Abstract
Fusarium keratoplasticum is the Fusarium species most commonly associated with human infections (fusariosis). Antifungal treatment of fusariosis is often hampered by limited treatment options due to resistance towards azole antifungals. The mechanisms of antifungal resistance and sterol biosynthesis in fusaria are poorly understood. Therefore, in this study we assessed the transcriptional response of F. keratoplasticum when exposed to voriconazole. Our results revealed a group of dramatically upregulated ergosterol biosynthesis gene duplicates, most notably erg6A (912-fold), cyp51A (52-fold) and ebp1 (20-fold), which are likely part of an alternative ergosterol biosynthesis salvage pathway. The presence of human cholesterol biosynthesis gene homologs in F. keratoplasticum (ebp1, dhcr7 and dhcr24_1, dhcr24_2 and dhcr24_3) suggests that additional sterol biosynthesis pathways may be induced in fusaria under other growth conditions or during host invasion. Voriconazole also induced the expression of a number of ABC efflux pumps. Further investigations suggested that the highly conserved master regulator of ergosterol biosynthesis, FkSR, and the pleiotropic drug resistance network that induces zinc-cluster transcription factor FkAtrR coordinate the response of FSSC species to azole antifungal exposure. In-depth genome mining also helped clarify the ergosterol biosynthesis pathways of moulds and provided a better understanding of antifungal drug resistance mechanisms in fusaria.
Keywords: FSSC, cyp51, erg6, ebp1, SR, AtrR, ABC transporters, azole, antifungal resistance, ergosterol biosynthesis, cholesterol biosynthesis
1. Introduction
Fusarium keratoplasticum belongs to the Fusarium solani species complex (FSSC), a genetically diverse mould genus within the fungal order Hypocreales and Sordariomycetes class. Fusaria can be found in various ecological niches such as soil, water, plants, and man-made habitats. Fusaria are plant pathogens that cause significant diseases in a variety of agriculturally important crops [1,2,3]. Fusaria also frequently cause invasive mould infections (IMIs) in humans [4], and invasive fusariosis (IF) has a high mortality rate (56%) [5], especially in neutropenic patients (up to 100%) [6]. Infections caused by FSSC are problematic as most are innately resistant to commonly prescribed azole antifungals such as voriconazole (VRC) [7,8] which is one of the recommended treatment options for IF in addition to liposomal amphotericin B formulations or a combination of both [9,10,11]. Excellent guidelines for the diagnosis and management of rare mould infections, including IF, have recently been provided by the One World-One Guideline initiative [11]. Most FSSC species are susceptible to amphotericin B [7,8], but the use of this antifungal can cause side effects. Thus, there are few treatment options for immunocompromised patients with IF [12,13,14,15]. The widespread application of azole fungicides in agriculture [16] has resulted in the spread of azole-resistant Aspergillus fumigatus isolates [17] from the environment into hospital settings [18,19,20]. Some of these isolates exhibit resistance to multiple antifungals [21] mainly associated with cyp51A TR34/L98H or TR46/Y121F/T289A mutations [22,23] and, rather concerningly, they are now spreading globally [24]. A similar trend of reduced azole susceptibility in isolates of the Fusarium solani [25] and Fusarium fujikuroi [26] species complexes of agriculture and human origin has been recently reported.
Sterols play important roles in many eukaryotes including the maintenance of membrane fluidity, the regulation and distribution of membrane proteins, and the control of the cell cycle [27]. Ergosterol is the major sterol of fungal cell membranes, while the counterpart in mammalian cell membranes is cholesterol. In fungi, ergosterol depletion caused by inhibition of ergosterol biosynthesis with azole antifungals results in the accumulation of toxic sterol intermediates, disruption of cell membrane integrity and cell death [28]. Therefore, the regulation of sterol biosynthesis is essential to maintain various cellular processes and for cell survival. There are three types of transcription factors known to regulate ergosterol biosynthesis in fungi: (i) the sterol regulatory element binding proteins (SREBPs) [29] that regulate ergosterol biosynthesis in Agaricomycotina, Taphrinomcyotina and the Eurotiomycetes, orthologs of which also regulate cholesterol biosynthesis in Metazoa, including humans [30]; (ii) the sterol uptake control (Upc2) Zn2Cys6 cluster transcription factor orthologs that regulate ergosterol biosynthesis in Saccharomycotina yeasts [31,32]; and (iii) the sterol regulator (SR) Zn2Cys6 cluster transcription factor orthologs recently discovered in Fusarium graminearum and related moulds of the Sordariomycetes and Leotiomycetes lineages [33]. Interestingly, orthologs of the Zn2Cys6 cluster transcription factor named ABC transporter regulator (AtrR) were found to be key determinants of azole resistance in both the Eurotiomycetes species A. fumigatus [34,35] as well as the Sordariomycetes species F. graminearum [36]. This is somewhat surprising because ergosterol biosynthesis is regulated by SREBP and SR orthologs in A. fumigatus and F. graminearum, respectively. In fungi, all these transcription factors have been implicated in virulence and antifungal resistance.
