Abstract
Background: a specific subset of metastatic triple-negative breast cancers (mTNBC) is characterized by homologous recombination deficiency (HRD), leading to enhanced sensitivity to platinum-based chemotherapy. Apart from mutations in BRCA1/2 genes, the evaluation of other HRD-related alterations has been limited to date. As such, we analyzed data from mTNBC patients enrolled in the ProfiLER-01 study to determine the prevalence of alterations in homologous recombination-related (HRR) genes and their association with platinum sensitivity. Methods: next-generation sequencing and promoter methylation of BRCA1 and RAD51C were performed on tumors from patients with mTNBC, using a panel of 19 HRR genes. Tumors were separated into three groups based on their molecular status: mutations in BRCA1/2, mutations in other HRR genes (BRCA1/2 excluded) or BRCA1/RAD51C promoter methylation and the absence of molecular alterations in HRR genes (groups A, B and C, respectively). Sensitivity to platinum-based chemotherapy was evaluated through the radiological response. Results: mutations in BRCA1/2 were detected in seven (13.5%) patients, while alterations in other HRR genes or hypermethylation in BRCA1 or RAD51C were reported in 16 (30.7%) patients; furthermore, no alteration was found in the majority of patients (n = 29; 55.8%). Among 27 patients who received platinum-based chemotherapy, the disease control rate was 80%, 55% and 18% (groups A, B and C, respectively; p = 0.049). Regarding group B, patients with disease control exhibited mutations in FANCL, FANCA and the RAD51D genes or RAD51C methylation; Conclusion: mutations in HRR genes and epimutations in RAD51C were associated with disease control through platinum-based chemotherapy. As such, apart from well-characterized alterations in BRCA1/2, a more comprehensive evaluation of HRD should be considered in order to enlarge the selection of patients with mTNBC that could benefit from platinum-based chemotherapy.
Keywords: triple-negative breast cancer, TNBC, platinum-based chemotherapy, DNA repair, homologous recombination, HRR genes, BRCA
1. Introduction
Breast cancer (BC) represents the leading cause of death among females in the world [1]. A specific subset of BC, triple-negative breast cancer (TNBC), is characterized by the lack of estrogen and progesterone receptors and human epidermal growth factor receptor 2 (HER2) amplification. TNBCs exhibit specific features such as higher prevalence in younger patients, high histologic grade and more aggressive disease than other BC subtypes, leading to a poorer clinical outcome [2]. Indeed, over 30% of patients with TNBC will relapse with metastatic disease within three years; furthermore, the median overall survival (OS) of metastatic TNBC (mTNBC) is around 18 months [2,3]. As such, in spite of accounting for 15% of all breast cancers, TNBC represents a challenge for clinicians, with the necessity to develop new treatment strategies for this distinct subgroup [4].
At the molecular scale, a germline mutation in BRCA1 or BRCA2 (gBRCA1/2) is found in 15–20% of patients with TNBC [5,6]. The disruption of BRCA1/2 genes leads to inefficient homologous recombination (HR) for double-strand break (DSB) processing, leading to the so-called homologous recombination deficiency (HRD) and subsequent specific features through genomic instability [7]. In the context of neoadjuvant chemotherapy, it has been shown that tumors with BRCA1/2 mutations exhibit an increased sensitivity to platinum salts [8,9]. In the metastatic setting, current guidelines from the European Society for Medical Oncology (ESMO) recommend platinum-based chemotherapies in the context of gBRCA1/2-mutated TNBC [10]. Furthermore, in the context of gBRCA1/2, TNBCs exhibit higher sensitivity to Poly(ADP-ribose) polymerase inhibitors (PARPi) through synthetic lethality [7]. In recent years, several trials have shown the efficiency of PARPi’s in the context of gBRCA1/2, both in the metastatic and adjuvant settings [11,12,13]. Apart from the well-characterized BRCA1/2, several other causes leading to HRD have been described, such as the promoter hypermethylation of BRCA1 or RAD51C or mutations in HR-related (HRR) genes, which are grouped under the concept of “BRCAness”, which represent any molecular alteration out of BRCA1/2 that leads to an HRD phenotype [2,7]. In spite of their putative effect on HR function, it remains unclear if their presence in TNBC is associated with enhanced sensitivity to platinum-based chemotherapy. To address this question, we analyzed a specific panel of HRR genes and the promoter methylation of BRCA1 or RAD51C of mTNBC from patients included in the ProfiLER-01 trial [14]. The objectives of the study were assessing the prevalence of alteration of HRR genes and their association with platinum-based chemotherapy.
