INTRODUCTION
During the early months of the SARS-CoV-2 pandemic, notable uncertainty emerged regarding the role of children in transmission dynamics1. With time, it became more clear that children were susceptible to infection with SARS-CoV-2, but that the vast majority of children experienced mild symptoms with lower incidence of severe disease2. This pattern remained consistent despite the later emergence of SARS-CoV-2 variants, including Delta and Omicron, even among children <5 ineligible for vaccination3. The relative lack of severe disease in the pediatric population raised questions regarding viral kinetics and infectivity in children versus adults.
We hypothesized that unique virologic features in children could explain this apparent decrease in symptoms and transmissibility early in the pandemic. Due to the challenges posed by measurement of infectious viral titers, the majority of work examining viral loads in clinical samples has measured viral RNA levels, as determined by RT-qPCR cycle threshold [CT]. A previous study using this technique reported no differences in viral RNA load in adults and children, when controlling for the presence of symptoms4. A different study reported both RNA viral load and level of infectious virus using a semi-quantitative method (TCID50) in pediatric clinical samples5. In contrast however, other work indicates that ancestral SARS-CoV-2 replicates less efficiently in both children and pediatric versus adult nasal epithelial cells, a defect that Omicron was able to abolish5–7. Finally, we and others have demonstrated a dynamic relationship between CT values and infectious viral titers with potential for significant discrepancies and a ratio dependent on both viral and host factors,8 but this work did not include children9.
Therefore, to further understand SARS-CoV-2 infection in children, we investigated the ratio of infectious virus titer to RNA viral load in children aged 0 to <18 years old. We hypothesized that the ratio of infectious virus to RNA viral load would be positively associated with age.
METHODS
Sample Selection
Banked SARS-CoV-2 positive nasopharyngeal specimens from children 0 to <18 years old collected and stored at Children’s Wisconsin, Milwaukee, Wisconsin between September 14, 2020 and May 17, 2021 were identified. Deidentified samples were binned into four age groups (<1, 1–5, 6–11, and 12–17) and stratified by clinical CT value (<20, 20–24, 25–29, and 30–34) to select a sample of children representing the full spectrum of both age and CT value . The study received an exempt determination for use of deidentified specimens from the University of Vermont (UVM) Institutional Review Board and the Children’s Wisconsin Institutional Review Board.
RNA extractions and RT-PCR
Total nucleic acid was extracted on the NucliSENS easyMAG or EMAG automated extraction instruments (bioMerieux). SARS-CoV-2 RNA was detected using previously published primers/probes for the SARS-CoV-2 E gene (Sarbeco10) on the 7500 Fast Real-Time PCR System or QuantStudio 7 Pro platforms.
Viral Titrations
SARS-CoV-2 viral titering was conducted under BSL-3 conditions at UVM using a microfocus forming unit (FFU) assay in VeroE6-TMPRSS2 cells, which increases assay sensitivity compared to standard VeroE6 cells, as previously described8.
Statistical Analysis
Viral titers were log-transformed for analysis. Linear regression was used to predict log titer as a function of CT, fitting separate models without age and to control for continuous and categorical age effects. Models were compared by F test. Data were analyzed and plotted with R. Code is available at https://github.com/emilybrucelab.
RESULTS
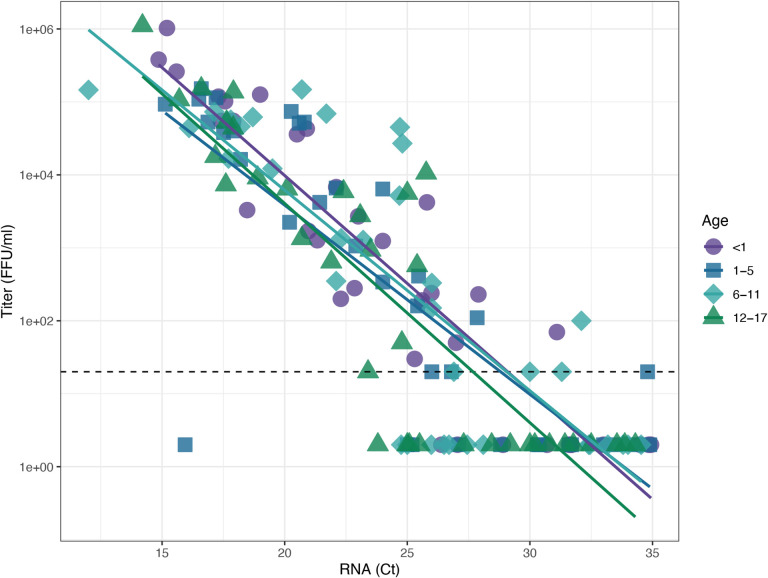
N=144 clinical specimens were selected to determine the relationship between the infectivity of SARS-CoV-2 in pediatric samples and RNA viral load. As expected, higher RNA viral load generally correlated with higher infectious virus titer, although as reported previously this ratio was somewhat variable8,9. In linear regression, the relationship between infectious viral titer and CT was not significantly modified by age (P=0.156) or age group (P=0.355 overall by F test). These data indicate that there is no difference in the infectiousness of SARS-CoV-2 produced by children, regardless of age.
