Abstract
Given the lack of effective vaccines to control Streptococcus suis infection and the lack of a rapid and reliable molecular diagnostic assay to detect its infection, a polyclonal antibody was raised against the whole-cell protein of S. suis type 2 and used to screen an S. suis gene library in an effort to identify protective antigen(s) and antigens of diagnostic importance. A clone that produced a 45-kDa S. suis-specific protein was identified by Western blotting. Restriction analysis showed that the gene encoding the 45-kDa protein was present on a 1.6-kb pair DraI region on the cloned chromosomal fragment. The nucleotide sequence contained an open reading frame that encoded a polypeptide of 448 amino acid residues with a calculated molecular mass of 48.8 kDa, in close agreement with the size observed on Western blots. A GenBank database search revealed that the derived amino acid sequence is homologous to the sequence of glutamate dehydrogenase (GDH) protein isolated from various sources, including conserved motifs and functional domains typical of the family 1-type hexameric GDH proteins, thus placing it in that family. Because of these similarities, the protein was designated the GDH of S. suis. Hybridization studies showed that the gene is conserved among the S. suis type 2 strains tested. Antiserum raised against the purified recombinant protein was reactive with a protein of the same molecular size as the recombinant protein in S. suis strains, suggesting expression of the gene in all of the isolates and antigenic conservation of the protein. The recombinant protein was reactive with serum from pigs experimentally infected with a virulent strain of S. suis type 2, suggesting that the protein might serve as an antigen of diagnostic importance to detect S. suis infection. Activity staining showed that the S. suis GDH activity is NAD(P)H dependent but, unlike the NAD(P)H-dependent GDH from various other sources, that of S. suis utilizes l-glutamate rather than α-ketoglutarate as the substrate. Highly virulent strains of S. suis type 2 could be distinguished from moderately virulent and avirulent strains on the basis of their GDH protein profile following activity staining on a nondenaturing gel. We examined the cellular location of the protein using a whole-cell enzyme-linked immunosorbent assay and an immunogold-labeling technique. Results showed that the S. suis GDH protein is exposed at the surface of intact cells.
Streptococcus suis type 2 is one of the causative agents of meningitis, arthritis, septicemia, endocarditis, encephalitis, abortions, polyserositis, bronchopneumonia, and sudden death in pigs (5, 6, 29, 36). S. suis has also been recovered from other animal species (5, 8). In humans, S. suis type 2 can cause meningitis, endocarditis, septicemia, and permanent deafness. All reported human cases of S. suis infection have been associated with slaughterhouse workers handling infected pork, and deaths have been reported among those workers (2, 27). For these reasons, attention has been focused on understanding the molecular mechanism of the disease process, identifying protective antigens that may be useful in the development of a subunit vaccine, and identifying antigens and the DNA region(s) that may be used in developing a rapid and sensitive molecular diagnostic assay for the detection of S. suis infection.
Strains of S. suis are divided into serotypes according to polysaccharide capsular antigens. Thirty-five capsular serotypes (types ½ and 1 through 34) have been identified (10, 11, 16, 25). Of the 35 serotypes, type 2 is the most frequently isolated serotype from pigs with disease, but strains belonging to other serotypes such as types ½, 1, 7, 9, and 14 can also cause disease (23, 36). However, not all strains of S. suis type 2 are virulent, and there is variation in the virulence of those strains that are virulent. The economic impact of S. suis infection on the swine industry is substantial due to a lack of effective means to control the infection (36), which, in turn, is largely due to a lack of understanding of the protective antigen(s) and pathogenic mechanism(s) and limited information on the genetics of the organism and the presence of many different serotypes with diverse genetic makeups (24, 36). A more thorough understanding of S. suis genetics will help in the development of new approaches to control S. suis infection.
We report the cloning, sequencing, and characterization of the gene encoding a 45-kDa antigen from a virulent strain of S. suis type 2 and show that the protein is surface exposed and conserved among S. suis type 2 strains tested and that it belongs to the NAD(P)H-dependent glutamate dehydrogenase (GDH) enzyme family. We also show that serum from pigs experimentally infected with a virulent strain of S. suis type 2 reacted with the 45-kDa recombinant protein, making it a good candidate for the development of a serological assay to detect S. suis infection. To our knowledge, this is the first report on the cloning and characterization of a major enzyme of S. suis involved in intermediary metabolism.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
S. suis type 2 strain 1933, a virulent field isolate recovered from a pig with meningitis, was obtained from M. M. Chengappa (Kansas State University, Manhattan, Kans.) and was used for the genomic library construction. Other S. suis isolates used in this study were obtained from the same source, and the degrees of pathogenicity of most of the isolates have been described previously (35). pUC19 propagated in Escherichia coli DH5α was used as the library expression vector. Plasmid, pGEM (Promega, Madison, Wis.) was used for DNA-sequencing purposes. Luria Berteni broth or agar was used to grow E. coli strains. Todd-Hewitt broth supplemented with 0.6% yeast (Difco Laboratories, Detroit, Mich.) was used to grow the S. suis strains. When appropriate, AMP was used at 60 μg/ml for E. coli. All cultures were grown at 37°C.
Chemicals and enzymes.
Restriction enzymes, T4 DNA ligase, and calf intestinal alkaline phosphatase were purchased either from Promega or New England Biolabs (Beverly, Mass.) and were used as recommended by the manufacturer. Molecular-grade chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.) or from Fisher Scientific Co. (Pittsburgh Pa.). The digoxigenin-labeled DNA molecular-weight marker II and digoxigenin-11-dUTP DNA-labeling kit and detection system were from Boehringer Mannheim (Indianapolis, Ind.). S. suis case serum samples were obtained from P. Halbur (Iowa State University, Ames, Iowa). The serum samples were from six pigs experimentally infected with a virulent strain of S. suis type 2 (ISU VDL isolate 40634/94).
