt and a cottony hyphal overgrowth with the reverse light yellowish. Prostrate hyphae were smooth, septate, hyaline, and 1.5 to 2.5 μm in diameter. Erect conidiophores usually arose from the aerial hyphae. Phialides were solitary or in groups of four. Phialides were 5.4 to 10.4 × 1.3 to 2.2 μm with a cylindrical or subellipsoidal basal portion and tapered into a short, distinct neck. Conidia were in chains, hyaline, ellipsoidal, one-celled, and 2.3 to 2.8 × 1.6 to 1.8 μm. The sexual state was not observed.
Host: Pupa (Lepidoptera).
Locality: Kaiyang County (27°5'12.138” N, 107°0'48.42” E), Guiyang, Guizhou Province, China.
Etymology: Referring to its location in Tiankeng.
Additional strain examined: China, Guizhou, Guiyang, Kaiyang County (27°5'12.138” N, 107°0'48.42” E). On a pupa (Lepidoptera), 16 November 2020, Wanhao Chen, KY11742.
Remarks: Samsoniella tiankengensis was easily identified as Samsoniella according to the phylogenetic analysis of the combined data sets (ITS, LSU, RPB2, TEF) (Fig. 1). When comparing the typical characteristics, S. tiankengensis was morphologically close to S. erucae and S. coleopterorum by its fusiform or ellipsoidal conidia, and it was distinguished from S. erucae and S. coleopterorum by its shorter phialides (5.4 to 10.4 × 1.3 to 2.2 μm), its bigger conidia (2.3 to 2.8 × 1.6 to 1.8 μm), and its pupa host. S. tiankengensis clustered into an independent subclade (Fig. 2) and was distinguished from other species.
DISCUSSION
Monkey-Ear Tiankeng is an important modern refugium. It contains cliffs, caves, and an underground river. Following a vertical distribution pattern, the bottom of Tiankeng is the center of the biodiversity, and the species distribution gradually increases from the pit mouth to the pit bottom, such that it can be used as a natural refuge for organisms in rocky karst, desert, and mountainous areas (26, 27). Previous studies have shown that many species of Ascomycota and Basidiomycota are present in the karst cave (an important refugium) (28, 29). Deng and Wu (30) showed that abundant species of Basidiomycota were found in Tiankeng of Leye County. However, few studies have addressed the species diversity of Ascomycota in Tiankeng.
Cordyceps-like fungi are important Ascomycota that originated in remote mountains and dense forests and have become closely intertwined with people’s lives (31). Cordyceps-like fungi were previously used as traditional medicinal fungi and as a type of health food, and their uses have expanded into other fields, such as biological materials, biochromes, and new bioactive substances, because of the expansion of their members and their host range. The physiological effects of extracts from Cordyceps-like fungi and their active components have been involved in promoting the production of cytokines, such as interleukin and tumor necrosis factor-α, and used for their antioxidant, anticancer, hypolipidemic, hypoglycemic, antifatigue, anti-aging, cholesterol-lowering, blood-pressure-lowering, antidepressant, aphrodisiac, and kidney-protecting properties (32). Cordyceps-like fungi also have broad application prospects in the fields of fungal feed additives, the environment, nano-materials, and biotransformation (33).
In the present study, the species diversity of a Cordyceps-like fungus in Monkey-Ear Tiankeng was investigated. 77 specimens were collected, and 51 strains were isolated. In a combined analysis of ITS sequences and morphological characteristics, 35 strains were identified: Beauveria bassiana (11 strains), B. pseudobassiana (5 strains), Cordyceps tenuipes (5 strains), C. fumosorosea (2 strains), C. cateniannulata (2 strains), C. cateniobliqua (9 strains), and C. javanica (1 strain). A total of 16 strains were clustered into the following genera: Akanthomyces (4 strains), Cordyceps (2 strains), and Samsoniellla (10 strains). More information is needed for further identification. Eight new species: A. araneosus, A. tiankengensis, C. tiankengensis, S. formicae, S. erucae, S. guizhouensis, S. neopupicola, and S. tiankengensis, were established and described according to a multilocus phylogenetic analysis and their morphological characteristics. Our results showed that Cordyceps-like fungi are abundant in Monkey-Ear Tiankeng. Interestingly, the month of the sampling was November, which is not an active period of growth and reproduction for Cordyceps-like fungi, revealing that unconventional time sampling should not be ignored, especially for a special eco-environment, such as Tiankeng. However, further research is needed to confirm whether the diversity of the Cordyceps-like fungi in Tiankeng, especially for the new species, is related to the sampling time and the environment.
Cordyceps-like fungi are all-rounders in their nutrition intake. The nutritional model of Cordyceps-like fungi was found to range from plants (including living plants and plant residues) to animals (especially insects) and even to fungi (34). The initial sources of nutrition of the Cordyceps-like fungi were plants and their residues. Beauveria bassiana is an important traditional medicinal fungus and was successfully applied in biocontrol. Previous studies have revealed that B. bassiana could cause endophytic colonization in plants, induce systemic resistance against plant pathogens, promote plant growth, and enhance the resistance of plants to insect pests (35, 36). Cordyceps fumosorosea is usually used for biocontrol and can enhance plant growth and prevent insect pests (37). Most Cordyceps-like fungi are soil-dwelling microbes (38), and they may play an important role in the cycling of carbon and nutrients in their habitats (39). Previous studies have found that plant diversity in the Tiankeng karst region is characterized by rich species diversity, ancient origins, and characteristics of secondary vegetation (3). The relationship among the Cordyceps-like fungi, plants, and Tiankeng is worthy of further research.
