ABSTRACT
Staphylococcus argenteus is a recently described member of the Staphylococcus aureus complex (SAC) and is associated with human disease. The frequency and intensity of infections caused by S. argenteus are similar to those of Staphylococcus aureus. S. argenteus can harbor antibiotic resistance genes and a variety of virulence factors analogous to methicillin-resistant S. aureus (MRSA). The aim of our study was to analyze a collection of isolates in the Dutch national MRSA surveillance from January 2008 until March 2021 that were nontypeable by multilocus variable-number tandem-repeat analysis (MLVA). Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-ToF MS) was used for identifying the S. argenteus isolates, and whole-genome sequencing and SeqSphere were used to generate an in-house whole-genome multilocus sequence typing (wgMLST) scheme for typing the isolates. Furthermore, the presence of antibiotic resistance genes, replicons, and virulence genes was determined. Of 52,467 isolates submitted as MRSA from January 2008 until March 2021, 64 isolates (0.12%) were nontypeable with MLVA, and 54 of them were identified with mass spectrometry (MALDI-ToF MS) as S. argenteus. It appeared in retrospect that the first methicillin-resistant S. argenteus (MRSArg) was already submitted in 2008. An in-house-developed S. argenteus wgMLST scheme revealed that S. argenteus isolates clustered in 5 genomic groups which were characterized by distinct MLST types, resistomes, plasmid replicon families, and virulence factors. All but one isolate carried the staphylococcal chromosomal cassette mec (SCCmec) type IV harboring the methicillin resistance gene mecA and represent MRSArg. Most of the isolates with SCCmec subtype IVc(2B) had a trimethoprim resistance gene, dfrG, and harbored a blaZ-carrying plasmid, and most MRSArg isolates have the immune-modulating genes scn and sak. Nine of the 47 isolates carried enterotoxin-encoding genes seg, sei, sem, seo, and seu, which might be able to cause food poisoning. In some persons there was long-term persistence of MRSArg, and there were several genetically related MRSArg isolates in people living in close proximity, suggesting direct human-human transmission.
IMPORTANCE We show that MRSArg has been circulating in the Netherlands since at least 2008. Although MRSArg is distinct from MRSA, it has a comparable population structure and carries similar resistance and virulence genes. The Dutch national MRSA surveillance has been expanded to include other methicillin-resistant members of the S. aureus complex, such as S. argenteus and Staphylococcus schweitzeri.
KEYWORDS: MRSArg, SCCmec type IV, SCCmec subtype IVc(2B), resistance genes, enterotoxins, Staphylococcus argenteus, virulence factors
INTRODUCTION
Staphylococcus argenteus belongs to the Staphylococcus aureus complex (SAC) together with Staphylococcus schweitzeri, S. aureus, and the recently described Staphylococcus roterodami (1) and Staphylococcus singaporensis (2). S. argenteus was first described in 2006 (3) and was shown to belong to multilocus sequence typing (MLST) clonal complex 75 (CC75) of S. aureus. However, a later study showed that these CC75 isolates were distinct from S. aureus in several ways, and therefore, they were designated a separate species in 2015 (4), namely, S argenteus. CC75 comprises several MLST types exclusively found in S. argenteus isolates, e.g., ST2250, ST1223, ST2198, and ST2854 (4–7). In 2011 it was found that isolates with other sequence types (STs) not belonging to CC75, e.g., ST2196, ST1594, and ST2793, were genetically very similar to S. argenteus when their genomes were compared (5). Nowadays, S. argenteus isolates belonging to more than 60 different STs, from five different clonal complexes, have been identified (6, 8). S. argenteus has been found in many countries all over the world, including in the Netherlands (3–20).
Initially it was thought that S. argenteus was less virulent than S. aureus, because it lacks staphyloxanthin (5, 14). However, a study in 2015 (9) demonstrated that S. argenteus can cause health issues and that the frequency of infections caused by S. argenteus is similar to that of S. aureus. S. argenteus can cause skin, bone, and joint infections (3, 6, 10), bloodstream infections (11), and toxin-mediated syndromes, like staphylococcal food poisoning (12). Other studies (7, 13–15) have shown that S. argenteus can be resistant to various classes of antibiotics due to the presence of antibiotic resistance genes, e.g., blaZ, fusA, tet(L), or aph(3′-III) (7, 8, 13, 15, 20), and that there are methicillin-susceptible S. argenteus (MSSArg) but also methicillin-resistant S. argenteus (MRSArg) isolates harboring SCCmec type IV containing mecA (8). A variety of virulence factors have been found in S. argenteus, including the Panton-Valentine leukocidin (PVL) gene and the five classical enterotoxins (6, 8, 12, 14–16, 20), which may have a clinical relevance.
In a previous study, we showed that a number of isolates that were submitted for the Dutch national methicillin-resistant S. aureus (MRSA) surveillance were MRSArg and that MRSArg had been detected in the Netherlands as early as 2008 (17).
From January 2008 until March 2021, 52,467 MRSA isolates were submitted for the Dutch national methicillin-resistant S. aureus (MRSA) surveillance that were subjected to multilocus variable-number tandem-repeat analysis (MLVA). However, 54 isolates from 47 different persons were determined as nontypeable for MLVA and were shown to be MRSArg.
In the study presented here, we performed next-generation sequencing (NGS) and third-generation sequencing (TGS) to assess the genetic relationships and to determine molecular characteristics of these Dutch MRSArg isolates.
RESULTS
Identification and geographic distribution of S. argenteus in the Netherlands.
Between January 2008 and March 2021, the National Institute for Public Health and the Environment (RIVM) received 52,467 presumed MRSA isolates of which 64 were nontypeable by MLVA (0.12%). All 64 isolates were analyzed by next-generation sequencing (NGS) and matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-ToF MS). In the mass spectrometry (MALDI-ToF MS), 54 isolates of the 64 nontypeable isolates (84%) appeared to belong to the Staphylococcus genus with the highest score being for S. argenteus, six isolates were classified as S. aureus, and four isolates were other species (Staphylococcus sciuri and three Gram-negative isolates). These results were corroborated by analysis of the NGS data. The six previously nontypeable S. aureus isolates yielded complete MLVA profiles in the MLVA of freshly made bacterial lysates. The 54 S. argenteus isolates were obtained from 47 different persons; from five persons two isolates were obtained and from one person three isolates were obtained (Table 1). For this study 47 unique isolates were used, the first isolate of each person.
TABLE 1.
