ABSTRACT
The marine bacterial genus Thalassospira has often been identified as an abundant member of polycyclic aromatic hydrocarbon (PAH)-exposed microbial communities. However, despite their potential usability for biotechnological applications, functional genes that are conserved in their genomes have barely been investigated. Thus, the goal of this study was to comprehensively examine the functional genes that were potentially responsible for aromatic hydrocarbon biodegradation in the Thalassospira genomes available from databases, including a new isolate of Thalassospira, strain GO-4, isolated from a phenanthrene-enriched marine bacterial consortium. Strain GO-4 was used in this study as a model organism to link the genomic data and their metabolic functions. Strain GO-4 growth assays confirmed that it utilized a common phenanthrene biodegradation intermediate 2-carboxybenzaldehyde (CBA) as the sole source of carbon and energy, but did not utilize phenanthrene. Consistently, strain GO-4 was found to possess homologous genes of phdK, pht, and pca that encode enzymes for biodegradation of CBA, phthalic acid, and protocatechuic acid, respectively. Further comprehensive genomic analyses for 33 Thalassospira genomes from databases showed that a gene cluster that consisted of phdK and pht homologs was conserved in 13 of the 33 strains. pca gene homologs were found in all examined genomes; however, homologs of the known PAH-degrading genes, such as the pah, phn, or nah genes, were not found. Possibly Thalassospira spp. co-occupy niches with other PAH-degrading bacteria that provide them with PAH degradation intermediates and facilitated their inhabitation in PAH-exposed microbial ecosystems.
IMPORTANCE Comprehensive investigation of multiple genomic data sets from targeted microbial taxa deposited in databases may provide substantial information to predict metabolic capabilities and ecological roles in different environments. This study is the first report that details the functional profiling of Thalassospira spp. that have repeatedly been found in polycyclic aromatic hydrocarbon (PAH)-exposed marine bacterial communities by using genomic data from a new isolate, Thalassospira strain GO-4, and other strains in databases. Through screening of functional genes potentially involved in aromatic hydrocarbon biodegradation across 33 Thalassospira genomes and growth assays for strain GO-4, it was suggested that Thalassospira spp. unexceptionally conserved the ability to metabolize single-ring, PAH biodegradation intermediates, while being incapable of utilizing PAHs. This expanded our understanding of this potentially important contributing member to PAH-degrading microbial ecosystems; such species are considered to be specialized in driving downstream reactions of PAH biodegradation.
KEYWORDS: aromatic hydrocarbons, biodegradation, genomics, marine bacteria
INTRODUCTION
The environmental fate of toxic and persistent organic pollutants that are released from anthropogenic activities has been of great concern. In marine environments, indigenous microorganisms play central roles in global carbon and nutrient cycling and thus are expected to be main drivers of restoration of natural environments when they are exposed to an accidental release of organic pollutants—such as in ocean oil spill disasters (1). Microbial function genes that were characterized as responsible for key steps in the biotransformation of specific hydrocarbons have been employed as functional biomarkers to evaluate the ability of microorganisms for removal of organic pollutants (2–4). At the same time, microbial (meta)genomic research has contributed to understanding complex microbial ecosystems that develop in polluted environments through identification of microbial taxa that play central roles in pollutant biodegradation and that may be applied to bioremediation (5–7).
The alphaproteobacterial genus Thalassospira (Rhodospirillales, Thalassospiraceae) was first characterized in 2002 (8), has been discovered globally in marine environments, and has repeatedly been identified as an abundant member of polycyclic aromatic hydrocarbon (PAH)-exposed marine microbial communities (9–12). Additionally, there are a few examples of isolates that were claimed to be capable of degrading PAHs and potentially growing on them as the carbon source (13–15). However, despite such unique metabolic capabilities and potential high usability for biotechnological applications, genomic information of Thalassospira strains has not been adequately discussed and characterized. Other well-studied bacterial groups that are capable of biodegrading and growing on aromatic hydrocarbons, such as Mycobacterium, Pseudomonas, or sphingomonad strains, are known to have acquired and conserved the specialized functional genes in their genomes that are responsible for their metabolisms: e.g., aromatic ring-hydroxylating dioxygenases (EC 1.14.12.x) and aromatic ring cleavage dioxygenases (EC 1.13.11.x) (16–20). In contrast, in Thalassospira, it remains largely unknown how these functional genes have been conserved evolutionally in different strains and how distinct they are from those in the known aromatic hydrocarbon-degrading bacterial groups.
In this study, comprehensive investigation of the genomes from Thalassospira spp. that were available in public databases was performed for the first time, in which the functional genes that were potentially involved in aromatic hydrocarbon biodegradation were screened across 33 Thalassospira genomes. Additionally, the complete genome of a new Thalassospira isolate, strain GO-4, which was obtained from a phenanthrene-enriched marine bacterial consortium (21), was sequenced and this organism was used as a model to link genomic data and metabolic functions. Through these investigations, this study aimed to predict the metabolic capability conserved in Thalassospira spp. and to understand their ecological roles in microbial ecosystems that drive aromatic hydrocarbon biodegradation.
RESULTS
Isolation of strain GO-4, which grows on CBA as the sole carbon source.
Thalassospira sp. strain GO-4 was isolated from a phenanthrene-enriched coastal marine bacterial consortium that has been maintained in the laboratory (see Materials and Methods and reference 21). Taxonomic analysis based on 16S rRNA gene sequencing indicated that strain GO-4 was most closely related to Thalassospira povalilytica strain Zumi 95T (99.93% identity) (Fig. 1), a polyvinyl alcohol-degrading organism that was isolated from Tokyo Bay (22). Although strain GO-4 was isolated from a phenanthrene-grown consortium, the isolate did not exhibit clear growth on phenanthrene or 1-hydroxy-2-naphthoic acid, a typically biodegraded downstream product of phenanthrene biodegradation (23), as a sole carbon source. When strain GO-4 cells were incubated with the putative further downstream compound of phenanthrene biodegradation, 2-carboxybenzaldehyde (CBA), cells showed an apparent increase in cell density, reaching stationary phase after around 24 h (Fig. 2A), and the spiral-rod-shaped cells multiplied in the culture (Fig. 2B).
FIG 1.
Neighbor-joining phylogenetic tree of 16S rRNA genes of strain GO-4 and reference Thalassospira strains. Strains with complete genomes available in databases are shown in boldface. Black diamonds indicate strains that possess the phdK-pht gene cluster in their genomes. The tree was created with 1,000 bootstrap iterations, and values below 50% are not reported.
FIG 2.
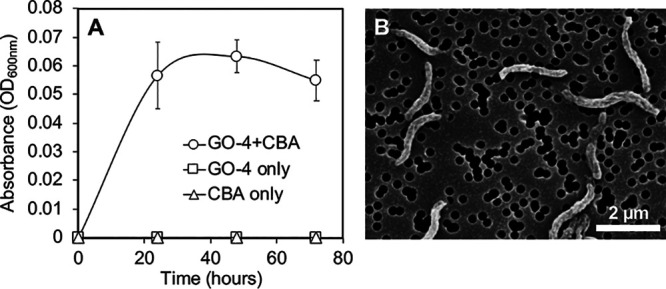
(A) Growth monitoring of strain GO-4 that grew on 100 mg L−1 2-carboxybenzaldehyde (CBA) as the sole carbon source. OD600 values of the cultures that contained bacterial cells with CBA (○; n = 3), bacterial cells only (□; n = 1), or CBA only (△; n = 1) are shown. (B) Scanning electron microscopic image of strain GO-4 cells that grew on CBA for 5 days.
The complete genome of strain GO-4 and taxonomic characterization.
