Abstract
Antibiotics at suboptimal doses promote biofilm formation and the development of antibiotic resistance. The underlying molecular mechanisms, however, were not investigated. Here, we report the effects of sub-minimum inhibitory concentrations (sub-MICs) of imipenem and colistin on genes associated with biofilm formation and biofilm-specific antibiotic resistance in a multidrug-tolerant clinical strain of Acinetobacter baumannii Sequence Type (ST) 1894. Comparative transcriptome analysis was performed in untreated biofilm and biofilm treated with sub-MIC doses of imipenem and colistin. RNA sequencing data showed that 78 and 285 genes were differentially expressed in imipenem and colistin-treated biofilm cells, respectively. Among the differentially expressed genes (DEGs), 48 and 197 genes were upregulated exclusively in imipenem and colistin-treated biofilm cells, respectively. The upregulated genes included those encoding matrix synthesis (pgaB), multidrug efflux pump (novel00738), fimbrial proteins, and homoserine lactone synthase (AbaI). Upregulation of biofilm-associated genes might enhance biofilm formation when treated with sub-MICs of antibiotics. The downregulated genes include those encoding DNA gyrase (novel00171), 30S ribosomal protein S20 (novel00584), and ribosome releasing factor (RRF) were downregulated when the biofilm cells were treated with imipenem and colistin. Downregulation of these genes affects protein synthesis, which in turn slows down cell metabolism and makes biofilm cells more tolerant to antibiotics. In this investigation, we also found that 5 of 138 small RNAs (sRNAs) were differentially expressed in biofilm regardless of antibiotic treatment or not. Of these, sRNA00203 showed the highest expression levels in biofilm. sRNAs regulate gene expression and are associated with biofilm formation, which may in turn affect the expression of biofilm-specific antibiotic resistance. In summary, when biofilm cells were exposed to sub-MIC doses of colistin and imipenem, coordinated gene responses result in increased biofilm production, multidrug efflux pump expression, and the slowdown of metabolism, which leads to drug tolerance in biofilm. Targeting antibiotic-induced or repressed biofilm-specific genes represents a new strategy for the development of innovative and effective treatments for biofilm-associated infections caused by A. baumannii.
Keywords: Acinetobacter baumannii, biofilm, colistin, imipenem, antibiotic resistance, RNA sequencing, small RNA, virulence
1. Introduction
Acinetobacter baumannii is an emerging global antibiotic-resistant gram-negative bacteria that primarily causes biofilm-associated infections such as ventilator-associated pneumonia and catheter-related infection, both of which are resistant to conventional antibiotics [1,2,3].The capacity of A. baumannii to form biofilms enhances its survival in adverse environments, making it a successful nosocomial pathogen [4,5,6,7,8]. Recently, several environmental reservoirs in hospital settings were reported to be the primary sources for outbreaks of multidrug resistance (MDR) A. baumannii [9,10]. Biofilm formation protects A. baumannii from antibacterial agents and allows the pathogen to survive on abiotic and biotic surfaces [11]. In our previous study, we identified a strong biofilm-former, A. baumannii ST1894 [12]. The strain was susceptible to imipenem and colistin in the planktonic state but was highly resistant in the biofilm state, and the minimum inhibitory concentrations (MICs) of imipenem and colistin were 2048 and 32 times higher than those required to eradicate the planktonic cells [12]. Imipenem and colistin are the most common antibiotics used to treat multidrug-resistant A. baumannii strains; a decrease in antibiotic susceptibility in biofilm creates a significant problem in the control of A. baumannii infections in clinical settings [13,14]. Moreover, the decrease in susceptibility renders the antibiotic dosage sub-optimal, studies have shown that exposure to sub-optimal antibiotic dosages triggered biofilm formation and expression of antibiotic resistance genes [15,16]. Thus, exposure of A. baumannii biofilm to sub-MICs of imipenem and colistin may trigger transcriptional and post-transcriptional changes in the biofilm cells, which might result in biofilm-specific antibiotic resistance in A. baumannii.
Despite the fact that biofilm formation in A. baumannii decreases susceptibility to imipenem and colistin, little is known about the molecular mechanisms involved in the expression of antibiotic resistance and virulence when A. baumannii biofilm is exposed to sub-optimal doses of the two antibiotics. Here, we have conducted a comparative transcriptome study to determine the transcriptional changes when biofilm cells were treated with sub-MICs of imipenem and colistin. This study lays the foundation for future research on the development of novel therapeutics for treatment of biofilm-associated infections caused by A. baumannii.
2. Results
2.1. Gene Expression Profile Associated with Biofilm Formation
Following the alignment of clean reads with the reference genome of A. baumannii ATCC17978, a list of differentially expressed genes (DEGs) in the untreated and antibiotic-treated biofilm cells was identified (Table S1). As shown in Table 1, 51.8% (1592/3075) of genes were differentially expressed in the untreated biofilm phase compared with their planktonic counterparts. In addition, 96 and 5 DEGs were classified as novel genes and sRNA, respectively. Of these 1592 genes, 20% (614/3075) were upregulated and 31.8% (978/3075) were downregulated in biofilm cells. For the imipenem-treated biofilm, 3.7% (106/2885) of the genes were differentially expressed. Seven and one DEGs were classified as novel genes and sRNA. Of the 106 genes, 45.3% (48/106) were upregulated and 54.7% (58/106) were downregulated in biofilm cells. For the colistin-treated biofilm, 12.6% (368/2912) of the genes were differentially expressed, 33 DEGs were classified as novel genes. Of the 368 genes, 64.9% (239/368) were upregulated and 35.1% (129/368) were downregulated in biofilm cells.
Table 1.
Summary of genes differentially expressed in untreated biofilm and biofilm exposed to imipenem and colistin at sub-MIC dosages.
| Group | Transcribed Genes | Differentially Expressed | |||
|---|---|---|---|---|---|
| Total No. (%) | Novel Genes | Genes Encoding Hypothetical Proteins | Small RNA | ||
| Biofilm vs. planktonic cells | 3075 | 1592 (51.8) | 96 | 431 | 5 |
| Imipenem-treated biofilm vs. untreated biofilm cells | 2885 | 106 (3.7) | 7 | 11 | 1 |
| Colistin-treated biofilm vs. untreated biofilm cells | 2912 | 368 (12.6) | 33 | 74 | 0 |
We classified the 1592 DEGs into different functional categories based on Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment. We focused on 211 genes that were linked to biofilms and antibiotic resistance for further analyses (Table S1 and Figure 1). To understand the effect of suboptimal doses of antibiotics, we subsequently identified 50 DEGs in biofilm cells upon exposure to a suboptimal dose of colistin or imipenem (Table 2).
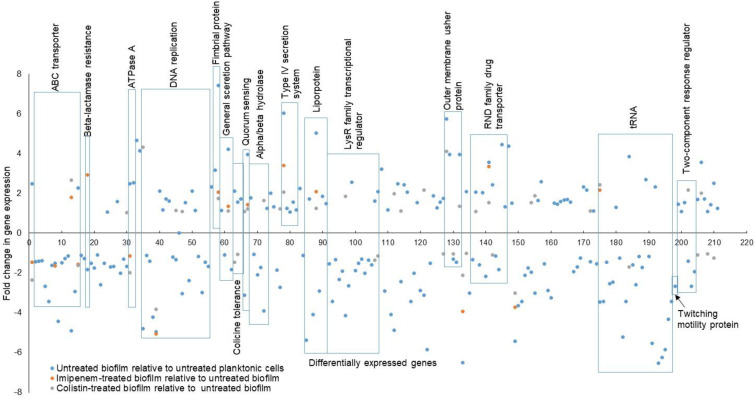
Figure 1.
Distribution of significantly up- or downregulated genes belonging to functional categories associated with virulence and antibiotic resistance in biofilm. The blue dots represent the DEGs between untreated biofilm and planktonic cells. The orange dots represent the DEGs between untreated and imipenem-treated biofilm cells. The grey dots represent the DEGs between untreated and colistin-treated biofilm cells.
Table 2.
Biofilm-specific genes induced or repressed by sub-MICs of imipenem and colistin.
