Abstract
The nucleotide sequence of the Anaplasma centrale 16S rRNA gene was determined and compared with the sequences of ehrlichial bacteria. The sequence of A. centrale was closely related to Anaplasma marginale by both level-of-similarity (98.08% identical) and distance analysis. A species-specific PCR was developed based upon the alignment data. The PCR can detect A. centrale DNA extracted from 10 infected bovine red blood cells in a reaction mixture. A. centrale DNA was amplified in the reaction, but not other related ehrlichial species.
Anaplasmosis is an arthropod-borne disease of cattle and other ruminants caused by the intraerythrocytic rickettsiae of the genus Anaplasma (18). Based upon location within the infected erythrocyte, two species of Anaplasma that infect cattle have been described, Anaplasma marginale and Anaplasma centrale. In addition to having differences in morphology, these species display differences in virulence and geographical distribution. A. marginale causes most severe hemolytic anemia and occurs in all six temperate continents (23). Although severe disease may also occur with A. centrale, it usually causes only a mild anemia in most cases (18). A. centrale is also widely distributed but has never been reported in North America. Despite differences in morphology, distribution, and virulence, A. marginale and A. centrale appear to be closely related as shown by comparison of protein and antigenic composition (5, 9, 11–14, 20, 26). Major surface protein 2 is the immunodominant outer membrane protein in both species and bears epitopes conserved among species of the Anaplasma group (13, 15, 16). Consequently these two species demonstrated serological cross-reactions in various studies: complement fixation assay, capillary tube agglutination test (3), and enzyme-linked immunosorbent assay (6–8). Cross-protective immunity to A. marginale in cattle can also be acquired by infecting cattle with A. centrale (1, 4, 12, 22). Although these observations support a phylogenetic relationship between A. centrale and A. marginale, the phylogenetic position of A. centrale has not yet been determined. In this paper, we report the results of a study of the 16S rRNA gene sequence of A. centrale. In addition, we developed a species-specific PCR that can amplify a partial sequence of A. centrale 16S rRNA gene.
MATERIALS AND METHODS
Ehrlichia strains.
A. centrale Aomori was first isolated from peripheral blood of infected cattle in the Aomori prefecture, Japan, in 1966. The isolate was maintained by the National Institute of Animal Health, Tsukuba, Japan. The heparinized peripheral blood was taken and kept in 10% glycerin at −80°C until used. The blood was thawed and fixed with 70% ethanol for international transfer of the infected blood. Genomic DNA of A. centrale was extracted by using the QIAamp blood kit procedure (QIAGEN GmbH, Hilden, Germany). Finally, DNA was extracted in 200 μl of Tris-EDTA buffer and stored at −20°C until used.
PCR amplification and sequence of A. centrale 16S rRNA gene.
The amplification of the 16S rRNA of A. centrale was performed by PCR with two sets of primers, fD1 (5′-AGA-GTT-TGA-TCC-TGG-CTC-AG-3′)-EHR16SR (5′- TAG-CAC-TCA-TCG-TTT-ACA-GC-3′) and EHR16SD (5′-GGT-ACC-YAC-AGA-AGA-AGT-CC-3′)-Rp2 (5′-ACG-GCT-ACC-TTG-TTA-CGA-CTT-3′), as described previously (17). EHR16SD and EHR16SR are Ehrlichia genus-specific primers, and fD1 and Rp2 are the universal primers. In each test, distilled water and DNA of the human granulocytic ehrlichia (HGE) agent were included as a negative and a positive control, respectively. The amplification products were visualized on a 1% agarose gel after electrophoretic migration of 8 μl of amplified material. The PCR products for DNA sequencing were purified with QIAquick PCR purification kits (QIAGEN GmbH). For DNA sequencing reactions, fluorescence-labeled dideoxynucleotide technology was used (Perkin-Elmer, Applied Biosystems Division, Foster City, Calif.). The sequencing fragments were separated, and data were collected on an ABI PRISM 310 Genetic Analyzer (Perkin-Elmer). The collected sequences were assembled and edited with the AutoAssembler (version 1.4; Perkin-Elmer). The obtained sequence was confirmed by performing the same PCR and sequence methods three times.
Data analysis.
