ABSTRACT
Uropathogenic Escherichia coli (UPEC) ecology-pathophysiology from the gut reservoir to its urothelium infection site is poorly understood, resulting in equivocal benefits in the use of cranberry as prophylaxis against urinary tract infections. To add further understanding from the previous findings on PAC antiadhesive properties against UPEC, we assessed in this study the effects of proanthocyanidins (PAC) rich cranberry extract microbial metabolites on UTI89 virulence and fitness in contrasting ecological UPEC’s environments. For this purpose, we developed an original model combining a colonic fermentation system (SHIME) with a dialysis cassette device enclosing UPEC and a 3D tissue-engineered urothelium. Two healthy fecal donors inoculated the colons. Dialysis cassettes containing 7log10 CFU/mL UTI89 were immersed for 2h in the SHIME colons to assess the effect of untreated (7-day control diet)/treated (14-day PAC-rich extract) metabolomes on UPEC behavior. Engineered urothelium were then infected with dialysates containing UPEC for 6 h. This work demonstrated for the first time that in the control fecal microbiota condition without added PAC, the UPEC virulence genes were activated upstream the infection site, in the gut. However, PAC microbial-derived cranberry metabolites displayed a remarkable propensity to blunt activation of genes encoding toxin, adhesin/invasins in the gut and on the urothelium, in a donor-dependent manner. Variability in subjects’ gut microbiota and ensuing contrasting cranberry PAC metabolism affects UPEC virulence and should be taken into consideration when designing cranberry efficacy clinical trials.
IMPORTANCE Uropathogenic Escherichia coli (UPEC) are the primary cause of recurrent urinary tract infections (UTI). The poor understanding of UPEC ecology-pathophysiology from its reservoir–the gut, to its infection site–the urothelium, partly explains the inadequate and abusive use of antibiotics to treat UTI, which leads to a dramatic upsurge in antibiotic-resistance cases. In this context, we evaluated the effect of a cranberry proanthocyanidins (PAC)-rich extract on the UPEC survival and virulence in a bipartite model of a gut microbial environment and a 3D urothelium model. We demonstrated that PAC-rich cranberry extract microbial metabolites significantly blunt activation of UPEC virulence genes at an early stage in the gut reservoir. We also showed that altered virulence in the gut affects infectivity on the urothelium in a microbiota-dependent manner. Among the possible mechanisms, we surmise that specific microbial PAC metabolites may attenuate UPEC virulence, thereby explaining the preventative, yet contentious properties of cranberry against UTI.
KEYWORDS: PAC, cranberry, gut metabolome, UPEC, urinary tract infections, urothelium, virulence genes
INTRODUCTION
Urinary tract infections (UTI) in human are generally caused by uropathogenic Escherichia coli (UPEC). More than 50% of women will experience symptomatic UTI during their lifetime. Among them, 1/3 will develop recurrent UTIs (1). UPEC life cycle starts in the intestinal niche, where the bacterium naturally cohabits with the gut microbiota (2, 3). The fecal-periurethral contamination promotes UPEC tropism and colonization through the urinary tract and bladder (3, 4). At this site, UPEC utilizes numerous virulence (e.g., adhesins, toxins) and fitness factors (e.g., iron acquisition system) to adhere, disseminate within the mucosa and ultimately cause disease (4). There is an evident lack of understanding of UPEC ecology-pathophysiology in the gut reservoir, with studies focusing mainly on the infection site. Elucidating the distinct UPEC lifestyles (e.g., survival, virulence genes expression) from the gut to the urinary tract is crucial to add knowledges from previous works that were dedicated to study UTI pathogenesis, recurrence mechanisms, and to improve treatment effectiveness.
So far, antibiotics remain the standard prophylaxis and treatment, contributing to the drastic rise of multidrug-resistant uropathogens and their concomitant health repercussions (5). Thus, there is a pressing need for the development of more effective prophylaxis to address the UTI societal burden and limit the use of antibiotics. Positioning cranberries consumption (Vaccinium macrocarpon Aiton) as a natural and nonantibiotic approach meets consumers’ demand for integral alternatives to prevent the development of the infections. Although cranberry is extensively recommended for UTIs prophylaxis (6), there is discrepancy and controversial results between studies with underlying mechanisms of action that are still open to debate. Cranberry is composed predominantly of water, followed by a complex mixture of organic acids, fructose and polyphenols including flavonoids, anthocyanidins, proanthocyanidins (PAC), catechins, and triterpenoids (7). The whole cranberry fruit or specific fractions have been studied for their potential role in inhibiting UPEC adhesion/invasion to the urinary tract, limiting biofilm formation, and/or in reducing clinical symptoms or UTI events (6, 8–15). In particular, many research evidence shows that cranberry PAC are the most clinically relevant component in preventing UTI in women (8). PAC display a limited absorption in the intestine and are extensively metabolized in the colon by the gut microbiota (12, 16). It is now believed that PAC antiadhesive properties on UPEC in bladder are more likely caused by their absorbed microbial metabolites, as well as for the reduction of biofilm formation or clinical symptoms (13, 14). Among the PAC microbial metabolites, conjugates (O-methyl ethers and sulfates) of phenolic acids (phenylpropionic, phenylvaleric, phenylacetic, benzoic, and cinnamic acids) (13), and other microbial metabolites, including phenyl-γ-valerolactones (17), have been associated with the preventive actions of cranberry flavonoids and phenolic acids on UTI.
To contribute to further understanding from the previous findings on PAC antiadhesive properties against UPEC, the present work aims to assess the effect of gut microbial metabolites of PAC-rich cranberry extract on UPEC virulence and fitness pathways. For this purpose, we developed an original model combining an artificial colonic fermentation system (SHIME) with a dialysis cassette device and a 3D tissue-engineered urothelium coculture model.
RESULTS
Metabolomes of donors A and B display a differential PAC catabolism in the colon.
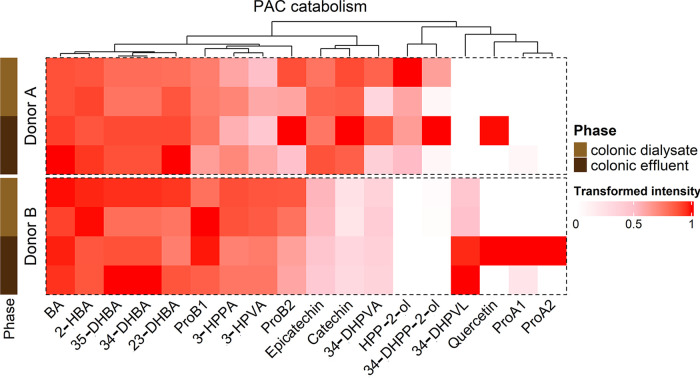
UPEC enclosed in dialysis cassettes were first exposed for 2-h to the metabolic environment of the SHIME transverse colon from two different donors after a control period and a 14-day PAC treatment (Fig. 1A and B). As proof of the efficacy of the dialysis device, most PAC metabolites present in colonic effluents were also found in colonic dialysate (Fig. 2). No PAC metabolites were found in the absence of microbiota. Differential production of PAC metabolites was seen between donors A and B. Donor B produced 34-DHPVL, ProA1 and ProA2, while donor A did not (Fig. 2). On the contrary, donor A produced HPP-2-ol and DHPP-2-ol. Finally, donor A predominantly produced catechin, epicatechin and 3,4-DHPVA compared to donor B.