| Gene_id | KEGG_ID | Fold Change in Gene Expression | Function/Product | ||
|---|---|---|---|---|---|
| Untreated Biofilm Relative to Untreated Planktonic | Imipenem-Treated Biofilm Relative to Untreated Biofilm | Colistin-Treated Biofilm Relative to Untreated Biofilm | |||
| Lipoprotein | |||||
| E5A72_RS12100 | acb: A1S_0938 | 5.0363 | 2.09 | 1.23 | Lipoprotein (biofilm matrix) |
| ATP-binding cassette (ABC) transporter | |||||
| E5A72_RS02170 | acb: A1S_2611 | 2.3183 | 2.14368 | 1.02 | Transport protein of outer membrane lipoproteins |
| E5A72_RS16335 | acb: A1S_1722 | −1.5054 | −1.639 | 0.095 | ABC transporter ATP-binding protein |
| E5A72_RS14510 | acb: A1S_1359 | −4.9058 | 1.7825 | 2.6702 | ABC-type Fe3+ transport system |
| E5A72_RS08785 | acb: A1S_0229 | 2.0395 | 1.5784 | −1.6349 | Lipopolysaccharide export system permease protein |
| E5A72_RS15155 | acb: A1S_1482 | −3.3293 | 1.0455 | 2.0257 | D-and L-methionine transport protein |
| β-lactam resistance | |||||
| E5A72_RS00940 | acb: A1S_2367 | −1.8362 | 2.92 | 1.16 * | ampC; β-lactamase |
| Novel00738 | acb: A1S_2736 | 3.5549 | 3.3532 | −1.013 | Resistance–nodulation–cell division (RND) family drug transporter |
| DNA replication and repair | |||||
| Novel00171 | acb: A1S_2626 | −4.9582 | −5.0584 | −3.8428 | DNA gyrase |
| E5A72_RS14680 | acb: A1S_1389 | −1.3449 | −0.0409 * | 1.1446 | DNA polymerase V component |
| E5A72_RS03815 | acb: A1S_2918 | −3.0414 | −0.08 * | 1.0746 | DNA repair protein |
| Fimbrial protein and csu operon | |||||
| E5A72_RS05130 | acb: A1S_3177 | 7.4033 | 2.0455 | 1.7527 | Fimbrial protein |
| E5A72_RS19200 | acb: A1S_2217 | 5.7238 | NA | 2.1677 | Protein CsuA |
| E5A72_RS19190 | acb: A1S_2215 | 1.3111 | −0.683 * | 1.0371 | Protein CsuC |
| E5A72_RS19180 | acb: A1S_2213 | 3.9517 | 0.2679 * | 4.1059 | Protein CsuE |
| Bacterial secretion system | |||||
| E5A72_RS14240 | acb: A1S_1310 | 6.0356 | 3.387 | 2.045 | Type VI secretion system protein |
| E5A72_RS15605 | acb: A1S_1564 | 4.2 | 1.3587 | 1.1006 | General secretion pathway protein J |
| E5A72_RS03500 | acb: A1S_2862 | 5.376 | 1.65783 | 2.349 | Preprotein translocase subunit SecA |
| Two-component regulatory system | |||||
| E5A72_RS09610 | acb: A1S_0399 | −4.1397 | −0.346 * | 1.8361 | LysR family transcriptional regulator |
| E5A72_RS12380 | acb: A1S_0992 | 1.617 | −0.132 * | −1.3517 | |
| E5A72_RS13515 | acb: A1S_1182 | 3.216 | 2.06 | 0.065 * | cAMP-activated global transcriptional regulator CRP |
| Novel00822 | acb: A1S_0685 | −1.4033 | 1.788 * | 2.4156 | Two-component response regulator |
| Quorum sensing or quenching | |||||
| E5A72_RS06870 | acb: A1S_0109 | 3.9355 | 1.43253 | 1.21081 | Acyl homoserine lactone (AHL) synthase (AbaI) |
| E5A72_RS16835 | acb: A1S_1809 | −3.9015 | 0.267 * | 1.6271 | Hydrolase transmembrane protein |
| Multidrug efflux pump | |||||
| E5A72_RS00640 | acb: A1S_2306 | 2.0527 | −0.083 * | 2.1244 | RND efflux transporter |
| E5A72_RS11940 | acb: A1S_0908 | 2.4322 | −0.4375 | 1.0937 | RND family multidrug resistance secretion protein |
| Transcription and translation | |||||
| E5A72_RS17735 | acb: A1S_1974 | −6.5036 | −3.9244 | −2.1244 | Ribosome releasing factor |
| Novel00584 | acb: A1S_1617 | −5.4437 | −3.7365 | −3.01 | 30S ribosomal protein S20 |
| Novel00490 | - | 7.0401 | 1.1587 | 0.415 | Transcription termination factor Rho OS |
| E5A72_RS04400 | acb: A1S_3029 | −3.4737 | 2.161 | 2.4156 | tRNA-Arg |
| Peptidoglycan biosynthesis | |||||
| E5A72_RS10790 | acb: A1S_1965 | 1.6823 | −0.1303 * | −1.6917 | UDP-N-acetylglucosamine 1-carboxyvinyltransferase |
| Novel00626 | acb: A1S_1965 | 3.5549 | NA | 2.1677 | UDP-N-acetylglucosamine acyltransferase |
| E5A72_RS05230 | acb: A1S_3203 | 1.0844 | −0217* | −1.032 | UDP-N-acetylmuramoylalanyl-D-glutamate-2, 6-diaminopimelate ligase |
| E5A72_RS17730 | acb: A1S_1973 | 2.513 | −0.0783 * | −1.2536 | Undecaprenyl pyrophosphate synthetase |
| E5A72_RS07480 | acb: A1S_2968 | −2.7334 | 0.4605 * | 1.2183 | Hypothetical protein |
| E5A72_RS05220 | acb: A1S_3200 | 1.7367 | 0.0003 * | −1.0371 | Phospho-N-acetylmuramoyl-pentapeptide transferase |
| Outer membrane protein | |||||
| E5A72_RS11985 | - | −4.8714 | −0.1244 * | 1.9751 | OprD family outer membrane porin |
| E5A72_RS18480 | acb: A1S_2076 | −2.4293 | 0.4473 * | 1.1074 | Outer membrane porin receptor for Fe (III)- coprogen |
| E5A72_RS01795 | acb: A1S_2538 | −3.1257 | 1.4605 * | 2.1677 | Outer membrane protein CarO precursor |
| Transcriptional regulators and others | |||||
| E5A72_RS13590 | acb: A1S_1197 | −2.5865 | −1.2664 | 0.240 * | Extracellular nuclease |
| E5A72_RS03015 | acb: A1S_2767 | −1.6789 | 0.7044 | 1.0387 | AraC family transcriptional regulator |
| E5A72_RS04330 | acb: A1S_3229 | 3.8412 | 0.25977 | −1.6917 | tRNA-i(6)A37 modification enzyme |
| E5A72_RS01785 | acb: A1S_2536 | 2.4782 | −1.1416 | −1.9789 | ATPase |
| E5A72_RS14655 | acb: A1S_1386 | −4.8062 | NA | 4.3282 | Catalase; K03781 catalase |
| E5A72_RS01830 | acb: A1S_2546 | −1.9701 | 0.9841 | 1.5253 | Secreted trypsin-like serine protease |
| E5A72_RS05375 | acb: A1S_3227 | 2.0827 | −0.02825 | −1.013 | RNA binding protein |
| E5A72_RS16050 | acb: A1S_1670 | −3.0262 | 0.598 * | −3.01 | Secretion protein HlyD |
| E5A72_RS02070 | acb: A1S_2592 | 1.5561 | 0.0036 | −1.0789 | Group A colicins tolerance protein |
| E5A72_RS02065 | acb: A1S_2591 | 2.0955 | −0.0461 | −1.4623 | Group A colicins tolerance protein |
| Non-coding RNA | |||||
| Gene encoding sRNA00203 | - | 5.3951 | 2.38935 | −0.389 * | |
Adjusted p (pad) < 0.004; * pad > 0.005; NA: not applicable. Minus sign (−): downregulated gene.
2.2. Gene Expression Profile of Biofilm Treated with Sub-MIC of Imipenem
Table 1 summarizes the number of genes expressed in biofilm cells of A. baumannii ST1894 when treated with imipenem at sub-MIC. Of the 2885 total expressed genes, 3.7% (106) were differentially expressed in the imipenem-treated biofilm cells and the details in listed in Table S1. Of these 106 DEGs, 78 were biofilm-specific, of which 48 were upregulated in sub-MIC of imipenem treated biofilm versus untreated biofilm cells. The upregulated genes with biofilm cells treated with sub-MIC of imipenem include genes encoding pgaB, genes encoding the fimbrial protein, AHL synthase, the T6SS protein ImpK, preprotein translocase subunit SecA, a cAMP-activated global transcriptional regulator and cyclic-AMP receptor protein (CRP), RND family drug transporter and sRNA00203, as shown in Table 2. Thirty genes were downregulated in treated versus untreated biofilm cells, including genes encoding the ATP-binding cassette (ABC) transporter, DNA gyrase, ribosome release factor (RRF), and 30S ribosomal protein S20 (Table 2).
2.3. Gene Expression Profile of Biofilm Treated with Sub-MIC of Colistin
Of the 2912 total expressed genes, 2.6% (368) were differentially expressed in the biofilm cells treated with Sub-MIC of colistin as illustrated in Table S1. Of the 368 DEGs, 285 were biofilm-specific genes, of which 197 were upregulated and 88 were downregulated compared to untreated biofilm cells. We selected 44 DEGs in the biofilm cells that were either induced or repressed when treated with colistin, as shown in Table 2. Of the 44 DEGs, 28 and 16 were upregulated and downregulated, respectively, when the biofilm cells were treated with sub-MICs of colistin. The colistin-induced biofilm-specific genes include those genes encoding ABC-type iron transport proteins, fimbrial protein, lipoprotein (biofilm matrix), AHL synthase (AbaI), multidrug efflux pumps, OMP protein, and catalase and the csu operon. The biofilm-specific genes repressed by sub-optimal concentrations of colistin included genes encoding the LPS export system permease, DNA gyrase, RRF, 30S ribosomal protein S20, tRNA-i(6)A37 modification enzyme, and ATPase. This indicates that a sub-MIC of colistin can either activate or suppress biofilm-specific genes to promote the survival of biofilm cells in the presence of antibiotics.