Metadata of the Dutch S. argenteus isolates used in this study
| Isolate | S. argenteus PCR | Persona | Age (yr) | Yr of sampling | Submitter of sample | Specimenb | Carrier or infection | Risk detail and/or reason for culturing | |
|---|---|---|---|---|---|---|---|---|---|
| RIVM_M003546 | + | Person 1 | 25 | 2008 | Swab (T) | Carrier | |||
| RIVM_M011864 | + | Person 1 | 28 | 2010 | Swab (N, P, T) | Carrier | |||
| RIVM_M025118 | + | Person 2 | 39 | 2014 | Other human material | Unknown | |||
| RIVM_M026823 | + | Person 2 | 40 | 2014 | Urine | Infection | |||
| RIVM_M031425 | + | Person 3 | 9 | 2015 | General practice | Pus | Infection | ||
| RIVM_M031426 | + | Person 3 | 9 | 2015 | General practice | Pus | Infection | ||
| RIVM_M040661 | + | Person 4 | 18 | 2017 | General practice | Swab (T) | Carrier | None | |
| RIVM_M040662 | + | Person 4 | 18 | 2017 | General practice | Swab (N) | Carrier | None | |
| RIVM_M040163 | + | Person 5 | 31 | 2017 | Hospital | Swab (N) | Carrier | Screening, was nursed in Australia | |
| RIVM_M041166 | + | Person 5 | 31 | 2017 | Rehabilitation | Swab (N) | Carrier | Screening, known MRSA carrier | |
| RIVM_M045471 | + | Person 5 | 32 | 2018 | General practice | Swab (T) | Carrier | ||
| RIVM_M046211 | + | Person 6 | 28 | 2019 | General practice | Swab (N) | Carrier | ||
| RIVM_M046343 | + | Person 6 | 28 | 2019 | Hospital | Swab (P) | Carrier | ||
| RIVM_M002983 | + | 60 | 2008 | Tumor thump | Unknown | ||||
| RIVM_M008822 | + | 61 | 2010 | Swab (N, P, T) | Carrier | ||||
| RIVM_M019208 | + | NAd | 2012 | Swab of ICDe pocket | Unknown | ||||
| RIVM_M020832 | + | 36 | 2013 | Swab (T) | Carrier | ||||
| RIVM_M023607 | + | 33 | 2013 | Wound fluid | Unknown | ||||
| RIVM_M024573 | + | 27 | 2014 | Wound fluid | Infection | ||||
| RIVM_M025002 | + | 49 | 2014 | Pus | Infection | ||||
| RIVM_M025606 | + | 29 | 2014 | Other human material | Unknown | ||||
| RIVM_M025618 | + | 28 | 2014 | Swab (N, P, T) | Carrier | ||||
| RIVM_M027397 | + | 56 | 2014 | Swab (N) | Carrier | ||||
| RIVM_M029687 | + | 70 | 2015 | Hospital | Swab (P, T) | Carrier | Screening, person was nursed in Belgiumc | ||
| RIVM_M029794 | + | 50 | 2015 | Sputum | Infection | ||||
| RIVM_M030565 | + | 42 | 2015 | Swab (N) | Carrier | ||||
| RIVM_M030644 | + | 20 | 2015 | Hospital | Wound fluid | Infection | Screening, vacation in Philippines | ||
| RIVM_M035744 | + | 69 | 2016 | General practice | Groin swab | Unknown | |||
| RIVM_M036020 | + | 25 | 2016 | Swab (N) | Carrier | ||||
| RIVM_M036044 | + | 62 | 2016 | Hospital | Wound fluid | Infection | |||
| RIVM_M036477 | + | 40 | 2016 | Hospital | Wound fluid | Infection | Coincidental finding in patient/client | ||
| RIVM_M036821 | + | 33 | 2016 | Hospital | Drain fluid | Unknown | Coincidental finding in patient/client | ||
| RIVM_M037020 | + | 34 | 2016 | Hospital | Swab (P, T) | Carrier | Screening, had had contact with MRSA+ person | ||
| RIVM_M037112 | + | 44 | 2016 | Hospital | Wound | Unknown | |||
| RIVM_M038289 | + | 48 | 2017 | Swab (T) | Carrier | ||||
| RIVM_M038319 | + | 46 | 2017 | Hospital | Wound fluid | Infection | Coincidental finding in patient/client | ||
| RIVM_M040187 | + | 19 | 2017 | Hospital | Swab (N) | Carrier | |||
| RIVM_M041614 | + | 37 | 2017 | General practice | Pus | Infection | |||
| RIVM_M042032 | + | 51 | 2018 | General practice | Swab (P) | Carrier | Screening, was nursed in Philippinesc | ||
| RIVM_M042169 | + | 40 | 2018 | Practice/clinic/treatment center | Swab (T) | Carrier | |||
| RIVM_M042294 | + | 22 | 2018 | Hospital | Swab (N) | Carrier | |||
| RIVM_M042580 | + | 10 | 2018 | General practice | Swab (T) | Carrier | Screening, from Somalia, here for family reunion | ||
| RIVM_M042731 | + | 29 | 2018 | General practice | Swab (R) | Carrier | Screening, had had contact with MRSA+ person | ||
| RIVM_M043825 | + | 43 | 2018 | Practice/clinic/treatment center | Swab (T) | Carrier | |||
| RIVM_M044970 | + | 65 | 2018 | Hospital | Swab (N) | Carrier | |||
| RIVM_M046950 | + | 25 | 2019 | Hospital | Swab (T) | Carrier | |||
| RIVM_M046968 | + | 66 | 2019 | Hospital | Blood | Infection | |||
| RIVM_M081107 | 2018 | ||||||||
| RIVM_M083837 | Not tested | 37 | 2020 | Hospital | Swab (N, P, T) | Carrier | |||
| RIVM_M085152 | Not tested | 0 | 2020 | Hospital swab (T) | Carrier | ||||
| RIVM_M085212 | Not tested | 39 | 2020 | Health service | Swab (N, T) | Carrier | |||
| RIVM_M087541 | Not tested | 64 | 2021 | Hospital, intensive care | Sputum | Infection | |||
| RIVM_M087650 | Not tested | 66 | 2021 | Hospital, intensive care | Swab (P) | Carrier | Screening | ||
| RIVM_M088004 | Not tested | 64 | 2021 | Hospital, nursing ward | Swab (T) | Carrier | Screening |
There were 6 persons with multiple isolates; these isolates are numbered person 1 to 6. All other isolates were obtained from different persons.
The different swab locations used for sampling (N, nose; P, perineum; R, rectum; T, throat).
The person was nursed in a health care facility in a foreign country, less than 2 months previously, for less than 24 h.
NA; not applicable.
ICD pocket, implantable cardioverter defibrillator pocket.
Twenty-three of the 47 isolates were obtained from persons living in the more densely populated provinces of the Netherlands, Utrecht and North and South Holland, 12 were from other provinces, three isolates were retrieved from the Dutch Caribbean island of St. Maarten, and for nine isolates this information was missing. The epidemiological questionnaires showed that the reason for sampling was screening for MRSA carriage based on risk factors (27/47; 57.5%) or clinical indication (11/47; 27.7%), and some isolates had an incomplete submission form (8/47; 17.0%). One isolate (1/47; 2.1%) was reported to be obtained from an implantable cardioverter defibrillator pocket. Most of the MRSA carriage screening samples were a swab from nose, perineum, rectum, or throat (29/47; 61.7%), and 34.0% (16/47) of the isolates came from other clinical materials such as blood, pus, sputum, urine, and wound fluid (Table 1).
wgMLST revealed distinct genogroups in the S. argenteus minimum spanning tree (MST).
