Abstract
The deepest marine ecosystem, the hadal zone, hosts endemic biodiversity resulting from geographic isolation and environmental selection pressures. However, the pan-ocean distribution of some fauna challenges the concept that the hadal zone is a series of isolated island-like habitats. Whether this remains true at the population genomic level is untested. We investigated phylogeographic patterns of the amphipod, Bathycallisoma schellenbergi, from 12 hadal features across the Pacific, Atlantic, Indian, and Southern oceans and analyzed genome-wide single-nucleotide polymorphism markers and two mitochondrial regions. Despite a cosmopolitan distribution, populations were highly restricted to individual features with only limited gene flow between topographically connected features. This lack of connectivity suggests that populations are on separate evolutionary trajectories, with evidence of potential cryptic speciation at the Atacama Trench. Together, this global study demonstrates that the shallower ocean floor separating hadal features poses strong barriers to dispersal, driving genetic isolation and creating pockets of diversity to conserve.
Population genomics of an amphipod supports that the ocean’s deepest ecosystems host isolated pockets of endemic biodiversity.
INTRODUCTION
Geographic isolation is a key mechanism in promoting biodiversity (1), and the deepest ocean ecosystems represent hotspots for endemic and undiscovered species (2–4). Extending from 6000 to ~11,000 m deep, the hadal zone accounts for nearly half of the ocean’s depth profile (4). Yet, the hadal zone represents a minor fraction of the seafloor, with only 47 known features extending to hadal depths in the Pacific, Indian, Atlantic, and Southern oceans (4, 5). Globally, hadal features create fragmented pockets of island-like habitats, often separated by thousands of kilometers of shallower abyssal plains (3, 5). Most of the features are trenches, formed at the subduction zone between tectonic plates, and are analogous to inverted continental mountain ranges such as the Andes and Himalayas. Two other geomorphic forms can reach hadal depths, transform faults/fracture zones, and troughs/basins (6). Hadal features are shaped by a suite of extrinsic and intrinsic factors making the environment, carbon flux, seismicity, and geomorphology of each trench, fracture zone, and trough unique (4). In general, the hadal environment can be defined as “extreme,” where life must tolerate immense hydrostatic pressures, the absence of natural light, scarcity of food, and near-freezing temperatures (4, 7).
Scientists did not think life could survive at hadal depths until the first hadal trawl at the Puerto Rico Trench in 1948 recovered holothurians, polychaetes, and isopods (8). We now know that the hadal zone is home to representatives from most major marine taxa, from bacteria to fishes, with some taxa present to full ocean depth (3). Hadal species have a suite of biochemical, physiological, morphological, and ecological adaptions to thrive in the challenging environment (e.g., 3, 4, 9). Furthermore, the hadal community has been consistently found to be distinct from shallower zones (3, 10). The inherent long-term geographical isolation and strong environmental selection pressures of hadal features are thought to follow island biogeography principles, whereby these features are inhabited by high levels of endemic biodiversity (3, 11, 12). The notion of endemic hadal diversity has been largely derived from amphipods, an order of crustaceans. Amphipods represent one of the best-sampled groups because of their relative ease in recovery through trawls and baited traps (e.g., 10, 13–15). Specifically, ~95% of hadal amphipod species are only known to inhabit a single trench or a group of adjacent trenches (3, 4). For example, the large amphipod Eurythenes atacamensis has only been found at the Peru-Chile Trench (16), while another large amphipod, Hirondellea gigas, is known to the deepest depths of the northwest Pacific Ocean trenches (17). However, many of these findings originated from the first hadal sampling campaigns, the RV Vityaz and Galathea expeditions of the 1950s and 1960s (3). The perception that hadal features are endemic hot spots has not been thoroughly tested, and large gaps remain in our understanding of the evolutionary processes that drive population-level differentiation and speciation across the hadal environment.
The remoteness of these features poses substantive technological and logistical challenges in conducting sampling expeditions due to high pressure at depths and distance from the surface (4). While the number of hadal expeditions has increased over the past 20 years, there is a bias toward the subduction trenches of the western Pacific Ocean and particularly the deepest point, Challenger Deep in the Mariana Trench (18). The focus on singular trenches along the Pacific Rim has left other hadal features, particularly nontrench and non–Pacific Ocean features, undersampled. As expeditions are often conducted by different research groups, the generation of an accessible global specimen collection has been difficult.
Genetic analyses of the limited number of specimens collected have primarily used single-gene mitochondrial markers, specifically DNA barcoding, to study fundamental taxonomy, uncover cryptic species, and assess biodiversity (14–16, 19), with few studies directly assessing patterns of population connectivity among features. One of the only studies using genome-wide markers found enough gene flow among four Pacific Ocean trench populations of the amphipod Paralicella spp. to conclude that trenches do not restrict dispersal (20). However, Paralicella spp. is not endemic to the hadal zone, predominately dwelling at the shallower abyssal depths (21). Thus, the degree to which hadal features act as isolated island habitats to promote, first, reproductive isolation and second, speciation, in hadal-specific organisms remains unresolved.
In this study, we use the amphipod Bathycallisoma schellenbergi (22) to examine the extent to which hadal features function as island habitats and host distinct and isolated populations. Bathycallisomsa schellenbergi is a large bentho-pelagic scavenger (length ~ 4.5 cm) and found between 5600 and 9400 m at 10 hadal features across the Pacific, Atlantic, Indian, and Southern oceans (23, 24) (Fig. 1). Common among Amphipoda, B. schellenbergi lacks a pelagic larval stage and juveniles emerge from the brood pouch ready for independent life. Dispersal of brooders is largely dependent on the adult’s swimming ability, often limiting wide distributions and accelerating endemism and the potential for cryptic speciation (25). Thus, this amphipod represents a model for examining how geographic isolation and topographical barriers limit the exchange of genetic material across the hadal zone.
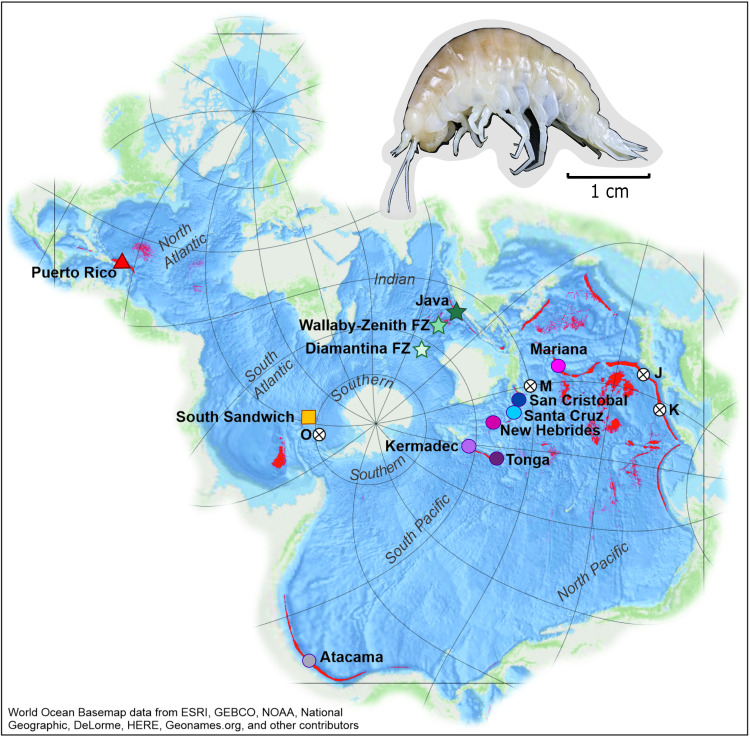
Fig. 1. Sample localities of the hadal amphipod B. schellenbergi.
The hadal zone (depths of >6000 m) is defined in bright red. Unsampled features with known records of B. schellenbergi are denoted with an X shape with the addition of “J” for the Japan Trench, “K” for the Kuril-Kamchatka Trench, “M” for the Massau Trench, and “O” for the Orkney Trench. Color and shape codes for each hadal-feature population are used consistently in the other figures. The map was created in ArcGIS Pro using the Spilhaus Ocean Map in Square projection. All elevation data are from Esri World Ocean Basemap. The analyses on the distribution of hadal zone were undertaken using data from the GEBCO Compilation Group (95). The map was created in ArcGIS Pro (version 2.9) using the Spilhaus Ocean Map in Square projection. FZ, Fracture Zone.
Here, we pair DNA barcoding with genome-wide single-nucleotide polymorphism (SNPs) markers generated by restriction site–associated DNA sequencing (RAD-seq) to assess the population genetic patterns among B. schellenbergi from 12 hadal features located in the Pacific, Atlantic, Indian, and Southern oceans. We aim to (i) uncover whether B. schellenbergi is a single species with a cosmopolitan distribution or a complex of cryptic species with narrower distributions, (ii) assess the degree of genetic differentiation among populations, and (iii) estimate whether gene flow and contemporary migration are occurring among features. By examining the population genetics of this broadly distributed hadal amphipod, we begin to unravel the interplay between speciation and tectonic-driven isolation at the deepest depths of the oceans and gain insights into the importance of conserving the hadal zone.
RESULTS
Overview of the global B. schellenbergi specimen and genetic datasets
During 13 sampling campaigns between 2011 and 2019, we amassed a global specimen collection from 10 subduction trenches and two fracture zones in the Pacific, Atlantic, Indian, and Southern oceans (Fig. 1 and Table 1). Bathycallisomsa schellenbergi were recovered from depths of 5920 to 8428 m during 31 baited lander deployments. The features are geographically isolated, with an average separation of 9743 km (table S1). The San Cristobal and Santa Cruz trenches are the closest two features, separated by only 288 km, whereas the Mariana and Puerto Rico trenches are separated by 25,990 km across the Atlantic, Indian, and Pacific oceans.
Table 1. Summary of the B. schellenbergi datasets.
