Abstract
Bacteria have evolved sophisticated regulatory circuits to modulate their gene expression in response to disparate environments. In order to monitor bacterial gene expression and regulation in the host, methods for direct transcript analysis from clinical specimens are needed. For most bacterial infections, amplification of the mRNAs of interest is necessary due to the low numbers of cells present and the low levels of specific transcripts. Here we compare two methods of quantitative reverse transcription-PCR (RT-PCR)—competitive RT-PCR using a one-tube system followed by standard gel analysis and the real-time detection of PCR product formation by fluorescence resonance energy transfer technology using the LightCycler unit. We isolated Staphylococcus aureus RNA directly from clinical specimens obtained from cystic fibrosis patients with chronic S. aureus lung infection and from an animal model of foreign-body infection with no further cultivation of the bacteria. Competitive RT-PCR and LightCycler RT-PCR were tested for their ability to quantify the transcription of a constitutively expressed gyrase gene (gyr) and a highly regulated α-toxin gene (hla) of S. aureus. Reproducible results were obtained with both methods. A sensitivity of 104 (gyr) and 103 (hla) copies, respectively, was reached, which was sufficient for the quantification of transcripts during bacterial infection. Overall, the competitive RT-PCR is a robust technique which does not need special RNA purification. On the negative side, it is labor intensive and time consuming, thus limiting the numbers of samples which can be analyzed at a given time. LightCycler RT-PCR is very susceptible to even traces of inhibitors, but it allows high-throughput processing of samples.
Over time, bacteria have evolved sophisticated regulatory circuits to modulate their gene expression in response to disparate environments (4). The sequential gene expression essential for host colonization and infection remains to be determined for most pathogens. Therefore, methods for direct quantitative transcript analysis during infection are needed (2). Hybridization techniques such as Northern analysis or chip technology for detecting specific mRNAs are limited to infections with very high bacterial numbers, e.g., Pseudomonas aeruginosa in cystic fibrosis (CF) patients (8). Amplifying the target mRNA is necessary for the quantification of specific transcripts during infections with low bacterial numbers. Reverse transcription (RT) followed by PCR is a powerful technique for the detection of low levels of mRNA. However, exact quantification by end-point measurement of product is cumbersome and requires an increased number of controls (including prior determination of the dynamic range, the exact PCR efficiencies, and the PCR plateau) in order to yield meaningful results. Quantification of transcripts can be achieved either by competitive RT-PCR followed by gel analysis or by real-time RT-PCR monitoring of product formation (6). Useful competitive RT-PCR is based on the coamplification of the target RNA and known amounts of a synthetic homologous competitor template, usually engineered to share the primer recognition site with the target sequence but to differ either in length or by a short heterologous sequence stretch, provided that the overall PCR efficiency is not affected by the modification (6, 9). The LightCycler (Roche Biochemicals) instrumentation allows detection of the PCR product during the entire course of amplification by hybridization of two internal probes labeled with two different fluorophores based on the fluorescence resonance energy transfer principle (5, 7). Thus, sequence-specific detection is ensured by the use of internal hybridization probes. The kinetics obtained during the exponential phase of PCR are used for quantification.
In addition, to monitor cell numbers in the specimens as well as the efficiency of the RNA extraction and the presence of PCR inhibitors, an ubiquitously expressed internal housekeeping gene is usually quantified at the same time, and the number of copies of the gene of interest is normalized against the number of copies of the housekeeping gene (6, 11). To determine temporal gene expression in bacteria, quantification of the 16S rRNA is often used as a reference (1, 3). Recently, we described the use of a constitutively expressed gene gyr, which codes for gyrase, as an internal control during bacterial infection (2); no alteration in gyr expression under different experimental conditions has been found in this laboratory so far.
Here we assess competitive RT-PCR and LightCycler RT-PCR for their value in studying bacterial gene expression in vivo. For the analysis we isolated Staphylococcus aureus RNA directly from clinical specimens from CF patients with chronic S. aureus lung infection and from an animal model of foreign-body infection. Both methods were evaluated for their usefulness in quantifying the transcription of the constitutively expressed gyr gene and a highly regulated α-toxin gene, hla.