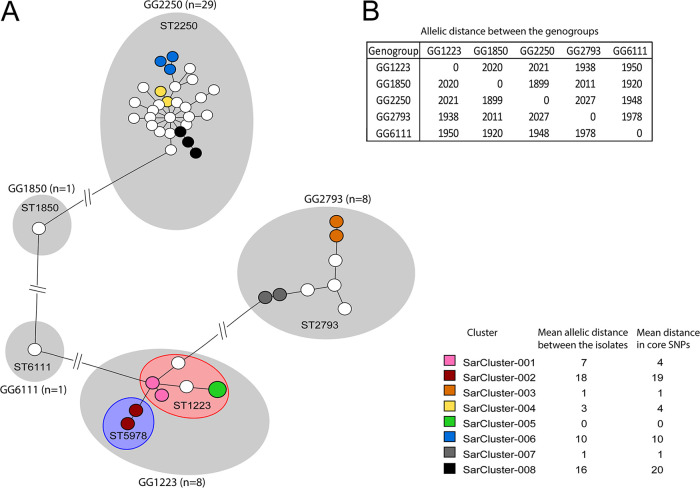
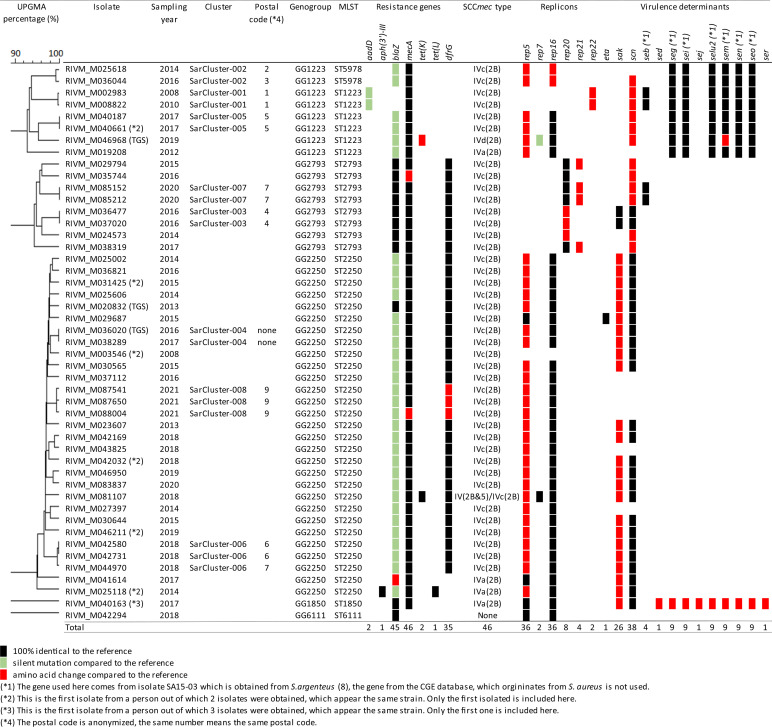
Whole-genome MLST (wgMLST) of the 47 Dutch S. argenteus isolates revealed that they were partitioned into five groups, designated as genogroups (GGs), with a pairwise difference between genogroups of more than 1,800 alleles (Fig. 1A and B) and with a pairwise distance of isolates within a genogroup of less than 145 alleles. The isolates within each genogroup had the same classical S. aureus MLST sequence type, except for genogroup GG1223, which comprised two sequence types, ST1223 and ST5978 (Fig. 1A). The isolates belonging to these two MLST types carried different alleles of the MLST gene tpi only, and this difference is based on a single mutation at position 186 in this gene, indicating that these two sequence types are very similar. One S. argenteus strain with a new allelic profile was found and was assigned MLST ST6111. The different S. argenteus GGs were characterized by distinct resistomes, plasmid replicon families, and virulence factors (Table 2).
FIG 1.
The wgMLST-based population structure of Dutch S. argenteus isolates. (A) MLST sequence types and wgMLST genogroups are indicated in the minimum spanning tree (MST). Each circle represents the first unique isolate per person, and the circle size indicates the number of isolates. The colors of the circles represent genetic clusters (right bottom). Genogroups (GG) are indicated with gray halos, while specific MLST STs are highlighted with colored halos. A genetic cluster of S. argenteus isolates is defined as ≥2 isolates that differ by ≤20 alleles. (B) Overview of the allelic distances between the genogroups from the wgMLST MST.
TABLE 2.
Genetic characteristics of the 47 unique Dutch S. argenteus isolates
The set of isolates was also analyzed using core single nucleotide polymorphism (SNP) typing (see Fig. S1 and Table S1 in the supplemental material). The distribution of genogroups and the genetic clusters are alike when comparing core SNPs with wgMLST. Only the genetic difference between the genogroups is much larger, more than 11,000 SNPs compared to an allelic difference of 1,800.
Evidence of long-term carriage of S. argenteus strains.
For six persons multiple isolates were submitted over time, which enabled intraperson analysis of the genetic variation of S. argenteus strains. The multiple isolates were obtained with different time periods between samplings, ranging from 0 days in person 4 to 834 days in person 1 (Table 3). In five of the six persons from whom multiple isolates were obtained, the wgMLST profiles within the isolate sets differed in a maximum of three alleles (3/2,555; 0.12%) independent of the time between the person’s samplings (Table 3). In addition, the resistomes and replicons within each isolate set were identical. The wgMLST profile of two isolates obtained from person 1 differed in 19 alleles (19/2,555; 0.74%), suggesting this person had acquired a different MRSArg strain. This idea was supported by the fact that the first isolate did not carry the blaZ resistance gene and the rep5 and rep16 replicons but did carry the immune-modulating genes sak and scn. The second isolate had a blaZ resistance gene and the rep5 and rep16 replicons but did not carry the immune-modulating genes sak and scn. However, the blaZ gene is located on a rep5-rep16 plasmid and the sak and scn genes are located on the Sa3 phage. Therefore, the second isolate may also represent the same MSRArg strain that had acquired a blaZ plasmid and had lost the Sa3 phage.
TABLE 3.
Genetic characteristics of duplicate Dutch S. argenteus isolates found in 6 persons
The multiple isolates from one person were also analyzed using core SNP typing (Table 3 and Table S1). The allelic differences found with wgMLST and the number of core SNPs are alike; only the two isolates from person 1 seemed to have a higher number of SNPs (30 SNPs) compared to the allelic differences found with wgMLST (19 wgMLST alleles).
Transmission of S. argenteus in the Netherlands.
Eight groups comprising 18 S. argenteus isolates were highly related and designated genetic clusters SarCluster-001 to SarCluster-008 (Fig. 1A). Within a genetic cluster the mean allelic distance between two isolates was 18 or less, with a maximum pairwise distance between two isolates of 19 alleles. The persons from whom S. argenteus isolates were obtained belonged to the same genetic cluster and lived within the same 4-digit postal code region, except for the two isolates of SarCluster-002 and the third isolate of SarCluster-006 (Table 3). Isolates from SarCluster-007 were obtained from a father and his newborn child, and SarCluster-008 isolates were retrieved from three men who shared a household. Isolates from SarCluster-003 were obtained from persons from the small Caribbean island of St. Maarten, but detailed residential data were lacking. Except for one isolate in SarCluster-008, with one single nucleotide polymorphism (SNP) in mecA, the isolates belonging to the same genetic cluster had identical resistance genes, replicons, SCCmec types, and subtypes as well as the same virulence factors (Table 2), suggesting the genetic clusters represent interperson S. argenteus transmissions.