Amphipods were collected from 13 sampling campaigns to 12 hadal features between 2011 and 2019 and used for the mtDNA (16S + COI regions) and RAD-seq datasets. SO261, Expedition SO261 on the RV Sonne; FDE, Five Deeps Expedition on the DSSV Pressure Drop; KAH1109/KAH1202/KAH1310 on the RV Kaharoa; Ho, observed heterozygosity; He, expected heterozygosity; FIS, inbreeding coefficient.
| Feature |
Depth stations (m) |
General location * |
Year | Expedition | n mtDNA | n RAD-seq |
mtDNA haplotype diversity |
SNP Ho | SNP He | F IS |
| Atacama Trench |
5920, 3025, and 6714 |
20°S, 71°W | 2018 | SO261 | 9 | 8 | 0.89 | 0.319 | 0.375 | 0.150 |
| Diamantina Fracture Zone† |
7009 | 33°S, 101°E | 2019 | FDE | 7* | 0 | – | – | – | – |
| Java Trench | 6957 and 7176 |
11°S, 114°E | 2019 | FDE | 15 | 9 | 0.63 | 0.214 | 0.419 | 0.491 |
| Kermadec Trench |
6709, 6879, and 7291 |
32°S, 177°W | 2011, 2012 | KAH1109, KAH1202 |
9 | 7 | 0.92 | 0.244 | 0.377 | 0.353 |
| Mariana Trench |
7094 and 7509 |
11°N, 142°E | 2019 | FDE | 5 | 4 | 0.40 | 0.267 | 0.384 | 0.305 |
| New Hebrides Trench |
6000, 6228, and 6948 |
20°S, 168°E | 2013 | KAH1310 | 8 | 7 | 1.00 | 0.289 | 0.352 | 0.179 |
| Puerto Rico Trench |
6954, 7505, and 8379 |
19°N, 67°W | 2018 | FDE | 6 | 10 | 0.60 | 0.274 | 0.372 | 0.263 |
| San Cristobal Trench |
6844, 7220, and 8407 |
11°S, 163°E | 2019 | FDE | 9 | 13 | 0.86 | 0.267 | 0.384 | 0.305 |
| Santa Cruz Trench |
7431 and 8428 | 11°S, 165°E | 2019 | FDE | 6 | 10 | 0.96 | 0.265 | 0.395 | 0.330 |
| South Sandwich Trench |
6640, 7400, 8100, and 8265 |
60°S, 25°W | 2019 | FDE | 11 | 11 | 0.89 | 0.268 | 0.348 | 0.229 |
| Tonga Trench |
6793, 7273, and 7928 |
23°S, 174°W | 2019 | FDE | 7 | 11 | 0.71 | 0.273 | 0.367 | 0.258 |
| Wallaby- Zenith Fracture Zone |
6537 and 6546 |
22°S, 102°E | 2017 | SO258 | 9 | 7 | 1.00 | 0.237 | 0.386 | 0.387 |
| Total | 94 | 97 |
*Number of depth stations represent in the study shown in brackets. See data S1 for full station details.
†Individuals recovered from Diamantina Fracture Zone were only successfully sequenced at the 16S mtDNA region.
Examined specimens from each feature were morphologically consistent with the diagnostic description of B. schellenbergi (24). In particular, the lacinia mobilis was a long, slender robust seta, and gnathopod 1 dactylus was small, simple, and highly modified with a blunt apical tip. Minor variation was observed on the angle of the distal lobe of the pereopod 7 basis, which varied from straight parallel with the body to slightly concave and angled upward 30o. However, this variation was not sufficient to indicate a different identification decision.
We extracted genomic DNA from 239 individuals and constructed two genetic datasets to investigate phylogenetic and population genomic relationships among B. schellenbergi populations living in the 12 hadal features and across their depth range at each feature, where possible. A challenge to working with deep-sea samples is often poor DNA quality due to a combination of shearing during depressurization and degradation during the time lag between recovery, onboard preservation, curation storage, and lastly DNA extraction (26). Hence, there was a level of attrition due to either poor DNA quality, no polymerase chain reaction (PCR) amplification, or low quality of reads.
The first dataset, “mtDNA” (mitochondrial DNA), was constructed by Sanger sequencing two partial mitochondrial regions, 16S ribosomal RNA [16S; 215 base pairs (bp)] and cytochrome c oxidase subunit I (COI; 559 bp). Amphipods from the Diamantina Fracture Zone had extremely poor DNA quality, most likely due to the lander being at the surface for >5 hours during recovery, and hence only seven were successfully sequenced at 16S (Table 1). From the other 11 features, 94 individuals were sequenced at both DNA barcoding regions.
The second dataset, “RAD-seq,” used the bestRAD protocol with the Sbf I enzyme to generate thousands of SNPs in nuclear loci (27). A total of 128 individuals from 11 hadal features (i.e., all features but the Diamantina Fracture Zone) met the high DNA quantity and integrity requirements for the bestRAD protocol. The final RAD-seq dataset matrix consisted of 97 individuals following forward read assembly and genotyping with ipryad, as well as the removal of poor-performing individuals (28, 29). The mean coverage per individual was 74.7×. All the 4611 recovered RADtag loci and 58,730 SNPs were used for the analysis with fineRADstucture (30), while a subset of 2933 SNPs was used for other analyses after filtering to retain 1 SNP per locus that was present in 70% of individuals (30) (table S2). The final RAD-seq sample size was uneven among features, ranging from 4 Mariana Trench to 13 San Cristobal Trench individuals (Table 1). Despite a small sample size in some features, this dataset has sufficient statistical power, as accurate estimates of genetic differentiation in highly structured populations can be obtained using ≥1500 SNPs and two to five individuals [e.g., (31)]. Outputs from each RAD-seq processing step are available in data S1. The DNA barcoding sequences and the demultiplexed bestRAD dataset are available via the National Center for Biotechnology Information (NCBI) (data S1).
Strong global phylogeographic structure and evidence of cryptic diversity at the Atacama Trench
As the occurrence of cryptic diversity is prevalent in the deep sea, particularly among amphipods (32, 33), we aimed for robust identification by pairing morphological identifications with molecular phylogenies. We applied three molecular species delimitation methods, namely, Assemble Species by Automatic Partitioning (ASAP) (34), Generalized Mixed Yule Coalescent (GMYC) (35), and Bayesian implementation of the Poisson Tree Processes (bPTP) (36) to test for evidence of cryptic speciation within the mtDNA alignments, and pairwise p-distances were calculated.
The mtDNA and RAD-seq phylogenies displayed broadly congruent patterns with six well-supported groups or clades (Fig. 2, A and C), grouped either by ocean or ocean basin. The first diverging clade was the Atacama Trench population [bayesian posterior probability (BPP): 1; bootstrap: 100]. The Atacama Trench population was the only clade consistently delineated as a separate species lineage across the three delimitation methods and the COI pairwise p-distances averaged 4.75% (Fig. 2B and table S3). The South Sandwich Trench population was the second divergent clade (BPP: 0.7; bootstrap: 1); however, this group was only inferred as a separate species by bPTP, and the mean COI pairwise p-distance for COI was 2.25%. The Atacama and South Sandwich trench individuals did not match an existing Barcode Index Number (BIN) on the global COI database, the Barcode of Life Data System (BOLD), suggestive of a separate molecular operational taxonomic unit or species. The western Pacific, Indian, and Atlantic ocean individuals were matched to the existing B. schellenbergi BIN (BOLD: ADD0645). The Puerto Rico Trench individuals formed a clade, Clade Bs3, and the mean pairwise p-distance for COI was 1.66%. Clade Bs4 included individuals from the Java Trench and the Wallaby-Zenith Fracture Zone in the Indian Ocean (BPP: 0.99; bootstrap: 100), with a mean p-distance of 0.62%. The Puerto Rico and Indian Ocean clades were monophyletic in the mtDNA phylogeny while paraphyletic in the RAD-seq phylogeny. Clades Bs5 and Bs6 were represented by western Pacific Ocean trench features. The Clade Bs5 was entirely composed of individuals from the Kermadec and Tonga trenches (BPP: 0.95; bootstrap: 100), with a COI p-distance of 0.48%. The Clade Bs6 was a mixture of individuals from the remaining trenches (BPP: 0.95: bootstrap: 100) with a mean pairwise p-distance of 0.76% for COI within the clade. In the mtDNA phylogeny, the comparative sequences from the Massau and Kuril-Kamchatka trenches were grouped with the Mariana Trench individuals (BPP: 1). While not present on the phylogeny, the 16S p-distance analysis placed the Diamantina Fracture Zone amphipods within this species (table S4). However, the 16S sequence length was too short to resolve intraspecific relationships across all individuals (21).
Fig. 2. The phylogenetic relationships among 11 hadal feature populations of the amphipod B. schellenbergi.
(A) The Bayesian phylogeny is based on a concatenated 16S (215 bp) and COI (559 bp) dataset. Branch nodes show Bayesian posterior probabilities greater than 0.70. (B) A collapsed 16S and COI phylogeny to illustrate results from morphology and three molecular species delimitation approaches, the bPTP, GMYC, and ASAP. (C) The maximum likelihood phylogeny generated by IQ-TREE is based on an RAD-seq dataset (2933 loci) of 11 feature populations. Branch nodes show standard bootstrap support values greater than 80.
Hadal features host highly structured populations of B. schellenbergi
Genetic structure is the variation of allele frequencies within and among populations, largely resulting from opposing forces of genetic drift and gene flow (1). To detect and quantify genetic structure among 11 hadal feature populations, we applied a complementary suite of six methods to the RAD-seq data, which included STRUCTURE (37), fineRADstructure (37), Discriminant Analysis of Principal Components (DAPC) (38), principal components analysis (PCA) (39), pairwise FST with the Hudson’s estimator (40), and analysis of molecular variance (AMOVA) (41). The RAD-seq patterns were compared to an mtDNA haplotype network. Furthermore, we investigated the extent to which geographic distance explains those differences and estimated heterozygosity and inbreeding within populations.
The suite of six analyses showed that these 11 hadal feature populations of B. schellenbergi are highly structured, as indicated by a global FST of 0.489, and uncovered three consistent patterns. The first pattern was that the Atacama Trench population was the most differentiated, which complements the delimitation results that infer that this lineage may be a separate genetic species. This degree of differentiation was illustrated by the low estimated coancestry in the fineRADstructure analysis (fig. S1), the main driver of variation in the first principal component (fig. S2), and a mean pairwise FST of 0.75 (table S5).
The second pattern was that populations separated by large geographical distances, or ocean basins, displayed strong patterns of genetic structure. In hierarchical analysis with STRUCTURE and DAPC, the populations were separated into the five respective ocean basins—Southern, Atlantic, Indian, west Pacific, and east Pacific (Fig. 3; KSTRUCTURE = 5; fig. S3, KDAPC = 6). The AMOVA showed that 48.3% (P = 0.001) of the variation was explained by hadal features. The second principal component, or 10.7% of the variation, was explained between west Pacific and non-Pacific Ocean populations (fig. S2). The fineRADstructure analysis also identified both the ocean basin separation and the Pacific and non-Pacific Ocean split (fig. S1). The degree of differentiation among oceans was high, where the mean pairwise FST among populations with the South Sandwich Trench, Puerto Rico Trench, and Indian Ocean populations were 0.531, 0.468, and 0.457, respectively (table S5).
Fig. 3. Bar plots showing the genetic clustering among 11 hadal feature populations of B. schellenbergi.
The hierarchical STRUCTURE analysis was conducted with the RAD-seq dataset. The colors correspond to each genetic cluster (K), in which each vertical bar represents an individual’s estimated membership probability to a genetic cluster.