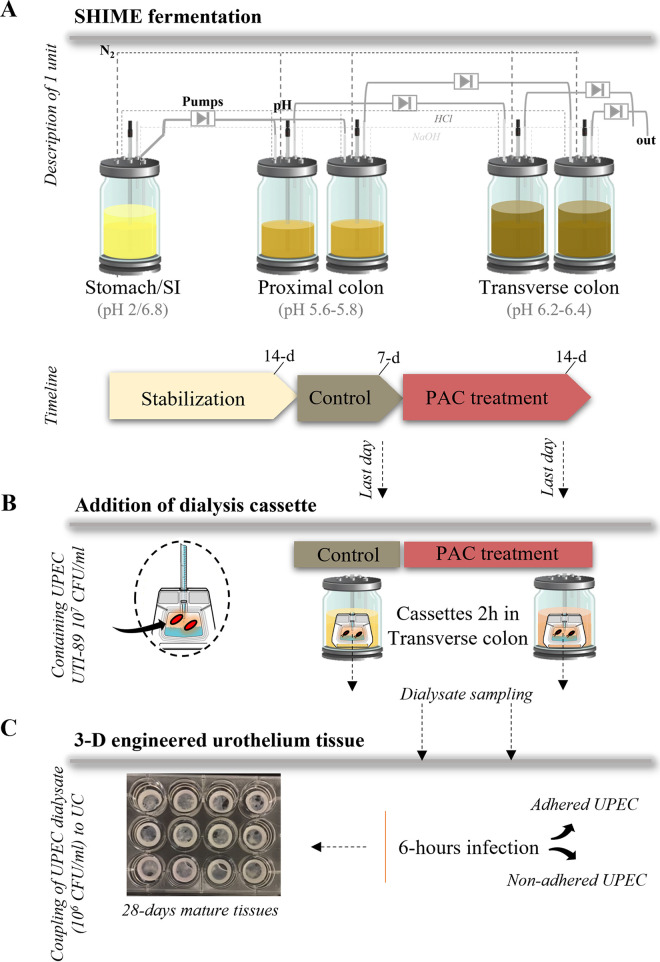
FIG 1.
Study design. (A) Illustration of one unit of the TWIN-SHIME fermentation system, including reactors in series from the stomach/small intestine (SI) to the transverse colon. The colonic reactors of each unit were inoculated with two different fecal donors, both in duplicates. The three phases and duration of fermentation are shown in the timeline. The treatment consisted of the addition of 86.8 mg PAC-rich cranberry extract/day in the SHIME stomach for 14 days, consecutively catabolized in the proximal and transverse colon. (B) The transverse colon was chosen as site of interest for UPEC reservoir and final PAC catabolism. Therefore, dialysis cassettes (10 kDa) containing 7log10 CFU/mL UTI89 were added for 2h in the transverse colon to assess the effect of untreated (control)/treated (PAC) metabolome on UPEC behavior. After exposure, dialysates containing UPEC were stored until processing. (C) Dialysates containing UPEC (adjusted concentration of 6log10 CFU/mL UTI89) were then combined to a 3D tissue-engineered urothelium (UC) to recreate a 6-h infection period.
FIG 2.
Hierarchical profiling of PAC catabolism between donors in gut effluents and dialysates after a 14-day treatment. Heatmap displaying the transformed intensity of the PAC catabolites remaining in gut effluents and dialysates after 14 days of PAC-rich extract treatment and following a 2-h UPEC exposure, as determined by UPLC-QToF in negative ionization. BA = Benzoic Acid; 3-HPVA = 5-(3′-hydroxyphenyl)valeric acid; 35-DHBA = 3,5-dihydroxybenzoic acid; ProA2 = Procyanidin A2; ProB2 = Procyanidin B2; HPP-2-ol = 1-(Hydroxyphenyl)-(2′,4′,6′-trihydroxyphenyl)-propan-2-ol; 2-HBA = 2-hydroxybenzoic acid; 3-HPPA = 3-(3′-hydroxyphenyl)propanoic acid;, 4-DHPVA = 5-(3′,4′-dihydroxyphenyl)valeric acid; ProA1 = Procyanidin A1; 34-DHPP-2-ol = 1-(Dihydroxyphenyl)-3-(2′,4′,6′-trihydroxyphenyl)-propan-2-ol; ProB1 = Procyanidin B1; 34-DHPVL = 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone; 34-DHBA = 3,4-dihydroxybenzoic acid; 4_HPAA = 2-(4′-hydroxyphenyl)acetic acid; 34-DHPPA = 3-(3′,4′-dihydroxyphenyl)propanoic acid; 4-HPPA = 3-(4′-hydroxyphenyl)propanoic acid.
PAC treatment significantly affects UPEC growth in a donor-dependent manner.
The gut metabolome of donor A caused a significant decrease in UPEC titer under PAC treatment (P ≤ 0.05) compared to donor B (Fig. 3A, Fig. S2 in the supplemental material). Such differential response might be explained by variability in PAC catabolism as shown in Fig. 2, and therefore, depends on microbiota composition (Fig. 3B). Although PAC treatment stimulated the growth of Akkermansia in both donors, donor A was characterized by an upsurge of Stenotrophomonas, Escherichia-Shigella, and a decrease of Megasphaera, while donor B displayed a decrease of Bacteroides under PAC treatment (Fig. 3B). The fermentation by-products were also measured under both conditions. PAC treatment induced (P ≤ 0.001) a sharp increase of butyrate and propionate in the transverse colon of both donors (Fig. 3C). Finally, in the absence of a microbiota, PAC treatment significantly stimulated UPEC growth (Fig. 3A). This suggests that the microbiota and microbial PAC metabolites blunt UPEC growth.
FIG 3.
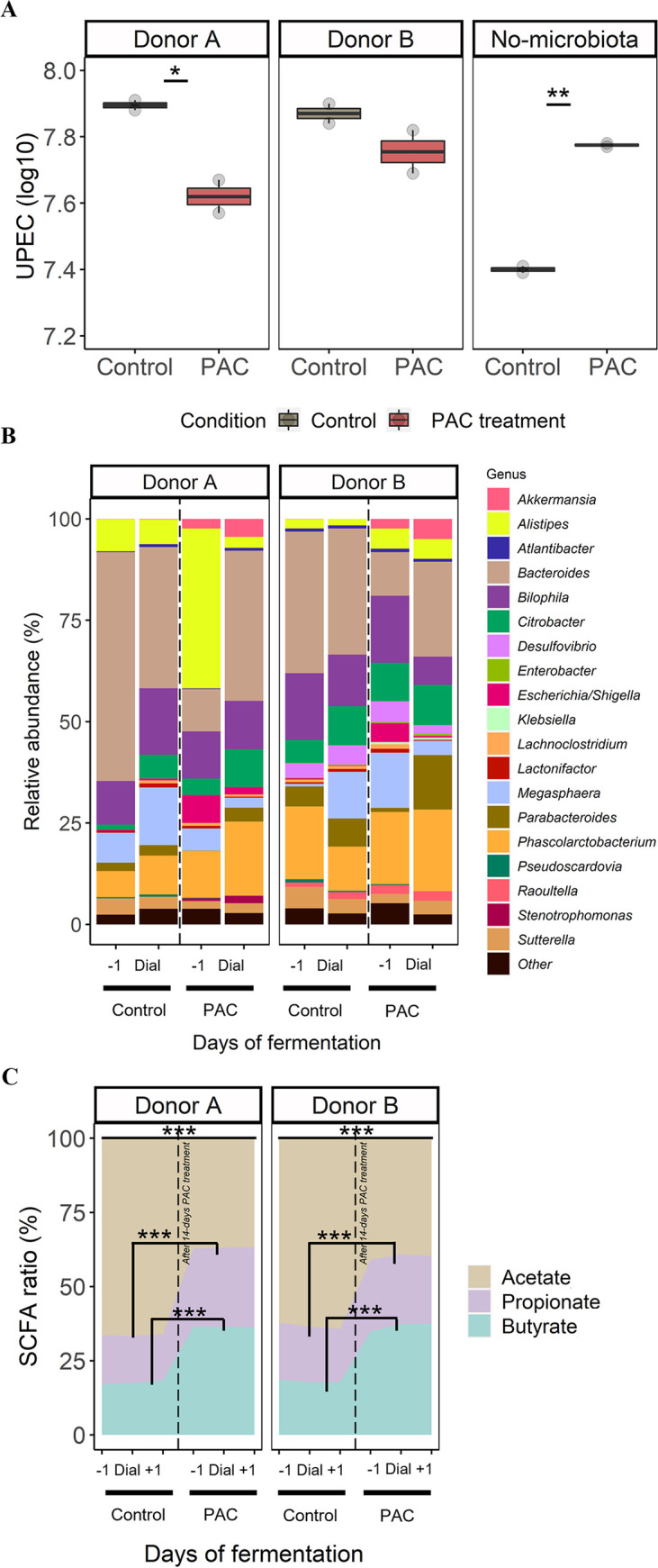
TWIN-SHIME combining dialysis cassette device as a model of short-term UPEC’s gut reservoir (A) Number of viable-culturable UPEC (log10 CFU/mL) remaining in the dialysate after a 2-h exposure with the PAC treated/non-treated metabolome of the transverse colon. A condition “no microbiota” was also performed by introducing dialysis cassettes in jars with digestive medium deprived of microbiota. Significant differences between PAC treatment and control are indicated with P ≤ 0.05 (*) or P ≤ 0.01 (**), as determined by the Friedman post hoc Wilcoxon test. (B) The microbial composition (%) of the 20 most abundant genera is shown. “Dial” represents the day of addition of dialysis cassettes containing UPEC in transverse colons of donors A and B, both during the control and PAC treatment. (C) Ratio-profile of short chain fatty acids (SCFA) for each treatment condition: before (-1 day), during addition of dialysis cassette “Dial,” and after addition (+1 day). Significant differences in production of acetate, butyrate and propionate were found between PAC treatment and control, P ≤ 0.001 (***), as determined by the Friedman post hoc Wilcoxon test.