Construction and screening of a recombinant DNA library and restriction mapping.
Genomic DNA was isolated by a previously described method (24) from a 30-ml culture of S. suis type 2 strain 1933 cells grown on Todd Hewitt broth containing 0.6% yeast. The DNA was digested with SpeI, and fragments were fractionated by agarose gel electrophoresis. Fragments in the size range of 2 to 23 kb were excised from the gel, purified by electroelution, and ligated into a pUC19 plasmid cloning vector that was digested to completion with XbaI. The recombinant plasmids were transformed into E. coli DH5α by electroporation. Transformed cells were plated onto Luria Bertani agar containing 60 μg of AMP (Ap60), isopropyl-β-d-thiogalactopyranoside (4 μl of a 20% solution), and X-Gal (40 μl of a 20-mg/ml solution), respectively, and grown at 37°C overnight. Resulting white colonies were transferred to a fresh plate and grown as above. One loopful of each colony was solubilized in 100 μl of 1× sodium dodecyl sulfate (SDS) sample buffer by heating for 5 min at 100°C. The preparation was centrifuged for 2 min to remove cellular debris, and the supernatant containing proteins was used for Western blot analysis with a 1:500 dilution of polyclonal antibody raised against whole-cell protein of S. suis type 2 as the primary antibody followed by a 1:1,000 dilution of anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase. Blots were developed with hydrogen peroxide and 4-chloro-1-naphthol (4CN). A colony designated DH5α(pOT401) was identified and characterized. The S. suis DNA insert in plasmid pOT401 was mapped by restriction analysis using standard protocols (28).
Nucleotide sequence determination and analysis.
The complete nucleotide sequences of both strands of the 1.6-kb DNA insert in plasmid pOT410 were determined by the dideoxy-chain termination method (30) using an automated nucleotide sequencer (Applied Biosystems, Foster City, Calif.). The nucleotide sequence and the deduced amino acid sequence were analyzed with the MacVector software (Oxford Molecular Group, Inc., Campbell, Calif.). Sequence similarity searches were performed with GenBank sequences by using the BLAST network service.
Electrophoresis and Southern blotting.
Genomic DNA (3 μg) from S. suis type 2 strains was digested with DraI restriction endonuclease. Restriction fragments were separated on a 0.8% agarose gel (Promega) in Tris-borate-EDTA buffer (90 mM Tris, 90 mM borate, 2 mM EDTA [pH 8.0]) and transferred to a positively charged nylon membrane (Boehringer Mannheim) using the method of Southern (34). Following DNA transfer, the nylon membrane was rinsed briefly in 6× SSC (1× SSC is 0.15 M NaCl, 0.015 M sodium citrate), air dried, and UV cross-linked to the DNA (GS gene linker; Bio-Rad, Richmond, Calif.).
Probe preparation and hybridization.
The 689-bp NcoI-PstI internal DNA fragment from pOT410 was labeled using a nonradioactive labeling system (Genius System; Boehringer Mannheim) that incorporated digoxigenin-11-dUTP by random priming as specified by the manufacturer. Prehybridization (2 h) and hybridization (16 h) were done at 65°C. Washes and hybrid detection were done according to the manufacturer's instruction with a Genius II nonradioactive labeling and detection kit (Boehringer Mannheim).
Overexpression of the recombinant protein.
The DNA insert in pOT410 was cloned in frame into a pBAD/Myc-His version B expression vector to create pOT411. pOT411 was transformed into E. coli TOP10-competent cells and overexpressed by following the manufacturer's protocol (Invitrogen).
Antigen and polyclonal antibody preparation.
Antigen was prepared as described elsewhere (14) with slight modifications. Following overexpression of protein in E. coli cells carrying plasmid pOT411, cells were solubilized in 1× SDS sample buffer by heating at 100°C for 5 min followed by centrifugation for 2 min to pellet cellular debris. Proteins in the supernatant were separated on SDS-10% polyacrylamide preparative gels, stained with Coomassie brilliant blue, and destained with a methanol-acetic acid-water mixture. The 45-kDa band then was excised from the gels and electroeluted. The protein concentration was determined using the method of Lowry et al. (21), with bovine serum albumen as a standard. Monospecific polyclonal antibody against the recombinant 45-kDa protein was obtained by immunizing New Zealand White rabbits (Shelton's Bunny Barn Rabbits, Waverly Hall, Ga.) subcutaneously at multiple sites with approximately 210 μg of purified protein emulsified 1:1 in Fruend complete adjuvant. The rabbits received one booster injection with the same antigen concentration emulsified 1:1 with Freund's incomplete adjuvant 14 days later and then were bled 7 days after the the booster was administered. Sera were filter sterilized and stored at −30°C until used.
GDH activity staining.
Cultures were grown overnight in 50 ml of Todd Hewitt or tryptic soy broth and centrifuged at 5,000 × g for 10 min at 4°C to pellet cells. Supernatant was discarded, and cells were resuspended in 1 ml of 50 mM Tris buffer (pH 8.0) and then disrupted by sonication (4°C) at a setting of 2.5, using two 30-s bursts with 10-s cooling intervals (Sonic Dismembrator, model F60; Fisher Scientific). Following sonication, samples were transferred to 1.5-ml Eppendorf tubes and centrifuged at 14,000 rpm (Eppendorf centrifuge 54-15C) for 2 min at room temperature. Fifteen microliters (approximately 165 μg) of the cell-free extract was subjected to nondenaturing polyacrylamide gel electrophoresis, modified by omitting SDS and β-mercaptoethanol. The acrylamide concentrations were 3.5 and 7.5% (wt/vol) in the stacking and resolving gels, respectively. Following electrophoresis, gels were incubated in 25 ml of staining solution consisting of 0.5 mM NAD(P)H or NAD, 50 mM Tris-HCl (pH 8.0), 20 mM l-glutamate, 0.3 mg of Nitro Blue Tetrazolium per ml, and 0.05 mg of phenazine methosulfate (1, 38) per ml. In another set of experiments, l-glutamate was replaced with 50 mM α-ketoglutarate. GDH activity appeared as a dark band against a clear background.