The third pattern was that genetic differentiation is weaker between hadal features located in the same region or subocean basin, specifically among the six western Pacific Ocean populations and between the two East Indian Ocean features (Fig. 3 and figs. S1 to S3). In the western Pacific Ocean, the tertiary STRUCTURE and DAPC analyses and fineRADstructure uncovered that the Mariana Trench population was distinctive (mean pairwise FST = 0.164). Low genetic differentiation was detected among the three central west Pacific Ocean features, the Santa Cruz, San Cristobal, and New Hebrides trenches (pairwise FST = 0.019 to 0.052; KSTRUCTURE = 1). Within the East Indian Ocean, the Java Trench and Wallaby-Zenith Fracture Zone populations lacked genetic structure (pairwise FST = 0.059; KSTRUCTURE = 1). A lack of genetic differentiation was detected between the two southwest Pacific Ocean features, the Kermadec and Tonga trenches (pairwise FST = 0.04; KSTRUCTURE = 1).
The isolation-by-distance analysis showed the overall trend that largest distances between features correlate to greater genetic differentiation (r2 = 0.44, P = 2.1 × 10−10; Mantel = 0.666, P = 0.001). This trend was consistent among all pairwise comparisons, except for the Atacama Trench. When Atacama Trench is compared with the other 10 features, genetic differences were irrespective of geographic distance (r2 = 0.00, P = 997, Mantel = 0.73, P = 0.001; Fig. 4A). When the Atacama Trench is excluded, the isolation-by-distance pattern was more significant among the remaining 10 features (r2 = 0.63, P = 6.8 × 10−13; Mantel = 0.791, P = 0.001; Fig. 4A). Among the western Pacific Ocean features, a pattern of isolation by distance was significant (r2 = 0.51, P = 0.042; Mantel = 0.714, P = 0.02; Fig. 4B).
Fig. 4. Linear relationships between genetic and geographic distance.
(A) All 11 hadal features, where the gray circles represent the pairwise comparisons between the Atacama Trench and the other features, while the black squares represent the pairwise comparisons among the other 10 sampled features. (B) A focus on the six western Pacific Ocean features. Asterisks represent *P < 0.05 and **P ≤ 0.001.
The RAD-seq patterns were mirrored in the mtDNA dataset, albeit to a lesser extent. Haplotype diversity was high (mean = 0.81; Table 1 and fig. S5), with no dominant haplotype by feature. Haplotypes were shared among three sets of features, specifically the Java Trench and Wallaby-Zenith Fracture Zone, the San Cristobal and Santa Cruz trenches, and the Kermadec and Tonga trenches, and each clustered together. Haplotypes from the South Sandwich and Atacama trenches formed district groups within the network and were separated by at least 7 and 25 mutational steps, respectively.
Within hadal features, no genetic substructure or pattern by depth was identified, which may be more indicative of insufficient sample size than lack of population structure. The inbreeding coefficient FIS was highly positive (mean = 0.295), suggestive of considerable levels of inbreeding with each feature. The Atacama Trench population had the lowest FIS value, 0.150, while the Java Trench population had the highest FIS value, 0.491 (Table 1).
Migration among hadal features in the central West Pacific Ocean
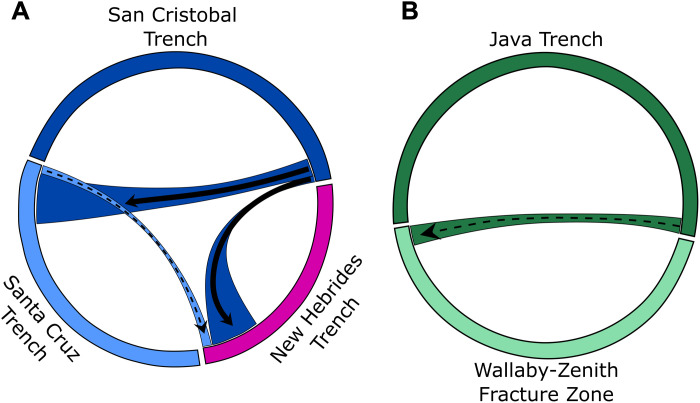
We used the program BayesAss to infer proportions of recent migrants among the two multifeature genetic populations identified through the hierarchical STRUCTURE analysis with pairwise FST > 0.05 (42, 43). The estimated probability of recent migration is summarized visually in Fig. 5 and shown in full in table S6. Good convergence among the Markov chains was achieved, with adequate acceptance rates for parameter estimations obtained (figs. S6 and S7). Asymmetrical migration was detected with migrants from the San Cristobal Trench to the Santa Cruz Trench (0.179) and the New Hebrides Trench (0.176). Between the Java Trench and Wallaby-Zenith Fracture Zone, the wide confidence intervals were too wide to infer contemporary migration.
Fig. 5. Contemporary gene flow among the two multifeature genetic clusters.
Estimates were derived from the program BayesAss based on the RAD-seq dataset. Colors correspond to the hadal feature: (A) San Cristobal (dark blue), Santa Cruz (light blue), and New Hebrides (pink) trenches in the central west Pacific Ocean and (B) Java Trench (dark green) and Wallaby-Zenith Fracture Zone (light green) in the East Indian Ocean. The width of the ribbon denotes the relative amount of gene flow. Solid arrows represent significant directionality based on nonoverlapping 95% confidence intervals, while dotted arrows are not significantly directional. See table S6 for exact gene flow estimates.
DISCUSSION
Bathycallisomsa schellenbergi is an extraordinary deep-ocean amphipod with a wide geographic and bathymetric distribution. Our global study revealed that despite its expansive distribution, populations were highly restricted to individual features, particularly among ocean basins, with only limited gene flow between spatially proximate and topographically connected features. We detected potential cryptic speciation in the Atacama Trench population, emphasizing that populations are on separate speciation paths. Together, this path-breaking study provides evidence that the shallower depths of the ocean floor separating hadal features function as a strong barrier to dispersal, leading to genetic isolation and creating islands of endemic biodiversity across the ocean’s deepest zone.
Cosmopolitan distribution but evidence of cryptic diversity at the Atacama Trench
Peracarid crustaceans, such as B. schellenbergi, are a highly diverse and abundant group in the deep ocean, yet the number of species present in three or more ocean basins is few (25). For the few taxa with wide distributions, molecular taxonomy tends to challenge this notion by uncovering that a species is a complex of cryptic species, morphological indistinguishable but genetically distinct (21, 33). One classic example is Eurythenes gryllus, a large amphipod historically presumed to live in all oceans between 550 and 8000 m but since 2013 has been known as a complex of at least 12 morphologically and genetically distinct species (16, 32). Given that the hadal zone is defined as fragmented habitats, we expected B. schellenbergi to have high levels of overlooked diversity, such as E. gryllus, with multiple species inhabiting individual or neighboring features. The question of whether B. schellenbergi is one or multiple species has been outstanding since initial descriptions, as Bathycallisoma pacifica Dahl, 1959 from the Kermadec Trench and B. schellenbergi (22) (originally described under Scopelocheirus) from the Kuril-Kamchatka Trench have been synonymized (24).
Unexpectedly, the species delimitation results strongly indicated that our B. schellenbergi specimens are the same species across the Atlantic, Southern, Indian, and west Pacific oceans (Fig. 2B). This finding supports the synonymization of multiple species to B. schellenbergi (24), adds new presence records to the San Cristobal and Santa Cruz trenches and the Diamantina Fracture Zone, and confirms records at the Java and South Sandwich trenches (24, 44). Although sequences were not available, we can presume that historical records at the Japan and Orkney trenches represent correct identifications (24) (Fig. 1). While this amphipod represents a rare example of a globally distributed hadal species, elevated diversity was uncovered, with the Atacama Trench population being consistently inferred as separate species-level lineage (Fig. 2). The interclade divergences did exceed the commonly applied 3% divergence threshold for crustaceans (45); however, other deep-ocean amphipods studies, including on Eurythenes, have proposed higher thresholds ranging from 4.5 to 12% [e.g., (21, 32)] (table S3). Here, if we assume two or even three distinct species, as suggested for the South Sandwich Trench population by bPTP (Fig. 2), the level of interspecific divergence is comparatively low and suggests relatively recent speciation events, of ~1.5 to 3.6 Ma as estimated by most adopted divergence rate of 1.4% per million years (46) (table S7). As our cursory morphological assessment did not indicate observable differences, future work is needed to identify diagnostic traits and confirm whether the Atacama Trench population is a distinct species.
This amphipod joins a short list of other species with wide distributions across the hadal zone, including the lophogastrid Eucopia sculpticauda (47), the amphipods Hirondellea dubia (48) and Alicellea gigantea (49), the cusk eel Abyssobrotula galatheae (12), the holothurian Prototrochus bruuni, and the sea star Eremicaster vicinus (3, 4). However, B. schellenbergi differs from these examples in two ways. First, apart from the holothurian and sea star, these species are found on the abyssal plains, whereas B. schellenbergi has yet to be found outside a trench or fracture zone. Although, it remains unconfirmed whether this is a true absence from the abyssal plains or is reflective of a sampling bias or limited sampling. Second, records of most hadal species, including the cusk eel, holothurian, and sea star, are based either on video images that lack the resolution for species-level identification or on morphological assessments of limited and often highly damaged material from the RV Vityaz and Galathea expeditions (3, 4, 44). These identifications have not been scrutinized with molecular approaches to rule out cryptic diversity. Other widespread hadal species may exist but have not yet been recovered and/or assessed at the global level because of undersampling and the dominant use of baited landers to sample hadal fauna (18), but this lack of representation may change with the expansion of hadal research with new submersible technologies.
Barriers to gene flow
The DNA barcoding provided compelling findings that B. schellenbergi is a single or potentially two recently diverged species with a global distribution across the inherently geographical isolated hadal zone. This emphasizes a key question: How did Bathycallisoma come to have a cosmopolitan distribution when it is restricted to the deep abyssal and hadal zones? The RAD-seq dataset allowed us to investigate this question by quantifying the extent to which these populations are independent of each other. From these data, three scenarios may explain how the species’ history of divergence interplays with the hadal zone geochronology: (i) dispersal between the hadal features across abyssal plains and mid-ocean ridges; (ii) independent immigration into the various hadal features (possibly without subsequent dispersal/migration) by a widely distributed abyssal ancestor; and (iii) an ancient immigration into the hadal zone and then ancient migration between hadal features when these were interconnected.
A key difference among the scenarios is the timing of hadal colonization. The age of the hadal community is a contentious subject since the first hadal organisms were recovered (3, 4, 7). While data are sparse on when hadal-dwelling lineages diverged, some evidence suggests that the contemporary hadal community is relatively young. Recent fossil-based time-calibrated Amphipoda phylogeny suggests that tectonic-induced climatic cooling and a more ventilated deep ocean in the Paleogene-Neogene promoted colonization and adaptive radiation of abyssal amphipods (50). In the study, the divergence time between Paracallisoma and B. schellenbergi was placed between 17 and 15 Ma in the Miocene (50), and our data align with that estimate using a standard estimated COI substitution rate for crustaceans of 1.4% per million year (46) (table S7). Applying a 1.4% substitution rate, preliminary divergence time estimations indicate that the Atacama Trench population may have diverged as early as 3.8 Ma in the Pliocene, and the west Pacific Ocean clades during the past ~300,000 to 500,000 years (table S7). In the following paragraphs, we will consider this timing estimate to examine the data at three scales—global, the west Pacific Ocean, and within-basin—by considering topographical barriers, geologic history, and geographic distance and then identify the most plausible scenario.