2. Materials and Methods
2.1. Patients and Tumors
All mTNBC patients included in the ProfiLER-01 trial (NCT01774409) from March 2013 to July 2017 were analyzed in the present study. Characteristics of the ProfiLER-01 trial have been previously published [14]. This study was approved by Ethics Committee of Lyon Sud-Est IV in agreement with the Good Clinical Practice guidelines of the International Conference on Harmonization and the Declaration of Helsinki, and with relevant French and European laws and directives. All patients provided written informed consent. Treatment response was assessed by clinical and radiological evaluations every 2 to 3 months, and radiological evaluation was based on RECIST1.1 criteria [15]. Stable disease was defined by the response or absence of signs of progression after three months of treatment. Disease control included complete response, partial response and stable disease. Relapse free survival (RFS) was defined as the time from date of diagnosis of early breast cancer to the time of loco-regional relapse or metastatic relapse. Overall survival (OS) was defined as the time from the diagnosis of incurable cancer (metastatic or locally advanced without curative intent) to the date of death or end of follow-up or cutoff date. Median OS (OS) was defined as the time from beginning of first-line treatment to the date of death (for any cause) or end of follow-up or cutoff date.
2.2. DNA Sequencing
DNA extraction was performed from Paraffin-embedded (FFPE) blocks (primary tumors or metastases). DNA-capture based next generation sequencing was performed using 50 ng of genomic DNA with a custom “HRR genes” panel of the following 19 selected genes: ATM, BARD1, BRCA1, BRCA2, BRIP1, CDK12, CHEK1, CHEK2, FANCA, FANCD2, FANCL, MRE11, NBN, PALB2, PPP2R2A, RAD51B, RAD51C, RAD51D and RAD54L (Sophia Genetics, Saint-Sulpice, Switzerland). The TP53 gene was used as internal control. The human genome Hg19 (GRCh37.p5) was used as the reference genome. Fastq files were analyzed with the Sophia DDM Platform version 5.1.9 (Sophia Genetics, Saint-Sulpice, Switzerland). SIFT, Polyphen-2e Clinvar, NNSPLICE, MaxEnt, SSF and guidelines from the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists were used to classify each variant as “pathogenic”, “likely pathogenic”, “uncertain significance”, “likely benign”, or “benign”. Variants were considered as “pathogenic” or “likely pathogenic” if at least three databases or tools predicted it [16]. Some patients underwent genetic counseling and germline analysis based on their familial and clinical characteristics; if applicable, germline results were collected.
2.3. Methylation Assays
300 ng of genomic DNA were amplified with ddPCR after treatment with bisulfite, using the EpiGenteck kit (EpiGenteck Inc., Farmingdale, NY, USA) according to the manufacturer’s instructions. Previously published specific primers and probes for methylated BRCA1 gene promoter analysis were used [17]. The RAD51C gene promoter was designed in-house. The ACTB gene was used as an internal amplification control as previously published [18]. Methylation percentage was defined as the ratio between levels of methylated BRCA1 (or RAD51C) and ACTB genes.
2.4. HRD Groups
Patients were divided into three groups according to their molecular alterations. Group A included patients with BRCA1/2 pathogenic or likely pathogenic mutations. Group B included patients without BRCA1/2 mutations and with pathogenic or likely pathogenic mutations in at least one of the other genes from the “HRR genes” panel described upwards or with the promoter methylation of BRCA1 or RAD51C genes. Group C included all of the remaining patients (i.e., without alterations detected).
2.5. Statistical Analyses
Between-group comparisons were performed using Fisher’s exact test for categorical data and a non-parametric Kruskall-Wallis test for continuous data. A p-value < 0.05 was considered significant. Survival data were performed using the Kaplan-Meier method. Missing data were censored to the date of latest news. SAS version 9.4 (SAS Institute, Cary, NC, USA) was used for all statistical analyses.
2.6. Study Endpoints
The primary endpoint was the description of the alteration of the genes of the HR pathway in mTNBC populations. Secondary endpoints were responses to treatment and the associations between molecular alterations and platinum efficacy.
3. Results
3.1. Patients’ and Treatments’ Characteristics
Fifty-nine patients from the ProFILER-01 trial exhibited mTNBC and were analyzed; seven patients were excluded due to the lack of genetic material or the poor quality of genetics samples. Fifty-two patients were divided into three groups with the following distribution: 7, 16 and 29 patients (groups A, B and C, respectively). Regarding clinical characteristics, all patients were females with median age at diagnosis significantly different (p = 0.006) with 37.1 years (range 27–40) in group A, 42.1 years (range 28–65) in group B and 48.2 years (range 34–65) in group C (Table 1).
Table 1.
Patient’s characteristics.