The isolates that form a genetic cluster were also analyzed using core SNP typing (Fig. 1, Fig. S1, and Table S1). The allelic differences found with wgMLST and the number of core SNPs are alike.
Genomic comparison of S. argenteus isolates from the Netherlands with isolates from other countries.
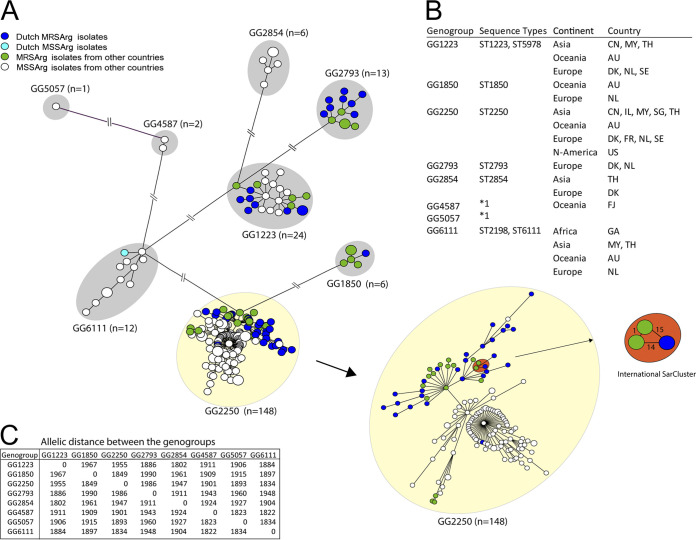
Comparison of the wgMLST profiles of the 47 S. argenteus isolates retrieved in the Netherlands with the wgMLST profiles of 167 isolates collected in other countries showed a highly similar distribution of genogroups (Fig. 2A). Eight genogroups were identified which differed by more than 1,800 alleles from each other (Fig. 2A and C), and isolates within a genogroup had a pairwise distance of less than 200 alleles. As with the Dutch S. argenteus isolates, most isolates from other countries belonged to GG2250 (Fig. 2A and B). A more detailed view of GG2250 showed two subgroups within the genogroup. Except for one isolate, all Dutch isolates resided in the upper branch of GG2250 with isolates from Denmark and Sweden and with two isolates sequenced in the United States. Most genogroups have a global composition, with isolates from several different continents, except for genogroup GG2793 (n = 13), which was comprised of European isolates only. Genogroups GG1850 and GG2854 carried isolates from two countries only. GG2793 and GG1850 contained only MRSArg isolates, whereas genogroups GG1223 and GG2250 have both MRSArg and MSSArg isolates. GG2250 was comprised of two subgroups, and the upper branch were mainly MRSArg isolates while the lower branch contained mainly MSSArg isolates. The other genogroups were comprised of only MSSArg isolates, and the only Dutch MSSArg isolate belongs to GG6111 together with other MSSArg isolates from other countries. In GG2250, a genetic cluster with a maximum allelic difference of 15 was identified with one Dutch isolate, RIVM_M025002, and two internationally obtained isolates (GenBank accession numbers JGHK00000000 and JGMK00000000, red circle in Fig. 2A). These isolates shared the same resistance genes, SCCmec type, and virulence factors. Isolate JGMK00000000 differed from the other two isolates because it harbored two replicons that were absent in the other isolates (data not shown). This was the only identified genetic cluster containing S. argenteus isolates from the Netherlands and other countries.
FIG 2.
International comparison of S. argenteus isolates by wgMLST. (A) In-house wgMLST MST of S. argenteus obtained in the Netherlands and from other countries, visualized in a minimum spanning tree (MST) with only the first unique isolate per person. GG indicates genogroups, which are encircled with gray halos, except for GG2250, which is light yellow. The Dutch MRSArg isolates are colored dark blue, and the Dutch MSSArg isolates are colored light blue. The MRSArg isolates from other countries are green, while the MSSArg isolates from other countries are white. An international genetic cluster with an S. argenteus isolate from the Netherlands and two isolates from the United States is depicted in red. (Inset) Closeup of GG2250 to provide greater detail of the international S. argenteus cluster. (B) Overview of the genogroups and isolates from different countries in the genogroups. The ISO 3166 code is used for country abbreviation. *1, these isolates had an incomplete MLST profile. The sequence type nearest the incomplete profile is used as the genogroup name. (C) Overview of the allelic distances between the genogroups.
The resistome of S. argenteus.
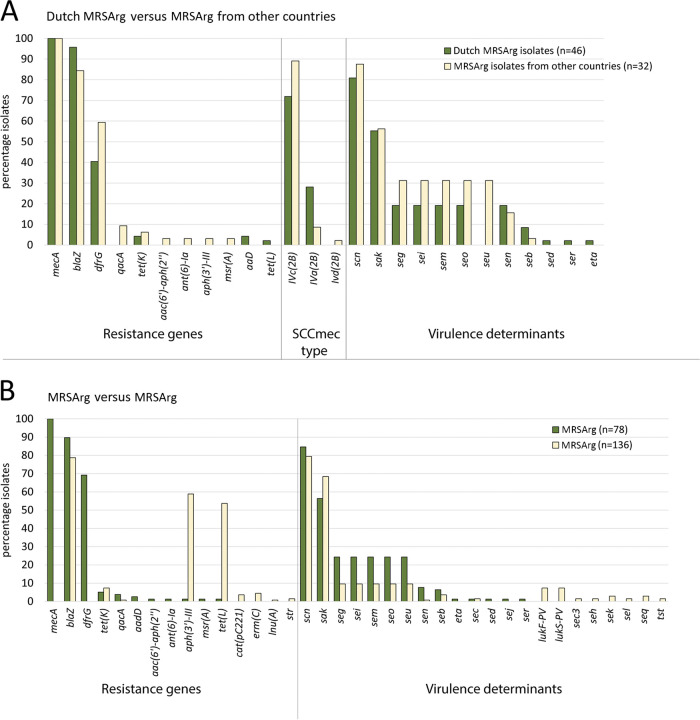
Forty-six of the 47 S. argenteus isolates (98%) from the Netherlands harbored a mecA gene and had the SCCmec type IV cassette (Fig. 3A). Thirty-five of the 41 isolates (85%) with the SCCmec type IVc(2B) also had a trimethoprim resistance gene, dfrG, embedded into the SCCmec, and this gene was found only in isolates with SCCmec type IVc(2B). Of all isolates studied in other countries only 32 of the 167 isolates (19.2%) were MRSArg. When comparing the Dutch MRSArg isolates with the MRSArg isolates from other countries, we observed that more isolates from other countries had the dfrG gene and the SCCmec type IVc(2B) (Fig. 3A). Next, we observed resistance genes aadD and tet(L) in a few Dutch MRSArg isolates while they were absent in isolates from other countries. Vice versa, the qacA, aac(6′)-aph(2′'), ant(6)-Ia, aph(3′)-III, and msr(A) genes were observed only in isolates from other countries. The tet(K) gene was found in MRSArg isolates in the Netherlands as well as in MRSArg isolates from other countries. When comparing the MRSArg isolates with MSSArg isolates, most MSSArg isolates had blaZ (78.7%), comparable to the MRSArg isolates (89.7%) (Fig. 3B). Two resistance genes, aph(3′)-III 3 and tet(L), were observed more often in MSSArg isolates (59% and 54%) than in MRSArg isolates (1% and 1%), respectively.
