Abstract
Bovine immunodeficiency virus (BIV) and Jembrana disease virus (JDV) are bovine lentiviruses that are closely related genetically. A recombinant fusion protein containing the capsid protein of BIV expressed in Escherichia coli was used to immunize mice and produce monoclonal antibodies. Six hybridomas specific for BIV capsid protein were identified, and one antibody, designated 10H1, was characterized further. Competitive binding assays were performed to analyze the topography of antigenic determinants by enzyme-linked immunosorbent assay and demonstrated the existence of at least three distinct antigenic determinants on capsid protein. The monoclonal antibody reacted specifically with both BIV capsid and the recombinant fusion protein in Western immunoblot analyses. However, it did not react with the recombinant capsid fusion protein of JDV, indicating that BIV contains at least one unique epitope in the capsid protein that is absent in JDV. Further mapping of the epitope by chemical cleavage analysis identified that the epitope is located at the 6.4-kDa N terminus of the 29-kDa capsid protein. This monoclonal antibody assay will be valuable for distinguishing the two closely related lentiviruses by Western blotting.
Bovine immunodeficiency virus (BIV) and Jembrana disease virus (JDV) are bovine lentiviruses. The BIV originally was isolated from cattle with lymphocytosis, lymphadenopathy, neuropathy, and progressive emaciation (11, 26). However, overt clinical disease in seropositive cattle is rare, and the infection is difficult to reproduce experimentally (5, 9, 25, 27, 29). Antibodies to BIV have been detected in beef and dairy cattle in the United States, some European countries, Australia, and New Zealand (1, 15, 16, 24, 28, 30). The JDV is a relatively new member of the Lentivirus family (6, 17). It causes Jembrana disease, an acute and sometimes fatal disease of domesticated banteng or Bali cattle that is endemic in parts of Indonesia (13). It also causes a milder disease syndrome in Bos taurus cattle (23).
Both BIV and JDV resemble human immunodeficiency virus in their structural, genomic, antigenic, and biological properties (6). Among the three major structural proteins—gag, pol, and env—gag protein developed the earliest and strongest antibodies in infected animals (27). The gag precursor of BIV has been shown to have a molecular mass of 53 kDa and can be processed into three smaller proteins, p17 (matrix), p26 (capsid), and p15 (nucleocapsid) (19, 20). Because the capsid protein is a major structural and immunodominant protein, the recombinant capsid protein can be used as an antigen source to detect animals infected by BIV.
BIV is closely related to JDV based upon nucleotide sequence homology (6, 10). The gag gene similarity was approximately 62% at the amino acid level, and the capsid protein had a high amino acid identity to that of JDV at 75% (7). Conservation of antigenic epitopes of this protein is broad within the lentiviruses, and cross-reactivity of sera from BIV-infected cattle against JVD recombinant capsid protein has been reported (7).
Monoclonal antibodies have been used successfully for detection of many viruses, including lentiviruses (8). Because each monoclonal antibody is made against a single epitope (14, 18, 22) a monoclonal antibody produced against a unique epitope possibly could be used to distinguish between two closely related lentiviruses. This study describes the production of such a monoclonal antibody against BIV recombinant capsid protein and mapping within the BIV capsid protein of the unique BIV antigenic epitope that is absent in JDV.
MATERIALS AND METHODS
Cell.
The myeloma cells P3X63Ag8.653 were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum, l-glutamine, nonessential amino acids, sodium pyruvate, vitamins (Gibco BRL, Grand Island, N.Y.), and antibiotics (penicillin [100 U/ml] and streptomycin [0.1 mg/ml]).
Expression and purification of recombinant BIV and JDV gag proteins.