Tiankeng acts as a refugium for biodiversity amid changing global climates, as has been shown in many studies (10, 17). A previous study showed that some ancient (Alsophila spinulosa) and unique plants (cool-adapted plants) are present in Tiankeng (23, 40). Interestingly, abundant Cordyceps-like fungi were found in Monkey-Ear Tiankeng, and the hosts of the new species were relatively simple. Akanthomyces species are often isolated from soil, insects, and spiders (41, 42), and Samsoniella species are often found initiated on a pupa or larva of Lepidoptera, beetles, bees, or ants (43, 44). Furthermore, sampling was performed in November, when the temperature of the environment was lower. Whether these fungi are more ancient than others, have adapted to the cold environment, or have coevolved with their hosts and have special metabolizing processes is worthy of further research.
MATERIALS AND METHODS
Specimen collection and identification.
77 infected insect and spider specimens (named KY1101 to KY1177) were collected from Monkey-Ear Tiankeng (27°5'12.138” N, 107°0'48.42” E), Kaiyang County, Guiyang, Guizhou Province, on 16 November 2020. The area belongs to a subtropical monsoon humid climate zone that receives an annual precipitation of 1,141 to 1,547 mm and has an annual average temperature of 10.6 to 15.3°C (26). The isolation of the strains was conducted as described by Chen et al. (45). Fungal colonies emerging from specimens were isolated and cultured at 25°C for 14 days under 12 h light/12 h dark conditions, following the protocol described by Zou et al. (46). The specimens and the isolated strains were deposited in the Institute of Fungus Resources, Guizhou University (formally Herbarium of Guizhou Agricultural College; code: GZAC), Guiyang City, Guizhou, China.
The macroscopic and microscopic morphological characteristics of the fungi were examined, especially for the arrangement, shape, and measurement of phialides and conidia. The growth rates were also determined from potato dextrose agar cultures incubated at 25°C for 14 days. The hyphae and conidiogenous structures were mounted in lactophenol cotton blue or a 20% lactate solution and observed with an optical microscope (OM, DM4 B, Leica, Germany).
DNA extraction, PCR amplification, and nucleotide sequencing.
DNA extraction was carried out using the Fungal Genomic DNA Extraction Kit (DP2033, BioTeke Corporation) in accordance with Liang et al. (47). The extracted DNA was stored at −20°C. The amplification of the internal transcribed spacer (ITS) region, the large subunit rRNA (LSU) loci, the small subunit rRNA (SSU), the RNA polymerase II largest subunit 2 (RPB2), and the translation elongation factor 1 alpha (TEF) was determined by polymerase chain reaction (PCR) as described by White et al. (48), Castlebury et al. (49), and van den Brink et al. (50), respectively. Primer sequence information is shown in Table S1. The PCR products were purified and sequenced at Sangon Biotech (Shanghai) Co. The resulting sequences were submitted to GenBank (Table 1).
TABLE 1.
Taxa included in the phylogenetic analyses
| Species | Strain no. | GenBank accession no. |
Reference | |||
|---|---|---|---|---|---|---|
| ITS | LSU | RPB2 | TEF | |||
| Akanthomyces aculeatus | HUA 186145 | MF416520 | MF416465 | 62 | ||
| Akanthomyces aculeatus | TS772 | KC519371 | KC519370 | KC519366 | 63 | |
| Akanthomyces araneogenum | GZUIFDX2 | KU893153 | MH978179 | MH978185 | MH978187 | 64 |
| Akanthomyces araneogenum | GZUIFDX1 | KU893152 | MH978178 | MH978184 | 64 | |
| Akanthomyces araneogenum | GZUIFSN1 | MH978177 | MH978180 | MH978186 | MH978188 | 64 |
| Akanthomyces araneosus | KY11341 | ON502826 | ON502832 | ON525442 | ON525443 | This study |
| Akanthomyces araneosus | KY11342 | ON502844 | ON502837 | ON525444 | ON525445 | This study |
| Akanthomyces attenuatus | CBS 402.78 | AJ292434 | AF339565 | EF468935 | EF468782 | 65 |
| Akanthomyces coccidioperitheciata | NHJ 6709 | JN049865 | EU369042 | EU369086 | EU369025 | 65 |
| Akanthomyces farinosa | CBS 541.81 | AY624180 | JQ425686 | 66 | ||
| Akanthomyces lecanii | CBS 101247 | JN049836 | AF339555 | DQ522466 | DQ522359 | 65 |
| Akanthomyces neocoleopterorum | GY11241 | MN093295 | MN093296 | MN097812 | MN097813 | 41 |
| Akanthomyces neocoleopterorum | GY11242 | MN093297 | MN093298 | MN097814 | MN097815 | 41 |
