Abstract
Candida albicans is a major human opportunistic pathogen causing infections, which range from cutaneous to invasive systemic infections. Herein, the antifungal and anti-biofilm potential of silver nanoparticles (AgNPs) green synthesized in the presence of Encephalartos laurentianus leaf extract (ELLE) were investigated. The bioactive chemicals of ELLE, including phenolics, flavonoids, and glycosides were identified and quantified for the first time. AgNPs showed minimum inhibitory concentration (MIC) values against C. albicans clinical isolates ranging from 8 to 256 µg/mL. In addition, AgNPs significantly decreased biofilm formation. The impact of AgNPs on the expression of the genes encoding biofilm formation was assessed using qRT-PCR. AgNPs had a beneficial role in the macroscopic wound healing, and they resulted in complete epithelization without any granulation tissue or inflammation. Treatment with AgNPs resulted in negative immunostaining of tumor necrosis factor-α. The levels of the inflammation markers, interleukin-6 and interleukin-1β, significantly decreased (p < 0.05) in the AgNPs-treated group. There was also a pronounced increase in the gene expression of fibronectin and platelet-derived growth factor in the wound tissues. Thus, AgNPs synthesized using ELLE may be a promising antifungal and wound healing agent.
Keywords: antifungal activity, LC-MS/MS, qRT-PCR, SEM, TEM, TNF-α
1. Introduction
Nanotechnology has yielded various beneficial applications in biotechnology and microbiology. This is due to its ability to develop, design, and manipulate nanostructures with a high ratio of surface area to volume [1].
Nanoparticles have different sizes, morphologies, and chemical characteristics that can be used in their classification. Metal nanoparticles are inorganic nanoparticles, such as silver nanoparticles (AgNPs), with distinctive characteristics [2]. Their sizes range from 1 to 100 nm with high porosity and surface charge. AgNPs have received special attention owing due to their high antimicrobial activity, resulting in numerous applications designed to control the spread of pathogenic microorganisms [3].
There are many methods for AgNPs synthesis including physical synthesis using laser ablation or chemical synthesis involving chemical reduction with organic and inorganic reducing agents. Most of these techniques have limitations including the high cost and the use of toxic or hazardous chemicals. To overcome such drawbacks, extensive research has been done to find alternative methods that enable eco-friendly, cheap, facile, and nontoxic synthesis of AgNPs [4].
Recently, green synthesis has used eco-friendly non-toxic reagents from plants and microorganisms to reduce silver ions. The use of natural extracts may be more advantageous than microorganisms for the synthesis of nanoparticles because complex and specific processes like culture maintenance and isolation are not needed [5].
Different plant parts can be utilized for the biosynthesis of nanoparticles, such as leaves, flowers, roots, and fruits. Encephalartos are among the plants that are not well studied. They belong to the Cycadales, the second-largest extant genus. Encephalartos laurentianus is the largest cycad and has recently been found to possess cytotoxic, antioxidant, and antibacterial characteristics [6]. We used the leaf extract from this plant to synthesize AgNPs as E. laurentianus contains a variety of active compounds, including flavonoids, flavonoid glycosides, and phenolic and organic acids [7].
Various reports of the biological activities of AgNPs led us to assess their activity against Candida albicans clinical isolates. C. albicans is among the most commonly encountered fungal pathogens and causes high morbidity and mortality, particularly among immunocompromised and critical care patients [8]. While C. albicans is a commensal of the oral cavity, gastrointestinal tract, and vagina, it is an opportunist able to produce severe systemic infections. In addition, C. albicans can infect wounds, which can result in invasive candidiasis [9].
There is an urgent need to identify new antifungal agents against C. albicans as the current antifungal drugs suffer from many disadvantages including drug resistance, narrow spectrum, and significant side effects [10]. This report describes the synthesis of AgNPs using ELLE, as a capping agent, and investigates their in vitro and in vivo antifungal activities against C. albicans. In addition, LC-MS/MS was used to identify the potential bioactive components of ELLE, involved in the synthesis of AgNPs.
2. Materials and Methods
2.1. Chemicals and Preparation of Plant Extract
Encephalartos laurentinus De Wild. leaves (Family Zamiaceae) were obtained from Al-Abid Farms on 13 January 2017. E. laurentianus was identified by Dr. Esraa Ammar, Tanta University. The leaves were dried at room temperature for two weeks and then ground. The powder (400 g) was extracted using the cold maceration method with methanol (4 L × four times till complete exhaustion, two-day intervals) and then concentrated under reduced pressure to yield 39.6 g of ELLE [7].
Betadine™ ointment was purchased from Mundi pharma, Cairo, Egypt. Culture media (including Sabouraud dextrose agar (SDA), brain heart infusion (BHI), and Sabouraud dextrose broth (SDB) were purchased from Oxoid, Hampshire, UK. All other chemicals used in the study were obtained from Merck, New Jersey, USA.
2.2. Animals
Thirty white male Wistar albino rats (180–210 g, eight weeks old) were obtained from the Faculty of Veterinary Medicine (Cairo University). They were housed in a pathogen-free environment at 25 ± 2 °C with a 12-h light/dark cycle. They were provided with a standard pellet diet (ad libitum feeding) and filtered water. The rats were acclimatized for one week prior to the in vivo experiments. The authorized standards of the Faculty of Pharmacy Research Ethical Committee (Tanta University) for the use of the laboratory animals, with code number TP/RE/3-22-P-006, were used.
2.3. Liquid Chromatography Analysis
Liquid chromatography-electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS) of the samples used an Exion LC AC system for separation and SCIEX Triple Quad 5500+ MS/MS system equipped with electrospray ionization (ESI) for detection. The separation was performed using ZORBAX SB-C18 Column (4.6 × 100 mm, 1.8 µm). The mobile phases were mainly from two eluents, A: 0.1% formic acid in water and B: acetonitrile (L.C. grade). The mobile phase was programmed as follows: 2% B from 0–1 min, 2–60% B from 1–21 min, 60% B from 21–25 min, and 2% B from 25.01–28 min. The flow rate was 0.8 mL/min, and the injected volume was 3 µL. For multiple reaction monitoring (MRM) analysis of the selected polyphenols, positive and negative ionization modes were applied in the same run with the following parameters: curtain gas: 25 psi; ion spray voltage: 4500 and 4500 V for positive and negative modes, respectively; source temperature: 400 °C; ion source gas 1 and 2 were 55 psi with a declustering potential (DP): 50; collision energy: 25 eV; collision energy spread: 10 times [11].
2.4. Green Synthesis of AgNPs
Aqueous silver nitrate solution (AgNO3, 1 mM) was stored in a cold, dark place. Then, ELLE (10 mL) was added to AgNO3 solution (90 mL), and the mixture was incubated overnight in a dark environment to reduce Ag+ ions. The formation of a yellowish-brown tint suggested the production of AgNPs. The resultant solution was assessed using a transmission electron microscope (TEM) and ultraviolet-visible spectroscopy (UV-Vis). AgNPs were purified via centrifugation at 11.000× g for 30 min followed by washing three times with distilled water and filtration with filter paper [12].
2.5. Characterization of AgNPs
2.5.1. UV-Vis Spectroscopy
The production of AgNPs was confirmed using a UV/Vis spectrophotometer (SHIMADZU, Kyoto, Japan) at a wavelength of 300–900 nm.
2.5.2. Fourier-Transform Infrared (FTIR)
FTIR analysis was used to determine the functional groups of the plant metabolites involved in the reduction process of silver. The attenuated total reflectance mode of a Jasco FTIR4100 spectrometer (SHIMADZU, Kyoto, Japan) in 4000–400 cm−1 was used [13].
2.5.3. High-Resolution (HR-TEM) and Scanning Electron Microscope (SEM)
HR-TEM (JEOL JEM-2100, Tokyo, Japan) and selected area electron diffraction (SAED) were used to determine the average particle size and the morphology of ELLE-capped AgNPs. SEM (TM1000, Hitachi, Tokyo, Japan) was used to examine the morphology of AgNPs [14].
2.5.4. Zeta Potentials and Dynamic Light Scattering (DLS)
DLS analysis was carried out using zeta-sizer (Malvern, UK) to determine the entire core size of the AgNPs and their charge.
2.5.5. X-ray Diffraction (XRD)
XRD analysis was used to confirm the occurrence of the crystal nature of AgNPs and determine their structure and size. XRD was performed on AgNPs using Cu-Kα1 X-ray diffractometer radiation (θ = 1.5406 Å) operating at 45 kV and 30 mA with 2θ in the range of 4.01–79.99° [15].
2.6. Determination of Total Flavonoids and Phenolics of ELLE
Colorimetric analysis of ELLE dilution series was carried out using the aluminum chloride procedure with Rutin as a reference to assess the total flavonoid concentration [15]. The content of polyphenols was evaluated using the Folin-Ciocalteu technique with gallic acid as the reference [16].
2.7. Antioxidant Activity of ELLE
2.7.1. ABTS Radical Scavenging Capacity
The 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) assay was conducted as previously described [17]. The results are expressed as µM Trolox equivalents (TE)/mg of samples using the linear regression equation derived from the calibration curve.