Two different BIV constructs expressing capsid proteins were used in this experiment: pATH and pQE32. The pATH capsid construct was used for the production of monoclonal antibody. The clone containing a 0.8-kb capsid gene from the R29 strain of BIV was provided kindly by B. Atkinson from the University of Nebraska, Lincoln (2). The capsid protein was expressed as a 67-kDa fusion protein to the TrpE protein. Another capsid expression vector, pQE32, was constructed recently in our laboratory (31) and contains the same 0.8-kb capsid insert as the pATH vector. This construct expressed a 29-kDa capsid protein with a small fusion of 13 amino acid residues at the N terminus. The JDV capsid construct, JCA, containing a 0.8-kb capsid insert (the same region as the BIV capsid), which expressed a 58-kDa fusion protein to glutathione-S-transferase, was provided kindly by E. J. Burkala from Murdoch University, Murdoch, Australia (4). Purification was achieved using affinity chromatography via immobilized reduced glutathione (4).
The procedure of Atkinson et al. (2) was used to express the capsid fusion protein from the pATH construct in Escherichia coli. Briefly, E. coli strain RR1 with a pATH expression vector containing BIV capsid gene was grown in a PATH medium with tryptophan for 10 h. The culture then was inoculated into a fresh PATH medium without tryptophan. After 1 h of growth with shaking, β-indoleacrylic acid was added, and the culture was grown for another 3 h with vigorous shaking to induce expression of the recombinant protein. Bacterial cells were pelleted and suspended in a solution containing lysozyme (2 mg/ml) and 10% Nonidet P-40. After incubation at room temperature for 20 min, the cell suspension was sonicated, pelleted, and resuspended in 10 mM Tris-HCl for storage at 4°C. Recombinant gag proteins were purified by electrophoresis and electroelution as described by Atkinson et al. (2). The expressed proteins were separated on a preparative sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel. A clear, thick, white protein band with a molecular mass of 67 kDa, visualized by soaking in 1 M potassium acetate, was excised and electroeluted using a Bio-Rad (Richmond, Calif.) Electro Eluter. The concentration of the eluted protein was determined by a dye-binding protein assay (Bio-Rad).
Expression and purification of the capsid protein from pQE32 were carried out as follows. The construct was maintained in E. coli M15, and expression of the gag protein was induced with 2 mM isopropylthio-β-d-galactoside (IPTG) for 4 h. E. coli strain M15 was transformed with a pQE32 expression vector containing the BIV gag gene or with plasmid pQE32 alone as negative control. Cells were grown overnight at 37°C in 1.5 ml of Luria-Bertani broth containing both ampicillin and kanamycin. The cultures were transferred to fresh Luria-Bertani broth (1:4 dilution) and incubated for 30 min at 37°C; then, IPTG was added to a final concentration of 2 mM for protein induction. After 4 h of induction, cells were harvested by centrifugation at 4,000 × g for 15 min and lysed by sonication in lysis buffer B (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl [pH 8.0]). The recombinant capsid proteins (29 kDa) were analyzed on an SDS–12.5% PAGE and purified on Ni-nitrilotriacetic acid columns (Pierce, Rockford, Ill.). The concentration of the purified protein was determined by a dye-binding protein assay (Pierce). The gag protein then was divided into aliquots and stored at −20°C until used.
Production of monoclonal antibodies.
Seven-week-old BALB/c mice were immunized intraperitoneally with 35 μg of purified recombinant capsid protein per mouse with RIBI adjuvant (RIBI Immunochem Research Inc., Hamiton, Mont.). A booster was given 2 weeks later. One week after the booster, mice were bled and tested for BIV capsid protein-specific antibody by a Western blot assay, and then antibody-positive mice received another booster. The mice were sacrified 3 days after the last injection, and their spleens were removed aseptically and minced to make single-cell suspensions. Spleen cells were fused with myeloma cells at a ratio of 2:1 using 50% polyethylene glycol 1450 (American Type Culture Collection [ATCC], Manassas, Va.). Cells were resuspended in myeloma cell culture medium supplemented with 10% fetal calf serum, 100 μmol of hypoxanthine per liter, 0.4 μmol of aminopterin per liter, and 16 μmol of thymidine per liter. Seven days after incubation, one half of this medium was removed and replaced with medium containing only hypoxanthine and thymidine. Supernatants from growing hybridoma cells were tested for reactivity to purified recombinant BIV gag protein by Western blotting. Positive hybridomas were recloned twice by limited dilution in medium, selected hybridoma cells were injected intraperitoneally into mice, and ascitic fluids were collected 10 days after injections.