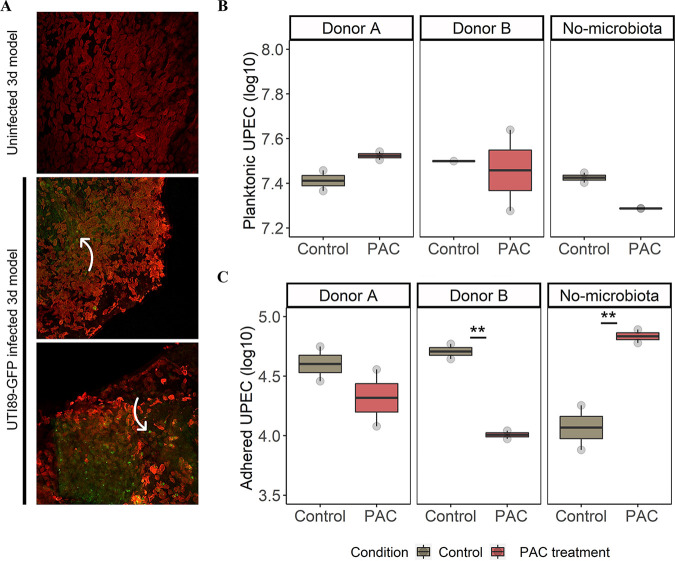
Next, UPEC dialysates were used to infect urothelial cells (UC) (Fig. 1C, Fig. 4A). After a 6-h infection, cells were washed, and two UPEC ecosystems were analyzed: planktonic and adhered UPEC (Fig. 4B and C). No change in the number of planktonic UPEC was observed between control versus PAC (Fig. 4B). Under all conditions, a low proportion of UPEC adhered persistently onto UC, approximately 4–5 log10 (Fig. 4C). In this fraction, UPEC previously exposed to PAC metabolites from donor B displayed a significantly lower adhesion onto UC (4 log10) compared to the control (4.5 log10) (P ≤ 0.01) (Fig. 4C). On the contrary, the lack of microbiota under PAC treatment stimulated the UPEC adhesion, while lowering the planktonic fraction.
FIG 4.
Engineered bladder mucosa as a model of acute urinary tract infection. (A) Bladder mucosa were reconstructed and infected by UPEC dialysates. Tissues were fixed and immunolabelled 6 h after infection to detect cytokeratins (red fluoresecnce, urothelial cells). UPEC were detected by GFP (green) fluorescence. Large number of bacteria at the surface or in the urothelial cells were visible and indicated by white arrows. (B) Number of viable-culturable UPEC (log10 CFU/mL) unattached to UC (planktonic) after a 6-h infection. (C) Number of viable-culturable UPEC (log10 CFU/mL) remaining attached to UC after a 6-h infection. A condition “no microbiota” was also performed by testing UPEC dialysate not exposed to the gut microbiota. Significant differences between PAC treatment and control are indicated with P ≤ 0.01 (**), as determined by the Friedman post hoc Wilcoxon test.
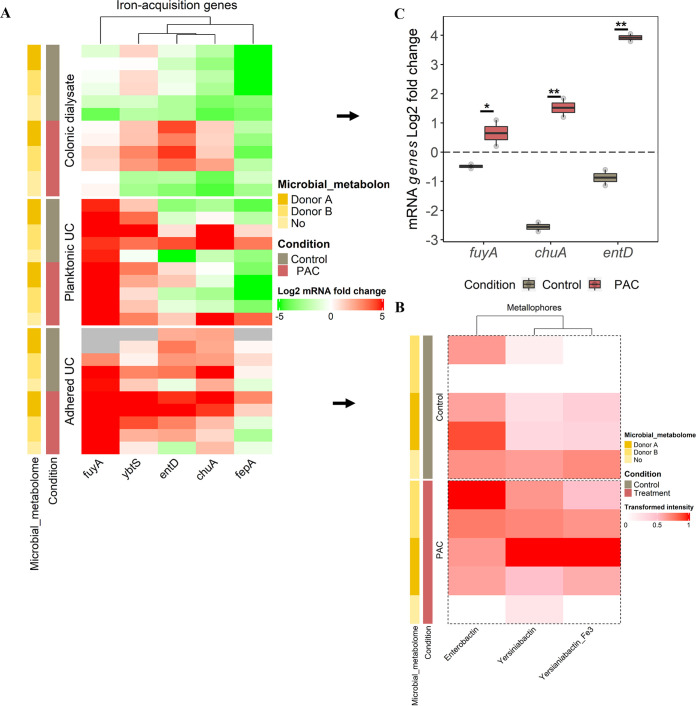
UPEC virulence genes are activated at the urothelium site and upstream in the gut reservoir.
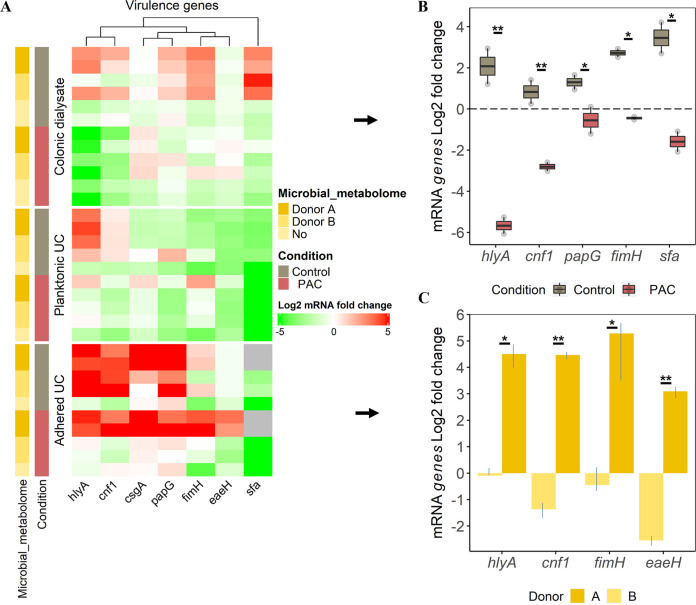
Key virulence genes specific to UPEC strain UTI89 were monitored at the end of the UPEC incubation period in each ecosystem studied (after 2 h in gut dialysate and after 6 h onto UC) under both control (PAC free) and PAC treatment. Results were expressed in log2 expression fold changes (FC). UPEC virulence genes expression signatures were specific to the ecosystem studied (e.g., gut dialysate, planktonic UC, adhered UC) (Fig. 5A). In the control condition, most of the virulence genes encoding for toxins production and adhesins were expressed at the expected site of action: when adhered onto UC. Strikingly, these genes were already activated at an earlier stage in the colonic dialysate (Fig. 5A). However, in the absence of a microbiota, no virulence genes were activated.
FIG 5.
Profiling of UPEC main virulence genes expression from the gut reservoir to urothelium. (A) Heatmap displaying the log2 fold change UPEC virulence genes expression according to the ecosystem exposure (colonic dialysate, planktonic and adhered bacteria to UC), the microbial metabolome origin and the treatment. Induction (log2 fold change expression ≥ 1) is denoted in shade of red, and repression (≤ -1) in shade of green, as determined by RT-qPCR. Samples with failed amplification are displayed in gray. (B) Selection of genes from dialysis cassettes remaining significantly different between control and PAC conditions are indicated with P ≤ 0.05 (*) or P ≤ 0.01 (**), as determined by the Friedman post hoc Wilcoxon test. (C) Selection of genes from adhered UPEC treated with PAC that are significantly different between donors A and B, as determined by the Friedman post hoc Wilcoxon test.
PAC treatment significantly impeded UPEC virulence genes activation in the gut reservoir.