Global
Population differentiation was most clearly structured among the four ocean basins (Fig. 4), expanding preliminary phylogeographic patterns identified between the amphipods at the Wallaby-Zenith Fracture Zone and among four western Pacific Ocean features (23). While we expected genetic differentiation between populations residing in two ocean basins (20), the strength of the differentiation, FST > 0.30, was unexpected. The near-complete differentiation contrasts findings of cross-directional gene flow among trench populations of Paralicella spp. across the Pacific Ocean (20), which was enough for that study to conclude that the populations in hadal features are not evolutionarily independent units. However, Paralicella spp., while present in the upper hadal depths, is predominantly distributed at the shallower abyssal depths and hence is not considered to have a restricted hadal distribution. B. schellenbergi has been found as shallow as 5600 m in the New Hebrides Trench, albeit only 13 of 792 individuals were found at the nonhadal sites (51); however, this amphipod has only been found at subduction trenches and fracture zones and not at adjacent abyssal-depth features or well-sampled abyssal plains, such as the Clarion-Clipperton Zone (52). Given the high levels of genetic structure between oceans and the time to accumulate difference, it is unlikely that gene flow is ongoing between oceans.
As lineages diverge, the barriers that limit genetic exchange among populations become stronger (1). Here, the strength of the differentiation among ocean basins is likely attributed to a combination of barriers. The most obvious barrier is the sheer extent of geographic distance, with almost 26,000 km separating the Puerto Rico and Mariana trenches (table S1). These vast distances are further subdivided by physical barriers of shallower topographic features such as abyssal plains, mid-ocean ridges, and continental landmasses. Mid-ocean ridges, such as the Mid-Atlantic Ridge, can be a biogeographic barrier, restricting dispersal for abyssal taxa based on a species dispersal strategy (53, 54). For example, the Mid-Atlantic Ridge is a minimal barrier for select active swimming Munnopsidae isopods while a near-absolute barrier for nonswimming Macrostylidae isopods (55). The third barrier is the environment of each hadal feature, which is distinctive with a combination of extrinsic factors, such as the flux of nutrients, oxygen supply, and temperature of the feature.
The largest differentiation was driven by the Atacama Trench population (Fig. 5 and fig. S2). As the species delimitation analyses are limited to the generally faster evolved maternally inherited mitochondrial genome (56), the nuclear genome divergence supports that the Atacama Trench population, as well as the South Sandwich population, is on a separate speciation trajectory, suggesting that strong isolating mechanisms are in action. The environmental conditions of both trenches are distinct. The Atacama Trench is situated below eutrophic surface waters, and the bottom water O2 concentrations are 25% lower than other trenches with data (57), and the South Sandwich Trench has subzero temperatures and receives large krill-based nutrient pulses (44). Coupled with genetic drift, these environmental conditions may act as selective pressures promoting adaptions to the Atacama Trench and South Sandwich Trench and as driving speciation, which has led to other endemic species such as E. atacamensis in the Atacama Trench (16) and several previously unidentified snailfishes in both trenches (12). While out of scope to confirm here, both trenches represent model settings for future studies to identify and elucidate protein-level differences and adaptive mechanisms across a range of taxa.
West Pacific Ocean
With the high levels of tectonic activity along the rim of the Pacific Plate, most of the hadal features are found in the western Pacific Ocean, giving an ideal opportunity to investigate the extent of genetic homogeneity between hadal populations in the same ocean basin. Previous research on the morphometric differences between three west Pacific trench populations of H. gigas suggested enough morphological distinctiveness within populations to indicate a reduction of gene flow among populations (17). Overall, our population structure findings, with a mean FST of 0.16 among the six west Pacific Ocean trenches, provide support for a reduced gene flow between hadal populations in the same region.
The population differentiation among the six west Pacific Ocean features showed more complexity to global-level patterns, where the differences between populations can be explained by a mix of geographic distance, topographical barriers, and evolutionary history. For example, the Kermadec Trench is ~4500 km farther from the Mariana Trench than the New Hebrides Trench, yet the Kermadec Trench population was similarly differentiated from the Mariana and New Hebrides trench populations (Figs. 3 and 4). The formation of these trenches and subsequent separation by the North Fiji Basin and the Melanesian Basin seafloor has been present since the Miocene Epoch (58), which is much earlier than the estimated ~1 Ma of estimated genetic divergence among these populations (table S7). This strongly suggests that either the trenches were later colonized or dispersal occurred after their separation. While our sampling was extensive and leveraged public DNA barcoding sequences, not all features B. schellenbergi are known to inhabit were sampled by this study, specifically the Massau, Japan, and Kuril-Kamchatka trenches (24) (Fig. 1). Additional sampling in the future and/or focused studies on other taxa may provide the opportunity to further assess how geologic complexity influences population dynamics.
Within-basin
While the dominant pattern uncovered by our data is that hadal features pose barriers to gene flow, three instances of lack of genetic differentiation among features were detected (Fig. 3) and for those with FST > 0.05 contrasting migration patterns emerged (Fig. 5). Connectivity differences can be explained, in part, by a combination of seabed topography, plate tectonics, and circulation patterns. The Tonga and Kermadec trenches are a part of the same subduction zone (59), separated by the Louisville Seamount Chain, which over millions of years has alternated between hadal continuity and abyssal partitioning as these seamounts subduct (60) (fig. S8A). This “leaky barrier” coupled with the strong swimming ability of B. schellenbergi may explain the lack of population divergence. Similarly, the San Cristobal, Santa Cruz, and New Hebrides trenches share a subduction zone complex, the Solomon Islands–Vanuatu subduction complex (61). The San Cristobal and Santa Cruz trenches are not currently connected at hadal depths, but they are only separated by a 100-km-wide topographic high of ~5000-m depth where the South Rennell Trough intersects the subduction zone (fig. S8B). Therefore, gene flow between San Cristobal and Santa Cruz trench populations seems plausible (Fig. 5A). Evidence of gene flow between the San Cristobal and New Hebrides populations is unexpected given that they are separated by ~1100 km of substantially shallower water depths and the island of Vanuatu, a result of the subduction of the North d’Entrecasteaux Ridge (fig. S8A). Connectivity may be explained by individuals transported by deep-ocean currents; however, deep-ocean circulation data for this region are scarce. The gene flow results are constrained by the sampling locations, where a different set of samples from within a trench and other unrepresented populations, such as the Massau Trench, are likely to modify the connectivity patterns. Yet, the pattern of a level of regional connectivity is a previously unidentified insight. Future investigations should aim to provide greater spatial resolution and sampling intensity to illuminate connectivity within and between adjacent trenches, both in these systems and in other multitrench systems with one continuous hadal habitat, such as the Izu-Bonin, Japan, and Kuril-Kamchatka trenches (6).
The contemporary migration pattern, or potential lack thereof, uncovered between the Wallaby-Zenith Fracture Zone and Java Trench contrasts those of the west Pacific Ocean populations (Figs. 3 and 5B). While in the same subocean basin, the main difference between these two features and the Pacific Ocean features is that the Java Trench and Wallaby-Zenith Fracture Zone do not share a convergent zone. The Java Trench is formed where the Indo-Australian Plate subducts beneath the Eurasian Plate (62) with convergence initiating in the early Paleogene, whereas the Wallaby-Zenith Fracture Zone sits within the Wharton Basin and formed as a transform fault during the opening of the Indian Ocean in the Early Cretaceous (63). The Wallaby-Zenith Fracture Zone, and other small nontrench hadal features, has been suggested to be spillover or stepping-stone populations from the Java Trench, with a corridor of connectivity between populations through the Wharton Basin (23). Yet, the lack of inferred migration and high inbreeding indicates that these two populations are not actively or directly connected. It is also potentially indicative of incomplete lineage sorting with insufficient evolutionary time for genetic differences to accumulate. More efforts are needed to directly test the hypothesis that small nontrench hadal features may function as stepping-stone habitats.
Scenarios for the descent of B. schellenbergi to the hadal zone
Through examining these data at the three different scales, we can evaluate the degree to which the data support each of the possible scenarios and speculate on the most likely: (1) dispersal between the hadal features, (2) a widely distributed abyssal species colonized the hadal zone, or (3) ancient immigration into the hadal zone and then ancient migration between hadal features. Our findings at both the global and regional levels provide strong evidence that abyssal plains, basins, and forearcs act as barriers to gene flow for B. schellenbergi. At the regional level, we see that these allopatric populations are highly restricted to respective features, except for minimal connectivity between adjoining features. On the basis of these time scales and that hadal features have been separated between longer periods, upward of 150 Ma (4), we find scenario (3) of an ancient lineage and ancient migration to be a less likely hypothesis. Between scenarios (1) and (2), we are unable to rule one out as they both likely work together and function more strongly at differing scales. Scenario (1) is the most plausible to explain the localized patterns between populations residing in topographically connected features, such as Tonga and Kermadec trenches, and some individuals may be able to actively swim and disperse between features. At the global and basin level, scenario (2) is more favorable to explain the global and basin level patterns of a species with a cosmopolitan distribution that is highly restricted to the disjunct hadal features.
Following scenario (2), the measured structure among the Bathycallisoma populations may reflect a signature of refugial bathymetric range truncation (64, 65), whereby populations are allopatrically separated—entirely isolated from each other or have limited genetic exchange between neighboring feature populations. During the Miocene and Pliocene Epochs, the deep-ocean temperatures are estimated to have been 3° to 6°C warmer than present-day conditions. For deep-ocean organisms, a warmer temperature could allow for both wider eurybathic and deeper distribution, as the temperature is less of a physiological limitation (53). Thus, it is plausible that a common ancestor of B. schellenbergi had an abyssal-centric cosmopolitan distribution, where the topographic barriers, such as the North Fiji Basin, were less disruptive. Following the Mid-Miocene Climatic Optimum, global-scale changes triggered a long-term period of cooling to current conditions, leading to a narrowing of vertical ranges in deep-ocean species (53, 66). While speculative, the ancestral Bathycallisoma may have had a suite of pressure adaptions necessary and the competitive advantage to restrictively survive in the hadal features. This hypothesis is an intriguing concept that highly warrants further sampling across the deep ocean and the inclusion of more taxa to resolve deep data gaps in our understanding of how and when life evolved into the hadal zone.