Biotin-labeled monoclonal antibody.
Biotin-labeled monoclonal antibodies were prepared by the method of Borrow and Oldstone (3). Briefly, monoclonal antibodies from ascitic fluid were purified on a protein A affinity column (Pierce). Then, 10 mg of monoclonal antibody was mixed with 2 mg of N-hydroxysuccinimidobiotin (Sigma Chemical Co., St. Louis, Mo.) in 0.01 M carbonate buffer (pH 9.6) and incubated overnight at 4°C. The mixture was dialyzed against 0.02 M phosphate-buffered saline (PBS). The biotin-conjugated antibody was separated from hydroxysuccinimidobiotin and from antibody that had not formed antibody-biotin complexes by passing the mixture through a Sephadex G-200 column.
Competitive binding assay.
The competitive binding assay was performed by enzyme-linked immunosorbent assay (ELISA). The purified recombinant capsid protein was used as an antigen. Serial dilutions of each competing antibody from ascitic fluid were added to the capsid-adsorbed wells in microplates. The ascitic fluid against bovine viral diarrhea virus was used as the control. After standing for 2 h at 37°C, the plates were washed four times with PBS-Tween 20, and biotinylated labeled monoclonal antibody, which had an optical density at 405 nm [OD405] of 1.0, was added. Plates again were washed four times with PBS-Tween buffer, a 1:5,000 dilution of avidin-peroxidase-conjugated goat anti-mouse immunoglobulin G (IgG) (Kirkegaard & Perry Laboratory, Gaithersburg, Md.) was added, and the plates were incubated at 37°C for 30 min. The plates then were washed, and ABTS [2,2′-azinobis (3-ethylbenzthiazolinesulfonic acid)] substrate (Kirkegaard & Perry Laboratory) was added to each well. The plates were incubated at 37°C for 30 min, and the absorbance was measured at 405 nm. The percentage of competitive inhibition was calculated as described by Kimura-Kuroda and Yasui (18) using the formula 100 × [(A − B)/(A − C)], where A is the OD in the absence of competitor monoclonal antibody, B is the OD in the presence of competitor monoclonal antibody, and C is the OD in the presence of homologous antibody.
Isotype characterization of monoclonal antibodies.
The immunoglobulin class and subclass were determined by ELISA using a Mouse Sub-isotyping kit (Bio-Rad). Monoclonal antibody at a constant concentration was adsorbed to the BIV capsid protein-coated wells of microtiter plates. Each class-specific antiserum was used to compete with the binding of peroxidase-conjugated antimouse gamma globulin. Isotypes of the monoclonal antibodies were determined by the reduction in OD405 compared with the value for an unblocked control.
Statistical analysis.
The competitive monoclonal antibody inhibition rates (Table 1) were analyzed using the Statistical Analysis System (PC-SAS; SAS Institute Inc., Cary, N.C.). Analysis of variance with means variance using least-significant difference determines which difference among the inhibition rates was considered to be real.
TABLE 1.
Epitope differentiation by ELISA
| Biotinylated MAba | Competitive MAb inhibition rate (%)
|
|||||
|---|---|---|---|---|---|---|
| 1H10 | 1B11 | 10F10 | 7G7 | 6B4 | 5G6 | |
| Group 1 | ||||||
| 1H10 | 100b | 85.8 | 13.5 | 9.2 | 10.1 | 5.0 |
| 1B11 | 82.1 | 100 | 12.0 | 11.4 | 14.2 | 2.8 |
| Group 2 | ||||||
| 10F10 | 15.0 | 10.0 | 100 | 60.0 | 20.4 | 10.0 |
| 7G7 | 10.1 | 14.9 | 64.5 | 100 | 10.9 | 12.9 |
| Group 3 | ||||||
| 6B4 | 8.9 | 16.4 | 15.8 | 14.2 | 100 | 64.5 |
| 5G6 | 3.6 | 11.2 | 15.4 | 13.6 | 59.2 | 100 |
MAb, monoclonal antibody.