Many antifungals target the ergosterol biosynthetic pathway. They include the azole class of antifungals that inhibit sterol 14α-demethylase encoded by cyp51, also known as ERG11 in many yeast species [27,37]. Yeast species usually have only one essential ERG11 gene, whereas moulds can have up to three cyp51 paralogs: Aspergillus fumigatus, A. niger and A. terreus, for example, have two (cyp51A, cyp51B) [37] and most Fusarium species have three cyp51 paralogs (cyp51A, cyp51B, cyp51C) [25,38]. An exception is Aspergillus flavus which also has three cyp51 paralogs including cyp51C [39]. The Cyp51C ortholog in Fusarium is required for invasion of plant tissue, but was suggested to have no sterol 14α-demethylase activity [38]. The yeast ortholog Erg11 (lanosterol-14α-demethylase) catalyses the C14-demethylation of lanosterol [27] while the two Cyp51 paralogs of moulds, Cyp51A and Cyp51B, are reported to use eburicol as their preferred substrate in A. fumigatus [40] and Mycosphaerella graminicola [41]. Alcazar-Fuoli and co-authors have reported that in A. fumigatus, lanosterol was first converted to eburicol by C24-methyltransferase, encoded by erg6, and then demethylated by Cyp51, which suggests that Cyp51 should instead be named eburicol-14α-demethylase [42,43]. It is evident that such misnomers often occur for mould genes because of their homology to genes in Saccharomyces cerevisiae, even though the ergosterol biosynthesis pathways in the two fungi differ [44].
As A. fumigatus is the most common cause of IMIs, it is to date the best studied mould in terms of antifungal resistance mechanisms. It has been shown that azole resistance involves modifications and/or overexpression of the azole drug target eburicol-14α-demethylase [37] and overexpression of ATP-binding cassette (ABC) drug efflux transporters [34,35,45]. In fusaria, however, azole resistance mechanisms are still poorly understood. We recently reported a close association between a deletion in the cyp51A promoter and VRC resistance in both clinical and environmental FSSC isolates from Malaysia. The association between the cyp51A promoter deletion and VRC resistance was independent of whether they were clinical or environmental isolates. Most intriguingly, however, this association held true across species boundaries with all VRC-resistant FSSC isolates (i.e., 3 F. keratoplasticum and 6 F. suttonianum isolates) possessing the 23 bp cyp51A promoter deletion [25]. We also recently reported a 372-fold induction of the mRNA levels of the pleiotropic drug resistance (PDR) transporter abc1 in response to VRC exposure [46]. When abc1 was expressed in S. cerevisiae it conferred up to 1024-fold increased resistance towards a range of xenobiotics including azoles [46].
The present study describes the transcriptional response of the clinically relevant FSSC species F. keratoplasticum to VRC exposure. The upregulation of a group of duplicated ergosterol biosynthesis genes and the annotation of all possible sterol biosynthesis homologs/orthologs in the sequenced genomes of related FSSC species enabled the proposal of an alternative ergosterol biosynthesis salvage pathway(s) that involve human cholesterol biosynthesis orthologs. Key players of the pleiotropic drug resistance network involved in azole antifungal drug resistance that seem conserved throughout the entire FSSC were also upregulated. The phylogenetic relationship of the manually curated ergosterol biosynthesis genes of the FSSC to their A. fumigatus and F. graminearum counterparts revealed possible alternative ergosterol biosynthesis pathways for these two model species as well.
2. Materials and Methods
2.1. Fungal Isolate and Culture Conditions
F. keratoplasticum strain 2781 is a clinical isolate obtained from a human nail as part of routine diagnostic procedures at the Hospital Canselor Tuanku Muhriz [25]. F. keratoplasticum 2781 showed high MICs to itraconazole (ITC; MIC > 32 mg/L) and VRC (MIC > 32 mg/L) [25]. Although antifungal clinical breakpoints have not yet been established for fusaria, epidemiological cut-off values have been defined using the CLSI method [7]. We have previously shown that exposure to high but sub-growth inhibitory concentrations of VRC (16 mg/L) caused increased expression of cyp51A [25] and the PDR transporter abc1 [46]. F. keratoplasticum strain 2781 was initially grown on potato dextrose agar, PDA (Merck & Co., Kenilworth, NJ, USA) at 28 °C for a week. The preparation of conidia suspensions, the growth and VRC induction, cell harvest and total RNA extraction were conducted as previously described [25]. In short, 50 mL potato dextrose broth, PDB (Merck & Co., Kenilworth, NJ, USA) was inoculated with 10 µL of F. keratoplasticum conidia suspension (∼1 × 108 cfu/mL) and cells were grown to mid-logarithmic phase for 21 h [47]. Then, cells were incubated for an additional 80 min in the presence or absence of VRC (16 mg/L) followed by cell harvest. VRC was added to the cell suspensions as a 100× stock (1.6 mg/mL) dissolved in DMSO and the control cells were incubated with an equal volume of DMSO only. The experiment included three biological replicates.
2.2. Total RNA Extraction and Sequencing
Total RNA extraction (hot phenol method), DNase treatment (PureLink DNase kit; Invitrogen Inc., Carlsbad, CA, USA) and quality checks of RNA samples by agarose gel electrophoresis were conducted as previously described [25]. Total RNA samples (3 each of VRC treated and untreated control samples) were submitted to Genewiz Biotechnology Co. Ltd. (now Azenta Life Science), Suzhou, China, for transcriptome sequencing. Sample quality control checks were performed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and a NanoDrop spectrophotometer (Thermo Fisher Scientific Inc. Waltham, MA, USA). RNA sequencing (RNA-seq) of 150 bp paired-end reads with a total of ~40 million reads was performed on NovaSeq 6000 (Illumina, San Diego, CA, USA).
2.3. De Novo Transcriptome Assembly and Analysis of Differentially Expressed Genes (DEGs)
Quality assessment of the sequencing data was evaluated using FastQC version 0.11.4 (Simon Andrew, Cambridge, UK, https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) [48] followed by the removal of adapter sequences using Cutadapt version 1.9.1 (Martin Marcel, https://cutadapt.readthedocs.io/en/v1.9.1/) [49]. The 5′ and 3′ end bases containing Ns or sequences with quality values < 20 were removed. Sequencing reads that were less than 75 bp long after trimming were also removed. De novo transcriptome assembly of the clean data was performed using Trinity version 2.2.0 (Brian J. Haas, https://github.com/trinityrnaseq/trinityrnaseq/releases) [50] followed by clustering into long non-redundant unique sequences. The prediction of coding regions within the assembled transcripts was performed using the TransDecoder version 3.0.0 (Brian J. Haas, https://github.com/TransDecoder/TransDecoder/releases?page=2) [51]. To determine their putative functions, transcripts were aligned with sequences in the following publicly available databases: NCBI non-redundant protein sequences (Nr), a manually annotated and reviewed protein sequence database (Swiss-Prot), Kyoto Encyclopedia of Genes and Genomes (KEGG), and Cluster of Orthologous Groups of proteins (COG).