2.7.2. Ferric Reducing Antioxidant Power (FRAP) Assay
FRAP assay was performed as previously reported. The results were expressed using the linear dose-response curve of Trolox [18].
2.8. Antifungal Potential (In Vitro)
2.8.1. Fungi
Thirteen C. albicans clinical isolates were collected from patients admitted to different departments of Tanta University Hospitals, Egypt. The samples were obtained from the patients for laboratory diagnosis and were not collected specifically for this study. The clinical specimens (skin swabs) were first cultured on SDA plates, then incubated overnight at 37 °C. The cultures were identified as C. albicans using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) (bioMerieux, Marcy-l’Étoile, France) with Candida albicans (MTCC 227) used as the standard isolate.
2.8.2. Antifungal Activity
The disc agar diffusion method for screening the antifungal potential of the green synthesized AgNPs was used as previously described [7,19]. In brief, sterile filter paper discs impregnated with AgNPs were added after spreading the fungal suspension on the SDA plates. Each plate included a positive control (fluconazole, 10 µg) and negative control (dimethyl sulfoxide, DMSO, 10%). The plates were then incubated at 37 °C for 24 h and inspected for the appearance of inhibition zones around the discs.
2.8.3. Estimation of Minimum Inhibitory Concentrations (MICs)
The MIC values of AgNPs against C. albicans isolates were estimated using the broth microdilution method in a 96-well microtitration plate using a two-fold serial dilution of AgNPs [20]. A positive control (untreated fungi) and a negative control (SDB only) were included in each microtitration plate. After incubation at 37 °C for 24 h, the minimum concentration of AgNPs, which caused a complete absence of the growth of C. albicans cells, was recorded as MIC [21,22].
2.8.4. Time Kill Assay
It was carried out as previously reported [23]. Overnight cultures of C. albicans isolates in BHI broth were adjusted to 106 CFU/mL. Then, AgNPs (0.5× MIC, 1× MIC, 2× MIC, 4× MIC, and 8× MIC values) were added and optical density (OD) values, at 530 nm, were determined after 4, 8, 24, 28, 32, and 48 h.
2.8.5. Biofilm Formation Assay
Overnight C. albicans cultures in SDB were adjusted to 1 × 106 CFU/mL, and 100 μL was added to the wells of the microtiter plates and incubated for 48 h at 37 °C. The wells were rinsed gently with phosphate-buffered saline (PBS) and the fungal cells that adhered to the wells were fixed using 100 μL methanol for 20 min. C. albicans cells were stained using 1% crystal violet for 20 min, the wells then gently washed with distilled water and allowed to dry. The bound dye was dissolved using glacial acetic acid (33%), and the OD values, at 540 nm, were measured using an ELISA reader (Sunrise Tecan, Zürich, Switzerland) [11]. The classification of C. albicans isolates according to their biofilm-forming ability is shown in Table S1.
2.8.6. Anti-Biofilm Activity
The anti-biofilm effect of AgNPs (at 0.5 MIC values) against C. albicans isolates moderately and strongly biofilm-forming was measured using a crystal violet assay as previously described [7]. The number of colony-forming units (CFU/mL) was used to assess the viability of the fungal cells forming biofilms. The cells forming biofilms were gently rinsed with PBS and then scraped off the wells using toothpicks after adding 200 μL PBS. The fungal suspensions were serially diluted and plated on SDA plates supplemented with chloramphenicol, and the number of CFU/mL was determined after overnight incubation at 37 °C.
2.8.7. SEM
Glass coverslips submerged with C. albicans isolates (before and after treatment with AgNPs) were overnight incubated at 37 °C to permit the fungal cells to form biofilms. The coverslips were gently rinsed with PBS, flooded with 2.5% glutaraldehyde solution, left overnight at 4 °C, and dehydrated using a series of ethanol concentrations (30% to 100%). They were allowed to air dry and coated with gold for examination using SEM (Hitachi, Tokyo, Japan).
2.8.8. Quantitative Real-Time PCR (qRT-PCR)
The expression of the biofilm encoding genes (BCR1, PLB2, ALS1, and SAP5) were evaluated, using qRT-PCR, after treatment with AgNPs [12]. After extracting the total RNA using an RNeasy mini kit (Qiagen, Hilden, Germany), cDNA was synthesized using the SensiFAST™ cDNA kit (Bioline, London, UK). The qRT-PCR was carried out using SensiFAST™ SYBR green PCR master mix (Bioline, London, UK) as described by the manufacturer. All reverse-transcription experiments included a negative control to test for contaminating genomic DNA. The negative control contained all components except for the template RNA. The used primers are presented in Table S2 using ACT1 as a housekeeping gene. The expression of the tested genes (BCR1, PLB2, ALS1, and SAP5) in C. albicans isolates before treatment with AgNPs was considered to have a value of one. Relative gene expression was calculated using the 2−ΔΔCT method [8]. Fold changes were considered to be statistically significant when there were two or more fold changes (either increased or decreased) [24].
2.9. In Vivo Antifungal Potential
2.9.1. Experimental Model
Rats were randomly grouped into three equal groups. These groups were: group I (control, 0.9% normal saline), group II (Betadine™ ointment), and group III (AgNPs solution, 5 μg/mL). After being anesthetized, a limited skin area on the backs of the rats was carefully shaved. Full thickness excisional wounds, with an initial size of 1.2 ± 0.2 cm2, were prepared and inoculated with C. albicans (C3 isolate, 106 CFU/mL). Betadine™ ointment, AgNPs, or 0.9% normal saline were applied topically on the wound’s surface daily for seven days.
2.9.2. Macroscopic Features of the Wound
From day zero (the day of wound creation), the process of wound healing was observed for seven days. Digital images of the wounds were recorded on days zero, three, and seven. The wound areas (cm2) were measured using a ruler on these days to assess healing efficacy [25]. The wound healing percentage was calculated using the following equation:
n represents day three or day seven.
2.9.3. Fungal Burden
The fungal burden was calculated on days three and seven post wounding. Excised wound tissues were homogenized in 10 mL PBS, serially diluted (10-fold) dilutions in SDB, and plated onto SDA plates, supplemented with chloramphenicol antibiotic to prevent bacterial growth. The plates were incubated at 37 °C for 24 h for quantification of CFU/mL.
2.9.4. Histopathological Examination
The entire wound, with a five-millimeter margin of the surrounding intact skin, was obtained from each rat at the end of the experiment for histological assessment [26]. The specimens were fixed with formalin solution (10%) and inserted into paraffin wax at 65 °C for block formation. The tissue blocks were sliced into thin sections, stained with hematoxylin and eosin (H&E) and Masson’s trichrome stain (for collagen staining) and inspected using a light microscope [27]. The percentage of the area of collagen fibers was assessed using image J software (National Institutes of Health, Bethesda, MD, USA).
2.9.5. Immunohistochemistry
The thin sections were dewaxed, rehydrated, and stained with tumor necrosis factor-alpha (TNF-α) antibody (ABclonal Technology, Woburn, MA, USA). The strength of staining was graded as negative (−) if there were not any positive cells, mild (+) if the positive cell percentage ranged from 1% to 10%, moderate (++) if the positive cell percentage ranged from 11% to 50%, and strong (+++) if the positive cell percentage was more than 50% [10].
2.9.6. ELISA
Interleukin-1β (IL-1β) and (IL-6) levels were assessed in the wound tissues (pg/mg tissues) using an ELISA kit (Abcam Co., Waltham, MA, USA).
2.9.7. Gene Expression of Fibronectin and Platelet-Derived Growth Factor (PDGF) Genes
qRT-PCR was carried out as previously described using the glyceraldehyde-phosphate dehydrogenase (GAPDH) gene as a reference gene. The relative expression of fibronectin and PDGF genes was determined (primers listed in Table S3) [25].
2.9.8. Statistical Analysis
All assays were performed in triplicate, and the results are presented as mean ± standard deviation (SD). The results were regarded as statistically significant at p < 0.05 using the GraphPad Prism 8 (USA) via ANOVA and a post hoc test.
3. Results
3.1. LC-ESI-MS/MS Analysis
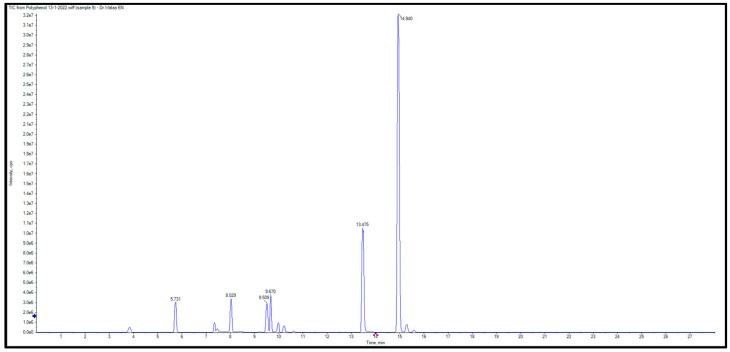
Liquid Chromatography with tandem mass spectrometry LC/MS was employed to identify and quantify phenolic chemicals in ELLE. The LC-ESI-MS/MS chromatogram for ELLE’s identified flavonoids and phenolics is shown in Figure 1. Ten phenolics were identified, which were (gallic, 3,4-dihydroxybenzoic, chlorogenic, methyl gallate, caffeic, syringic, coumaric, vanillic, ellagic, and ferulic acid). In addition, seven flavonoids were detected (rutin, luteolin, quercetin, naringenin, apigenin, kaempferol, and hesperetin). The LC-MS/MS analysis of E. laurentianus found in Table 1 explored the presence of naringenin at a concentration of 66,647 μg/g as the major flavonoid, followed by luteolin (486 μg/g). The main phenolic acid was 3,4-dihydroxybenzoic acid (767 μg/g), followed by chlorogenic acid (106 μg/g).