| Group A n = 7 |
Group B n = 16 |
Group C n = 29 |
All n = 52 |
p-Value ** | |
|---|---|---|---|---|---|
| Age at diagnosis | |||||
| Median (min–max) | 37.1 (27–40) | 42.1 (28–65) | 48.2 (34–65) | 42.1 (27–65) | 0.006 |
| Personal history of breast cancer n, (%) | 0 | 0 | 4 (14%) | 4 (14%) | |
| Family history of cancer * | |||||
| 1st and/or 2nd degree | 4 (57%) | 7 (44%) | 7 (25%) | 18 (35%) | 0.16 |
| No history | 3 (43%) | 9 (56%) | 22 (75%) | 34 (65%) | |
| gBRCA1/2 testing n (%) | 7 (100%) | 8 (50%) | 7 (24%) | 22 (42%) | |
| Histology, n (%) | |||||
| NOS | 7 (100%) | 13 (81%) | 25 (86%) | 45 (87%) | |
| Lobular | 0 | 0 | 1 (4%) | 1 (2%) | |
| other | 0 | 3 (19%) | 3 (10%) | 6 (11%) | |
| Grade, n (%) | |||||
| 1 | 0 | 0 | 2 (7%) | 2 (4%) | |
| 2 | 0 | 3 (20%) | 7 (24%) | 10 (20%) | |
| 3 | 6 (100%) | 12 (80%) | 20 (69%) | 38 (76%) | |
| Unknown | 1 | 1 | 0 | 2 | |
| Number of metastatic sites | |||||
| 1 | 1 (14%) | 5 (31.3%) | 12 (41.4%) | 18 (34.6%) | |
| 2–3 | 6 (86%) | 11 (68.8%) | 13 (44.8%) | 30 (57.7%) | |
| >3 | 0 | 0 | 4 (13.8%) | 4 (7.7%) | p = 0.21 |
| Sites of metastasis, n (%) | |||||
| Bones | 1 (14%) | 5 (31%) | 10 (34%) | 16 (31%) | |
| CNS | 0 | 2 (12%) | 2 (7%) | 4 (8%) | |
| Skin | 2 (29%) | 5 (31%) | 5 (17%) | 12 (23%) | |
| Liver | 0 | 3 (19%) | 8 (28%) | 11 (21%) | |
| Lung | 5 (71%) | 7 (44%) | 14 (50%) | 26 (51%) | |
| Regional lymph nodes | 5 (71%) | 6 (37%) | 17 (58%) | 28 (54%) |
Abbreviations: gBRCA1/2: germline mutation of BRCA1/2; NOS: not otherwise specified. * Ovary, breast, prostate or uterus. ** Kruskal-Wallis test or Fisher exact test.
Germline BRCA testing was performed in seven patients in group A, eight patients in group B, and 7 patients in group C. There were no significant differences between groups in terms of family history of cancer, tumor grades or number of metastatic sites. Forty-nine (94.2%) patients underwent surgery of the primary breast tumor; and 44 (84.6%) patients received chemotherapy in the (neo)-adjuvant setting (Table 2). Initially, local or locally advanced disease at diagnosis were as follows: five (71%), 14 (88%) and 25 (86%) patients in group A, B, and C, respectively. Upon neoadjuvant treatment, responses to therapy were evaluated in three, eight and nine patients in group A, B and C, respectively. One patient in group A had pathological complete response to neoadjuvant treatment, while two patients in group B and one patient in group C progressed on therapy. No difference was seen regarding PFS from localized therapy (p = 0.91). No patients received platinum-based treatment in this setting.
Table 2.
Treatment’s Characteristics.
| Group A n = 7 |
Group B n = 16 |
Group C n = 29 |
All n = 52 |
p-Value * | |
|---|---|---|---|---|---|
| Disease stage at diagnosis | |||||
| Local/Locoregional | 5 (71%) | 14 (88%) | 25 (86%) | 44 (85%) | |
| Metastatic | 2 (29%) | 2 (12%) | 4 (14%) | 8 (15%) | |
| Neo/adjuvant chemotherapy | n = 5 (71%) | n = 14 (88%) | n = 25 (86%) | n = 44 (85%) | |
| Cyclophosphamide | 5 (100%) | 13 (92%) | 25 (100%) | 43 (97%) | |
| Anthracycline | 5 (100%) | 11 (78%) | 24 (96%) | 40 (90%) | |
| Taxane | 5 (100%) | 13 (92%) | 24 (96%) | 42 (95%) | |
| Breast surgery | 7 (100%) | 14 (87%) | 28 (97%) | 49 (94%) | |
| Nodes involvement | 1 (14%) | 11 (68%) | 17 (59%) | 29 (55%) | |
| Response to neoadjuvant therapy | n = 3 | n = 8 | n = 9 | n = 20 | |
| pCR | 1 (33%) | 0 | 0 | 1 (5%) | |
| PR/SD | 2 (67%) | 6 (75%) | 8 (89%) | 16 (80%) | |
| PD on therapy | 0 | 2 (25%) | 1 (11%) | 3 (15%) | |
| Adjuvant RT | 5 (71%) | 14 (88%) | 21 (72%) | 40 (76%) | |
|
RFS from localized therapy (months) Median (min–max) |
16.0 (11–17) |
14.3 (7–53) |
13.6 (3–202) |
14.2 (3–202) |
p = 0.91 |
|
Number of lines in metastatic setting Median (min–max) |
4 (2–6) | 6 (1–9) | 3 (1–9) | 4 (1–9) | |
|
OS (months) Median (min–max) |
19.2 (8.0–44.0) |
23.3 (14.5–31.2) |
16.6 (10.2–28.4) |
17.9 (8.0–44.0) |
p = 0.86 |
Abbreviations: CNS: Central Nervous System ET: endocrine therapy; gBRCA1/2: germline BRCA1/2 mutation; NOS: not otherwise specified; OS: overall survival; pCR: pathological complete response; PD: progression disease; PR: partial response; RFS: Response Free Survival; RT: Radiation therapy; SD: Stable disease. * Fisher exact test or Kruskall-Wallis test.