| Akanthomyces lepidopterorum | SD05151 | MT705971 | MT705973 | MT727044 | 52 | |
| Akanthomyces lepidopterorum | SD05152 | MT705972 | MT705974 | MT727045 | 52 | |
| Akanthomyces noctuidarum | BCC 36265 | MT356072 | MT356084 | MT477987 | MT477978 | 42 |
| Akanthomyces kanyawimiae | TBRC 7244 | MF140752 | MF140716 | MF140836 | 67 | |
| Akanthomyces pissodis | CBS 118231 | KM283799 | KM283864 | KM283822 | 52 | |
| Akanthomyces pyralidarum | BCC28816 | MT356080 | MT356091 | MT478007 | MT477982 | 42 |
| Akanthomyces sulphureus | TBRC 7248 | MF140758 | MF140722 | MF140812 | MF140843 | 67 |
| Akanthomyces thailandicus | TBRC 7245 | MF140754 | MF140809 | MF140839 | 67 | |
| Akanthomyces tiankengensis | KY11571 | ON502848 | ON502825 | ON525446 | ON525447 | This study |
| Akanthomyces tiankengensis | KY11572 | ON502821 | ON502827 | ON525448 | ON525449 | This study |
| Akanthomyces tortricidarum | BCC72638 | MT356076 | MT356088 | MT477992 | MT478004 | 42 |
| Akanthomyces tuberculatus | OSC 111002 | JN049830 | DQ518767 | DQ522435 | DQ522338 | 34 |
| Akanthomyces waltergamsii | TBRC 7252 | MF140748 | MF140714 | MF140834 | 67 | |
| Ascopolyporus polychrous | P.C. 546 | DQ118737 | DQ118745 | 68 | ||
| Ascopolyporus villosus | ARSEF 6355 | AY886544 | DQ118750 | 68 | ||
| Beauveria bassiana | ARSEF 1564 | HQ880761 | HQ880905 | HQ880974 | 69 | |
| Beauveria brongniartii | ARSEF 617 | HQ880782 | HQ880926 | HQ880991 | 69 | |
| Beauveria brongniartii | BCC 16585 | JN049867 | JF415967 | JF415991 | JF416009 | 65 |
| Beauveria caledonica | ARSEF 2567 | HQ880817 | AF339520 | HQ880961 | EF469057 | 63 |
| Blackwellomyces cardinalis | OSC 93609 | AY184962 | DQ522422 | DQ522325 | 63 | |
| Blackwellomyces cardinalis | OSC 93610 | JN049843 | AY184963 | EF469106 | EF469059 | 65 |
| Blackwellomyces pseudomilitaris | NBRC 101409 | JN943305 | JN941393 | 70 | ||
| Blackwellomyces pseudomilitaris | NBRC 101410 | JN943307 | JN941394 | 70 | ||
| Cordyceps amoene-rosea | CBS 107.73 | AY624168 | MG665224 | MG665234 | 67 | |
| Cordyceps amoene-rosea | CBS 729.73 | AY624169 | MG665225 | MG665235 | HM161732 | 67 |
| Cordyceps bifusispora | EFCC 5690 | EF468806 | EF468909 | EF468746 | 51 | |
| Cordyceps bifusispora | EFCC 8260 | EF468807 | EF468910 | EF468747 | 51 | |
| Cordyceps cateniannulata | CBS 152.83 | AY624172 | MG665226 | JQ425687 | 67 | |
| Cordyceps cateniobliqua | CBS 153.83 | AY624173 | MG665236 | JQ425688 | 67 | |
| Cordyceps chiangdaoensis | TBRC 7274 | KT261393 | MF140732 | KT261403 | 67 | |
| Cordyceps coleopterorum | CBS 110.73 | AY624177 | JF415988 | JF416006 | JF416028 | 66 |
| Cordyceps farinosa | CBS 111113 | AY624181 | FJ765253 | GU979973 | GQ250022 | 67 |
| Cordyceps fumosorosea | CBS 107.10 | AY624184 | MG665227 | MG665237 | HM161735 | 67 |
| Cordyceps fumosorosea | CBS 337.52 | EF411219 | MG665228 | MG665233 | 67 | |
| Cordyceps javanica | CBS 134.22 | AY624186 | MG665231 | JQ425683 | 67 | |
| Cordyceps kyusyuensis | EFCC 5886 | EF468813 | EF468917 | EF468754 | 51 | |
| Cordyceps militaris | OSC 93623 | JN049825 | AY184966 | DQ522332 | 51 | |
| Cordyceps morakotii | TBRC 7275 | KT261388 | MF140730 | KT261398 | 67 | |
| Cordyceps morakotii | TBRC 7276 | KT261390 | MF140731 | KT261400 | 67 | |
| Cordyceps ninchukispora | EFCC 5197 | EF468820 | EF468760 | 51 | ||
| Cordyceps ninchukispora | EFCC 5693 | EF468821 | EF468762 | 51 | ||
| Cordyceps oncoperae | AFSEF 4358 | AF339532 | EF468936 | EF468785 | 51 | |
| Cordyceps tiankengensis | KY11141 | ON502831 | ON502824 | ON525440 | This study | |
| Cordyceps tiankengensis | KY11142 | ON502829 | ON502836 | ON525441 | This study | |
| Cordyceps piperis | CBS 116719 | AY466442 | EU369083 | DQ118749 | 71 | |
| Cordyceps pruinosa | ARSEF 5413 | JN049826 | AY184968 | DQ522451 | DQ522351 | 65 |
| Cordyceps tenuipes | ARSEF 5135 | AY624196 | JF415980 | JF416000 | JF416020 | 65 |
| Gibellula longispora | NHJ 12014 | EU369075 | EU369017 | 71 | ||
| Gibellula pulchra | NHJ 10808 | EU369035 | EU369076 | EU369018 | 71 | |
| Gibellula ratticaudata | ARSEF 1915 | DQ518777 | DQ522467 | DQ522360 | 71 | |
| Hevansia arachnophila | NHJ 10469 | EU369031 | EU369008 | 71 | ||
| Hevansia cinerea | NHJ 3510 | EU369070 | EU369009 | 71 | ||
| Hevansia novoguineensis | NHJ 11923 | EU369032 | EU369072 | EU369013 | 71 | |
| Leptobacillium coffeanum | COAD 2057 | MF066034 | MF066032 | 72 | ||