The obtained sequence of A. centrale was compared with 16S rRNA gene sequences of related ehrlichial species and Rickettsia rickettsii deposited in GenBank. Pairwise percent identities of the sequences with all gaps omitted were calculated by a program designed by H. Ogata, IGS CNRS-UMR, Marseilles, France. Multiple alignment analysis, distance matrix calculation, and construction of a phylogenetic tree were performed with the CLUSTAL W program (21), version 1.8, available in the DNA Data Bank of Japan (Mishima, Japan [http://www.ddbj.nig.ac.jp/htmls/E-mail/clustalw-e.html]). The distance matrices for the aligned sequences with all gaps ignored were calculated using the Kimura two-parameter method (2), and the neighbor-joining method was used for constructing a phylogenetic tree (19). The stability of the tree obtained was estimated by bootstrap analysis for 1,000 replications using the same program. Tree figures were generated using the TREEVIEW program, version 1.61 (10).
Species-specific PCR.
A forward primer, CENTRALE (5′-CAA-ATC-TGT-AGC-TTG-CTA-CGG-A-3′), was designed based upon the alignment data and used with the Ehrlichia genus-specific reverse primer GA1UR (5′-GAG-TTT-GCC-GGG-ACT-TCT-TCT-3′) (24) to amplify a partial 16S rRNA gene of A. centrale specifically. PCR conditions were the same as above with an annealing temperature at 55°C and 40 cycles. The sensitivity of the PCR was evaluated using a dilution of DNA in water. The specificity of the reaction was also tested with DNA extracted from related ehrlichial species: the HGE agent strain Webster (J. S. Dumler), Ehrlichia equi strain California (J. E. Madigan), Ehrlichia phagocytophila strain 1602 (A. Garcia-Perez), A. marginale strains Florida and South Idaho (G. H. Palmer), Ehrlichia platys strain Okinawa (H. Inokuma), Ehrlichia canis strain Oklahoma (J. Dawson), Ehrlichia chaffeensis strain Arkansas (J. Dawson), Ehrlichia muris (M. Kawahara), Cowdria ruminantium (C. E. Yunker), Wolbachia pipientis (M. Taylor), Ehrlichia risticii (ATCC), Ehrlichia sennetsu strain Miyayama (G. Dash), and Neorickettsia helminthoeca (Y. Rikihisa).
Sequences for phylogenetic trees.
The species and GenBank accession numbers of the 16S rRNA gene sequences used to construct phylogenetic trees are as follows: E. chaffeensis, M73222; the new Ehrlichia sp. found in Ixodes ovatus strain Yamaguchi, AF260591; E. muris, U15527; Ehrlichia ewingii, M73227; E. canis, M73221; C. ruminantium, AF069758; E. phagocytophila, M73224; E. equi, M73223; HGE agent, U02521; Ehrlichia bovis, U03775; E. platys, M82801; A. marginale, M60313, W. pipientis, AF179630; E. sennetsu, M73225; E. risticii, M21290; N. helminthoeca, U12457; and R. rickettsii, U11021.
Nucleotide sequence accession numbers.
The sequence of the partial 16S rRNA gene of A. centrale has been deposited in the GenBank data library under accession number AF283007.
RESULTS AND DISCUSSION
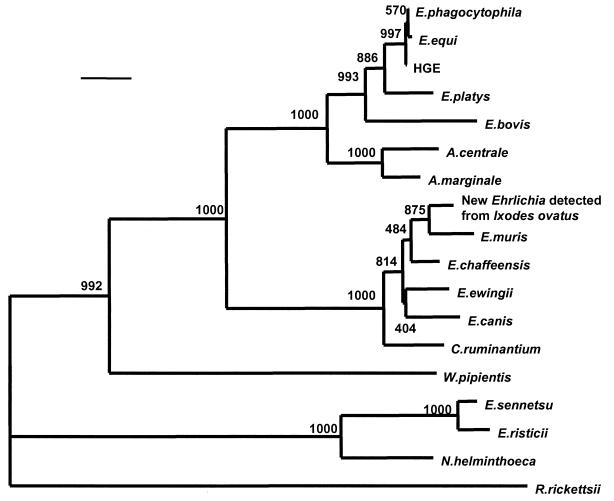
PCR using two sets of primers yielded a 1,425-bp nucleotide sequence. The level of similarity and evolutionary distance between 16S rRNA gene sequences are shown in Table 1. The sequence of 16S rRNA A. centrale was most similar to that of A. marginale, with 98.08% nucleotide identity. It also had the closest evolutionary distance to A. marginale (0.019). The related group species, including E. bovis, E. platys, E. phagocytophila, E. equi, and the HGE agent, had similar sequences, with levels of sequence similarity of more than 95% and an evolutionary distance of 0.041 to 0.054. All other species in the family were only distantly related phylogenically (levels of sequence similarity, 84 to 92%; evolutionary distance, 0.082 to 0.178). The 16S rRNA gene sequence of A. centrale was included in a phylogenetic tree, and the organism was demonstrated to be close to A. marginale and the E. phagocytophila group (Fig. 1). The phylogenetic relationship between A. centrale and A. marginale was significantly independent, with the bootstrap value of 1,000.