3.2. Molecular Alterations
From the 52 tumor samples, variant analysis, large rearrangement analysis of HRR genes and assessment of promoter methylation of BRCA1 and RAD51C were performed on 52, 50 and 47 samples, respectively. Seven samples were discarded due to technical failure (n = 2) or insufficient tissue (n = 5). Tumor DNA was extracted from 40 primary tumors and 12 metastases. Among the seven patients in group A, six patients had BRCA1 mutations and one had a BRCA2 mutation; all of these mutations were also found constitutively. In group B, the mutated genes were as follows: FANCA (n = 2), FANCL (n = 1), RAD51D (n = 1; also detected as a germline mutation), NBN (n = 1) and CHEK2 (n = 1). Promoters of BRCA1 and RAD51C were found to be methylated in eight and two patients, respectively.
3.3. Response to Platinum-Based Chemotherapy
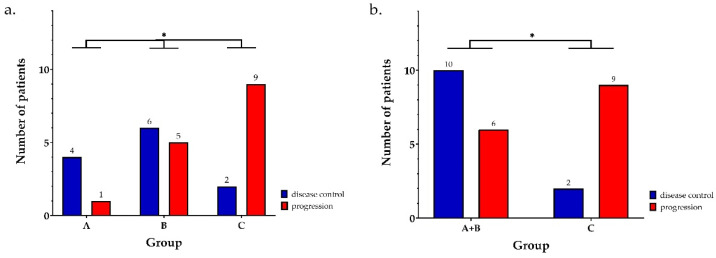
Among the 27 patients who received platinum-based chemotherapy, disease control rate at three months was 80% in group A, 55% in group B, and 18% in group C (p = 0.049; Figure 1a). When combining groups A and B versus group C (corresponding to presence or absence of molecular alterations, respectively), a statistically significant differential disease control remained (p = 0.047; Figure 1b).
Figure 1.
Response to platinum-based therapy according to molecular status phenotype. Responses are given for each group taken separately (a), or by comparing patients with or without molecular alteration (groups A + B versus group C respectively) (b). * represents differences with statistical significance (p < 0.05).
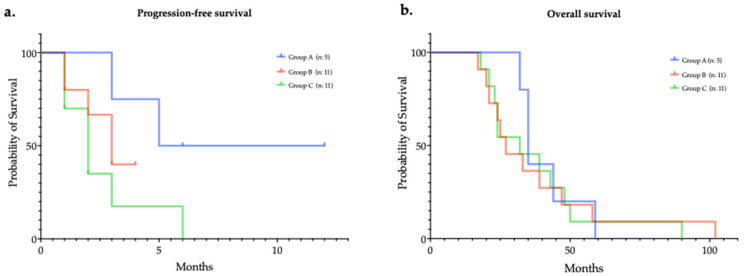
Regarding survival associated with platinum-based chemotherapy, the median PFS were 5.3, 3.0, and 2.1 months (groups A, B and C, respectively), nevertheless without reaching a statistically significant difference (p = 0.36; Figure 2a). Similarly, median OS were as follows: 35.0, 27.2, and 32.4 months (groups A, B and C, respectively; p = 0.77; Figure 2b).
Figure 2.
Progression-free (a) and overall (b) survivals of patients challenged with platinum-based chemotherapy, according to molecular status.
3.4. Exploratory Analysis at the Individual Scale
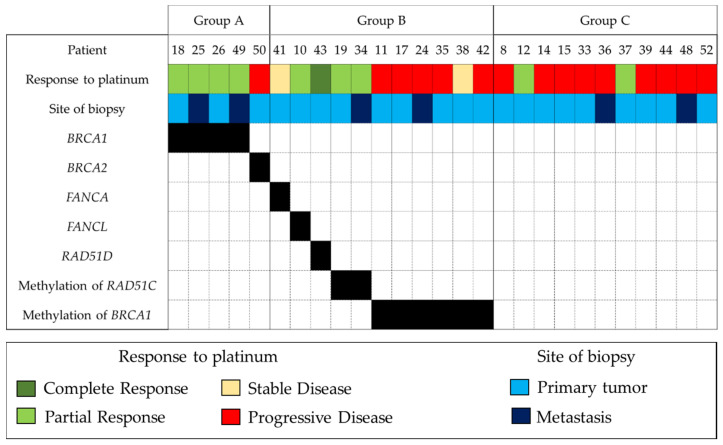
Individual responses are described in Figure 3. It is worthy of note that patient 50 carried a two-base germline mutation of BRCA2 (c.3545_3546delTT) in exon 11, causing a frameshift variant and a premature termination codon. A second deletion c.3509_3547del was found on tumor analysis, which restored the reading frame and BRCA2 function.