DISCUSSION
Consistent with previous findings, we found no significant differences in the relationship between SARS-CoV-2 infectious virus titer and RNA viral load in children across the pediatric age spectrum4,7. Our findings suggest equal levels of viral infectivity in children and in adults with similar RNA viral loads. Limitations of this study include lack of access to viral sequencing and individual level metadata, which could reveal differences in infectivity as a result of viral genetic background, days post-symptom onset, host immune status, and vaccination status. Furthermore, there was no direct comparison with adult samples, although we did include samples in older teens who would closely resemble adults biologically.
Figure 1. SARS-CoV-2 viral infectivity does not vary by age in a pediatric population.
A set of 144 clinical samples from children infected with SARS-CoV-2 was used to examine the relationship between infectious virus titer and RNA viral load as a function of patient age. Individual specimen measurements of E gene RNA levels (CT) on the x-axis are plotted against viral titer, as measured in focus forming units (FFU/mL) on the y-axis. Dashed line indicates the limit of detection for infectious titer (20 FFU/mL). Samples for which we could not measure a viral titer were assigned fixed values of one-tenth the limit of detection (2 FFU/mL). Lines of best fit were generated by linear regression on log-transformed titer data as a function of CT and age group.
ACKNOWLEDGEMENTS
We thank Ms. Kubinski and Dr. Oetjen for technical assistance.
Funding/Support:
This study was supported by NIH grant P30GM118228-04 (to E.A.B., J.W.C. and B.L.)
Role of Funder:
The NIH had no role in the design and conduct of the study.
Abbreviations:
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- COVID-19
coronavirus disease 2019
- CT
cycle threshold
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- RNA
ribonucleic acid
- TMPRSS2
transmembrane protease, serine 2
- FFU
focus forming unit
Footnotes
Conflict of Interest Disclosures: The authors have no conflicts to disclose.
REFERENCES
- 1.Lee B, Raszka WV Jr. COVID-19 Transmission and Children: The Child Is Not to Blame. Pediatrics. 2020;146(2). [DOI] [PubMed] [Google Scholar]
- 2.Lu X, Zhang L, Du H, et al. SARS-CoV-2 Infection in Children. New England Journal of Medicine. 2020;382(17):1663–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang L, Berger NA, Kaelber DC, Davis PB, Volkow ND, Xu R. Incidence Rates and Clinical Outcomes of SARS-CoV-2 Infection With the Omicron and Delta Variants in Children Younger Than 5 Years in the US. JAMA Pediatrics. 2022;176(8):811–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung E, Chow EJ, Wilcox NC, et al. Comparison of Symptoms and RNA Levels in Children and Adults With SARS-CoV-2 Infection in the Community Setting. JAMA Pediatrics. 2021;175(10):e212025–e212025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonker LM, Boucau J, Regan J, et al. Virologic Features of Severe Acute Respiratory Syndrome Coronavirus 2 Infection in Children. J Infect Dis. 2021;224(11):1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones TC, Biele G, Mühlemann B, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373(6551). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu Y, Chew KY, Wu M, et al. Ancestral SARS-CoV-2, but not Omicron, replicates less efficiently in primary pediatric nasal epithelial cells. PLOS Biology. 2022;20(8):e3001728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Despres HW, Mills MG, Shirley DJ, et al. Measuring infectious SARS-CoV-2 in clinical samples reveals a higher viral titer:RNA ratio for Delta and Epsilon vs. Alpha variants. Proc Natl Acad Sci U S A. 2022;119(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puhach O, Adea K, Hulo N, et al. Infectious viral load in unvaccinated and vaccinated patients infected with SARS-CoV-2 WT, Delta and Omicron. medRxiv. 2022:2022.2001.2010.22269010. [DOI] [PubMed] [Google Scholar]
- 10.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 (3). [DOI] [PMC free article] [PubMed] [Google Scholar]