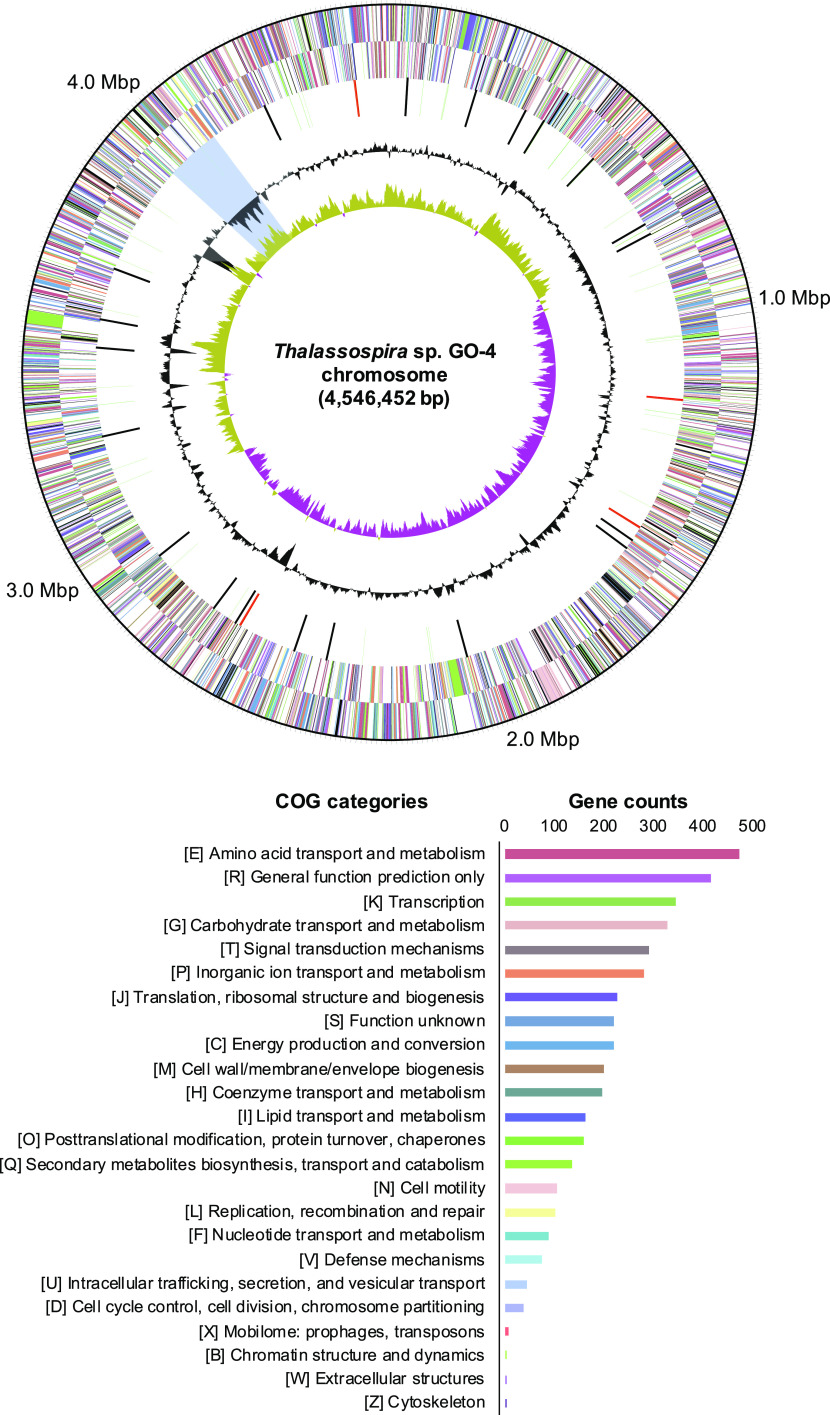
The complete genome sequence of strain GO-4 was successfully obtained through the hybrid assembly technique using the short-read (DNBSEQ) and long-read (GridION) sequencing data (see Table S1 in the supplemental material) (21). The genome of strain GO-4 consisted of a single circular chromosome with a size of 4,546,452 bp without plasmids (Fig. 3), and 4,046 coding genes (CDSs) were detected on the chromosome (according to the Prokaryotic Genome Annotation Pipeline [PGAP]). The COG categorization of these coding genes and their location on the chromosome are summarized in Fig. 3. The genome of strain GO-4 showed 97.7% average nucleic acid identity (ANI) to T. povalilytica Zumi 95T (draft genome; 21 contigs), 84.0% ANI to Thalassospira indica PB8BT (complete genome; 1 chromosome), 83.9% ANI to Thalassospira profundimaris WP0211T (draft genome; 28 contigs), and 83.0% ANI to Thalassospira australica NP 3b2T (draft genome; 32 contigs).
FIG 3.
Circular map of the strain GO-4 chromosome with functional annotation. Rings from outside to the center represent genes on the forward strand and reverse strand (colored by COG annotation categories, as listed in the bar graph with gene counts), RNA genes (tRNAs, green; rRNAs, red; other RNAs, black), GC content (gray), and GC skew (yellow and purple). The putative foreign gene-rich region with lower GC contents is shown with a blue background.
Functional genes of aromatic hydrocarbon degradation conserved in Thalassospira genomes.
As the potential marker genes responsible for aromatic hydrocarbon biodegradation, genes that were annotated as homologous genes of the known aromatic ring-hydroxylating dioxygenase genes or aromatic ring cleavage dioxygenase genes were screened in the genomes of strain GO-4 and the selected 32 Thalassospira strains in databases. Strain GO-4 and other 12 strains possessed homologous genes that encode the subunits of phthalate 4,5-dioxygenase Pht2/Pht3 (EC 1.14.12.7) in their genomes. However, genes that were annotated as other known aromatic ring-hydroxylating dioxygenase genes (EC 1.14.12.x) were not found in all genomes (Table 1). Regarding aromatic ring cleavage dioxygenase genes, all the selected 33 genomes possessed homologous genes for alpha/beta subunits of protocatechuate 3,4-dioxygenase PcaGH (EC 1.13.11.3), while genes for other known aromatic ring cleavage dioxygenases (EC 1.13.11.x) were not found except for catechol 2,3-dioxygenase XylE (EC 1.13.11.2) (Table 1); xylE was found in four reference genomes, although other xyl genes responsible for the downstream reactions of catechol degradation were not.
TABLE 1.
List of the representative known functional gene products for aromatic hydrocarbon degradation and number of the selected 33 Thalassospira genomes that were found to possess each gene
| Gene product | No. of Thalassospira genomes that have gene encoding the product shown |
|---|---|
| Aromatic ring-hydroxylating dioxygenase | |
| Benzene 1,2-dioxygenase (EC 1.14.12.3) | 0/33 |
| Phthalate 4,5-dioxygenase (EC 1.14.12.7) | 13/33 |
| Benzoate 1,2-dioxygenase (EC 1.14.12.10) | 0/33 |
| Toluene 2,3-dioxygenase (EC 1.14.12.11) | 0/33 |
| Naphthalene 1,2-dioxygenase (EC 1.14.12.12) | 0/33 |
| Biphenyl 2,3-dioxygenase (EC 1.14.12.18) | 0/33 |
| Aromatic ring cleavage dioxygenase | |
| Catechol 1,2-dioxygenase (EC 1.13.11.1) | 0/33 |
| Catechol 2,3-dioxygenase (EC 1.13.11.2) | 4/33 |
| Protocatechuate 3,4-dioxygenase (EC 1.13.11.3) | 33/33 |
| Protocatechuate 4,5-dioxygenase (EC 1.13.11.8) | 0/33 |
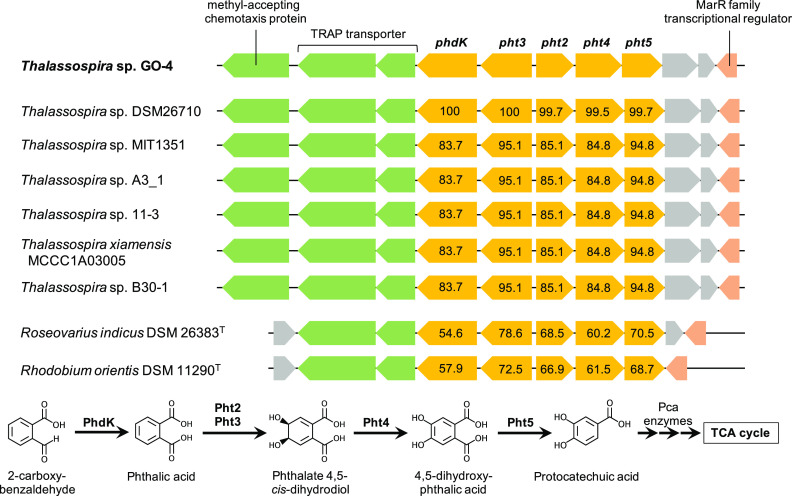
The pht2/pht3 gene homologs in the strain GO-4 genome were present as a putative pht gene cluster (Fig. 4), that shall be responsible for biotransformation of phthalic acid to protocatechuic acid (24, 25), and the pcaGH homologs were also found to be clustered with other pca genes that shall catalyze the ring cleavage of protocatechuic acid and downstream degradation to the tricarboxylic acid (TCA) cycle (26, 27) (Table 2). Degradation of phthalic acid shall be initiated from the addition of two oxygen atoms to the benzene ring by phthalate 4,5-dioxygenase Pht2/Pht3 and phthalate 4,5-cis-dihydrodiol dehydrogenase Pht4 (EC 1.3.1.64), and then subsequently decarboxylated by 4,5-dihydroxyphthalate decarboxylase Pht5 (EC 4.1.1.55) resulting in the generation of protocatechuic acid (Fig. 4). Furthermore, a gene that was located next to pht genes in the strain GO-4 genome was annotated as the phdK gene, which encodes 2-carboxybenzaldehyde dehydrogenase (EC 1.2.1.78) (28). This putative phdK-pht gene cluster, including the neighboring genes that potentially encode a methyl-accepting chemotaxis protein, tripartite ATP-independent periplasmic (TRAP) transporter—known to typically be involved in phthalate uptake (29)—and a MarR family transcriptional regulator, were found to be conserved in genomes of other Thalassospira strains available in databases (Fig. 4 and Table 3), throughout different phylogenetic branches (Fig. 1). The amino acid sequences of these putative PhdK and Pht enzymes showed 83 to 100% identities among Thalassospira strains, and the enzymes in Thalassospira sp. strain DSM26710 were most closely related to those in strain GO-4. This gene set was also found in the genomes of Roseovarius indicus DSM 26383T (IMG genome ID 2619618997; 54 to 79% amino acid identities), a Rhodobacteraceae bacterium that was isolated from a PAH-degrading marine bacterial consortium (30), and Rhodobium orientis DSM 11290T (IMG genome ID 2887621230; 57 to 73% amino acid identities), a phototrophic bacterium that was isolated from coastal seawater (31) (Fig. 4). Considering downstream metabolic processes, an aromatic ring cleavage dioxygenase, protocatechuate 3,4-dioxygenase PcaGH, and other Pca enzymes that are putatively encoded by the clustered pca gene homologs in the strain GO-4 genome should be responsible for biodegradation of protocatechuic acid into the TCA cycle. The pca gene cluster was found in all 33 Thalassospira genomes in the databases (Table 3).