Implications for hadal research
Previously limited by a lack of physical samples, this study exemplifies how hadal biology may at last be able to test these decades-old ecoevolutionary questions and showcases that global hadal-depth datasets are now possible. Our study demonstrates that hadal biology and ecology are progressing from being a largely observational science. Here, we have provided an in-depth investigation into the complex geological, biological, and environmental drivers of distributional patterns of a key hadal species across a geographical extent never before attempted. Although this study focuses on Bathycallisoma, our finding that hadal features act as isolating island habitats may explain distribution patterns in other hadal taxa, particularly snailfishes (12) and less mobile taxa, such as holothurians. It is increasingly clear that the large-scale geomorphology of the hadal zone comprises distinct habitats that affect biodiversity and drives speciation. With mounting evidence that these remote ecosystems and the communities they contain are the final receptors for Anthropocene impacts (18), we suggest its inclusion in future global ocean conservation strategies.
MATERIALS AND METHODS
Study locations and design: Amphipods from 12 hadal features during 13 sampling campaigns
Amphipods were recovered from 12 hadal features using autonomous, free-fall lander vehicles (67) between 2011 and 2019 (Fig. 1 and Table 1). Each lander was equipped with a bespoke invertebrate funnel trap that was baited with whole mackerel (Scombridae). Landers remained on the seafloor for 7 to 12 hours.
Upon recovery of landers and following initial sorting on deck, amphipods were preserved using 70% ethanol. In the laboratory, amphipods were morphologically identified following Kilgallen and Lowry (24) using a stereomicroscope (Wild Heerbrugg M8). For a subset of individuals from each feature, appendages, particularly the mandible and gnathopods 1 and 2, were dissected, temporarily mounted with glycerol, and viewed with a Leica DMi8 inverted microscope.
DNA extraction
We selected individuals to represent the full recovered depth range at each feature, with representatives from minimum and maximum depths, where possible. Total genomic DNA was extracted from 239 amphipods following a magnetic bead–based protocol (68). The head was dissected and added to reaction tubes with a solution of low-salt detergent buffer, Proteinase K (Thermo Fisher Scientific), and ribonuclease A [New England Biolabs (NEB)]. Following incubation at 50°C overnight, DNA was precipitated and cleaned in a series of washes with Sera-Mag SpeedBeads Carboxylate-Modified Magnetic Particles (SpeedBeads) using isopropanol and fresh 80% ethanol. The SpeedBeads were air-dried for approximately 10 min before elution with 45 μl of low TE (pH 7).
DNA quantity and integrity were measured with a 4200 TapeStation (Agilent Technologies). Samples with concentrations of <35 ng/μl and/or the TapeStation assigned DNA Integrity Number < 4.0, an indication of high fragmentation, were excluded from the RAD-seq library. All amphipods from the Diamantina Fracture Zone failed to meet the DNA quantity and integrity mark and were excluded from RAD-seq library preparation.
DNA barcoding dataset
Morphological-based identification was cross-validated with DNA barcoding at two mitochondrial regions. The partial regions of 16S (260 bp) and COI (658 bp) were amplified using published PCR primers and protocols (14). PCR products were enzymatically cleaned and sequenced with an ABI 3730XL sequencer (Eurofins Genomics, Germany). Electropherograms were manually trimmed in MEGA X (69). The COI sequences were translated to assess for stop codon presence. Sequence quality, absence of contamination, and initial species identification were verified on the NCBI BLASTn and BOLD v4 websites (70).
The mtDNA phylogenetic and phylogeographic relationships among the hadal-feature populations were investigated by constructing two alignments, specifically a 16S (215 bp) and a COI (559 bp) alignments. In addition to the sequences generated here, nine sequence sets were retrieved from GenBank for individuals identified as either B. schellenbergi or the synonymized name Scopelocheirus schellenbergi from the Wallaby-Zenith Fracture Zone (23) and Tonga, Kermadec, New Hebrides, Massau, and Kuril-Kamchatka trenches (table S8). Three species were selected as outgroups: Paracallisoma sp. as a species from the same family, Scopelocheiridae, and H. dubia and H. gigas as hadal fauna in a separate family (Hirondellidae) (table S8). Nucleotide sequences were aligned with the MAFFT v7 (71) webserver with the FFT-NS-1 strategy and trimmed to equal length.
The phylogenetic and phylogeographic relationships were inferred using the Bayesian Evolutionary Analysis by Sampling Trees (BEAST) software package v1.10.4 (72) for each gene and concatenated genes with partitions. The optimal evolutionary models were identified in MEGA X (69) as the HKY model for 16S and the TN93 model for COI, both with gamma distributions. For each combination, two independent runs were performed for 40,000,000 generation sampling every 10,000 generations with an uncorrelated relaxed clock. Outputs were assessed with Tracer v1.7 to ensure convergence [effective sample size (ESS) > 200] and combined in LogCombiner v1.8.4. The first 4,000,000 states were discarded as burn-in, and the maximum clade credibility tree was generated in TreeAnnotator v1.8.4. The final trees were viewed in FigTree v1.4.3. The concatenated 16S + COI tree is used as the graphical presentation of the phylogenetic and molecular species delimitation results and annotated in Inkscape v0.92.2.
Three molecular species delimitation approaches were applied to test for evidence of cryptic speciation, with one distance-based and two tree-based methods. The distance-based method was ASAP, which uses pairwise genetic distances to make groups and a scoring system to estimate species partitions (34). The ASAP method was applied to the 16S and COI alignments separately on the ASAP webserver using the Jukes-Cantor (JC69) substitution model (34). The two tree-based approaches were the GMYC and the bPTP methods (35, 36). GYMC delimits species based on the inter- and intraspecies branching patterns of an ultrametic tree (35), whereas bPTP models speciation based on the number of substitutions (36). The input trees of the COI and concatenated dataset were constructed in BEAST for both approaches. For GMYC, the ultrametric trees were constructed on the basis of a normalized exponential relaxed clock and a Yule process of speciation. For the GYMC, the number of delimited species was determined through the “gymc” function in the splits v1.0-20 package (73) in the program R v3.6.3 (74). For the bPTP method, the unrooted tree was analyzed on the bPTP webserver for 200,000 generations, a thinning of 100, and a burn-in of 25%.
Pairwise p-distances (transition and transversion substitution included and complete deletion) between study-generated and comparative B. schellenbergi individuals were calculated separately on the 16S and COI alignments using MEGA X. For this analysis, representatives from Diamantina Fracture Zone were included in the 16S alignment. The p-distances were summarized by the hadal feature population.
A 16S + COI concatenated alignment (774 bp) was used to infer a haplotype network using the statistical parsimony method (75) in PopART v1.7 (76). Haplotype diversity was calculated with the package pegas v0.14 (77) in R.
Nuclear dataset using RAD-seq
bestRAD library preparation
RAD-seq libraries were constructed for 128 individuals from 11 features (Table 1) using the bestRAD protocol (27). Samples were normalized to 250 ng in 10 μl of 1× TE. The genomic DNA was digested with Sbf I–HF restriction enzyme (NEB). Following ligation of the biotinylated bestRAD adapters, samples were multiplexed into two libraries consisting of 96 and 32 samples. Pooled DNA was sheared in a Bioruptor NGS sonicator (Diagenode) with three cycles of 30 s on/90 s off and then visualized on a fragment analyzer to ensure DNA fragments ranged from 100 to 500 bp.
The bestRAD adapters contain biotinylated ends, which bind to Dynabeads M-280 streptavidin magnetic beads (Life Technologies) to physically isolate the RADtagged DNA fragments. The RADtags were freed from the Dynabeads with Sbf I–HF digestion and resuspended in 55.5 μl of low TE. The NEBNext Ultra DNA Library Prep Kit for Illumina was used to repair blunt ends and ligate adapters. The RADtags were size-selected for 500 bp. A test library enrichment PCR was run with 5 μl of library for 15 cycles to determine the optimal number of PCR cycles. The final library enrichment PCR was run for 13 cycles with 15 μl of library. Paired-end sequencing of 150 bp was carried out across two lanes on an Illumina HiSeq 4000 at the UC Davis Genome Center.
Processing and filtering of reads
MultiQC reports were examined for each library before demultiplexing and quality filtering with the Stacks v2.54 pipeline (78). The RADtags were processed using “process_radtags” with the rescue feature activated and allowing for three mismatches between barcodes. The reads were trimmed to 130 bp. Following “process_radtags,” a total of 697,111,355 reads were retained from the initial 1,038,164,688 raw reads (67.1%), with an average of 5,445,430 reads per individual. Six individuals had <500,000 retained reads and were excluded from subsequent processing (data S1).
We tested two parallel approaches for sequence assembly and SNP discovery, specifically the Stacks v2 “de novo” pipeline and ipryad v.0.9.68 (28), with a 22-subsample dataset (two individuals per feature). From the Stacks v2 pipeline, few loci were shared between individuals. This problem was not observed in the ipyrad pipeline, as indicated by an average sample coverage of 74.7×, and thus deemed a suitable approach. A similar SNP discovery problem has been documented with a coral-associated amphipod ddRAD dataset and overcome with ipyrad (79). As ipyrad is not developed for paired-end data from the bestRAD protocol, only the forward reads were used. The “clustering threshold [14]” was first optimized with the 22-subsample dataset, testing at 0.85, 0.87, 0.90, and 0.93. The clustering threshold was selected at 0.87 as the balance between under- and oversplitting loci. The parameters, “max cluster depth within samples [13]”, “max N’s (uncalled bases) in consensus [19]”, “max H’s (heterozygotes) in consensus [20]”, and “max # SNPs per locus [22]”, were further optimized with the full dataset through branching. We selected the combination of parameters that maximized the number of total filtered loci at the end of the ipyrad pipeline. The optimized parameters (nondefault) were maximum depth within samples of 100 and a clustering threshold of 0.87, and the minimum number of individuals per locus was set to 70%. The final parameter file is provided (table S9).
RAD-seq datasets require strict filtering to ensure that the final genotypes represent independent markers and do not lead to misleading patterns (30). Following de novo assembly, we filtered the dataset using VCFtools 0.1.16 (80) to remove loci with a low read depth (<6x), excessively high read depth (>35×), missing from 30% or more of individuals, less than 0.05 minimum allele frequency, and more than one variable site per RADtag to remove short-distance linkage disequilibrium between SNPs (filtering details and results are in table S2) (81). After locus filtering, 25 individuals had >50% missingness and a dataset of 498 SNPs. These poor-performing individuals had <3.5M retained reads following “process_radtags.” The ipryad and filtering pipeline was rerun with the poor-performing individuals removed (29). The locus recovery improved, with the final thinned dataset consisting of 97 individuals and 2933 SNPs. This high-quality dataset was used for the subsequent phylogenetic, population genetic structure, and contemporary migration analyses that require single SNPs per loci input. One exception is fineRADstructure, which combines all SNPs for each locus into haplotypes and then performs structure analyses on these haplotypes (37). For fineRADstructure, we used the prefiltered output, direct from ipyrad, of 4611 loci and 58,730 SNPs.
