ABSTRACT
VlsE (variable major protein-like sequence, expressed) is an outer surface protein of the Lyme disease pathogen (Borreliella species) responsible for its within-host antigenic variation and a key diagnostic biomarker of Lyme disease. However, the high sequence variability of VlsE poses a challenge to the development of consistent VlsE-based diagnostics and therapeutics. In addition, the standard diagnostic protocols detect immunoglobins elicited by the Lyme pathogen, not the presence of the pathogen or its derived antigens. Here, we described the development of recombinant monoclonal antibodies (rMAbs) that bound specifically to conserved epitopes on VlsE. We first quantified amino-acid sequence variability encoded by the vls genes from 13 B. burgdorferi genomes by evolutionary analyses. We showed broad inconsistencies of the sequence phylogeny with the genome phylogeny, indicating rapid gene duplications, losses, and recombination at the vls locus. To identify conserved epitopes, we synthesized peptides representing five long conserved invariant regions (IRs) on VlsE. We tested the antigenicity of these five IR peptides using sera from three mammalian host species including human patients, the natural reservoir white-footed mouse (Peromyscus leucopus), and VlsE-immunized New Zealand rabbits (Oryctolagus cuniculus). The IR4 and IR6 peptides emerged as the most antigenic and reacted strongly with both the human and rabbit sera, while all IR peptides reacted poorly with sera from natural hosts. Four rMAbs binding specifically to the IR4 and IR6 peptides were identified, cloned, and purified. Given their specific recognition of the conserved epitopes on VlsE, these IR-specific rMAbs are potential novel diagnostic and research agents for direct detection of Lyme disease pathogens regardless of strain heterogeneity.
IMPORTANCE Current diagnostic protocols of Lyme disease indirectly detect the presence of antibodies produced by the patient upon infection by the bacterial pathogen, not the pathogen itself. These diagnostic tests tend to underestimate early-stage bacterial infections before the patients develop robust immune responses. Further, the indirect tests do not distinguish between active or past infections by the Lyme disease bacteria in a patient sample. Here, we described novel monoclonal antibodies that have the potential to become the basis of direct and definitive diagnostic detection of the Lyme disease pathogen, regardless of its genetic heterogeneity.
KEYWORDS: Borrelia burgdorferi, C6 peptide, Lyme disease, VlsE, monoclonal antibodies
INTRODUCTION
Lyme disease is a multistage, tick-transmitted infection caused by spirochetes of the bacterial species complex Borrelia burgdorferi sensu lato (Bbsl), known more concisely (albeit controversially) as a new genus Borreliella (1, 2). Lyme disease is the most common tick-borne disease in regions of North America, Europe, and Asia (3, 4). In the United States, approximately 476,000 cases are diagnosed annually (5). Most Lyme disease cases in the US are caused by the single species B. burgdorferi and transmitted by the hard-bodied Ixodes scapularis or I. pacificus ticks, although the same tick vectors carry other Borreliella species as well as Borrelia species closely related to relapsing fever spirochetes (6–8). B. burgdorferi causes multisystemic manifestations in humans including erythema migrans (EM) at early stages, arthritis, carditis, neuroborreliosis in late stages, and chronic symptoms associated with persistent infections (4, 9, 10).
Antigenic variation via continuously altering the sequences of surface antigens during infection is a common strategy that microbial pathogens employ to escape the adaptive immune responses of vertebrate hosts (11, 12). In the two sister spirochetal groups – Borrelia causing relapsing fever and Borreliella causing Lyme disease, two homologous but distinct molecular systems have evolved facilitating continuous antigenic variation through recombination between an expressed locus and silent archival loci during persistent infection within the vertebrate hosts (13). In B. burgdorferi, the molecular system able to generate antigenic variation consists of one expression site (vlsE, variable major protein-like sequence, expressed) and a set of tandemly arranged silent cassettes that share more than 90% similarities to the central cassette region of vlsE (14–17) (Fig. 1). During mammalian infection, vlsE continuously expresses and undergoes random segmental recombination with the silent cassettes, generating a considerable number of new VlsE antigen variants to prolong spirochete infection in hosts (13, 16).
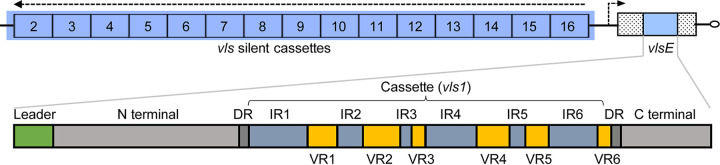
FIG 1.
Genomic and gene structures of the vls locus in B. burgdorferi strain B31. (Top) The vls locus is located close to the telomere of the linear plasmid lp28-1 (GenBank accession AE000794) in the B31 genome, consisting of cassettes of silent (unexpressed) open reading frames (ORFs) (vls2 through vls16) and an expressed ORF (vlsE) containing the vls1 cassette introduced by recombination (14). Dashed arrows indicate the direction of coding strands. (Bottom) The VlsE protein consisted of a leader peptide, an N terminus domain, a cassette flanked by two direct repeats (DRs), and a C terminus domain. The central cassette consisted of interspersed variables (VR1-6) and invariant regions (IR1-6).
The vlsE gene encodes a 36 kDa lipoprotein that is anchored to the outer membrane on the cell surface. The primary structure of VlsE comprises the N- and C-terminal domains, as well as the central cassette, which consists of six highly variable regions (VR1-VR6), interspersed with six conserved invariant regions (IR1-IR6) (Fig. 1). The N- and C-terminal regions do not undergo antigenic variation and are thought to be important in maintaining the functional structure of the molecule (15). The cassette sequences undergo antigenic variation during infection (18). The crystal structure of recombinant VlsE protein revealed that the six VRs constitute loop structures and form a “dome” on the membrane distal surface exposed to the host environment, which may shield the IRs from antibody binding (19).
VlsE elicits strong humoral responses that can be detected throughout Lyme disease, making it a powerful antigen in serologic assays of Lyme disease diagnosis (18–21). Contrary to the established paradigm of weak immunogenicity of the conserved regions of bacterial surface proteins, the conserved IR6 elicits immunodominant antibody responses during human infection despite the region being largely inaccessible on the intact VlsE molecule (18, 22, 23). The surprising finding of immunodominance of IR6 in human patients is hypothesized to be a result of antigen processing of the VlsE proteins in nonreservoir host species (24).
A 26-amino acid peptide that reproduces the IR6 sequence, known as the C6 peptide, is used in commercial diagnostic tests for Lyme disease (18, 25). The standardized two-tiered testing (STTT) for Lyme disease diagnosis includes a screening enzyme immunoassay (EIA) with the whole-cell sonicate and a subsequent confirmatory Western blot assay for the presence of both IgM and IgG antibodies against 10 Borreliella antigens (26, 27). Recently, a modified two-tiered testing (MTTT) protocol using two sequential EIAs with C6 peptide or the whole VlsE protein has been developed. MTTT improved sensitivity and specificity relative to STTT, especially in Lyme patients with early-stage manifestations (28). Nevertheless, the overall sensitivity for early-stage diagnosis remains low, ranging from 36% to 54%, even with MTTT (29). In addition, both diagnostic assays are indirect tests and do not distinguish between active infection and past exposure. In summary, there is a need to simplify the testing protocol for Lyme disease, improve testing sensitivity in the early infection stage, and detect the presence of the Lyme pathogen or its derivative antigens directly.
During the transmission cycle of B. burgdorferi, the vls locus is expressed during the late-stage persistent infection within the mammalian host, in contrast to genes like ospA (encoding outer surface protein A) expressed within the ticks, and genes like ospC expressed exclusively during a short window of time when the spirochetes begin to migrate from the tick to the mammalian host (30–32). As a multicopy gene family and driven by diversifying natural selection, the silent vls cassettes exhibit high sequence variability not only between B. burgdorferi strains but also within the same genome (33–35). In the present study, we developed a bioinformatics workflow to facilitate the automated identification of vls sequences from the sequenced Borreliella genomes. We quantified evolutionary rates at individual amino acid sites of the vls coding sequences identified from 13 B. burgdorferi genomes. Extending the previous analysis of mechanisms of evolution at the vls locus (8, 34), we explored the evolution mechanisms by comparing the vls gene phylogeny with the genome-derived strain phylogeny. Our experimental investigations of the immunogenicity of the VlsE protein confirmed the immunodominance of the IR6 peptide and discovered a similar immunodominance of the IR4 peptide in human patients and immunized rabbits but not the reservoir hosts. Furthermore, we identified, cloned, and purified four recombinant IR-specific monoclonal antibodies (rMAbs) that are promising theragnostic agents for the direct assay of B. burgdorferi infection in clinical samples and model organisms of Lyme disease.
RESULTS
Phylogenetic inconsistencies indicated duplications and losses, sequence divergence, and recombination at the vls locus.
We identified 194 vls cassette sequences from 13 B. burgdorferi strains and inferred a maximum likelihood tree of the cassette (Fig. 2). These B. burgdorferi strains have been classified into four phylogenetic groups (A to D) based on chromosomal single-nucleotide polymorphisms (SNPs) (36). The vls gene phylogeny consists of eight major clades and is consistent with a previously published vls cassette phylogeny (34). Here, we analyzed the vls gene phylogeny in the broader context of strain phylogeny. Phylogenetic inconsistencies between gene and strain trees may result from – and thus indicate the occurrence of – horizontal gene transfers between strains, ancestral gene duplications followed by the loss of duplicated copies, and incomplete lineage sorting when strains rapidly diverge from one other (37, 38).