TABLE 1.
Levels of similarity and evolutionary distances between 16S rRNA gene sequences
| Organism | Level of similarity (%) or evolutionary distancea
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. centrale | A. marginale | E. bovis | E. platys | HGE agent | E. equi | E. phagocytophila | E. muris | New Ehrlichia sp. | E. chaffeensis | E. ewingii | E. canis | C. ruminantium | W. pipientis | E. sennetsu | E. risticii | N. helminthoeca | R. rickettsii | |
| A. centrale | 0.019 | 0.054 | 0.043 | 0.041 | 0.043 | 0.042 | 0.087 | 0.082 | 0.087 | 0.088 | 0.086 | 0.088 | 0.129 | 0.176 | 0.178 | 0.175 | 0.182 | |
| A. marginale | 98.08 | 0.046 | 0.037 | 0.036 | 0.038 | 0.037 | 0.083 | 0.082 | 0.083 | 0.085 | 0.083 | 0.086 | 0.132 | 0.177 | 0.177 | 0.172 | 0.184 | |
| E. bovis | 94.80 | 95.51 | 0.034 | 0.033 | 0.033 | 0.032 | 0.092 | 0.094 | 0.094 | 0.097 | 0.097 | 0.095 | 0.138 | 0.184 | 0.186 | 0.177 | 0.194 | |
| E. platys | 95.58 | 96.15 | 96.58 | 0.015 | 0.014 | 0.014 | 0.086 | 0.087 | 0.085 | 0.087 | 0.089 | 0.087 | 0.125 | 0.172 | 0.174 | 0.168 | 0.191 | |
| HGE agent | 96.09 | 96.37 | 96.73 | 98.50 | 0.001 | 0.001 | 0.082 | 0.079 | 0.077 | 0.079 | 0.081 | 0.079 | 0.126 | 0.172 | 0.174 | 0.166 | 0.181 | |
| E. equi | 95.94 | 96.22 | 96.72 | 98.57 | 99.86 | 0.001 | 0.082 | 0.081 | 0.078 | 0.081 | 0.082 | 0.081 | 0.127 | 0.173 | 0.175 | 0.167 | 0.183 | |
| E. phagocytophila | 96.01 | 96.29 | 96.79 | 98.65 | 99.93 | 99.93 | 0.081 | 0.080 | 0.077 | 0.080 | 0.081 | 0.080 | 0.126 | 0.172 | 0.174 | 0.166 | 0.182 | |
| E. muris | 91.88 | 92.23 | 91.45 | 91.80 | 92.39 | 92.38 | 92.46 | 0.014 | 0.019 | 0.023 | 0.028 | 0.029 | 0.135 | 0.181 | 0.186 | 0.170 | 0.197 | |
| New Ehrlichia sp. | 92.02 | 92.30 | 91.38 | 91.73 | 92.67 | 92.53 | 92.60 | 98.65 | 0.013 | 0.019 | 0.022 | 0.027 | 0.132 | 0.182 | 0.186 | 0.166 | 0.194 | |
| E. chaffeensis | 91.95 | 92.09 | 91.24 | 91.94 | 92.89 | 92.74 | 92.81 | 98.09 | 98.72 | 0.017 | 0.018 | 0.024 | 0.132 | 0.181 | 0.184 | 0.164 | 0.190 | |
| E. ewingii | 91.73 | 91.79 | 90.73 | 91.43 | 92.52 | 92.38 | 92.45 | 97.44 | 97.87 | 98.15 | 0.019 | 0.023 | 0.134 | 0.185 | 0.189 | 0.171 | 0.196 | |
| E. canis | 91.22 | 92.07 | 90.94 | 91.42 | 92.30 | 92.16 | 92.23 | 97.02 | 97.59 | 98.01 | 98.08 | 0.028 | 0.135 | 0.181 | 0.186 | 0.169 | 0.191 | |
| C. ruminantium | 91.66 | 91.94 | 91.38 | 91.79 | 92.67 | 92.52 | 92.59 | 97.09 | 97.23 | 97.51 | 97.58 | 97.15 | 0.129 | 0.178 | 0.184 | 0.170 | 0.184 | |
| W. pipientis | 88.11 | 87.67 | 87.47 | 88.66 | 88.47 | 88.39 | 88.46 | 87.96 | 87.82 | 87.74 | 87.59 | 87.58 | 88.10 | 0.178 | 0.181 | 0.171 | 0.186 | |
| E. sennetsu | 84.25 | 84.53 | 83.91 | 84.87 | 84.97 | 84.89 | 84.96 | 84.18 | 84.04 | 84.18 | 83.66 | 84.00 | 84.38 | 84.41 | 0.010 | 0.050 | 0.195 | |
| E. risticii | 84.11 | 84.53 | 83.76 | 84.80 | 84.82 | 84.74 | 84.81 | 83.82 | 83.35 | 83.97 | 83.44 | 83.72 | 83.95 | 84.19 | 99.00 | 0.044 | 0.197 | |
| N. helminthoeca | 83.76 | 84.11 | 83.63 | 84.60 | 84.69 | 84.54 | 84.61 | 84.26 | 84.55 | 84.55 | 84.03 | 84.37 | 84.18 | 84.27 | 94.28 | 94.78 | 0.188 | |