Conducting phylogenetic analysis
A phylogeny was constructed using the maximum likelihood approach implemented by IQ-TREE v1.6.12 (82). The analysis was run on the W-IQ-TREE Web Service provided by Los Alamos National Laboratory, USA (83), where ModelFinder (84) applied the best model, TVM + F + R2, and an ascertainment bias correction (+ASC) model. Branch support was generated by the standard bootstrap method with 100 replicates. The final tree was viewed in FigTree and annotated in Inkscape.
Performing population genetic structure analyses
We used a suite of six analyses to examine the degree to which the 11 hadal features are population genetically structured or differentiated and the extent geographic distance explains those differences according to isolation by distance. These analysis approaches invoke a variety of different assumptions and underlying models (e.g., Hardy-Weinberg equilibrium). By using a complementary suite of methods, we can develop a robust understanding of patterns of population structure.
The first analysis was STRUCTURE, which is a model-based clustering method that uses a Bayesian approach to detect allele frequency differences and assign individuals to subpopulations on the basis of an analysis of likelihoods. We implemented STRUCTURE v2.3.4 (85) by the parallelized and automated structure_threader (86). For all runs, an admixture model was selected with correlated allele frequencies. Population information was not used to preassign individuals to clusters, i.e., USEPOPINFO 0. The Markov chain Monte Carlo (MCMC) runs were completed with a burn-in of 10,000 and 100,000 repeats. Ten independent runs were performed for each value of K (assumed genetic cluster). STRUCTURE HARVESTER Web v0.6.94 (87) collated the results and inferred the most likely number of clusters by using the maximum log probability approach [ln Pr(X|K)] (85) and the ΔK statistic (88). We applied a hierarchical approach to the clustering analysis. The first level included all 11 a priori populations and assumed 1 to 12 genetic clusters (K). The second level was the six a priori population situated in the western Pacific Ocean with K ranging from one to eight. The third level was the four a priori western central Pacific Ocean populations with K varying from one to six. The fourth level was three sets of populations [i.e., (i) the Kermadec and Tonga trenches; (ii) the Santa Cruz, San Cristobal, and New Hebrides trench; and (iii) the Java Trench and Wallaby-Zenith Fracture Zone] with K ranging from one to two plus the number of a priori populations. The log probabilities and the ΔK statistic results are presented in figs. S9 and S10.
The second structure analysis was fineRADstructure, which, similar to STRUCTURE, uses a Bayesian clustering approach. Instead of a single SNP per locus, fineRADstructure treats all SNPs within a locus as a haplotype to infer coancestry (37). To assign individuals to populations, the MCMC analysis was run with a burn-in of 100,000 iterations, 1 × 106 sample iterations, and thinning to every 1000th iteration.
The third structure analysis was DAPC, applied with the package adegenet v2.1.3 (38) in R. In contrast to STRUCTURE and fineRADstructure, DAPC is a model-free multivariate ordination method that uses a sequential K-means and model selection to infer genetic clusters (39). As with STRUCTURE methods, we applied a hierarchical approach. For each dataset, the optimal K was selected using the find.clusters function and was based on the number of clusters with the smallest Bayesian information criterion.
The fourth structure analysis was a PCA, which is a multivariate analysis that reduces dimensionality by calculating principal components from allele frequencies. We performed a PCA using the package ade4 v1.7-16 (39) in R.
The fifth structure analysis was pairwise FST, which is a measure of population differentiation based on the variance of allele frequencies between populations. To account for the unequal sample sizes, pairwise FST was calculated among the 11 sampled features with the Hudson’s FST estimator using the package popkinsuppl v1.0.17.9000 in R (89).
The sixth structure analysis was an AMOVA, which tests for the hierarchical population structure with the calculation of F-statistics. Conducted in GenoDive 3.04 (41), we evaluated variation in the “all populations” dataset at three levels: (i) individual, (ii) hadal feature, and (iii) ocean. An infinite allele model was used with a significance test using 999 permutations.
Patterns of isolation by distance were evaluated among the 11 sampled features using the calculated pairwise FST and pairwise geographical distance between each feature. For the sampled features that included individuals from more than one sampled depth, the sampling location for the deepest site was used as the geographic location to represent the feature (table S1). The geographic distance between sampled features was calculated with the Geographic Distance Matrix Generator (version 1.2.3), using the semimajor axis of the WGS84 reference system as the default radius of Earth (90). Where a continental landmass separated any sampled features, an oceanographic link was determined, and the distance was measured directly in a three-dimensional global scene generated in ArcGIS Pro (version 2.9). A Mantel test was applied to test for a linear correlation between the two distance matrixes using the mantel.randtest function in adegenet v2.1.3 (38) at four levels (α = 0.05): (i) all features, (ii) all features except Atacama Trench, (iii) only Atacama Trench, and (iv) the six western Pacific Ocean features. Linear models were fit using the package stats v3.6.3 in R.
Estimating heterozygosity and inbreeding
To minimize biases in our estimates of heterozygosity, we constructed feature-specific datasets with a modified filtering approach (91). For each feature population, four random individuals were selected to account for differences in sample size between populations and run through the ipyrad pipeline. SNPs were retained with a minor allele count of 1 and had to be genotyped at all four individuals (e.g., 0% missingness). Heterozygosity was computed using the packages adegenet and hierfstat for two versions of each dataset: first, thinned to one SNP per RADtag, and second, an unthinned dataset with all SNPs per RADtag that met the other filtering criteria. The FIS (inbreeding coefficient) was calculated following Nei (1987) in the package hierfstat v0.5-7 (92) in R.
Determining patterns of gene flow among genetic populations
Directionality of gene flow between features was inferred using BayesAss v3.04, a Bayesian-based program that infers the proportion of recent migrants among populations (42, 93). We estimated the rates of gene flow among the two multifeature genetic clusters with pairwise FST > 0.05 (43), namely, (i) among the Santa Cruz, San Cristobal, and New Hebrides trenches and (ii) between the Java Trench and Wallaby-Zenith Fracture Zone. For each dataset, trials were conducted by adjusting the three mixing parameters (i.e., migration rate, allele frequency, and inbreeding coefficient) to bring the acceptance rates into the recommended range of 20 to 60% (42). For the Java Trench and Wallaby-Zenith Fracture Zone, mixing parameters were set at dM = 0.3, dA = 0.9, and dF = 0.1. For the Santa Cruz, San Cristobal, and New Hebrides trenches, mixing parameters were set at dM = 0.45, dA = 1, and dF = 0.1. For the Santa Cruz, San Cristobal, and New Hebrides trenches, five runs were conducted with a different seed for 40,000,000 iterations, a burn-in of the first 4,000,000 iterations, and a sampling interval of 5000. For the Java Trench and Wallaby-Zenith Fracture Zone, five runs were conducted for 31,000,000 iterations, a burn-in of the first 1,000,000 iterations, and a sampling interval of 5000. The log outputs were assessed in Tracer, and the Bayesian deviance was calculated (94). The best run was selected as the run with ESS > 200 and the lowest Bayesian deviance. Tracer plots are provided in figs. S6 and S7. Significant differences in proportions of immigrant ancestry were assessed using nonoverlapping 95% confidence intervals.
Acknowledgments
Amphipods were recovered over 11 years onboard several vessels and funding schemes. We thank the captains, crew, and company of the New Zealand RV Kaharoa (KAH1109, KAH1202, and KAH1310), the German RV Sonne (SO261 and SO258), and the DSSV Pressure Drop (Five Deeps Expeditions) for their work to make specimen recovery possible. We thank A. G. Rivera-Colon (University of Illinois-Champaign), M. Wenzel (Aberdeen University), K. Dicks and H. Ritchie (RZSS Wildgenes Laboratory), A. Haupt (California State University, Monterrey Bay), and P. Leary (University of Zurich) for support and demystification of RAD-seq methods. We are thankful for science writing advice from A. Pineda (I focus and write). We are grateful to S. O’Rouke (University of California, Davis) for conducting the RAD-seq library construction, especially during the COVID-19 pandemic. We thank J. Nelson (Esri) for showing us that a map projection can change how we see the hadal zone. H.A.S. publishes with permission of the Executive Director of the British Geological Survey (UKRI). We thank the reviewers and editor for their constructive comments and insights that greatly improved this body of work.
Funding: The KAH1109, KAH1202, and KAH1310 expeditions were part of HADEEP II-IV, which were funded by the TOTAL Foundation (France) through the “Multi-disciplinary investigations of the deepest scavengers on Earth” (2010–2012) and “Trench Connection” (2013–2015). The SO258 Expedition was funded by the German Federal Ministry of Education and Research and GEOMAR. The SO261 Expedition was funded by the HADES-ERC Advanced Grant “Benthic diagenesis and microbiology of hadal trenches” (grant #669947). The Five Deeps Expedition (sea time and logistics) was privately funded by Victor Vescovo of Caladan Oceanic LLC (USA). Participation of A.J.J. on these expeditions was supported by Newcastle University’s Research Infrastructure Fund, Exploration of Extreme Ocean Environments, awarded to A.J.J. Molecular work and sequencing were funded by the “Hadal Zones of our Overseas Territories” by the Darwin Initiative funded by the U.K. Government (grant DPLUS093), awarded to H.A.S., and the 2017 Systematics Research Fund, awarded to J.N.J.W. The Illumina sequencing was carried out by the DNA Technologies and Expression Analysis Core at the UC Davis Genome Center, supported by the NIH Shared Instrumentation Grant 1S10OD010786-01. The bioinformatics was conducted with the Rocket High-Performance Computing Service provided by Newcastle University.
Author contributions: Conceptualization: J.N.J.W. and J.J.N.K. Specimen collection: A.J.J. Methodology: J.N.J.W. and J.J.N.K. Investigation: J.N.J.W. Formal analysis: J.N.J.W., E.L.J., and M.S.R.H. Visualization: J.N.J.W., E.L.J., and H.A.S. Supervision: A.J.J. and E.L.J. Writing (original draft): J.N.J.W. Writing (review and editing): J.N.J.W., E.L.J., H.A.S., and A.J.J.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All genetic data produced in this study are available through the NCBI. 16S accession numbers for the seven individuals from the Diamantina Fracture Zone are OK480969 to OK480975. 16S accession numbers for the other 94 individuals from 11 hadal features are OK480978 to OK481071. COI accession numbers are OK483380 to OK483473. The demultiplexed bestRAD data (truncated to 130 bp) are part of BioProject PRJNA772153 with BioSamples SAMN22366349 to SAMN22366476 and SRA SRR16470572 to SRR16470699 (all details included in data S1).