To generate read densities for gene expression analysis, sequencing reads were aligned to the assembled transcripts using Bowtie2 version 2.2.6 (Langmead et al., https://sourceforge.net/projects/bowtie-bio/files/) [52] with default settings. The level of gene expression normalized to read density was calculated using RNA-seq by Expectation Maximization (RSEM) version 1.2.4 (Li et al., https://github.com/deweylab/RSEM/releases) [53] and quantified as fragments per kilobase of transcripts per million mapped reads (FPKM). The analysis of DEGs between the two groups of RNA samples was performed using the Bioconductor package DESeq2 version 1.6.3 (Love et al., https://bioconductor.org/packages/release/bioc/html/DESeq2.html) [54]. The results were further analysed to determine significant DEGs based on the criteria of fold change > 2 and a q-value (false discovery rate adjusted p-value) < 0.05.
In order to classify the biological functions of significant DEGs, GO functional enrichment analysis was performed using GOSeq [55]. Gene length and read count biases were included in the GO analysis using a filtering threshold for overrepresented sequences by applying a p-value ≤ 0.05.
We further compared the log2 fold-change values of F. keratoplasticum 2781 in response to VRC to F. graminearum PH-1 strains (PH-1 is a well-studied strain for azole resistance and transcriptional regulation) deleted in the transcription factor gene ΔFgSR [33] or ΔFgAtrR [36]. These data were obtained from Supplementary Data 3 (∆FgSR) in [33] and Supplementary Table S2 (∆FgAtrR) in [36], respectively. We used these log2 fold-change values to plot column clustered heatmaps by using the gplots package version 3.1.3 [56] on R version 4.2.0 (R Core Team, Vienna, Austria, https://cran.r-project.org/bin/windows/base/) [57].
2.4. Search for Homologs of F. keratoplasticum Genes in Other Fungi
We identified and manually curated all possible ergosterol and cholesterol biosynthesis homologs of A. fumigatus and F. graminearum and the three FSSC species F. vanettenii, F. solani FSSC5 and F. keratoplasticum 2781 by searching their entire genome or, in the case of F. keratoplasticum 2781, the entire transcriptome of cells grown either in the presence or absence of VRC using all known ergosterol or cholesterol biosynthesis protein sequences of A. fumigatus and S. cerevisiae or H. sapiens as query sequences (see Supplementary File S1).
Coding sequences (CDSs) within the F. keratoplasticum mRNA transcripts were used as query sequences with the online NCBI BLASTx to search the non-redundant protein sequence database (Nr) of Fusarium taxa (taxid: 5506). CDSs conserved across the Fusarium taxa were identified from the alignments and translated into protein sequences. The BLAST+ software version 2.13.0 [58] was used to conduct a local BLASTp search against the protein sequences of Fusarium solani FSSC 5 [59], F. vanettenii 77-13-4 [60], F. graminearum PH-1 [61], A. fumigatus Af293 [62] and S. cerevisiae S288C [63] downloaded from the publicly available MycoCosm [64] database (accessed on 29 June 2022) of the Joint Genome Institute (JGI). F. solani FSSC 5 and F. vanettenii 77-13-4 were selected because they are the closest relatives to F. keratoplasticum within the FSSC clade. The F. keratoplasticum genome has yet to be sequenced. F. graminearum is a more distant relative belonging to a neighbour clade Fusarium sambunicum species complex. A. fumigatus was included as a pathogenic mould for which azole resistance mechanisms have been well studied. S. cerevisiae was included as a well characterised yeast species.
2.5. Validation of RNA-Seq DEGs Using qPCR
First strand cDNA synthesis of 1 µg total RNA and the quantification of mRNA expression levels of individual genes was conducted by real-time qPCR as previously described [25,46]. The three cyp51 paralogs (cyp51A, cyp51B and cyp51C) and abc1 were selected for validation of the RNA-seq results [25,46]. The erg6A and erg6B paralogs encoding sterol C24-methyltransferases were also included. qPCR of three biological replicates with three technical replicates each was performed to confirm the RNA-seq results.
3. Results
3.1. De Novo Transcriptome Assembly and Annotation of Transcripts
De novo transcriptome assembly of sequencing reads obtained from all six RNA samples generated a total of 9,011,979 contigs, of which 37,274 were identified as unique transcripts. The sequence length distribution of the 37,274 transcripts (Figure 1a) revealed that the majority (49.51%) were 200–500 nucleotides in length. Out of the 37,274 transcripts, 2752 had matches in all four databases Nr, Swiss-prot, KEGG and COG (Figure 1b). The Nr database had the most matches to the transcripts (22,830), and 10,180 of these transcripts were not present in any of the other three databases (Figure 1b). A similar number of transcripts were found in the Swissprot (12,357) and COG (10,454) databases, while KEGG only had matches to 4062 unique transcripts.
Figure 1.
Transcript analysis. (a) Pie chart showing the size distribution (length in nucleotides) of the 37,274 F. keratoplasticum transcripts. (b) Venn diagram of the number of unique transcripts found in the Nr, Swissprot, KEGG and COG, databases (Nr = 22,830; Swissprot = 12,357; KEGG = 4062; COG = 10,454).