| R. rickettsii | 83.56 | 83.33 | 82.56 | 82.90 | 83.64 | 83.49 | 83.56 | 82.55 | 82.69 | 83.05 | 82.52 | 82.80 | 83.32 | 83.13 | 82.22 | 82.15 | 81.88 | |
The values on the upper right are the evolutionary distance calculated by using the Kimura two-parameter method in the CLUSTAL W program. The values on the lower left are the levels of fractional nucleotide identity between sequences.
New Ehrlichia species detected from l. ovatus tick.
FIG. 1.
Phylogenetic relationship between A. centrale and various Ehrlichia spp. based on the 16S rRNA gene sequences. The neighbor-joining method was used to construct the phylogenetic tree using the CLUSTAL W program. The scale bar represents 1% divergence. The numbers at nodes are the proportions of 1,000 bootstrap resamplings that support the topology shown.
Since Weisburg et al. (25) showed the usefulness of 16S rRNA gene sequence analysis in phylogenetic determination, this approach has been widely used to both identify newly discovered bacteria as well as redefine existing taxonomy. A. centrale has been thought to be close to A. marginale on the basis of morphological, protein structural, and immunological studies. The present results confirm that A. centrale is an independent species and most closely related to A. marginale by both level-of-similarity and distance analysis.
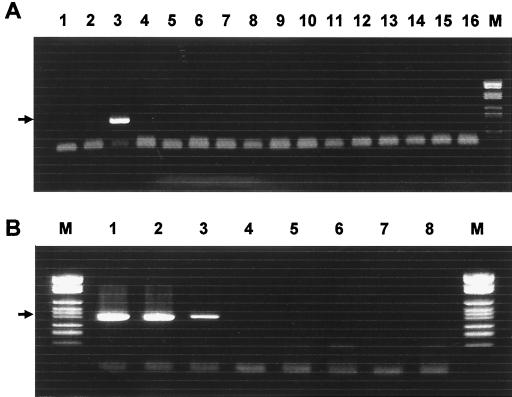
Based upon the alignment data of the 16S rRNA gene of ehrlichial species, a new PCR primer was designed to amplify the A. centrale DNA specifically. The PCR produced a 403-bp fragment of the 16S rRNA gene with A. centrale DNA extracted from infected bovine blood cells. No amplification was observed in the specificity test using other DNA from related ehrlichial species, including most closely related A. marginale strains (Fig. 2A). Nonspecific bands were not observed in a reaction with A. centrale, although there were primer dimer bands in negative controls. With this set of primers, the PCR can detect A. centrale DNA from 10 infected red blood cells (Fig. 2B). These findings suggest that the PCR can be used to detect A. centrale DNA specifically from a small amount of cattle blood. Only one strain of A. centrale was used in the present study for PCR. The positive PCR detection of A. centrale might be strain specific. Studies with additional strains of A. centrale and other species may confirm the sensitivity and specificity of the PCR. PCR is a powerful tool for epidemiological or diagnostic purposes because of its high sensitivity and specificity; however, there have been few molecular tools available for A. centrale identification until now. Most of the epidemiological data on A. centrale are based on immunological or hematological examination, and fewer findings have been reported on the epidemiology of A. centrale than on that of A. marginale. The present study revealed the possibility of developing a PCR to detect A. centrale. Such a PCR would be a useful tool for epidemiological studies and the diagnosis of A. centrale in cattle.