Under PAC treatment, most of the virulence genes were downregulated in UPEC dialysate and planktonic UC compared to the control, regardless of the microbial metabolome origin (Fig. 5A). However, significant differences between control and treatment were noted for UPEC dialysate only (Fig. 5B). The genes encoding for toxins production including alpha hemolysin (hlyA) and cytotoxic necrotizing factor 1 (cnf1) were highly repressed (P ≤ 0.01) under PAC with -5.8 log2 FC and -3 log2 FC versus 2 log2 FC and 1 log2 FC under control, respectively (Fig. 5B). Still in UPEC dialysate, genes encoding for adhesins/invasins, such as pili associated pyelonephritis (papG), type 1 fimbrial adhesin (fimH) and S fimbriae (Sfa), were significantly downregulated (P ≤ 0.05). For instance, a 2.5 log2 FC expression was observed for fimH under control versus -0.5 log2 FC under PAC condition (Fig. 5B).
Adhered UPEC onto UC displayed a particular expression pattern under PAC treatment that appeared dependent on the microbial metabolome origin (Fig. 5A). Indeed, UPEC previously exposed to the PAC-treated metabolome of donor A (Fig. 2) showed significant overexpression of hlyA, cnf1, fimH and the gene encoding for attaching and effacing protein (eaeH) when adhering to urothelium, compared to donor B (Fig. 5C).
PAC treatment significantly activates UPEC iron-acquisition genes in the gut reservoir.
Genes encoding for UPEC iron-acquisition were also studied, including the outer membrane ferric yersiniabactin/enterobactin importers (fuyA/fepA), the yersiniabactin (ybtS) and enterobactin synthesis (entD) and the heme transport (chuA) (Fig. 6A). Regardless of the condition, most of the genes were upregulated (except fepA) at the UC site for the planktonic and adhered UPEC. This is due to a large amount of iron present in the UC medium. Per the gene’s expression, their corresponding metabolites (e.g., yersiniabactin, enterobactin) were found in UC medium under both treatment and control conditions (Fig. 6B).
FIG 6.
Profiling of UPEC iron-acquisition genes expression from the gut reservoir to urothelium. (A) Heatmap displaying the log2 fold change UPEC iron-acquisition genes expression according to the ecosystem exposure (colonic dialysate, planktonic and adhered bacteria to UC), the microbial metabolome origin and the treatment. Induction (log2 fold change expression ≥ 1) is denoted in shade of red and repression (≤ -1) in shade of green, as determined by RT-qPCR. Samples with failed amplification are displayed in gray. (B) Heatmap displaying the transformed intensity of the metallophores metabolites remaining in UC medium after 6 h of UPEC infection, as determined by Reverse-Phase Liquid Chromatography (RPLC) positive. (C) Selection of genes from dialysis cassettes remaining significantly different between control and PAC conditions are indicated with P ≤ 0.05 (*) or P ≤ 0.01 (**), as determined by the Friedman post hoc Wilcoxon test.
A significant overexpression of fuyA (P ≤ 0.05), chuA and entD (P ≤ 0.01) genes were found in gut dialysate under PAC treatment, while genes remained downregulated under the control condition (Fig. 6C). This was true in the presence of the microbial metabolome, while in its absence, they remained downregulated like the control condition (Fig. 6A). However, the metallophores were not detected in UPEC dialysate.
DISCUSSION
UPEC display remarkable capacity to metabolically adapt to diverse nutritional/physicochemical environments as found in the gut and the bladder (18, 19). Considering the limited information available on UPEC spatial ecology-pathophysiology, this study explored, in a unique fashion, the effects of metabolized PAC-rich cranberry extract on growth and virulence genes expression of UTI89 in contrasting ecosystems (i.e., transverse colon and urothelium). These ecosystems were mimicked in a novel model combining a colonic fermentation system interacting with a dialysis cassette device containing UPEC and a 3D tissue-engineered urothelium.
Tracking UPEC virulence in its different ecological niches.
To our knowledge, our study reveals for the first time that UPEC virulence genes activation is independent of the urothelium infection site “urovirulence” (20). Remarkably, most of the studied genes encoding adhesins and toxins were already overexpressed upstream in the colonic non-treated microbiota. At this site, UPEC is recognized as being inoffensive (2, 3). However, our work, supports the idea that virulence factors cannot directly predict UTI symptoms but presumably confer multiple other functions (21). Since the SHIME only mimics the microbial interactions and physicochemical parameters of the luminal part of the colon but does not include intestinal epithelial cells, we cannot truly claim UPEC as enterovirulent. Such concept of UPEC enterovirulence should be specifically tested in the presence of intestinal epithelial cells, as recently discussed in the work of Schultz et al., using an intestinal model of Caco-2 cells (22). The authors have shown that the strain UPEC 536, positive for the hemolysin toxin production (HlyA+), induced epithelial barrier dysfunction by compromising tight junctions as potentiator of the leaky gut syndrome (22). Our work concords with that of Schulz et al. (22), highlighting the importance of studying UPEC upstream the famous urovirulent stage.
Cranberry PAC microbial-derived metabolites impede UPEC virulence genes activation: dealing with donor-dependent responses for a more personalized approach.
The European Association of Urology has not yet formally recommended the use of PAC-rich cranberry extracts against UTI due to inconsistencies among human clinical trials and knowledge gaps in the mechanisms of action of PAC in the urinary tract (15, 23). Our study attempted therefore to provide new insights on the presumed mode of actions, and specifically assess the molecular mechanisms at play in the gut reservoir in the presence of a whole and natural microbiota. We demonstrated that PAC-rich cranberry extract microbial metabolites significantly impede the activation of UPEC virulence genes at early stage in the gut reservoir (hlyA, cnf1, papG, fimH, Sfa). Such gene inhibitions under PAC treatment tended also to be seen in the urothelium habitat (planktonic and adhered UPEC), although the effect was not significant. The lack of statistical difference was due to controversial response between donors in UPEC genes expression. The genes hlyA, cnf1, fimH, and eaeH remained upregulated in donor A under both control and PAC treatment, while an opposite profile was observed in donor B. Hence, the interindividual variability in genes expression at the urothelium site might be attributed to upstream differences in intestinal microbiota composition (e.g., age, gender, race, diet, health status, environmental exposure, etc.) among individuals (6) and its ensuing PAC metabolism. Indeed, cranberry PAC-polyphenols underwent extensive microbial metabolism during colonic digestion, triggering a wide range of phenolic metabolites, as observed here and in previous studies (12–17). However, contrasting microbiota between individuals can lead to the selective release of specific bioavailable microbial-derived PAC metabolites (14, 24–27). In our study, among the spectrum of PAC metabolites produced under colonic fermentation, donor B was producing 5-(3′,4′-dihydroxyphenyl)-γ-valerolactone contrary to donor A. Such variability between donors, therefore, indicates the need to profile individuals’ gut metabolomes as metabotypes when conducting clinical interventions, to define responder(s) class(es) in a perspective of targeted personalized nutrition (28).
Interestingly, compared with donor A, donor B exhibited an overall better response to the PAC treatment, that is, a marked virulence genes downregulation and a reduced UPEC adhesion onto UC. The antiadhesive activity of phenyl-γ-valerolactone against UPEC was previously reported in bladder epithelial cells (17), but no studies have assessed its role on the anti-virulent activity of UPEC. Most studies only assessed the ability of PAC and its corresponding metabolites to inhibit bacterial adherence/invasion to UC, biofilm formation, or clinical symptoms reduction (6, 12–15). They briefly described the anti-virulent properties of PAC in simple static culture medium (29–31). For instance, one study reported that cranberry compounds could inhibit UPEC motility via downregulation of the fliC gene (30). In a more extensive microarray study, UPEC transcriptomic profiles were assessed in simple cultures supplemented with cranberry PAC alone or in combination with propolis (31). Both the PAC treatment and the combination downregulated expression of genes involved in UPEC adhesion, motility, and biofilm formation (31). Although these results are in accordance with our observations, our work is the only one assessing UPEC virulence in a natural luminal microbiota environment with its full metabolome complement, therefore reproducing more accurately the true human gut conditions. Finally, the observed modulation of virulence genes expression under PAC treatment does not necessarily indicates a modulation at protein level. Indeed, post-transcriptional and post-translational regulations are additional key steps that we must consider in future studies by using proteomic tools.
Additional mode of action of cranberry PAC on UPEC: metallophore encoding genes interaction.