3.2. Genes Differentially Expressed in the Presence of VRC
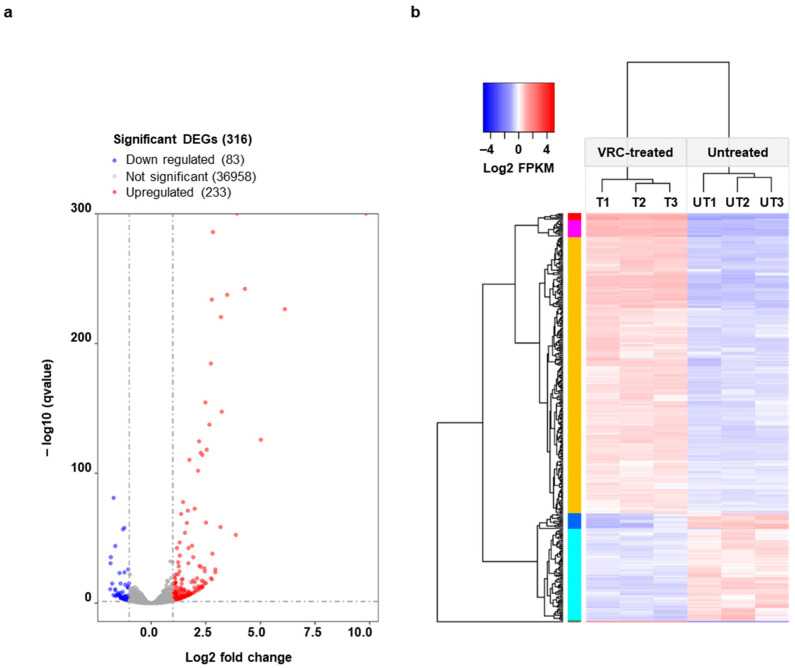
A total of 316 genes were significantly differently expressed (fold-change > 2 and q-value < 0.05) in the presence of VRC; 233 were upregulated and 83 were downregulated (Figure 2a). To assess the reproducibility of the 316 significant DEGs in each sample from the two treatment groups (VRC-treated and untreated), we compared their FPKM profiles. Figure 2b shows that the triplicate repeats for cells grown either in the presence (T1, T2, T3) or absence (UT1, UT2, UT3) of VRC clearly grouped into two distinct clusters, confirming good reproducibility and the reliability of the results produced by the biological replicates.
Figure 2.
Genes differentially expressed in the presence of VRC. (a) Volcano plot of differentially expressed genes (DEGs) showing the log10 q-value (y-axis) vs. log2 fold change (x-axis). The cut-off values (dotted grey lines) used to define significant DEGs were a q-value of < 0.05 and a log2 fold change > |1|, i.e., larger than 2-fold. (b) Heatmap dendrogram (blue = down-regulation, red = up-regulation) for the normalized mRNA expression levels (log2 of FPKMs) of the 316 DEGs relative to their combined average expression levels for each of the three biological replicates grown in the absence (UT1, UT2, UT3) or presence (80 min; T1, T2, T3) of sub-growth inhibitory concentrations of VRC (16 mg/L). The hierarchical clustering indicates biologically meaningful results. Colours of the vertical bar next to the y-axis dendrogram indicate different levels of down-regulation (cyan-blue) or up-regulation (orange-magenta-red) of groups of transcripts.
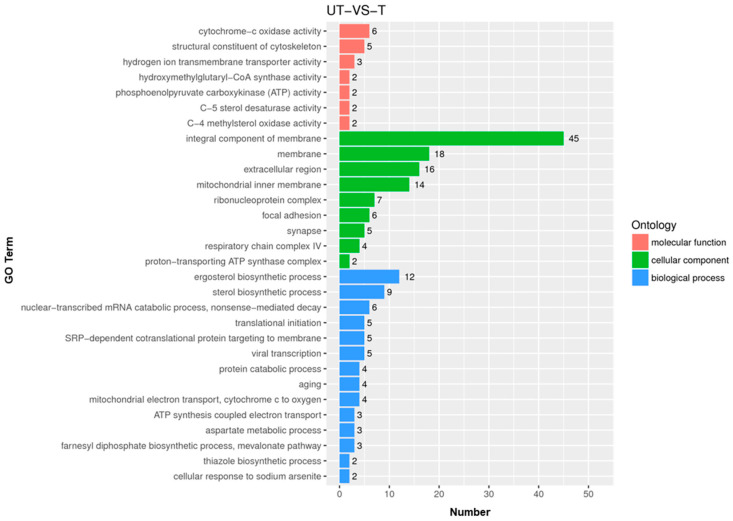
Of the 316 DEGs, GO enrichment analysis revealed 117 genes were associated with various cellular components, 67 were involved in biological process and 22 were involved in molecular functions as shown in Figure 3.
Figure 3.
Gene ontology (GO) annotations of the 30 most prominent GO terms enriched in DEGs grouped into three main GO categories: cellular component (117), biological process (67), and molecular function (22).