FIG. 2.
(A) A. centrale-specific PCR was evaluated for its specificity with DNA from A. marginale strains South Idaho (lane 1) and Florida (lane 2), A. centrale (lane 3), HGE agent (lane 4), E. equi (lane 5), E. phagocytophila (lane 6), E. platys (lane 7), W. pipientis (lane 8), E. muris (lane 9), E. chaffeensis (lane 10), E. canis (lane 11), C. ruminantium (lane 12), E. risticii (lane 13), E. sennetsu (lane 14), N. helminthoeca (lane 15), and distilled water (lane 16). (B) The PCR was also evaluated for its sensitivity with diluted DNA extracted from A. centrale-infected bovine red blood cells. Lane 1 contains DNA equivalent to that extracted from 103 infected cells. Consequently, DNA equivalent to that extracted from 102, 101, 100, 10−1, 10−2, and 10−3 infected cells was used in lane 2 to 7, respectively. Lane M, DNA ladder. Arrows show the 412-bp target band.
ACKNOWLEDGMENTS
We thank H. Ogata for analyzing the sequence data and G. H. Palmer for reviewing the manuscript.
H. Inokuma was supported by a grant from the EGIDE, Paris, France.
REFERENCES
- 1.Abdala A A, Pipano E, Aguirre D H, Grido A B, Zurbriggen M A, Mangold A J, Gugielmone A A. Frozen and fresh Anaplasma centrale vaccines in the protection of cattle against Anaplasma marginale infection. Rev Elev Med Vet Pays Trop. 1990;43:155–158. [PubMed] [Google Scholar]
- 2.Kimura M. A simple method for estimating evolutional rates of base substitutions through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 3.Kuttler K L. Serological relationship of Anaplasma marginale and Anaplasma centrale as measured by the complement fixation and capillary tube agglutination test. Res Vet Sci. 1967;8:207–211. [PubMed] [Google Scholar]
- 4.Kuttler K L. A study of the immunological relationship of Anaplasma marginale and Anaplasma centrale. Res Vet Sci. 1967;8:467–471. [PubMed] [Google Scholar]
- 5.McGuire T L, Palmer G H, Goff W L, Johnson M I, Davis W C. Common and isolated-restricted antigens of Anaplasma marginale detected with monoclonal antibodies. Infect Immun. 1984;45:697–700. doi: 10.1128/iai.45.3.697-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Molloy J B, Bowles P M, Knowles D P, McElwain T F, Bock R E, Kingston T G, Blight G W, Dalgleish R J. Comparison of a competitive inhibitor ELISA and the card agglutination test for detection of antibodies to Anaplasma marginale and Anaplasma centrale in cattle. Aust Vet J. 1999;77:245–249. doi: 10.1111/j.1751-0813.1999.tb11712.x. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura Y, Shimizu S, Minami T, Ito S. Enzyme-linked immunosorbent assay using solubilised antigen for detection of antibodies to Anaplasma marginale. Trop Anim Health Prod. 1988;20:259–266. doi: 10.1007/BF02239994. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura Y, Shimizu S, Minami T, Ito S. Enzyme-linked immunosorbent assay for detection of antibodies to Anaplasma centrale. J Vet Med Sci. 1988;50:933–935. doi: 10.1292/jvms1939.50.933. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura Y, Kawazu S, Minami T. Analysis of protein comparisons and surface protein epitopes of Anaplasma marginale and Anaplasma centrale. J Vet Med Sci. 1991;53:73–79. doi: 10.1292/jvms.53.73. [DOI] [PubMed] [Google Scholar]
- 10.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Applic Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 11.Palmer G H, McGuire T L. Immune serum against Anaplasma marginale initial bodies neutralizes infectivity for cattle. J Immunol. 1984;133:1010–1015. [PubMed] [Google Scholar]