Figure 3.
Site of biopsy, molecular status and radiological response for each patient challenged with platinum-based chemotherapy. A black square indicates the presence of a given molecular alteration.
4. Discussion
Beyond BRCA1/2 mutations, this retrospective analysis shows that specific alterations of HR genes are associated with platinum sensitivity in mTNBC patients, namely FANCL mutation, FANCA mutation, RAD51D mutation and promoter methylation of RAD51C. In our study, we tested tumor DNA by NGS with a small HR-gene panel, as well as BRCA1 and RAD51C promoter methylation. We found that 44% of TNBC tumors had HR gene alterations; and 30.7% had alterations in HRR genes other than BRCA1/2, in accordance with the literature. Indeed, HRD score evaluated with MyChoice® (Myriad Genetics Inc., Salt Lake City, UT, USA) was considered high for 41–71% of selected patients with early and metastatic cancer, but it remains unknown whether these patients may be eligible for PARPi TNBC [9,19,20,21]. In these studies, BRCA1/2 mutations account for only 15–20% of tumors with high HRD score. In another study, the HRDetect mutational-signature-based algorithm score was high in 59% of patients with early TNBC [22].
Early TNBC with a high HRD score and especially BRCA1/2 mutations showed higher pCR rates after neoadjuvant chemotherapy [19,20,23,24]. In the metastatic setting, the TNT trial failed to demonstrate the benefit of platinum-based chemotherapy for the whole cohort of TNBC patients, as well as for patients with high-HRD scores. Nevertheless, this trial showed that gBRCA1/2 gene mutations were significantly associated with better tumor response to carboplatin than docetaxel treatment (68% versus 33%) [9]. In the present study, putative HRD tumors (with BRCA 1/2 mutation or other HR gene alteration) presented a higher disease control rate than non-HRD patients after platinum-based chemotherapy. Patients with gBRCA1 gene mutation responded to platinum-based chemotherapy, consistent with previous studies [9,25]. The only patient with a gBRCA2 mutation who did not respond harbored a reverse mutation of the BRCA2 gene. This resistance mechanism to platinum-based chemotherapy has been previously described [26]. In the context of putative HRD without BRCA1/2 mutations, patients who responded to platinum-based chemotherapy harbored the following alterations: the promoter methylation of RAD51C, and mutations of the RAD51D, FANCA and FANCL genes. Each alteration represented less than 2% of our entire TNBC cohort, in concordance with existing data [27,28]. The methylation of RAD51C has been associated with “signature 3” like BRCA1/2 mutated tumors and has been shown to exhibit the features of HRD tumors [22,27]. Regarding the 6 patients with promoter methylation of BRCA1, only one presented a stable disease with platinum-based chemotherapy in our cohort. Although this epimutation is considered as a marker of HRD, notably with its association with “signature 3”, its theragnostic impact has been controversial [27]. In the TNT trial, although a majority of tumors with BRCA1 promoter methylation had a high HRD score (based on the MyChoice® assay), they did not exhibit a better response to carboplatin treatment [9]. The same absence of impact has been reported in the TBCR009 trial [25]. Nevertheless, this epimutation was associated with response to olaparib in untreated TNBC [29]. Several limits may explain this apparent discordance. Firstly, there is not an absolute correlation between promoter methylation and the absence of expression at the RNA level [17]. Secondly, we performed methylation assays on primary tumors for all patients but one; as such, based on the dynamic nature of methylation, we cannot exclude a different BRCA1 promoter methylation status between the initial biopsy and the pre-platinum time points. Indeed, the reversion of BRCA1 promoter methylation has already been reported, leading to HR pathway restoration [30].
Our study has several limits. Firstly, although it is an almost “real-life” trial regarding enrolled patients, the sample size is quite modest, at both the whole cohort and platinum-challenged patient’s scales. This may have led to the absence of statistical significance regarding the different PFS observed after platinum-based chemotherapy. Secondly, as previously discussed, the vast majority of molecular analyses were performed on tissues obtained from initial biopsies; as such, the molecular alterations observed do not necessarily perfectly reflect the actual status upon platinum-challenge. Nevertheless, assessing the predictive value of initial biomarkers allowed us to decipher more robust ones (i.e., less sensitive to evolutionary fluctuations); furthermore, iterative biopsies with multiple molecular assays are not necessarily compatible with real-life practice. Thirdly, it is now well-known that HRD status may be assessed through diverse companion diagnostic assays; they nevertheless do not overlap perfectly [7]. The MyChoice® assay has been shown to represent the current gold standard in ovarian cancer as a prerequisite to PARPi challenge [31]. As such, it would have been interesting to analyze the potent correlation between the molecular alterations observed in our study and the associated HRD score. Regarding perspectives, PARPi have already demonstrated their interest in breast tumors with gBRCA1/2 [11,12]. Results from the phase II RUBY trial, which assessed rucaparib in patients with an HRD status evaluated with the Foundation Medicine® T5 assay (Foundation Medicine, Cambridge, MA, USA) in mTNBC showed encouraging data regarding larger populations than gBRCA1/2 patients [32].