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Tables S1 to S9
References
Other Supplementary Material for this manuscript includes the following:
Data file S1
REFERENCES AND NOTES
- 1.Sobel J. M., Chen G. F., Watt L. R., Schemske D. W., The biology of speciation. Evolution 64, 295–315 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Mora C., Tittensor D. P., Adl S., Simpson A. G. B., Worm B., How many species are there on Earth and in the Ocean? PLOS Biol. 9, e1001127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.G. M. Belyaev, Deep-Sea Ocean Trenches and Their Fauna (Nauka, 1989). [Google Scholar]
- 4.A. J. Jamieson, The Hadal Zone: Life in the Deepest Oceans (Cambridge Univ. Press, 2015). [Google Scholar]
- 5.Stewart H. A., Jamieson A. J., Habitat heterogeneity of hadal trenches: Considerations and implications for future studies. Prog. Oceanogr. 161, 47–65 (2018). [Google Scholar]
- 6.Jamieson A. J., Stewart H. A., Hadal zones of the Northwest Pacific Ocean. Prog. Oceanogr. 190, 102477 (2021). [Google Scholar]
- 7.Wolff T., The hadal community, an introduction. Deep Sea Res. 6, 95–124 (1959). [Google Scholar]
- 8.Jamieson A. J., Stewart H. A., Nargeolet P. H., Exploration of the Puerto Rico Trench in the mid-twentieth century: Today’s significance and relevance. Endeavour 44, 100719 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Yancey P. H., Cellular responses in marine animals to hydrostatic pressure. J. Exp. Zool. A Ecol. Integr. Physiol. 333, 398–420 (2020). [DOI] [PubMed] [Google Scholar]
- 10.Lacey N. C., Rowden A. A., Clark M. R., Kilgallen N. M., Linley T., Mayor D. J., Jamieson A. J., Community structure and diversity of scavenging amphipods from bathyal to hadal depths in three South Pacific Trenches. Deep Sea Res. Part I Oceanogr. Res. Pap. 111, 121–137 (2016). [Google Scholar]
- 11.Eustace R. M., Ritchie H., Kilgallen N. M., Piertney S. B., Jamieson A. J., Morphological and ontogenetic stratification of abyssal and hadal Eurythenes gryllus sensu lato (Amphipoda: Lysianassoidea) from the Peru-Chile Trench. Deep-Sea Res. I Oceanogr. Res. Pap. 109, 91–98 (2016). [Google Scholar]
- 12.Jamieson A. J., Linley T. D., Eigler S., Macdonald T., A global assessment of fishes at lower abyssal and upper hadal depths (5000 to 8000 m). Deep-Sea Res. I Oceanogr. Res. Pap. 178, 103642 (2021). [Google Scholar]
- 13.Dahl E., Amphipoda from depths exceeding 6000 m. Galathea Rep. 1, 211–240 (1959). [Google Scholar]
- 14.Ritchie H., Jamieson A. J., Piertney S. B., Phylogenetic relationships among hadal amphipods of the Superfamily Lysianassoidea: Implications for taxonomy and biogeography. Deep-Sea Res. I Oceanogr. Res. Pap. 105, 119–131 (2015). [Google Scholar]
- 15.Jażdżewska A. M., Mamos T., High species richness of Northwest Pacific deep-sea amphipods revealed through DNA barcoding. Prog. Oceanogr. 178, 102184 (2019). [Google Scholar]
- 16.Weston J. N. J., Espinosa-Leal L., Wainwright J. A., Stewart E. C. D., González C. E., Linley T. D., Reid W. D. K., Hidalgo P., Oliva M. E., Ulloa O., Wenzhöfer F., Glud R. N., Escribano R., Jamieson A. J., Eurythenes atacamensis sp. nov. (Crustacea: Amphipoda) exhibits ontogenetic vertical stratification across abyssal and hadal depths in the Atacama Trench, eastern South Pacific Ocean. Mar. Biodivers. 51, 51 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.France S. C., Geographic variation among three isolated populations of the hadal amphipod Hirondellea gigas (Crustacea: Amphipoda: Lysianassoidea). Mar. Ecol. Prog. Ser. 92, 277–287 (1993). [Google Scholar]
- 18.Jamieson A. J., A contemporary perspective on hadal science. Deep-Sea Res. II Top. Stud. Oceanogr. 155, 4–10 (2018). [Google Scholar]
- 19.Taylor M. L., Roterman C. N., Invertebrate population genetics across Earth’s largest habitat: The deep-sea floor. Mol. Ecol. 26, 4872–4896 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Ritchie H., Jamieson A. J., Piertney S. B., Population genetic structure of two congeneric deep-sea amphipod species from geographically isolated hadal trenches in the Pacific Ocean. Deep-Sea Res. I Oceanogr. Res. Pap. 119, 50–57 (2017). [Google Scholar]
- 21.Jazdzewska A. M., Horton T., Hendrycks E. A., Mamos T., Driskel A., Brix S., Arbizu P., Pandora’s box in the deep sea–Intraspecific diversity patterns and distribution of two congeneric scavenging amphipods. Front. Mar. Sci. 8, 750180 (2021). [Google Scholar]
- 22.Birstein J. A., Vinogradov M. E., Pelagicheskie gammaridy (Amphipoda, Gammaridea) severo-zapadnoi chasti Tikhogo Okeana. [Pelagic Gammaridea from the northwestern Pacific Ocean]. Trudy Instituta Okeanologii Akademija Nauk S.S.S.R. 27, 219–257 (1958). [Google Scholar]
- 23.Weston J. N. J., Peart R. A., Stewart H. A., Ritchie H., Piertney S. B., Linley T. D., Jamieson A. J., Scavenging amphipods from the Wallaby-Zenith Fracture Zone: Extending the hadal paradigm beyond subduction trenches. Mar. Biol. 168, 1–14 (2021). [Google Scholar]
- 24.Kilgallen N. M., Lowry J. K., A review of the scopelocheirid amphipods (Crustacea, Amphipoda, Lysianassoidea), with the description of new taxa from Australian waters. Zoosyst. Evol. 91, 1–43 (2015). [Google Scholar]
- 25.Brandt A., Błazewicz-Paszkowycz M., Bamber R. N., Mühlenhardt-Siegel U., Malyutina M. V., Kaiser S., de Broyer C., Havermans C., Are there widespread peracarid species in the deep sea (crustacea: Malacostraca)? Polish Polar Res. 33, 139–162 (2012). [Google Scholar]
- 26.Dixon D. R., Pruski A. M., Dixon L. R. J., The effects of hydrostatic pressure change on DNA integrity in the hydrothermal-vent mussel Bathymodiolus azoricus: Implications for future deep-sea mutagenicity studies. Mutat. Res-Fund. Mol. Mech. Mutagen. 552, 235–246 (2004). [DOI] [PubMed] [Google Scholar]
- 27.Ali O. A., O’Rourke S. M., Amish S. J., Meek M. H., Luikart G., Jeffres C., Miller M. R., Rad capture (Rapture): Flexible and efficient sequence-based genotyping. Genetics 202, 389–400 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eaton D. A. R., Overcast I., ipyrad: Interactive assembly and analysis of RADseq datasets. Bioinformatics 36, 2592–2594 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Cerca J., Maurstad M. F., Rochette N. C., Rivera-Colón A. G., Rayamajhi N., Catchen J. M., Struck T. H., Removing the bad apples: A simple bioinformatic method to improve loci-recovery in de novo RADseq data for non-model organisms. Methods Ecol. Evol. 12, 805–817 (2021). [Google Scholar]
- 30.O’Leary S. J., Puritz J. B., Willis S. C., Hollenbeck C. M., Portnoy D. S., These aren’t the loci you’e looking for: Principles of effective SNP filtering for molecular ecologists. Mol. Ecol. 27, 3193–3206 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Nazareno A. G., Bemmels J. B., Dick C. W., Lohmann L. G., Minimum sample sizes for population genomics: An empirical study from an Amazonian plant species. Mol. Ecol. Resour. 17, 1136–1147 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Havermans C., Sonet G., d’Udekem d’Acoz C., Nagy Z. T., Martin P., Brix S., Riehl T., Agrawal S., Held C., Genetic and morphological divergences in the cosmopolitan deep-sea amphipod Eurythenes gryllus reveal a diverse abyss and a bipolar species. PLOS ONE 8, e74218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brasier M. J., Wiklund H., Neal L., Jeffreys R., Linse K., Ruhl H., Glover A. G., DNA barcoding uncovers cryptic diversity in 50% of deep-sea Antarctic polychaetes. R. Soc. Open Sci. 3, 160432 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puillandre N., Brouillet S., Achaz G., ASAP: Assemble species by automatic partitioning. Mol. Ecol. Resour. 21, 609–620 (2021). [DOI] [PubMed] [Google Scholar]
- 35.Fujisawa T., Barraclough T. G., Delimiting species using single-locus data and the generalized Mixed Yule Coalescent Approach: A revised method and evaluation on simulated data sets. Syst. Biol. 62, 707–724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang J., Kapli P., Pavlidis P., Stamatakis A., A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29, 2869–2876 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malinsky M., Trucchi E., Lawson D. J., Falush D., RADpainter and fineRADstructure: Population inference from RADseq data. Mol. Biol. Evol. 35, 1284–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jombart T., Ahmed I., adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J. Thioulouse, S. Dray, A.-B. Dufour, A. Siberchicot, T. Jombart, S. Pavoine, Multivariate Analysis of Ecological Data with ade4 (Springer, 2018). [Google Scholar]
- 40.Bhatia G., Patterson N., Sankararaman S., Price A. L., Estimating and interpreting FST: The impact of rare variants. Genome Res. 23, 1514–1521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meirmans P. G., genodive version 3.0: Easy-to-use software for the analysis of genetic data of diploids and polyploids. Mol. Ecol. Resour. 20, 1126–1131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilson G. A., Rannala B., Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163, 1177–1191 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faubet P., Waples R. S., Gaggiotti O. E., Evaluating the performance of a multilocus Bayesian method for the estimation of migration rates. Mol. Ecol. 16, 1149–1166 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Jamieson A. J., Stewart H. A., Weston J. N. J., Bongiovanni C., Hadal fauna of the South Sandwich Trench, Southern Ocean: Baited camera survey from the five deeps expedition. Deep-Sea Res. II Top. Stud. Oceanogr. 194, 104987 (2021). [Google Scholar]
- 45.Hebert P. D. N., Ratnasingham S., DeWaard J. R., Barcoding animal life: Cytochromecoxidase subunit 1 divergences among closely related species. Proc. R. Soc. B: Biol. Sci. 270, S96–S99 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Knowlton N., Weigt L. A., New dates and new rates for divergence across the Isthmus of Panama. Proc. R. Soc. B Biol. Sci. 265, 2257–2263 (1998). [Google Scholar]
- 47.Kou Q., Meland K., Li X., He L., Wang Y., Deepest record of Eucopia sculpticauda (Crustacea: Lophogastrida: Eucopiidae) and the order, with new insights into the distribution and genetic diversity of the species. Bull. Mar. Sci. 95, 327–335 (2019). [Google Scholar]