Metallophore encoding genes are additional key survival genes for UPEC, standing at midway between virulence and fitness pathways. UPEC encounter iron limitation within the host, such as in the urinary tract. Cranberry is also known as an iron chelator strongly limiting bacterial availability (30, 32). To ensure adequate intracellular iron levels, UPEC upregulates the expression of genes involved in iron acquisition (31, 33). Such iron-acquisition genes upregulation mechanism is confirmed in our study reproducing a complex colonic environment under PAC treatment compared to the control. Indeed, UPEC display a capacity to adapt to the presence of cranberry PAC by upregulating iron acquisition systems and reducing iron storage, as reported by Ranfaing et al. (31). Finally, in the urothelium, most of the iron-acquisition genes were upregulated under both conditions. The urothelium was however not the best model to study metallophore action since the medium was already supplemented with iron to maintain UC growth.
Beyond PAC microbial-derived metabolites.
In addition to the PAC metabolites which appear to alter UPEC virulence genes, one should not overlook the role of SCFA as key fermentation metabolites affecting bacterial physiology. In our study, the 14-day treatment shaped the gut microbiota in a donor-dependent way. Yet, such changes resulted in a clear shift of SCFA production marked by a significant increase of propionate and butyrate for both donors. Such SCFA increase can be partly explained through the bloom of Akkermansia muciniphila in the two donors, a species recognized as propionate and butyrate producer (34, 35). Several studies have reported the role of SCFA in the modulation of enteric pathogens by lowering intraluminal pH and the capacity of SCFA to prime the virulence genes (36). However, there is so far no consensus regarding the SCFA role as virulence enhancer or inhibitor, which appear to be pathogen specific (37, 38). In our experimental conditions, the change of colonic pH cannot be observed since the pH was controlled. In addition, it is unlikely that the UPEC virulence genes downregulation under PAC treatment was associated to the increased production of propionate/butyrate. Indeed, in the control condition performed without microbiota, and thus without SCFA, no change in the expression profiles was seen compared to the treated microbiota.
Conclusion.
Cranberry PAC microbial-derived metabolites could be of great public health significance and may represent a nonantibiotic alternative for UTI prophylaxis. In addition to reducing UPEC growth, we unraveled that microbial cranberry PAC metabolites display a remarkable capacity to blunt activation of genes encoding toxin production and adhesin/invasins in the gut reservoir and in coculture on the urothelium, in a donor-dependent manner. We should obviously consider interindividual variability in PAC microbial metabolism to identify responsive metabotypes in the patients and stratify them according to the specific PAC metabolizing microbiota (39). This approach would be of great interest to design UPEC anti-virulence prophylaxis in a personalized fashion, based on microbial or metabolomic markers of response. Further investigations integrating multi-stage microenvironments such as the coculturing of urothelium with bladder microvascular endothelial cells exposed to urine as well as microbiome and urobiome interactions would also provide more insights on the PAC metabolites underlying mechanisms of action.
MATERIALS AND METHODS
Strain.
UPEC strain used in this study was SLC-719 O18:K1:H7, an isolate derivative of the human cystitis strain UTI89, carrying the fluorescent protein vsfGFP-9 (40, 41) which possesses virulence genes encoding for the type 1 fimbrial adhesin (fimH), attaching and effacing protein (eaeH), S fimbriae (sfa), pili associated pyelonephritis (papG), curlin major subunit (csgA), as well as for the toxins alpha hemolysin (hlyA) and cytotoxic necrotizing factor 1 (cnf1). UPEC was routinely grown under agitation (37°C, 150 rpm, overnight) in Luria Bertani (LB) broth (BD Difco, USA). The number of cultivable UPEC was determined by direct plating onto LB agar and expressed in log10 CFU (CFU)/mL.
Fermentation system.
The TWIN-SHIME (Prodigest, Belgium) is a simulator of the human intestinal microbial ecosystem with two anaerobic SHIME units operated in parallel, in semi-continuous mode (42). In this study, a SHIME unit consisted of a stomach/small intestine vessel to reproduce gastric digestion of a standardized nutritional medium pancreatic/bile juice delivery, followed by a proximal colon and a transverse colon vessel. Two healthy fecal donors, one in each SHIME unit, inoculated the proximal and transverse colons to capture inter-individual variability in microbiota composition and PAC metabolism. Consent for fecal donation was obtained under registration number 2019-312 (Laval University, Canada). Procedure for fecal inoculum preparation was previously described (43). After a 2-week stabilization period, a 7-day control diet followed by a 14-day treatment was applied to the system (Fig. 1A). Treatment consisted in the supplementation of a PAC-rich cranberry extract, containing 86.8 mg PAC/day/donor (Urophenol-Diana Food, France) in the SHIME stomach for 14 days. Extract characterization is provided in Table S1 in the supplemental material.
Microbial community analysis.
Following the DNA extraction of the SHIME samples as previously described (44), next-generation 16S rRNA gene amplicon sequencing of the V3-V4 region (341F-805R) was performed at IBIS (Institut de Biologie Intégrative et des Systèmes, Laval University, Canada), on an Illumina MiSeq platform with Illumina V3 chemistry using the 600-cycle reagent kit (Illumina, USA). The Divisive Amplicon Denoising Algorithm (DADA2) workflow implemented in the dada2 R package (version 1.18.0) was employed to identify Amplicons Sequences Variants (ASVs) (45). The advantage of using dada2 over traditional clustering methods is that it determines the exact sequences based on an error model for the sequencing run, resolving as little as one nucleotide difference. The reads were merged if they overlapped precisely, and an ASVs table was constructed, recording the number of times each ASV was observed in each sample. Default parameters were used to estimate error parameters using learnErrors, and chimeras were removed using removeBimeraDenova (method = "consensus”). ASVs sequences were assigned taxonomy using the most recent SILVA taxonomic database (SILVA SSURef 138.1 NR, March 2021) as a reference data set (46). A phyloseq data object was created using the phyloseq package in R (47). Unassigned taxa and singletons were removed. Rarefaction curves were constructed to ensure that the samples were sequenced at sufficient depth (48). Relative abundances of microbial genera were plotted using the ggplot2 packageafter transforming abundance data into relative abundances.
Short Chain Fatty Acids (SCFA) analysis.
Colonic samples from SHIME were centrifuged 8 min at 14000g at 4°C. Supernatants were stored at –20°C until analysis. Aliquots of 125 μL of sample were diluted with H2O Milli-Q water, spiked with a solution containing an internal standard (4-methylvaleric acid, Sigma, Canada) and H3PO4 (Fisher, Canada), 10% to obtain a pH of 2. A volume of methyl tert-butyl ether (Sigma, Canada), equivalent to the volume of diluted sample was added to extract SCFA by vortexing2min. Samples were then centrifuged 10min at 14000g at 4°C and organic phases were transferred to a glass vial. SCFA analysis was performed on a GC-FID system (Shimadzu, Japan), consisting of a GC2010 Plus gas chromatograph equipped with an AOC-20s auto-sampler, an AOC-20i auto-injector and a flame ionization detector. The system was controlled by GC solution software. One microliter of organic phase was injected in a split mode into a Nukol capillary GC column (30m ×0.25mm id, 0.25 μM film thickness, Supelco analytical) and hydrogen was used as carrier gas. The injector and detector were set at 250°C. The oven temperature was initially programmed at 60°C, then increased to 200°C at 12° C/min and hold at this temperature for 2 min. SCFA were quantified using a 5-points calibration curve prepared with a mix of standards (acetic acid, propionic acid, butyric acid, isobutyric acid, valeric acid and isovaleric acid, Sigma, Canada) extracted following the same procedure as samples.
Dialysis cassette confined UPEC exposure to SHIME metabolome.
Slide-A-Lyzer Dialysis Cassettes 10 kDa pores (ThermoFisher Scientific, Canada) were used to test UPEC short-term exposure to the colonic metabolome of the TWIN-SHIME. This device maintains UPEC confined within the cassette and avoids the contamination of the whole reactor while allowing recovery of the pathogen for further analysis. The semipermeable membrane contains 10 kDa pores that are large enough to let colonic metabolic compounds pass through but restrict the passage of bacteria. UPEC pre-culture was resuspended in saline buffer and injected into the cassette at a dose of 7log10 CFU/3 mL (Fig. 1B). Cassettes containing UPEC were introduced in the transverse colon treated with or without PAC extract until the endpoint of diffusion of 2 h. The transverse colon was chosen for UPEC reservoir and PAC catabolism (49–51). An additional condition without microbiota was tested separately in anaerobic jars containing only the nutritional medium and pancreatic juice adjusted at the pH of the transverse colon. In each condition the cassettes were removed from the reactors after a 2-h incubation. Dialysates containing UPEC were collected with a syringe and aliquoted. To count the number of UPEC cells remaining after dialysis, 100 μL of dialysates were taken apart and serially diluted in saline water and plated onto LB agar (overnight incubation at 37°C). In parallel, the remaining 2.9 mL of dialysate samples were divided in two tubes and both pelleted 10 min at 14000g at 4°C. One tube of UPEC pellet was resuspended in 500 μL RNAlater (Thermo Fisher Scientific, USA) and stored at –80°C prior to RNA extraction, while the second tube of UPEC pellet was stored and preserved at –80°C in glycerol 10% until cell culture.