MATERIALS AND METHODS
Specimens.
Sputum samples from CF patients were collected as described in an earlier paper (2). Further specimens were obtained from an animal model of foreign-body infection (10). Tissue cages were infected with 105 CFU of S. aureus/ml, and fluid (exudate) was aspirated at 48 and 96 h after infection and stored immediately in liquid nitrogen.
RNA preparation.
Frozen samples were thawed rapidly and 200-μl aliquots were used for RNA isolation. S. aureus cells were lysed directly in 1 ml of Trizol LS reagent (Gibco BRL, Karlsruhe, Germany) with 0.5 ml of zirconia/silica beads (0.1-mm diameter) in a high-speed homogenizer (Savant Instruments, Farmingdale, N.Y.) at 6,500 rpm for 20 s. RNA was isolated as described in the instructions of the manufacturer (Gibco BRL). In order to remove PCR inhibitors, the RNA was further purified with the viral nucleic acid kit (Roche Biochemicals, Mannheim, Germany) by following the manufacturer's instructions. Contaminating DNA was degraded by digesting RNA samples with DNase as previously described (2).
Construction of specific RNA standards.
Sequence-modified RNA standards specific for gyr and hla were engineered as previously described (2).
Quantification of specific transcripts with competitive RT-PCR.
Competitive RT-PCR for the quantification of gyr and hla was performed as previously described (2). Briefly, serial dilutions of RNA standards were spiked with equal amounts of total sample RNA and subjected to RT-PCR using the TITAN One-Tube RT-PCR system (Roche Biochemicals). Aliquots of the amplified products were separated on a 3% agarose gel and visualized after ethidium bromide staining. The agarose gels were digitalized and subjected to quantitative densitometry. The absolute amounts of target products were calculated by determination of the competition equivalence point of target and standard amplicons (11).
Quantification of specific transcripts with LightCycler RT-PCR.
LightCycler RT-PCR was carried out using the LightCycler RNA amplification kit for hybridization probes (Roche Biochemicals). Master mixes were prepared by following the manufacturer's instructions, using the primers for gyr and hla listed in Table 1. After RT for 20 min at 50°C, the following temperature profile was utilized for amplification: denaturation for 1 cycle at 95°C for 30 s and 45 cycles at 95°C for 1 s (temperature transition, 20°C/s), 55 to 50°C (step size, 1°C; step delay, 1 cycle) for 15 s (temperature transition, 20°C/s), and 72°C for 15 s (temperature transition, 2°C/s) with fluorescence acquisition at 55 to 50°C in single mode. Melting-curve analysis was done at 45 to 90°C (temperature transition, 0.2°C/s) with stepwise fluorescence acquisition. Sequence-specific standard curves were generated using 10-fold serial dilutions (102 to 108 copies/μl) of the specific RNA standards. The number of copies of each sample transcript was then determined with the aid of the LightCycler software. The specificity of the PCR reaction was verified by ethidium bromide staining on 3% agarose gels.
TABLE 1.
Oligonucleotide primers and LightCycler hybridization probes
| Target gene | GenBank accession no. | Primer | Nucleotide positions | Sequencea | Source |
|---|---|---|---|---|---|
| gyr | D10489 | gyrU | 219–239 | TTATGGTGCTGGGCAAATACA | 2 |
| gyrL | 536–556 | CACCATGTAAACCACCAGATA | 2 | ||
| gyrFL1 | 484–512 | ATTTTAACTGTTTTACATGCTGGTGGTAA-F | This study | ||
| gyrLC1 | 514–532 | Red640-TTTGGCGGTGGCGGATACA-ph | This study | ||
| hla | X01645 | hlaU | 498–518 | AGAAAATGGCATGCACAAAAA | 2 |
| hlaL | 874–895 | TATCAGTTGGGCTCTCTAAAA | 2 | ||
| hlaFL1 | 794–813 | CAGGAAAAATTGGCGGCCTT-F | This study | ||
| hlaLC1 | 815–836 | Red640-TTGGTGCAAATGTTTCGATTGG-ph | This study |
F, fluorescein; Red640, LightCycler-Red 640-N-hydroxysuccinimide ester; ph, 3′-phosphate.