| Leptobacillium coffeanum | COAD 2061 | MF066035 | MF066033 | 72 | ||
| Leptobacillium filiforme | URM 7918 | MH979338 | MH979399 | 73 | ||
| Purpureocillium lilacinum | CBS 284.36 | EF468941 | EF468792 | 65 | ||
| Purpureocillium lilacinum | CBS 431.87 | HQ842812 | EF468844 | EF468940 | EF468791 | 51 |
| Samsoniella alboaurantium | CBS 240.32 | JF415979 | JF415999 | JF416019 | 65 | |
| Samsoniella alboaurantium | CBS 262.58 | AB080087 | MF416448 | MF416497 | 67 | |
| Samsoniella alpina | YFCC 5818 | MN576809 | MN576923 | MN576979 | 74 | |
| Samsoniella alpina | YFCC 5831 | MN576810 | MN576924 | MN576980 | 74 | |
| Samsoniella antleroides | YFCC 6016 | MN576803 | MN576917 | MN576973 | 74 | |
| Samsoniella antleroides | YFCC 6113 | MN576804 | MN576918 | MN576974 | 74 | |
| Samsoniella aurantia | TBRC 7271 | MF140728 | MF140818 | MF140846 | 67 | |
| Samsoniella aurantia | TBRC 7272 | MF140727 | MF140817 | MF140845 | 67 | |
| Samsoniella cardinalis | YFCC 5830 | MN576788 | MN576902 | MN576958 | 74 | |
| Samsoniella cardinalis | YFCC 6144 | MN576786 | MN576900 | MN576956 | 74 | |
| Samsoniella coleopterorum | A19501 | MT626376 | MN101585 | MN101586 | 43 | |
| Samsoniella coleopterorum | A19502 | MT626625 | MN101587 | MT642602 | 43 | |
| Samsoniella cristata | YFCC 6021 | MN576791 | MN576905 | MN576961 | 74 | |
| Samsoniella cristata | YFCC 6023 | MN576792 | MN576906 | MN576962 | 74 | |
| Samsoniella erucae | KY11121 | ON502828 | ON502835 | ON525424 | ON525425 | This study |
| Samsoniella erucae | KY11122 | ON502847 | ON502822 | ON525426 | ON525427 | This study |
| Samsoniella formicae | KY11041 | ON502852 | ON525420 | ON525421 | This study | |
| Samsoniella formicae | KY11042 | ON502842 | ON525422 | ON525423 | This study | |
| Samsoniella guizhouensis | KY11161 | ON502823 | ON502830 | ON525428 | ON525429 | This study |
| Samsoniella guizhouensis | KY11162 | ON502845 | ON502846 | ON525430 | ON525431 | This study |
| Samsoniella hepiali | ICMM 82-2 | MN576794 | MN576908 | MN576964 | 74 | |
| Samsoniella hepiali | YFCC 661 | MN576795 | MN576909 | MN576965 | 74 | |
| Samsoniella hymenopterorum | A19521 | MN128224 | MT642604 | MN101588 | 43 | |
| Samsoniella hymenopterorum | A19522 | MN128081 | MN101590 | MN101591 | 43 | |
| Samsoniella inthanonensis | TBRC 7915 | MF140761 | MF140815 | MF140849 | 67 | |
| Samsoniella inthanonensis | TBRC 7916 | MF140760 | MF140814 | MF140848 | 67 | |
| Samsoniella kunmingensis | YHH 16002 | MN576802 | MN576916 | MN576972 | 74 | |
| Samsoniella lanmaoa | YFCC 6148 | MN576789 | MN576903 | MN576959 | 74 | |
| Samsoniella lanmaoa | YFCC 6193 | MN576790 | MN576904 | MN576960 | 74 | |
| Samsoniella lepidopterorum | DL10071 | MN128076 | MN101593 | MN101594 | 43 | |
| Samsoniella lepidopterorum | DL10072 | MN128084 | MT642605 | MT642606 | 43 | |
| Samsoniella neopupicola | KY11321 | ON502843 | ON502839 | ON525432 | ON525433 | This study |
| Samsoniella neopupicola | KY11322 | ON502834 | ON502833 | ON525434 | ON525435 | This study |
| Samsoniella pseudogunii | GY407201 | MZ827470 | MZ827010 | MZ855239 | MZ855233 | 44 |
| Samsoniella pseudogunii | GY407202 | MZ831863 | MZ831865 | MZ855240 | MZ855234 | 44 |
| Samsoniella pupicola | DY101681 | MZ827085 | MZ827009 | MZ855237 | MZ855231 | 44 |
| Samsoniella pupicola | DY101682 | MZ827008 | MZ827635 | MZ855238 | MZ855232 | 44 |
| Samsoniella ramose | YFCC 6020 | MN576805 | MN576919 | MN576975 | 74 | |
| Samsoniella tiankengensis | KY11741 | ON502840 | ON502838 | ON525436 | ON525437 | This study |
| Samsoniella tiankengensis | KY11742 | ON502849 | ON502841 | ON525438 | ON525439 | This study |
| Samsoniella tortricidae | YFCC 6013 | MN576807 | MN576921 | MN576977 | 74 | |
| Samsoniella tortricidae | YFCC 6131 | MN576806 | MN576920 | MN576976 | 74 | |
| Samsoniella yunnanensis | YFCC 1527 | MN576812 | MN576926 | MN576982 | 74 | |
| Samsoniella yunnanensis | YFCC 1824 | MN576813 | MN576927 | MN576983 | 74 | |
| Simplicillium formicae | MFLUCC 18–1379 | MK766511 | MK766512 | MK926451 | 75 | |
| Simplicillium lamellicola | CBS 116.25 | AJ292393 | AF339552 | DQ522462 | DQ522356 | 51 |
| Simplicillium lanosoniveum | CBS 101267 | AJ292395 | AF339554 | DQ522463 | DQ522357 | 51 |
| Simplicillium lanosoniveum | CBS 704.86 | AF339553 | DQ522464 | DQ522358 | 51 | |
| Simplicillium obclavatum | CBS 311.74 | AF339517 | EF468798 | 51 | ||
Sequence alignment and phylogenetic analyses.