FIG 4.
phdK-pht gene cluster that is potentially responsible for 2-carboxybenzaldehyde biotransformation and is conserved in the genomes of Thalassospira strains, Roseovarius indicus DSM 26383T, and Rhodobium orientis DSM 11290T. Amino acid sequence similarities (percentages) of PhdK and Pht enzymes to those of strain GO-4 are presented.
TABLE 2.
List of functional genes that were found as the two gene clusters in the strain GO-4 genome and are responsible for biodegradation of 2-carboxybenzaldehyde via phthalic acid and protocatechuic acid
| Gene, product | EC no. | COG category | aa length | Locus | IMG gene ID |
|---|---|---|---|---|---|
| phdK, 2-carboxybenzaldehyde dehydrogenase | 1.2.1.78 | C | 492 | 4025393–4026871 | 2963527656 |
| pht3, phthalate 4,5-dioxygenase oxygenase subunit | 1.14.12.7 | P | 425 | 4023933–4025210 | 2963527655 |
| pht2, phthalate 4,5-dioxygenase reductase subunit | 1.14.12.7 | C | 316 | 4022846–4023796 | 2963527654 |
| pht4, phthalate 4,5-cis-dihydrodiol dehydrogenase | 1.3.1.64 | R | 382 | 4021639–4022862 | 2963527653 |
| pht5, 4,5-dihydroxyphthalate decarboxylase | 4.1.1.55 | 330 | 4020650–4021642 | 2963527652 | |
| pcaB, 3-carboxy-cis,cis-muconate cycloisomerase | 5.5.1.2 | F | 445 | 1138719–1140056 | 2963525014 |
| pcaF, acetyl-CoA C-acyltransferase | 2.3.1.16 | I | 401 | 1140209–1141414 | 2963525015 |
| pcaJ, 3-oxoadipate CoA-transferase beta subunit | 2.8.3.6 | I | 257 | 1141438–1142211 | 2963525016 |
| pcaI, 3-oxoadipate CoA-transferase alpha subunit | 2.8.3.6 | I | 284 | 1142226–1143080 | 2963525017 |
| pcaG, protocatechuate 3,4-dioxygenase alpha subunit | 1.13.11.3 | Q | 195 | 1143223–1143810 | 2963525018 |
| pcaH, protocatechuate 3,4-dioxygenase beta subunit | 1.13.11.3 | Q | 233 | 1143814–1144515 | 2963525019 |
| pcaC, 4-carboxymuconolactone decarboxylase | 4.1.1.44 | R | 130 | 1144521–1144913 | 2963525020 |
| pcaD, 3-oxoadipate enol-lactonase | 3.1.1.24 | H | 266 | 1144906–1144706 | 2963525021 |
TABLE 3.
Information of selected Thalassospira genomes in the databases
| Organism | IMG genome ID | NCBI accession no. | Completeness (no. of contigs) | Presence of: |
|
|---|---|---|---|---|---|
| phdK-pht cluster | pca cluster | ||||
| Thalassospira sp. strain GO-4 | 2963523905 | ASM2358896 | Complete (1) | + | + |
| Thalassospira povalilytica Zumi 95T | ASM284423 | Draft (21) | + | ||
| Thalassospira australica NP3b2T | 2617271273 | ASM76329 | Draft (32) | + | |
| Thalassospira sp. strain HF15 | 2895203324 | ASM1180652 | Draft (25) | + | |
| Thalassospira indica PB8BT | 2840662270 | ASM340309 | Complete (1) | − | + |
| Thalassospira sp. strain MCCC 1A01148 | 2724679112 | ASM161379 | Draft (24) | + | |
| Thalassospira sp. strain MIT1351 | 2681813577 | Complete (1) | + | + | |
| Thalassospira sp. strain A40-3 | ASM1587148 | Complete (1) | − | + | |
| Thalassospira tepidiphila 1-1BT | 2829832560 | ASM1192768 | Draft (5) | + | |
| Thalassospira profundimaris WP0211T | 2529292925 | ASM30027 | Draft (28) | + | |
| Thalassospira sp. strain MCCC 1A02491 | 2724679110 | ASM161378 | Draft (33) | + | + |
| Thalassospira sp. strain MCCC 1A02803 | ASM198342 | Draft (22) | + | + | |
| Thalassospira sp. strain HJ | 2648501770 | ASM94841 | Draft (22) | + | |
| Thalassospira sp. strain DSM 26710 | 2728369743 | ASM361021 | Draft (12) | + | + |
| Thalassospira profundimaris S25-3-2 | 2808606712 | ASM332675 | Draft (85) | + | |
| Thalassospira marina CSC3H3T | 2775506933 | ASM284437 | Complete (2) | − | + |
| Thalassospira sp. strain TSL5-1 | ASM190769 | Draft (27) | + | ||
| Thalassospira xianhensis P-4T | ASM332647 | Draft (100) | + | ||
| Thalassospira sp. strain A3_1 | 2893749381 | ASM1774421 | Draft (11) | + | + |
| Thalassospira permensis NBRC 106175T | 2585427703 | Draft (72) | + | ||
| Thalassospira sp. strain MIT1004 | 2839174240 | ASM185854 | Draft (47) | + | + |
| Thalassospira sp. strain 11-3 | 2744054425 | ASM320199 | Draft (28) | + | + |
| Thalassospira xiamenensis M-5T | 2681813519 | ASM30023 | Complete (2) | − | + |
| Thalassospira xiamenensis IOP_1 | ASM2102127 | Draft (65) | + | + | |
| Thalassospira xiamenensis S27-11 | ASM332671 | Draft (56) | + | + | |
| Thalassospira xiamenensis MCCC 1A03005 | 2744054535 | ASM161816 | Draft (62) | + | + |
| Thalassospira sp. strain B30-1 | ASM1576757 | Complete (1) | + | + | |
| Thalassospira sp. strain KO164 | 2751185779 | Draft (2) | + | + | |
| Thalassospira lucentensis HF2 | 2915113161 | ASM1180642 | Draft (13) | + | |
| Thalassospira lohafexi 139Z-12T | ASM284427 | Draft (23) | + | ||
| Thalassospira lucentensis DSM 14000T | 2526164517 | ASM42126 | Draft (22) | + | |
| Thalassospira alkalitolerans JCM 18968T | 2791355065 | ASM211574 | Draft (72) | + | |
| Thalassospira mesophila JCM 18969T | 2791355063 | ASM211575 | Draft (94) | + | |
The phdK-pht gene cluster was located on a foreign gene-rich region of the strain GO-4 genome.
Sequence alignment comparison between genomes of strain GO-4 and the closest strain, T. povalilytica Zumi 95T, indicated that the lack of the phdK-pht gene cluster in the strain Zumi95T draft genome was not due to sequencing gaps between different contigs (Fig. 5). In fact, the phdK-pht gene cluster of strain GO-4 was located on the less-conserved genomic region (approximately genome positions 3.98 to 4.06 Mb) (Fig. 5), which showed lower GC content than other regions (Fig. 3), suggesting that this region was inserted and enriched with horizontally acquired genes (32, 33). This region also included functional gene operons for nitrate reductase, nitric oxide reductase, isoquinoline oxidoreductase, and ribose transport systems (Table S2). Through further alignment comparison against the genome of strain DSM 26710, which possessed the phdK-pht gene cluster most closely related to that of strain GO-4 (Fig. 4), the phdK-pht gene cluster and surrounding operons were found to be distributed at different locations and in different orientations in the strain DSM 26710 genome, which may indicate that these operons were individually mobilized and transposed (Fig. 5).