Numbers in boldface type indicate percent inhibition between antibodies from the same group.
Western blotting.
The purified capsid protein was loaded onto an SDS–12% PAGE gel (Bio-Rad) and electrophoresed in a Tris-glycine buffer (0.025 M Tris base, 0.192 M glycine, 0.1% SDS) or SDS–16% PAGE gel in a Tricine buffer at 30 mA/gel for 1 h. The 16% gel was used to separate smaller proteins after chemical cleavage. The protein then was transferred onto a nitrocellulose membrane (0.45-μm pore size) with a Transblot apparatus (Bio-Rad) at 259 mA for 1 h. The membrane was blocked with 2% bovine albumin, 0.02 M Tris base, 0.385 M NaCl, and 0.1% Tween 20 (pH 7.5) (TTBS) at room temperature for 2 h and rinsed three times with TTBS. The first antibody was diluted with TTBS (monoclonal antibody diluted 1:1,000 and bovine serum diluted 1:50) and incubated overnight at 4°C. After being washed three times with TTBS, the membranes were incubated with horseradish peroxidase-labeled goat anti-bovine IgG (heavy and light chain) for bovine serum or anti-mouse IgG for monoclonal antobody (1:3,000) (Kirkegaard & Perry Laboratory) at room temperature for another 2 h and washed twice with TTBS and once with TBS (0.02 M Tris base, 0.385 M Nacl [pH 7.5]). Finally, color was developed with a 4-chloro-naphthol substrate solution (100 ml of TBS, 60 μl of H2O2, 20 ml of ice-cold methanol, and 60 μg of of 4-chloro-naphthol [Pierce]) at room temperature for 15 min.
Chemical cleavage of capsid protein by acid and cyanogen bromide.
The purified capsid protein (29 kDa) from pQE32 construct was cleaved chemically with mild acid and cyanogen bromide (CNBr) to define the region of the unique epitope for BIV (14). Five micrograms of the purified capsid protein was dissolved in 50 μl of 75% formic acid and incubated overnight at 37°C. The acid was evaporated under a nitrogen stream, and the peptides were resuspended and dried three times, with water being used to remove residual acid. Another 5 μg of the protein was resuspended in 50 μl of 70% formic acid containing CNBr (10 mg/ml), and the sample was incubated overnight at room temperature. The CNBr was evaporated under a stream of nitrogen, and the protein was washed with water as described above. The cleaved samples were resuspended in 1X SDS-PAGE sample buffer and separated on a 16% discontinuous Tricine gel (21). The gel was stained with 0.1% Coomassie brilliant blue in 11.9% ethanol and 5% glacial acetic acid and transferred to membrane for Western blotting.
RESULTS
Formation of antibody-producing hybridoma.
At 10 to 14 days after fusion, culture fluids from 41 of 216 wells of microplates showed positive activity by ELISA (19%). After recloning and freezing, six stable hybridomas were selected and intraperitonelly injected into BALB/c mice to produce monoclonal antibodies in ascitic fluid. ELISA showed that the average antibody titer in ascitic fluid was 1:105, whereas that in cell culture supernatants ranged from 1:102 to 1:103. All monoclonal antibodies reacted positively with the recombinant capsid protein as well as native capsid protein from BIV in Western blots. All antibodies were determined to be of κ light chain and IgG1 heavy chain.
Topographical analysis of antigenic determinants on capsid protein.