- 48.Weston J. N. J., Jamieson A. J., The multi-ocean distribution of the hadal amphipod, Hirondellea dubia Dahl, 1959 (Crustacea, Amphipoda). Front. Mar. Sci. 9, 155 (2022). [Google Scholar]
- 49.Jamieson A. J., Lacey N. C., Lörz A. N., Rowden A. A., Piertney S. B., The supergiant amphipod Alicella gigantea (Crustacea: Alicellidae) from hadal depths in the Kermadec Trench, SW pacific ocean. Deep-Sea Res. II Top. Stud. Oceanogr. 92, 107–113 (2013). [Google Scholar]
- 50.Copilaş-Ciocianu D., Borko Š., Fišer C., The late blooming amphipods: Global change promoted post-Jurassic ecological radiation despite Palaeozoic origin. Mol. Phylogenet. Evol. 143, 106664 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Lacey N. C., Mayor D. J., Linley T. D., Jamieson A. J., Population structure of the hadal amphipod Bathycallisoma (Scopelocheirus) schellenbergi in the Kermadec Trench and New Hebrides Trench, SW Pacific. Deep-Sea Res. II Top. Stud. Oceanogr. 155, 50–60 (2018). [Google Scholar]
- 52.Mohrbeck I., Horton T., Jażdżewska A. M., Martínez Arbizu P., DNA barcoding and cryptic diversity of deep-sea scavenging amphipods in the Clarion-Clipperton Zone (Eastern Equatorial Pacific). Mar. Biodiver. 51, 1–15 (2021). [Google Scholar]
- 53.McClain C. R., Hardy S. M., The dynamics of biogeographic ranges in the deep sea. Proc. R. Soc. B Biol. Sci. 277, 3533–3546 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riehl T., Kaiser S., Brandt A., Vema-TRANSIT – An interdisciplinary study on the bathymetry of the Vema-Fracture Zone and Puerto Rico Trench as well as abyssal Atlantic biodiversity. Deep-Sea Res. II Top. Stud. Oceanogr. 148, 1–6 (2018). [Google Scholar]
- 55.Riehl T., Lins L., Brandt A., The effects of depth, distance, and the Mid-Atlantic Ridge on genetic differentiation of abyssal and hadal isopods (Macrostylidae). Deep-Sea Res. II Top. Stud. Oceanogr. 148, 74–90 (2018). [Google Scholar]
- 56.Avise J. C., Arnold J., Ball R. M., Bermingham E., Lamb T., Neigel J. E., Reeb C. A., Saunders N. C., Intraspecific phylogeography: The mitochondrial DN a bridge between population genetics and systematics. Annu. Rev. Ecol. Evol. Syst. 18, 489–522 (2003). [Google Scholar]
- 57.Glud R. N., Berg P., Thamdrup B., Larsen M., Stewart H. A., Jamieson A. J., Glud A., Oguri K., Sanei H., Rowden A. A., Wenzhöfer F., Hadal trenches are dynamic hotspots for early diagenesis in the deep sea. Commun. Earth Environ. 2, 1–8 (2021). [Google Scholar]
- 58.Schellart W. P., Lister G. S., Jessell M. W., Analogue modeling of arc and backarc deformation in the New Hebrides arc and North Fiji Basin. Geology 30, 311–314 (2002). [Google Scholar]
- 59.R. J. Wysoczanski, M. R. Clark, Southern Kermadec Arc–Havre Trough geohabitats and biological communities, in Seafloor Geomorphology as Benthic Habitat: GeoHab Atlas of Seafloor Geomorphic Features and Benthic Habitats, P. T. Harris, E. K. Baker, Eds. (Elsevier Insights Series, Elsevier, 2012), pp. 853–867. [Google Scholar]
- 60.A. J. Jamieson, H. A. Stewart, A. A. Rowden, M. R. Clark, Geomorphology and benthic habitats of the Kermadec Trench, Southwest Pacific Ocean, in Seafloor Geomorphology as Benthic Habitat, P. T. Harris, E. Baker, Eds. (Elsevier, ed. 2, 2019), pp. 949–966. [Google Scholar]
- 61.Neely J. S., Furlong K. P., Evidence of displacement-driven maturation along the San Cristobal Trough transform plate boundary. Earth Planet. Sci. Lett. 485485, 88–98 (2018). [Google Scholar]
- 62.DeMets C., Gordon R. G., Argus D. F., Geologically current plate motions. Geophys. J. Int. 181, 1–80 (2010). [Google Scholar]
- 63.Olierook H. K. H., Merle R. E., Jourdan F., Sircombe K., Fraser G., Timms N. E., Nelson G., Dadd K. A., Kellerson L., Borissova I., Age and geochemistry of magmatism on the oceanic Wallaby Plateau and implications for the opening of the Indian Ocean. Geology 43, 971–974 (2015). [Google Scholar]
- 64.Lau S. C. Y., Wilson N. G., Silva C. N. S., Strugnell J. M., Detecting glacial refugia in the Southern Ocean. Ecography 43, 1639–1656 (2020). [Google Scholar]
- 65.Stewart J. R., Lister A. M., Barnes I., Dalén L., Refugia revisited: Individualistic responses of species in space and time. Proc. R. Soc. B Biol. Sci. 277, 661–671 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Herold N., Huber M., Müller R. D., Seton M., Modeling the Miocene climatic optimum: Ocean circulation. Paleoceanography 27, P1209 (2012). [Google Scholar]
- 67.Jamieson A. J., Fujii T., Solan M., Priede I. G., HADEEP: Free-falling landers to the deepest places on earth. Mar. Technol. Soc. J. 43, 151–160 (2009). [Google Scholar]
- 68.Oberacker P., Stepper P., Bond D. M., Höhn S., Focken J., Meyer V., Schelle L., Sugrue V. J., Jeunen G.-J., Moser T., Hore S. R., von Meyenn F., Hipp K., Hore T. A., Jurkowski T. P., Bio-on-magnetic-beads (BOMB): Open platform for high-throughput nucleic acid extraction and manipulation. PLOS Biol. 17, e3000107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ratnasingham S., Hebert P. D. N., A DNA-based registry for all animal species: The barcode index number (BIN) system. PLoS One. 8, e66213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh K., Rozewicki J., Yamada K. D., MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suchard M. A., Lemey P., Baele G., Ayres D. L., Drummond A. J., Rambaut A., Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.T. Ezard, T. Fujisawa, T. Barraclough, splits: SPecies’ LImits by threshold statistics (2021); https://R-Forge.R-project.org/projects/splits/.
- 74.R Core Team, R: A language and environment for statistical computing (2020); www.R-project.org/.
- 75.Templeton A., Hedges S., Kumar S., Tamura K., Stoneking M., Human origins and analysis of mitochondrial DNA sequences. Science 255, 737–739 (1992). [DOI] [PubMed] [Google Scholar]
- 76.Leigh J. W., Bryant D., popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 6, 1110–1116 (2015). [Google Scholar]
- 77.Paradis E., pegas: An R package for population genetics with an integrated–Modular approach. Bioinformatics 26, 419–420 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Rochette N. C., Rivera-Colón A. G., Catchen J. M., Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol. Ecol. 28, 4737–4754 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Schwentner M., Lörz A. N., Population genetics of cold-water coral associated Pleustidae (Crustacea, Amphipoda) reveals cryptic diversity and recent expansion off Iceland. Mar. Ecol. 42, e12625 (2021). [Google Scholar]
- 80.Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., DePristo M. A., Handsaker R. E., Lunter G., Marth G. T., Sherry S. T., McVean G., Durbin R., The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jensen E. L., Edwards D. L., Garrick R. C., Miller J. M., Gibbs J. P., Cayot L. J., Tapia W., Caccone A., Russello M. A., Population genomics through time provides insights into the consequences of decline and rapid demographic recovery through head-starting in a Galapagos giant tortoise. Evol. Appl. 11, 1811–1821 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen L.-T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trifinopoulos J., Nguyen L.-T., von Haeseler A., Minh B. Q., W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44, W232–W235 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., Jermiin L. S., ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pritchard J. K., Stephens M., Donnelly P., Inference of population structure using multilocus genotype data. Genetics 155, 945–959 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pina-Martins F., Silva D. N., Fino J., Paulo O. S., Structure_threader: An improved method for automation and parallelization of programs structure, fastStructure and MavericK on multicore CPU systems. Mol. Ecol. Resour. 17, e268–e274 (2017). [DOI] [PubMed] [Google Scholar]
- 87.Earl D. A., vonHoldt B. M., STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359–361 (2012). [Google Scholar]
- 88.Evanno G., Regnaut S., Goudet J., Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 14, 2611–2620 (2005). [DOI] [PubMed] [Google Scholar]
- 89.A. Ochoa, J. D. Storey, popkinsuppl: Supplement to popkin package (2022); https://github.com/OchoaLab/popkinsuppl.
- 90.P. J. Ersts, Geographic Distance Matrix Generator (version 1.2.3) (American Museum of Natural History, Center for Biodiversity and Conservation); http://biodiversityinformatics.amnh.org/open_source/gdmg.
- 91.Schmidt T. L., Jasper M. E., Weeks A. R., Hoffmann A. A., Unbiased population heterozygosity estimates from genome-wide sequence data. Meth. Ecol. Evol. 12, 1888–1898 (2021). [Google Scholar]
- 92.Goudet J., hierfstat: A package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186 (2005). [Google Scholar]
- 93.Mussmann S. M., Douglas M. R., Chafin T. K., Douglas M. E., BA3-SNPs: Contemporary migration reconfigured in BayesAss for next-generation sequence data. Meth. Ecol. Evol. 10, 1808–1813 (2019). [Google Scholar]
- 94.Meirmans P. G., Nonconvergence in Bayesian estimation of migration rates. Mol. Ecol. Resour. 14, 726–733 (2014). [DOI] [PubMed] [Google Scholar]
- 95.GEBCO Compilation Group, GEBCO 2021 Grid (2021); 10.5285/c6612cbe-50b3-0cff-e053-6c86abc09f8f. [DOI]
- 96.Loeza-Quintana T., Carr C. M., Khan T., Bhatt Y. A., Lyon S. P., Hebert P. D. N., Adamowicz S. J., Recalibrating the molecular clock for Arctic marine invertebrates based on DNA barcodes. Genome 62, 200–216 (2019). [DOI] [PubMed] [Google Scholar]
- 97.Ritchie H., Jamieson A. J., Piertney S. B., Genome size variation in deep-sea amphipods. R. Soc. Open Sci. 4, 170862 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chan J., Pan B., Geng D., Zhang Q., Zhang S., Guo J., Xu Q., Genetic diversity and population structure analysis of three deep-sea amphipod species from geographically isolated hadal trenches in the Pacific Ocean. Biochem. Genet. 58, 157–170 (2020). [DOI] [PubMed] [Google Scholar]
- 99.Blankenship L. E., Yayanos A. A., Universal primers and PCR of gut contents to study marine invertebrate diets. Mol. Ecol. 14, 891–899 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Tables S1 to S9
References
Data file S1