3.3. Inventory of All F. keratoplasticum Sterol Biosynthesis Gene Transcripts and Their Orthologs in F. solani, F. vanettenii, F. graminearum, A. fumigatus and S. cerevisiae
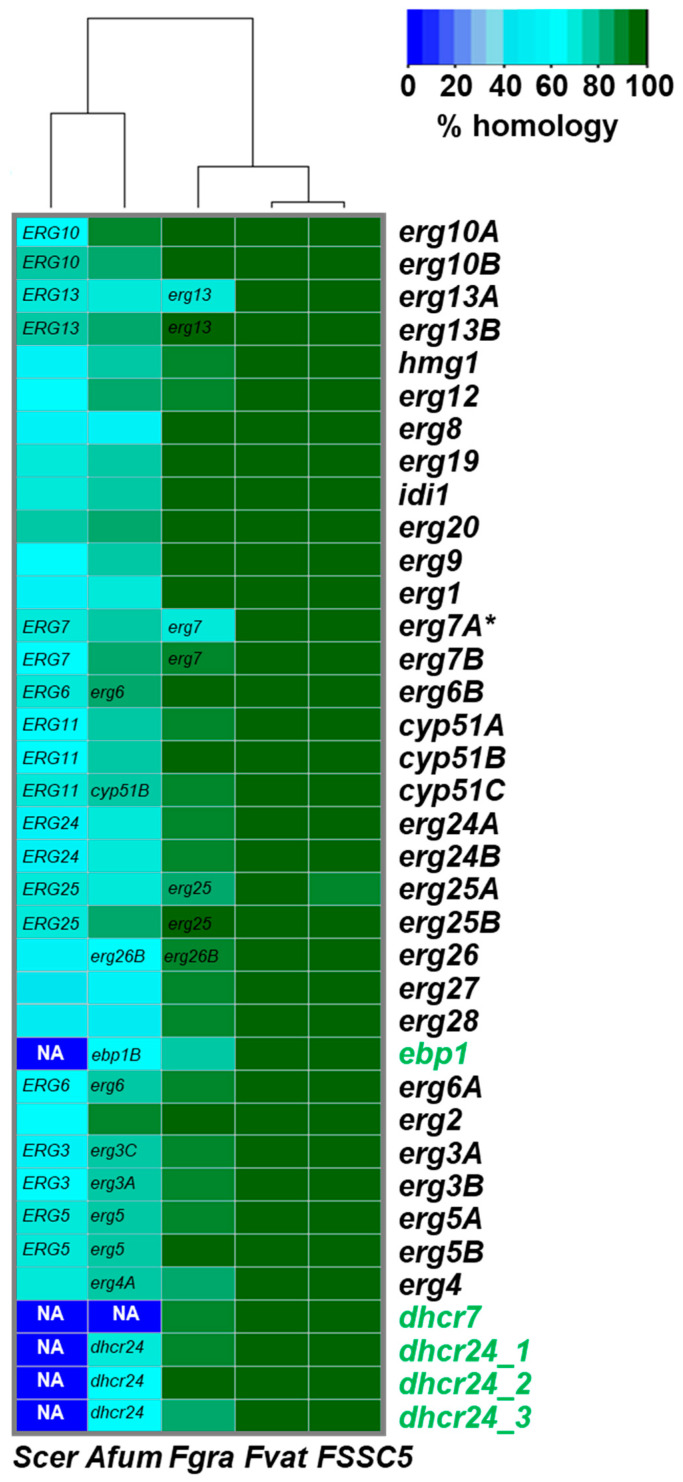
The search for all possible fungal ergosterol biosynthesis gene orthologs in F. keratoplasticum mRNA transcripts revealed the presence of eight duplicated genes (erg10, erg13, erg7, erg6, erg24, erg25, erg3 and erg5) in addition to the azole drug target, cyp51 which is known to have three paralogs in Fusarium [25]. In order to ‘correctly’ assign ‘A’ and ‘B’ orthologs (e.g., cyp51A or cyp51B) for the paralogs of erg10, erg13, erg7, erg6, cyp51, erg24, erg25, erg3 and erg5, we compared F. keratoplasticum protein sequences with all homologous proteins from three different Fusarium species (F. solani FSSC 5, F. vanettenii, and F. graminearum), A. fumigatus and S. cerevisiae (Figure 4). The two FSSC species F. solani and F. vanettenii had the same eight duplicates as F. keratoplasticum and three cyp51 genes. A. fumigatus had nine ergosterol biosynthesis gene duplicates (erg10, erg13, hmg, erg7, cyp51, erg24, erg25, erg26 and erg4) and it had three erg3 paralogs. F. graminearum had seven gene duplicates (erg10, erg6, cyp51, erg24, erg26, erg3 and erg5) and S. cerevisiae had just one (HMG). After careful annotation of all ORF sequences (some introns and ATG start codons were clearly misannotated; see Supplementary File S1 for further details) their phylogenetic relationships were determined to help identify the corresponding orthologs for all six fungal species. To remain consistent with the commonly accepted nomenclature for mould cyp51 orthologs (i.e., ‘A’ orthologs are usually expressed at low levels under normal growth conditions and are less conserved but they are upregulated in response to azole stress), ‘B’ was assigned to the more conserved ortholog across all species and ‘A’ was assigned to the less conserved gene duplicate. Thus, the F. keratoplasticum ‘B’ orthologs of all duplicated genes had the highest homology to their orthologs in F. graminearum, A. fumigatus and S. cerevisiae (Figure 4). For example, of the two Fk-erg13 paralogs, Fk-erg13B had a much higher homology to F. graminearum erg13 (95%) than did Fk-erg13A (69%). A. fumigatus also had two erg13 paralogs, Af-erg13A and Af-erg13B. As expected, Fk-erg13B was significantly more conserved (81%) than Fk-erg13A (71%) when compared to their respective A. fumigatus orthologs or to S. cerevisiae ERG13 (‘A’ 68% and ‘B’ 75%). The phylogenetic relationships between all F. keratoplasticum genes in the later stages of the ergosterol biosynthesis pathway, beginning with erg7 (lanosterol synthase), and their homologs in the remaining five fungal species are presented in Supplementary Figures S1a,b. Phylogenies of the human cholesterol gene (dhcr7 and dhcr24) homologs and the mevalonate pathway genes (erg10, erg13, hmg1) are available in Supplementary Figure S1c,d, respectively. The percentage identities and homologies for all genes across all species analysed in this study can be found in the ‘Fker FSSC2 (protein)’ sheet in Supplementary File S1.