FIG 2.
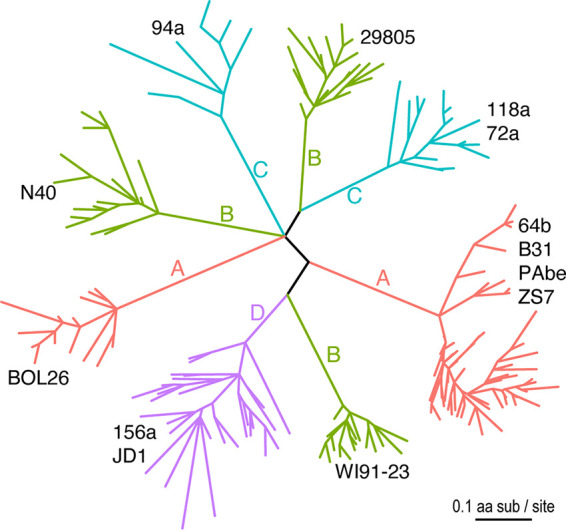
Sequence diversity of vls cassettes of B. burgdorferi strains. Eight major clades of vls alleles were identified based on the codon alignment of 194 cassette sequences from 13 North American and European B. burgdorferi genomes (strain names shown by the clades) (35). The maximum likelihood tree was inferred using IQ-TREE (version 1.6.1) (71). All branches were supported by ≥80% bootstrap values. The tree was rendered using the R package ggtree (Version 2.2.4) (72). Branches were colored according to the four phylogenetic groups (A through D) identified based on genome-wide single-nucleotide polymorphisms (SNPs). SNP groups A, B, and C split into multiple clades, indicating rapid vls sequence divergence between closely related strains (34).
The vls sequences from the two SNP group D strains (JD1 and 156a) formed a monophyletic group consistent with the strain phylogeny. However, within this major clade, the vls sequences did not separate into two strain-specific clades. This phylogenetic inconsistency could be caused by a mixture of paralogous and orthologous gene copies as a result of random gene duplications and losses. Another possible cause of the mixed paralogs from two closely related strains is incomplete lineage sorting, by which descendant strains stochastically inherited gene copies from a common ancestor (39). Horizontal gene exchanges, on the other hand, are unlikely the reason for this inconsistency because recombination would have introduced vls sequences from outside the SNP group D.
In contrast, the vls sequences from the strains belonging to the SNP groups A, B, and C all formed paraphyletic groups, each of which contained multiple clades highly divergent from one another than one would expect from the strain phylogeny (Fig. 2). In the SNP group A, the vls sequences from the European strain BOL26 formed a clade highly divergent from the vls sequences from the North American strains B31, PAbe and 64B and the European strain ZS7. In the SNP group B, the vls sequences from three strains (WI91-23, N40, and 29805) formed three strain-specific clades. The vls sequences from strains belonging to the SNP group C were split into two clades, one consisting of the sequences from strain 94a and the other consisting of sequences from strains 72a and 118a.
As in group D, the vls sequences within the SNP groups A, B, and C did not sort into strain-specific clades, indicating similar evolutionary processes including frequent gene duplications, rapid gene losses, and fast sequence divergence within each group. Indeed, it has been shown that the rapid sequence evolution of the vls cassettes was driven by adaptive differentiation evidenced by the accelerated nonsynonymous nucleotide substitutions (i.e., a high dN/dS ratio) (34).
Evolutionary rates and molecular structure of vls cassettes.
Rates of amino-acid substitutions are not uniform along the translated vls sequence, which consists of mostly fast-evolving variant regions (VRs) interspersed with six short conserved invariant regions (IR1-6) (14). Here, we quantified vls variability at individual amino-acid sites among the 13 B. burgdorferi strains using the 194 vls sequences including both the expressed and unexpressed cassettes. Conserved regions were detected by computing the relative evolutionary rate of each amino-acid site in the multiple sequence alignment, with the average variability score scaled to zero (Fig. 3). Most residues in the IRs showed negative variability scores, indicating below-average evolutionary rates. The mean variability score for each IR was shown in Table 1.
FIG 3.
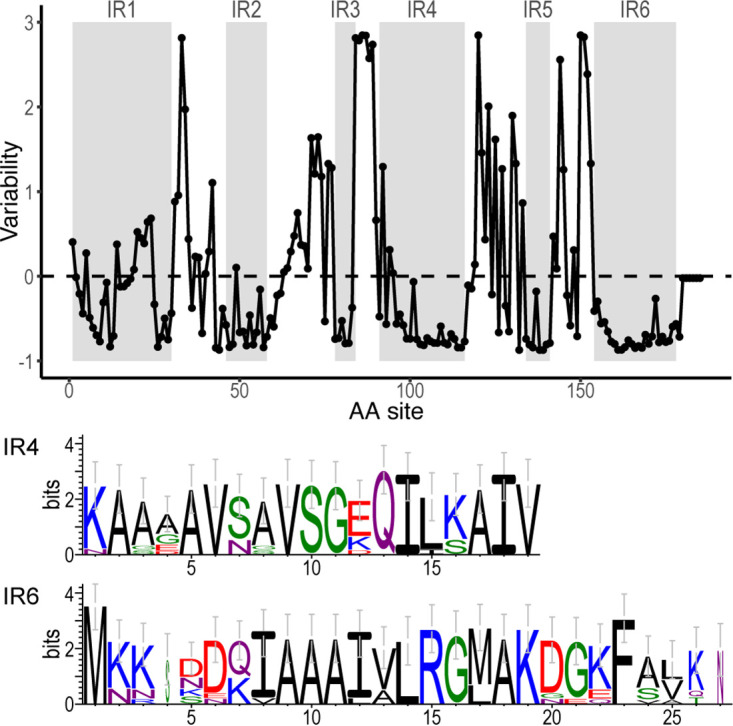
Site-specific evolutionary rates of vls cassettes. (Top) Evolutionary rate, denoted as variability score (in the unit of standard deviation, y-axis), at each amino acid site was estimated by Rate4Site (Version 3.0.0) (73) based on an alignment of translated sequences of 194 vls cassettes and the maximum-likelihood tree (Fig. 2). The dashed line at 0 indicates the average evolutionary rate. The six IRs, showing generally lower-than-average rates, were shaded in gray. VlsE of the B31-5A3 clone (GenBank accession U76405) was used as the reference for computation and annotation. (Bottom) SeqLogo images of IR4 and IR6 sequences, constructed based on 12 representative vls alleles (translated) (35). Amino acid residues were colored according to physiochemistry. Letter heights correspond to information content in bits, a measure of site conservation (74).
TABLE 1.
Peptides used to screen for IR-specific monoclonal antibodies
| Peptidea | Sequenceb | Length | Variability (z-score)d |
|---|---|---|---|
| IR1 | EVSELLDKLVKAVKTAEGASSG | 22 | −0.1746 |
| IR2 | (ASVK)GIAKGIKEIVEAA(GGSE) | 21 | −0.6066 |
| IR3c | AGKLFVK | 7 | −0.6585 |
| IR4 | KAAGAVSAVSGEQILSAIV(TAA) | 22 | −0.5393 |
| IR5 | (AEEAD)NPIAAAIG(TTNEDA) | 19 | −0.7378 |
| IR6 | MKKDDQIAAAIALRGMAKDGKFAVK | 25 | −0.6932 |
IRs: Invariant regions. Six IRs were identified from an alignment of VlsE proteins from three strains representing two species (18).
IR sequences were based on the VlsE protein of the B31 strain (GenBank accession U76405). Flanking residues (in parentheses) were padded to the IR2 and IR4 to facilitate ELISA. For IR5, the padded residues were the most common residues flanking this conserved region.
Antigenicity of IR3, the shortest IR, was not tested in the present study.
Standard deviation from the mean amino-acid substitution rate of zero, with a lower score indicating a higher level of sequence conservation (see Materials and Methods).
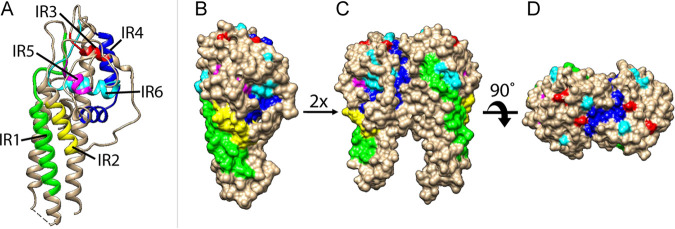
We further mapped the IRs to a published three-dimensional structure of the VlsE protein (from the strain B31) (19) (Fig. 4). All the IRs formed alpha helixes, as the ribbon model showed (Fig. 4A). The space-filled model showed that IR1, IR2, and IR4 were partially surface exposed while IR3, IR5, and IR6 exhibited limited surface exposure (Fig. 4B). The VlsE molecules likely form dimers on the spirochete cell surface (16, 19, 40, 41), which would further shield the invariant regions located on the monomer-monomer interface (Fig. 4C and D). Nevertheless, IR4 is partially exposed at the membrane distal surface even in a dimerized form (Fig. 4D).