Competitive binding assays were used to analyze the topography of the capsid protein to which the monoclonal antibodies bound. If two epitopes are close each other, the binding of antibody to one of the epitopes will sterically hinder binding of the competitive antibody to the other epitope. Competition of antibody binding was caused by another antibody, which defined the antigenic determinant. To analyze epitopes of capsid protein, a competitive binding assay was performed with six biotin-labeled monoclonal antibodies: clones 1H10, 1B11, 10F10, 7G7, 6B4, and 5G6. The six monoclonal antibodies were tested for competition with the binding of each biotin-labeled antibody. The binding assays were repeated three times, and the average competitive monoclonal antibody inhibition rates are summarized in Table 1. The monoclonal antibodies were classified into three antigenic groups based on the binding assay. The group 1 clones, 1H10 and 1B11, were nearly completely blocked by each other (82 to 85%). Antibodies from other groups cannot compete as well; only between 2.8 to 12% competition by the other antibodies was detected. The group 2 clones, 10F10 and 7G7, were blocked reciprocally (60%) and were not blocked by other antibodies tested. The group 3 clones, 6B4 and 5G6, also were blocked reciprocally (about 60%) (Table 1). A statistical analysis was performed on the inhibition rates in Table 1 using least-significant difference on variance with means separation. The inhibition rates were found to be significantly different at the 0.01 level among the three groups, and no significant difference was observed within each group. None of the monoclonal antibodies reacted with the recombinant capsid protein containing only the 87 C-terminal amino residues of the capsid, indicating that all three antigenic sites were located at the N terminus and not at the C terminus of the capsid protein.
Immunological assay of JDV recombinant proteins with BIV monoclonal antibody.
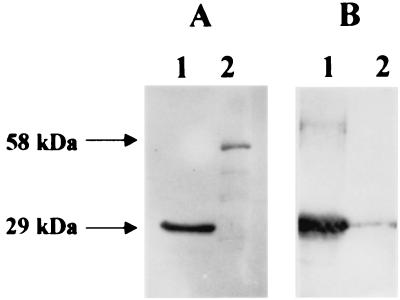
The recombinant BIV capsid (29 kDa) and JDV capsid (58 kDa) proteins were both detected with BIV polyclonal antisera in Western blots (Fig. 1A). No immunoreactive band was observed in the proteins prepared from the vector alone, indicating that the antisera reacted specifically to the recombinant proteins. The BIV monoclonal antibody, 1H10, detected only the recombinant BIV capsid protein and not the JDV recombinant capsid protein (Fig. 1B, lane 2) even though the two expressed proteins were from the same region. Another antibody in the same group, 1B11, reacted with the same pattern with 1H10. This result suggests that the monoclonal antibody was raised against an antigenic epitope that is unique to BIV capsid and is absent in the JDV capsid.
FIG. 1.
Differentiation of BIV and JDV gag protein by immunoblotting. The recombinant capsid proteins of BIV and JDV were reacted with polyclonal antibodies of BIV (A) and monoclonal antibody against BIV capsid (B). The capsid BIV and JDV were expressed as 29- and 58-kDa proteins, respectively (lanes 1 and 2).
Localization of a unique epitope site in BIV capsid protein.
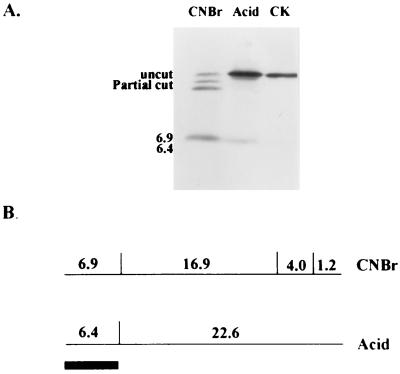
To locate the unique epitope for BIV, the purified 29-kDa recombinant capsid protein was subjected to cleavage by mild acid and CNBr treatments, followed by Tricine gel electrophoresis and Western blotting. The peptide bond that links aspartic and proline residues is susceptible to cleavage under mild acidic conditions. The capsid contains a single aspartate-proline bond at amino acid 60 (from the capsid N terminus), and cleavage at that site within the capsid protein yielded an amino-terminal 6.4-kDa fragment and a 22.6-kDa carboxyl-terminal fragment (Fig. 2B). Only the 6.4-kDa fragment at the N terminus of the protein reacted positively with the monoclonal antibody (Fig. 2A). The cleavage with CNBr occurred at three methionine residues in the capsid protein resulting in four bands: 6.9, 16.9, 4.0, and 1.2 kb (from the N terminus to C terminus) (Fig. 2B). However, the CNBr digestion gave two extra partially digested bands: 23.8 and 27.8 kb. Both bands contain a 6.9-kb N-terminal fragment. Figure 2A shown that only the 6.9-kDa fragment at the N terminus and the two partially digested bands which contain the 6.9-kDa fragment reacted positively with the monoclonal antibody. The apparent molecular mass values agreed well with those predicted on the basis of the known BIV sequence (12). The cleavage results confirmed that the unique epitope site is within the 6.4-kDa fragment located at the N terminus of the BIV capsid protein. Comparison of the amino acid sequences (6, 12) in capsid proteins of BIV and JDV has revealed that the identity at the first 60-amino-acid region is only 25%, compared to 75% in the rest of the capsid region (Fig. 3).