RESULTS AND DISCUSSION
Use of in vitro-transcribed RNA standards.
Synthetic, quantified RNA molecules are necessary to quantify specific sample transcripts. The standard RNAs used in this study were specifically designed to allow identical experimental conditions for both standard RNA and samples during RT-PCR with identical primer pairs. This was achieved by generating transcription-competent DNA fragments using the gene-specific primer for gyr or hla with a 5′-extension encompassing the T7 phage promoter sequence. The standard RNAs were then synthesized by in vitro transcription using T7 polymerase. In competitive RT-PCR, a sequence-modified standard RNA allowed us to distinguish between competitor and target products. Such standards were constructed by a deletion mutagenesis PCR technique using oligonucleotides composed of two distinctly spaced target sites, thus generating deletions in the final sequence (2). This modification of the RNA standard is not necessarily required for the LightCycler RT-PCR since the standard is run separately. The synthesis of an unmodified standard is less time-consuming and can be easily achieved within 1 day. In the work described here, the same modified standard RNAs were used for both competitive RT-PCR and LightCycler RT-PCR.
The yield of an in vitro transcription ranged from 1012 to 1013 copies of synthetic RNA. Aliquots of >1010 copies were stored at −70°C with the addition of M2 phage RNA for stabilization. This allowed the storage of synthetic transcripts for up to 1 year. Working dilutions containing 108 copies and below are less stable and cannot be stored for quantitative experiments. Due to the high yield of the in vitro transcription process and the stability of concentrated synthetic RNAs, one batch of standard RNA is sufficient even for extensive research projects.
Range and sensitivity.
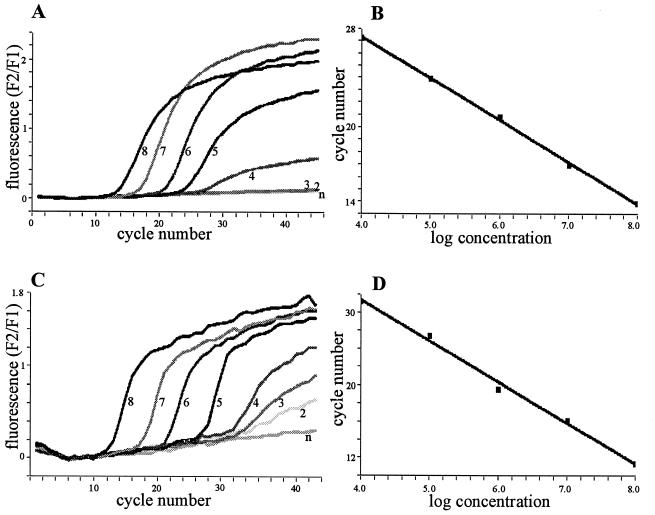
For competitive RT-PCR, constant amounts of samples are spiked with serial dilutions of the RNA standard. The amount of expected transcript must be roughly estimated prior to the actual experiment (for instance in reference to bacterial counts or total RNA) to stay within the appropriate working range of the assay. The LightCycler RT-PCR allowed the quantification of transcripts differing by over five orders of magnitude within one run (Fig. 1B and D). This broad range permitted analysis without the need for testing several dilutions of the sample.
FIG. 1.
Detection of the synthetic homologous RNA standards for gyr (A and B) and for hla (C and D) using the LightCycler technique. (A and C) The amplification of 108 (8), 107 (7), 106 (6), 105 (5), 104 (4), 103 (3), and 102 (2) copies of the RNA standard and of the negative control (n). (B and D) The generated standard curves.