Lasergene software (version 6.0, DNASTAR) was applied for the assembly and editing of the DNA sequences in this study. ITS, LSU, RPB2, and TEF sequences were downloaded from GenBank, based on Sung et al. (51), Chen et al. (41, 43, 44, 52), Li et al. (53), Aini et al. (42), and others selected on the basis of Basic Local Alignment Search Tool (BLAST) algorithm-based searches in GenBank (Table 1). A single-locus data set was aligned and edited using multiple alignment using fast Fourier transform (MAFFT) v7.037b (54) and Molecular Evolutionary Genetics Analysis version 6 (55). Combined sequences of ITS, LSU, RPB2, and TEF were performed in SequenceMatrix v.1.7.8 (56). The model for the Bayesian analysis was selected using ModelFinder (57) with PhyloSuite software (58).
The combined loci were analyzed using Bayesian inference (BI) and maximum likelihood (ML) methods. For the BI, a Markov chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities for the combined sequence data sets using MrBayes v.3.2 (59). The Bayesian analysis resulted in 20,001 trees after 10,000,000 generations. The first 4,000 trees, representing the burn-in phase of the analyses, were discarded, while the remaining 16,001 trees were used to calculate the posterior probabilities for the majority rule consensus tree. After the analysis was finished, each run was examined using the Tracer v1.5 program (60) to determine the burn-in and to confirm that both runs had converged. The ML analysis was designed with IQ-TREE (61), and the model was automatically selected by the software.
ACKNOWLEDGMENTS
This work was funded by the National Natural Science Foundation of China (31860002), High-level Innovative Talents Training Object in Guizhou Province (Qiankehepingtairencai [2020]6005), Science and Technology Foundation of Guizhou Province (Qiankehejichu [2020]1Y060), Program of Innovative Scientific and technological Talent Team of Guizhou Province (2020-5010), Construction Program of Guizhou Engineering Research Center (Qian Fa Gai Gao Ji 2020-896), and Guizhou Science and Technology Support Project (Qiankehezhicheng [2019]2776).
Footnotes
Supplemental material is available online only.
Contributor Information
Yan-Feng Han, Email: swallow1128@126.com.
Matthew Zack Anderson, The Ohio State University.
REFERENCES
- 1.Zhu XW. 2001. China’s karst Tiankeng and its value for science and tourism. Sci Technol Rev 10:60–65. [Google Scholar]
- 2.Zhu XW, Waltham T. 2005. Tiankeng: definition and description. Cave and Karst Sci 32:75–79. [Google Scholar]
- 3.Pu GZ, Lv YN, Xu GP, Zeng DJ, Huang Y. 2017. Research progress on karst tiankeng ecosystems. Bot Rev 83:5–37. doi: 10.1007/s12229-017-9179-0. [DOI] [Google Scholar]
- 4.Su Y, Tang Q, Mo F, Xue Y. 2017. Karst tiankengs as refugia for indigenous tree flora amidst a degraded landscape in southwestern China. Sci Rep 7:1–10. doi: 10.1038/s41598-017-04592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ozkan K, Gulsoy S, Mert A, Ozturk M, Muys B. 2010. Plant distribution-altitude and landform relationships in karstic sinkholes of Mediterranean region of Turkey. J Environ Biol 31:51–60. [PubMed] [Google Scholar]
- 6.Palmer AN, Palmer MV. 2005. Hydraulic processes in the origin of Tiankengs. Cave and Karst Sci 32:101–106. [Google Scholar]
- 7.Klimchouk A. 2006. Cave un-roofing as a large-scale geomorphic process. Speleogenesis Evolution Karst Aquifers 4:1–11. [Google Scholar]
- 8.Waltham T. 2008. Fengcong, fenglin, cone karst and tower karst. Cave Karst Sci 35:77–88. [Google Scholar]
- 9.De Waele J, Gutiérrez F, Parise M, Plan L. 2011. Geomorphology and natural hazards in karst areas: a review. Geomorphology 134:1–8. doi: 10.1016/j.geomorph.2011.08.001. [DOI] [Google Scholar]
- 10.Shui W, Chen YP, Wang YW, Su ZA, Zhang S. 2015. Origination, study progress and prospect of karst tiankeng research in China. Acta Geogr Sin 70:431–446. doi: 10.11821/dlxb201503007. [DOI] [Google Scholar]
- 11.Jiang ZG. 2012. Biodiversity and its conservation in the Tiankeng-Difeng region, 1st ed, Chinese Forestry Press, Beijing, China. [Google Scholar]
- 12.Jian XM, Shui W, Wang YN, Wang QF, Chen YP, Jiang C, Xiang ZY. 2018. Species diversity and stability of grassland plant community in heavily-degraded karst tiankeng: a case study of Zhanyi Tiankeng in Yunnan. China Acta Ecol Sin 38:4704–4714. doi: 10.5846/stxb201706281163. [DOI] [Google Scholar]
- 13.Fan BB. 2014. Master thesis. The study on characteristics and succession of karst Tiankeng community in Dashiwei. Guangxi Normal University, Guilin, Guangxi. [Google Scholar]