FIG. 2.
Chemical cleavage of the capsid BIV protein and identification of the positive reaction bands by monoclonal antibody. (A) Western blot of acid and CNBr-cleaved capsid BIV. Five micrograms of capsid BIV protein was incubated overnight with 75% formic acid at 37°C. Another 5 μg was incubated overnight at room temperature with CNBr (10 mg/ml) in 70% formic acid. All samples were solubilized in SDS-PAGE sample buffer and analyzed by Tricine gel electrophoresis. The molecular mass (in kilodaltons) of each of the peptides is indicated. CK, uncut control; uncut, uncleaved capsid protein; Partial cut, partially digested fragments. (B) Physical map of the 29-kDa BIV capsid protein. The location of the single Asp-Pro linkage cleavable by acid and the molecular masses (in kilodaltons) of the two fragments are indicated; so are the locations of three methionine residues and the molecular masses of the four fragments generated by the CNBr cleavage. The positions of the peptides that reacted positively are indicate by the filled box.
FIG. 3.
Alignment of amino acid sequences of BIV and JDV capsid proteins. Peptide sequences were aligned by using the Genetics Computer Group GAP program. Vertical lines indicate amino acids that are conserved in the two sequence.
DISCUSSION
The amino acid sequences of capsid proteins are highly similar among all lentiviruses. Finding an epitope that is unique to only one virus has been difficult. The recombinant capsid protein of JDV has been found to cross-react with BIV polyclonal antisera. This result was expected, because previous Western immunoblot analyses using native JDV proteins showed antigenic cross-reactivity with BIV antisera and vice versa, indicating that the capsid proteins of these two bovine lentiviruses share common antigenic epitopes (17). The lentivirus capsid proteins contain a conserved epitope, the major homology region, to which the antigenic cross-reactivity of lentiviruses can be attributed.
The present study produced monoclonal antibodies to detect virus-specific capsid protein. Three antigenic epitopes within the capsid protein were identified using these monoclonal antibodies. Negative reactions of C-terminal amino residues of the capsid protein with all the antibodies further indicated that the antigenic sites were located on the N terminus of the protein. Previous studies using capsid deletion mutant and polyclonal antibody identified one major epitope located near the carboxyl terminus of the capsid protein (2). Thus, the capsid protein appears to have at least four epitopes. Because as few as 18 amino acid can make up one epitope, the 240-amino-acid capsid protein could potentially have more than 10 epitopes.
The monoclonal antibody produced in this study reacts specifically to BIV, but not JDV, and the unique epitope identified was mapped to the N terminus of the capsid protein. This monoclonal antibody provides a useful tool for distinguishing between these two closely related lentiviruses, BIV and JDV. To our knowledge, this is the first report to demonstrate the production of monoclonal antibody against a unique epitope at the N terminus of the BIV capsid protein that is absent in JDV.