FIG 3.
International comparison of SCCmec type, resistance genes, and virulence determinants of S. argenteus. (A) Resistance genes, virulence factors, and SCCmec cassette types as identified through ResFinder, VirulenceFinder, and SCCmecFinder found in the Dutch MRSArg isolates compared with the findings in MRSArg isolates from other countries. (B) Resistance genes and virulence factors as identified through ResFinder and VirulenceFinder found in all the MRSArg isolates compared with the findings in all MSSArg isolates.
Virulence determinants of MRSArg and MSSArg.
When comparing the Dutch MRSArg isolates with the MRSArg isolates from other countries, we observed that 55% of the isolates had a sak gene (Fig. 3A). The scn gene was found in 80.6% of the Dutch MRSArg isolates and 87.5% of the MRSArg isolates from other countries. MRSArg isolates from other countries contained more putative enterotoxin-encoding genes seg, sei, sem, seo, and seu than did the Dutch MRSArg isolates. The percentage identity of the seg, sei, sem, sen, seo, and seu genes in the Dutch MRSArg isolates were less than 97%, suggesting these represent novel variants. The six putative enterotoxin genes found in the Dutch MRSArg isolates were analyzed by BLAST, revealing 100% similarity with enterotoxin genes found in the S. argenteus strain MSHR1132 (GenBank accession number FR821777) described by Wakabayashi et al. (12), but they used the name selu2 instead of seu. It appears that the enterotoxin-encoding genes in the only Dutch MSSArg isolate are slightly different from those found in the MRSArg isolates. Furthermore, both MRSArg and MSSArg groups contained the scn gene in 80% of the isolates (Fig. 3B). The MSSArg isolates contained the sak gene (68.3%) more often than did MRSArg isolates (56.4%). MRSArg contained enterotoxin-encoding genes seg, sei, sem, seo, and seu (24.4%) more often than did MSSArg (9.6%). The PVL-encoding genes were identified in the MSSArg isolates (7.4%), but these were absent in MRSArg isolates.
DISCUSSION
Here, we report the identification and genomic characterization of methicillin-resistant S. argenteus isolates that were misidentified and submitted as MRSA to the Dutch national MRSA surveillance in the period from January 2008 until March 2021.
MRSArg has been circulating in the Netherlands since at least 2008 and is different from MRSA; however, it has a comparable population structure and harbors similar resistance and virulence genes.
We cannot estimate the prevalence of MRSArg in the Netherlands, but nevertheless our study shows that MRSArg carriage and transmission do occur, which merits further study.
Using an in-house wgMLST scheme for S. argenteus, we found a division of the Dutch MRSArg isolates into five different genogroups, which differed in MLST sequence types, resistance genes, replicon families, and virulence determinants. A similar division was found in the S. argenteus isolates from other countries obtained from the NCBI and SRA databases, suggesting global spread of S. argenteus strains. Previously the identification of six clusters of S. argenteus was described with phylogenetic analysis (14), and later seven S. argenteus clades were described with the use of SNP typing with 52,329 SNPs (6). The identified genogroups in this study have a distribution similar to those in the work of Kaden et al. (14), so it seems that the two typing methods yield comparable results for determining genetic relationships between isolates. We also compared the wgMLST analyses with a core SNP typing method but found no significant differences, except for 2 isolates obtained from one person, but there was a long time between the sample collection dates. The person could still carry the same strain that has adapted and changed or perhaps the person encountered a genetically highly related strain and became reinfected.
In this study, we defined a genetic cluster cutoff value of ≥2 isolates that differ by ≤20 alleles to determine whether the isolates were genetically related. We cannot conclude whether two isolates that form a genetic cluster are the same strain if we rely only on genetic distance. Additional information on the isolates in the genetic cluster is needed, including the number and variety of resistance genes, replicon families, virulence determinants, and also epidemiological and clinical data. Until now, only a few Dutch S. argenteus genetic clusters have been investigated; thus, possibly the cluster cutoff may vary in the future.
From six persons multiple S. argenteus isolates were obtained over a period of time which revealed that each person carried her or his own unique strain. In other studies, it was also found that S. argenteus can persist in a person for longer periods of time (18, 19). The wgMLST showed that some isolates that were genetically closely related were obtained from persons living in the same postal area, suggesting transmission. For two such genetic clusters, SarCluster-007 and SarCluster-008, the persons from whom the MRSArg isolates were obtained were living in the same household, making transmission highly likely.
When comparing the Dutch MRSArg isolates with the international MRSArg isolates, only one genetic cluster was identified with an isolate retrieved from the Netherlands and two isolates sequenced in the United States. One Dutch MRSArg isolate was found in genogroup GG1850 next to Australian MRSArg isolates. There was only one Dutch isolate with this sequence type, and since this person was hospitalized in Australia in the past, an international transmission is likely.
The studied MRSArg isolates from the Netherlands resembled MRSArg isolates from other countries, when their resistance genes were compared. Only a few genes encoding resistance to methicillin, trimethoprim, and beta-lactam were identified, indicating that these MRSArg isolates were not highly resistant. MSSArg isolates were not methicillin resistant and lacked the dfrG gene, but >50% of the MSSArg isolates carried the aminoglycoside resistance gene aph(3)-III (58.8%) and tetracycline resistance gene tet(L) (53.7%). When the virulence determinants were compared between MRSArg isolates from the Netherlands and MRSArg isolates from other countries, only small differences were found. Most of the Dutch MRSArg isolates carried the immunomodulating genes sac and scn, suggesting that the isolates are as virulent as the MRSArg and MSSArg isolates from other countries. The enterotoxins identified in S. argenteus are different from those in S. aureus but similar to the enterotoxins reported in S. argenteus by Wakabayashi et al. (12). These enterotoxin-encoding genes occur in 25% of the MRSArg isolates but in only 9.6% of the MSSArg isolates. Some studies reported PVL-encoding genes in MSSArg isolates (3, 8, 11, 16, 19) but not in MRSArg isolates (3, 18); since we studied only MRSArg isolates, we did not observe these.
The isolates were identified as S. argenteus by mass spectrometry (MALDI-ToF MS), but since the discrimination between the different species belonging to the S. aureus complex is not very reliable, we also used the core genes present in the S. argenteus and S. aureus wgMLST scheme to confirm the species identification. While doing so, we noticed that the core genes found in S. argenteus differed considerably from those found in S. aureus. Also, the other members of the SAC, S. schweitzeri, S. roterodami, and S. singaporensis, seemed to have different core genes when they were analyzed with the S. aureus and S. argenteus wgMLST scheme. This shows that the five species belonging to the S. aureus complex differ significantly in their core genes and that the wgMLST scheme designed for S. argenteus is highly specific for S. argenteus. This finding puts some doubt on the grouping of these five species in a single complex. For S. roterodami only one reference strain was available (EMCR19), so this is a limitation. When more S. roterodami sequences are available, it may be interesting to analyze them as well to see if they have the same number of core genes for the two schemes as EMCR19.