FIG 4.
IR-highlighted three-dimensional structures of VlsE. Structure diagrams of VlsE protein from B31 were prepared in Chimera (Version 1.15) (77) based on the PDB file (accession 1L8W) (19). IRs were highlighted in different colors (IR1 in green, IR2 in yellow, IR3 in red, IR4 in dark blue, IR5 in magenta, and IR6 in cyan). (A) Ribbon diagram showing that IRs tend to form alpha helices. (B) Surface-filled diagram showing membrane surface exposure of IRs in monomeric form. (C) and (D) Dimerized structure models. The structures were oriented to show the membrane-proximal part at the bottom (A, B, and C).
Antigenicity of IRs against host sera.
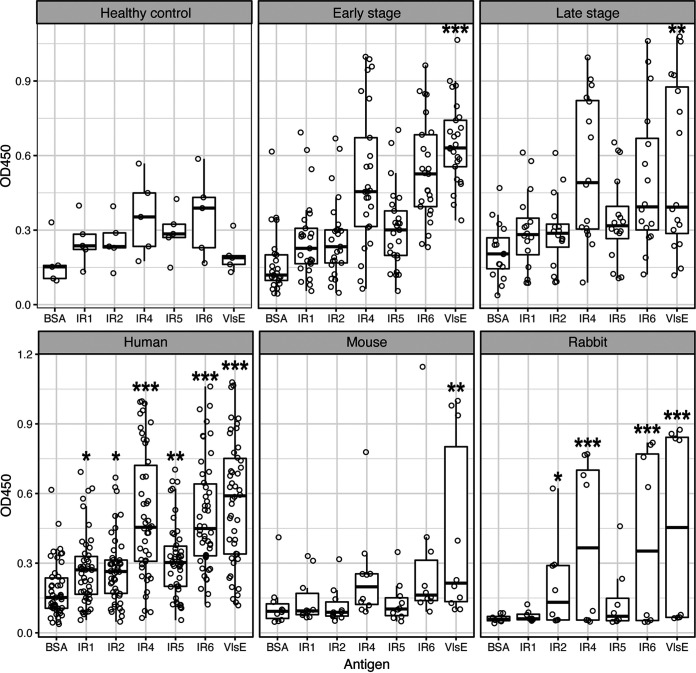
We measured the antigenicity of IRs with sera from Lyme disease patients, white-footed mice, and recombinant VlsE-immunized rabbits using an enzyme-linked immunosorbent assay (ELISA). For the 46 human sera, reactivities of the IR4 and IR6 peptides with the human sera were significantly higher than that of BSA (P = 2.87e−13 and 3.06e−13 by analysis of variance [ANOVA], respectively), while reactivities of IR1, IR2, and IR5 were less significant (P = 0.034, 0.034, and 0.0019 by ANOVA, respectively) (Fig. 5A and B, left). Reactivity of VlsE recombinant protein was the strongest (P < 2.2e−16 by ANOVA). In addition, reactivities of IR4 and IR6 with the human sera were weakly although significantly correlated with those of the VlsE (P = 7.6e−4 and R2 = 0.212 for IR4, P = 3.6e−3 and R2 = 0.158 for IR6, both by linear regression). Reactivities of both early and late-stage samples were significantly higher than the non-Lyme control samples (P = 8.49e−5 and P = 2.63e−4, respectively, by t test) but there was no significant difference in reactivity between the early and late-stage patient samples (P = 0.8654 by t test).
FIG 5.
Antigenicity of conserved epitopes against host sera. The antigenicity of five IRs (x-axis) was quantified with ELISA (see Materials and Methods). Bovine serum albumin (BSA) was used as the negative control and the purified recombinant VlsE protein (of strain B31) as the positive control. Each IR peptide was tested for reactivity (optical density at 450 nm [OD450], y-axis) with host sera (represented by dots). (Top 3) Reactivity with 46 human sera, including those from four healthy controls (left), 25 early-stage Lyme disease patients (middle), and 17 late-stage Lyme disease patients (right). The antigens reacted significantly stronger with the patient sera than with the control sera for both the early-stage and late-stage samples (see Results for t test results). Reactivity of individual IRs, however, was not significant between the patient and control. VlsE reacted strongly with both the early and late-stage patient samples relative to the control sera. (Bottom) Reactivity with 46 patients (left), 10 naturally infected reservoir hosts (white-footed mouse, Peromyscus leucopus) (middle), and 4 New Zealand rabbits (Oryctolagus cuniculus) immunized with recombinant VlsE (right). Asterisks indicate levels of statistical significance: *, 0.01 < P < 0.05; **, 0.001 < P < 0.01; ***, P < 0.001.
For sera from 10 naturally infected white-footed mice, the natural reservoir host of B. burgdorferi, the IR peptides showed weakly significant differences in antigenicity among the antigens (P = 0.0159 by ANOVA), with only VlsE showing a significant difference from the BSA control (P = 2.9e−3 by ANOVA) (Fig. 5B, middle). These results are consistent with the findings of an earlier study that showed low antigenicity of the IR6 peptide in natural hosts relative to its antigenicity in humans (18).
The gross anti-VlsE polyclonal antibodies were extracted from the serum of four immunized rabbits. Reactivities of the IRs against the rabbit polyclonal antibodies showed a similar pattern as those against the naturally infected human (Fig. 5B, right). For example, VlsE, IR4, and IR6 displayed the highest antigenicity (P = 6.8e−10, 1.2e−7, and 2.1e−8, respectively, with an overall P = 2.2e−10 by ANOVA). Antigenicity of the IR1, IR2, and IR5 peptides against the rabbit polyclonal antibodies did not differ or differed weakly from that of BSA, the negative control (P = 0.855, 0.011, and 0.236 by ANOVA, respectively). The preimmunized rabbit sera did not react with any antigens (unpublished data; P = 1.76e−6 by paired t test between the preimmunized and postimmunized sera).
In sum, these ELISA results suggested that (i) anti-VlsE antibodies were present in patients throughout different stages of Lyme disease, (ii) antibodies against the VlsE IRs were strongly present in naturally infected or artificially immunized nonreservoir hosts but minimally present in reservoir hosts, and (iii) the IR4 and IR6 peptides were highly immunogenic conserved epitopes on the VlsE molecule in nonreservoir hosts relative to the IR1, IR2, and IR5 peptides. These results were consistent with the conclusions of earlier studies on the antigenicity of VlsE and conserved epitopes, which established the use of VlsE and the C6 peptide (derived from IR6) in both the standard and modified diagnostics tests for Lyme disease (18, 28, 29, 42–46).
Here, we established that the IR4 peptide was as antigenic as the IR6 peptide. Indeed, both IR peptides reacted at a level similar to the reactivity of the whole VlsE protein with the sera from naturally infected and immunized hosts (Fig. 5). The use of the highly conserved IR4 and IR6 as targets for theragnostic agents has the advantage that they are expected to exhibit antigenicity against a broad set of B. burgdorferi strains, with the potential to mitigate the challenge of strain-specific antigenicity of the highly variable antigens including VlsE and OspC (47, 48).
Identification and characterization of recombinant IR-specific monoclonal antibodies.
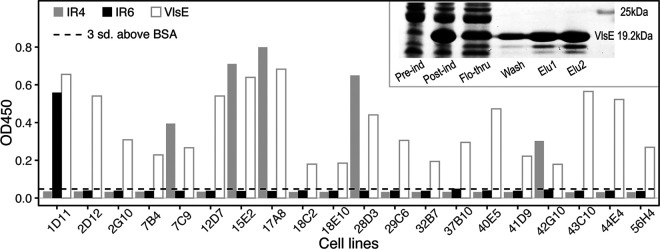
Recombinant VlsE of the strain B31 was overexpressed, purified, and used to immunize New Zealand rabbits (Fig. 6, gel image). IR-specific antibodies were identified via B cell sorting and by testing the reactivity of the supernatant of the 20 B cell lines against the five IR peptides with ELISA. We found that one cell line (1D11) bound specifically to the IR6 peptide and five cell lines (7C9, 15E2, 17A8, 28D3, and 42G10) specifically to the IR4 peptide in addition to their binding to the recombinant VlsE protein (Fig. 6, bar plots). Supernatants of the remaining 14 B cell lines reacted with VlsE but not with the IR peptides, suggesting that most B cell lines in the immunized rabbit expressed antibodies recognizing epitopes located on the variable but not the conserved regions.
FIG 6.
Identification of rabbit B cells producing IR-specific antibodies. (Bar plot) Twenty VlsE-positive cell lines (x-axis) from rabbits immunized with recombinant VlsE were selected with the B cell sorting technique and tested against purified antigens with ELISA. Bovine serum albumin (BSA) was used as the negative control and the purified recombinant VlsE protein (of strain B31) as the positive control. An OD450 value (y-axis) greater than 3 standard deviations above the mean BSA reactivity (dashed lines) was considered to show significant antibody-antigen reactivity. Five cell lines expressing anti-IR4 antibodies and one cell line expressing anti-IR6 antibodies were identified. (Inset) SDS-PAGE image of induction and purification of the recombinant B31 VlsE. Elution 1 and Elution 2 were combined into a single preparation with an estimated purity of ~65% and a concentration of ~5.0 mg/mL, which was subsequently used to immunize New Zealand rabbits. “Pre-ind,” pre-IPTG induction; “Post-ind,” postinduction; “Flo-through,” flowthrough sample; “Wash,” wash sample; “Elu1” and “Elu2,” elution samples.