2.4. Verification of Genes Induced or Repressed by the Sub-MICs of Imipenem and Colistin
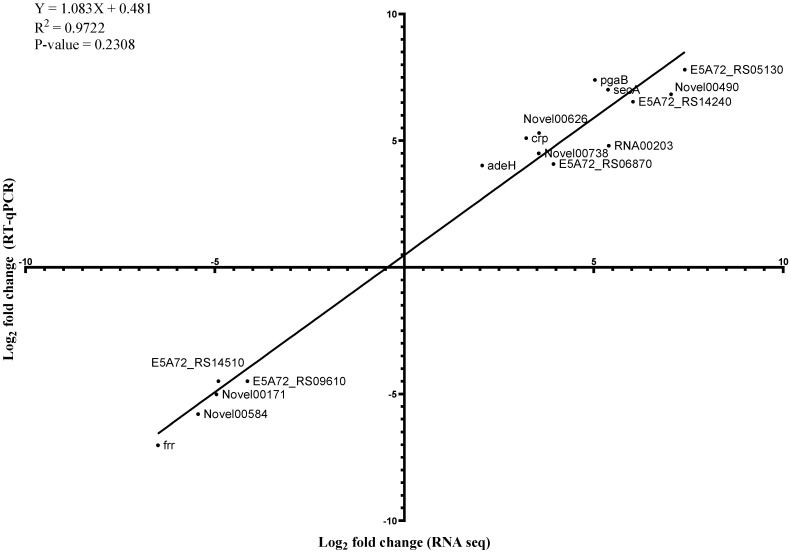
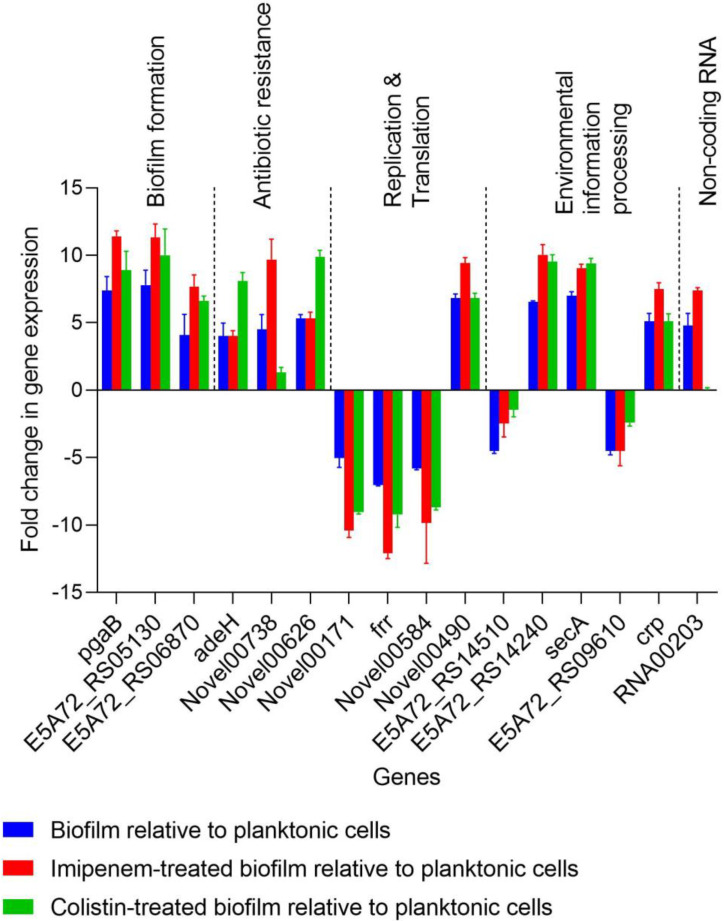
To verify the RNA-seq results, 16 genes that were induced or suppressed by the sub-MICs of imipenem and colistin were selected for verification with qRT-PCR experiments (Figure 2 and Figure 3). These DEGs were associated with adherence, biofilm matrix synthesis, QS, β-lactam resistance, multidrug efflux pumps, replication and translation, environmental information processing, and ncRNAs (Figure 2 and Figure 3). A correlation coefficient of 0.98 was obtained from the linear regression plotted between the RT-qPCR and RNA-seq data, suggesting a strong positive correlation (Figure 2). The changes in gene expression measured using RT-qPCR displayed a pattern similar to the one seen in the RNA-seq data (p ≥ 0.05), suggesting that RNA-seq can be used interchangeably to describe transcriptional changes observed in biofilm and planktonic cells.
Figure 2.
Relationship between gene expression fold changes obtained from RNA sequencing and RT-qPCR. A total of 16 genes associated with virulence and antibiotic resistance in biofilm were compared. The measured log2 fold change in gene expression of biofilm cells relative to planktonic cells are plotted against the RNA sequencing data (statistical goodness of fit value provided).
Figure 3.
Effect of imipenem and colistin at sub-MICs on genes associated with virulence and antibiotic resistance in A. baumannii ST1894 biofilm. pgaB: biofilm matrix synthesis; E5A72_RS05130: fimbrial protein; E5A72_RS06870: quorum sensing; adeH: multidrug efflux pump; Novel00738: resistance–nodulation–cell division (RND) efflux pumps; Novel00626: peptidoglycan biosynthesis; Novel00171: DNA replication; frr, Novel00584, Novel00490: translation; E5A72_RS14510: ATP-binding cassette (ABC) transporter; E5A72_RS14240, secA: bacterial secretion system; E5A72_RS09610, crp: two-component system; RNA00203: non-coding RNA.
3. Discussion
The capacity of A. baumannii to form biofilms enhances its survival in adverse environments, making it a successful nosocomial pathogen [5,17,18,19]. In our previous study, we identified the non-MDR and strong biofilm-forming strain A. baumannii ST1894, the biofilm cells of which exhibited reversible antibiotic tolerance to colistin, imipenem, and ciprofloxacin [12]. This finding indicated that biofilms play a substantial role in the survival of A. baumannii by modifying its responses to antibiotics [12]. The phenotypic changes observed in this strain could be a result of alterations in gene expression in the biofilm cells and not irreversible genetic mutations. In this study, we analyzed the transcriptomic profiles between biofilm and planktonic cells, untreated biofilm cells and imipenem-treated or colistin-treated biofilms of A. baumannii ST1894. Upon conducting RNA-seq of the biofilm and biofilm cells treated sub-optimal doses antibiotics, we identified the genes involved in biofilm formation and biofilm specific antibiotic resistances.
In this study, an increase in the expression levels of the pgaB gene and genes encoding LPS biosynthesis components was observed in biofilm cells relative to planktonic cells. Other research groups have reported that deletion of the pga locus led to the loss of the strong biofilm formation phenotype of A. baumannii, demonstrating that this gene is essential for biofilm formation [20,21]. The production of EPS indicates that biofilm cells have reached the stage of irreversible adherence to the surface. The pgaB gene is involved in the biosynthesis of EPS, which is a significant component of the biofilm matrix [22].
Components of the biofilm matrix can limit the penetration of antimicrobial agents, such as bleach and antibiotics, into biofilm cells by binding to or consuming the antimicrobial agent [23,24]. However, when we observed antibiotic-treated biofilm cells using confocal imaging, we observed that most of the biofilm cells were killed by treatment with high concentrations of colistin, imipenem, and ciprofloxacin [12]. This observation suggests that the antibiotics can be passed through the matrix of the biofilm. Although the biofilm matrix might have reduced the penetration of antibiotics into the biofilm cells, this possibility needs to be experimentally verified.
We also observed the increased expression of pgaB and the gene coding for UDP-N-acetylglucosamine acyltransferase (novel00626) in biofilm cells exposed to sub-MICs of imipenem and colistin. This observation explains how sub-MICs of antimicrobials enhance biofilm formation which was described in a previous in-vitro study [16,25]. The biofilm matrix formation triggers stress-induced metabolic or transcriptional changes that increase resistance in cells exposed to sub-MICs of antibiotics [26,27].
We also observed that expression of the pilA gene, which encodes the fimbrial protein (A1S_3177) or type IV pilus assembly protein, was upregulated 222-fold in biofilm cells compared with planktonic cells. The expression of pilA also increased when the biofilm cells were treated with sub-MICs of imipenem and colistin.
A previous study reported that this type IV pilus assembly protein, commonly found in pathogenic A. baumannii strains, plays an essential role in host cell adhesion, biofilm formation, microcolony formation, and horizontal gene transfer [28,29]. In this study, the upregulated expression of type IV pili genes upon exposure of the cells to sub-MICs of imipenem and colistin might have activated signaling cascades associated with pathogenicity and antibiotic resistance in A. baumannii. When A. baumannii cells form biofilms in clinical settings, overexpression of the type IV pilus assembly protein upon exposure to sub-optimal concentrations of imipenem and colistin can increase the rates of conjugative gene transfer, thereby increasing the likelihood of developing irreversible antibiotic resistance and consequent therapeutic failure. Sub-MICs of antibiotics also promote mutation, leading to the emergence of antibiotic resistance [30,31]. This sequence of events can eventually result in the evolution of sensitive strains into resistant strains.
We observed that the csu operon (proteins CsuA, CsuC, and CsuE) was upregulated in biofilm cells. The csu operon encodes proteins involved in the chaperon-usher pili assembly mechanism, which is essential for the assembly of pili and the formation of biofilms [32]. The csu operon has been identified in pathogenic strains of A. baumannii, indicating its role as a virulence factor [33,34]. However, we observed that exposure to imipenem and colistin at sub-MICs did not significantly affect the expression of the csu operon.
QS is a regulatory mechanism that allows bacteria to communicate cell density information through the diffusion of small molecules and adjust their gene expression profiles accordingly [35,36]. All QS bacteria generate and release chemical signal molecules called autoinducers (AIs) that increase in concentration as a function of cell density [21,35,37]. AHLs are a class of AI signal used by Gram-negative bacteria to regulate various physiological processes such as conjugation, virulence factor production and biofilm formation [35,38,39,40,41,42].
AHLs have been identified as major components of biofilm formation in A. baumannii cells [43,44,45,46]. In this study, the genes encoding AHL synthase (E5A72_RS06870) were upregulated by 16.9-fold in biofilm cells compared with planktonic cells. The exposure of biofilm cells to sub-MICs of imipenem and colistin also induced genes encoding AHL synthase, which were upregulated by 12.2-fold and 5.8-fold, respectively. At a particular threshold, the binding of AHL to receptors within the cell promotes a signal transduction cascade that eventually changes the expression levels of specific genes involved in virulence and antibiotic resistance [42,45,47]. These changes in gene expression enable this pathogen to survive in adverse environmental conditions by promoting biofilm formation. When treated with sub-MICs of imipenem and colistin, the increased expression of genes encoding AHL synthase in biofilm cells suggests that a higher concentration of AIs is required to counteract adverse conditions encountered in the biofilm state.