FIG 5.
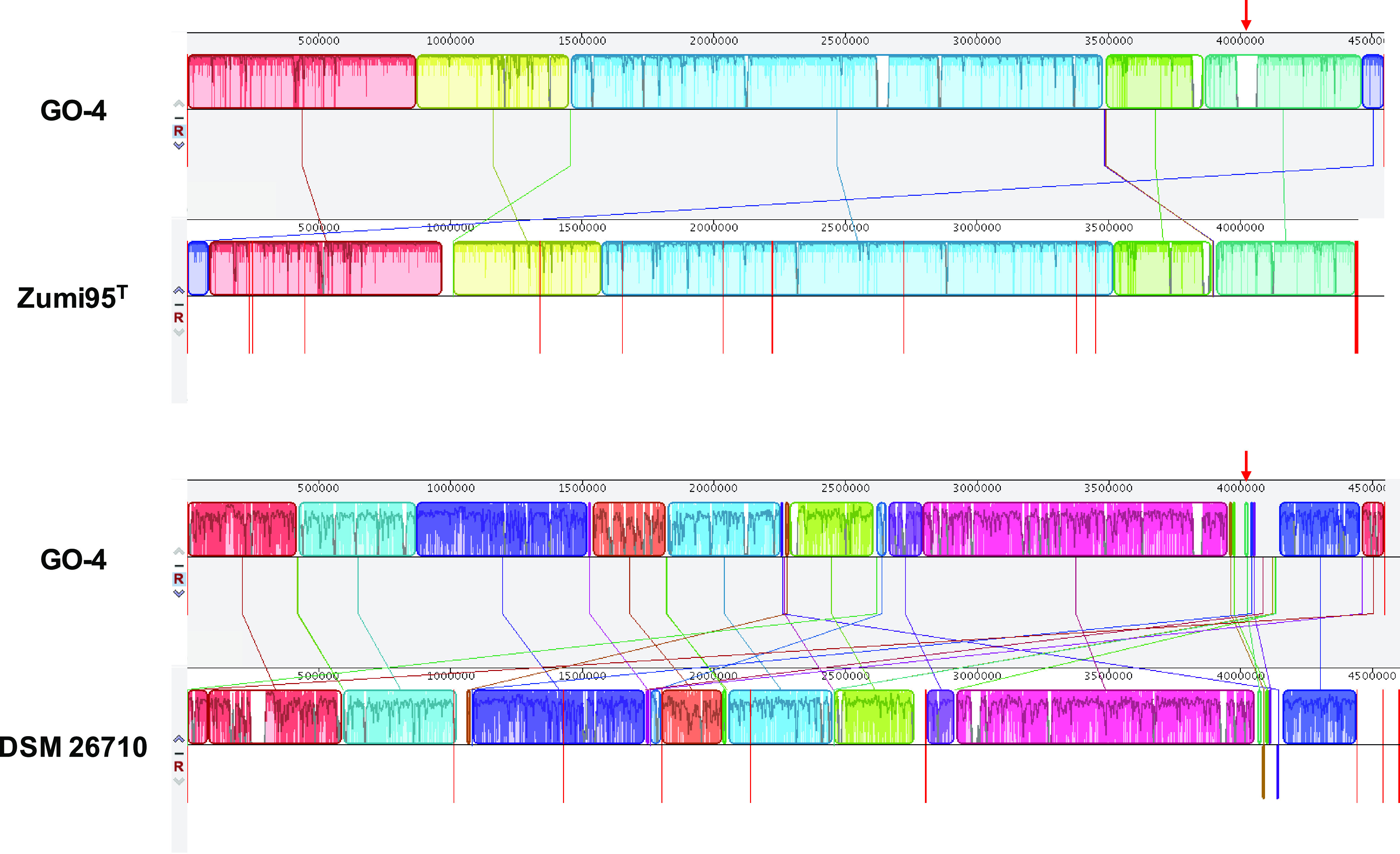
Sequence alignment comparisons of the strain GO-4 genome against the genomes of T. povalilytica Zumi 95T (above) and Thalassospira sp. strain DSM 26710 (below). Red arrows indicate the location of phdK-pht gene cluster that was located on the putative foreign gene-rich genomic region.
Identification of biotransformation products of CBA by liquid chromatography-electrospray ionization tandem mass spectrometry.
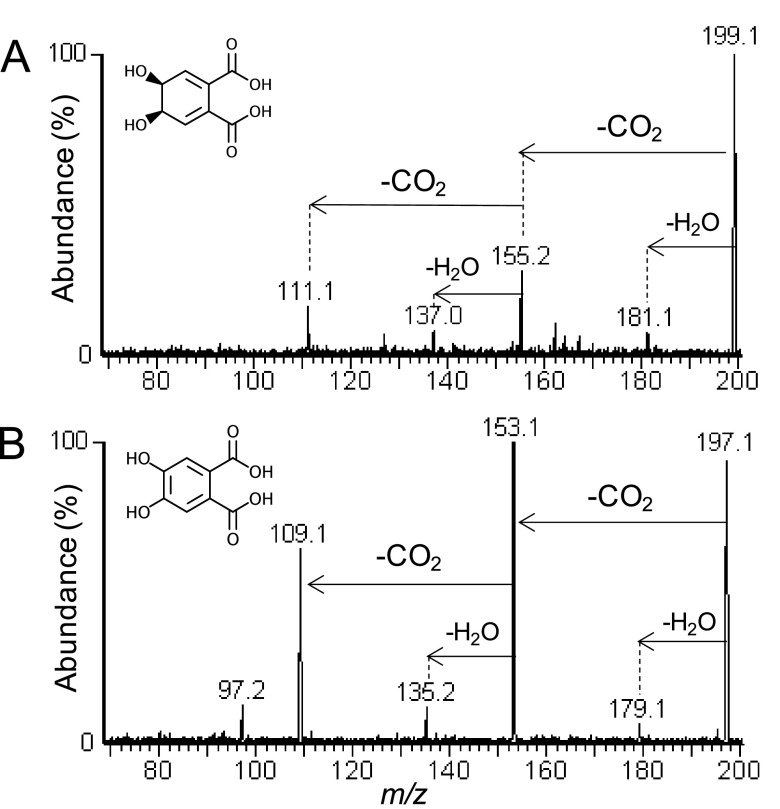
When strain GO-4 was grown on CBA as the sole carbon source, two biodegradation products were detected by liquid chromatography-electrospray ionization tandem mass spectrometry [LC-ESI(−)-MS/MS] analyses that corresponded to the deprotonated molecules [M – H]− = 199 and [M – H]− = 197. As shown in Fig. 6A and B, results of product ion scan analyses of these compounds revealed similar mass spectra in that two consecutive losses of 44 Da each as CO2 were detected as the most abundant fragments during collision-induced dissociation (CID) of both compounds. In the case of the compound that corresponded to [M – H]− = 199, m/z 155 and m/z 111 were observed (Fig. 6A). The fragment m/z 181 represented a loss of water as 18 Da from the parent deprotonated molecule and the fragment m/z 137 represented losses of CO2 and water (Fig. 6A). When these results were considered in combination with a molar mass of 200 Da and a molecular formula of C8H8O6, they provided evidence that this compound was a phthalate-dihydrodiol. As shown in Fig. 6B, results of CID of the compound that corresponded to [M – H]− = 197 revealed losses of 44 Da (CO2) and 88 Da (2CO2) from the parent deprotonated molecule as m/z 153 and m/z 109, respectively; losses of water occurred identically to those described above. Taken together with a molar mass of 198 and a molecular formula of C8H6O6, these results provided evidence that this compound was a dehydroxylated phthalic acid. Shown in Fig. 6 are the compounds phthalate 4,5-cis-dihydrodiol (Fig. 6A) and 4,5-dihydroxyphthalic acid (Fig. 6B), which were reported as the biotransformation products of phthalic acid by the enzymes Pht2/Pht3 and Pht4 (Fig. 4) (25). When strain GO-4 was incubated with a mixture of 100 mg L−1 each of CBA and phenanthrene, phenanthrene loss was not detected after 8 days of incubation, which confirmed that strain GO-4 was unable to biodegrade this compound, even though cells grew by consuming CBA (Fig. S1).
FIG 6.