One pair of the most abundant heavy chain and light chain variable region (VH and VL) sequences in each of four IR-specific cell lines – including the anti-IR6 1D11 cell line and three top anti-IR4 cell lines – were identified by pyrosequencing and subsequently cloned and overexpressed. Specificities of the purified recombinant monoclonal antibodies (rMAb) were validated using ELISA. The initial rMAb cloned from the 1D11 cell line based on the most abundant VH and VL sequences was not reactive to the IR6 peptide as the supernatant of the cell line did. A new rMAb – based on the second most abundant VH and VL sequences – was recloned and overexpressed and reacted with the IR6 peptide strongly and specifically. The VH and VL sequences of the four IR-specific rMAbs are shown in Table 2 and their binding characteristics were obtained by titration experiments (Fig. 7).
TABLE 2.
Specificity and sequences of IR-specific MAbs
| MAb | Specificity | EC50 (ng/mL)a | Variable heavy chain (VH) sequenceb | Variable light chain (VL) sequence |
|---|---|---|---|---|
| 1D11-4 | IR6 | 59.07 (1.75) | MGWSCIILFLVATATGVHSQSLVESGGGLVQPEGSLTLTCKASGFSFSSGYDMCWVRQAPGKGLEYIACIDAGDDITHYASWVKGRFTVSKTSSTTVTLQLNSLTVADTATYFCGRFWDLWGPGTLVTVSS (131 aa) | MGWSCIILFLVATATGVHSSVLTQTPSPVSAAVGGTVTINCQSSQSVYDSTWLGWYQQKPGQPPKLLIYKASNLASGVPSRFKGSGSGTHFTLTISDLECDDAATYYCVGGYSGSVDNWAFGGGTEVVVK (130 aa) |
| 15E2-1 | IR4 | 125.17 (5.10) | METGLRWLLLVAVLKGVQCQSLEESGGGLFKPTDTLTLTCTVSGIDLSSYAMIWVRQAPGKGLEWIGYIWSSGRIWYASWAKGRFTISRTSTTVDLKLASPTTEDTATYFCARLWDIWGPGTLVTVSS (128 aa) | MDTRAPTQLLGLLLLWLPGATFAQVLTQTPSSVSAAVGGTVTISCQASQSLYNGVNLAWYQQKPGQPPKLLIFGASNLESGVSSRFRGSGSGTQFTLTISGVQCDDAATYYCLGEFSCSSADCLAFGGGTEVVVK (135 aa) |
| 17A8-1 | IR4 | 86.14 (2.44) | METGLRWLLLVAVLKGVQCQSVEESGGRLVTPGTPLTLTCTVSGFPLSSYSMAWVRQAPGKGLEYIGFINTDGSAYYASWAKGRITISKTSTTVELKITSPTTEDTATYFCGTGNIWGPGTLVTVSS (127 aa) | MDTRAPTQLLGLLLLWLPGATFAQVLTQTPSSVSAAVGGTVTINCQASQSVSNNNVLAWFQQKPGQPPKRLIYSALTLDSGVPSRFKGSGSGTHFTLTISGVQCDDAATYYCAGGYDCSSNDCIAFGGGTEVVVK (135 aa) |
| 28D3-1 | IR4 | 245.91 (7.93) | METGLRWLLLVAVLKGVQCQSVEESGGRLVTPGTPLTLTCTVSGFSLSSYSMGWVRQAPGKGLEYIGMIISNNSTYYASWAKGRITISKTSTTVELKITSPTTEDTATYFCGTGNIWGPGTLVTVSS (127 aa) | MDTRAPTQLLGLLLLWLPGATFAQVLTQTPASVSAAVGGTVTINCQASQSTSNNNALAWFQQKPGQPPKRLIYSALTLDSGVPSRFKGSGSGTHFTLTISGVQCDDAATYYCAGGYDCSSNDCITFGGGTEVVVK (135 aa) |
EC50, effective concentration of 50% response level (and standard error) based on titration by ELISA. A lower EC50 indicates effective binding at a lower concentration (Fig. 7).
Peptide sequences from the topmost (or 2nd topmost) abundant sequences identified in the sorted single B cells producing IR-specific antibodies. The B cells originated from rabbits immunized with recombinant VlsE proteins (see Materials and Methods).
FIG 7.
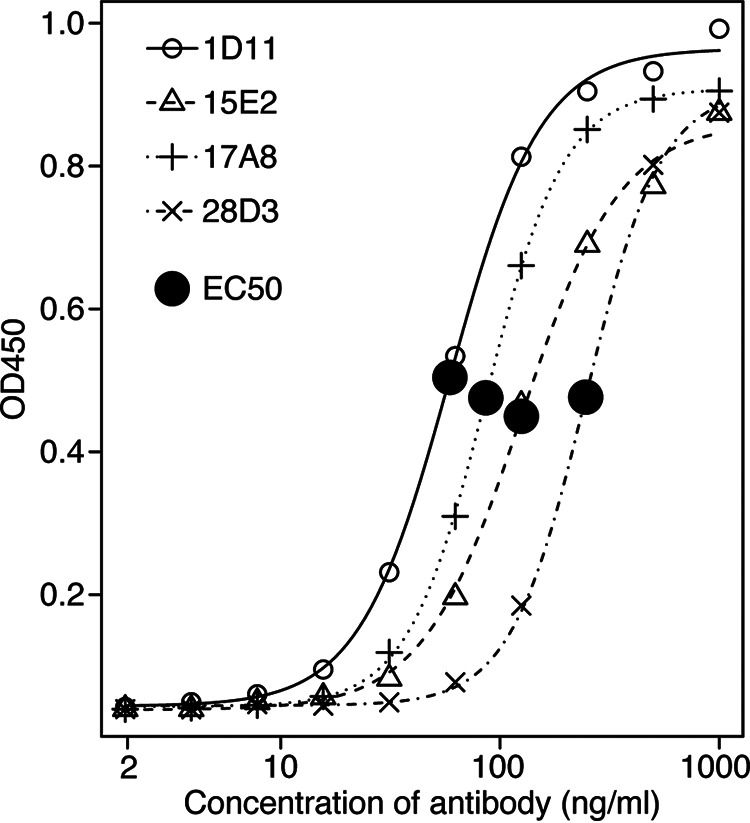
Binding characteristics of IR-specific monoclonal antibodies. Serially diluted preparations of the four affinity-purified IR-specific rMAbs cloned from rabbits immunized with recombinant VlsE were tested with ELISA against their respective IRs (1D11 against IR6; 15E2, 17A8, and 28D3 against IR4). The R package drc (Version 3.0-1) (79) was used to estimate the effective concentration and to plot the titration curves. EC50 (effective concentration at 50% of the maximum activity) values were estimated from the fitted curves, with a lower EC50 indicating stronger antigen affinity.
DISCUSSION
Rapid adaptive diversification of vls cassettes.
The vls gene system in Borreliella was discovered based on gene sequence homology with the vsp/vlp (variable small and large proteins) system in Borrelia spirochetes causing relapsing-fever (13, 14). Since then, the molecular mechanism of segmental recombination between the expression site and the archival cassettes has been well characterized in B. burgdorferi B31, the strain type (16, 40, 41, 49–51). In parallel, genome-based comparative analysis of the vls system among Borreliella species and strains of the same species uncovered rapid evolution in sequence, copy number, and genomic location of the vls cassettes (8, 33, 34).
In the present study, we showed pervasive phylogenetic inconsistencies between the vls gene tree and the genome-based strain tree, suggesting frequent gene duplications, gene losses, and gene exchanges, in addition to adaptive sequence evolution at the locus (Fig. 2). The highly divergent vls cassette sequences between phylogenetic sister strains are reminiscent of the rapid amino-acid sequence diversification at the locus encoding the outer surface protein C (ospC), another immunodominant antigen of B. burgdorferi (52). Protein sequences of major ospC alleles diverge in a strain-specific fashion with an average sequence identity of ~75.9% among B. burgdorferi strains in the Northeast US, due to a history of recombination among coexisting strains and diversifying selection driven by host immunity and possibly host-species preferences (53–56). In contrast, the coding sequences of the vls cassettes vary at a significantly higher level between the eight major sequence clusters (~56.3% average sequence identity), while varying at a high level between copies within the same genome as well (e.g., 81.0% for B31, 76.5% for N40, and 76% for JD1) (Fig. 2). Such a high sequence variability among the vls cassettes is caused by elevated intrinsic mutation and recombination rates mediated by site-specific genetic mechanisms including error-prone repair and frequent gene conversion (41, 49, 57). Nevertheless, positive natural selection, by which new VlsE sequence variants are advantageous to survive the host adaptive immunity, is fundamental and essential for maintaining the high sequence variability among vls cassettes (34). locus
As more Borreliella genomes are sequenced, the bioinformatics workflow including the customized web-based tool (http://borreliabase.org/vls-finder) established in the present study will facilitate large-scale automated identification of vls sequences and quantification of the rates of gene duplication, losses, exchanges, and sequence divergence in this key adaptive molecular system in Borreliella.