Efflux pumps actively eliminate antimicrobial agents from intracellular targets, facilitating the reduced antibiotic susceptibility of biofilm and planktonic cells [46,48,49]. In the current study, we observed that 12 RND family MDR genes were differentially expressed in biofilm cells. Among these genes, novel00738 was upregulated in untreated biofilms and biofilm cells exposed to sub-MICs of imipenem. The product of this gene is functionally similar to the MexB-AcrB efflux pump, which confers resistance to β-lactams and cationic antimicrobial peptides. Poole et al. (1993) previously reported that deleting the genes encoding the MexAB-OprM efflux pathway in Pseudomonas aeruginosa resulted in hypersensitivity to many antimicrobial compounds [50,51,52,53,54].
The gene novel00738 encodes as a multidrug efflux pump that is active in β-lactam resistance pathways and is overexpressed when treated with sub-MICs of imipenem. The overexpression of novel00738 can result in tolerance to imipenem when biofilm cells are treated with high concentrations of β-lactam antibiotics. This finding is similar to one reported by He et al. [55], who demonstrated the role of the AdeFGH efflux pump in biofilm formation in response to low-concentration antimicrobial therapy. The increased expression of genes encoding efflux systems could reduce the cytoplasmic concentrations of bactericidal antibiotics to below the threshold required for antibacterial activity.
One of the main mechanisms underlying the development of antibiotic resistance is the pumping of antibiotics out of cells by efflux systems [55,56,57]. Efflux pumps can not only provide resistance to antibiotics used in clinical therapy but can also drive bacterial pathogenicity and persistence [58,59]. The increased expression of novel00738 in biofilm cells might be responsible for antibiotic tolerance and not resistance, as this strain was seen to revert to the susceptible form after treatment with a high concentration of antibiotics.
Analysis of the cationic antimicrobial resistance peptide path also revealed that novel00738 is functionally similar to acrB. The expression of acrB gene affects the expression of a group of efflux pump genes such as tolC, which is believed to confer tolerance to cationic peptides. The tolC gene is expressed at significantly higher levels in persister cells than in normal viable cells [59,60,61].
We also found that the adeH (acb: A1S 2306) gene was upregulated in biofilm cells and was further induced by treatment with sub-MICs of colistin. AdeH can pump colistin out of the cell and thereby confer tolerance to this antibiotic. Overexpression of AdeFGH has also been reported to facilitate the synthesis and transport of AHLs in A. baumannii during biofilm production [55,57,62]. We further observed a positive correlation between the expression of AIs and the upregulation of adeH, indicating that this gene might be involved in the transport of QS molecules, in addition to the expulsion of antibiotics.
We also observed that novel00626, which encodes UDP-N acetylglucosamine O-acyltransferase, appears to play a role similar to that of lpxA. This novel gene was highly upregulated in biofilm cells compared with their planktonic counterparts, and its expression level also significantly increased when biofilm cells were treated with sub-MICs of colistin. Previous studies have demonstrated that strains mutant for LOS production cannot survive desiccation, implying that the development of desiccation resistance is dependent on the composition of the outer membrane [4,63,64]. However, the processes that mediate desiccation resistance have not been studied and are currently being characterized. The upregulation of genes encoding UDP-N acetyl glucosamine O-acyltransferase during biofilm formation and in cells treated with sub-MICs of colistin is attributable to the role of LPS in the synthesis of the biofilm matrix.
We found that genes involved in DNA replication, transcription, and translation were differentially expressed in biofilm cells compared with planktonic cells. The genes involved in DNA replication were significantly downregulated in biofilm cells. The novel00171 gene, thought to encode DNA gyrase, was downregulated 32.4-fold in biofilm cells compared with planktonic cells. When treated with sub-MICs of imipenem, the expression of novel00171 decreased by a 42.2-fold relative to untreated biofilm cells. Such downregulation of DNA gyrase genes during biofilm formation has never been reported previously.
DNA gyrase is necessary for the replication and transcription of DNA. Reductions in intracellular gyrase proteins by >50% have been shown to affect cell growth [65,66]. The altered supercoiling of DNA due to gyrase depletion causes subsequent changes in the density of RNA polymerase in the transcription units, thereby altering transcription. The consequently reduced transcriptional activity generates a high number of slow-growing biofilm cells, which can also occur due to the limited availability of nutrients. The presence of slow-growing cells in biofilms may lead to the development of decreased susceptibility to antibiotics [67]. We also observed no substantial changes in the expression level of the novel00171 gene when the biofilm cells were exposed to sub-MICs of colistin.
The expression level of the gene encoding the RRF (frr) was reduced by 129.7-fold in biofilm cells. When the biofilm cells were treated with sub-MICs of imipenem and colistin, the expression level of frr decreased by 33.3-fold and 4.5-fold, respectively. The primary purpose of RRF is to recycle ribosomes for subsequent rounds of protein synthesis; it is thus essential for bacterial growth [68,69]. The downregulation of genes encoding RRFs in untreated biofilms and in antibiotic-treated biofilms might be due to the presence of slow-growing cells, which can confer antibiotic tolerance.
The expression of novel00490, which encodes the transcription termination factor Rho OS, was 113.7-fold higher in biofilm cells compared with planktonic cells. Expression of novel00490 increased significantly by 6.1-fold when the biofilm cells were treated with sub-MICs of imipenem. The transcriptional changes observed in both untreated biofilm cells and those exposed to sub-MICs of imipenem and colistin may shut down metabolic activity. Such a shutdown could directly inhibit other vital cellular activities and inactivate antibiotic targets.
We further observed that all of the DEGs involved in the citric acid cycle were downregulated, while all of those involved in glycolysis were upregulated. The downregulation of the citric acid cycle suggests that biofilm cells have lower metabolic rates than planktonic cells. This metabolic quiescence might contribute to the reduced susceptibility to antibiotics observed in hyper biofilm-forming strains such as A. baumannii ST1894. It also implies that the persisters observed in the biofilms of A. baumannii ST1894 after treatment with high concentrations of bactericidal antibiotics might have emerged a result of reduced metabolic activity.
The transcriptomic analysis revealed that genes encoding acinetobactin biosynthesis proteins were downregulated in biofilm cells relative to their planktonic counterparts. The identified genes encode iron-induced proteins, such as the iron storage protein Bfr, metabolic proteins, such as AcnA, AcnB, GlyA, SdhA and SodB, and lipid biosynthesis proteins [70]. The reduced expression of these genes can further downregulate the expression of genes involved in aerobic respiration. Similar patterns were observed in our current study: the downregulation of genes involved in the citric acid cycle suggests slower metabolic rates in biofilm cells. This reduced metabolic activity could further increase the antibiotic tolerance of biofilm cells compared with planktonic cells.
In addition to the novel protein-coding transcripts, the total RNA-seq data showed that 5 of 138 sRNAs were differentially expressed in biofilm cells relative to planktonic cells. Of these sRNAs, sRNA00203 exhibited the highest expression levels in biofilm cells that were untreated or treated with sub-MICs of imipenem. sRNAs regulate protein expression by complementing target mRNAs and interacting with mRNA transcripts at or near the RBS [71,72,73]. sRNA00203, an ncRNA, is believed to play roles in various cellular processes, including the regulation of gene expression. sRNAs are genetic regulators that enable biofilm cells to recognize environmental signals and relay information in the form of metabolic changes with significant physiological effects during biofilm formation [45,74,75].
Overall, we observed that the hyper biofilm-producing strain A. baumannii ST1894 demonstrated a high degree of reduced susceptibility to antibiotics. The capacity of A. baumannii ST1894 to survive the effects of bactericidal antibiotic exposure during biofilm formation might emanate from genetic changes that arise due to this exposure in the biofilm state.
This is the first study to characterize transcriptional changes in biofilm cells in response to treatment with sub-MICs of imipenem and colistin. Several novel findings of this study are reported here. First, the consistent upregulation of genes involved in biofilm matrix synthesis (pgaB), multidrug efflux pump (novel00738) and LPS synthesis (novel00626) in A. baumannii in response to treatment with sub-MICs (half of the MIC) of imipenem and colistin may lead to increased biofilm production. This finding illustrates the possible relationship between low-dose antimicrobial therapy and enhanced biofilm production, which can occur during Acinetobacter infections. Second, this study showed a reduced expression of genes linked to acinetobactin biosynthesis and protein synthesis (novel00171, RRF, novel00584) during biofilm formation, which might slow down metabolism. Such changes enhance biofilm-specific antibiotic resistance and environmental resilience when the cells are exposed to antibiotics. Third, an upregulation of sRNA00203 in the biofilm cells was observed. As sRNAs are involved in the regulation of many cellular processes, activation of sRNA gene expression may in turn affect the expression of biofilm-specific antibiotic resistance. Based on our findings of the transcriptome study, multiple genetic factors account for the decreased susceptibility of biofilm cells to antibiotics, the precise interactions between various factors warrant further investigation.
4. Materials and Methods
4.1. Bacterial Strain and Culture Conditions
The A. baumannii strain used for comparative transcriptome analysis was A. baumannii ST1894, which was a clinical strain isolated from the sputum of a patient suffering from a lower respiratory tract infection. A. baumannii ST1894 has been characterized in our previous investigation and shown to be a hyper biofilm-former and was susceptible to colistin and imipenem when grown in the planktonic phase. However, the strain exhibited a high degree of resistance to colistin and imipenem when grown in the biofilm phase [12]. The strain was grown in LB broth at 37 °C with shaking (180 rpm) and stored at −80 °C in LB broth containing 20% glycerol until being used.