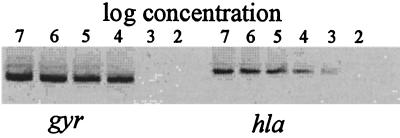
The sensitivity of both methods was comparable; for the synthetic RNAs, 104 copies of gyr (Fig. 1A and Fig. 2) and 103 copies of hla (Fig. 1C and Fig. 2) were still detected. The sensitivity of the conventional RT-PCR is limited by the ability to stain the products with ethidium bromide in the agarose gel. For the LightCycler RT-PCR, a sensitivity limit of 10 copies of human cytokine RNA is asserted by the manufacturer. The discrepancy observed may be due to the AT-rich RNA derived from S. aureus, which made it necessary to run the PCR reaction with low annealing temperatures. In contrast, the detection limit for some viral or human RNA was 10 copies (unpublished data).
FIG. 2.
Detection of the specific RNA standards for gyr and hla using competitive RT-PCR. Serial dilutions (107 to 102 copies) of each RNA standard were amplified.
Direct analysis of bacterial transcripts during infection.
For the direct analysis of transcripts during infection, S. aureus RNA was prepared from specimens without subculturing of the bacteria. Sputa from CF patients chronically infected with S. aureus were chosen to monitor bacterial gene expression and regulation during the infection process in the human host. Using competitive RT-PCR, we detected the specific expression of virulence genes in vivo (2). Transcription of the highly regulated hla gene ranged from 1.5 to 6 copies of hla/copy of gyr in the sputa. Analyzing the samples with LightCycler RT-PCR yielded the same relative results (0.3 to 5.5 copies of hla/copy of gyr).
In order to investigate a different type of infection, we also determined S. aureus gene expression in an animal model of foreign-body infection. Tissue cages were infected with 105 CFU of S. aureus/ml, and the exudate was aspirated. In our standard method of RNA preparation, bacteria are disrupted in a phenol reagent, and RNA is precipitated after chloroform extraction (2). Whereas this procedure was sufficient to detect low amounts of sample RNA using competitive RT-PCR, we found that especially the exudates containing blood inhibited the RT-PCR using the LightCycler. Consequently, an additional purification step using a silica matrix had to be incorporated into the RNA preparation. In general the LightCycler RT-PCR is more susceptible even to traces of inhibitors, such as hemine, ethanol, etc. Therefore, RNA has to be purified carefully and thoroughly. However, when highly purified RNA samples were used, both methods yielded comparable results in the foreign-body infection as well.
Reproducibility.
Competitive RT-PCR was successfully applied to the analysis of in vivo transcription with reproducible results (2). In order to analyze the reproducibility of the LightCycler RT-PCR method, we quantified sample RNA from exudates of the foreign-body infection independently on four different occasions. For example, determination of gyr in a given sample resulted in 1.4 × 106, 2.1 × 106, 2.4 × 106, and 1.7 × 106 copies (mean, 1.9 × 106 ± 4.7 × 105 copies). Reliable determination is ensured, with less than a twofold difference between the different RT-PCRs in the number of copies detected. However, we recommend repeating each run at least once based on our observation that even small variations in the RT-PCR set-up can result in profound discrepancies.
For further validation we also prepared RNA from the same exudates independently four times. For example, the concurrent determination of gyr in four different preparations from one specimen resulted in 5.6 × 106, 5.2 × 106, 6.0 × 106, and 5.2 × 106 copies (mean, 5.5 × 106 ± 4.0 × 105 copies). Hence, the variation between different RT-PCR runs was higher than between different RNA preparations quantified in one run. In summary, transcript quantification by LightCycler can be successfully applied to gain insight into temporal bacterial gene expression in vivo if performed carefully.
Practicability.
Both methods are useful for direct quantification of bacterial transcripts during infection, but both need experienced personnel since commercial kits for expression analysis in bacteria are not yet available. The need to analyze serial dilutions in the competitive RT-PCR considerably limits the number of samples which can be processed on any given day. The whole procedure takes at least 8 h of hands-on time, including agarose gels and evaluation of the results. In contrast, 25 samples (plus 5 standard RNA dilutions and 2 controls) can be analyzed in the LightCycler in one run, and the results are available within 2 h.