Immunogenicity of the IRs in nonreservoir hosts.
The VlsE and its derivative C6 peptide (based on IR6) are key diagnostic antigens in serological tests of Lyme disease (21, 28, 58). In the present study, we confirmed the predominant immunogenicity of IR6 in serum samples from human patients and VlsE-immunized rabbits (Fig. 5). In contrast to an earlier study but consistent with another one (18, 23), the IR4 peptide showed as a similar level of antigenicity as the IR6 peptide in all three host species. Indeed, epitopes on IR4 might be more immunodominant than the IR6 epitopes in rabbits, as we obtained five anti-IR4 cell lines and only one anti-IR6 cell line out of a total of 20 randomly selected VlsE-reactive B cell lines (Fig. 6). The IR1, IR2, and IR5 appeared to be barely immunogenic in reservoir hosts as well as nonreservoir hosts (Fig. 6).
Epitope mapping studies suggested that the IR6 may function as a single conformational epitope (43). On an intact VlsE molecule (or its dimerized structure), the IR6 is almost entirely buried underneath the membrane surface and immunofluorescence assays demonstrated that the IR6 was inaccessible to antibodies on intact spirochetes (22, 24, 59). It appears paradoxical that IR4 and IR6, two highly conserved and mostly buried regions on VlsE, contain immunodominant epitopes in human patients. Evolutionary arms races drive codiversification of the antigen sequences in microbial pathogens along with the sequences of antigen-recognition proteins in vertebrate hosts through population mechanisms like negative frequency-dependent selection (55, 60, 61). Regions on antigen molecules shielded from host immune systems, like the IRs on VlsE, are not under such diversifying selection and thereby expected to be conserved in molecule sequences. The paradox resolves itself however when one considers that the IRs were indeed weakly immunogenic in the infected mice that belong to the natural reservoir species of B. burgdorferi (Fig. 6). Indeed, as the whole VlsE molecule elicits significant antibody responses in the infected P. leucopus mice, such immunogenicity is likely due to epitopes on the variable regions as expected from the pathogen-host coevolutionary arms race (44) (Fig. 6).
Likely, B. burgdorferi is well adapted to natural hosts and able to maintain a high level of cell integrity including intact VlsE molecules on the cell surfaces. Indeed, B. burgdorferi expresses cell surface proteins binding specifically to proteins of the host complement system to downregulate innate and adaptive host immunity (31, 62–64). On an intact spirochete cell surface, the VlsE molecules can shield other surface antigens from being recognized by antibodies (46).
In nonnatural hosts, such as humans and rabbits to which B. burgdorferi is poorly adapted, the pathogen may lose or diminish its ability to inhibit host immune responses and is thus more easily recognized by the host immune system. Upon cell disintegration and degradative processing of the surface antigens including VlsE by the major histocompatibility complex (HMC), the IR6 would be exposed along with other epitopes and elicit strong antibody responses. Because the IRs are conserved among the vls alleles and, unlike the VRs, their total amount remains stable during antigenic shift during infection, the IRs would result in stronger and more long-lasting host responses and become immunodominant in nonnatural hosts including humans.
Potential of rMAbs as diagnostic agents.
B. burgdorferi infection is characterized by a low number of colonizing spirochetes. It is difficult to directly detect the pathogen through culture or PCR approaches due to the extreme scarcity of the organism in infected hosts (58). Further, current standard diagnostic assays of Lyme disease, targeting the anti-VlsE or anti-C6 antibodies, do not distinguish between active and past infections (25, 29). The VlsE-recognizing recombinant monoclonal antibodies are potential diagnostic agents for the direct detection of B. burgdorferi infections. For example, we plan to label the IR-specific rMAbs with a radioactive isotope, such as zirconium-89, and perform a positron emission tomography (PET) for the sensitive detection of trace quantities of spirochetes in experimentally infected mice. Clinical immunoPET has been shown to detect very small lesions (~1 mm3) bearing cancer antigens with very low density (<500 antigen copies per cell) (65). As a result, we are hopeful that these rMAbs can be harnessed for the immunoPET of Lyme disease despite the scarcity of spirochetes during human infection, but only rigorous preclinical experiments will show us whether this is possible.
To validate the utility of these rMAbs as diagnostic agents, it is necessary to perform in vitro testing using cultured B. burgdorferi cells followed by in vivo testing using a mouse model of Lyme disease. We anticipate several biological and technical challenges during in vitro and in vivo validation testing of the rMAbs. For in vitro testing using cultured spirochetes, first, it is unclear if the rMAbs would bind VlsE anchored on the surface of live B. burgdorferi cells because of limited surface accessibility of the IRs at native conformations, even though the rMAbs reacted strongly with VlsE molecules fixed on an ELISA plate (Fig. 2) (43, 44). Molecular conformation may also differ between the IR peptides used in ELISA and the IRs as a part of VlsE molecules anchored to the outer membrane of spirochete cells. Second, B. burgdorferi does not constitutively express a large quantity of VlsE during in vitro culture, and supplementing the standard media with human tissue cells may be necessary to increase VlsE expression for in vitro validation of rMAb binding (66, 67). Third, both the IR4 and IR6 sequences vary slightly among B. burgdorferi strains despite high sequence conservation (Fig. 3). The affinity of these rMAbs, which were raised using a single allelic variant (the B31 VlsE), is expected to vary among the B. burgdorferi strains. Effects of sequence variability to rMAb affinity could be quantified with ELISA using synthetic peptides representing the IR variants. Ideally, amino acid residues essential for the rMAb binding could be accurately pinpointed with systematic epitope mapping (23).
MATERIALS AND METHODS
Identification of vls cassette sequences and evolutionary analysis.
We downloaded the whole-genome sequences of 13 B. burgdorferi strains from NCBI GenBank (35). The silent cassette sequences of the B31-5A3 clone (GenBank accession U76406) (14) were used as the queries to search for sequences homologous to the vls cassette sequences using HMMER (version 3.3.2) (68). A customized web-based software tool was developed to identify and extract individual vls sequences given a B. burgdorferi replicon sequence (http://borreliabase.org/vls-finder). Sequences of the silent cassettes and vlsE were translated, aligned, and converted into a codon alignment using MUSCLE (version 3.8.31) (69) and the bioaln utility (–dna2pep method) of the BpWrapper (version 1.13) toolkit (70). A maximum likelihood tree was subsequently inferred using IQ-TREE (version 1.6.1) with the best-fit nucleotide substitute model KOSI07 and 1000 bootstrap replicates (71). Branches with lower than 80% bootstrap support were collapsed using the biotree utility (-D method) of the BpWrapper toolkit (70). The tree was rendered using the R package ggtree (Version 2.2.4) (72). To quantify the sequence conservation, evolutionary rates at individual amino acid positions were estimated using Rate4Site (version 3.0.0) with the protein alignment and the phylogenetic tree as inputs and the B31-5A3 VlsE sequence (GenBank accession U76405) as the reference (73). Sequence conservation at the IRs was further quantified and visualized with WebLogo (74).
Synthesis of peptides representing conserved epitopes of VlsE.
The preparation of the peptides was based on the annotation of the B31-5A3 VlsE protein sequence (14). Five invariant regions, IR1, IR2, IR4, IR5, and IR6, were tested for antigenicity using sera from three host species. IR3 (AGKLFVK), the shortest IR, was excluded from the antigenicity test. Extra flanking amino acids were added to IR2, IR4, and IR5 to meet the minimum length for peptide synthesis. Peptides were commercially synthesized and biotin-labeled on the N terminus using Fmoc chemistry (GenScript, Piscataway, NJ, USA). Sequences of these peptides are shown in Table 1.
Sera collection from naturally infected hosts.
The 56 serum samples, consisting of Lyme patient and control sera provided by the US Centers for Disease Control and Prevention (CDC; n = 40), Lyme patient sera provided by Maria Gomes-Solecki (University of Tennessee Health Science Center; n = 6), and sera from naturally infected individuals of the reservoir host white-footed mouse (Peromyscus leucopus) originated from Millbrook, New York (n = 10), have been used and described in previous publications (53, 75, 76). Briefly, among the human samples, 25 serum samples were derived from patients with early-stage Lyme disease including those diagnosed as having the skin symptom erythema migran (EM) or as EM convalescence. Seventeen human serum samples were from patients displaying late-stage Lyme disease symptoms including arthritic, cardiac, and neurological Lyme diseases. Four human serum samples were from healthy individuals as controls.
Cloning, overexpression, and purification of recombinant VlsE protein.