LC-ESI(−)-MS/MS product ion scan mass spectra of the biotransformation products that corresponded to (A) [M – H]− = 199, which was assigned as phthalate 4,5-cis-dihydrodiol, and (B) [M – H]− = 197, which was assigned as 4,5-dihydroxyphthalic acid.
DISCUSSION
The putative CBA-degrading gene cluster that was conserved in Thalassospira.
2-Carboxybenzaldehyde (CBA), phthalic acid, and protocatechuic acid are known as the intermediate products in one of the typical phenanthrene biodegradation pathways, the so-called “phthalate pathway,” which was previously reported in both Gram-negative (Pseudomonas spp.) and Gram-positive (Mycobacterium and Nocardioides spp.) bacteria (23, 34, 35). The key reactions for the phthalate pathway shall be the transformation of 1-hydroxy-2-naphthoic acid to phthalic acid via CBA, instead of generating 1,2-dihydroxynaphthalene that is further degraded via salicylic acid (the so-called “salicylate pathway”). Functional genes that are responsible for those key reactions were previously characterized in Mycobacterium and Nocardioides spp. as the phd gene cluster: 1-hydroxy-2-naphthoate dioxygenase (PhdI; EC 1.13.11.38) and cis-2′-carboxybenzalpyruvate aldolase (PhdJ; EC 4.1.2.34) convert 1-hydroxy-2-naphthoic acid to CBA, which is further transformed to phthalic acid by CBA dehydrogenase (PhdK) (28, 36, 37). The phdK gene had so far only been found and characterized in Nocardioides strain KP7 (28), in which the phdK gene was clustered with phdI and phdJ genes but not with pht genes for phthalic acid degradation. The amino acid sequences of the putative PhdK enzyme in strain GO-4 (492 amino acids [aa]) and PhdK in Nocardioides strain KP7 (485 aa) shared 40.6% sequence similarity, and both consisted of one domain of the aldehyde dehydrogenase (IPR015590). In Thalassospira, homologous genes of phdI or phdJ genes were not found in their genomes in databases, which is consistent with the fact that strain GO-4 did not grow on 1-hydroxy-2-naphthoic acid. The putative phdK-pht gene cluster that was found in Thalassospira spp. can be considered to be responsible for their ability to grow on CBA as the sole carbon source. To the best of our knowledge, this study is the first report of this putative novel functional gene cluster, whereas further transcriptomic and/or enzymatic examinations are required to fully prove their functions. Although CBA has often been identified as an intermediate product of PAH biodegradation, the bacterial ability to utilize CBA as the sole growth substrate has not been much studied. A previous study reported a Thalassospira isolate that was capable of degrading and growing on phthalic acid (38); however, the functional genes that were responsible for the organism’s metabolism were not adequately characterized until the current study. The phdK-pht gene cluster was located in a putative foreign gene-rich region of the strain GO-4 genome (Fig. 5) and was found to be shared among genomes of at least 12 other Thalassospira strains and even at least two other bacterial genera, Roseovarius and Rhodobium (Fig. 4). Thus, this gene cluster had likely been acquired in the genomes of these strains by horizontal gene transfers and may have benefited them to inhabit aromatic hydrocarbon-exposed environments.
Potential of Thalassospira to biotransform PAHs.
Comparative genomic analyses of the Thalassospira genomes in the current study indicated that homologous genes for an aromatic ring-hydroxylating dioxygenase Pht2/Pht3 and an aromatic ring cleavage dioxygenase PcaGH were commonly found in Thalassospira in addition to other genes necessary for further downstream reactions that were clustered with them. However, any related genes of the well-studied pah, phn, or nah genes, which are reported to be responsible for the upstream reactions of phenanthrene/naphthalene degradation and thus are the functional marker genes for PAH degradation (4, 39, 40), were not found in the genomes of strain GO-4 and all other 32 Thalassospira strains that were investigated, including strains TSL5-1, T. xianhensis P-4T, and T. tepidiphila 1-1BT, which were previously claimed to be potential PAH-degrading strains (13–15). In the literature, there has been no discussion regarding the functional genes or enzymes that were responsible for PAH biotransformation in these strains, and thus these are still open questions. Genomes of these strains are currently all incomplete (27, 100, and 5 uncirculated contigs were deposited for the genomes of strains TSL5-1, P-4T, and 1-1BT, respectively); therefore, the possibility that other considerable functional genes were overlooked cannot be eliminated. However, according to the genomic overview of Thalassospira strains obtained in the current study, members of Thalassospira had more likely evolved by adapting to metabolize the downstream, single-ring aromatic acids such as CBA, phthalic acid, and protocatechuic acid. Even if Thalassospira strains did not possess the known PAH degradation genes and thus were incapable of growing on PAHs as the sole carbon sources, these PAHs may have been biotransformed by Thalassospira through cometabolic reactions (41–43), whereas this phenomenon was not observed in strain GO-4 (see Fig. S1 in the supplemental material).
Insights into the ecological niches of Thalassospira in aromatic hydrocarbon-exposed environments.
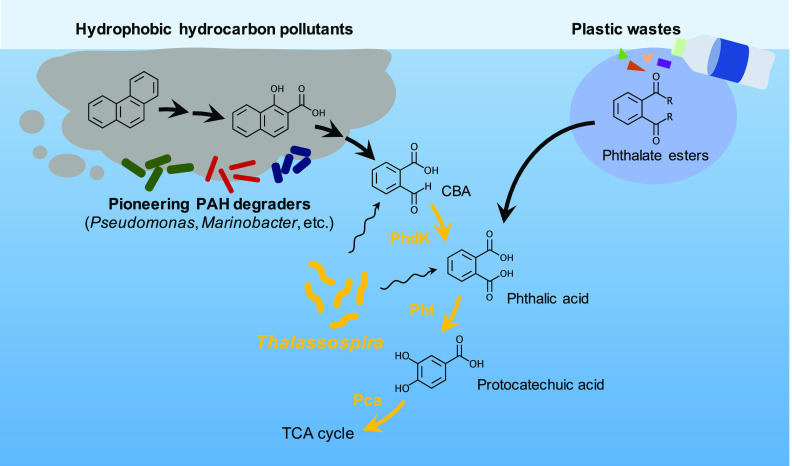
Certain microbial groups that are capable of utilizing complex, hydrophobic aromatic compounds like PAHs for their growth and degrading them into more bioavailable compounds are considered to play important roles in nutrient flows in microbial ecosystems, in which other coexisting (secondary) microorganisms grow on metabolites of these pioneers and thus are metabolically dependent upon them (7, 44, 45). Although Thalassospira has often been detected as an abundant member of PAH-exposed microbial communities in previous studies (9–12), Thalassospira species were not characterized as the pioneering players of PAH degradation. For example, in a previous study by Wang et al. that worked on a phenanthrene-degrading marine bacterial consortium (9), Thalassospira was characterized as one of the contributing members that potentially degraded protocatechuate and/or gentisate produced by the pioneer phenanthrene-degrading Marinobacter and Martelella. Consistent with these previous reports, the current study suggested that members of Thalassospira have conserved the ability to grow on single-ring aromatic acids like CBA, phthalic acid, and protocatechuic acid, while lacking known PAH degradation genes; thus, they were more likely metabolically dependent on the coexisting pioneering PAH degraders to grow in PAH-exposed communities. Some members of Thalassospira seemed to have acquired the phdK-pht genes for CBA and phthalic acid degradation via horizontal gene transfers, in addition to pca genes for the downstream protocatechuic acid degradation, which was unexceptionally conserved in Thalassospira. Acquisition of the ability to utilize CBA as a growth substrate shall have advantaged Thalassospira by allowing them to scavenge these aromatic acids after they became bioavailable from more hydrophobic aromatic hydrocarbon sources (Fig. 7), and hypothetically facilitated Thalassospira to commonly inhabit the PAH-exposed environments by outcompeting other microbial groups. Furthermore, the ability to utilize phthalic acid may allow Thalassospira to adapt to environments that are polluted with phthalate esters that are derived from plastic wastes (46–48). Strain GO-4 was found to possess at least six different types of esterase-encoding genes; thus, Thalassosopira may even contribute to converting phthalate esters to phthalic acid, although no previous study has reported this. Thalassospira was also reported to have the abilities of biosurfactant production (49, 50) and biofilm formation (51, 52), which are known as typical strategies to utilize aromatic hydrocarbons (53) and thus also may allow them to dominate aromatic hydrocarbon-degrading microbial communities. Furthermore, Thalassospira potentially indirectly contributes to PAH degradation in microbial communities, by mediating synergistic PAH degradation (54) and/or suppressing growth inhibitors of the PAH-degrading pioneers (55) as the other bacterial groups were reported to do in PAH-degrading communities.