The evaluation of the results from competitive RT-PCR depends on the accurate quantification of band intensity, either visually or densitometrically. This quantification is hampered by variabilities caused by the individual interpreter. In contrast, with LightCycler RT-PCR, the results are calculated directly by the integrated software. However, one is tempted to overlook principal sources of error such as unspecific amplification. Additional amplicons may be present which do not hybridize to the fluorescence-labeled probes but compete with the specific PCR. Therefore, the PCR products have to be analyzed on agarose gels and, in cases of multiple bands, the PCR conditions should be optimized to avoid false priming.
One major drawback of the LightCycler system using the hybridization probe format is that there is no way to discriminate between accumulated PCR products derived either from contaminating standard RNA or from the target RNA. Therefore, extreme care must be taken to avoid any contamination of the samples with concentrated RNA standard solutions. In this study the described modified RNA standards allowed us to visualize a suspected contamination by gel analysis of the LightCycler amplicons.
ACKNOWLEDGMENTS
We thank U. Flückiger for providing the specimens from the animal model of foreign-body infection and D. Blaurock for critically reading the manuscript.
This work was supported by grants from Fortüne (No. 688-0-0) and the Deutsche Forschungsgemeinschaft (Wo 578/3-1 and Wo 578/3-2).
REFERENCES
- 1.Cheung A L, Eberhardt K J, Fischetti V A. A method to isolate RNA from gram-positive bacteria and mycobacteria. Anal Biochem. 1994;222:511–514. doi: 10.1006/abio.1994.1528. [DOI] [PubMed] [Google Scholar]
- 2.Goerke C, Campana S, Bayer M G, Döring G, Botzenhart K, Wolz C. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun. 2000;68:1304–1311. doi: 10.1128/iai.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathews S A, Volp K M, Timms P. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of sigma factors. FEBS Lett. 1999;458:354–358. doi: 10.1016/s0014-5793(99)01182-5. [DOI] [PubMed] [Google Scholar]
- 4.Mekalanos J J. Environmental signals controlling expression of virulence determinants in bacteria. J Bacteriol. 1992;174:1–7. doi: 10.1128/jb.174.1.1-7.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mergny J L, Boutorine A S, Garestier T, Belloc F, Rougee M, Bulychev N V, Koshkin A A, Bourson J, Lebedev A V, Valeur B, et al. Fluorescence energy transfer as a probe for nucleic acid structures and sequences. Nucleic Acids Res. 1994;22:920–928. doi: 10.1093/nar/22.6.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlando C, Pinzani P, Pazzagli M. Developments in quantitative PCR. Clin Chem Lab Med. 1998;36:255–269. doi: 10.1515/CCLM.1998.045. [DOI] [PubMed] [Google Scholar]
- 7.Selvin P R. Fluorescence resonance energy transfer. Methods Enzymol. 1995;246:300–334. doi: 10.1016/0076-6879(95)46015-2. [DOI] [PubMed] [Google Scholar]
- 8.Storey D G, Ujack E E, Rabin H R. Population transcript accumulation of Pseudomonas aeruginosa exotoxin A and elastase in sputa from patients with cystic fibrosis. Infect Immun. 1992;60:4687–4694. doi: 10.1128/iai.60.11.4687-4694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang A M, Doyle M V, Mark D F. Quantitation of mRNA by the polymerase chain reaction [published erratum appears in Proc Natl Acad Sci USA 1990 Apr;87(7):2865] Proc Natl Acad Sci USA. 1989;86:9717–9721. doi: 10.1073/pnas.86.24.9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerli W. Experimental models in the investigation of device-related infections. J Antimicrob Chemother. 1993;31(Suppl D):97–102. doi: 10.1093/jac/31.suppl_d.97. [DOI] [PubMed] [Google Scholar]
- 11.Zimmermann K, Mannhalter J W. Technical aspects of quantitative competitive PCR. BioTechniques. 1996;21:268. doi: 10.2144/96212rv01. [DOI] [PubMed] [Google Scholar]