Figure 1.
Liquid chromatography-electrospray ionization–tandem mass spectrometry (LC-ESI-MS/MS) of ELLE.
Table 1.
Chemical composition analysis of ELLE’s compounds.
| Number | Retention Time (RT) | Compound | Conc. (μg/g) |
|---|---|---|---|
| 1 | 3.85 | Gallic acid | 61.24 |
| 2 | 5.74 | 3.4-Dihydroxybenzoic acid | 767.72 |
| 3 | 7.26 | Catechin | ND |
| 4 | 7.35 | Chlorogenic acid | 106.25 |
| 5 | 7.46 | Methyl gallate | 4.18 |
| 6 | 8.04 | Caffeic acid | 63.53 |
| 7 | 8.4 | Syringic acid | 17.10 |
| 8 | 9.52 | Coumaric acid | 37.46 |
| 9 | 9.56 | Vanillin | 59.76 |
| 10 | 9.69 | Rutin | 81.99 |
| 11 | 9.89 | Ellagic acid | 8.59 |
| 12 | 10.23 | Ferulic acid | 108.53 |
| 13 | 11.79 | Myricetin | ND |
| 14 | 12.9 | Daidzein | ND |
| 15 | 13.49 | Luteolin | 486.28 |
| 16 | 13.56 | Quercetin | 6.94 |
| 17 | 14.18 | Cinnamic acid | ND |
| 18 | 14.95 | Naringenin | 66,647.08 |
| 19 | 15.01 | Apigenin | 356.88 |
| 20 | 15.32 | Kaempferol | 4.05 |
| 21 | 15.6 | Hesperetin | 14.37 |
3.2. Characterization of the AgNPs
3.2.1. UV-Vis Spectroscopy
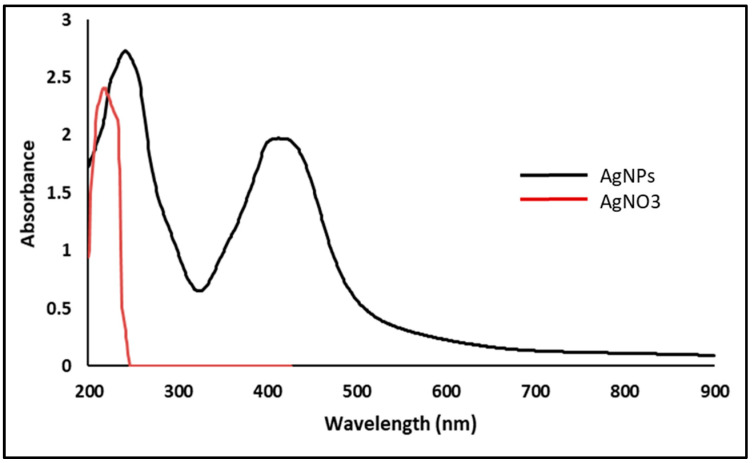
It was utilized to demonstrate AgNPs creation because of UV’s selectivity for the produced AgNPs. AgNPs interact strongly with specific wavelengths of light due to their distinctive optical reflectivity. For example, free electrons create a surface plasmon resonance (SPR) absorption band in AgNPs due to collective electron oscillation [28]. An UV absorbance peak in the ELLE-capped nanoparticles was detected at 432 nm (Figure 2).
Figure 2.
UV spectrum of AgNPs.
3.2.2. FTIR
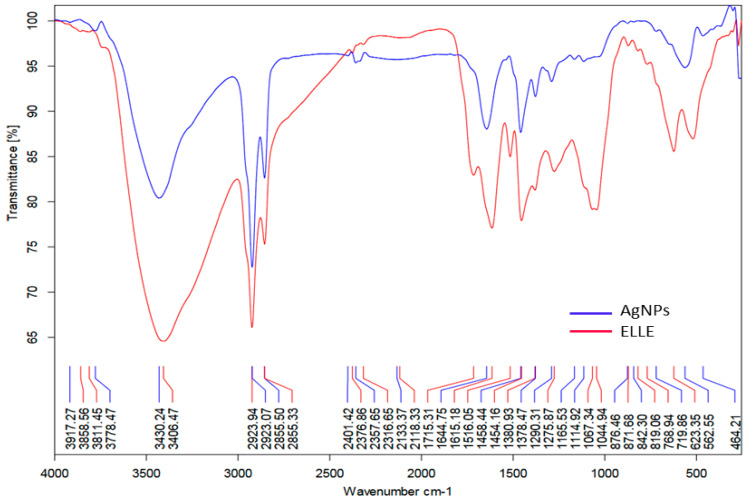
The presence of an OH group in the phenolic compounds was confirmed via FTIR data for ELLE (Figure 3), which showed multiple bands at 3430 cm−1. The medium peak shows a stretch alkane group at 2923 cm−1 (CH). CH bending vibrations and C-C or C=O stretch in the phenolic components caused the bands in the ELLE spectra in 1713, 1624, 1516, and 1378 cm−1. Another intense band at 842 cm−1 was characteristic at the aromatic ring. Owing to the phenolic and amino groups attachment, these bands’ frequencies reduced in strength attachment via AgNPs compared with ELLE [29].
Figure 3.
FTIR spectrum of AgNPs and ELLE.
ELLE contains flavonoids, glycosides, phenolics, and organic acids components that might cap the AgNPs. From the phytochemical results of LC-ESI-MS/MS, we suggest that -OH (hydroxyl), -C=O (Carbonyl) of ELLE also had a role in AgNPs production.
3.2.3. TEM and SEM
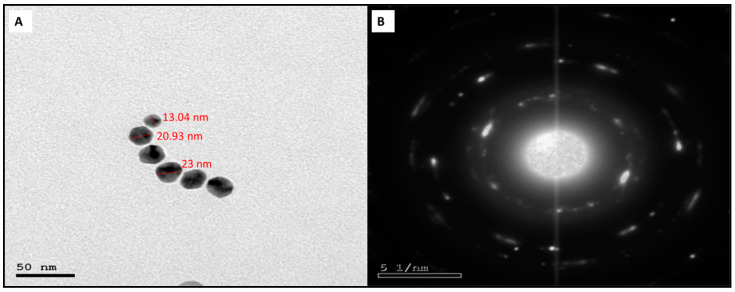
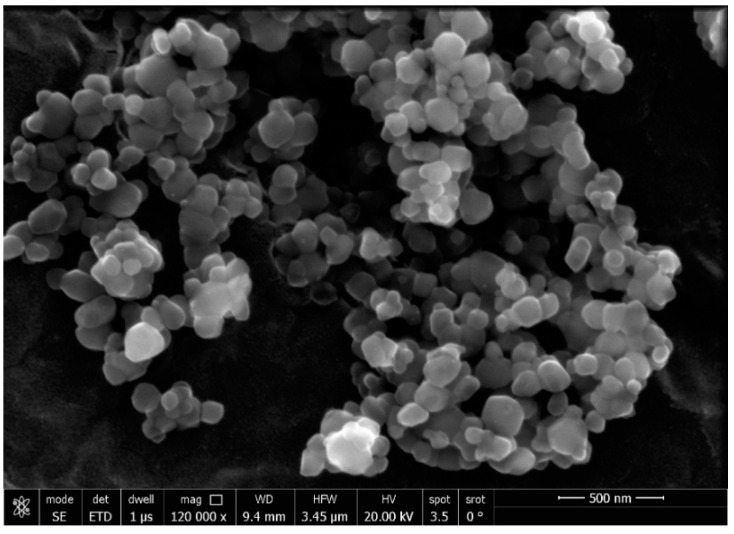
The production of AgNPs with a spherical shape and a particle size range of 13.04–23.0 nm, with an average size of 18 ± 5 nm, was observed employing HR-TEM on AgNPs (Figure 4). The crystallinity of the AgNPs was confirmed by the sharp ring diffraction pattern detected with SAED. SEM analysis showed the AgNPs had a spherical shape and a tendency to aggregate (Figure 5).
Figure 4.
Electron micrographs of the green synthesized AgNPs capped with ELLE. (A) TEM and (B) SAED.
Figure 5.
SEM of AgNPs capped with ELLE.