Western immunoblotting.
Cell-free extracts prepared as described above were vacuum concentrated (15-fold), and 15 μl of each sample was used for Western blot analysis (20, 28). The proteins were combined in a reaction mixture with a 1:500 dilution of the polyclonal antibody raised against the purified 45-kDa GDH protein, followed by a horseradish peroxidase-conjugated goat anti-rabbit IgG antibody (ICN) diluted 1:1,000. Bound antibodies were detected colorimetrically with hydrogen peroxide and 4CN. To screen pig antisera for antibody against the 45-kDa antigen, a 1:100 dilution of the pig sera was used as the primary antibody followed by a 1:1,000 dilution of alkaline phosphatase-conjugated affinity-purified anti-swine IgG (Rockland Immunochemicals, Gilbertsville, Pa.). Blots were developed with a 5-bromo-4-chloro-indolyl phosphate–nitroblue tetrazolium salt mixture.
Whole-cell ELISA and immunogold labeling.
A whole-cell enzyme-linked immunosorbent assay (ELISA) was performed essentially as described elsewhere (26) with plates coated with whole cells of S. suis type 2 strain 1933. Whole cells were combined in reaction mixtures with a 1:500 dilution of the polyclonal antibody raised against the purified 45-kDa GDH protein followed by a 1:1,000 dilution of horseradish peroxidase-conjugated goat anti-rabbit immunoglobulins (ICN Immunochemicals, Inc., Irvine, Calif.). Peroxidase activity was measured by the addition of 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonic acid) peroxidase substrate (Sigma) to each well. Following incubation for 1.5 h at room temperature, the optical density at 405 nm was read with an ELISA microplate reader. Negative controls included wells containing preimmune rabbit serum as the primary antibody, wells without antibody, and wells without antigen. Values were considered positive when the optical density at 405 nm was twice or more the reading obtained for control wells.
For immunogold labeling postembedding, S. suis cells were first fixed for 30 min at room temperature in 1% paraformaldehyde and 0.2% glutaraldehyde made in 0.1 M phosphate buffer (pH 7.4) and stored in the same fixative solution overnight at 4°C. After four washes, cells were postfixed in 1% osmium tetroxide for 2 h at room temperature. Washed cells were dehydrated in increased concentrations of ethanol and embedded in Durcupan ACM resin. Ultrathin sections (70 nm) were cut, placed on nickel grids, and etched with 3% hydrogen peroxide for 5 min. After lysin treatment, grids were floated on a blocking solution of phosphate-buffered saline (PBS [pH 7.4]) containing 5% nonfat dried milk and 0.1% bovine serum albumin for 30 min and incubated at 4°C overnight in a 1:50 dilution of the polyconal antibody raised against the purified 45-kDa GDH protein. For the control, the primary antibody was replaced with preimmune rabbit serum. A second control that omitted the primary antibody was also used. After washing, grids were incubated for 2 h in a 1:40 dilution of goat anti-rabbit antibody conjugated to 10-nm gold particles (Sigma) that had been centrifuged to eliminate aggregates. The grids were washed twice in PBS (5× concentration) and then in regular PBS (three times, 5 min per wash), incubated in 2% glutaraldehyde, and washed again in PBS and distilled water, respectively (five times, 2 min each). The sections were stained for 35 min in 4% aqueous uranyl acetate and for 3 min in 0.4% aqueous lead. The grids then were Formvar coated, and sections were visualized with a Philips 301 transmission electron microscope (FEI Co., Hillsboro, Oreg.).
RESULTS
Identification and localization of the gdh gene.
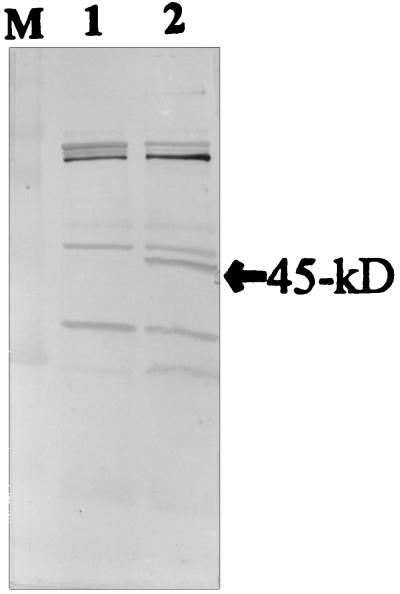
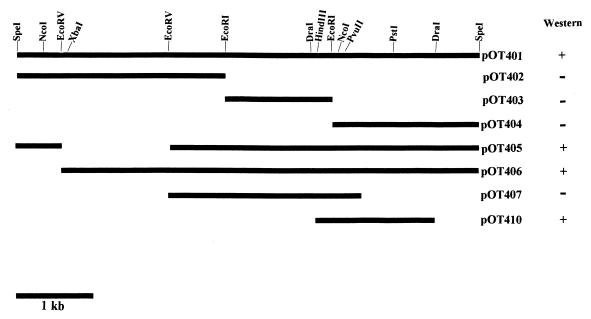
The gene library yielded a colony designated E. coli DH5α(pOT401) that produced a 45-kDa protein which reacted to the polyclonal antibody raised against whole-cell protein of S. suis type 2. The protein was not present in the negative control, indicating that it originated from the cloned S. suis DNA fragment. Other reactive bands were considered to be E. coli protein bands that cross-reacted with the antibody as judged by the result in the negative control lane (Fig. 1). On these bases, the 45-kDa protein was chosen and characterized. A series of deletion subclones was generated in order to define the location of the gdh structural gene and the promoter elements for transcriptional initiation. Subcloning localized the gene to within a 1.6-kb DraI region on the cloned chromosomal fragment (Fig. 2). The gene encoding the protein was well expressed from both the original recombinant plasmid pOT401 and the plasmid pOT410, which contains the 1.6-kb DraI insert DNA in the reverse orientation in pUC19. This indicated that the gdh gene was expressed from promoter elements located within the cloned DNA fragment.