4.2. Biofilm Preparation and Exposure to Antibiotics
A single colony was picked from a pure culture grown on LB agar plates and then inoculated into 20 mL of LB broth to obtain a planktonic culture. Similarly, five colonies were selected and inoculated into a larger Petri dish (150 mm diameter) containing 100 mL of LB broth. The planktonic and biofilm cultures were incubated at 37 °C for 48 h [76]. After 47 h of incubation, 62.5 mg/mL of imipenem (half of the MIC) and 250 mg/mL of colistin (half the MIC) were added to the biofilm incubated at 37 °C for another hour. After 48 h of cultivation, all of the single cells were carefully washed three times with maximum recovery diluent (1 g peptone and 9 g of NaCl per liter of distilled water) without disturbing the biofilm.
Subsequently, the biofilm cells attached to the Petri dish surface were removed using a plastic cell scraper and then resuspended in maximum recovery diluent. The cells were placed on ice and then washed thrice with 1 mL of cold phosphate-buffered saline (PBS). After washing, the biofilm and planktonic cultures were centrifuged for 15 min at 3500× g and 4 °C. The pellets obtained from the biofilm cells, antibiotic-treated biofilm cells, and planktonic cells were stored at −80 °C until they were processed for RNA extraction.
4.3. RNA Extraction
Cell pellets prepared from the planktonic, biofilm, imipenem-treated (half of the MIC), and colistin-treated biofilm cells (half of the MIC) were processed for total RNA isolation using a PureLink RNA Mini Kit (Ambion, Life Science Technologies, Carlsbad, CA, USA). The isolated RNA was treated with DNase to eliminate contaminating genomic DNA using TURBO DNase (Life Science Technologies).
4.4. RNA Sequencing
Library preparation and RNA-seq were performed by Groken Bioscience Ltd, Hong Kong. Three micrograms of input RNA were used to construct the libraries using the NEBNext UltraTM Directional RNA Library Prep Kit for Illumina (NEB, Northborough, MA, USA). The libraries were sequenced on an Illumina HiSeq2500 platform (San Diego, CA, USA) with 150PE reads.
4.5. Bioinformatics Analysis
The clean reads were obtained by quality filtering and trimming of adapters by using custom Perl scripts and Trimmomatic v0.3032 (Cambridge, UK) with the default parameters.
The reference genome of A. baumannii ATCC17978 with RefSeq assembly accession GCF_004797155.2 (latest) was retrieved from the National Center for Biotechnology Information (NCBI) website. The reads were mapped to the reference genome using Bowtie 2 v2.2.3 with default parameters [77]. HTSeq v0.6.1 was used to count the number of reads mapped to each gene, and the fragments per kilobase of transcript sequence per million base pairs (FPKM) were calculated to quantify the level of gene expression. Simultaneously, the log2 (FPKM biofilm/FPKM planktonic), which accounts for the effects of sequencing depth and gene length on the read count, was determined [78]. The results show the number of genes with different expression levels and the expression levels of single genes. An FPKM value of 0.1 or 1 was set as the criterion for determining the expression levels of the target groups. The raw sequences were deposited in the NCBI Sequence Read Archive (SRA) under the BioProject ID PRJNA892543 and BioSample accession numbers SAMN31387122, SAMN31387123, SAMN31387124, SAMN31387125.
The sequencing reads were annotated using the R Bioconductor package [79]. Normalization of gene expression was performed using the edge program package and a single scaling factor. The DEGseq R package (1.18.0) was used to estimate the DEGs between biofilm and planktonic cells, untreated biofilms, imipenem-treated biofilm cells, and untreated biofilms and colistin-treated biofilm cells. The fold change and level of significance, which indicate differential expression, were evaluated using a model based on the binomial distribution (which could be approximated by a Poisson distribution) [80]. Genes with an expression level of log2 fold change > 1 and a corrected p (q) ≤ 0.005 were considered to be differentially expressed. Genes were considered to be differentially expressed if they fulfilled the following criteria: (1) the biofilm cells had a normalized gene expression value > 2-fold that of the planktonic cells; and (2) the antibiotic-treated biofilm cells had a normalized expression value > 2-fold that of untreated biofilm cells. The DEGs were then used for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway analyses.
4.5.1. Novel Gene and Gene Structure Analysis
The RNA-seq reads were mapped to the reference genome of A. baumannii ATCC17978 using Rockhopper, and the novel genes were compared to known gene structures [81]. The novel transcripts were BLASTx (cut-off: e value < 1 × 10−5) against non-redundant protein database. Novel transcripts with NR protein sequence annotations were considered to be novel potential protein-coding transcripts. The transcription start sites (TSSs) and termination sites of operons were predicted based on the positions of the reads in the reference genome using Rockhopper. A 700-bp sequence in the upstream TSS was extracted and used to identify the promoter according to the time-delay neural network (TDNN) method.
4.5.2. The GO and KEGG Enrichment Analyses
The GO seq R package was used to perform the GO enrichment study of the DEGs; GO terms with an adjusted p < 0.05 were considered to be enriched [82]. KEGG enrichment analysis was conducted to identify significantly enriched metabolic pathways or signal transduction pathways across antibiotic-treated biofilms, untreated biofilms, and planktonic cells. KOBAS (Dalian, China) was used to evaluate the statistical enrichment of DEGs in KEGG pathways. KEGG pathways with an adjusted p < 0.05 were considered to be significantly enriched in DEGs.
4.6. Verification of Genes of Induced or Repressed by Exposure to Sub-MICs of Imipenem and Colistin
RT-qPCR was used to verify the differentially expressed genes obtained from the RNA-seq data [81,83]. Sixteen genes associated with biological functions such as matrix formation, QS, β-lactam resistance, cationic antimicrobial peptide resistance, bacterial secretion system, and the two-component system were selected for RT-qPCR verification (Table 3). The same RNA samples (technical replicates) used for transcriptomic analysis were reversely transcribed to cDNA followed by RT-qPCR. In addition, RNA samples prepared from two more independent experiments of antibiotic exposure and prepared under the same biological conditions were used as biological replicas. Thus, for each group (planktonic, untreated biofilm, imipenem-treated or colistin-treated biofilm), three replica RNA samples were subjected to RT-qPCR.
Table 3.
Lists of genes induced or repressed by exposure to sub-MICs of imipenem and colistin and selected for RT-qPCR verification.
| mRNA_ID | KEGG_ID | KEGG Annotation | Strand | Start | End | Length (bp) | Pathway |
|---|---|---|---|---|---|---|---|
| E5A72_RS12100 | acb: A1S_0938 | PgaB | + | 2520896 | 2522890 | 1995 | Biofilm matrix |
| E5A72_RS05130 | acb: A1S_3177 | Fimbrial protein | − | 1078917 | 1079381 | 2538 | Two-component system |
| E5A72_RS06870 | acb: A1S_0109 | Homoserine lactone synthase | + | 1427610 | 1428176 | 567 | Quorum sensing |
| E5A72_RS00640 | acb: A1S_2306 | RND2 efflux transporter | + | 116590 | 118041 | 555 | Multidrug efflux system |
| Novel00738 | acb: A1S_2736 | RND family drug transporter | + | 617454 | 623989 | 6536 | β-lactam resistance |
| Novel00626 | acb: A1S_1965 | UDP-N acetylglucosamine acyltransferase | − | 3676375 | 3677683 | 1309 | Cationic antimicrobial peptide resistance |
| Novel00171 | acb: A1S_2626 | DNA gyrase | − | 483250 | 484821 | 1572 | DNA replication |
| E5A72_RS17735 | acb: A1S_1974 | Ribosome releasing factor | − | 3685962 | 3686516 | 555 | Translation |
| Novel00584 | acb: A1S_1617 | 30S ribosomal protein S20 | − | 3284582 | 3284881 | 300 | |
| Novel00490 | - | Transcription termination factor Rho OS | − | 2214580 | 2215974 | 1395 | Transcription |
| E5A72_RS14510 | acb: A1S_1359 | ABC3-type Fe3+ transport system | + | 3025424 | 3026461 | 1038 | ABC transporters |
| E5A72_RS14240 | acb: A1S_1310 | K11892 type VI secretion system protein ImpK | + | 2971603 | 2972409 | 807 | Bacterial secretion system |
| E5A72_RS03500 | acb: A1S_2862 | Preprotein translocase subunit SecA | − | 756066 | 758789 | 2724 | |
| E5A72_RS09610 | acb: A1S_0399 | LysR family transcriptional regulator | + | 1993774 | 1994670 | 897 | Two-component system |
| E5A72_RS13515 | acb: A1S_1182 | CRP4 transcriptional regulator | − | 2818333 | 2819040 | 708 | |
| sRNA00203 | - | - | − | 1245743 | 1245795 | 53 | Non-coding RNA |
The sequences of the 16 selected genes were retrieved from the A. baumannii ST1894 transcriptome data and used as references for the design of primers and probes for qPCR. The primers and probes shown in Table 4 were designed using Primer3 plus (Boston, USA). A two-step protocol was used to perform RT-qPCR, 500 ng of pure RNA samples were reverse-transcribed using a Luna Script RT Supermix Kit (NEB, Massachusetts, USA). The prepared cDNA and no-RT control reactions were diluted at 1:100 in nuclease-free water to be used for RT-qPCR reactions. The SYBR Green RT-qPCR reaction was prepared as a 20-µL mixture by adding 10 µL of Luna Universal qPCR Master mix (NEB, Massachusetts, USA), 0.5 µL of forward primer (10 µM), 0.5 µL of reverse primer (10 µM), 2 µL of 1:10 diluted cDNA and 7 µL of nuclease-free water. A no-RT control was run in parallel.
Table 4.