3D tissue-engineered urothelium culture and infection.
Bladder mesenchymal primary cells (BMC) and urothelial cells (UC) were extracted from a bladder biopsy (52). BMC and UC were cultured in media as previously described (53) and incubated at 37°C in a humidified 8% CO2 atmosphere. A suspension of 1,500,000 BMCs in 3 mL BMC medium was mixed with 3 mL of 5 mg/mL PureCol-EZ collagen solution (Advanced Biomatrix, USA) and distributed in 6 wells (including a paper anchorage) to produce 1 cm3 of cellularized collagen gel. The day after, 200,000 UCs were seeded on top of the collagen gel. Constructs were cultivated for 7 days in UC medium supplemented with 50 μ/mL ascorbate (Sigma, Canada). Engineered tissues were then put at the air/liquid interface using a specific device (54) and cultivated in UC medium for another 21 days with ascorbate (Fig. S1 in the supplemental material). On the day of infection, an antibiotic-free UC medium was used. UC were infected with 1 mL UPEC dialysates containing 6log10 CFU/mL, in duplicate for each condition (Fig. 1C). After 6 h, the medium containing non-adhered (planktonic) bacteria was collected, and tissues were rinsed three times using 2 mL PBS. Tissues were fixed 10 min in pure acetone at –20°C before performing immunolabelling. 100 μL of UC medium (planktonic UPEC) and rinsed tissue (adhered UPEC) were serially diluted in saline water and plated onto LB agar (overnight incubation at 37°C). The number of cultivable UPEC was expressed in log10 CFU (CFU)/mL.
Immunolabelling.
Punches of 4 mm from fixed tissues were rinsed in PBS and incubated 1 h with PBS/BSA 1% before sequential 24 h incubation with a primary antibody AE1/AE3 diluted 1/200 and a mix of secondary antibodies Alexa Fluor 594 (Abcam, Canada) diluted 1/400 with Hoechst 33258 diluted 1/200 (Sigma, Canada). Between each step, tissues were washed several times with PBS, and finally covered with mounting medium and coverslips. The slides were observed with a Zeiss LSM 800 laser-scanning confocal microscopy system (Zeiss, Canada).
Isolation of UPEC RNA and qRT-PCR.
RNA was isolated from UPEC dialysates, adherent and planktonic UPEC in UC, using TRIzol method (Invitrogen, USA) (43) and MICROBEnrich kit (Ambion, USA). UPEC mRNA was converted to cDNA using PrimeScript RT Kit (TaKaRa, Japan). Expression of UPEC genes was quantified on StepOnePlus real-time PCR system (Applied Biosystems, USA) and normalized to the housekeeping genes arcA and gapA. Primers used are described in Table S2 in the supplemental material.
Semi-targeted metabolomics for relative quantification of PAC secondary metabolites and metallophores.
Samples preparation consisted in the addition of 50 μL of cold methanol (LC-MS grade) to 50 μL colonic or UC samples to stop the metabolic activity. Ten microliters of internal standards solution (in 1:1 methanol/water) were added at a final concentration of 10 ppm for gallic acid-d2, succinic acid-d6, trans-cinnamic acid-d5, L-tryptophan-d5, glycocholic acid-d4, l-leucine-d7, 4-hydroxybenzoic acid-d4, methyl 4-hydroxybenzoate-d4; except for caffeine methyl-d3 (1 ppm) and n-dodecylphosphocholine-d38 (0,1 ppm). Samples were then filtered using micro-spin filter tubes (Nylon 0.22 μm, Canadian Life Science, Canada) by centrifuging them for 10 min at 10,000 g before LC-MS analysis. Relative quantification of PAC secondary metabolites and metallophores was performed on UPLC-Q-ToF (Acquity I-Class coupled with a Synapt G2-Si, Waters, USA).
The chromatographic method was adapted from Mena et al., 2017 (17). Briefly, 5 μL of samples were injected onto an ACQUITY UPLC HSS T3 column (2.1 × 100 mm, 1.8 μm) (Waters, USA) with an ACQUITY UPLC HSS T3 VanGuard pre-column (2.1 × 5 mm, 1.8 μm) (Waters, USA) heated to 30°C. The mobile phases were water (A) and acetonitrile (B), both acidified with 0.1% formic acid, and the flow rate was 0.3 mL/min. The gradient was as follows: 0–0.5 min: 2% B, 0.5–9 min: 2–45% B, 9–9.5 min: 45–80% B, 9.5–15.5 min: 80% B, 15.5–16 min: 80–2% B and 16–22 min: 2% B. Samples were kept at 4°C in the autosampler compartment. For the MS analysis, data were acquired by MSE in both positive and negative electrospray ionization and resolution mode (resolution ≈ 25 000) with a scan time of 0.2 s. Each function was collected with a scan range (m/z) of 50 to 1200. A collision energy ramp of 20 to 50 V was applied in the high energy function. The source parameters were as follows: capillary voltage, +1,20 kV in positive ionization and -2,40 kV in negative ionization; source temperature, 120°C; desolvation temperature, 400°C; cone gas flow, 50 L/h and desolvation gas, 800 L/h. Leucine-enkephaline (200 pg/μL) was infused at a flow rate of 10 μL/min for use as internal mass standard. Each sample was injected in triplicate.
Metabolite identification was confirmed using an UHPLC-Orbitrap (Vanquish Flex coupled with Orbitrap Tribrid Fusion, Thermo Scientific, USA) (Tables S3 and S4 in the supplemental material). Chromatographic conditions were the same as for analysis with UPLC-QToF. Data-dependant MS2 with an inclusion list was performed to confirm the identification of metabolites. The method started with a survey scan in the Orbitrap analyzer in positive ionization mode with a resolution of 120 000. The scan range (m/z) was 100 to 1,000, and the RF lens was set to 60%. After, ions were isolated by the quadrupole with a window size of 1.6 m/z and fragmented in the HCD cell with stepped collision energy (20, 40 and 65%). Fragments were analyzed in the Orbitrap with a resolution of 30 000. For each MS survey scan, three MS2 scans were acquired. The source parameters were as follows: spray voltage, +3,500 V; sheath gas flow, 50 Arb; aux gas flow, 10 Arb; sweep gas flow, 1 Arb; ion transfer tube temperature, 325°C and vaporizer temperature, 350°C. All acquired data were corrected using the internal mass calibrant (EASY-ICTM). Data acquired with the UHPLC-Orbitrap were processed using Compound Discoverer. Progenesis QI and Skyline were used to process data from the UPLC-Q-ToF.
Statistical analyses.
Statistical analyses were performed in R studio, version 4.0.4 (55), using the PMCMR package version 4.3. All formal hypothesis using non-parametric tests were conducted on the 5% significance level (P ≤ 0.05). Friedman test with post hoc Wilcoxon test was used to compare control versus PAC treatment conditions.
ACKNOWLEDGMENTS
We thank Swaine L. Chen from the Infectious Diseases Group, Genome Institute of Singapore for the generous gift of the UPEC strain used in this work, Dominique Mayrand for her help with confocal microscopy and Quebec Cell, Tissue and Gene Therapy Network-ThéCell (a thematic network supported by the FRQS), and Pier-Luc Plante from the metabolomic platform of INAF for his support with the metabolomic analysis.
We declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper. Y.D. hold the NSERC-DianaFood Industrial Research Chair (IRC) on the prebiotic effect of polyphenols in fruits and vegetables. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Footnotes
Supplemental material is available online only.
Contributor Information
Yves Desjardins, Email: yves.desjardins@fsaa.ulaval.ca.