Figure 4.
A heatmap showing the percentage homology of F. keratoplasticum ergosterol biosynthesis pathway protein sequences to S. cerevisiae (Scer), A. fumigatus (Afum), F. graminearum (Fgra), F. vanettenii (Fvat) and Fusarium solani sensu stricto (FSSC5). For genes with no obvious orthologs in other fungal species the gene most homologous to the F. keratoplasticum gene is indicated inside the rectangle. NA indicates a homolog was not found. Genes (ebp1, dhcr7 and dhcr24) with characteristics similar to human cholesterol biosynthesis [65] genes are highlighted in green font. Asterisk (*) indicates only a partial gene transcript was assembled/sequenced.
To our surprise, we also found five genes in all three fusaria (Figure 4, green font) that were homologs of human cholesterol biosynthesis genes [65]: the ∆8,∆7-sterol isomerase homolog of emopamil binding protein (ebp1) [66], the 7-dehydrocholesterol reductase (dhcr7) and three paralogs of 24-dehydrocholesterol reductase (dhcr24) which suggests that fusaria can also synthesize cholesterol or cholesterol-like compounds [67]. Further investigations revealed two ebp1 homologs and one dhcr24 homolog, ortholog of dchr24_1, in A. fumigatus and the same five epb1, dhcr7 and three dhcr24 orthologs in F. graminearum. Phylogenetic trees demonstrating the evolutionary relationship between these human cholesterol biosynthesis homologs are presented in Supplementary Figure S1b,c.
3.4. Identification of F. keratoplasticum ABC Transporters and Transcription Factors Differentially Expressed in VRC Treated Cells
Among the 316 F. keratoplasticum significant DEGs, seven encoded proteins (DN33072, DN3265, DN5825, DN16847, DN36936, DN21770 and DN29261) that belonged to the ABC transporter superfamily. These proteins had homologs in other fusaria (Table 1) with percentage identities between 75–100%.
Table 1.
Fungal orthologs or closest homologs of proteins encoded by F. keratoplasticum DEGs. Results of BLASTp searches with F. keratoplasticum gene products as queries against protein sequences of F. solani FSSC5, F. vanettenii 77-13-4, F. graminearum PH-1, A. fumigatus Af293 and S. cerevisiae S288C obtained from the Joint Genome Institute (JGI) MycoCosm database.
| F. keratoplasticum 2781 | JGI Protein ID (% Identity/% Homology) 1, 2 | |||||
|---|---|---|---|---|---|---|
| Gene | Fusarium solani FSSC 5 |
F. vanettenii 77-13-4 |
F. graminearum PH-1 | A. fumigatus Af293 | S. cerevisiae S288C | |
| ABC transporters | ||||||
| ABCB-1 | DN33072 | 511443 (100/100) | 96330 (98/99) | 6063 (80/88) | 3538 (54/72) | 3617 (28/47) |
| ABCB-2 | DN3265 | 543122 (100/100) | 92751 (99/100) | 7949 (75/85) | 8558 (47/66) | 6078 (28/48) |
| ABCC-yor1 | DN5825 | 393338 (100/100) | 63546 (98/100) | 8483 (85/92) | 5371 (59/74) | 2864 (37/56) |
| ABCG-abc1 | DN16847 | 537064 (100/100) | 63187 (98/99) | 3418 (81/89) | 1300 (60/73) | 1445 (46/64) |
| ABCG-abc3 | DN36936 | 417245 (100/100) | 35467 (99/100) | 9970 (86/91) | 1300 (63/76) | 5806 (49/67) |
| ABCG-H1 | DN21770 | 439905 (100/100) | 35868 (99/100) | 9973 (84/91) | 6736 (64/78) | 6299 (37/56) |
| ABCG-F | DN29261 | 57426 (99/99) | 68948 (99/99) | 12739 (81/90) | 4105 (56/72) | 5566 (38/58) |
| Transcription factors | ||||||
| atrR | DN25544 | 393104 (100/100) | 96223 (98/99) | 7871 (84/90) | 1901 (63/73) | NA |
| moc3 | DN12705 | 489853 (98/99) | 1518 (97/98) | 8308 (66/76) | NA | NA |
| atF | DN14162 | 502182 (96/98) | 84321 (100/100) | 9383 (36/55) | NA | NA |
| SR | DN37099 | 384730 (100/100) | 103092 (99/100) | 1375 (87/93) | NA | NA |
| yap5 | DN16099 | 42136 (99/100) | 74330 (97/98) | 1660 (73/82) | 1198 (46/59) | NA |
| SREBP1 | DN13837 | 554245 (100/100) | 67823 (99/100) | 5412 (84/91) | NA | NA |
F. keratoplasticum abc1 (DN16847) and abc3 (DN36936) were the closest homologs of A. fumigatus PDR transporter abcG1, also known as abcC or cdr1B, a major efflux pump involved in azole resistance in A. fumigatus [34,35] (JGI protein ID: 1300; Supplementary File S1), with percentages identity/homology of 60/73 and 63/76 (Table 1), respectively. F. keratoplasticum DEGs also included transcription factors. All six (DN25544, DN12705, DN14162, DN37099, DN16099 and DN13837) differentially expressed transcription factors were also present in all other fusaria. F. keratoplasticum atF (DN14162) orthologs were present in F. solani FSSC5 and F. vanettenii but the F. graminearum atF homolog was not as highly conserved as the other five TFs, suggesting that Fg-atF is possibly not an ortholog of Fk-atF and has evolved its own unique function in F. graminearum. Homologs of moc3 (DN12705), atF (DN14162), SR (DN37099) and SREBP1 (DN13837) were not found in A. fumigatus. No homologs of the F. keratoplasticum transcription factors were found in S. cerevisiae.