Recombinant VlsE protein from the B31 strain was cloned, overexpressed, and purified using a protocol described previously (53). Briefly, the 585-bp vlsE cassette region (including the IR1 through VR6 regions) of the B31-5A3 clone was codon-optimized, synthesized, and cloned into the pET24 plasmid vector which then transfected Escherichia coli BL21 cells. A10 × Histidine-tag was added on the N terminus of the construct to facilitate the downstream purification. All cloning work was performed by a commercial service (GeneImmune Biotechnology Corp., Rockville, MD, USA). The E. coli strain that contained a cloned vlsE cassette was cultured in Luria-Bertani (LB) broth containing 0.4% glucose and 50 μg/mL Ampicillin. When the culture reached exponential growth, we induced the expression of the cloned vlsE cassette by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.25 mM and by incubation overnight at 25°C. Cells were collected and then lysed by lysozyme and sonication. The lysate supernatant, containing the recombinant VlsE protein, was purified using nickel Sepharose beads (Ni-NTA, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocol. The identity and concentration of the purified protein were examined and quantified using the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and the Pierce Bradford Protein assay kit (Thermo Fisher Scientific, Waltham, MA, USA).
Immunization of rabbits and preparation of polyclonal and monoclonal antibodies.
Antibody preparation was conducted with a commercial service GenScript (Piscataway, NJ, USA). Briefly, the project consisted of four stages. In stage 1, animals were immunized, and polyclonal antibodies were obtained. Specifically, four New Zealand rabbits (Oryctolagus cuniculus) were immunized with 100 μg purified recombinant VlsE protein on days 1, 14, and 28. The rabbits were bled for antiserum collection 1 week after the third immunization. The antisera were subsequently purified by affinity chromatography to obtain polyclonal antibodies (pAbs), which were assayed for anti-VlsE activity. In stage 2, monoclonal antibodies (MAbs) were identified via single B cell sorting. Peripheral blood mononuclear cells (PBMC) were collected from the two selected immunized rabbits 1 week after a booster dose with the recombinant VlsE. Plasma B cells (CD138+) were isolated and enriched using a commercial kit. B cells were transformed by a proprietary process and then cloned by limiting dilution. The supernatants of positive cell lines were used to test for binding with VlsE and positive cell lines were chosen to produce monoclonal antibodies. In Stage 3, the variable domains of the light and heavy chains of the VlsE-binding antibodies were sequenced. Total RNA was isolated from the VlsE-binding B cell lines and reverse-transcribed into cDNA using universal primers. DNA sequences encoding variable domains of the heavy chain and the light chain were amplified and sequenced. In Stage 4, the amplified antibody variable fragments were cloned into plasmid vector pcDNA3.4, which was then transfected into mouse cells for expression. Supernatants of cell cultures were harvested continuously. The recombinant monoclonal antibodies (rMAbs) were purified using protein A/G affinity chromatography (with immobilized protein A and G from Staphylococcus aureus) followed by size exclusion chromatography (SEC).
Identification of IR-specific MAbs with ELISA.
Sera from naturally infected humans and P. leucopus were tested to evaluate reactivity to the IR peptides (Table 1) and the recombinant VlsE protein with ELISA using a protocol described previously (53). Briefly, a 96-well MICROLON 600 plate (USA Scientific, Inc., Ocala, FL, USA) was incubated with 10 μg/mL of antigen overnight at 4°C. Serum samples diluted between 1:100 to 1:1000 were applied after blocking with 5% milk and were incubated for 2 h at 37°C, followed by the application of horseradish peroxidase (HRP)-conjugated secondary antibodies. We used the Goat Anti-Human IgG/IgM (H+L) (Abcam, Cambridge, UK) 1:40,000 for assays of human sera and the Goat anti-P. leucopus IgG (H+L) (SeraCare Life Sciences, MA, USA) 1:1000 for assays of P. leucopus sera. The antigen-antibody reaction was probed by TMB ELISA Substrate Solution (Invitrogen eBioscience) and was terminated with 1 M sulfuric acid after 15 min. Binding intensities were measured at the 450 nm wavelength using a SpectraMax i3 microplate reader (Molecular Devices, LLC, CA, USA).
The same ELISA protocol was followed to test against binding with the rabbit-originated antibodies as well, including the purified anti-VlsE pAbs, the supernatants of selected B cell cultures, and the purified rMAbs. Mouse Anti-Rabbit IgG Fr secondary antibody (GenScript, Piscataway, NJ) 1:30,000 was used for assays of these rabbit-derived samples. Serial dilutions of MAbs by factors from 1,000 to 512,000 were tested with ELISA to quantify the binding activities.
Protein structure visualization.
The PDB file of the VlsE protein structure (accession no. 1L8W) was downloaded from the protein data bank (PDB) (19). The PDF file describes a tetramer of VlsE. We used Chimera (version 1.15) (77) to visualize the protein structure in ribbon and surface-filled formats and to color the six invariable regions (IR1-6).
Animal care.
Antibody production from the New Zealand rabbits followed the protocols approved by the Office of Laboratory Animal Welfare (OLAW) Assurance and the Institutional Animal Care and Use Committee (IACUC) of the vendor (GenScript, Piscataway, NJ).
Data availability.
Data visualization and statistical analysis were performed in the R statistical computing environment (78) accessed with RStudio. The alignment of translated vls sequences, ELISA readings, and R scripts are publicly available on GitHub at https://github.com/weigangq/vls-mabs.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Public Health Service awards AI139782 (WGQ) from the National Institute of Allergy and Infectious Diseases (NIAID) and EB030275 (B.M.Z. and W.Q.) from the National Institute of Bio-medical Imaging and Bioengineering (NIBIB) of the US National Institutes of Health (NIH). The content of the manuscript is solely the responsibility of the authors and does not necessarily represent the official views of NIH. Li Li and Saymon Akther were supported in part by the Doctoral Program in Biology of the Graduate Center, the City University of New York.
We thank Christopher Sexton and Jeannine Petersen of the Centers for Disease Control and Prevention (CDC) and Maria Gomes-Solecki (University of Tennessee Health Science Center) for providing serum samples from human patients and reservoir mice, respectively.
Li Li performed evolutionary analysis, protein purification, and ELISA. Li Li composed the initial draft. Lia Di developed the bioinformatics pipeline and web tool and participated in protein purification and ELISA. Saymon Akther participated in protein purification and ELISA and edited the manuscript. Brian M. Zeglis and Weigang Qiu conceived, obtained funding for, and supervised the project. Weigang Qiu and Brian M. Zeglis revised the manuscript.
We declare no conflicts of interest.
Contributor Information
Weigang Qiu, Email: wqiu@hunter.cuny.edu.
Catherine Ayn Brissette, University of North Dakota.
REFERENCES
- 1.Barbour AG, Qiu W. 2019. Borreliella, p 1–22. In Bergey’s Manual of Systematics of Archaea and Bacteria. John Wiley & Sons, Inc., in association with Bergey’s Manual Trust. [Google Scholar]
- 2.Margos G, Castillo-Ramirez S, Cutler S, Dessau RB, Eikeland R, Estrada-Peña A, Gofton A, Graña-Miraglia L, Hunfeld K-P, Krause A, Lienhard R, Lindgren P-E, Oskam C, Rudolf I, Schwartz I, Sing A, Stevenson B, Wormser GP, Fingerle V. 2020. Rejection of the name Borreliella and all proposed species comb. nov. placed therein. Int J Syst Evol Microbiol 70:3577–3581. doi: 10.1099/ijsem.0.004149. [DOI] [PubMed] [Google Scholar]
- 3.Kilpatrick AM, Dobson ADM, Levi T, Salkeld DJ, Swei A, Ginsberg HS, Kjemtrup A, Padgett KA, Jensen PM, Fish D, Ogden NH, Diuk-Wasser MA. 2017. Lyme disease ecology in a changing world: consensus, uncertainty and critical gaps for improving control. Philos Trans R Soc B 372:20160117. doi: 10.1098/rstb.2016.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanek G, Wormser GP, Gray J, Strle F. 2012. Lyme borreliosis. Lancet 379:461–473. doi: 10.1016/S0140-6736(11)60103-7. [DOI] [PubMed] [Google Scholar]
- 5.Kugeler KJ, Schwartz AM, Delorey MJ, Mead PS, Hinckley AF. 2021. Estimating the frequency of lyme disease diagnoses, United States, 2010–2018. Emerg Infect Dis 27:616–619. doi: 10.3201/eid2702.202731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbour AG, Bunikis J, Travinsky B, Hoen AG, Diuk-Wasser MA, Fish D, Tsao JI. 2009. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am J Trop Med Hyg 81:1120–1131. doi: 10.4269/ajtmh.2009.09-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritt BS, Respicio-Kingry LB, Sloan LM, Schriefer ME, Replogle AJ, Bjork J, Liu G, Kingry LC, Mead PS, Neitzel DF, Schiffman E, Hoang Johnson DK, Davis JP, Paskewitz SM, Boxrud D, Deedon A, Lee X, Miller TK, Feist MA, Steward CR, Theel ES, Patel R, Irish CL, Petersen JM. 2016. Borrelia mayonii sp. nov., a member of the Borrelia burgdorferi sensu lato complex, detected in patients and ticks in the upper midwestern United States. Int J Syst Evol Microbiol 66:4878–4880. doi: 10.1099/ijsem.0.001445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwartz I, Margos G, Casjens SR, Qiu W-G, Eggers CH. 2021. Multipartite Genome of Lyme Disease Borrelia: Structure, Variation and ProphagesLyme Disease and Relapsing Fever Spirochetes: Genomics, Molecular Biology, Host Interactions and Disease Pathogenesis. Caister Academic Press. [Google Scholar]