FIG 7.
Schematic model summarizing the putative ecological features of Thalassospira in aromatic hydrocarbon-exposed marine environments. Thalassospira occupied niches that utilize single-ring aromatic acids (CBA, phthalic acid, and protocatechuic acid) that are released from complex, hydrophobic aromatic hydrocarbon pollutants. In PAH-exposed environments, Thalassospira likely grew through metabolic dependencies on the pioneering members that possessed specialized enzymes for PAH degradation and thus were responsible for upstream biotransformations. Acquisition of the ability to utilize CBA in Thalassospira may have advantaged them to become abundant in PAH-exposed communities, by allowing them to scavenge the relatively less hydrophobic aromatic acids (CBA and phthalic acid) right after becoming available from the more hydrophobic aromatic hydrocarbon sources that originated from ocean oil spill or plastic waste pollution.
In summary, this study provided the first comprehensive functional profile of Thalassospira spp. that expanded scientific insight into their potential ecological roles in aromatic hydrocarbon-exposed environments. Further growth assays and metabolite profiling of a new Thalassospira isolate strain GO-4 supported the linkage of the metabolic capabilities of strain GO-4 and the results of genomic analyses. In PAH-exposed environments, Thalassospira spp. may be adapted to utilize the single-ring aromatic acids (CBA, phthalic acid, and protocatechuic acid) that are released from the degradation of more hydrophobic aromatic substrates, rather than playing a pioneering role in PAH biodegradation. Thus, Thalassospira spp. likely co-occupy niches with other pioneer PAH-degrading players that provide them with biodegradation intermediates that may have allowed them to be commonly found as abundant members of microbial communities that developed under exposure to PAHs (9–12). The high culturability of Thalassospira in laboratories shall benefit future studies using their pure cultures, which may allow us to discover unknown metabolic functions and to evaluate their potential for biotechnological usages.
MATERIALS AND METHODS
Chemicals.
2-Carboxybenzaldehyde (>98% purity) was purchased from Tokyo Chemical Industries (Tokyo, Japan). Phthalic acid (98% purity), N,N-dimethylformamide (DMF) (>99% purity), methanol (LC-MS grade), and ethyl acetate (high-performance liquid chromatography [HPLC] grade) were purchased from Wako Chemical (Osaka, Japan). 1-Hydroxy-2-naphthoic acid (>97% purity) was purchased from Kanto Chemical Co. (Tokyo, Japan). Phenanthrene (98% purity) was purchased from Sigma-Aldrich (St. Louis, MO, USA).
Bacterial isolation and growth conditions.
Thalassospira sp. strain GO-4 was isolated from a phenanthrene-enriched marine bacterial consortium that originated from the coast of Nojima, Yokohama, Japan (21). The sampling site was not exposed to excess hydrocarbon pollutions such as oil spills, but was surrounded by urban beaches that may face continuous inputs of hydrophobic hydrocarbon pollutants and plastic debris (56). The bacterial consortium was grown on 50 mg L−1 phenanthrene as the sole carbon source in Artificial Sea Water medium (57), and bacterial cells were repeatedly transferred to a new medium once per month. Strain GO-4 was isolated from this consortium through dilution-to-extinction techniques using 10 mM glucose as the carbon source, and its purity was confirmed by sequencing analysis of 16S rRNA gene (Macrogen Japan Co., Tokyo, Japan) and microscopic observation using a light microscope (Eclipse E800; Nikon, Tokyo, Japan) or a scanning electron microscope (JSM-6000 NeoScope; JEOL, Tokyo, Japan) after collecting cells on a 0.2-μm-pore polycarbonate membrane filter (Isopore, Merck Millipore, Tullagreen, Ireland). To evaluate the growth capability of strain GO-4 on different carbon sources (phenanthrene, 1-hydroxy-2-naphthoic acid, or 2-carboxybenzaldehyde dissolved in DMF prior to addition), cells were incubated under aerobic conditions with 50 to 100 mg L−1 of the selected substrates for ~8 days by rotary shaking at 120 rpm in the dark at 30°C. Bacterial growth was evaluated by visual inspection of turbidity and by measuring the optical density of the cultures at 600 nm (OD600).
Complete genome sequencing of strain GO-4 and comparative genomic analyses.
The complete genome sequence of strain GO-4 was recently announced with detailed methodological information regarding the sequencing analyses and data processing (21). In brief, genomic DNA of strain GO-4 was extracted from a glucose-grown culture by using the NucleoBond high-molecular-weight DNA kit (Macherey-Nagel, Düren, Germany) and was subjected to complete genome sequencing that employed a hybrid assembly of short-read (DNBSEQ-G400; MGI Tech, Shenzhen, China) and long-read (GridION X5; Oxford Nanopore Tech, Oxford, United Kingdom) sequencing technologies. Gene annotation was performed by using the annotation pipelines of NCBI (PGAP v.6.1) and JGI (IMG annotation pipeline v.5.1.6). Comparative genomic analyses using the reference Thalassospira genomes deposited in public databases (NCBI and IMG) were conducted as described in a previous work (58); average nucleic acid identity (ANI) among Thalassospira strains was determined using FastANI (v.1.32) (59), and sequence alignment comparisons between Thalassospira genomes were performed by using Mauve (v.2.4.0) (60).
Profiling of biotransformation products of strain GO-4 by LC-ESI(−)-MS/MS.
Metabolic products released by strain GO-4 cells that grew on 50 mg L−1 CBA were extracted and analyzed by LC-ESI(−)-MS/MS according to Tomiyama et al. (61). Briefly, cultures were extracted with equal volumes of ethyl acetate at pH 7 and pH 2 (pH adjustment by HCl) by liquid-liquid extraction, after which organic phases were filtered through anhydrous sodium sulfate and concentrated in vacuo at 28°C via rotary evaporation (Eyela model N-1000; Tokyo Rikakikai Co., Ltd., Tokyo, Japan). Residues were resuspended in 1 mL of methanol and analyzed using a Waters 2690 separations module delivery system in line with a Quattro Ultima triple-stage quadrupole mass spectrometer (Micromass, Manchester, United Kingdom). Extracts were eluted isocratically in 77% methanol/water at a flow rate of 0.3 mL/min and separated on a Shimadzu Shim-pack XR-ODS column (150 mm by 3.0-mm inside diameter, 2.2-μm particle size) that was in line with a Security Guard cartridge system precolumn fitted with a Widepore C18 cartridge (Phenomenex, CA). Data were processed by MassLynx software v.4.1.
Data availability.
The genome sequence of strain GO-4 has been deposited in NCBI GenBank under accession no. CP097807 and in the IMG/MER database under accession no. 2963523905. The raw sequences are available from SRA accession no. SRR19369649 and SRR19369650 under BioProject no. PRJNA841389 and BioSample no. SAMN28591689.
ACKNOWLEDGMENT
This research work was funded by the Japanese Society for the Promotion and Science (JSPS) KAKENHI grant 19K15738 and 22K14813 to J.F.M.
Footnotes
Supplemental material is available online only.
Contributor Information
Jiro F. Mori, Email: morij@yokohama-cu.ac.jp.