FIG. 1.
Immunoblot (Western blot) analysis with the polyclonal antibody raised against whole-cell proteins of S. suis type 2. Lanes: M, rainbow molecular weight marker (Amersham); 1, E. coli DH5α(pUC19) negative control; 2, E. coli DH5α(pOT401). The location of the 45-kDa GDH protein is indicated by the arrow.
FIG. 2.
Restriction map and characteristics of subclones derived from pOT401. Restriction endonuclease sites are indicated above the top line. Clones were analyzed for expression of recombinant protein by Western immunoblot with the polyclonal antibody raised against whole-cell proteins of S. suis type 2 as the primary antibody. Subclones in which recombinant GDH was detected are denoted by a plus sign. Clones with no detectable recombinant GDH are denoted by a minus sign.
Nucleotide sequence analysis.
Analysis of the nucleotide sequence of the cloned gene revealed that it contains an open reading frame that codes for a protein of 448 amino acid residues with a calculated molecular mass of 48.8 kDa (data not shown). This is in close agreement with the 45-kDa molecular mass observed by Western blot analysis (Fig. 1). Potential promoter sequences and a ribosome binding site were located upstream from the open reading frame on the basis of homology with published concensus sequences (9, 12, 32). The protein lacks a signal sequence (31, 37) and has an isoelectric point of 5.3. A hydrophilicity plot (19) revealed several regions of high hydrophilicity and few short hydrophilic regions compared with regions of hydrophobicity (data not shown). The calculated G+C content of the gdh gene was 41.5%, in agreement with the G+C content reported for S. suis (38 to 42% [18]). An 11-bp inverted repeat was found downstream of the stop codon between positions 1692 and 1703 and positions 1711 and 1721.
A GenBank database search revealed that the derived amino acid sequence shared identity (48 to 59%) with GDH proteins isolated from a variety of organisms, all of which fall into family 1. Highly conserved domains characteristic of the family-1 hexameric GDHs such as KFLGFEQ (amino acid positions 108 through 114), RPEATGY (positions 208 through 214), GSGNVAQYAVQKATELG (positions 240 through 256), DSNG (positions 274 through 277), and EKYD (positions 418 through 421) were present, but several amino acid substitutions were apparent (3). Because of these similarities the protein was designated the GDH of S. suis. On the nucleotide level, the gdh gene of S. suis is not homologous with similar genes or any bacterial gene in the GenBank data base.
Construction and overexpression of the GDH protein.
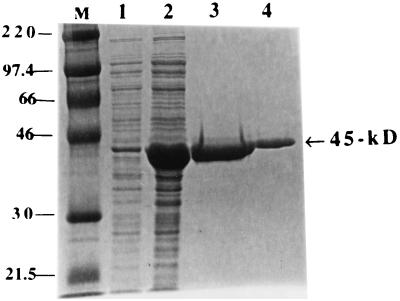
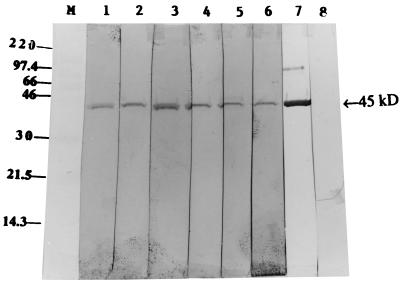
To produce a sufficient quantity of the GDH protein for antibody production, S. suis DNA inserted in plasmid pOT410 was cloned in frame into the pBAD/Myc-His version B expression vector, overexpressed, and gel purified. The purified protein gave a prominent 45-kDa band (Fig. 3) and was used to generate polyclonal antibody in rabbit cells.
FIG. 3.
Overexpression and purification of the 45-kDa recombinant GDH protein. The protein was overexpressed and purified as described in Materials and Methods. An SDS-10% polyacrylamide gel stained with Coomassie brilliant blue R-250 is shown. Lanes: M, rainbow molecular size marker in kilodaltons; 1, whole-cell lysate of pOT411 transformant of E. coli TOP10 uninduced; 2, whole-cell lysate of E. coli transformed with pOT411 and induced with arabinose; 3 and 4, different amounts of the recombinant protein purified from pOT411 transformant of E. coli TOP10.
Conservation of the gdh gene and its product.