5. Conclusions
Our present study showed that 30% of TNBC tumors had at least one mutation in our HRR-gene panel (BRCA1/2 genes excluded), and these specific alterations were associated with an objective response to platinum-based chemotherapy. As such, a more comprehensive approach should be considered when assessing the HRD status for TNBC patients, with the possible effect of enlarging the selection of patients that could benefit from platinum-based treatments. Further studies are needed regarding the impact of HRR-genes in mTNBC and their predictive impact regarding sensitivity to PARPi.
Author Contributions
Conceptualization, E.B., O.T. and J.-Y.B.; supervision, project administration and funding acquisition: O.T. and J.-Y.B.; methodology, software, validation, data curation and formal analysis: V.H., A.L.-C., O.T., V.A., Q.W., V.C., D.P. (Daniel Pissaloux), D.P. (David Pérol), I.T., A.B., V.B., C.L., S.P., A.V. and E.S.; investigation: P.-E.H., T.B., A.D., L.E., P.T., O.T., J.-Y.B. and I.R.-C.; writing—original draft preparation, S.Q., K.-A.B. and E.B.; writing—review and editing: S.Q., K.-A.B., E.B., O.T. and J.-Y.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Characteristics of the ProfiLER-01 (NCT01774409) trial, including detailed information regarding Institutional Review Board Statement have been previously published [14]. The study protocol was approved by the Ethics Committee of Lyon Sud-Est IV (protocol code 12/114; date of approval 13 December 2012), in agreement with the Good Clinical Practice guidelines of the International Conference on Harmonization and the Declaration of Helsinki, and with relevant French and European laws and directives.
Informed Consent Statement
Written informed consent has been obtained from all the patients enrolled in this study.
Data Availability Statement
Datasets are available upon request to corresponding author.
Conflicts of Interest
J.-Y.B.: received research support and honoraria from Astrazeneca, GSK, Novartis; P.-E.H. received grants and non-financial support from AstraZeneca, Roche, Novartis, Pfizer, personal fees from Mylan, personal fees and non-financial support from EISAI, outside the submitted work; I.R.-C.: received honoraria from AstraZeneca, Clovis, Tesaro and PharmaMar; Consulting/advisory board fees from AstraZeneca, Roche, Clovis, Tesaro, Genmab, PharmaMar, MSD and Pfizer, research funding from MSD and BMS, travel expenses from AstraZeneca, GSK and Roche; O.T.: received grants from Roche, MSD-Merck, BMS; personal fees from Roche, MSD-Merck, Novartis-Sandoz, Pfizer, Lilly, Astra-Zeneca, Daiichi Sankyo, Eisai, Pierre Fabre; D.P. (David Pérol): received honoraria from Astrazeneca, BMS, ELI-Lilly, IPSEN, Roche, Novartis, Pierre Fabre, MSD and Takeda, research funding from MSD Avenir, travel expenses from Astrazeneca; T.B.: received grants, advisory board fees and non-financial support from Novartis, AstraZeneca and Pfizer, advisory board fees and non-financial support from Roche, advisory board fees from SeattleGenetics. E.B., V.H., S.Q., K.-A.B., A.L.-C., Q.W., L.E., L.C., V.B., I.T., V.A., V.C., C.L., A.D., E.S., D.P. (Daniel Pissaloux), A.V., S.P., A.B. and P.T. declare that they have no conflict of interest.