Jeffrey A. Gralnick, University of Minnesota
REFERENCES
- 1.Head IM, Jones DM, Röling WFM. 2006. Marine microorganisms make a meal of oil. Nat Rev Microbiol 4:173–182. doi: 10.1038/nrmicro1348. [DOI] [PubMed] [Google Scholar]
- 2.Martin F, Malagnoux L, Violet F, Jakoncic J, Jouanneau Y. 2013. Diversity and catalytic potential of PAH-specific ring-hydroxylating dioxygenases from a hydrocarbon-contaminated soil. Appl Microbiol Biotechnol 97:5125–5135. doi: 10.1007/s00253-012-4335-2. [DOI] [PubMed] [Google Scholar]
- 3.DeBruyn JM, Chewning CS, Sayler GS. 2007. Comparative quantitative prevalence of mycobacteria and functionally abundant nidA, nahAc, and nagAc dioxygenase genes in coal tar contaminated sediments. Environ Sci Technol 41:5426–5432. doi: 10.1021/es070406c. [DOI] [PubMed] [Google Scholar]
- 4.Liang C, Huang Y, Wang H. 2022. pahE, a functional marker gene for polycyclic aromatic hydrocarbon-degrading bacteria. Appl Environ Microbiol 85:e02399-18. doi: 10.1128/AEM.02399-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tremblay J, Fortin N, Elias M, Wasserscheid J, King TL, Lee K, Greer CW. 2019. Metagenomic and metatranscriptomic responses of natural oil degrading bacteria in the presence of dispersants. Environ Microbiol 21:2307–2319. doi: 10.1111/1462-2920.14609. [DOI] [PubMed] [Google Scholar]
- 6.Handley KM, Piceno YM, Hu P, Tom LM, Mason OU, Andersen GL, Jansson JK, Gilbert JA. 2017. Metabolic and spatio-taxonomic response of uncultivated seafloor bacteria following the Deepwater Horizon oil spill. ISME J 11:2569–2583. doi: 10.1038/ismej.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mori JF, Kanaly RA. 2020. Multispecies diesel fuel biodegradation and niche formation are ignited by pioneer hydrocarbon-utilizing proteobacteria in a soil bacterial consortium. Appl Environ Microbiol 87:e02268-20. doi: 10.1128/AEM.02268-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.López-López A, Pujalte MJ, Benlloch S, Mata-Roig M, Rosselló-Mora R, Garay E, Rodríguez-Valera F. 2002. Thalassospira lucentensis gen. nov., sp. nov., a new marine member of the alpha-Proteobacteria. Int J Syst Evol Microbiol 52:1277–1283. doi: 10.1099/00207713-52-4-1277. [DOI] [PubMed] [Google Scholar]
- 9.Wang C, Huang Y, Zhang Z, Wang H. 2018. Salinity effect on the metabolic pathway and microbial function in phenanthrene degradation by a halophilic consortium. AMB Express 8:67. doi: 10.1186/s13568-018-0594-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo G, He F, Tian F, Huang Y, Wang H. 2016. Effect of salt contents on enzymatic activities and halophilic microbial community structure during phenanthrene degradation. Int Biodeterior Biodegradation 110:8–15. doi: 10.1016/j.ibiod.2016.02.007. [DOI] [Google Scholar]
- 11.Plotnikova EG, Anan’ina LN, Krausova VI, Ariskina EV, Prisyazhnaya NV, Lebedev AT, Demakov VA, Evtushenko LI. 2011. Thalassospira permensis sp. nov., a new terrestrial halotolerant bacterium isolated from a naphthalene-utilizing microbial consortium. Microbiology 80:703–712. doi: 10.1134/S0026261711050109. [DOI] [PubMed] [Google Scholar]
- 12.Gallego S, Vila J, Tauler M, Nieto JM, Breugelmans P, Springael D, Grifoll M. 2014. Community structure and PAH ring-hydroxylating dioxygenase genes of a marine pyrene-degrading microbial consortium. Biodegradation 25:543–556. doi: 10.1007/s10532-013-9680-z. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H, Wang H, Huang Y, Fang T. 2016. Characterization of pyrene degradation by halophilic Thalassospira sp. strain TSL5-1 isolated from the coastal soil of Yellow Sea, China. Int Biodeterior Biodegradation 107:62–69. doi: 10.1016/j.ibiod.2015.10.022. [DOI] [Google Scholar]
- 14.Zhao B, Wang H, Li R, Mao X. 2010. Thalassospira xianhensis sp. nov., a polycyclic aromatic hydrocarbon-degrading marine bacterium. Int J Syst Evol Microbiol 60:1125–1129. doi: 10.1099/ijs.0.013201-0. [DOI] [PubMed] [Google Scholar]
- 15.Kodama Y, Stiknowati LI, Ueki A, Ueki K, Watanabe K. 2008. Thalassospira tepidiphila sp. nov., a polycyclic aromatic hydrocarbon-degrading bacterium isolated from seawater. Int J Syst Evol Microbiol 58:711–715. doi: 10.1099/ijs.0.65476-0. [DOI] [PubMed] [Google Scholar]
- 16.Stingley RL, Brezna B, Khan AA, Cerniglia CE. 2004. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR-1. Microbiology (Reading) 150:3749–3761. doi: 10.1099/mic.0.27263-0. [DOI] [PubMed] [Google Scholar]
- 17.Moser R, Stahl U. 2001. Insights into the genetic diversity of initial dioxygenases from PAH-degrading bacteria. Appl Microbiol Biotechnol 55:609–618. doi: 10.1007/s002530000489. [DOI] [PubMed] [Google Scholar]
- 18.Pinyakong O, Habe H, Omori T. 2003. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J Gen Appl Microbiol 49:1–19. doi: 10.2323/jgam.49.1. [DOI] [PubMed] [Google Scholar]
- 19.Maeda AH, Nishi S, Hatada Y, Ohta Y, Misaka K, Kunihiro M, Mori JF, Kanaly RA. 2020. Chemical and genomic analyses of polycyclic aromatic hydrocarbon biodegradation in Sphingobium barthaii KK22 reveals divergent pathways in soil sphingomonads. Int Biodeterior Biodegradation 151:104993. doi: 10.1016/j.ibiod.2020.104993. [DOI] [Google Scholar]
- 20.Parales RE, Resnick SM. 2004. Aromatic hydrocarbon dioxygenases, p 175–195. In Singh A, Ward OP (ed), Biodegradation and bioremediation. Soil biology, vol 2. Springer, Berlin, Germany. [Google Scholar]
- 21.Kayama G, Kanaly RA, Mori JF. 2022. Complete genome sequence of Thalassospira sp. strain GO-4, a marine bacterium isolated from a phenanthrene-enriched bacterial consortium. Microbiol Resour Announc 11:e00532-22. doi: 10.1128/mra.00532-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogi Y, Yoshizumi M, Miyazaki M. 2014. Thalassospira povalilytica sp. nov., a polyvinyl-alcohol-degrading marine bacterium. Int J Syst Evol Microbiol 64:1149–1153. doi: 10.1099/ijs.0.058321-0. [DOI] [PubMed] [Google Scholar]
- 23.Samanta SK, Chakraborti AK, Jain RK. 1999. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Appl Microbiol Biotechnol 53:98–107. doi: 10.1007/s002530051621. [DOI] [PubMed] [Google Scholar]
- 24.Batie CJ, LaHaie E, Ballou DP. 1987. Purification and characterization of phthalate oxygenase and phthalate oxygenase reductase from Pseudomonas cepacia. J Biol Chem 262:1510–1518. doi: 10.1016/S0021-9258(19)75664-6. [DOI] [PubMed] [Google Scholar]
- 25.Nomura Y, Nakagawa M, Ogawa N, Harashima S, Oshima Y. 1992. Genes in PHT plasmid encoding the initial degradation pathway of phthalate in Pseudomonas putida. J Ferment Bioeng 74:333–344. doi: 10.1016/0922-338X(92)90028-S. [DOI] [Google Scholar]
- 26.Fujisawa H, Hayaishi O. 1968. Protocatechuate 3,4-dioxygenase. I. Crystallization and characterization. J Biol Chem 243:2673–2681. doi: 10.1016/S0021-9258(18)93425-3. [DOI] [PubMed] [Google Scholar]
- 27.Buchan A, Collier LS, Neidle EL, Moran MA. 2000. Key aromatic-ring-cleaving enzyme, protocatechuate 3,4-dioxygenase, in the ecologically important marine Roseobacter lineage. Appl Environ Microbiol 66:4662–4672. doi: 10.1128/AEM.66.11.4662-4672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iwabuchi T, Harayama S. 1997. Biochemical and genetic characterization of 2-carboxybenzaldehyde dehydrogenase, an enzyme involved in phenanthrene degradation by Nocardioides sp. strain KP7. J Bacteriol 179:6488–6494. doi: 10.1128/jb.179.20.6488-6494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boll M, Geiger R, Junghare M, Schink B. 2020. Microbial degradation of phthalates: biochemistry and environmental implications. Environ Microbiol Rep 12:3–15. doi: 10.1111/1758-2229.12787. [DOI] [PubMed] [Google Scholar]
- 30.Lai Q, Zhong H, Wang J, Yuan J, Sun F, Wang L, Zheng T, Shao Z. 2011. Roseovarius indicus sp. nov., isolated from deep-sea water of the Indian Ocean. Int J Syst Evol Microbiol 61:2040–2044. doi: 10.1099/ijs.0.023168-0. [DOI] [PubMed] [Google Scholar]