3.2.4. Zeta Potential and DLS
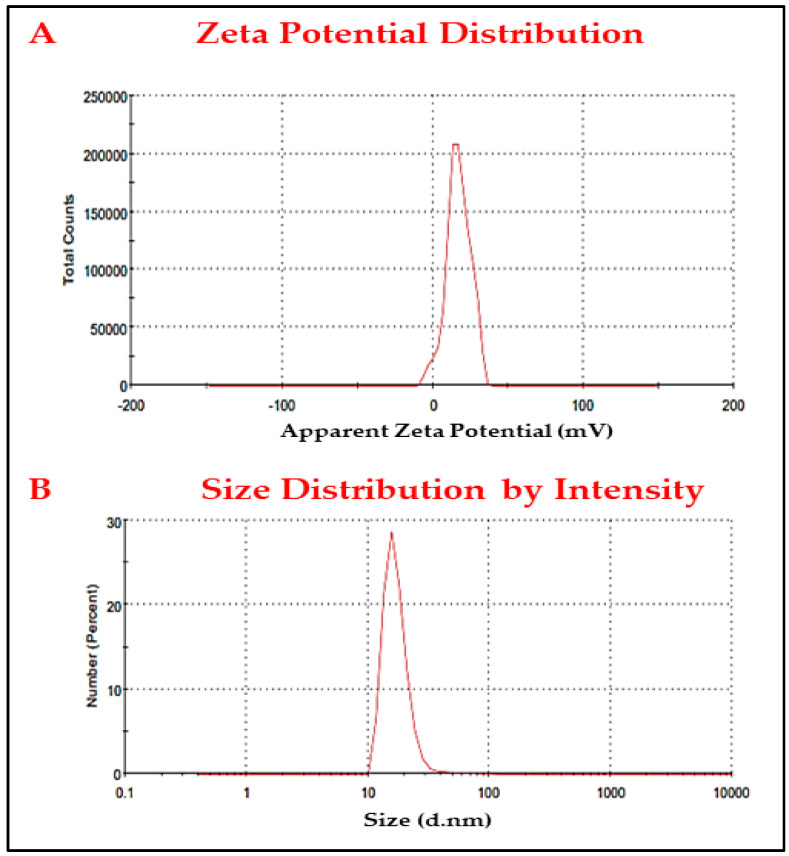
The surface charge of the synthesized AgNPs was determined using the ζ-potential approach. AgNPs have a mean ζ-potential of −7.04 ± 0.7 mV, with the negative charge highlighting the nanoparticles’ stability (Figure 6A). The DLS technique was used to measure the AgNPs size distribution, which included the metallic shell of the nanoparticles. AgNPs had a mean size of 78.4 ± 3.8 nm and a PDI value of 0.406 (Figure 6B).
Figure 6.
(A) ζ−potential analysis and (B) DLS of the synthesized AgNPs using ELLE.
3.2.5. X-ray Diffraction
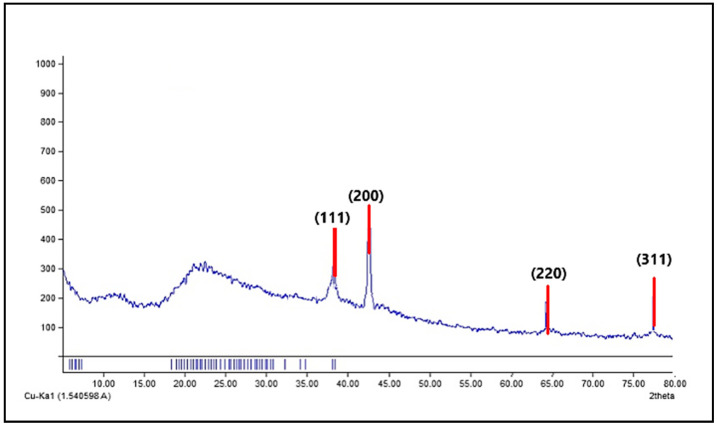
The intense diffraction peaks were observed at the 2θ values of 38.44, 44.47, 64.35, and 77.94, corresponding to (111), (200), (220), and (311) planes for AgNPs, respectively (Figure 7). Remarkably, there are no other peaks, which implies that the AgNPs were pure.
Figure 7.
X-ray diffraction (XRD) pattern of the green synthesized AgNPs by ELLE.
3.3. Flavonoids and Phenolics’ Total Content
The total flavonoids content of ELLE was 17.38 ± 0.95 µg rutin equivalent/mg sample, and the total polyphenols content was 95.89 ± 2.42 µg gallic acid/mg sample. These findings indicate that ELLE has high contents of phenols and flavonoids.
3.4. Antioxidant Activity of ELLE
The radical scavenging and metal-reducing assays were used to explore ELLE’s antioxidant effects. According to ABTS, ELLE has an antioxidant activity of 530.73 µM Trolox equivalents (TE)/mg. The FRAP test was utilized as a metal-reduction assay, and the ELLE activity was 189.87 µM TE/mg.
3.5. In Vitro Antifungal Potential
AgNPs showed antifungal potential against C. albicans isolates with formation of inhibition zones like fluconazole. The MIC values of AgNPs against the tested C. albicans isolates ranged from 8 to 256 µg/mL (Table S4).
3.5.1. Time Kill Curve
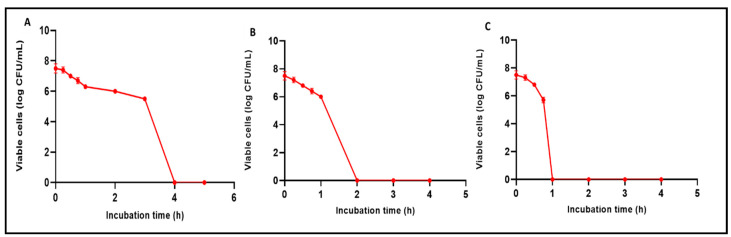
The CFU/mL of C. albicans isolates decreased by at least three log units after incubation with AgNPs at concentrations of 2× MIC for four hours, 4× MIC for two hours, and 8× MIC for one hour for 69.23%, 53.85%, and 30.77% of C. albicans isolates, respectively. An illustrative example is presented in Figure 8.
Figure 8.
Charts representing the decrease in the number of CFU/mL of C. albicans isolates after incubation with AgNPs: (A) with concentrations of 2× MIC for four hours, (B) 4× MIC for two hours, and (C) 8× MIC for one hour.
3.5.2. Antibiofilm Activity
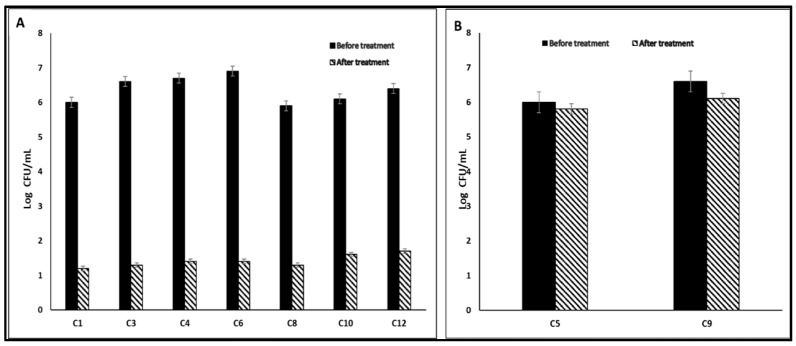
The impact of AgNPs on the biofilms of C. albicans isolates was assessed via a crystal violet test. The AgNPs reduced the percentage of C. albicans isolates, which form biofilm strongly and moderately (C1, C3, C4, C5, C6, C8, C9, C10, and C12), from 69.23% to 30.77% (Table 2). Furthermore, AgNPs decreased significantly (p < 0.05) the number of CFU/mL in 53.85% of C. albicans isolates (Figure 9).
Table 2.
Impact of AgNPs on biofilm formation by C. albicans isolates.
| Biofilm Formation | No. of Isolates | |
|---|---|---|
| Before Treatment | After Treatment | |
| No formation | 1 | 3 |
| Weak | 3 | 6 |
| Moderate | 5 | 3 |
| Strong | 4 | 1 |
Figure 9.
Bar chart showing (A) a significant decrease (p < 0.05) and (B) non-significant change (p > 0.05) in the number of CFU/mL after AgNPs treatment.
3.5.3. SEM
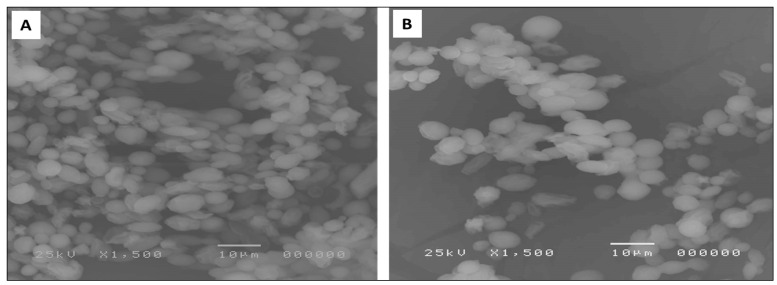
SEM was used to study the effect of the biosynthesized nanoparticles on the biofilm morphology. A remarkable decrease in the biofilms formed by C. albicans isolates was found after treatment with AgNPs in five isolates (38.46% of isolates). An illustration of the visible inhibition of the biofilm formation is shown in Figure 10.