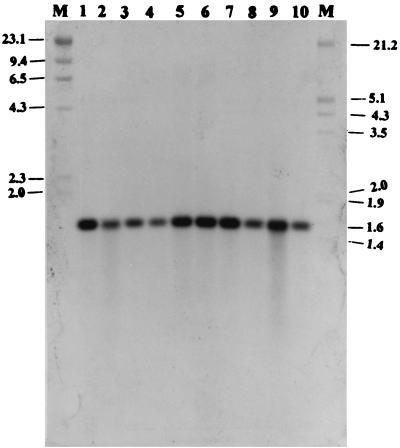
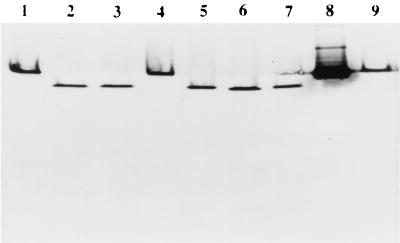
Because genetic diversity and antigenic variation have been shown to exist among isolates of the same serotype (24), we wanted to determine whether the gene and its product are conserved among S. suis type 2 strains. When 10 isolates were used in Southern blot analysis with a 689-bp NcoI-PstI fragment probe derived from within the gdh gene, the probe hybridized to a 1.6-kb fragment in all of the strains, suggesting that the gene originated from S. suis and is conserved in the strains tested (Fig. 4). When the genomic DNA of S. suis type 2 strain 1933 was digested with several restriction enzymes that do not cut within the gdh gene, followed by Southern analysis with the 689-bp probe, only one band was visible, indicating that the gene is present in the chromosome of S. suis as a single copy (data not shown).
FIG. 4.
Southern blot of chromosomal DNA from different strains of S. suis type 2 digested with DraI and hybridized to the 689-bp NcoI-PstI fragment internal to the GDH gene. Lanes: M, digoxigenin- labeled molecular weight markers II and III; 1, S. suis 1933; 2, S. suis 86-3977B; 3, S. suis AAH6; 4, S. suis 95-13626; 5, S. suis DH5; 6, S. suis 89-1591; 7, S. suis D930; 8, S. suis 95-7220; 9, S. suis 0891; 10, S. suis 88–5955.
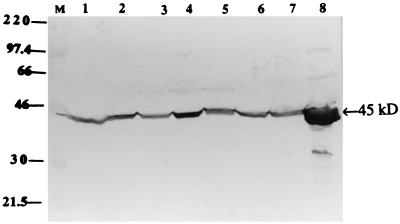
To determine whether S. suis type 2 strains encode antigenically similar GDH proteins, seven different strains were used in Western blot analysis with the polyclonal antibody raised against the purified 45-kDa GDH protein. Results showed that a protein in S. suis strains and in E. coli cells carrying the gdh gene reacted specifically to the antibody, giving a band of identical molecular mass (Fig. 5). This indicated that the gene is expressed by all of the isolates and that the protein is antigenically conserved in the strains tested.
FIG. 5.
Western immunoblot analysis of cell-free extracts of S. suis type 2 strains with the polyclonal antibody raised against the purified 45-kDa GDH recombinant protein. Lanes: M, rainbow molecular size marker (Amersham) in kilodaltons; 1, S. suis 1933; 2, S. suis AAH6; 3, S. suis DH5; 4, S. suis type strain; 5, S. suis 88-5955; 6, S. suis 95-13626; 7, S. suis 89-1591, and 8, E. coli DH5α(pOT410).
GDH activity of recombinant and native GDH.
Because the recombinant protein was identified as a GDH enzyme by amino acid homology, it was of interest to analyze the recombinant and native GDH proteins for biological activity. GDH zymograms with l-glutamate and α-ketoglutarate as substrates, respectively, were used to characterize the biological activity of the recombinant and native GDH molecules. Cell-free extracts of E. coli carrying the gdh gene [DH5α(pOT410)] and seven different strains of S. suis type 2 with known degrees of virulence were electrophoresed in a nondenaturing polyacrylamide gel and stained as described in Materials and Methods. A GDH zymogram with l-glutamate as the substrate and NAD(P)H as the cofactor showed a single band signifying the S. suis GDH in samples containing recombinant GDH or native GDH (Fig. 6). When NAD was supplied as the cofactor, no apparent GDH activity was detected in any of the samples. Substitution of α-ketoglutarate for l-glutamate with either NAD(P)H or NAD as the cofactor also did not result in apparent GDH activity (Table 1), suggesting that a single GDH was present. In the protein profile, the migration distance of native GDH from S. suis strain 1933, the source of the cloned fragment, and S. suis type culture strain including E. coli carrying the recombinant gdh gene were the same (Fig. 6, lanes 1, 4, 8, and 9), whereas the remaining isolates tested had a different migration pattern (Fig. 6, lanes 2, 3, and 5 to 7), suggesting the existence of two conformational types of GDH among the strains tested. It is interesting that isolates in lanes 1, 4, and 9 are highly virulent, whereas isolates in lanes 2, 3, and 5 to 7 are either weakly virulent or nonvirulent (35). This analysis, although not exhaustive, indicated that highly virulent strains of S. suis type 2 may be distinguished from moderately virulent and avirulent isolates on the basis of their GDH protein profile following activity staining on a nondenaturing gel.
FIG. 6.
Activity staining of GDH showing NADPH-dependent GDH activity on acrylamide gel. l-Glutamate was used as substrate for detection of GDH activity. Lanes: 1, S. suis 1933; 2, S. suis 88-5955; 3, S. suis DH5; 4, S. suis type strain; 5, S. suis 95-13626; 6, S. suis 89-1591; 7, S. suis AAH6; 8, E. coli DH5α(pOT410); and 9, S. suis 1933.
TABLE 1.
Results of GDH activity staining assaya
| Source | NAD(P)H l-glutamate | NAD l-glutamate | NAD(P)H α-ketoglutarate | NAD α-ketoglutarate |
|---|---|---|---|---|
| Clone pOT410 | +++ | ++ | − | − |
| S. suis strain 1933 | +++ | ± | − | − |
+++, very strong; ++, strong; ±, faint; −, negative.