- 31.Hiraishi A, Urata K, Satoh T. 1995. A new genus of marine budding phototrophic bacteria, Rhodobium gen. nov., which includes Rhodobium orientis sp. nov. and Rhodobium marinum comb. nov. Int J Syst Evol Microbiol 45:226–234. [DOI] [PubMed] [Google Scholar]
- 32.Daubin V, Lerat E, Perrière G. 2003. The source of laterally transferred genes in bacterial genomes. Genome Biol 4:R57. doi: 10.1186/gb-2003-4-9-r57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gomes ALC, Johns NI, Yang A, Velez-Cortes F, Smillie CS, Smith MB, Alm EJ, Wang HH. 2020. Genome and sequence determinants governing the expression of horizontally acquired DNA in bacteria. ISME J 14:2347–2357. doi: 10.1038/s41396-020-0696-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun S, Wang H, Chen Y, Lou J, Wu L, Xu J. 2019. Salicylate and phthalate pathways contributed differently on phenanthrene and pyrene degradations in Mycobacterium sp. WY10. J Hazard Mater 364:509–518. doi: 10.1016/j.jhazmat.2018.10.064. [DOI] [PubMed] [Google Scholar]
- 35.Iwabuchi T, Inomata-Yamauchi Y, Katsuta A, Harayama S. 1998. Isolation and characterization of marine Nocardiodes capable of growing and degrading phenanthrene at 42°C. J Mar Biotechnol 6:86–90. [Google Scholar]
- 36.Pagnout C, Frache G, Poupin P, Maunit B, Muller J-F, Férard J-F. 2007. Isolation and characterization of a gene cluster involved in PAH degradation in Mycobacterium sp. strain SNP11: expression in Mycobacterium smegmatis mc2155. Res Microbiol 158:175–186. doi: 10.1016/j.resmic.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Stingley RL, Khan AA, Cerniglia CE. 2004. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR-1. Biochem Biophys Res Commun 322:133–146. doi: 10.1016/j.bbrc.2004.07.089. [DOI] [PubMed] [Google Scholar]
- 38.Iwaki H, Nishimura A, Hasegawa Y. 2012. Isolation and characterization of marine bacteria capable of utilizing phthalate. World J Microbiol Biotechnol 28:1321–1325. doi: 10.1007/s11274-011-0925-x. [DOI] [PubMed] [Google Scholar]
- 39.Goyal AK, Zylstra GJ. 1997. Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni. J Ind Microbiol Biotechnol 19:401–407. doi: 10.1038/sj.jim.2900476. [DOI] [PubMed] [Google Scholar]
- 40.Laurie AD, Lloyd-Jones G. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J Bacteriol 181:531–540. doi: 10.1128/JB.181.2.531-540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Herwijnen R, Wattiau P, Bastiaens L, Daal L, Jonker L, Springael D, Govers HAJ, Parsons JR. 2003. Elucidation of the metabolic pathway of fluorene and cometabolic pathways of phenanthrene, fluoranthene, anthracene and dibenzothiophene by Sphingomonas sp. LB126. Res Microbiol 154:199–206. doi: 10.1016/S0923-2508(03)00039-1. [DOI] [PubMed] [Google Scholar]
- 42.Nzila A. 2013. Update on the cometabolism of organic pollutants by bacteria. Environ Pollut 178:474–482. doi: 10.1016/j.envpol.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 43.Izawa M, Sakai M, Mori JF, Kanaly RA. 2021. Cometabolic benzo[a]pyrene biotransformation by Sphingobium barthaii KK22 proceeds through the kata-annelated ring and 1-pyrenecarboxylic acid to downstream products. J Hazard Mater Adv 4:100018. doi: 10.1016/j.hazadv.2021.100018. [DOI] [Google Scholar]
- 44.Louvado A, Gomes NCM, Simões MMQ, Almeida A, Cleary DFR, Cunha A. 2015. Polycyclic aromatic hydrocarbons in deep sea sediments: microbe-pollutant interactions in a remote environment. Sci Total Environ 526:312–328. doi: 10.1016/j.scitotenv.2015.04.048. [DOI] [PubMed] [Google Scholar]
- 45.Gao Y, Guo S, Wang J, Li D, Wang H, Zeng D-H. 2014. Effects of different remediation treatments on crude oil contaminated saline soil. Chemosphere 117:486–493. doi: 10.1016/j.chemosphere.2014.08.070. [DOI] [PubMed] [Google Scholar]
- 46.Giam CS, Atlas E, Powers MA, Jr, Leonard JE. 1984. Phthalic acid esters, p 67–142. In Hutzinger O (ed), Anthropogenic compounds. The handbook of environmental chemistry, vol 3. Springer, Berlin, Germany. [Google Scholar]
- 47.Taylor BF, Curry RW, Corcoran EF. 1981. Potential for biodegradation of phthalic acid esters in marine regions. Appl Environ Microbiol 42:590–595. doi: 10.1128/aem.42.4.590-595.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benson NU, Fred-Ahmadu OH. 2020. Occurrence and distribution of microplastics-sorbed phthalic acid esters (PAEs) in coastal psammitic sediments of tropical Atlantic Ocean, Gulf of Guinea. Sci Total Environ 730:139013. doi: 10.1016/j.scitotenv.2020.139013. [DOI] [PubMed] [Google Scholar]
- 49.Morales-Guzmán D, Martínez-Morales F, Bertrand B, Rosas-Galván NS, Curiel-Maciel NF, Teymennet-Ramírez KV, Mazón-Román LE, Licea-Navarro AF, Trejo-Hernández MR. 2021. Microbial prospection of communities that produce biosurfactants from the water column and sediments of the Gulf of Mexico. Biotechnol Appl Biochem 68:1202–1215. doi: 10.1002/bab.2042. [DOI] [PubMed] [Google Scholar]
- 50.Santisi S, Zoccali M, Catania V, Quatrini P, Mondello L, Genovese M, Cappello S. 2022. Biodegradation potential of oil-degrading bacteria related to the genus Thalassospira isolated from polluted coastal area in Mediterranean Sea. Soil Sediment Contam Int J 31:316–332. doi: 10.1080/15320383.2021.1937935. [DOI] [Google Scholar]
- 51.Liu N, Dopffel N, Hovland B, Alagic E, Vik BF, Bødtker G. 2020. High osmotic stress initiates expansion and detachment of Thalassospira sp. biofilms in glass microchannels. J Environ Chem Eng 8:104525. doi: 10.1016/j.jece.2020.104525. [DOI] [Google Scholar]
- 52.Chen S, Zhang D. 2019. Corrosion behavior of Q235 carbon steel in air-saturated seawater containing Thalassospira sp. Corros Sci 148:71–82. doi: 10.1016/j.corsci.2018.11.031. [DOI] [Google Scholar]
- 53.Johnsen AR, Karlson U. 2004. Evaluation of bacterial strategies to promote the bioavailability of polycyclic aromatic hydrocarbons. Appl Microbiol Biotechnol 63:452–459. doi: 10.1007/s00253-003-1265-z. [DOI] [PubMed] [Google Scholar]
- 54.Sun S, Wang H, Yan K, Lou J, Ding J, Snyder SA, Wu L, Xu J. 2021. Metabolic interactions in a bacterial co-culture accelerate phenanthrene degradation. J Hazard Mater 403:123825. doi: 10.1016/j.jhazmat.2020.123825. [DOI] [PubMed] [Google Scholar]
- 55.Ogawa N, Kato H, Kishida K, Ichihashi E, Ishige T, Yoshikawa H, Nagata Y, Ohtsubo Y, Tsuda M. 2019. Suppression of substrate inhibition in phenanthrene-degrading Mycobacterium by co-cultivation with a non-degrading Burkholderia strain. Microbiology (Reading) 165:625–637. doi: 10.1099/mic.0.000801. [DOI] [PubMed] [Google Scholar]
- 56.Hirai H, Takada H, Ogata Y, Yamashita R, Mizukawa K, Saha M, Kwan C, Moore C, Gray H, Laursen D, Zettler ER, Farrington JW, Reddy CM, Peacock EE, Ward MW. 2011. Organic micropollutants in marine plastics debris from the open ocean and remote and urban beaches. Mar Pollut Bull 62:1683–1692. doi: 10.1016/j.marpolbul.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Kester DR, Duedall IW, Connors DN, Pytkowicz RM. 1967. Preparation of artificial seawater. Limnol Oceanogr 12:176–179. doi: 10.4319/lo.1967.12.1.0176. [DOI] [Google Scholar]
- 58.Mori JF, Kanaly RA. 2022. Natural chromosome-chromid fusion across rRNA operons in a Burkholderiaceae bacterium. Microbiol Spectr 10:e02225-21. doi: 10.1128/spectrum.02225-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2018. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun 9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomiyama Y, Takeshita T, Mori JF, Kanaly RA. 2021. Functionalization of the model asphaltene 1-dodecylnaphthalene by Pseudomonas aeruginosa KK6 through subterminal metabolism. J Pet Sci Eng 205:108870. doi: 10.1016/j.petrol.2021.108870. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.03149-22-s0001.pdf, PDF file, 0.1 MB (138.5KB, pdf)
Data Availability Statement
The genome sequence of strain GO-4 has been deposited in NCBI GenBank under accession no. CP097807 and in the IMG/MER database under accession no. 2963523905. The raw sequences are available from SRA accession no. SRR19369649 and SRR19369650 under BioProject no. PRJNA841389 and BioSample no. SAMN28591689.