Figure 10.
SEM micrograph showing biofilm formation by C. albicans (C6) isolate (A) before and (B) after AgNPs treatment.
3.5.4. qRT-PCR
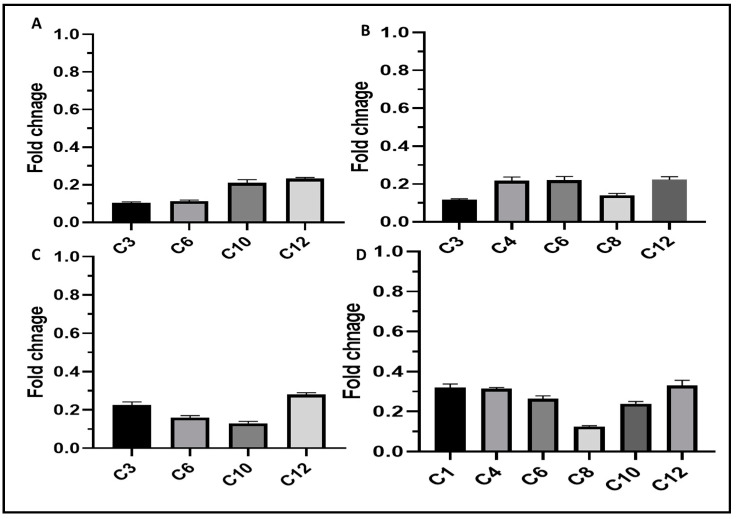
Relative gene expression of BCR1, PLB2, ALS1, and SAP5 biofilm genes was determined via qRT-PCR in C. albicans isolates to assess the influence of AgNPs on biofilm formation. As shown in Figure 11, a significant downregulation of the expression of BCR1, PLB2, ALS1, and SAP5 was detected in 30.77%, 38.46%, 30.77%, and 46.15% of the isolates, respectively.
Figure 11.
Bar charts showing the downregulation caused by AgNPs in the expression of (A) BCR1, (B) PLB2, (C) ALS1, and (D) SAP5 genes in C. albicans isolates.
3.6. In Vivo Antifungal Potential
3.6.1. Macroscopic Wound Healing and Fungal Cell Viability
The Betadine™- and AgNPs-treated groups exhibited an entire and prominent wound healing process relative to the control group (Figure S1).
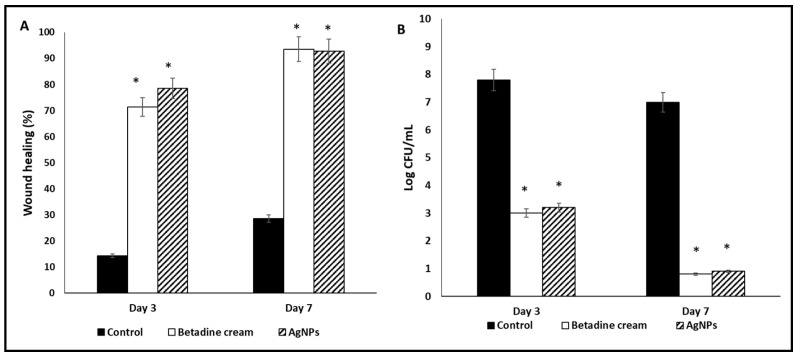
The rats of Betadine™- and AgNPs-treated groups showed remarkable wound healing on day three (percentages of wound healing were 71.43% and 78.57%, respectively) in comparison with the control group. The two groups revealed complete wound healing on day seven, with percentages of wound healing of 93.57% and 92.86%, respectively (Figure 12A). Moreover, the number of viable C. albicans cells (CFU/mL) were several orders of magnitude lower in the Betadine™- and AgNPs-treated groups compared to the control group (Figure 12B).
Figure 12.
Characters of the wounds on days 3 and 7: (A) wound healing percentage and (B) log CFU/mL. The symbol (*) represents a significant change (p < 0.05) in comparison with the control group.
3.6.2. Histopathology
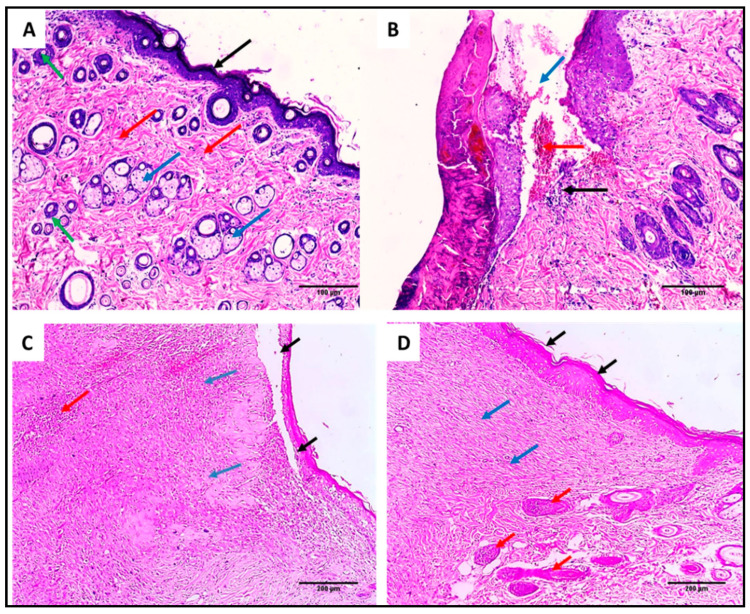
Normal skin sections revealed normal epidermal thickness, with the underlying dermis showing thick collagen bands, hair follicles, and sebaceous glands (Figure 13A). In contrast, the wound sections of the control group revealed the presence of ulcers filled with hemorrhage accompanied by acute and chronic inflammatory cellular infiltrate (Figure 13B). The wound section of the Betadine™ group exhibited a complete epithelization with underlying granulation tissue without any inflammatory cellular infiltrate (Figure 13C). Wound sections of the AgNPs-treated group also showed a complete epithelization with underlying dermis containing thick collagen bundles and proliferating hair follicles without any granulation tissue or inflammation (Figure 13D).
Figure 13.
H&E-stained wound sections of (A) section in normal skin showed normal epidermal thickness (black arrow), with underlying dermis showing thick collagen bands (red arrows), hair follicles (green arrows), and sebaceous glands (blue arrows) (×200). (B) Section in the wound of the control group showed an ulcer (blue arrow) filled with hemorrhage (red arrow) and acute and chronic inflammatory cellular infiltrate (black arrow) (×200). (C) Section in the Betadine™-treated group showed partial epithelization (black arrow) with underlying granulation tissue (blue arrows) with little inflammatory cellular infiltrate (red arrow) (×200). (D) Section in the AgNPs-treated group showed complete epithelization (black arrows) with underlying focal granulation tissue formation (blue arrows) with adjacent proliferating hair follicles (red arrows) without inflammation (×200).
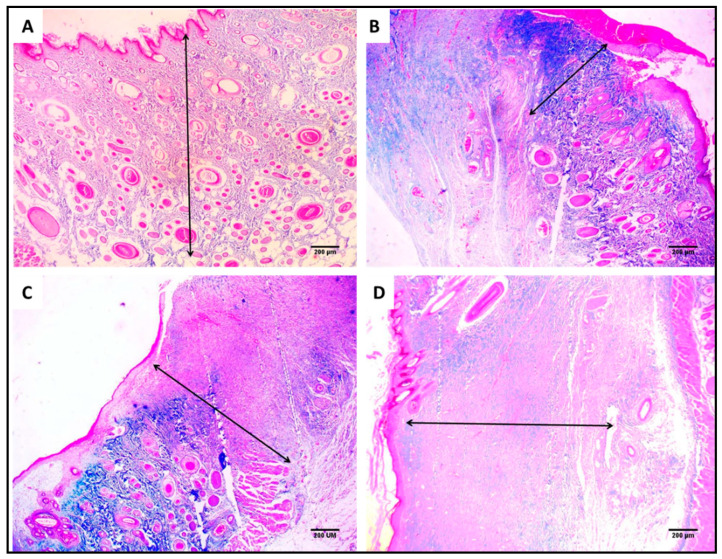
Light microscopy of Masson’s trichrome-stained wound sections was used to inspect the collagen fibers (Figure 14). The percentages of the collagen fibers in the Betadine™- and AgNPs-treated groups were significantly higher than the control group.
Figure 14.
Masson’s trichrome staining of the wound showing the area of collagen fibers (%) using image J software of (A) section in normal skin (negative control) showing 28.2% area of collagen fiber (×40). (B) Section in the wound of the control group showing 8.2% area of collagen fiber (×40). (C) Section in the Betadine™-treated group showing 14.7% area of collagen fiber (×40). (D) Section in the AgNPs-treated group showing 22.4% area of collagen fiber (×40).