In conclusion, MRSArg appears to be a unique pathogen sharing important resistance and virulence characteristics with MRSA. In order to gain insights in the prevalence and clinical relevance of MRSArg in the Netherlands, the Dutch national MRSA surveillance has been expanded to include other methicillin-resistant members of the S. aureus complex, including S. schweitzeri and S. argenteus.
MATERIALS AND METHODS
Bacterial isolates.
The National Institute for Public Health and the Environment (RIVM) performs the national MRSA surveillance in the Netherlands. Dutch medical microbiology laboratories (MMLs) submit MRSA isolates to the RIVM, where they are typed by MLVA (21). Some of the isolates are not Staphylococcus aureus and cannot be typed using MLVA, since they lack more than two of the eight required variable-number tandem repeat loci. mass spectrometry (MALDI-ToF MS) mass spectrometry (MS) microflex LT (Bruker, Billerica, MA, USA) with the December 2018 updated database including the S. argenteus spectrum (BDAL version 8.0.0.0 & SR 1.0.0.0 & in-house; Bruker, Billerica, MA, USA) was performed on 64 isolates.
The submitted isolates were accompanied with an epidemiological questionnaire containing patient characteristics, but for some of the older isolates the questionnaire was only partially filled in (Table 1).
Whole-genome sequencing.
NGS was performed using a NovaSeq 6000 machine (Illumina, San Diego, CA, USA) yielding 150-base-long paired-end reads. The NGS data were used to create contigs, using CLC Genomics Workbench v20.0.3 (Qiagen Bioinformatics, Aarhus, Denmark), and only contigs with a minimum length of 500 bases and a read coverage of >30 were used for analysis. Three isolates, RIVM_M020832, RIVM_M036020, and RIVM_M046968, were also sequenced with Nanopore long-read sequencing (Oxford Nanopore Technologies, Oxford, United Kingdom). Chromosomes and plasmids were reconstructed using Unicycler hybrid assembly, and the resulting contigs were annotated with Prokka as described previously (22) and submitted to NCBI.
Genomic analyses.
An S. argenteus-specific wgMLST scheme was designed in SeqSphere v3.5.0 (Ridom, Münster, Germany) using the annotated chromosome sequence of isolate CP086572 as the seed sequence and the two other isolates CP086570 and CP086574 as query genomes. This yielded a wgMLST scheme comprising 2,168 core genes and 387 accessory genes. All S. argenteus isolates were analyzed with this wgMLST scheme, and each isolate yielded more than 95% good targets, which means that 95% of the genes from the core genome were present in each isolate.
Ten MRSA isolates from our collection were analyzed with the in-house S. argenteus wgMLST scheme, and this showed only 34.9% of the S. argenteus core genes present in S. aureus. Nine available S. schweitzeri sequences ware analyzed and yielded 77.0% of the core genes present in the S. argenteus wgMLST scheme. And the recently described S. roterodami EMCR19, which is only one isolate, and 6 available sequences of the newly described S. singaporensis both yielded 87.3% of the core genes present in the S. argenteus wgMLST scheme. The same sequences were also analyzed with the wgMLST scheme for S. aureus. Here, we found that the S. aureus isolates scored more than 96.4% of the core genes present in the wgMLST scheme for S. aureus, but S. schweitzeri yielded 47.0%, S. roterodami yielded 39.3%, and S. singaporensis yielded only 37.7% of the core genes present in the S. aureus wgMLST scheme.
Apart from the Dutch isolates, we used the NCBI (129 isolates) and SRA (38 isolates) databases to obtain NGS data from S. argenteus isolates studied in other countries. Of 129 NCBI isolates 30 were MRSArg and 99 were methicillin-sensitive S. argenteus (MSSArg). Of 38 SRA isolates two were MRSArg and 36 were MSSArg. The in-house S. argenteus wgMLST scheme was used to compare international S. argenteus isolates with the isolates retrieved in the Netherlands. Next, the allelic profiles were imported in BioNumerics (v7.6.3; Applied Maths, Sint-Martens-Latem, Belgium) and used to assess genetic relationships between isolates, which were visualized in a minimum spanning tree (MST). Missing alleles were ignored and not counted as allelic differences. A genetic cluster is defined as ≥2 isolates that differ by ≤20 alleles, and genogroups differ by ≥1,800 alleles from each other.
All 54 Dutch isolates were typed using an SNP typing tool of CLC Genomics Workbench v20.0.3, using CP086572 as a reference. The core genome was annotated and used for core SNP typing, excluding the variable regions.
ResFinder, PlasmidFinder, VirulenceFinder, and SCCmec Finder databases from the Center for Genomic Epidemiology (https://bitbucket.org/genomicepidemiology, downloaded on 1 April 2020) were used for the identification of resistance genes, replicons, virulence determinants, and the SCCmec type, respectively. A threshold of 95% was used for identity and 60% for the minimum length for ResFinder and PlasmidFinder. Targets that had less than 100% identity with the ResFinder, VirulenceFinder, or PlasmidFinder database reference gene for the specific resistance genes, virulence genes, or replicons were further analyzed by mapping Illumina reads to the specific reference gene to determine whether the found differences were silent mutations or caused amino acid changes.
Ethics statement.
The bacterial isolates belong to the medical microbiological laboratories participating in the Dutch national MRSA surveillance program and were obtained as part of routine clinical care in the past years. Since no identifiable personal data were collected and data were analyzed and processed anonymously, written or verbal patient consent was not required. According to the Dutch Medical Research Involving Human Subjects Act (WMO), this study was exempt from review by an Institutional Review Board.
Data availability.
The Illumina (NGS) sequence data set generated and analyzed in this study is available in the Sequence Read Archive (SRA) in project PRJNA794761 (accession numbers SRR17445809 through SRR17445863). The plasmid and chromosome sequences are deposited in GenBank of the National Center for Biotechnology Information (NCBI) and available through the accession numbers CP086570, CP086572, and CP086574. We confirm that all supporting data, code, protocols, and accession numbers have been provided within the article and through supplemental material.
ACKNOWLEDGMENTS
We thank the Dutch medical microbiology laboratories and hospitals for sending in the Staphylococcus aureus isolates for the Dutch national MRSA surveillance, which turned out to be Staphylococcus argenteus. Without their contribution, this study could not have been performed. We also thank Romy Zwittink for critical reading of the manuscript.