Reactivity of the recombinant GDH protein with serum from pigs.
Figure 7 shows the reactivity of serum samples from pigs experimentally infected with S. suis type 2 to the recombinant 45-kDa GDH protein on immunoblot. All samples produced detectable signal against the recombinant antigen. This result illustrates the potential use for the recombinant GDH protein in the development of a serodiagnostic assay for S. suis disease.
FIG. 7.
Immunoblot analysis of pig sera and their reactivities against the purified recombinant GDH protein. Lanes: M, molecular size marker in kilodaltons; 1 through 6, serum from pigs infected with S. suis type 2; 7, polyclonal antibody raised in a rabbit against the purified recombinant GDH protein (positive control); and 8, preimmune serum from the rabbit used to raise antibody against the purified protein (negative control). The location and size of the GDH protein are indicated by the arrow.
Cellular location of the Streptococcus GDH protein.
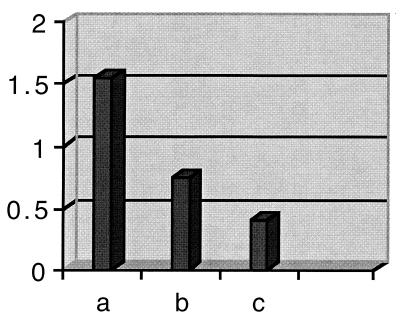
Whole-cell ELISA and immunogold-labeling techniques were performed to determine whether the polyclonal antibody generated against the purified streptococcal GDH protein was able to recognize its specific epitopes at the surface of the organism. For negative control, the primary antibody was replaced with preimmune rabbit serum. When examined using a whole-cell ELISA technique, the antibody bound strongly to the epitope at the surface of the organism, with a value of 1.55. Absorption of the antibody with whole homologous cells clearly diminished the reaction of the antibody with the antigen (value of 0.88) indicating that the GDH antibodies had been removed by absorption. Preimmune serum gave a value of 0.414 (Fig. 8). These results suggest that the GDH protein is exposed on the surface of intact cells.
FIG. 8.
Binding properties of the anti-S. suis GDH polyclonal antibody to intact cells as determined by the whole-cell ELISA technique. Live intact S. suis type 2 strain 1933 cells were incubated with (a) the polyclonal antibody raised against the purified GDH protein, (b) polyclonal antibody absorbed with whole-cell extracts of S. suis type 2 strain 1933, and (c) preimmune rabbit antiserum.
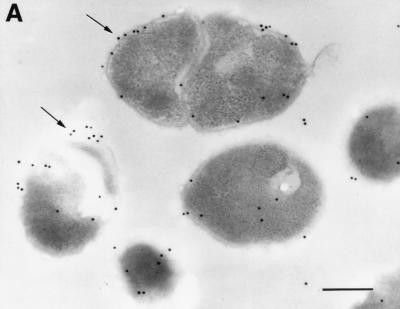
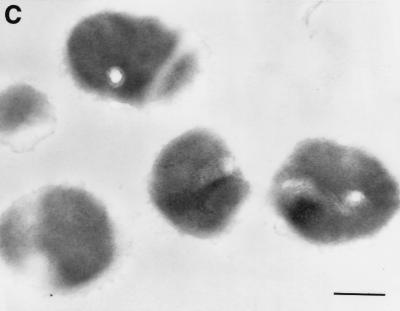
To confirm the ELISA results, an immunogold-labeling technique was employed. An electron micrograph showed that the gold particles were associated with the surface of the cells. Additionally, the gold particles were also associated with the interior region of the cells. Preimmune serum prepared from the rabbit used for the production of anti-S. suis GDH antibody (negative control) failed to label the thin section (Fig. 9). No labeling was also noted when primary antibody was omitted (data not shown). Consistent with the whole-cell ELISA technique results, these results indicated that the GDH protein is exposed on the surface of S. suis cells.
FIG. 9.
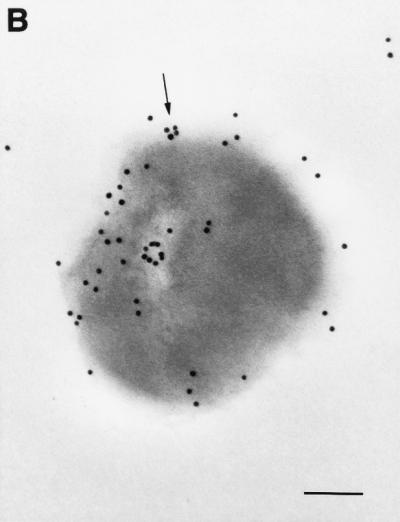
Visualization of GDH proteins with an immunogold ultrathin-section transmission electron microscope. Thin sections were prepared from S. suis cells and probed with primary antibodies to GDH protein (A and B) or preimmune rabbit serum (C). Representative gold particles on the surface of the cells are indicated by arrows; the bar equals 200 nm.
Nucleotide sequence accession number.
The GenBank accession number for the sequence reported in this paper is AF229683.
DISCUSSION
GDH has been shown previously to be an important diagnostic antigen, and it has also been correlated to virulence of certain bacteria (15, 22). Additionally, GDHs have been shown to play a key role in nitrogen metabolism (33). This is the first enzyme of S. suis type 2 involved in intermediary metabolism to be cloned, sequenced, and characterized.
The gdh gene is located within a 1.6-kb DraI DNA fragment and appears to be transcribed from a promoter located within the cloned DNA fragment, since expression is not dependent on insertional orientation. In agreement with this notion, sequences typical of concensus promoters are present upstream of the translation start point.