3.5. Transcriptional Response of F. keratoplasticum Cells to VRC Exposure Suggests the Presence of an Alternative Route in the Ergosterol Biosynthesis Pathway
Among the 37,274 transcripts (Figure 1a), 32 were genes from the ergosterol biosynthesis pathway of which 22 were significantly upregulated (Figure 5a, red shading in the rectangles on the right column of the heatmap; and Table 2) in response to VRC. In fact, erg6A, cyp51A and, most interestingly, the human cholesterol biosynthesis ortholog, ebp1 (Table 2), were the three most upregulated DEGs in response to VRC; 912-fold, 52-fold and 20-fold upregulated, respectively. To determine the transcription factors possibly involved in F. keratoplasticum 2781 ergosterol pathway gene regulation, we compared the log2 fold change values to those caused by transcription factor deletion in F. graminearum—strains PH-1 ΔFgSR [33] and ΔFg-atrR [36] (Figure 5a). We found that 16 out of the 22 genes in F. keratoplasticum upregulated in response to VRC were downregulated in F. graminearum ΔFgSR suggesting that FkSR is also the main transcription factor involved in the upregulation of the ergosterol biosynthesis genes in F. keratoplasticum upon azole antifungal treatment. The unchanged FkSR expression level upon VRC exposure (Table 2) is not unexpected because the FgSR expression levels also remained unchanged upon tebuconazole exposure of F. graminearum PH-1 cells [33]. The F. keratoplasticum atrR ortholog of Fg-atrR, however, was the most significantly upregulated (4.5-fold) transcription factor upon VRC exposure (Table 2). This too was expected as Fg-atrR transcript levels were also increased in response to tebuconazole treatment of F. graminearum cells [68]. The F. graminearum ΔFg-atrR expression profile demonstrates that FgAtrR is also involved in the upregulation of some ergosterol biosynthesis genes, but it has a smaller target range as only six genes (erg1, cyp51A, erg26A, ebp1, erg6A and erg5A) were downregulated upon its deletion (Figure 5a).
Figure 5.
VRC exposure causes the upregulation of an alternative ergosterol biosynthesis pathway in F. keratoplasticum. (a) A heatmap showing log2 fold-change values for F. keratoplasticum 2781 when exposed to VRC, and effects of F. graminearum PH-1 ΔFgSR [33] and ΔFgAtrR [36] mutations. Genes existing as a single copy in either F. graminearum or FSSC are indicated in blue font. Genes that are normally involved in cholesterol biosynthesis are highlighted in green. * = Fg-erg3, Fg-erg5, Fg-erg6 and Fg-erg24 ‘A’ and ‘B’ paralogs were renamed to be consistent with the F. keratoplasticum nomenclature (see text for further details). (b) Proposed alternative pathway upon azole exposure is indicated by the dashed arrow line (X = to be experimentally confirmed sterol intermediates). Differential expression of genes is indicated by log2 fold-change values in the green boxes beside the genes. Dashed line (---) in green boxes indicates log2 fold change values < 1.00 and/or p-value > 0.05. The expression levels of erg6A, erg6B, cyp51A, cyp51B, highlighted in bold, were verified by qPCR. Genes in orange font do not take part in ergosterol biosynthesis under normal growth conditions nor in the cells’ response to azole exposure. Log2 fold change values of more than 3 are highlighted in bold numerals.
Table 2.
Expression levels of DEGs involved in ergosterol biosynthesis, drug efflux and gene regulation which could contribute to F. keratoplasticum azole resistance. Log2 fold-change and mean FPKM values of all other significant DEGs are available in Supplementary File S2.
| Gene ID | Gene Description 1 | Gene | Log2 Fold Change 2, 3, 4 | Mean FPKM Values | |
|---|---|---|---|---|---|
| VRC-Treated (n = 3) | Untreated (n = 3) | ||||
| Ergosterol biosynthesis 5 | |||||
| DN33281 | Acetyl CoA thiolase | erg10A | −0.41 | 99 | 119 |
| DN17604 | Acetyl CoA thiolase | erg10B | 2.49 | 1165 | 180 |
| DN37027 | HMG CoA synthase | erg13A | 2.44 | 209 | 25 |
| DN8815 | HMG CoA synthase | erg13B | 2.78 | 1199 | 151 |
| DN33169 | HMG CoA reductase | hmg1 | 1.64 | 208 | 59 |
| DN18060 | Mevalonate kinase | erg12 | 2.28 | 110 | 20 |
| DN33074 | Phosphomevalonate kinase | erg8 | * −0.10 | 30 | 29 |
| DN29440 | Mevalonate pyrophosphate kinase | erg19 | * −0.03 | 90 | 83 |
| DN29378 | Isopentenyl pyrophosphate isomerase | idi1 | * −0.16 | 171 | 172 |
| DN10425 | Geranyl pyrophosphate synthase | erg20 | * 0.17 | 20 | 16 |
| DN17806 | Squalene synthase | erg9 | 0.88 | 343 | 166 |
| DN18090 | Squalene epoxidase | erg1 | 1.11 | 78 | 31 |
| DN26925 | Lanosterol synthase | erg7A | * 0.08 | 2 | 0 |
| DN37088 | Lanosterol synthase | erg7B | 2.25 | 59 | 11 |
| DN33059 | Sterol C-24 methyltransferase | erg6B | 1.11 | 1694 | 207 |