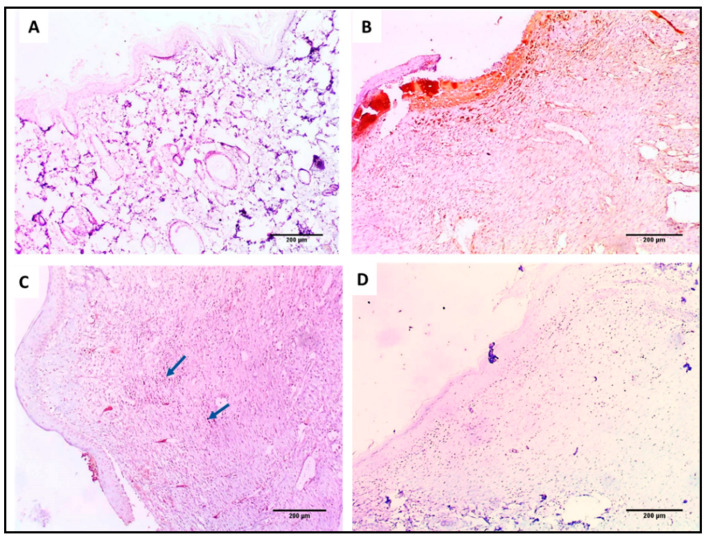
3.6.3. Immunohistochemistry
Light microscopy of the TNF-α-immunostained skin sections is shown in Figure 15. The AgNPs-treated group showed negative TNF-α immunostaining.
Figure 15.
TNF-α immunohistochemical staining of (A) section in normal skin (negative control) showing negative TNF-α immunostaining (−) as no wound healing area (×100). (B) Section in the wound of the control group showing strong positive immunostaining (+++) in the wound healing area (×100). (C) Section in the Betadine™-treated group showing mild positive TNF-α immunostaining (+) in the wound healing area (×100). (D) Section in the AgNPs-treated group showing negative TNF-α immunostaining (−) in the wound healing area (×100).
3.6.4. ELISA
The pro-inflammatory cytokines IL-6 and IL-1β significantly decreased (p < 0.05) in the Betadine™- and AgNPs-treated groups relative to the control (Table 3).
Table 3.
The levels of IL-6 and IL-1β of the wound tissues of the different groups.
| Parameters | Control Group | Betadine Group | AgNPs Group |
|---|---|---|---|
| IL-6 (pg/mg tissues) |
395.6 ± 4.2 | 72.8 ± 4.2 | 70.5 ± 5.5 |
| IL-1β (pg/mg tissues) |
471.3 ± 5.4 | 132.7 ± 6.7 | 148.8 ± 5.4 |
3.6.5. qRT-PCR
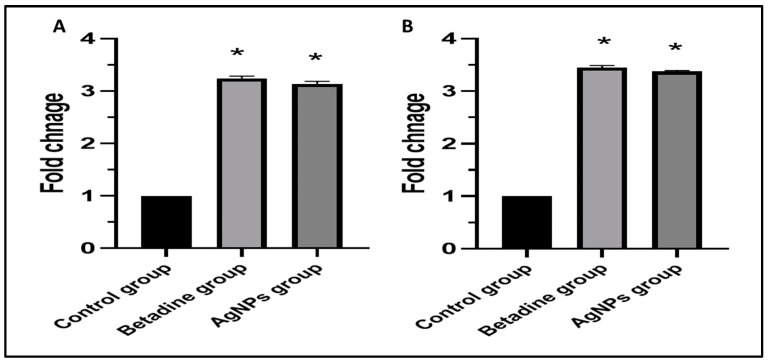
Both groups treated with Betadine™ and AgNPs demonstrated a pronounced increase (p < 0.05) in fibronectin and PDGF gene expression compared to the control group, as shown in Figure 16.
Figure 16.
Bar charts represent the fold change in the gene expression of (A) fibronectin and (B) PDGF genes. The symbol (*) denotes a significant increase (p < 0.05) relative to the control group.
4. Discussion
C. albicans pathogenicity is primarily attributed to numerous virulence factors and their escalating resistance to the current antifungals [30]. Thus, the discovery of novel antifungals with activity against both planktonic and biofilm-forming cells is needed to overcome the problematic infections caused by this pathogen. Herein, we synthesized AgNPs from ELLE and investigated their antifungal activity against C. albicans.
Using 21 reference chemicals, LC-ESI/MS assessed and quantified the flavonoids and phenolic acids in ELLE. The bioactive chemicals in distinct Gymnosperm species have been documented in a few publications [6,31,32]. The LC-MS/MS analysis detected phenolic compounds claimed to have antimicrobial, antioxidant, and anticancer properties via several pathways [33].
Based on our observations of the strong antioxidant potential of ELLE, it not only scavenges free radicals but also breaks them down and reduces ferric ions. E. laurentianus have phenolic and flavonoid compounds that possess the ability to form and stabilize AgNPs. These may also be responsible for ELLE-dependent properties.
AgNPs demonstrated antifungal activity via an agar diffusion assay against C. albicans isolates and broth microdilution assay with a range of MICs of 8–256 µg/mL. However, other researchers have assessed the antifungal activity of AgNPs. Xia et al. [34] reported that AgNPs have antifungal activity against Trichosporon asahii. Jalal et al. [35] noted the antifungal potential of AgNPs against C. albicans, C. krusei, C. tropicalis, and C. parapsilosis. Herein, the AgNPs produced using ELLE have been shown, for the first time, to have antifungal activity both in vitro and in vivo. Time-kill curves provide a relationship between kill rate and MIC values [36,37]. Increased concentrations of AgNPs decrease the time required for killing fungi.
The formation of biofilm is an important virulence factor of C. albicans which helps in its colonization in human tissues. In addition, it provides the fungal cells with a protective environment in tissues that increases its survival rate compared with planktonic cells [36,37]. The AgNPs reduced the percentage of C. albicans isolates, which are capable of forming biofilm strongly and moderately, from 69.23% to 30.77%. SEM analysis showed that the morphology of some C. albicans isolates was altered due to treatment with AgNPs. A limited number of studies reported the antibiofilm potential of AgNPs on the C. albicans biofilms. Some attempt has been made to explore the mechanism of the antibiofilm action. Różalska et al. [38] proposed that the antibiofilm action was due to the ability of AgNPs to bind and penetrate the biofilm structure, which leads to disrupting the cell membranes. The same study also suggested that AgNPs inhibit biofilm formation by disrupting yeast morphogenesis [38,39]. Several studies have assessed the antibiofilm activity of plant extracts. One reported that biofilm formation was inhibited in Candida spp clinical isolates by 65% [8].
We tried to reveal the possible mechanisms of the antibiofilm action of AgNPs. The use of qRT-PCR to study the impact on the expression of the genes encoding biofilm formation showed that AgNPs exposure led to the downregulation of the biofilm genes to different extents in the tested isolates. Many genes in C. albicans are involved in biofilm development. Some of these genes encode transcription factors that function indirectly to control biofilm formation, such as BCR1 [40,41]. Others are related to adhesion, which is the first step in biofilm formation, such as surface adhesins ALS1 [42]. Furthermore, the genes encoding phospholipases, such as PLB2, and aspartyl proteases, such as SAP5, have an important role in fungal colonization [43]. The mode of action of AgNPs against C. albicans isolates was elucidated in previous research. It was reported that AgNPs affect the membrane of the fungal cells and possibly the integrity of the membrane due to increasing the cell permeability and outflow of the cellular proteins, DNA, and RNA [44]. Other researchers reported that the antibiofilm activity is mainly attributed to the inhibition of the adherence of the planktonic cells of fungi to different surfaces [45]. Future studies are needed to elucidate the exact mode of action of AgNPs.
Animal models can allow efficient assessment of the progression of fungal pathogenesis and host immune responses. Moreover, they can be used to investigate the antifungal properties of certain drug candidates [46]. AgNPs exhibited a promising wound healing potential for the infected rat wounds, as they induced complete epithelization and decreased the TNF-α immunostaining. TNF-α usually increases in non-healing wounds due to oxidative stress and inflammation [47]. Furthermore, the synthesis of collagen fibers was significantly higher in the case of treatment with AgNPs. This is an important finding as the wound healing can be improved via deposition of new collagen into the wound tissues [48]. In addition, it significantly reduced (p < 0.05) the levels of the pro-inflammatory cytokines, which cause inflammation and retardation of the wound healing process [49]. In addition, there was a considerable upregulation (p < 0.05) of the fibronectin and PDGF genes. Fibronectin and PDGF are essential in improving wound repair and inducing tissue regeneration [50].
5. Conclusions
ELLE has a significant content of phenols and flavonoids including compounds such as 3,4-dihydroxybenzoic acid, chlorogenic acid, naringenin, and luteolin. These bioactive constituents are likely to be responsible for forming and stabilizing AgNPs. The antifungal potential of AgNPs green synthesized from ELLE was elucidated. They exhibited in vitro antifungal activity with MIC values of 8 to 256 µg/mL for C. albicans clinical isolates.
Significant antibiofilm activity was revealed using crystal violet and SEM. Furthermore, AgNPs substantially down-regulated mRNAs important for biofilm formation in some clinical isolates. Furthermore, AgNPs exhibited a wound healing potential as they induced a complete epithelization, significantly decreased IL-6 and IL-1β levels, and markedly upregulated the genes encoding fibronectin and PDGF. This study opens opportunities to explore the value of AgNPs in the treatment of infections caused by pathogenic fungi like C. albicans. Further experimental and clinical studies can now be performed to elucidate the potential of AgNPs as a practical antifungal agent.