- 9.Feng J, Li T, Yee R, Yuan Y, Bai C, Cai M, Shi W, Embers M, Brayton C, Saeki H, Gabrielson K, Zhang Y. 2019. Stationary phase persister/biofilm microcolony of Borrelia burgdorferi causes more severe disease in a mouse model of Lyme arthritis: implications for understanding persistence, Post-treatment Lyme Disease Syndrome (PTLDS), and treatment failure. Discov Med 27:125–138. [PubMed] [Google Scholar]
- 10.Sharma B, Brown AV, Matluck NE, Hu LT, Lewis K. 2015. Borrelia burgdorferi, the causative agent of lyme disease, forms drug-tolerant persister cells. Antimicrob Agents Chemother 59:4616–4624. doi: 10.1128/AAC.00864-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palmer GH, Bankhead T, Seifert HS. 2016. Antigenic variation in bacterial pathogens. Microbiol Spectr 4:4-1. doi: 10.1128/microbiolspec.VMBF-0005-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vink C, Rudenko G, Seifert HS. 2012. Microbial antigenic variation mediated by homologous DNA recombination. FEMS Microbiol Rev 36:917–948. doi: 10.1111/j.1574-6976.2011.00321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norris SJ. 2006. Antigenic variation with a twist - The Borrelia story. Mol Microbiol 60:1319–1322. doi: 10.1111/j.1365-2958.2006.05204.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang JR, Hardham JM, Barbour AG, Norris SJ. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275–285. doi: 10.1016/s0092-8674(00)80206-8. [DOI] [PubMed] [Google Scholar]
- 15.Norris SJ. 2014. vls antigenic variation systems of lyme disease borrelia: eluding host immunity through both random, segmental gene conversion and framework heterogeneity. Microbiol Spectr 2:471–489. doi: 10.1128/microbiolspec.MDNA3-0038-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhey TB, Castellanos M, Chaconas G. 2019. Antigenic variation in the Lyme spirochete: detailed functional assessment of recombinational switching at vlsE in the JD1 strain of Borrelia burgdorferi. Mol Microbiol 111:750–763. doi: 10.1111/mmi.14189. [DOI] [PubMed] [Google Scholar]
- 17.Chaconas G, Castellanos M, Verhey TB. 2020. Changing of the guard: how the Lyme disease spirochete subverts the host immune response. J Biol Chem 295:301–313. doi: 10.1074/jbc.REV119.008583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang FT, Alvarez AL, Gu Y, Nowling JM, Ramamoorthy R, Philipp MT. 1999. An immunodominant conserved region within the variable domain of VlsE, the variable surface antigen of Borrelia burgdorferi. J Immunol Baltim Md 1950 163:5566–5573. [PubMed] [Google Scholar]
- 19.Eicken C, Sharma V, Klabunde T, Lawrenz MB, Hardham JM, Norris SJ, Sacchettini JC. 2002. Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia burgdorferi. J Biol Chem 277:21691–21696. doi: 10.1074/jbc.M201547200. [DOI] [PubMed] [Google Scholar]
- 20.Lawrenz MB, Hardham JM, Owens RT, Nowakowski J, Steere AC, Wormser GP, Norris SJ. 1999. Human antibody responses to VlsE antigenic variation protein of Borrelia burgdorferi. J Clin Microbiol 37:3997–4004. doi: 10.1128/JCM.37.12.3997-4004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bacon RM, Biggerstaff BJ, Schriefer ME, Gilmore R, Philipp MT, Steere AC, Wormser GP, Marques AR, Johnson BJ. 2003. Serodiagnosis of Lyme disease by kinetic enzyme-linked immunosorbent assay using recombinant VlsE1 or peptide antigens of Borrelia burgdorferi compared with 2-tiered testing using whole-cell lysates. J Infect Dis 187:1187–1199. doi: 10.1086/374395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elzbieta J, Tang KS, Komorowski L, Ajamian M, Probst C, Stevenson B, Wormser GP, Marques AR, Alaedini A. 2016. Epitope-specific evolution of human B cell responses to Borrelia burgdorferi VlsE protein from early to late stages of Lyme disease. J Immunol 196:1036–1043. doi: 10.4049/jimmunol.1501861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandra A, Latov N, Wormser GP, Marques AR, Alaedini A. 2011. Epitope mapping of antibodies to VlsE protein of Borrelia burgdorferi in post-Lyme disease syndrome. Clin Immunol 141:103–110. doi: 10.1016/j.clim.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Embers ME, Jacobs MB, Johnson BJ, Philipp MT. 2007. Dominant epitopes of the C6 diagnostic peptide of Borrelia burgdorferi are largely inaccessible to antibody on the parent VlsE molecule. Clin Vaccine Immunol 14:931–936. doi: 10.1128/CVI.00075-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wormser GP, Schriefer M, Aguero-Rosenfeld ME, Levin A, Steere AC, Nadelman RB, Nowakowski J, Marques A, Johnson BJB, Dumler JS. 2013. Single-tier testing with the C6 peptide ELISA kit compared with two-tier testing for Lyme disease. Diagn Microbiol Infect Dis 75:9–15. doi: 10.1016/j.diagmicrobio.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.CDC. 1995. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. MMWR Surveill Summ 44:590–591. [PubMed] [Google Scholar]
- 27.Moore A, Nelson C, Molins C, Mead P, Schriefer M. 2016. Current guidelines, common clinical pitfalls, and future directions for laboratory diagnosis of Lyme disease, United States. Emerg Infect Dis 22:1169–1177. doi: 10.3201/eid2207.151694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Branda JA, Strle K, Nigrovic LE, Lantos PM, Lepore TJ, Damle NS, Ferraro MJ, Steere AC. 2017. Evaluation of modified 2-tiered serodiagnostic testing algorithms for early Lyme disease. Clin Infect Dis 64:1074–1080. doi: 10.1093/cid/cix043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pegalajar-Jurado A, Schriefer ME, Welch RJ, Couturier MR, MacKenzie T, Clark RJ, Ashton LV, Delorey MJ, Molins CR. 2018. Evaluation of modified two-tiered testing algorithms for Lyme disease laboratory diagnosis using well-characterized serum samples. J Clin Microbiol 56:e01943-17. doi: 10.1128/JCM.01943-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crother TR, Champion CI, Whitelegge JP, Aguilera R, Wu X-Y, Blanco DR, Miller JN, Lovett MA. 2004. Temporal analysis of the antigenic composition of Borrelia burgdorferi during infection in rabbit skin. Infect Immun 72:5063–5072. doi: 10.1128/IAI.72.9.5063-5072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuels DS. 2011. Gene regulation in Borrelia burgdorferi. Annu Rev Microbiol 65:479–499. doi: 10.1146/annurev.micro.112408.134040. [DOI] [PubMed] [Google Scholar]
- 32.Tilly K, Bestor A, Rosa PA. 2013. Lipoprotein succession in Borrelia burgdorferi: similar but distinct roles for OspC and VlsE at different stages of mammalian infection. Mol Microbiol 89:216–227. doi: 10.1111/mmi.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glöckner G, Lehmann R, Romualdi A, Pradella S, Schulte-Spechtel U, Schilhabel M, Wilske B, Sühnel J, Platzer M. 2004. Comparative analysis of the Borrelia garinii genome. Nucleic Acids Res 32:6038–6046. doi: 10.1093/nar/gkh953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graves CJ, Ros VID, Stevenson B, Sniegowski PD, Brisson D. 2013. Natural selection promotes antigenic evolvability. PLoS Pathog 9:e1003766. doi: 10.1371/journal.ppat.1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schutzer SE, Fraser-Liggett CM, Casjens SR, Qiu W-G, Dunn JJ, Mongodin EF, Luft BJ. 2011. Whole-genome sequences of thirteen isolates of Borrelia burgdorferi. J Bacteriol 193:1018–1020. doi: 10.1128/JB.01158-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mongodin EF, Casjens SR, Bruno JF, Xu Y, Drabek EF, Riley DR, Cantarel BL, Pagan PE, Hernandez YA, Vargas LC, Dunn JJ, Schutzer SE, Fraser CM, Qiu WG, Luft BJ. 2013. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics 14:693. doi: 10.1186/1471-2164-14-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kundu S, Bansal MS. 2018. On the impact of uncertain gene tree rooting on duplication-transfer-loss reconciliation. BMC Bioinformatics 19:290. doi: 10.1186/s12859-018-2269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rogers J, Fishberg A, Youngs N, Wu Y-C. 2017. Reconciliation feasibility in the presence of gene duplication, loss, and coalescence with multiple individuals per species. BMC Bioinformatics 18:292. doi: 10.1186/s12859-017-1701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu L, Yu L, Kubatko L, Pearl DK, Edwards SV. 2009. Coalescent methods for estimating phylogenetic trees. Mol Phylogenet Evol 53:320–328. doi: 10.1016/j.ympev.2009.05.033. [DOI] [PubMed] [Google Scholar]
- 40.Verhey TB, Castellanos M, Chaconas G. 2018. Analysis of recombinational switching at the antigenic variation locus of the Lyme spirochete using a novel PacBio sequencing pipeline. Mol Microbiol 107:104–115. doi: 10.1111/mmi.13873. [DOI] [PubMed] [Google Scholar]