Cezar M. Khursigara, University of Guelph
REFERENCES
- 1.Geerlings SE. 2016. Clinical presentations and epidemiology of urinary tract infections. Microbiol Spectr 4:4.5.03. doi: 10.1128/microbiolspec.UTI-0002-2012. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen KL, Stegger M, Kiil K, Godfrey PA, Feldgarden M, Lilje B, Andersen PS, Frimodt-Moller N. 2017. Whole-genome comparison of urinary pathogenic Escherichia coli and faecal isolates of UTI patients and healthy controls. Int J Med Microbiol 307:497–507. doi: 10.1016/j.ijmm.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamamoto S, Tsukamoto T, Terai A, Kurazono H, Takeda Y, Yoshida O. 1997. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. J Urol 157:1127–1129. doi: 10.1016/S0022-5347(01)65154-1. [DOI] [PubMed] [Google Scholar]
- 4.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Y-H, Ko W-C, Hsueh P-R. 2013. Emerging resistance problems and future perspectives in pharmacotherapy for complicated urinary tract infections. Expert Opin Pharmacother 14:587–596. doi: 10.1517/14656566.2013.778827. [DOI] [PubMed] [Google Scholar]
- 6.González de Llano D, Moreno-Arribas MV, Bartolomé B. 2020. Cranberry polyphenols and prevention against urinary tract infections: relevant considerations. Molecules 25:3523. doi: 10.3390/molecules25153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pappas E, Schaich KM. 2009. Phytochemicals of cranberries and cranberry products: characterization, potential health effects, and processing stability. Crit Rev Food Sci Nutr 49:741–781. doi: 10.1080/10408390802145377. [DOI] [PubMed] [Google Scholar]
- 8.Sihra N, Goodman A, Zakri R, Sahai A, Malde S. 2018. Nonantibiotic prevention and management of recurrent urinary tract infection. Nat Rev Urol 15:750–776. doi: 10.1038/s41585-018-0106-x. [DOI] [PubMed] [Google Scholar]
- 9.Maki KC, Kaspar KL, Khoo C, Derrig LH, Schild AL, Gupta K. 2016. Consumption of a cranberry juice beverage lowered the number of clinical urinary tract infection episodes in women with a recent history of urinary tract infection [published correction appears in Am J Clin Nutr. 2017 Aug;106(2):708]. Am J Clin Nutr 103:1434–1442. doi: 10.3945/ajcn.116.130542. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Howell AB, Zhang DJ, Khoo C. 2019. A randomized, double-blind, placebo-controlled pilot study to assess bacterial anti-adhesive activity in human urine following consumption of a cranberry supplement. Food Funct 10:7645–7652. doi: 10.1039/C9FO01198F. [DOI] [PubMed] [Google Scholar]
- 11.Jensen HD, Struve C, Christensen SB, Krogfelt KA. 2017. Cranberry juice and combinations of its organic acids are effective against experimental urinary tract infection. Front Microbiol 8:542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scharf B, Schmidt TJ, Rabbani S, Stork C, Dobrindt U, Sendker J, Ernst B, Hensel A. 2020. Antiadhesive natural products against uropathogenic E. coli: what can we learn from cranberry extract? J Ethnopharmacol 257:112889. doi: 10.1016/j.jep.2020.112889. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, Deering RW, Peng Z, Najia L, Khoo C, Cohen PS, Seeram NP, Rowley DC. 2019. Pectic oligosaccharides from cranberry prevent quiescence and persistence in the uropathogenic Escherichia coli CFT073. Sci Rep 9:19590. doi: 10.1038/s41598-019-56005-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feliciano RP, Mills CE, Istas G, Heiss C, Rodriguez-Mateos A. 2017. Absorption, metabolism and excretion of cranberry (poly)phenols in humans: a dose response study and assessment of inter-individual variability. Nutrients 9:268. doi: 10.3390/nu9030268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peron G, Sut S, Pellizzaro A, Brun P, Voinovich D, Castagliuolo I, Dall'Acqua S. 2017. The antiadhesive activity of cranberry phytocomplex studied by metabolomics: Intestinal PAC-A metabolites but not intact PAC-A are identified as markers in active urines against uropathogenic Escherichia coli. Fitoterapia 122:67–75. doi: 10.1016/j.fitote.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Clifford MN. 2004. Diet-derived phenols in plasma and tissues and their implications for health. Planta Med 70:1103–1114. doi: 10.1055/s-2004-835835. [DOI] [PubMed] [Google Scholar]
- 17.Mena P, González de Llano D, Brindani N, Esteban-Fernández A, Curti C, Moreno-Arribas MV, Del Rio D, Bartolomé B. 2017. 5-(3′,4′-Dihydroxyphenyl)-γ-valerolactone and its sulphate conjugates, representative circulating metabolites of flavan-3-ols, exhibit anti-adhesive activity against uropathogenic Escherichia coli in bladder epithelial cells. J Functional Foods 29:275–280. doi: 10.1016/j.jff.2016.12.035. [DOI] [Google Scholar]
- 18.Mann R, Mediati DG, Duggin IG, Harry EJ, Bottomley AL. 2017. Metabolic adaptations of uropathogenic E. coli in the urinary tract. Front Cell Infect Microbiol 7:241. doi: 10.3389/fcimb.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alteri CJ, Mobley HLT. 2015. Metabolism and Fitness of Urinary Tract pathogens. Microbiol Spectr 3. doi: 10.1128/microbiolspec.MBP-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connolly JPR, O'Boyle N, Turner NCA, Browning DF, Roe AJ. 2019. Distinct intraspecies virulence mechanisms regulated by a conserved transcription factor. Proc Natl Acad Sci USA 116:19695–19704. doi: 10.1073/pnas.1903461116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garretto A, Miller-Ensminger T, Ene A, Merchant Z, Shah A, Gerodias A, Biancofiori A, Canchola S, Canchola S, Castillo E, Chowdhury T, Gandhi N, Hamilton S, Hatton K, Hyder S, Krull K, Lagios D, Lam T, Mitchell K, Mortensen C, Murphy A, Richburg J, Rokas M, Ryclik S, Sulit P, Szwajnos T, Widuch M, Willis J, Woloszyn M, Brassil B, Johnson G, Mormando R, Maskeri L, Batrich M, Stark N, Shapiro JW, Montelongo Hernandez C, Banerjee S, Wolfe AJ, Putonti C. 2020. Genomic survey of E. coli from the bladders of women with and without lower urinary tract symptoms. Front Microbiol 11:2094. doi: 10.3389/fmicb.2020.02094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulz E, Schumann M, Schneemann M, Dony V, Fromm A, Nagel O, Schulzke JD, Bucker R. 2021. Escherichia coli alpha-hemolysin HlyA induces host cell polarity changes, epithelial barrier dysfunction and cell detachment in human colon carcinoma caco-2 cell model via PTEN-dependent dysregulation of cell junctions. Toxins 13:520. doi: 10.3390/toxins13080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Professionals S-O. EAU guidelines: urological infections. Uroweb. https://uroweb.org/guideline/urological-infections/. [Google Scholar]
- 24.Rodriguez-Mateos A, Vauzour D, Krueger CG, Shanmuganayagam D, Reed J, Calani L, Mena P, Del Rio D, Crozier A. 2014. Bioavailability, bioactivity and impact on health of dietary flavonoids and related compounds: an update. Arch Toxicol 88:1803–1853. doi: 10.1007/s00204-014-1330-7. [DOI] [PubMed] [Google Scholar]
- 25.Mena P, Ludwig IA, Tomatis VB, Acharjee A, Calani L, Rosi A, Brighenti F, Ray S, Griffin JL, Bluck LJ, Del Rio D. 2019. Inter-individual variability in the production of flavan-3-ol colonic metabolites: preliminary elucidation of urinary metabotypes. Eur J Nutr 58:1529–1543. doi: 10.1007/s00394-018-1683-4. [DOI] [PubMed] [Google Scholar]
- 26.Monagas M, Urpi-Sarda M, Sánchez-Patán F, Llorach R, Garrido I, Gómez-Cordovés C, Andres-Lacueva C, Bartolomé B. 2010. Insights into the metabolism and microbial biotransformation of dietary flavan-3-ols and the bioactivity of their metabolites. Food Funct 1:233–253. doi: 10.1039/c0fo00132e. [DOI] [PubMed] [Google Scholar]