Acknowledgments
The authors would like to thank Princess Nourah bint Abdulrahman University Researchers Supporting Project (number PNURSP2022R322), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/jof8101005/s1: Table S1. Categories of biofilm-forming C. albicans isolates. Table S2. Sequences of primer used in qRT-PCR in vitro. Table S3. Sequences of primers utilized in qRT-PCR in vivo. Table S4. MIC values. Figure S1. The wound healing process of the different groups on days 0, 3, and 7.
Author Contributions
Conceptualization, W.A.N. and E.E.; Methodology, W.A.N., E.E., M.J.A. and I.A.H.; formal analysis, F.A.A., W.A.N., E.E., T.A.E.-M., M.J.A. and I.A.H.; investigation, F.A.A., W.A.N., E.E., T.A.E.-M., E.M.H., M.J.A. and I.A.H.; funding, F.A.A. and T.A.E.-M.; resources, F.A.A., E.M.H. and T.A.E.-M.; writing—original draft preparation, W.A.N., E.M.H. and E.E.; writing—review and editing, F.A.A., W.A.N. and E.E.; supervision, T.A.E.-M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This experiment was carried out in accordance with Research Ethical Committee (Tanta University) approval to use laboratory animals with the code number (TP/RE/3-22-P-006).
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by Princess Nourah bint Abdulrahman University Researchers Supporting Project (number PNURSP2022R322), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jeevanandam J., Barhoum A., Chan Y.S., Dufresne A., Danquah M.K. Review on nanoparticles and nanostructured materials: History, sources, toxicity and regulations. Beilstein J. Nanotechnol. 2018;9:1050–1074. doi: 10.3762/bjnano.9.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X.-F., Liu Z.-G., Shen W., Gurunathan S. Silver Nanoparticles: Synthesis, Characterization, Properties, Applications, and Therapeutic Approaches. Int. J. Mol. Sci. 2016;17:1534. doi: 10.3390/ijms17091534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qing Y., Cheng L., Li R., Liu G., Zhang Y., Tang X., Wang J., Liu H., Qin Y. Potential antibacterial mechanism of silver nanoparticles and the optimization of orthopedic implants by advanced modification technologies. Int. J. Nanomed. 2018;13:3311–3327. doi: 10.2147/IJN.S165125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jeevanandam J., Krishnan S., Hii Y.S., Pan S., Chan Y.S., Acquah C., Danquah M.K., Rodrigues J. Synthesis approach-dependent antiviral properties of silver nanoparticles and nanocomposites. J. Nanostructure Chem. 2022;12:1–23. doi: 10.1007/s40097-021-00465-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jain A.S., Pawar P.S., Sarkar A., Junnuthula V., Dyawanapelly S. Bionanofactories for Green Synthesis of Silver Nanoparticles: Toward Antimicrobial Applications. Int. J. Mol. Sci. 2021;22:11993. doi: 10.3390/ijms222111993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Negm W.A., Abo El-Seoud K.A., Kabbash A., Kassab A.A., El-Aasr M. Hepatoprotective, cytotoxic, antimicrobial and antioxidant activities of Dioon spinulosum leaves Dyer Ex Eichler and its isolated secondary metabolites. Nat. Prod. Res. 2021;35:5166–5176. doi: 10.1080/14786419.2020.1789636. [DOI] [PubMed] [Google Scholar]
- 7.Negm W.A., El-Aasr M., Attia G., Alqahtani M.J., Yassien R.I., Abo Kamer A., Elekhnawy E. Promising Antifungal Activity of Encephalartos laurentianus de Wild against Candida albicans Clinical Isolates: In Vitro and In Vivo Effects on Renal Cortex of Adult Albino Rats. J. Fungi. 2022;8:426. doi: 10.3390/jof8050426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 9.Santos G.C.D.O., Vasconcelos C.C., Lopes A.J.O., Cartágenes M.D.S.D.S., Filho A.K.D.B., Nascimento F.R.F.D., Ramos R., Pires E.R.R.B., Andrade M., Rocha F.M.G., et al. Candida Infections and Therapeutic Strategies: Mechanisms of Action for Traditional and Alternative Agents. Front. Microbiol. 2018;9:1351. doi: 10.3389/fmicb.2018.01351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang D., Liu Y., Jiang F., Ji F., Wang H., Han X. Synergistic antibacterial activity of designed Trp-containing antibacterial peptides in combination with antibiotics against multidrug-resistant Staphylococcus epidermidis. Front. Microbiol. 2019;10:2719. doi: 10.3389/fmicb.2019.02719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elekhnawy E., Negm W.A., El-Aasr M., Kamer A.A., Alqarni M., Batiha G.E.-S., Obaidullah A.J., Fawzy H.M. Histological assessment, anti-quorum sensing, and anti-biofilm activities of Dioon spinulosum extract: In vitro and in vivo approach. Sci. Rep. 2022;12:1–15. doi: 10.1038/s41598-021-03953-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Barros P.P., Rossoni R.D., Garcia M.T., Kaminski V.D.L., Loures F.V., Fuchs B.B., Mylonakis E., Junqueira J.C. The Anti-Biofilm Efficacy of Caffeic Acid Phenethyl Ester (CAPE) In Vitro and a Murine Model of Oral Candidiasis. Front. Cell. Infect. Microbiol. 2021;11:700305. doi: 10.3389/fcimb.2021.700305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Attallah N.G., Elekhnawy E., Negm W.A., Hussein I.A., Mokhtar F.A., Al-Fakhrany O.M. In Vivo and In Vitro Antimicrobial Activity of Biogenic Silver Nanoparticles against Staphylococcus aureus Clinical Isolates. Pharmaceuticals. 2022;15:194. doi: 10.3390/ph15020194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Attallah N.G., Al-Fakhrany O.M., Elekhnawy E., Hussein I.A., Shaldam M.A., Altwaijry N., Alqahtani M.J., Negm W.A. Anti-Biofilm and Antibacterial Activities of Cycas media R. Br Secondary Metabolites: In Silico, In Vitro, and In Vivo Approaches. Antibiotics. 2022;11:993. doi: 10.3390/antibiotics11080993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attallah N.G.M., El-Kadem A.H., Negm W.A., Elekhnawy E., El-Masry T.A., Elmongy E.I., Altwaijry N., Alanazi A.S., Al-Hamoud G.A., Ragab A.E. Promising Antiviral Activity of Agrimonia pilosa Phytochemicals against Severe Acute Respiratory Syndrome Coronavirus 2 Supported with In Vivo Mice Study. Pharmaceuticals. 2021;14:1313. doi: 10.3390/ph14121313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Attard E. A rapid microtitre plate Folin-Ciocalteu method for the assessment of polyphenols. Open Life Sci. 2013;8:48–53. doi: 10.2478/s11535-012-0107-3. [DOI] [Google Scholar]
- 17.Arnao M.B., Cano A., Acosta M. The hydrophilic and lipophilic contribution to total antioxidant activity. Food Chem. 2001;73:239–244. doi: 10.1016/S0308-8146(00)00324-1. [DOI] [Google Scholar]
- 18.Nallusamy S., Mannu J., Ravikumar C., Angamuthu K., Nathan B., Nachimuthu K., Ramasamy G., Muthurajan R., Subbarayalu M., Neelakandan K. Exploring phytochemicals of traditional medicinal plants exhibiting inhibitory activity against main protease, Spike glycoprotein, RNA-dependent RNA polymerase and non-structural proteins of SARS-CoV-2 through virtual screening. Front. Pharmacol. 2021;12:1704. doi: 10.3389/fphar.2021.667704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alqahtani M.J., Elekhnawy E., Negm W.A., Mahgoub S., Hussein I.A. Encephalartos villosus Lem. Displays a Strong In Vivo and In Vitro Antifungal Potential against Candida glabrata Clinical Isolates. J. Fungi. 2022;8:521. doi: 10.3390/jof8050521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pa W. 30th ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2020. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. [Google Scholar]