- 41.Verhey TB, Castellanos M, Chaconas G. 2018. Antigenic variation in the Lyme spirochete: insights into recombinational switching with a suggested role for error-prone repair. Cell Rep 23:2595–2605. doi: 10.1016/j.celrep.2018.04.117. [DOI] [PubMed] [Google Scholar]
- 42.Liang FT, Philipp MT. 1999. Analysis of antibody response to invariable regions of VlsE, the variable surface antigen of Borrelia burgdorferi. Infect Immun 67:6702–6706. doi: 10.1128/IAI.67.12.6702-6706.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang FT, Philipp MT. 2000. Epitope mapping of the immunodominant invariable region of Borrelia burgdorferi VlsE in three host species. Infect Immun 68:2349–2352. doi: 10.1128/IAI.68.4.2349-2352.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDowell JV, Sung S-Y, Hu LT, Marconi RT. 2002. Evidence that the variable regions of the central domain of VlsE are antigenic during infection with lyme disease spirochetes. Infect Immun 70:4196–4203. doi: 10.1128/IAI.70.8.4196-4203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lone AG, Bankhead T. 2020. The Borrelia burgdorferi VlsE lipoprotein prevents antibody binding to an arthritis-related surface antigen. Cell Rep 30:3663–3670.e5. doi: 10.1016/j.celrep.2020.02.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhatia B, Hillman C, Carracoi V, Cheff BN, Tilly K, Rosa PA. 2018. Infection history of the blood-meal host dictates pathogenic potential of the Lyme disease spirochete within the feeding tick vector. PLoS Pathog 14:e1006959. doi: 10.1371/journal.ppat.1006959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bockenstedt LK, Hodzic E, Feng S, Bourrel KW, de Silva A, Montgomery RR, Fikrig E, Radolf JD, Barthold SW. 1997. Borrelia burgdorferi strain-specific OspC-mediated immunity in mice. Infect Immun 65:4661–4667. doi: 10.1128/iai.65.11.4661-4667.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang JR, Norris SJ. 1998. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun 66:3698–3704. doi: 10.1128/IAI.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, Botkin DJ, Norris SJ. 2003. Characterization of the vls antigenic variation loci of the Lyme disease spirochaetes Borrelia garinii Ip90 and Borrelia afzelii ACAI. Mol Microbiol 47:1407–1417. doi: 10.1046/j.1365-2958.2003.03386.x. [DOI] [PubMed] [Google Scholar]
- 51.Coutte L, Botkin DJ, Gao L, Norris SJ. 2009. Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog 5:e1000293. doi: 10.1371/journal.ppat.1000293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbour AG, Travinsky B. 2010. Evolution and distribution of the ospC gene, a transferable serotype determinant of Borrelia burgdorferi. mBio 1:e00153-10. doi: 10.1128/mBio.00153-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di L, Akther S, Bezrucenkovas E, Ivanova L, Sulkow B, Wu B, Mneimneh S, Gomes-Solecki M, Qiu WG. 2021. Maximum antigen diversification in a lyme bacterial population and evolutionary strategies to overcome pathogen diversity. ISME J 16:447–464. doi: 10.1038/s41396-021-01089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brisson D, Dykhuizen DE. 2004. ospC diversity in Borrelia burgdorferi different hosts are different niches. Genetics 168:713–722. doi: 10.1534/genetics.104.028738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haven J, Vargas LC, Mongodin EF, Xue V, Hernandez Y, Pagan P, Fraser-Liggett CM, Schutzer SE, Luft BJ, Casjens SR, Qiu W-G. 2011. Pervasive recombination and sympatric genome diversification driven by frequency-dependent selection in Borrelia burgdorferi, the Lyme disease bacterium. Genetics 189:951–966. doi: 10.1534/genetics.111.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang I-N, Dykhuizen DE, Qiu W, Dunn JJ, Bosler EM, Luft BJ. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15–30. doi: 10.1093/genetics/151.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin T, Gao L, Edmondson DG, Jacobs MB, Philipp MT, Norris SJ. 2009. Central role of the Holliday junction helicase RuvAB in vlsE recombination and infectivity of Borrelia burgdorferi. PLoS Pathog 5:e1000679. doi: 10.1371/journal.ppat.1000679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marques AR. 2015. Laboratory diagnosis of Lyme disease: advances and challenges. Infect Dis Clin North Am 29:295–307. doi: 10.1016/j.idc.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang FT, Nowling JM, Philipp MT. 2000. Cryptic and exposed invariable regions of VlsE, the variable surface antigen of Borrelia burgdorferi sl. J Bacteriol 182:3597–3601. doi: 10.1128/JB.182.12.3597-3601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papkou A, Guzella T, Yang W, Koepper S, Pees B, Schalkowski R, Barg M-C, Rosenstiel PC, Teotónio H, Schulenburg H. 2019. The genomic basis of Red Queen dynamics during rapid reciprocal host-pathogen coevolution. Proc Natl Acad Sci USA 116:923–928. doi: 10.1073/pnas.1810402116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schierup MH, Mikkelsen AM, Hein J. 2001. Recombination, balancing selection and phylogenies in MHC and self-incompatibility genes. Genetics 159:1833–1844. doi: 10.1093/genetics/159.4.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hallström T, Siegel C, Mörgelin M, Kraiczy P, Skerka C, Zipfel PF. 2013. CspA from Borrelia burgdorferi inhibits the terminal complement pathway. mBio 4:e00481-13. doi: 10.1128/mBio.00481-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammerschmidt C, Koenigs A, Siegel C, Hallström T, Skerka C, Wallich R, Zipfel PF, Kraiczy P. 2014. Versatile roles of CspA orthologs in complement inactivation of serum-resistant Lyme disease spirochetes. Infect Immun 82:380–392. doi: 10.1128/IAI.01094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kraiczy P, Rossmann E, Brade V, Simon MM, Skerka C, Zipfel PF, Wallich R. 2006. Binding of human complement regulators FHL-1 and factor H to CRASP-1 orthologs of Borrelia burgdorferi. Wien Klin Wochenschr 118:669–676. doi: 10.1007/s00508-006-0691-1. [DOI] [PubMed] [Google Scholar]
- 65.Sharma SK, Pourat J, Abdel-Atti D, Carlin SD, Piersigilli A, Bankovich AJ, Gardner EE, Hamdy O, Isse K, Bheddah S, Sandoval J, Cunanan KM, Johansen EB, Allaj V, Sisodiya V, Liu D, Zeglis BM, Rudin CM, Dylla SJ, Poirier JT, Lewis JS. 2017. Noninvasive interrogation of DLL3 expression in metastatic small cell lung cancer. Cancer Res 77:3931–3941. doi: 10.1158/0008-5472.CAN-17-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson CR, Frye JG, Quinn FD, Gherardini FC. 2001. Increased expression of Borrelia burgdorferi vlsE in response to human endothelial cell membranes. Mol Microbiol 41:229–239. doi: 10.1046/j.1365-2958.2001.02511.x. [DOI] [PubMed] [Google Scholar]
- 67.Bykowski T, Babb K, von Lackum K, Riley SP, Norris SJ, Stevenson B. 2006. Transcriptional regulation of the Borrelia burgdorferi antigenically variable VlsE surface protein. J Bacteriol 188:4879–4889. doi: 10.1128/JB.00229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eddy SR. 2011. Accelerated profile HMM searches 2011. PLoS Comp Biol 7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hernández Y, Bernstein R, Pagan P, Vargas L, McCaig W, Ramrattan G, Akther S, Larracuente A, Di L, Vieira FG, Qiu WG. 2018. BpWrapper: BioPerl-based sequence and tree utilities for rapid prototyping of bioinformatics pipelines. BMC Bioinformatics 19:76. doi: 10.1186/s12859-018-2074-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yu G, Smith D, Zhu H, Guan Y, Lam T. 2017. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol 8:28–36. doi: 10.1111/2041-210X.12628. [DOI] [Google Scholar]
- 73.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. 2002. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 18:S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.S71. [DOI] [PubMed] [Google Scholar]
- 74.Crooks GE, Hon G, Chandonia J-M, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ivanova L, Christova I, Neves V, Aroso M, Meirelles L, Brisson D, Gomes-Solecki M. 2009. Comprehensive seroprofiling of sixteen B. burgdorferi OspC: implications for Lyme disease diagnostics design. Clin Immunol 132:393–400. doi: 10.1016/j.clim.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molins CR, Sexton C, Young JW, Ashton LV, Pappert R, Beard CB, Schriefer ME. 2014. Collection and characterization of samples for establishment of a serum repository for Lyme disease diagnostic test development and evaluation. J Clin Microbiol 52:3755–3762. doi: 10.1128/JCM.01409-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 78.R Core Team. 2013. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. [Google Scholar]
- 79.Ritz C, Baty F, Streibig JC, Gerhard D. 2015. Dose-Response Analysis Using R. PLoS One 10:E0146021. doi: 10.1371/journal.pone.0146021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data visualization and statistical analysis were performed in the R statistical computing environment (78) accessed with RStudio. The alignment of translated vls sequences, ELISA readings, and R scripts are publicly available on GitHub at https://github.com/weigangq/vls-mabs.