Members of the Dutch MRSA Surveillance Study Group include the following: B. Wintermans, ADRZ medisch centrum, Department of Medical Microbiology, Goes; M. A. Leversteijn-van Hall, Alrijne Hospital, Department of Medical Microbiology, Leiden; W. van den Bijllaardt, Amphia Hospital, Microvida Laboratory for Microbiology, Breda; I. J. B. Spijkerman, Amsterdam UMC—location AMC, Department of Medical Microbiology, Amsterdam; K. van Dijk, Amsterdam UMC—location Vumc, Department of Medical Microbiology and Infection Control, Amsterdam; T. Halaby, Analytical Diagnostic Center N.V. Curaçao, Department of Medical Microbiology, Willemstad (Curaçao); B. Zwart, Atalmedial, Department of Medical Microbiology, Amsterdam; B. M. W. Diederen, Bravis Hospital/ZorgSaam Hospital Zeeuws-Vlaanderen, Department of Medical Microbiology, Roosendaal/Terneuzen; A. Voss, Canisius Wilhelmina Hospital, Department of Medical Microbiology and Infectious Diseases, Nijmegen; J. W. Dorigo-Zetsma, CBSL, Department of Medical Microbiology, Hilversum; A. Ott, CERTE, Department of Medical Microbiology Groningen/Drenthe, Groningen; K. Waar, CERTE, Department of Medical Microbiology Friesland/NOP, Leeuwarden; J. H. Oudbier, Comicro, Department of Medical Microbiology, Hoorn; M. van der Vusse, Deventer Hospital, Department of Medical Microbiology, Deventer; A. L. M. Vlek, Diakonessenhuis, Department of Medical Microbiology and Immunology, Utrecht; A. G. M. Buiting, Elisabeth-TweeSteden (ETZ) Hospital, Department of Medical Microbiology and Immunology, Tilburg; L. G. M. Bode, Erasmus MC, University Medical Center Rotterdam, Department of Medical Microbiology and Infectious Diseases, Rotterdam; S. Paltansing, Franciscus Gasthuis and Vlietland, Department of Medical Microbiology and Infection Control, Rotterdam; A. J. van Griethuysen, Gelderse Vallei Hospital, Department of Medical Microbiology, Ede; M. den Reijer, Gelre Hospitals, Department of Medical Microbiology and Infection prevention, Apeldoorn; M. van Trijp, Groene Hart Hospital, Department of Medical Microbiology and Infection Prevention, Gouda; M. Wong, Haga Hospital, Department of Medical Microbiology, The Hague; A. E. Muller, HMC Westeinde Hospital, Department of Medical Microbiology, The Hague; M. P. M. van der Linden, IJsselland hospital, Department of Medical Microbiology, Capelle a/d Ijssel; M. van Rijn, Ikazia Hospital, Department of Medical Microbiology, Rotterdam; M. J. H. M. Wolfhagen, Isala Hospital, Laboratory of Medical Microbiology and Infectious Diseases, Zwolle; E. Kolwijck, Jeroen Bosch Hospital, Department of Medical Microbiology and Infection Control, 's-Hertogenbosch; N. al Naiemi, LabMicTA, Regional Laboratory of Microbiology Twente Achterhoek, Hengelo; T. Schulin, Laurentius Hospital, Department of Medical Microbiology, Roermond; M. Damen, Maasstad Hospital, Department of Medical Microbiology, Rotterdam; S. Dinant, Maasstad Hospital, Department of Medical Microbiology, Rotterdam; S. P. van Mens, Maastricht University Medical Centre, Department of Medical Microbiology, Maastricht; D. C. Melles, Meander Medical Center, Department of Medical Microbiology, Amersfoort; J. W. T. Cohen Stuart, Noordwest Ziekenhuisgroep, Department of Medical Microbiology, Alkmaar; P. Gruteke, Onze Lieve Vrouwe Gasthuis, Department of Medical Microbiology, Amsterdam; I. T. M. A. Overdevest, PAMM, Department of Medical Microbiology, Veldhoven; A. P. van Dam, Amsterdam Health Service, Public Health Laboratory, Amsterdam; H. Wertheim, Radboud University Medical Center, Department of Medical Microbiology, Nijmegen; B. Maraha, Albert Schweitzer Hospital, Department of Medical Microbiology, Dordrecht; J. C. Sinnige, Regional Laboratory of Public Health, Department of Medical Microbiology, Haarlem; E. E. Mattsson, Reinier de Graaf Groep, Department of Medical Microbiology, Delft; R. W. Bosboom, Rijnstate Hospital, Laboratory for Medical Microbiology and Immunology, Velp; A. Stam, Saltro Diagnostic Centre, Department of Medical Microbiology, Utrecht; E. de Jong, Slingeland Hospital, Department of Medical Microbiology, Doetinchem; N. Roescher, St Antonius Hospital, Department of Medical Microbiology and Immunology, Nieuwegein; E. Heikens, St Jansdal Hospital, Department of Medical Microbiology, Harderwijk; R. Steingrover, St. Maarten Laboratory Services, Department of Medical Microbiology, Cay Hill (St. Maarten); A. Troelstra, University Medical Center Utrecht, Department of Medical Microbiology, Utrecht; E. Bathoorn, University of Groningen, Department of Medical Microbiology, Groningen; T. A. M. Trienekens, VieCuri Medical Center, Department of Medical Microbiology, Venlo; D. W. van Dam, Zuyderland Medical Centre, Department of Medical Microbiology and Infection Control, Sittard-Geleen; E. I. G. B. de Brauwer, Zuyderland Medical Centre, Department of Medical Microbiology and Infection Control, Heerlen; F. S. Stals, Zuyderland Medical Centre, Department of Medical Microbiology and Infection Control, Heerlen.
Conceptualization and methodology, S.W., A.P.A.H., and L.M.S.; visualization, S.W.; data curation, F.L., S.W., and M.G.V.S.-V.; formal analysis, S.W. and L.M.S.; funding, not applicable; sample collection, Dutch MRSA Surveillance Study Group; laboratory experiments, F.L., A.D.H., and M.G.V.S.-V.; supervision, A.P.A.H. and L.M.S.; manuscript preparation – original draft, S.W.; review and editing, S.W., A.P.A.H., and L.M.S.; review and approval of final manuscript, all authors.
We declare that there are no conflicts of interest.
This work received no specific grant from any funding agency.
Footnotes
Supplemental material is available online only.
Contributor Information
Sandra Witteveen, Email: sandra.witteveen@rivm.nl.
Susan Realegeno, Quest Diagnostics.