Primers and probes used for verification of expression levels of the 16 selected DEGs.
| Gene ID | Amplicon Size (bp) | Primer/Probe | Sequence (5′ to 3′) |
|---|---|---|---|
| E5A72_RS12100 (pgaB) | 105 | E5A72_RS12100-F | CGGATGCGAATGGTTCTGC |
| E5A72_RS12100-R | GCGTACGGGTTTGAATTTGC | ||
| E5A72_RS05130 | 217 | E5A72_RS05130_4_F | CCGAAGGTACAGCTAACAGTG |
| E5A72_RS05130_4_R | CCACCCACATTTGCATTTACT | ||
| E5A72_RS06870 | 121 | E5A72_RS06870_F | GCCAGACTACTACCCACCAC |
| E5A72_RS06870_R | CTACGGCTGAAAACCTTGAT | ||
| E5A72_RS00640 | 108 | E5A72_RS00640_F | TCAGGCTTCACGTGCACTAC |
| E5A72_RS00640_R | AAACCGAGTGAAGCTGGAGA | ||
| Novel00738 | 79 | Novel00738_F | GCTGCCATTACTCGTTTACCT |
| Novel00738_R | CAGGACGGCTCTCAACAAC | ||
| Novel00738_IN | FAM-GGCAAGCTGTAGCGATGCTTGTTAAT-TAMRA | ||
| Novel00626 | 110 | Novel00626_F | CGCATCGTTACCCATTCTT |
| Novel00626_R | GAAATGCCCTTGTAGGAACTCT | ||
| Novel00626_IN | FAM-TTGGTTGATCGTGTGACTGAAGTTACTGA-TAMRA | ||
| Novel00171 | 109 | Novel00171_F | CATTGCCGGATGTGAGAG |
| Novel00171_R | ACACGAGCAGATTTCTTGTAGG | ||
| E5A72_RS17735 (Frr) | 98 | E5A72_RS17735_F | GCGAAAGTTGCTATCCGTAA |
| E5A72_RS17735_R | GCACGACGCTCATCATCT | ||
| Novel00584 | 114 | Novel00584_F | TGCGTTCTATGGTTCGTACTT |
| Novel00584_R | GCACGACGCTCATCATCT | ||
| Novel00490 | 109 | Novel00490_F | TTAGCCCGTGCATACAACAC |
| Novel00490_R | TAGCCCGTGCATACAACAC | ||
| Novel00490_IN | FAM-TGGTGTGGATGCACATGCTTTAGAAC-TAMRA | ||
| E5A72_RS14510 | 103 | E5A72_RS14510_F | AGGTTTAGGCTGGGAAATGG |
| E5A72_RS14510_R | ATTTGCTGCTTTGCTTACCG | ||
| E5A72_RS14240 | 110 | E5A72_RS14240_1_F | GCACGAGTAGGCGATGAA |
| E5A72_RS14240_1_R | AAAGGTAGCTCACGATGGATAA | ||
| E5A72_RS03500 (secA) | 107 | E5A72_RS03500_F | GACATTATTGCTCAGGCAGGT |
| E5A72_RS03500_R | GCAAGTTTCGCTTTCCAGTT | ||
| E5A72_RS09610 | 86 | E5A72_RS09610_3_F | AAGGTGGAACTGTGATGATGG |
| E5A72_RS09610_3_R | AATTCCCAAACCTGCACAAG | ||
| E5A72_RS13515 | 118 | E5A72_RS13515_3_F | ATCGACCTATCTTCACAACCAG |
| E5A72_RS13515_3_R | ATACACGGCCAACCATTTC | ||
| sRNA00203 | 76 | sRNA00203_4_F | GCATAAAAACCTCTTGAAACTGTTC |
| sRNA00203_4_R | AGCGTTCATTTCAACCGATA | ||
| sRNA00203_4_IN | TCAAGTTCCTTATGATCTCTTCCTTGA | ||
| gyrB | 93 | gyrB-F | ACGATTTACCGTGCTGGTC |
| gyrB-R | GGTATTATCCGTTTCACCAATC | ||
| gyrB_IN | FAM-TATCATCATGGTGATCCGCAATATCC-TAMRA |
The SYBR Green qPCRs were performed on a ViiA 7 Real-Time PCR system (Applied Biosystems, USA) using the following parameters: 1 min at 95 °C; 40 cycles of 95 °C for 15 s and 60 °C for 30 s; and 1 min at 72 °C. A melting curve analysis was added to ensure the specificity of the PCR product.
TaqMan assays were performed for the novel transcripts and sRNAs using the Luna Universal Probe qPCR Master mix (NEB). The reaction parameters set on the ViiA 7 Real-Time PCR were: 1 min at 95 °C and 40 cycles of 95 °C for 15 s and 60 °C for 30 s. For the SYBR Green and TaqMan qPCR experiments, no-RT controls were used for each target gene. The expression levels of three housekeeping genes (rpoD, gyrB, and E5A72 RS18355) were evaluated across all of the RNA-seq data. gyrB displayed the least variability across different sample groups, and therefore, the cycle threshold (CT) values of all 16 target genes were normalized using gyrB as an internal control.
5. Conclusions
In this investigation, we demonstrated that exposure of A. baumannii ST1894 to suboptimal doses of imipenem and colistin increased the expression of genes involved in biofilm formation and antibiotic resistance while decrease the expression of genes involved in protein synthesis in the biofilm state. This confirmed our previous observation that exposure of A. baumannii ST1894 biofilm to suboptimal antibiotic doses induced biofilm formation and antibiotic resistance. We also showed that expression of non-coding sRNA00203 was highly induced in either untreated or imipenem-treated biofilm cells. As sRNAs play critical regulatory roles in many cellular processes, this adds to the complexity and versatility of biofilm regulatory mechanisms and confers a survival advantage to the pathogen.
Acknowledgments
We are grateful to Barry Kin Chung Wong of United Christian Hospital, Hong Kong for providing the Acinetobacter baumannii strains for phenotypic and genetic investigations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms232012705/s1.
Author Contributions
Conceptualization, A.M.S. and P.H.M.L.; methodology, A.M.S. and P.H.M.L.; software A.M.S.; validation, A.M.S., M.Y. and P.H.M.L.; formal analysis, A.M.S.; investigation, A.M.S., J.Z. and P.H.M.L.; resources, P.H.M.L.; writing—original draft preparation, A.M.S.; writing—review and editing, P.H.M.L., J.Z., F.W.N.C. and D.A.; visualization, J.Z., P.H.M.L. and F.W.N.C.; supervision, P.H.M.L.; project administration, P.H.M.L.; funding acquisition, P.H.M.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw RNA sequencing datasets used for this study were deposited at the NCBI Sequence Read Archive; https://submit.ncbi.nlm.nih.gov/subs/sra under Bioproject ID number PRJNA892543; BioSample accession numbers SAMN31387122, SAMN31387123, SAMN31387124, SAMN31387125.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
A.M. Shenkutie is supported by the Hong Kong PhD Fellowship Scheme (PH16-01625) of the Research Grant Council, the Government of the Hong Kong Special Administrative Region and the Department of Health Technology and Informatics, The Hong Kong Polytechnic University.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gedefie A., Demsis W., Ashagrie M., Kassa Y., Tesfaye M., Tilahun M., Bisetegn H., Sahle Z. Acinetobacter Baumannii Biofilm Formation and Its Role in Disease Pathogenesis: A Review. Infect. Drug Resist. 2021;14:3711. doi: 10.2147/IDR.S332051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Espinal P., Martí S., Vila J. Effect of Biofilm Formation on the Survival of Acinetobacter Baumannii on Dry Surfaces. J. Hosp. Infect. 2012;80:56–60. doi: 10.1016/j.jhin.2011.08.013. [DOI] [PubMed] [Google Scholar]
- 3.Yang C.H., Su P.W., Moi S.H., Chuang L.Y. Biofilm Formation in Acinetobacter Baumannii: Genotype-Phenotype Correlation. Molecules. 2019;24:1849. doi: 10.3390/molecules24101849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harding C.M., Hennon S.W., Feldman M.F. Uncovering the Mechanisms of Acinetobacter Baumannii Virulence. Nat. Rev. Microbiol. 2018;16:91–102. doi: 10.1038/nrmicro.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding C.M., Tracy E.N., Carruthers M.D., Rather P.N., Actis L.A., Munson R.S. Acinetobacter Baumannii Strain M2 Produces Type IV Pili Which Play a Role in Natural Transformation and Twitching Motility but Not Surface-Associated Motility. MBio. 2013;4:e00360-13. doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 7.Penesyan A., Nagy S.S., Kjelleberg S., Gillings M.R., Paulsen I.T. Rapid Microevolution of Biofilm Cells in Response to Antibiotics. NPJ Biofilms Microbiomes. 2019;5:34. doi: 10.1038/s41522-019-0108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ababneh Q., Abu Laila S., Jaradat Z. Prevalence, Genetic Diversity, Antibiotic Resistance and Biofilm Formation of Acinetobacter Baumannii Isolated from Urban Environments. J. Appl. Microbiol. 2022. In press . [DOI] [PubMed]
- 9.Cheng V.C.C., Wong S.C., Chen J.H.K., So S.Y.C., Wong S.C.Y., Ho P.L., Yuen K.Y. Control of Multidrug-Resistant Acinetobacter Baumannii in Hong Kong: Role of Environmental Surveillance in Communal Areas after a Hospital Outbreak. Am. J. Infect. Control. 2018;46:60–66. doi: 10.1016/j.ajic.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Jung J.Y., Park M.S., Kim S.E., Park B.H., Son J.Y., Kim E.Y., Lim J.E., Lee S.K., Lee S.H., Lee K.J. Risk Factors for Multi-Drug Resistant Acinetobacter Baumannii bacteremia in Patients with Colonization in the Intensive Care Unit. BMC Infect. Dis. 2010;10:228. doi: 10.1186/1471-2334-10-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Kheloui Raja E.M.S., Asma L., Rachida M., Fatima H. Acinetobacter Baumannii Extracellular Matrix as An Antibiofilm and Anti-Infection Target. World J. Pharm. Res. 2022;11:10–35. [Google Scholar]