- 14.Feng HZ. 2015. Master thesis. The studied on origin and evolution of karst tiankeng flora in Dashiwei. Guangxi Normal University, Guilin, Guangxi. [Google Scholar]
- 15.Sheng MY, Xiong KN, Cui GY, Liu Y. 2015. Plant diversity and soil physical-chemical properties in karst rocky desertification ecosystem of Guizhou, China. Acta Ecol Sin 35:434–448. doi: 10.5846/stxb201303220488. [DOI] [Google Scholar]
- 16.Jiang C, Feng J, Zhu SF, Shui W. 2021. Characteristics of the soil microbial communities in different slope positions along an inverted stone slope in a degraded karst Tiankeng. Biology 10:474. doi: 10.3390/biology10060474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pu GZ, Lv YN, Dong L, Zhou LW, Huang KC, Zeng DJ, Mo L, Xu GP. 2019. Profiling the bacterial diversity in a typical karst Tiankeng of China. Biomolecules 9:187. doi: 10.3390/biom9050187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long Y, Nong Q, Xie L, Zhang W, Chen Y, Zhang Y. 2022. Tiankengomelania guangxiense, gen. et sp. nov., a dark septate endophytic fungus, promotes the growth of the medicinal orchid Dendrobium officinale. Fungal Biol 126:333–341. doi: 10.1016/j.funbio.2022.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Zhu XW, Chen WH. 2006. Tiankeng in the karst of China. Carsol Sin 25:7–24. [Google Scholar]
- 20.Ao LM. 2007. Master thesis. Distribution, sources and transportation of polycyclic aromatic hydrocarbons and organochlorine pesticides in tiankeng group, south China. China University Geosciences, Beijing. [Google Scholar]
- 21.Kong XS. 2012. Master thesis. Environmental behaviour of persistent organic pollutants in a typical karst sinkhole-a case in Dashiwei Tiankeng group in Guangxi. China University Geoscience, Beijing. [Google Scholar]
- 22.Bátori Z, Erdős L, Morschhauser T, Török P, Körmöczi L. 2009. Vegetation of the dolines in Mecsek Mountains (South Hungary) in relation to the local plant communities. AC 38:237–252. doi: 10.3986/ac.v38i2-3.125. [DOI] [Google Scholar]
- 23.Bátori Z, Vojtkó A, Farkas T, Szabó A, Havadtői K, Vojtkó AE, Tölgyesi C, Cseh V, Erdős L, Maák IE, Keppel G. 2017. Large-and small-scale environmental factors drive distributions of cool-adapted plants in karstic microrefugia. Ann Bot 119:301–309. doi: 10.1093/aob/mcw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang Z, Bing LU, Chen B, Qin LU. 2014. Screening, identification and enzyme-producing conditions of strains with high keratinase yield. Guizhou Agric Sci 42:139–141. [Google Scholar]
- 25.Lan TJ, Chen YL, Huang CM, Zhang WL, Xie L, Shi GY, Qin LP, Zhang Y, Nong Q. 2017. Community constituent of dark septate endophytic fungi in Dashiwei doline group and their effects on pioneer plants’ drought resistance capability. J Microbiol 37:26–34. [Google Scholar]
- 26.Li CY, Zhang CH, Wu J, Li XF. 2019. Distribution pattern of liverwort community in relation to environmental factors of caves in karst Tiankeng: a case study of Monkey-Ear Tiankeng of Guizou Province. Chin J Ecol 38:744–752. doi: 10.13292/j.1000-4890.201903.007. [DOI] [Google Scholar]
- 27.Li XF, Zhang CH, Li CY, Wu J, Wang ZH. 2018. Diversity of bryophytes in underground forest of Monkey-Ear Tiankeng. Acta Bot Boreali-Occident Sin 38:2324–2333. [Google Scholar]
- 28.Zhang ZF, Liu F, Zhou X, Liu XZ, Liu SJ, Cai L. 2017. Culturable mycobiota from Karst caves in China, with descriptions of 20 new species. Persoonia 39:1–31. doi: 10.3767/persoonia.2017.39.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang ZF, Zhou SY, Eurwilaichitr L, Ingsriswang S, Raza M, Chen Q, Zhao P, Liu F, Cai L. 2021. Culturable mycobiota from Karst caves in China II, with descriptions of 33 new species. Fungal Divers 106:29–136. doi: 10.1007/s13225-020-00453-7. [DOI] [Google Scholar]
- 30.Deng CY, Wu XL. 2014. The component and assessment of macro-fungi in leye county, Guangxi autonomous region. China Guizhou Sci 32:1–18. [Google Scholar]
- 31.Chen WH, Liang JD, Han YF, Zou X, Zhang YJ, Liang ZQ. 2021. Research overviews of Cordyceps: past, present and future. Mycosystema 40:2894–2905. doi: 10.13346/j.mycosystema.210164. [DOI] [Google Scholar]
- 32.Das G, Shin H-S, Leyva-Gómez G, Prado-Audelo MLD, Cortes H, Singh YD, Panda MK, Mishra AP, Nigam M, Saklani S, Chaturi PK, Martorell M, Cruz-Martins N, Sharma V, Garg N, Sharma R, Patra JK. 2020. Cordyceps spp.: a review on its immune-stimulatory and other biological potentials. Front Pharmacol 11:602364. doi: 10.3389/fphar.2020.602364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litwin A, Nowak M, Różalska S. 2020. Entomopathogenic fungi: unconventional applications. Rev Environ Sci Biotechnol 19:23–42. doi: 10.1007/s11157-020-09525-1. [DOI] [Google Scholar]