Funding Statement
Lyric (DGOS-INCa-4664), Bpifrance Financement abounded by European Community (E8983—PREDICTIV), LabEx DevweCAN (ANR-10-LABX-0061), Ligue contre le Cancer, Ligue de l’Ain contre le Cancer, la Fondation ARC and EURACAN (EU project 739521).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G., De Angelis C., Licata L., Gianni L. Treatment Landscape of Triple-Negative Breast Cancer—Expanded Options, Evolving Needs. Nat. Rev. Clin. Oncol. 2022;19:91–113. doi: 10.1038/s41571-021-00565-2. [DOI] [PubMed] [Google Scholar]
- 3.Zagami P., Carey L.A. Triple Negative Breast Cancer: Pitfalls and Progress. NPJ Breast Cancer. 2022;8:95. doi: 10.1038/s41523-022-00468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahtani R., Kittaneh M., Kalinsky K., Mamounas E., Badve S., Vogel C., Lower E., Schwartzberg L., Pegram M., Breast Cancer Therapy Expert Group (BCTEG) Advances in Therapeutic Approaches for Triple-Negative Breast Cancer. Clin. Breast Cancer. 2021;21:383–390. doi: 10.1016/j.clbc.2020.12.011. [DOI] [PubMed] [Google Scholar]
- 5.Belli C., Duso B.A., Ferraro E., Curigliano G. Homologous Recombination Deficiency in Triple Negative Breast Cancer. Breast. 2019;45:15–21. doi: 10.1016/j.breast.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Howard F.M., Olopade O.I. Epidemiology of Triple-Negative Breast Cancer: A Review. Cancer J. 2021;27:8–16. doi: 10.1097/PPO.0000000000000500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quesada S., Fabbro M., Solassol J. Toward More Comprehensive Homologous Recombination Deficiency Assays in Ovarian Cancer, Part 1: Technical Considerations. Cancers. 2022;14:1132. doi: 10.3390/cancers14051132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Telli M.L., Jensen K.C., Vinayak S., Kurian A.W., Lipson J.A., Flaherty P.J., Timms K., Abkevich V., Schackmann E.A., Wapnir I.L., et al. Phase II Study of Gemcitabine, Carboplatin, and Iniparib As Neoadjuvant Therapy for Triple-Negative and BRCA1/2 Mutation-Associated Breast Cancer with Assessment of a Tumor-Based Measure of Genomic Instability: PrECOG 0105. J. Clin. Oncol. 2015;33:1895–1901. doi: 10.1200/JCO.2014.57.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tutt A., Tovey H., Cheang M.C.U., Kernaghan S., Kilburn L., Gazinska P., Owen J., Abraham J., Barrett S., Barrett-Lee P., et al. Carboplatin in BRCA1/2-Mutated and Triple-Negative Breast Cancer BRCAness Subgroups: The TNT Trial. Nat. Med. 2018;24:628–637. doi: 10.1038/s41591-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gennari A., André F., Barrios C.H., Cortés J., de Azambuja E., DeMichele A., Dent R., Fenlon D., Gligorov J., Hurvitz S.A., et al. ESMO Clinical Practice Guideline for the Diagnosis, Staging and Treatment of Patients with Metastatic Breast Cancer. Ann. Oncol. 2021;32:1475–1495. doi: 10.1016/j.annonc.2021.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Robson M., Im S.-A., Senkus E., Xu B., Domchek S.M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 12.Litton J.K., Rugo H.S., Ettl J., Hurvitz S.A., Gonçalves A., Lee K.-H., Fehrenbacher L., Yerushalmi R., Mina L.A., Martin M., et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018;379:753–763. doi: 10.1056/NEJMoa1802905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tutt A.N.J., Garber J.E., Kaufman B., Viale G., Fumagalli D., Rastogi P., Gelber R.D., de Azambuja E., Fielding A., Balmaña J., et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. N. Engl. J. Med. 2021;384:2394–2405. doi: 10.1056/NEJMoa2105215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trédan O., Wang Q., Pissaloux D., Cassier P., de la Fouchardière A., Fayette J., Desseigne F., Ray-Coquard I., de la Fouchardière C., Frappaz D., et al. Molecular Screening Program to Select Molecular-Based Recommended Therapies for Metastatic Cancer Patients: Analysis from the ProfiLER Trial. Ann. Oncol. 2019;30:757–765. doi: 10.1093/annonc/mdz080. [DOI] [PubMed] [Google Scholar]
- 15.Eisenhauer E.A., Therasse P., Bogaerts J., Schwartz L.H., Sargent D., Ford R., Dancey J., Arbuck S., Gwyther S., Mooney M., et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1) Eur. J. Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibragimova I., Cairns P. Assays for Hypermethylation of the BRCA1 Gene Promoter in Tumor Cells to Predict Sensitivity to PARP-Inhibitor Therapy. Poly(ADP-Ribose) Polym. 2011;780:277–291. doi: 10.1007/978-1-61779-270-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ida C.M., Butz M.L., Jenkins R.B., Sarkaria J.N., Kitange G.J., Giannini C., Kipp B.R. Real-Time Methylation-Specific Polymerase Chain Reaction for MGMT Promoter Methylation Clinical Testing in Glioblastoma: An Alternative Detection Method for a Heterogeneous Process. Am. J. Clin. Pathol. 2017;148:296–307. doi: 10.1093/ajcp/aqx073. [DOI] [PubMed] [Google Scholar]