- 12.Shenkutie A.M., Yao M.Z., Siu G.K.H., Wong B.K.C., Leung P.H.M. Biofilm-Induced Antibiotic Resistance in Clinical Acinetobacter Baumannii Isolates. Antibiotics. 2020;9:817. doi: 10.3390/antibiotics9110817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Quintanilla M., Pulido M.R., López-Rojas R., Pachón J., McConnell M.J. Emerging Therapies for Multidrug Resistant Acinetobacter Baumannii. Trends Microbiol. 2013;21:157–163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 14.Paul M., Carrara E., Retamar P., Tängdén T., Bitterman R., Bonomo R.A., De Waele J., Daikos G.L., Akova M., Harbarth S. European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Guidelines for the Treatment of Infections Caused by Multidrug-Resistant Gram-Negative Bacilli (Endorsed by European Society of Intensive Care Medicine) Clin. Microbiol. Infect. 2022;28:521–547. doi: 10.1016/j.cmi.2021.11.025. [DOI] [PubMed] [Google Scholar]
- 15.Høiby N., Bjarnsholt T., Givskov M., Molin S., Ciofu O. Antibiotic Resistance of Bacterial Biofilms. Int. J. Antimicrob. Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan J.B. Antibiotic-Induced Biofilm Formation. Int. J. Artif. Organs. 2011;34:737–751. doi: 10.5301/ijao.5000027. [DOI] [PubMed] [Google Scholar]
- 17.Tarín-Pelló A., Suay-García B., Pérez-Gracia M.-T. Antibiotic Resistant Bacteria: Current Situation and Treatment Options to Accelerate the Development of a New Antimicrobial Arsenal. Expert Rev. Anti. Infect. Ther. 2022;20:1095–1108. doi: 10.1080/14787210.2022.2078308. [DOI] [PubMed] [Google Scholar]
- 18.Jolivet-Gougeon A., Bonnaure-Mallet M. Biofilms as a Mechanism of Bacterial Resistance. Drug Discov. Today Technol. 2014;11:49–56. doi: 10.1016/j.ddtec.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 19.Hathroubi S., Mekni M.A., Domenico P., Nguyen D., Jacques M. Biofilms: Microbial Shelters against Antibiotics. Microb. Drug Resist. 2017;23:147–156. doi: 10.1089/mdr.2016.0087. [DOI] [PubMed] [Google Scholar]
- 20.Choi A.H.K., Slamti L., Avci F.Y., Pier G.B., Maira-Litrán T. The PgaABCD Locus of Acinetobacter Baumannii Encodes the Production of Poly-β-1-6-N-Acetylglucosamine, Which Is Critical for Biofilm Formation. J. Bacteriol. 2009;191:5953–5963. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Law S.K.K., Tan H.S. The Role of Quorum Sensing, Biofilm Formation, and Iron Acquisition as Key Virulence Mechanisms in Acinetobacter Baumannii and the Corresponding Anti-Virulence Strategies. Microbiol. Res. 2022;260:127032. doi: 10.1016/j.micres.2022.127032. [DOI] [PubMed] [Google Scholar]
- 22.Vu B., Chen M., Crawford R.J., Ivanova E.P. Bacterial Extracellular Polysaccharides Involved in Biofilm Formation. Molecules. 2009;14:2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mah T.F. Biofilm-Specific Antibiotic Resistance. Future Microbiol. 2012;7:1061–1072. doi: 10.2217/fmb.12.76. [DOI] [PubMed] [Google Scholar]
- 24.Branda S.S., Vik Å., Friedman L., Kolter R. Biofilms: The Matrix Revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Kaushik V., Tiwari M., Tiwari V. Interaction of RecA Mediated SOS Response with Bacterial Persistence, Biofilm Formation, and Host Response. Int. J. Biol. Macromol. 2022;217:931–943. doi: 10.1016/j.ijbiomac.2022.07.176. [DOI] [PubMed] [Google Scholar]
- 26.Jefferson K.K., Goldmann D.A., Pier G.B. Use of Confocal Microscopy to Analyze the Rate of Vancomycin Penetration through Staphylococcus Aureus Biofilms. Antimicrob. Agents Chemother. 2005;49:2467–2473. doi: 10.1128/AAC.49.6.2467-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R., Ray P., Das A., Sharma M. Penetration of Antibiotics through Staphylococcus Aureus and Staphylococcus Epidermidis Biofilms. J. Antimicrob. Chemother. 2010;65:1955–1958. doi: 10.1093/jac/dkq257. [DOI] [PubMed] [Google Scholar]
- 28.Ronish L.A., Lillehoj E., Fields J.K., Sundberg E.J., Piepenbrink K.H. The Structure of PilA from Acinetobacter Baumannii AB5075 Suggests a Mechanism for Functional Specialization in Acinetobacter Type IV Pili. J. Biol. Chem. 2019;294:218–230. doi: 10.1074/jbc.RA118.005814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamabe K., Arakawa Y., Shoji M., Miyamoto K., Tsuchiya T., Minoura K., Akeda Y., Tomono K., Onda M. Enhancement of Acinetobacter Baumannii Biofilm Growth by Cephem Antibiotics via Enrichment of Protein and Extracellular DNA in the Biofilm Matrices. J. Appl. Microbiol. 2022;133:2002–2013. doi: 10.1111/jam.15712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laureti L., Matic I., Gutierrez A. Bacterial Responses and Genome Instability Induced by Subinhibitory Concentrations of Antibiotics. Antibiotics. 2013;2:100–114. doi: 10.3390/antibiotics2010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baharoglu Z., Garriss G., Mazel D. Multiple Pathways of Genome Plasticity Leading to Development of Antibiotic Resistance. Antibiotics. 2013;2:288–315. doi: 10.3390/antibiotics2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomaras A.P., Dorsey C.W., Edelmann R.E., Actis L.A. Attachment to and Biofilm Formation on Abiotic Surfaces by Acinetobacter Baumannii: Involvement of a Novel Chaperone-Usher Pili Assembly System. Microbiology. 2003;149:3473–3484. doi: 10.1099/mic.0.26541-0. [DOI] [PubMed] [Google Scholar]
- 33.Peleg A.Y., de Breij A., Adams M.D., Cerqueira G.M., Mocali S., Galardini M., Nibbering P.H., Earl A.M., Ward D.V., Paterson D.L., et al. The Success of Acinetobacter Species; Genetic, Metabolic and Virulence Attributes. PLoS ONE. 2012;7:e46984. doi: 10.1371/journal.pone.0046984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costa P.S., Rezende I.M., Luehring T.A.F.M., Vieira C.D., Drumond B.P., de Macêdo Farias L., Nobre V., Gonçalves R., dos Santos S.G. Sub-Inhibitory Concentrations of Polymyxin B Modulates Pathogenicity Factors and Transcriptional Regulators Genes in Multi-Resistant Acinetobacter Baumannii Strains. Braz. J. Dev. 2021;7:8463–8480. doi: 10.34117/bjdv7n1-574. [DOI] [Google Scholar]
- 35.Antunes L.C.M., Ferreira R.B.R., Buckner M.M.C., Finlay B.B. Quorum Sensing in Bacterial Virulence. Microbiology. 2010;156:2271–2282. doi: 10.1099/mic.0.038794-0. [DOI] [PubMed] [Google Scholar]
- 36.Breijyeh Z., Jubeh B., Karaman R. Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It. Molecules. 2020;25:1340. doi: 10.3390/molecules25061340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raju D.V., Nagarajan A., Pandit S., Nag M., Lahiri D., Upadhye V. Effect of Bacterial Quorum Sensing and Mechanism of Antimicrobial Resistance. Biocatal. Agric. Biotechnol. 2022;43:102409. doi: 10.1016/j.bcab.2022.102409. [DOI] [Google Scholar]
- 38.Papenfort K., Bassler B.L. Quorum Sensing Signal–Response Systems in Gram-Negative Bacteria. Nat. Rev. Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y.-H., Tian X. Quorum Sensing and Bacterial Social Interactions in Biofilms. Sensors. 2012;12:2519–2538. doi: 10.3390/s120302519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nazzaro F., Fratianni F., Coppola R. Quorum Sensing and Phytochemicals. Int. J. Mol. Sci. 2013;14:12607–12619. doi: 10.3390/ijms140612607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passos da Silva D., Schofield M.C., Parsek M.R., Tseng B.S. An Update on the Sociomicrobiology of Quorum Sensing in Gram-Negative Biofilm Development. Pathogens. 2017;6:51. doi: 10.3390/pathogens6040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang K., Zhang X.-H. Quorum Quenching Agents: Resources for Antivirulence Therapy. Mar. Drugs. 2014;12:3245–3282. doi: 10.3390/md12063245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brelles-Mario G., Bedmar E.J. Detection, Purification and Characterisation of Quorum-Sensing Signal Molecules in Plant-Associated Bacteria. In Proceedings of the J. Biotechnol. 2001;91:197–209. doi: 10.1016/S0168-1656(01)00330-3. [DOI] [PubMed] [Google Scholar]
- 44.Pompilio A., Scribano D., Sarshar M., Di Bonaventura G., Palamara A.T., Ambrosi C. Gram-Negative Bacteria Holding Together in a Biofilm: The Acinetobacter Baumannii Way. Microorganisms. 2021;9:1353. doi: 10.3390/microorganisms9071353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kröger C., Kary S.C., Schauer K., Cameron A.D.S. Genetic Regulation of Virulence and Antibiotic Resistance in Acinetobacter Baumannii. Genes. 2016;8:12. doi: 10.3390/genes8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ayoub Moubareck C., Hammoudi Halat D. Insights into Acinetobacter Baumannii: A Review of Microbiological, Virulence, and Resistance Traits in a Threatening Nosocomial Pathogen. Antibiotics. 2020;9:119. doi: 10.3390/antibiotics9030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rao R., Karthika R., Singh S., Shashikala P., Kanungo R., Jayachandran S., Prashanth K. Correlation between Biofilm Production and Multiple Drug Resistance in Imipenem Resistant Clinical Isolates of Acinetobacter Baumannii. Indian J. Med. Microbiol. 2008;26:333–337. doi: 10.1016/S0255-0857(21)01809-0. [DOI] [PubMed] [Google Scholar]