- 34.Spatafora JW, Sung GH, Sung JM, Hywel-Jones NL, White JF. 2007. Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol Ecol 16:1701–1711. doi: 10.1111/j.1365-294X.2007.03225.x. [DOI] [PubMed] [Google Scholar]
- 35.Ownley BH, Griffin MR, Klingeman WE, Gwinn KD, Moulton JK, Pereira RM. 2008. Beauveria bassiana: endophytic colonization and plant disease control. J Invertebr Pathol 98:267–270. doi: 10.1016/j.jip.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Shaalan RS, Gerges E, Habib W, Ibrahim L. 2021. Endophytic colonization by Beauveria bassiana and Metarhizium anisopliae induces growth promotion effect and increases the resistance of cucumber plants against Aphis gossypii. J Plant Prot Res 61:358–370. [Google Scholar]
- 37.Sun T, Shen Z, Shaukat M, Du C, Ali S. 2020. Endophytic isolates of Cordyceps fumosorosea to enhance the growth of Solanum melongena and reduce the survival of whitefly (Bemisia tabaci). Insects 11:78. doi: 10.3390/insects11020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu X, Xie W, Zhang J, Hu Q. 2019. Biodiversity of entomopathogenic fungi in the soils of South China. Microorganisms 7:311. doi: 10.3390/microorganisms7090311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman DC, Callaham MA, Crossley DA. 2018. Fundamentals of Soil Ecology, 3rd ed, Academic Press, Salt Lake City, USA. [Google Scholar]
- 40.Shen LN, Hou MF, Xu WB, Huang YF, Liang SC, Zhang YH, Jiang ZC, Chen WH. 2020. Research on flora of seed plants in Dashiwei Karst Tiankeng group of Leye. Guangxi Guihaia 40:751–764. doi: 10.11931/guihaia.gxzw201902015. [DOI] [Google Scholar]
- 41.Chen WH, Han YF, Liang JD, Liang ZQ. 2020. Akanthomyces neocoleopterorum, a new verticillium-like species. Phytotaxa 432:119–124. doi: 10.11646/phytotaxa.432.2.2. [DOI] [Google Scholar]
- 42.Aini AN, Mongkolsamrit S, Wijanarka W, Thanakitpipattana D, Luangsa-ard JJ, Budiharjo A. 2020. Diversity of Akanthomyces on moths (Lepidoptera) in Thailand. MycoKeys 71:1–22. doi: 10.3897/mycokeys.71.55126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen WH, Han YF, Liang JD, Tian WY, Liang ZQ. 2020. Morphological and phylogenetic characterizations reveal three new species of Samsoniella (Cordycipitaceae, Hypocreales) from Guizhou, China. MycoKeys 74:1–15. doi: 10.3897/mycokeys.74.56655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen WH, Liang JD, Ren XX, Zhao JH, Han YF, Liang ZQ. 2021. Cryptic diversity of Isaria-like species in Guizhou, China. Life 11:1093. doi: 10.3390/life11101093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen WH, Liu C, Han YF, Liang JD, Tian WY, Liang ZQ. 2019. Three novel insect-associated species of Simplicillium (Cordycipitaceae, Hypocreales) from Southwest China. MycoKeys 58:83–102. doi: 10.3897/mycokeys.58.37176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zou X, Liu AY, Liang ZQ, Han YF, Yang M. 2010. Hirsutella liboensis, a new entomopathogenic species affecting Cossidae (Lepidoptera) in China. Mycotaxon 111:39–44. doi: 10.5248/111.39. [DOI] [Google Scholar]
- 47.Liang JD, Han YF, Zhang JW, Du W, Liang ZQ, Li ZZ. 2011. Optimal culture conditions for keratinase production by a novel thermophilic Myceliophthora thermophila strain GZUIFR-H49-1. J Appl Microbiol 110:871–880. doi: 10.1111/j.1365-2672.2011.04949.x. [DOI] [PubMed] [Google Scholar]
- 48.White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p 315–322. In Innis MA, Gelfand DH, Sninsky JJ, White TJ (ed), PCR protocols: a guide to methods and applications. Academic Press, New York. [Google Scholar]
- 49.Castlebury LA, Rossman AY, Sung GH, Hyten AS, Spatafora JW. 2004. Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol Res 108:864–872. doi: 10.1017/s0953756204000607. [DOI] [PubMed] [Google Scholar]
- 50.van den Brink J, Samson RA, Hagen F, Boekhout T, de Vries RP. 2012. Phylogeny of the industrial relevant, thermophilic genera Myceliophthora and Corynascus. Fungal Divers 52:197–207. doi: 10.1007/s13225-011-0107-z. [DOI] [Google Scholar]
- 51.Sung GH, Hywel-Jones NL, Sung JM, Luangsa-ard JJ, Shrestha B, Spatafora JW. 2007. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud Mycol 57:5–64. doi: 10.3114/sim.2007.57.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen WH, Han YF, Liang JD, Liang ZQ. 2020. Akanthomyces lepidopterorum, a new lecanicillium-like species. Phytotaxa 459:117–123. doi: 10.11646/phytotaxa.459.2.3. [DOI] [Google Scholar]
- 53.Li YP, Chen WH, Han YF, Liang JD, Liang ZQ. 2020. Cordyceps yinjiangensis, a new ant-pathogenic fungus. Phytotaxa 453:284–292. doi: 10.11646/phytotaxa.453.3.10. [DOI] [Google Scholar]