Genetic heterogeneity has been shown to exist among isolates of the same S. suis serotype (24). It was therefore of interest to note that all of the strains tested by DNA-DNA hybridization studies contained the gdh gene (Fig. 4) and that the product of the gene is similar both antigenically and in molecular mass in the strains tested by Western immunoblot under denaturing conditions (Fig. 5). Under nondenaturing conditions, activity staining also showed that all strains tested contained the same type of GDH. However, two different patterns as judged by the electrophoretic mobility of the proteins on the nondenaturing gel were observed (Fig. 6). Because the protein sizes are identical when analyzed in a denatured compared to nondenatured state, we believe that the observed difference may be the result of differences in conformation (folding) of the proteins in their native form or amino acid substitutions. S. suis strains have been classified into three groups (highly virulent, moderately virulent, and avirulent) based on their degree of pathogenicity (35). The pathogenicity of the strains used in this study are known (35). Although a limited number of strains were tested, highly virulent strains (Fig. 6, lanes 1, 4, and 9) have banding patterns that differ from those of moderately virulent or avirulent strains. We do not know whether this enzyme contributes to certain metabolic properties that distinguish highly virulent from moderately virulent and avirulent strains of S. suis type 2. It is interesting that group 1 highly virulent Clostridium botulinum strains responsible for most cases of human botulism are distinguishable from group II strains, which are nonvirulent or weakly virulent, on the basis of their NAD-dependent l-glutamate dehydrogenase activity (15). It is therefore possible that such might be the case with S. suis type 2.
GDHs are of interest because they are highly conserved and exhibit an extremely low rate of point mutations relative to many other proteins (7). Additionally, GDHs have been successfully used in the diagnosis of certain bacterial infection. For example, infection of Clostridium difficile, an important nosocomial pathogen which causes pseudomembranous colitis, can be diagnosed by means of the GDH-based latex agglutination tests marketed by Becton Dickinson Microbiology Systems (Culturette CDT) and Meridian Diagnostics (Meritec-C. difficile) (22). The GDH of S. suis is antigenic, is conserved in the strains tested, and reacts with serum from animals with S. suis type 2 infection, thus making it a potential candidate for development of a serological assay to detect S. suis infection.
In general, bacterial GDH enzymes are located in the cytoplasm or the cytoplasmic membrane of the cell (4, 33). However, the GDH of Porphyromonas gingivalis has been shown to be surface associated (17). We have shown in this work that the GDH protein of S. suis is also surface exposed. This finding may explain why rabbits and pigs inoculated with log phase live whole-cell S. suis elicited a strong immune response to the GDH protein within a few days.
Glutamate and α-ketogluterate are the substrates of two different reactions (forward or reverse) performed by GDH enzymes. In the forward reaction, GDH utilizes α-ketogluterate as the substrate and NAD(P)H as a cofactor to produce l-glutamate. In the reverse reaction, GDH utilizes l-glutamate as the substrate and NAD as a cofactor to produce α-ketogluterate (4, 33). In contrast to the NAD(P)H-dependent GDHs from various sources which utilize α-ketoglutarate as the substrate, the S. suis GDH is active only when l-glutamate is used as the substrate with NAD(P)H as the cofactor (Table 1). This suggest that the GDH of S. suis may be regulated in a different way from the GDH from other sources and that a different l-glutamate pathway may exist in S. suis. Griffith and Carlsson (13) previously demonstrated that the GDH of streptococci is regulated in a way not previously described for bacteria.
In conclusion, we have cloned and characterized the gene encoding the GDH of S. suis type 2. Availability of the cloned gene will make it possible to construct mutant strains expressing little or no GDH, which could be used to determine whether this protein could be correlated to S. suis pathogenicity as in the case with C. botulinum types A, B, and F (group 1) (15). It will also facilitate studies on the factors affecting the gene regulation and expression.
ACKNOWLEDGMENTS
This work was supported by the Department of Health and Human Services, Health Research Service Administration, Center for Excellence and Minority Veterinary Medical Education grant 5D34MB00001-12.
REFERENCES
- 1.Abrahams G L, Abratt V R. The NADH-dependent glutamate dehydrogenase enzyme of Bacteroides fragilis Bfl is induced by peptides in the growth medium. Microbiology. 1998;144:1659–1667. doi: 10.1099/00221287-144-6-1659. [DOI] [PubMed] [Google Scholar]
- 2.Arends J P, Zanen H C. Meningitis caused by Streptococcus suis in humans. Rev Infect Dis. 1988;10:131–137. doi: 10.1093/clinids/10.1.131. [DOI] [PubMed] [Google Scholar]
- 3.Benachenhou-Lahfa N, Forterre P, Labedan B. Evolution of glutamate dehydrogenase genes: evidence for two paralogous protein families and unusual branching patterns of the archaebacteria in the universal tree of life. J Mol Evol. 1993;36:335–346. doi: 10.1007/BF00182181. [DOI] [PubMed] [Google Scholar]
- 4.Brunhuber N M W, Blanchard J S. The biochemistery and enzymology of amino acid dehydrogenases. Crit Rev Biochem Mol Biol. 1994;29:415–467. doi: 10.3109/10409239409083486. [DOI] [PubMed] [Google Scholar]
- 5.Chanter N, Jones P W, Alexander T J L. Meningitis in pigs caused by Streptococcus suis—a speculative review. Vet Microbiol. 1993;36:39–55. doi: 10.1016/0378-1135(93)90127-s. [DOI] [PubMed] [Google Scholar]