- 48.Poole K. Efflux Pumps as Antimicrobial Resistance Mechanisms. Ann. Med. 2007;39:162–176. doi: 10.1080/07853890701195262. [DOI] [PubMed] [Google Scholar]
- 49.Subhadra B., Kim D.H., Woo K., Surendran S., Choi C.H. Control of Biofilm Formation in Healthcare: Recent Advances Exploiting Quorum-Sensing Interference Strategies and Multidrug Efflux Pump Inhibitors. Materials. 2018;11:1676. doi: 10.3390/ma11091676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poole K., Krebes K., McNally C., Neshat S. Multiple Antibiotic Resistance in Pseudomonas Aeruginosa: Evidence for Involvement of an Efflux Operon. J. Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaillancourt M., Limsuwannarot S.P., Bresee C., Poopalarajah R., Jorth P. Pseudomonas Aeruginosa MexR and MexEF Antibiotic Efflux Pump Variants Exhibit Increased Virulence. Antibiotics. 2021;10:1164. doi: 10.3390/antibiotics10101164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Impey R.E., Hawkins D.A., Sutton J.M., Soares da Costa T.P. Overcoming Intrinsic and Acquired Resistance Mechanisms Associated with the Cell Wall of Gram-Negative Bacteria. Antibiotics. 2020;9:623. doi: 10.3390/antibiotics9090623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou G., Shi Q.-S., Huang X.-M., Xie X.-B. The Three Bacterial Lines of Defense against Antimicrobial Agents. Int. J. Mol. Sci. 2015;16:21711–21733. doi: 10.3390/ijms160921711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scoffone V.C., Trespidi G., Barbieri G., Irudal S., Perrin E., Buroni S. Role of RND Efflux Pumps in Drug Resistance of Cystic Fibrosis Pathogens. Antibiotics. 2021;10:863. doi: 10.3390/antibiotics10070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He X., Lu F., Yuan F., Jiang D., Zhao P., Zhu J., Cheng H., Cao J., Lu G. Biofilm Formation Caused by Clinical Acinetobacter Baumannii Isolates Is Associated with Overexpression of the AdeFGH Efflux Pump. Antimicrob. Agents Chemother. 2015;59:4817–4825. doi: 10.1128/AAC.00877-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vrancianu C.O., Gheorghe I., Czobor I.B., Chifiriuc M.C. Antibiotic Resistance Profiles, Molecular Mechanisms and Innovative Treatment Strategies of Acinetobacter Baumannii. Microorganisms. 2020;8:935. doi: 10.3390/microorganisms8060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aurilio C., Sansone P., Barbarisi M., Pota V., Giaccari L.G., Coppolino F., Barbarisi A., Passavanti M.B., Pace M.C. Mechanisms of Action of Carbapenem Resistance. Antibiotics. 2022;11:421. doi: 10.3390/antibiotics11030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abel Zur Wiesch P., Abel S., Gkotzis S., Ocampo P., Engelstädter J., Hinkley T., Magnus C., Waldor M.K., Udekwu K., Cohen T. Classic Reaction Kinetics Can Explain Complex Patterns of Antibiotic Action. Sci. Transl. Med. 2015;7:287ra73. doi: 10.1126/scitranslmed.aaa8760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pu Y., Ke Y., Bai F. Active Efflux in Dormant Bacterial Cells–New Insights into Antibiotic Persistence. Drug Resist. Updat. 2017;30:7–14. doi: 10.1016/j.drup.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Pu Y., Zhao Z., Li Y., Zou J., Ma Q., Zhao Y., Ke Y., Zhu Y., Chen H., Baker M.A.B., et al. Enhanced Efflux Activity Facilitates Drug Tolerance in Dormant Bacterial Cells. Mol. Cell. 2016;62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis K. Riddle of Biofilm Resistance. Antimicrob. Agents Chemother. 2001;45:999–1007. doi: 10.1128/AAC.45.4.999-1007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alav I., Sutton J.M., Rahman K.M. Role of Bacterial Efflux Pumps in Biofilm Formation. J. Antimicrob. Chemother. 2018;73:2003–2020. doi: 10.1093/jac/dky042. [DOI] [PubMed] [Google Scholar]
- 63.Boll J.M., Tucker A.T., Klein D.R., Beltran A.M., Brodbelt J.S., Davies B.W., Trent M.S. Reinforcing Lipid a Acylation on the Cell Surface of Acinetobacter Baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. MBio. 2015;6:e00478-15. doi: 10.1128/mBio.00478-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mengiste B., Lulie S., Getachew B., Gebrelibanos M., Mekuria A., Masresha B. In-vitro Antibacterial Activity of Extracts from Aerial Parts of Verbena Officinalis. Adv. Biol. Res. 2015;9:53–57. [Google Scholar]
- 65.Guha S., Udupa S., Ahmed W., Nagaraja V. Rewired Downregulation of DNA Gyrase Impacts Cell Division, Expression of Topology Modulators, and Transcription in Mycobacterium Smegmatis. J. Mol. Biol. 2018;430:4986–5001. doi: 10.1016/j.jmb.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 66.Willmott C.J.R., Critchlow S.E., Eperon I.C., Maxwell A. The Complex of DNA Gyrase and Quinolone Drugs with DNA Forms a Barrier to Transcription by RNA Polymerase. J. Mol. Biol. 1994;242:351–363. doi: 10.1006/jmbi.1994.1586. [DOI] [PubMed] [Google Scholar]
- 67.Rovinskiy N., Agbleke A.A., Chesnokova O., Pang Z., Higgins N.P. Rates of Gyrase Supercoiling and Transcription Elongation Control Supercoil Density in a Bacterial Chromosome. PLOS Genet. 2012;8:e1002845. doi: 10.1371/journal.pgen.1002845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kiel M.C., Kaji H., Kaji A. Ribosome Recycling: An Essential Process of Protein Synthesis. Biochem. Mol. Biol. Educ. 2007;35:40–44. doi: 10.1002/bmb.6. [DOI] [PubMed] [Google Scholar]
- 69.Pavlov M.Y., Freistroffer D.V., MacDougall J., Buckingham R.H., Ehrenberg M. Fast Recycling of Escherichia Coli Ribosomes Requires Both Ribosome Recycling Factor (RRF) and Release Factor RF3. EMBO J. 1997;16:4134–4141. doi: 10.1093/emboj/16.13.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nwugo C.C., Gaddy J.A., Zimbler D.L., Actis L.A. Deciphering the Iron Response in Acinetobacter Baumannii: A Proteomics Approach. J. Proteomics. 2011;74:44–58. doi: 10.1016/j.jprot.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Storz G., Vogel J., Wassarman K.M. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papenfort K., Vogel J. Multiple Target Regulation by Small Noncoding RNAs Rewires Gene Expression at the Post-Transcriptional Level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 73.Roncarati D., Scarlato V., Vannini A. Targeting of Regulators as a Promising Approach in the Search for Novel Antimicrobial Agents. Microorganisms. 2022;10:185. doi: 10.3390/microorganisms10010185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bak G., Lee J.Y.J., Suk S., Kim D., Lee J.Y.J., Kim K.S., Choi B.S., Lee Y. Identification of Novel sRNAs Involved in Biofilm Formation, Motility, and Fimbriae Formation in Escherichia Coli. Sci. Rep. 2015;5:15287. doi: 10.1038/srep15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pandey R.P., Mukherjee R., Chang C.-M. Emerging Concern with Imminent Therapeutic Strategies for Treating Resistance in Biofilm. Antibiotics. 2022;11:476. doi: 10.3390/antibiotics11040476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Di Bonaventura G., Spedicato I., D’Antonio D., Robuffo I., Piccolomini R. Biofilm Formation by Stenotrophomonas Maltophilia: Modulation by Quinolones, Trimethoprim-Sulfamethoxazole, and Ceftazidime. Antimicrob. Agents Chemother. 2004;48:151–160. doi: 10.1128/AAC.48.1.151-160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B., Salzberg S.L. Fast Gapped-Read Alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., Van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript Assembly and Quantification by RNA-Seq Reveals Unannotated Transcripts and Isoform Switching during Cell Differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu B.H., Yu H., Tu K., Li C., Li Y.X., Li Y.Y. DCGL: An R Package for Identifying Differentially Coexpressed Genes and Links from Gene Expression Microarray Data. Bioinformatics. 2010;26:2637–2638. doi: 10.1093/bioinformatics/btq471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McClure R., Balasubramanian D., Sun Y., Bobrovskyy M., Sumby P., Genco C.A., Vanderpool C.K., Tjaden B. Computational Analysis of Bacterial RNA-Seq Data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bharti R., Siebert D., Blombach B., Grimm D.G. Systematic Analysis of the Underlying Genomic Architecture for Transcriptional–Translational Coupling in Prokaryotes. NAR Genom. Bioinform. 2022;4:lqac074. doi: 10.1093/nargab/lqac074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw RNA sequencing datasets used for this study were deposited at the NCBI Sequence Read Archive; https://submit.ncbi.nlm.nih.gov/subs/sra under Bioproject ID number PRJNA892543; BioSample accession numbers SAMN31387122, SAMN31387123, SAMN31387124, SAMN31387125.