- 21.Elekhnawy E., Sonbol F., Abdelaziz A., Elbanna T. An investigation of the impact of triclosan adaptation on Proteus mirabilis clinical isolates from an Egyptian university hospital. Braz. J. Microbiol. 2021;52:927–937. doi: 10.1007/s42770-021-00485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alotaibi B., Negm W.A., Elekhnawy E., El-Masry T.A., Elseady W.S., Saleh A., Alotaibi K.N., El-Sherbeni S.A. Antibacterial, Immunomodulatory, and Lung Protective Effects of Boswelliadalzielii Oleoresin Ethanol Extract in Pulmonary Diseases: In Vitro and In Vivo Studies. Antibiotics. 2021;10:1444. doi: 10.3390/antibiotics10121444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemos A.S.O., Florêncio J.R., Pinto N.C.C., Campos L.M., Silva T.P., Grazul R.M., Pinto P.F., Tavares G.D., Scio E., Apolônio A.C.M., et al. Antifungal Activity of the Natural Coumarin Scopoletin Against Planktonic Cells and Biofilms From a Multidrug-Resistant Candida tropicalis Strain. Front. Microbiol. 2020;11:1525. doi: 10.3389/fmicb.2020.01525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesti R., Kontra L., Paul K., Vass I., Csorba T., Havelda Z., Várallyay É. Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLoS ONE. 2019;14:e0216618. doi: 10.1371/journal.pone.0216618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alotaibi B., Negm W.A., Elekhnawy E., El-Masry T.A., Elharty M.E., Saleh A., Abdelkader D.H., Mokhtar F.A. Antibacterial activity of nano zinc oxide green-synthesised from Gardenia thailandica triveng. Leaves against Pseudomonas aeruginosa clinical isolates: In vitro and in vivo study. Artif. Cells Nanomed. Biotechnol. 2022;50:96–106. doi: 10.1080/21691401.2022.2056191. [DOI] [PubMed] [Google Scholar]
- 26.Attallah NG E.-S.S., El-Kadem A.H., Elekhnawy E., El-Masry T.A., Elmongy E.I., Altwaijry N., Negm W.A. Elucidation of the Metabolite Profile of Yucca gigantea and Assessment of its Cytotoxic, Antimicrobial, and Anti-Inflammatory Activities. Molecules. 2022;16:1329. doi: 10.3390/molecules27041329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alotaibi B., Mokhtar F.A., A El-Masry T., Elekhnawy E., A Mostafa S., Abdelkader D.H., E Elharty M., Saleh A., A Negm W. Antimicrobial Activity of Brassica rapa L. Flowers Extract on Gastrointestinal Tract Infections and Antiulcer Potential Against Indomethacin-Induced Gastric Ulcer in Rats Supported by Metabolomics Profiling. J. Inflamm. Res. 2021;14:7411–74301. doi: 10.2147/JIR.S345780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ali A., Said D., Khayyat M., Boustimi M., Seoudi R. Improving the efficiency of the organic solar cell (CuPc/C60) via PEDOT: PSS as a photoconductor layer doped by silver nanoparticles. Results Phys. 2020;16:102819. doi: 10.1016/j.rinp.2019.102819. [DOI] [Google Scholar]
- 29.Bala N., Saha S., Chakraborty M., Maiti M., Das S., Basu R., Nandy P. Green synthesis of zinc oxide nanoparticles using Hibiscus subdariffa leaf extract: Effect of temperature on synthesis, anti-bacterial activity and anti-diabetic activity. RSC Adv. 2015;5:4993–5003. doi: 10.1039/C4RA12784F. [DOI] [Google Scholar]
- 30.Talapko J., Juzbašić M., Matijević T., Pustijanac E., Bekić S., Kotris I., Škrlec I. Candida albicans—The Virulence Factors and Clinical Manifestations of Infection. J. Fungi. 2021;7:79. doi: 10.3390/jof7020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negm W.A., Ibrahim A.E.-R.S., El-Seoud K.A., Attia G.I., Ragab A.E. A new cytotoxic and antioxidant Amentoflavone Monoglucoside from Cycas revoluta Thunb growing in Egypt. J. Pharm. Sci. Res. 2016;8:343. [Google Scholar]
- 32.Negm W.A., El-Aasr M., Kamer A.A., Elekhnawy E. Investigation of the Antibacterial Activity and Efflux Pump Inhibitory Effect of Cycas thouarsii R. Br. Extract against Klebsiella pneumoniae Clinical Isolates. Pharmaceuticals. 2021;14:756. doi: 10.3390/ph14080756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuchinda P., Saiai A., Pohmakotr M., Yoosook C., Kasisit J., Napaswat C., Santisuk T., Reutrakul V. Anti-HIV-1 cycloartanes from leaves and twigs of Gardenia thailandica. Planta Med. 2004;70:366–370. doi: 10.1055/s-2004-818951. [DOI] [PubMed] [Google Scholar]
- 34.Xia Z.-K., Ma Q.-H., Li S.-Y., Zhang D.-Q., Cong L., Tian Y.-L., Yang R.-Y. The antifungal effect of silver nanoparticles on Trichosporon asahii. J. Microbiol. Immunol. Infect. 2014;49:182–188. doi: 10.1016/j.jmii.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Jalal M., Ansari M.A., Alzohairy M.A., Ali S.G., Khan H.M., Almatroudi A., Siddiqui M.I. Anticandidal activity of biosynthesized silver nanoparticles: Effect on growth, cell morphology, and key virulence attributes of Candida species. Int. J. Nanomed. 2019;14:4667–4679. doi: 10.2147/IJN.S210449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendoza E.J., Manguiat K., Wood H., Drebot M. Two detailed plaque assay protocols for the quantification of infectious SARS-CoV-2. Curr. Protoc. Microbiol. 2020;57:cpmc105. doi: 10.1002/cpmc.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Velusamy P., Das J., Pachaiappan R., Vaseeharan B., Pandian K. Greener approach for synthesis of antibacterial silver nanoparticles using aqueous solution of neem gum (Azadirachta indica L.) Ind. Crops Prod. 2015;66:103–109. doi: 10.1016/j.indcrop.2014.12.042. [DOI] [Google Scholar]
- 38.Różalska B., Sadowska B., Budzyńska A., Bernat P., Różalska S. Biogenic nanosilver synthesized in Metarhizium robertsii waste mycelium extract—As a modulator of Candida albicans morphogenesis, membrane lipidome and biofilm. PLoS ONE. 2018;13:e0194254. doi: 10.1371/journal.pone.0194254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das J., Das M.P., Velusamy P. Sesbania grandiflora leaf extract mediated green synthesis of antibacterial silver nanoparticles against selected human pathogens. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013;104:265–270. doi: 10.1016/j.saa.2012.11.075. [DOI] [PubMed] [Google Scholar]
- 40.Almukainzi M., El-Masry T.A., Negm W.A., Elekhnawy E., Saleh A., Sayed A.E., Ahmed H.M., Abdelkader D.H. Co-delivery of Gentiopicroside and Thymoquinone Using Electrospun m-PEG/PVP Nanofibers: In-Vitro and In Vivo Studies for Antibacterial Wound Dressing in Diabetic Rats. Int. J. Pharm. 2022;625:122106. doi: 10.1016/j.ijpharm.2022.122106. [DOI] [PubMed] [Google Scholar]
- 41.Das J., Velusamy P. Antibacterial effects of biosynthesized silver nanoparticles using aqueous leaf extract of Rosmarinus officinalis L. Mater. Res. Bull. 2013;48:4531–4537. doi: 10.1016/j.materresbull.2013.07.049. [DOI] [Google Scholar]
- 42.Finkel J.S., Mitchell A.P. Genetic control of Candida albicans biofilm development. Nat. Rev. Genet. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samaranayake Y.H., Cheung B.P.K., Yau J.Y.Y., Yeung S.K.W., Samaranayake L.P. Human Serum Promotes Candida albicans Biofilm Growth and Virulence Gene Expression on Silicone Biomaterial. PLoS ONE. 2013;8:e62902. doi: 10.1371/journal.pone.0062902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim K.-J., Sung W.S., Suh B.K., Moon S.-K., Choi J.-S., Kim J.G., Lee D.G. Antifungal activity and mode of action of silver nano-particles on Candida albicans. Biometals. 2009;22:235–242. doi: 10.1007/s10534-008-9159-2. [DOI] [PubMed] [Google Scholar]
- 45.Miškovská A R.M., Michailidu J., Masák J., Čejková A., Lorinčík J., Maťátková O. A ntibiofilm activity of silver nanoparticles biosynthesized using viticultural waste. PLoS ONE. 2022;17:e0272844. doi: 10.1371/journal.pone.0272844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oshiro K., Rodrigues G., Monges B., Cardoso M., Franco O. Bioactive peptides against fungal biofilms. Front. Microbiol. 2019;10:2169. doi: 10.3389/fmicb.2019.02169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaikh-Kader A., Houreld N.N., Rajendran N.K., Abrahamse H. Levels of cyclooxygenase 2, interleukin-6, and tumour necrosis factor-α in fibroblast cell culture models after photobiomodulation at 660 nm. Oxidative Med. Cell. Longev. 2021;2021:6667812. doi: 10.1155/2021/6667812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng P., Luo Y., Ke C., Qiu H., Wang W., Zhu Y., Hou R., Xu L., Wu S. Chitosan-based functional materials for skin wound repair: Mechanisms and applications. Front. Bioeng. Biotechnol. 2021;9:111. doi: 10.3389/fbioe.2021.650598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Juráňová J.A.-L., J., Boumediene K., Baugé C., Biedermann D., Ulrichová J., Franková J. Modulation of Skin Inflammatory Response by Active Components of Silymarin. Molecules. 2019;24:123. doi: 10.3390/molecules24010123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Schaik P.E., Zuhorn I.S., Baron W. Targeting Fibronectin to Overcome Remyelination Failure in Multiple Sclerosis: The Need for Brain-and Lesion-Targeted Drug Delivery. Int. J. Mol. Sci. 2022;23:8418. doi: 10.3390/ijms23158418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are contained within the article and Supplementary Materials.