Footnotes
Contribution 00-362-J from the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Amborski G F, Lo J L, Seger C L. Serological detection of multiple retroviral infection in cattle: bovine leukemia virus, bovine syncytial virus and bovine visna virus. Vet Microbiol. 1989;20:247–253. doi: 10.1016/0378-1135(89)90048-5. [DOI] [PubMed] [Google Scholar]
- 2.Atkinson B, Liu Z Q, Wood C. Use of bacterial trpE fusion vectors to express and characterize the bovine immunodeficiency like virus core protein. J Virol Methods. 1992;36:35–39. doi: 10.1016/0166-0934(92)90155-7. [DOI] [PubMed] [Google Scholar]
- 3.Borrow P, Oldstone B A. Characterization of lymphocytic choriomeningitis virus-binding protein(s): a candidate cellular receptor for virus. J Virol. 1992;66:70–81. doi: 10.1128/jvi.66.12.7270-7281.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burkala E J, Narayani I, Hartaningsih N, Kertayadnya G, Berryman D, Wilcox G E. Recombinant Jembrana disease virus proteins as antigens for the detection of antibody to bovine lentiviruses. J Virol Methods. 1998;74:39–46. doi: 10.1016/s0166-0934(98)00066-4. [DOI] [PubMed] [Google Scholar]
- 5.Carpenter S, Miller L D, Alexanderson S, Whetstone C A, Van Der Maaten M J, Viuff B, Wannemuehler Y, Miller J M, Roth J A. Characterization of early pathogenic effects after experimental infection of calves with bovine immunodeficiency-like virus: J. Virol. 1992;66:1074–1083. doi: 10.1128/jvi.66.2.1074-1083.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chadwick B J, Coelen R J, Sammels L M, Kertayadnya G, Wilcox G E. Genomic sequence analysis identifies Jembrana disease virus as a new bovine lentivirus. J Gen Virol. 1995;76:189–192. doi: 10.1099/0022-1317-76-1-189. [DOI] [PubMed] [Google Scholar]
- 7.Chadwick B J, Coelen R J, Wilcox G E, Sammels L M, Kertayadnya G. Nucleotide sequence analysis of Jembrana disease virus: a bovine lentivirus associated with an acute disease syndrome. J Gen Virol. 1995;76:1637–1650. doi: 10.1099/0022-1317-76-7-1637. [DOI] [PubMed] [Google Scholar]
- 8.Chesebro B, Wehrly K. Development of sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaming K, Van Der Maaten M, Whetstone C, Carpenter S, Frank D, Roth J. Effect of bovine immunodeficiency-like virus infection on immune function in experimentally infected cattle. Vet Immunol Immunopathol. 1993;36:91–105. doi: 10.1016/0165-2427(93)90100-i. [DOI] [PubMed] [Google Scholar]
- 10.Garvey K J, Oberste M S, Elser J E, Braun M J, Gonda M A. Nucleotide sequence and genome organization of biologically active proviruses of the immunodeficiency-like virus. Virology. 1990;175:391–409. doi: 10.1016/0042-6822(90)90424-p. [DOI] [PubMed] [Google Scholar]
- 11.Gonda M A. Bovine immunodeficiency virus. AIDS. 1992;6:759–776. doi: 10.1097/00002030-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 12.Gonda M A, Brau M J, Carter S G, Kost T A, Bess Jr J W, Arthur L O, Van Der Maaten M J. Characterization and molecular cloning of a bovine lentivirus related to human immunodeficiency virus. Nature. 1987;330:388–391. doi: 10.1038/330388a0. [DOI] [PubMed] [Google Scholar]
- 13.Hartaningsih N, Wilcox G E, Kertayadnya G, Astawa M. Distribution of Jembrana disease in cattle in Indonesia. Vet Microbiol. 1993;38:23–29. doi: 10.1016/0378-1135(93)90072-f. [DOI] [PubMed] [Google Scholar]
- 14.Higman M A, Niles E G. Location of the S-adenosyl-l-methionine binding region of the vaccinia virus mRNA (guanine-7-) methyltransferase. J Biol Chem. 1994;269:14982–14987. [PubMed] [Google Scholar]
- 15.Horner G W. Serological evidence of bovine immunodeficiency-like virus and bovine syncytial virus in New Zealand. Surveillance. 1991;18:9–14. [Google Scholar]