- 6.Chengappa M M, Maddux R L, Kadel W L, Greer S C, Herrin C E. Proceedings of the Annual Meeting of the American Association of Veterinary Diagnosticians. Brookings, S.Dak: Inc. Reynolds Printing Co.; 1986. Streptococcus suis infection in pigs: incidence and experimental reproduction of the syndrome; pp. 25–38. [Google Scholar]
- 7.Creighton T E. Proteins. Structures and molecular principles. New York, N.Y: W. H. Freeman & Co.; 1984. [Google Scholar]
- 8.Devries L A, Sustronck B, Maenhouth T, Haesebrouck F. Streptococcus suis meningitis in a horse. Vet Rec. 1990;127:68. [PubMed] [Google Scholar]
- 9.Ferretti J J, Curtis III R, editors. Streptococcal genetics. Washington, D.C.: American Society for Microbiology; 1987. Compilation of nucleotide sequences that signal the initiation of transcription and translation in streptococci; p. 293. [Google Scholar]
- 10.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichson J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–2594. doi: 10.1128/jcm.29.11.2590-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottschalk M, Higgins R, Jacques M, Mittal K R, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graves M C, Rabinowitz J C. In vivo and in vitro transcription of the Clostridium pasteurianum ferrodoxin gene. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 13.Griffith C J, Carlsson J. Mechanism of ammonia assimilation in streptococci. J Gen Microbiol. 1974;82:253–260. doi: 10.1099/00221287-82-2-253. [DOI] [PubMed] [Google Scholar]
- 14.Haase E M, Zmuda J L, Scannapieco F A. Identification and molecular analysis of rough-colony-specific outer membrane proteins of Actinobacillus actinomycetemcomitans. Infect Immun. 1999;67:2901–2908. doi: 10.1128/iai.67.6.2901-2908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammer B A, Johnson E A. Purification, properties, and metabolic roles of NADH-glutamate dehydrogenase in Clostridium botulinum 113B. Arch Microbiol. 1988;150:460–464. doi: 10.1007/BF00422287. [DOI] [PubMed] [Google Scholar]
- 16.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. Description of six new capsular types (29–34) of Streptococcus suis. J Vet Diagn Investig. 1995;7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 17.Joe A, Murray C S, McBride B C. Nucleotide sequence of a Porphyromonasgingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect Immun. 1994;62:1358–1368. doi: 10.1128/iai.62.4.1358-1368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilper-Balz R, Schleifer K H. Streptococcus suis sp. nov., nom. rev. Int J Syst Bacteriol. 1987;37:160–162. [Google Scholar]
- 19.Kyte I, Dolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–695. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Lowry O H, Rosebrough N J, Farr A R, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 22.Lyerly D M, Barroso L A, Wilkins T D. Identification of the latex test-reactive protein of Clostridium difficile as glutamate dehydrogenase. J Clin Microbiol. 1991;29:2639–2642. doi: 10.1128/jcm.29.11.2639-2642.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLennan M, Foster G, Dick K, Smith W J, Nielsen B. Streptococcus suis serotypes 7, 8, and 14 from diseased pigs in Scotland. Vet Rec. 1996;139:423–424. doi: 10.1136/vr.139.17.423. [DOI] [PubMed] [Google Scholar]
- 24.Okwumabua O, Staats J, Chengappa M M. Detection of genomic heterogeneity in Streptococcus suis isolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping) J Clin Microbiol. 1995;33:968–972. doi: 10.1128/jcm.33.4.968-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perch B, Pedersen K B, Henrichsen J. Serology of capsulated streptococci pathogenic for pigs:six new serotypes of Streptococcus suis. J Clin Microbiol. 1983;19:993–996. doi: 10.1128/jcm.17.6.993-996.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plante M, Cadieux N, Rioux C R, Hamel J, Brodeur B R, Martin D. Antigenic and molecular conservation of the gonococcal NspA protein. Infect Immun. 1999;67:2855–2861. doi: 10.1128/iai.67.6.2855-2861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson I D, Blackmore D K. Occupational exposure to Streptococcus suis type 2. Epidemiol Infect. 1989;103:157–164. doi: 10.1017/s0950268800030454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanford S E, Higgins R. Streptococcal diseases. In: Lemon A, et al., editors. Diseases of swine. 7th ed. Ames, Iowa: Iowa University Press; 1992. pp. 588–590. [Google Scholar]
- 30.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarvas M. Protein secretion in bacilli. Curr Top Microbiol Immunol. 1986;125:103–125. doi: 10.1007/978-3-642-71251-7_8. [DOI] [PubMed] [Google Scholar]
- 32.Shine J, Dalgarno J. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementary to non-sense triplets and ribosome binding sites. Proc Natl Acad Sci USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith E L, Austen B M, Blumenthal K M, Nyc J F. Glutamate dehydrogenases. In: Boyer P D, editor. The Enzymes. Vol. 2. New York, N.Y: Academic Press; 1975. pp. 293–367. [Google Scholar]
- 34.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 35.Staats J J, Plattner B L, Nietfeld J, Dritz S, Chengappa M M. Use of ribotyping and hemolysin activity to identify highly virulent Streptococcus suis type 2 isolates. J Clin Microbiol. 1998;36:15–19. doi: 10.1128/jcm.36.1.15-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staats J J, Feder I, Okwumabua O, Chengappa M M. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/a:1005870317757. [DOI] [PubMed] [Google Scholar]
- 37.Watson M E E. Compilation of published signal sequences. Nucleic Acids Res. 1984;12:5145–5159. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wen Z, Morrison M. The NAD(P)H-dependent glutamate dehydrogenase activities of Prevotella ruminicola B14 can be attributed to one enzyme (GdhA), and gdhA expression is regulated in response to the nitrogen source available for growth. Appl Environ Microbiol. 1996;62:3826–3833. doi: 10.1128/aem.62.10.3826-3833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]