- 54.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vaidya G, Lohman DJ, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27:171–180. doi: 10.1111/j.1096-0031.2010.00329.x. [DOI] [PubMed] [Google Scholar]
- 57.Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour 20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 59.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Drummond A, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kepler RM, Luangsa-ard JJ, Hywel-Jones NL, Quandt CA, Sung GH, Rehner SA, Aime MC, Henkel TW, Sanjuan T, Zare R, Chen M, Li Z, Rossman AY, Spatafora JW, Shrestha B. 2017. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8:335–353. doi: 10.5598/imafungus.2017.08.02.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sanjuan T, Tabima J, Restrepo S, Læssøe T, Spatafora JW, Franco-Molano AE. 2014. Entomopathogens of Amazonian stick insects and locusts are members of the Beauveria species complex (Cordyceps sensu stricto). Mycologia 106:260–275. doi: 10.3852/13-020. [DOI] [PubMed] [Google Scholar]
- 64.Chen WH, Liu C, Han YF, Liang JD, Liang ZQ. 2018. Akanthomyces araneogenum, a new Isaria-like araneogenous species. Phytotaxa 379:66–72. doi: 10.11646/phytotaxa.379.1.6. [DOI] [Google Scholar]
- 65.Kepler RM, Sung GH, Ban S, Nakagiri A, Chen MJ, Huang B, Li Z, Spatafora JW. 2012. New teleomorph combinations in the entomopathogenic genus Metacordyceps. Mycologia 104:182–197. doi: 10.3852/11-070. [DOI] [PubMed] [Google Scholar]
- 66.Luangsa-ard JJ, Hywel-Jones NL, Manoch L, Samson RA. 2005. On the relationships of Paecilomyces sect. Isarioidea species. Mycol Res 109:581–589. doi: 10.1017/s0953756205002741. [DOI] [PubMed] [Google Scholar]
- 67.Mongkolsamrit S, Noisripoom W, Thanakitpipattana D, Wutikhun T, Spatafora JW, Luangsa-Ard J. 2018. Disentangling cryptic species with Isaria-like morphs in Cordycipitaceae. Mycologia 110:230–257. doi: 10.1080/00275514.2018.1446651. [DOI] [PubMed] [Google Scholar]
- 68.Chaverri P, Bischoff JF, Evans HC, Hodge KT. 2005. Regiocrella, a new entomopathogenic genus with a pycnidial anamorph and its phylogenetic placement in the Clavicipitaceae. Mycologia 97:1225–1237. doi: 10.1080/15572536.2006.11832732. [DOI] [PubMed] [Google Scholar]
- 69.Rehner SA, Minnis AM, Sung GH, Luangsa-ard JJ, Devotto L, Humber RA. 2011. Phylogeny and systematics of the anamorphic, entomopathogenic genus Beauveria. Mycologia 103:1055–1073. doi: 10.3852/10-302. [DOI] [PubMed] [Google Scholar]
- 70.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque A, Chen W, Fungal Barcoding Consortium . 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johnson D, Sung GH, Hywel-Jones NL, Luangsa-ard JJ, Bischoff JF, Kepler RM, Spatafora JW. 2009. Systematics and evolution of the genus Torrubiella (Hypocreales, Ascomycota). Mycol Res 113:279–289. doi: 10.1016/j.mycres.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 72.Gomes AA, Pinho DB, Cardeal ZL, Menezes HC, De Queiroz MV, Pereira OL. 2018. Simplicillium coffeanum, a new endophytic species from Brazilian coffee plants, emitting antimicrobial volatiles. Phytotaxa 333:188–198. doi: 10.11646/phytotaxa.333.2.2. [DOI] [Google Scholar]
- 73.Crous PW, Luangsa-ard JJ, Wingfield MJ, Carnegie AJ, Hernández-Restrepo M, Lombard L, Roux J, Barreto RW, Baseia IG, Cano-Lira JF, Martín MP, Morozova OV, Stchigel AM, Summerell BA, Brandrud TE, Dima B, García D, Giraldo A, Guarro J, Gusmão LFP, Khamsuntorn P, Noordeloos ME, Nuankaew S, Pinruan U, Rodríguez-Andrade E, Souza-Motta CM, Thangavel R, van Iperen AL, Abreu VP, Accioly T, Alves JL, Andrade JP, Bahram M, Baral HO, Barbier E, Barnes CW, Bendiksen E, Bernard E, Bezerra JDP, Bezerra JL, Bizio E, Blair JE, Bulyonkova TM, Cabral TS, Caiafa MV, Cantillo T, Colmán AA, Conceição LB, Cruz S, Cunha AOB, et al. 2018. Fungal Planet description sheets: 785–867. Persoonia 41:238–417. doi: 10.3767/persoonia.2018.41.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang YB, Wang Y, Fan Q, Duan DE, Zhang GD, Dai RQ, Dai YD, Zeng WB, Chen ZH, Li DD, Tang DX, Xu ZH, Sun T, Nguyen T, Tran N, Dao V, Zhang CM, Huang LD, Liu YJ, Zhang XM, Yang DR, Sanjuan T, Liu XZ, Yang ZL, Yu H. 2020. Multigene phylogeny of the family Cordycipitaceae (Hypocreales): new taxa and the new systematic position of the Chinese cordycipitoid fungus Paecilomyces hepiali. Fungal Divers 103:1–46. doi: 10.1007/s13225-020-00457-3. [DOI] [Google Scholar]
- 75.Wei D-P, Wanasinghe DN, Hyde KD, Mortimer PE, Xu J, Xiao Y-P, Bhunjun CS, To-Anun C. 2019. The genus Simplicillium. MycoKeys 60:69–92. doi: 10.3897/mycokeys.60.38040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Download spectrum.01975-22-s0001.pdf, PDF file, 0.3 MB (276.9KB, pdf)