- 16.Horzinek M, Keldermans L, Stuurman T, Black J, Herrewegh A, Sillekens P, Koolen M. Bovine immunodeficiency virus: immunochemical characterization and serological survey. J Gen Virol. 1991;72:2923–2928. doi: 10.1099/0022-1317-72-12-2923. [DOI] [PubMed] [Google Scholar]
- 17.Kertayadnya G, Wilcox G E, Soeharsono S, Hartaningsih N, Coelen R J, Cook R D, Collins M E, Brownlie J. Characteristics of a retrovirus associated with Jembrana disease in Bali cattle. J Gen Virol. 1993;74:1765–1773. doi: 10.1099/0022-1317-74-9-1765. [DOI] [PubMed] [Google Scholar]
- 18.Kimura-Kuroda J, Yasui K. Topographical analysis of antigenic determinants on envelope glycoprotein V3 (E) of Japanese encephalitis virus, using monoclonal antibodies. J Virol. 1983;45:124–132. doi: 10.1128/jvi.45.1.124-132.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lillehoj E P, Salazar H R, Mervis R J, Raum M G, Chan H W, Ahmad N, Venkatesan S. Purification and structural characterization of the putative gag-pol protease of human immunodeficiency virus. J Virol. 1988;62:3053–3058. doi: 10.1128/jvi.62.8.3053-3058.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mervis R J, Ahmad N, Lillehoj N. The gag gene products of human immunodeficiency virus type I: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schagger H, Jagow G V. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Smirnov I U A, Lipatov A S, Okuno I, Gitel'man A K. A common antigenic epitope in influenza A virus (H1, H2, H5, H6) hemagglutinin. Vopr Virusol. 1999;44:111–115. [PubMed] [Google Scholar]
- 23.Soeharsono S, Wilcox G E, Dharma D M N, Hartaningsih N, Kertayadnya G, Budiantono A. Species differences in the reaction of cattle to Jembrana disease virus infection. J Comp Pathol. 1995;112:391–402. doi: 10.1016/s0021-9975(05)80020-9. [DOI] [PubMed] [Google Scholar]
- 24.St. Cyr Coats K, Pruett S F, Nash J W, Cooper C R. Bovine immunodeficiency virus: incidence of infection in Mississippi dairy cattle. Vet Micorbiol. 1994;49:132–141. doi: 10.1016/0378-1135(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 25.Suarez D L, Van Der Maaten M J, Wood C, Whetstone C A. Isolation and characterization of new wild-type isolates of bovine lentivirus. J Virol. 1993;67:5051–5055. doi: 10.1128/jvi.67.8.5051-5055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Der Maaten M J, Boothe A D, Seger C L. Isolation of virus from cattle with persistent lymphocytosis. J Nat Cancer Inst. 1972;49:1649–1657. doi: 10.1093/jnci/49.6.1649. [DOI] [PubMed] [Google Scholar]
- 27.Whetstone C A, Van Der Maaten M J, Black J W. Humoral immune response to the bovine immunodeficiency-like virus in experimentally and naturally infected cattle. J Virol. 1990;64:3557–3561. doi: 10.1128/jvi.64.7.3557-3561.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whetstone C A, Van Der Maaten M J, Miller J M. A western blot assay for the detection of antibodies to bovine immunodeficiency-like virus in experimentally inoculated cattle, sheep, and goats. Arch Virol. 1991;116:119–131. doi: 10.1007/BF01319236. [DOI] [PubMed] [Google Scholar]
- 29.Zhang S, Wood C, Xue W, Krukenberg S M, Minocha H C. Immune suppression in calves with bovine immunodeficiency virus. Clin Diagn Lab Immunol. 1997;4:232–235. doi: 10.1128/cdli.4.2.232-235.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang S, Xue W, Wood C, Kapil S, Minocha H C. Detection of bovine immunodeficiency virus antibodies in cattle by western blot assay with recombinant gag protein. J Vet Diagn Investig. 1997;9:347–351. doi: 10.1177/104063879700900401. [DOI] [PubMed] [Google Scholar]
- 31.Zheng L, Swanson M, Liao J, Wood C, Kapil S, Snider R, Loughin T A, Minocha H C. Cloning of bovine immunodeficiency virus gag gene and development of the recombinant protein-based enzyme-linked immunosorbent assay. Clin Diagn Lab Immunol. 2000;7:557–562. doi: 10.1128/cdli.7.4.557-562.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]