- 19.Telli M.L., Timms K.M., Reid J., Hennessy B., Mills G.B., Jensen K.C., Szallasi Z., Barry W.T., Winer E.P., Tung N.M., et al. Homologous Recombination Deficiency (HRD) Score Predicts Response to Platinum-Containing Neoadjuvant Chemotherapy in Patients with Triple-Negative Breast Cancer. Clin. Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loibl S., Weber K.E., Timms K.M., Elkin E.P., Hahnen E., Fasching P.A., Lederer B., Denkert C., Schneeweiss A., Braun S., et al. Survival Analysis of Carboplatin Added to an Anthracycline/Taxane-Based Neoadjuvant Chemotherapy and HRD Score as Predictor of Response-Final Results from GeparSixto. Ann. Oncol. 2018;29:2341–2347. doi: 10.1093/annonc/mdy460. [DOI] [PubMed] [Google Scholar]
- 21.Sharma P., Barlow W.E., Godwin A.K., Pathak H., Isakova K., Williams D., Timms K.M., Hartman A.R., Wenstrup R.J., Linden H.M., et al. Impact of Homologous Recombination Deficiency Biomarkers on Outcomes in Patients with Triple-Negative Breast Cancer Treated with Adjuvant Doxorubicin and Cyclophosphamide (SWOG S9313) Ann. Oncol. 2018;29:654–660. doi: 10.1093/annonc/mdx821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staaf J., Glodzik D., Bosch A., Vallon-Christersson J., Reuterswärd C., Häkkinen J., Degasperi A., Amarante T.D., Saal L.H., Hegardt C., et al. Whole-Genome-Sequencing of Triple Negative Breast Cancers in a Population-Based Clinical Study. Nat. Med. 2019;25:1526–1533. doi: 10.1038/s41591-019-0582-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Von Minckwitz G., Schneeweiss A., Loibl S., Salat C., Denkert C., Rezai M., Blohmer J.U., Jackisch C., Paepke S., Gerber B., et al. Neoadjuvant Carboplatin in Patients with Triple-Negative and HER2-Positive Early Breast Cancer (GeparSixto; GBG 66): A Randomised Phase 2 Trial. Lancet Oncol. 2014;15:747–756. doi: 10.1016/S1470-2045(14)70160-3. [DOI] [PubMed] [Google Scholar]
- 24.Sikov W.M., Berry D.A., Perou C.M., Singh B., Cirrincione C.T., Tolaney S.M., Kuzma C.S., Pluard T.J., Somlo G., Port E.R., et al. Impact of the Addition of Carboplatin and/or Bevacizumab to Neoadjuvant Once-per-Week Paclitaxel Followed by Dose-Dense Doxorubicin and Cyclophosphamide on Pathologic Complete Response Rates in Stage II to III Triple-Negative Breast Cancer: CALGB 40603 (Alliance) J. Clin. Oncol. 2015;33:13–21. doi: 10.1200/JCO.2014.57.0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isakoff S.J., Mayer E.L., He L., Traina T.A., Carey L.A., Krag K.J., Rugo H.S., Liu M.C., Stearns V., Come S.E., et al. TBCRC009: A Multicenter Phase II Clinical Trial of Platinum Monotherapy With Biomarker Assessment in Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2015;33:1902–1909. doi: 10.1200/JCO.2014.57.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobalina L., Armenia J., Irving E., O’Connor M.J., Forment J.V. A Meta-Analysis of Reversion Mutations in BRCA Genes Identifies Signatures of DNA End-Joining Repair Mechanisms Driving Therapy Resistance. Ann. Oncol. 2021;32:103–112. doi: 10.1016/j.annonc.2020.10.470. [DOI] [PubMed] [Google Scholar]
- 27.Polak P., Kim J., Braunstein L.Z., Karlic R., Haradhavala N.J., Tiao G., Rosebrock D., Livitz D., Kübler K., Mouw K.W., et al. A Mutational Signature Reveals Alterations Underlying Deficient Homologous Recombination Repair in Breast Cancer. Nat. Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Castéra L., Harter V., Muller E., Krieger S., Goardon N., Ricou A., Rousselin A., Paimparay G., Legros A., Bruet O., et al. Landscape of Pathogenic Variations in a Panel of 34 Genes and Cancer Risk Estimation from 5131 HBOC Families. Genet. Med. 2018;20:1677–1686. doi: 10.1038/s41436-018-0005-9. [DOI] [PubMed] [Google Scholar]
- 29.Eikesdal H.P., Yndestad S., Elzawahry A., Llop-Guevara A., Gilje B., Blix E.S., Espelid H., Lundgren S., Geisler J., Vagstad G., et al. Olaparib Monotherapy as Primary Treatment in Unselected Triple Negative Breast Cancer. Ann. Oncol. 2021;32:240–249. doi: 10.1016/j.annonc.2020.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Prieske K., Prieske S., Joosse S.A., Trillsch F., Grimm D., Burandt E., Mahner S., Schmalfeldt B., Milde-Langosch K., Oliveira-Ferrer L., et al. Loss of BRCA1 Promotor Hypermethylation in Recurrent High-Grade Ovarian Cancer. Oncotarget. 2017;8:83063–83074. doi: 10.18632/oncotarget.20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quesada S., Fabbro M., Solassol J. Toward More Comprehensive Homologous Recombination Deficiency Assays in Ovarian Cancer Part 2: Medical Perspectives. Cancers. 2022;14:1098. doi: 10.3390/cancers14041098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Patsouris A., Diop K., Tredan O., Nenciu D., Gonçalves A., Arnedos M., Sablin M.-P., Jézéquel P., Jimenez M., Droin N., et al. Rucaparib in Patients Presenting a Metastatic Breast Cancer with Homologous Recombination Deficiency, without Germline BRCA1/2 Mutation. Eur. J. Cancer. 2021;159:283–295. doi: 10.1016/j.ejca.2021.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets are available upon request to corresponding author.