REFERENCES
- 1.Schutte AHJ, Strepis N, Zandijk WHA, Bexkens ML, Bode LGM, Klaassen CHW. 2021. Characterization of Staphylococcus roterodami sp. nov., a new species within the Staphylococcus aureus complex isolated from a human foot infection. Int J Syst Evol Microbiol 71. doi: 10.1099/ijsem.0.004996. [DOI] [PubMed] [Google Scholar]
- 2.Chew KL, Octavia S, Lai D, Lin RTP, Teo JWP. 2021. Staphylococcus singaporensis sp. nov., a new member of the Staphylococcus aureus complex, isolated from human clinical specimens. Int J Syst Evol Microbiol 71. doi: 10.1099/ijsem.0.005067. [DOI] [PubMed] [Google Scholar]
- 3.McDonald M, Dougall A, Holt D, Huygens F, Oppedisano F, Giffard PM, Inman-Bamber J, Stephens AJ, Towers R, Carapetis JR, Currie BJ. 2006. Use of a single-nucleotide polymorphism genotyping system to demonstrate the unique epidemiology of methicillin-resistant Staphylococcus aureus in remote aboriginal communities. J Clin Microbiol 44:3720–3727. doi: 10.1128/JCM.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tong SYC, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. 2015. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the non-human primate-associated Staphylococcus schweitzeri sp. nov. Int J Syst Evol Microbiol 65:15–22. doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holt DC, Holden MTG, Tong SYC, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. 2011. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soderquist B, Wildeman P, Stenmark B, Stegger M. 2020. Staphylococcus argenteus as an etiological agent of prosthetic hip joint infection: a case presentation. J Bone Jt Infect 5:172–175. doi: 10.7150/jbji.44848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang D-F, Zhi X-Y, Zhang J, Paoli GC, Cui Y, Shi C, Shi X. 2017. Preliminary comparative genomics revealed pathogenic potential and international spread of Staphylococcus argenteus. BMC Genomics 18:808. doi: 10.1186/s12864-017-4149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen TA, Bartels MD, Høgh SV, Dons LE, Pedersen M, Jensen TG, Kemp M, Skov MN, Gumpert H, Worning P, Westh H. 2017. Whole genome sequencing of Danish Staphylococcus argenteus reveals a genetically diverse collection with clear separation from Staphylococcus aureus. Front Microbiol 8:1512. doi: 10.3389/fmicb.2017.01512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thaipadungpanit J, Amornchai P, Nickerson EK, Wongsuvan G, Wuthiekanun V, Limmathurotsakul D, Peacock SJ. 2015. Clinical and molecular epidemiology of Staphylococcus argenteus infections in Thailand. J Clin Microbiol 53:1005–1008. doi: 10.1128/JCM.03049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rigaill J, Grattard F, Grange S, Forest F, Haddad E, Carricajo A, Tristan A, Laurent F, Botelho-Nevers E, Verhoeven PO. 2018. Community-acquired Staphylococcus argenteus sequence type 2250 bone and joint infection, France, 2017. Emerg Infect Dis 24:1958–1961. doi: 10.3201/eid2410.180727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupieux C, Blonde R, Bouchiat C, Meugnier H, Bes M, Laurent S, Vandenesch F, Laurent F, Tristan A. 2015. Community-acquired infections due to Staphylococcus argenteus lineage isolates harbouring the Panton-Valentine leucocidin, France, 2014. Euro Surveill 20:21154. doi: 10.2807/1560-7917.es2015.20.23.21154. [DOI] [PubMed] [Google Scholar]
- 12.Wakabayashi Y, Umeda K, Yonogi S, Nakamura H, Yamamoto K, Kumeda Y, Kawatsu K. 2018. Staphylococcal food poisoning caused by Staphylococcus argenteus harboring staphylococcal enterotoxin genes. Int J Food Microbiol 265:23–29. doi: 10.1016/j.ijfoodmicro.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 13.Moradigaravand D, Jamrozy D, Mostowy R, Anderson A, Nickerson EK, Thaipadungpanit J, Wuthiekanun V, Limmathurotsakul D, Tandhavanant S, Wikraiphat C, Wongsuvan G, Teerawattanasook N, Jutrakul Y, Srisurat N, Chaimanee P, Eoin West T, Blane B, Parkhill J, Chantratita N, Peacock SJ. 2017. Evolution of the Staphylococcus argenteus ST2250 clone in northeastern Thailand is linked with the acquisition of livestock-associated staphylococcal genes. mBio 8:e00802-17. doi: 10.1128/mBio.00802-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaden R, Engstrand L, Rautelin H, Johansson C. 2018. Which methods are appropriate for the detection of Staphylococcus argenteus and is it worthwhile to distinguish S. argenteus from S. aureus? Infect Drug Resist 11:2335–2344. doi: 10.2147/IDR.S179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aung M, San T, Aye M, Mya S, Maw W, Zan K, Htut W, Kawaguchiya M, Urushibara N, Kobayashi N. 2017. Prevalence and genetic characteristics of Staphylococcus aureus and Staphylococcus argenteus isolates harboring Panton-Valentine leukocidin, enterotoxins, and TSST-1 genes from food handlers in Myanmar. Toxins (Basel) 9:241. doi: 10.3390/toxins9080241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker K, Schaumburg F, Kearns A, Larsen AR, Lindsay JA, Skov RL, Westh H. 2019. Implications of identifying the recently defined members of the Staphylococcus aureus complex S. argenteus and S. schweitzeri: a position paper of members of the ESCMID Study Group for Staphylococci and Staphylococcal Diseases (ESGS). Clin Microbiol Infect 25:1064–1070. doi: 10.1016/j.cmi.2019.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Bank LEA, Bosch T, Schouls LM, Weersink AJL, Witteveen S, Wolffs PFG, Nijhuis RHT. 2021. Methicillin-resistant Staphylococcus argenteus in the Netherlands: not a new arrival. Eur J Clin Microbiol Infect Dis 40:1583–1585. doi: 10.1007/s10096-021-04204-7. [DOI] [PubMed] [Google Scholar]
- 18.Jauneikaite E, Pichon B, Mosavie M, Fallowfield JL, Davey T, Thorpe N, Nelstrop A, Sriskandan S, Lamb LE. 2021. Staphylococcus argenteus transmission among healthy Royal Marines: a molecular epidemiology case-study. J Infect 83:550–553. doi: 10.1016/j.jinf.2021.08.040. [DOI] [PubMed] [Google Scholar]
- 19.Senok A, Nassar R, Kaklamanos EG, Belhoul K, Abu Fanas S, Nassar M, Azar AJ, Müller E, Reissig A, Gawlik D, Monecke S, Ehricht R. 2020. Molecular characterization of Staphylococcus aureus isolates associated with nasal colonization and environmental contamination in academic dental clinics. Microb Drug Resist 26:661–669. doi: 10.1089/mdr.2019.0318. [DOI] [PubMed] [Google Scholar]
- 20.Argudín MA, Dodémont M, Vandendriessche S, Rottiers S, Tribes C, Roisin S, de Mendonça R, Nonhoff C, Deplano A, Denis O. 2016. Low occurrence of the new species Staphylococcus argenteus in a Staphylococcus aureus collection of human isolates from Belgium. Eur J Clin Microbiol Infect Dis 35:1017–1022. doi: 10.1007/s10096-016-2632-x. [DOI] [PubMed] [Google Scholar]
- 21.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, van der Heide HGJ, Grundmann H, Heck MEOC, de Neeling AJ. 2009. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa-typing. PLoS One 4:e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendrickx APA, Landman F, de Haan A, Witteveen S, van Santen-Verheuvel MG, Schouls LM, The Dutch Cpe Surveillance Study Group . 2021. bla OXA-48-like genome architecture among carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the Netherlands. Microb Genom 7:000512. doi: 10.1099/mgen.0.000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01035-22-s0001.pdf, PDF file, 0.3 MB (316.1KB, pdf)
Data Availability Statement
The Illumina (NGS) sequence data set generated and analyzed in this study is available in the Sequence Read Archive (SRA) in project PRJNA794761 (accession numbers SRR17445809 through SRR17445863). The plasmid and chromosome sequences are deposited in GenBank of the National Center for Biotechnology Information (NCBI) and available through the accession numbers CP086570, CP086572, and CP086574. We confirm that all supporting data, code, protocols, and accession numbers have been provided within the article and through supplemental material.