- 12.Palmer G H, Barbet A F, Davis W C, McGuire T L. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science. 1986;231:1299–1302. doi: 10.1126/science.3945825. [DOI] [PubMed] [Google Scholar]
- 13.Palmer G H, Barbet A F, Muske A J, Katende J M, Rurangirna F, Stikap V, Pipano E, Davis W C, McGuire T L. Recognition of conserved surface protein epitopes on Anaplasma centrale and Anaplasma marginale isolates from Israel, Kenya and the United States. Int J Parasitol. 1988;18:33–38. doi: 10.1016/0020-7519(88)90033-1. [DOI] [PubMed] [Google Scholar]
- 14.Palmer G H, Oberle S M, Barbet A F, Davis W C, Goff W L, McGuire T L. Immunization with a 36-kilodalton surface protein induced protection against homologous and heterologous Anaplasma marginale challenge. Infect Immun. 1988;56:1526–1531. doi: 10.1128/iai.56.6.1526-1531.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer G H, Eid G, Barbet A F, McGuire T L, McElwain T F. The immunoprotective Anaplasma marginale major surface protein 2 (MSP-2) is encoded by a polymorphic multigene family. Infect Immun. 1994;62:3808–3816. doi: 10.1128/iai.62.9.3808-3816.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer G H, Abbott J R, French D M, McElwain T F. Persistence of Anaplasma ovis infection and conservation of the msp-2 and msp-3 multigene families within the genus Anaplasma. Infect Immun. 1998;66:6035–6039. doi: 10.1128/iai.66.12.6035-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parola P, Roux V, Camicas J-L, Baradji I, Brouqui P, Raoult D. Detection of ehrlichiae in African ticks by PCR. Trans R Soc Trop Med Hyg. 2000;94:707–708. doi: 10.1016/s0035-9203(00)90243-8. [DOI] [PubMed] [Google Scholar]
- 18.Ristic M, Kreier J P. Genus Anaplasma. In: Kreig N R, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 1. Baltimore, Md: Wiliams and Wilkins; 1984. pp. 720–722. [Google Scholar]
- 19.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Med Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 20.Shkap V, Pipano E, McGuire T L, Palmer G H. Identification of immunodominant polypeptides common between Anaplasma centrale and Anaplasma marginale. Vet Immunol Immunopathol. 1991;29:31–40. doi: 10.1016/0165-2427(91)90050-m. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turton J A, Katsande T C, Matingo M B, Jorgensen W K, Ushewokunze-Obatolu U, Dalgliesh R J. Observation on the use of Anaplasma centrale for immunization of cattle against anaplasmosis in Zimbabwe. Onderstepoort J Vet Res. 1998;65:81–86. [PubMed] [Google Scholar]
- 23.Wannduragala L, Ristic M. Anaplasmosis. In: Woldehiwet Z, Ristic M, editors. Rickettsial and chlamydial diseases of domestic animals. Oxford, United Kingdom: Pergamon Press; 1993. pp. 65–87. [Google Scholar]
- 24.Warner C K, Dawson J E. Genus- and species-level identification of Ehrlichia species by PCR and sequencing. In: Persing D H, editor. PCR protocols for emerging infectious diseases. Washington, D.C.: ASM Press; 1996. pp. 100–105. [Google Scholar]
- 25.Weisburg W G, Barnes S, Pelletier D, Lane D. 16S rRNA ribosome DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zakimi S, Tsuji N, Fujisaki K. Protein analysis of Anaplasma marginale and Anaplasma centrale by two-dimentional polyacrylamide gel electrophoresis. J Vet Med Sci. 1994;56:1025–1027. doi: 10.1292/jvms.56.1025. [DOI] [PubMed] [Google Scholar]