- 27.Feliciano RP, Boeres A, Massacessi L, Istas G, Ventura MR, Nunes dos Santos C, Heiss C, Rodriguez-Mateos A. 2016. Identification and quantification of novel cranberry-derived plasma and urinary (poly)phenols. Arch Biochem Biophys 599:31–41. doi: 10.1016/j.abb.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 28.Riedl A, Gieger C, Hauner H, Daniel H, Linseisen J. 2017. Metabotyping and its application in targeted nutrition: an overview. Br J Nutr 117:1631–1644. doi: 10.1017/S0007114517001611. [DOI] [PubMed] [Google Scholar]
- 29.Samarasinghe S, Reid R, AL-Bayati M. 2019. The anti-virulence effect of cranberry active compound proanthocyanins (PACs) on expression of genes in the third-generation cephalosporin-resistant Escherichia coli CTX-M-15 associated with urinary tract infection. Antimicrob Resist Infect Control 8:181. doi: 10.1186/s13756-019-0637-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hidalgo G, Chan M, Tufenkji N. 2011. Inhibition of Escherichia coli CFT073 fliC expression and motility by cranberry materials. Appl Environ Microbiol 77:6852–6857. doi: 10.1128/AEM.05561-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ranfaing J, Dunyach-Remy C, Lavigne JP, Sotto A. 2018. Propolis potentiates the effect of cranberry (Vaccinium macrocarpon) in reducing the motility and the biofilm formation of uropathogenic Escherichia coli. PLoS One 13:e0202609. doi: 10.1371/journal.pone.0202609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin B, Johnson BJ, Rubin RA, Malanoski AP, Ligler FS. 2011. Iron chelation by cranberry juice and its impact on Escherichia coli growth. Biofactors 37:121–130. doi: 10.1002/biof.110. [DOI] [PubMed] [Google Scholar]
- 33.Garcia EC, Brumbaugh AR, Mobley HLT. 2011. Redundancy and specificity of Escherichia coli iron acquisition systems during urinary tract infection. Infect Immun 79:1225–1235. doi: 10.1128/IAI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Hage R, Hernandez-Sanabria E, Calatayud Arroyo M, Props R, Van de Wiele T. 2019. Propionate-producing consortium restores antibiotic-induced dysbiosis in a dynamic in vitro model of the human intestinal microbial ecosystem. Front Microbiol 10:1206. doi: 10.3389/fmicb.2019.01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Herreweghen F, De Paepe K, Roume H, Kerckhof FM, Van de Wiele T. 2018. Mucin degradation niche as a driver of microbiome composition and Akkermansia muciniphila abundance in a dynamic gut model is donor independent. FEMS Microbiol Ecol 94. doi: 10.1093/femsec/fiy186. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, O'Riordan MX. 2013. Regulation of bacterial pathogenesis by intestinal short-chain Fatty acids. Adv Appl Microbiol 85:93–118. doi: 10.1016/B978-0-12-407672-3.00003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobson A, Lam L, Rajendram M, Tamburini F, Honeycutt J, Pham T, Van Treuren W, Pruss K, Stabler SR, Lugo K, Bouley DM, Vilches-Moure JG, Smith M, Sonnenburg JL, Bhatt AS, Huang KC, Monack D. 2018. A gut commensal-produced metabolite mediates colonization resistance to salmonella infection. Cell Host Microbe 24:296–307. doi: 10.1016/j.chom.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jubelin G, Desvaux M, Schüller S, Etienne-Mesmin L, Muniesa M, Blanquet-Diot S. 2018. Modulation of enterohaemorrhagic Escherichia coli survival and virulence in the human gastrointestinal tract. Microorganisms 6:115. doi: 10.3390/microorganisms6040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mena P, Favari C, Acharjee A, Chernbumroong S, Bresciani L, Curti C, Brighenti F, Heiss C, Rodriguez-Mateos AM, Del Rio D. 2022. Metabotypes of flavan-3-ol colonic metabolites after cranberry intake: elucidation and statistical approaches. Eur J Nutr 61:1299–1317. doi: 10.1007/s00394-021-02692-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen SL, Hung C-S, Xu J, Reigstad CS, Magrini V, Sabo A, Blasiar D, Bieri T, Meyer RR, Ozersky P, Armstrong JR, Fulton RS, Latreille JP, Spieth J, Hooton TM, Mardis ER, Hultgren SJ, Gordon JI. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: A comparative genomics approach. Proc Natl Acad Sci USA 103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eshaghi M, Mehershahi KS, Chen SL. 2016. Brighter fluorescent derivatives of UTI89 utilizing a monomeric vGFP. Pathogens 5:3. doi: 10.3390/pathogens5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van de Wiele T, Van den Abbeele P, Ossieur W, Possemiers S, Marzorati M. 2015. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME®), 305–17. In Verhoeckx K, Cotter P, López-Expósito I, Kleiveland C, Lea T, Mackie A (ed), The impact of food bioactives on health: in vitro and ex vivo models. Springer International Publishing, Cham, France. [PubMed] [Google Scholar]
- 43.Roussel C, De Paepe K, Galia W, De Bodt J, Chalancon S, Leriche F, Ballet N, Denis S, Alric M, Van de Wiele T, Blanquet-Diot S. 2020. Spatial and temporal modulation of enterotoxigenic E. coli H10407 pathogenesis and interplay with microbiota in human gut models. BMC Biol 18:141. doi: 10.1186/s12915-020-00860-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geirnaert A, Wang J, Tinck M, Steyaert A, Van den Abbeele P, Eeckhaut V, Vilchez-Vargas R, Falony G, Laukens D, De Vos M, Van Immerseel F, Raes J, Boon N, Van de Wiele T. 2015. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol Ecol 91:fiv054. [DOI] [PubMed] [Google Scholar]
- 45.Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AA, Holmes SP. 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McLaren MR, Callahan BJ. 2021. Silva 138.1 prokaryotic SSU taxonomic training data formatted for DADA2 [Data set]. Zenodo, Geneva, Switzerland. [Google Scholar]
- 47.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oksanen J, Blanchet G, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H. 2020. Vegan: community ecology package (R package version 2.5-7). https://cran.r-project.org/web/packages/vegan/vegan.pdf.
- 49.Spaulding CN, Klein RD, Ruer S, Kau AL, Schreiber HL, Cusumano ZT, Dodson KW, Pinkner JS, Fremont DH, Janetka JW, Remaut H, Gordon JI, Hultgren SJ. 2017. Selective depletion of uropathogenic E. coli from the gut by a FimH antagonist. Nature 546:528–532. doi: 10.1038/nature22972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sánchez-Patán F, Barroso E, Van de Wiele T, Jiménez-Girón A, Martín-Alvarez PJ, Moreno-Arribas MV, Martinez-Cuesta MC, Pelaez C, Requena T, Bartolomé B. 2015. Comparative in vitro fermentations of cranberry and grape seed polyphenols with colonic microbiota. Food Chem 183:273–282. doi: 10.1016/j.foodchem.2015.03.061. [DOI] [PubMed] [Google Scholar]
- 51.Barroso E, Van de Wiele T, Jiménez-Girón A, Muñoz-González I, Martín-Alvarez PJ, Moreno-Arribas MV, Bartolomé B, Pelaez C, Martinez-Cuesta MC, Requena T. 2014. Lactobacillus plantarum IFPL935 impacts colonic metabolism in a simulator of the human gut microbiota during feeding with red wine polyphenols. Appl Microbiol Biotechnol 98:6805–6815. doi: 10.1007/s00253-014-5744-1. [DOI] [PubMed] [Google Scholar]
- 52.Chabaud S, Rousseau A, Marcoux T-L, Bolduc S. 2017. Inexpensive production of near-native engineered stromas. J Tissue Eng Regen Med 11:1377–1389. doi: 10.1002/term.2036. [DOI] [PubMed] [Google Scholar]
- 53.Ringuette Goulet C, Bernard G, Chabaud S, Couture A, Langlois A, Neveu B, Pouliot F, Bolduc S. 2017. Tissue-engineered human 3D model of bladder cancer for invasion study and drug discovery. Biomaterials 145:233–241. doi: 10.1016/j.biomaterials.2017.08.041. [DOI] [PubMed] [Google Scholar]
- 54.Chabaud S, Bolduc S. 2016. Production of a self-aligned scaffold, free of exogenous material, from dermal fibroblasts using the self-assembly technique. Dermatol Res Pract 2016:5397319. doi: 10.1155/2016/5397319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Core Team . 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.02432-21-s0001.pdf, PDF file, 0.5 MB (476.8KB, pdf)