| DN25638 | Eburicol 14-α demethylase | cyp51A | 5.70 | 2982 | 2 |
| DN21545 | Eburicol 14-α demethylase | cyp51B | 2.46 | 899 | 104 |
| DN8317 | Claimed not to encode sterol 14-α demethylase 6 | cyp51C | 1.88 | 20 | 5 |
| DN29591 | Sterol C-14 reductase | erg24A | 3.20 | 160 | 15 |
| DN32963 | Sterol C-14 reductase | erg24B | 2.12 | 290 | 58 |
| DN8081 | Sterol C-4 methyl oxidase | erg25A | 1.87 | 2 | 0 |
| DN17697 | Sterol C-4 methyl oxidase | erg25B | 2.11 | 1543 | 248 |
| DN29406 | Sterol C-3 dehydrogenase | erg26 | 0.35 | 82 | 58 |
| DN3112 | 3-keto sterol reductase | erg27 | 2.20 | 149 | 28 |
| DN22157 | Sterol C-4 demethylase | erg28 | * 0.06 | 68 | 58 |
| DN33250 | Sterol C-8 isomerase | ebp1 | 4.30 | 152 | 6 |
| DN32910 | Sterol C-24 methyltransferase | erg6A | 9.83 | 4186 | 3 |
| DN25482 | Sterol C-8 isomerase | erg2 | 2.97 | 1039 | 109 |
| DN15167 | Sterol C-5 desaturase | erg3A | 2.55 | 364 | 52 |
| DN37320 | Sterol C-5 desaturase | erg3B | 1.06 | 103 | 24 |
| DN16841 | Sterol C-22 desaturase | erg5A | * 0.30 | 0 | 0 |
| DN10857 | Sterol C-22 desaturase | erg5B | 1.76 | 575 | 151 |
| DN21765 | Sterol C-24 reductase | erg4 | 0.98 | 170 | 77 |
| DN12846 | C-7 sterol reductase | dhcr7 | NA | 0 | 0 |
| DN10241 | C-24 sterol reductase | dhcr24-1 | NA | 9 | 8 |
| DN18330 | C-24 sterol reductase | dhcr24-2 | NA | 27 | 29 |
| DN14443 | C-24 sterol reductase | dhcr24-3 | NA | 1 | 1 |
| ABC transporters 7 | |||||
| DN3265 | ABCB family | - | 1.38 | 4 | 1.4 |
| DN33072 | ABCB family | - | 1.25 | 20 | 5.4 |
| DN16847 | ABCG family (PDR transporter; cluster B) | abc1 | 2.29 | 1 | 0.05 |
| DN36936 | ABCG family (PDR transporter; cluster B) | abc3 | 1.48 | 21 | 3.3 |
| DN21770 | ABCG family (PDR transporter; cluster H1) | - | 1.24 | 17 | 6.4 |
| DN29261 | ABCG family (PDR transporter; cluster F) | - | –1.86 | 3 | 11.9 |
| Transcription factors | |||||
| DN25544 | Zn2-Cys6 transcription factor 8 | atrR | 2.15 | 34 | 7 |
| DN12705 | Transcriptional regulatory protein | moc3 | 1.32 | 5 | 2 |
| DN14162 | Terreic acid cluster-specific transcription factor | atF | 1.05 | 3 | 1 |
| DN37099 | Ergosterol biosynthesis regulator in fusaria 9 | SR | 0.13 | 33 | 27 |
| DN16099 | AP-1-like transcription factor | yap5 | –1.08 | 0 | 2 |
| DN13837 | Sterol regulatory element-binding protein 1 | SREBP1 | –1.16 | 4 | 15 |
1 Gene descriptions are based on the Uniprot descriptions except for genes with citations. 2 Negative values are the log2 values for downregulated genes. 3 DEGs with log2 fold changes above 2.00 are highlighted with a grey background. 4 Asterisk (*) indicates no significant change in expression (log2 fold changes with a q-value of > 0.05). 5 Genes are listed according to their predicted position in the ergosterol biosynthesis pathway (see text for further details). 6 Fan et al. [38] claimed that F. graminearum cyp51C does not encode sterol 14α-demethylase. 7 The cluster grouping of the ABCG family is based on previous reports by Lamping et al. [69] and James et al. [46]. 8 AtrR transcription factor orthologs have been reported to mediate azole resistance in A. fumigatus [34,35] and F. graminearum [36]. 9 The novel zinc-cluster transcription factor, FgSR, is the master regulator of ergosterol biosynthesis in F. graminearum [33].
The final clue for the proposed alternative ergosterol biosynthesis pathway in Figure 5b came with the discovery of the human ebp1 ortholog which was present in all fusaria species investigated (Supplementary File S1 and Supplementary Figure S1b). Human EBP orthologs perform the same ∆8,∆7-sterol isomerase reaction as fungal Erg2 orthologs [66]. Thus, like its human counterpart FkEbp1 most likely acts on sterol intermediates that are not methylated at their C24 position. Methylation at the C24 position, a characteristic feature differentiating ergosterol from cholesterol, is catalyzed by Erg6. In S. cerevisiae, Erg6 is thought to act after the Erg25/26/27/28 complex converting zymosterol to fecosterol which is the substrate for Erg2. However, mould Erg6B orthologs prefer lanosterol as a substrate [38,43] which places them before the Cyp51B orthologs in the ergosterol biosynthesis pathway (Figure 5b). These observations together with the facts that (i) Fk-erg6A, Fk-cyp51A and Fk-ebp1 were the most dramatically upregulated genes in response to VRC; (ii) they were the only three genes, apart from erg5A, that were significantly downregulated upon the deletion of both FgSR and Fg-atrR; and (iii) the likelihood that ebp1 acts on non-C24 methylated sterol intermediates led to the proposed alternative ergosterol biosynthesis pathway shown in Figure 5b. Fk-erg7A, Fk-erg25A and Fk-erg5A may not take part in either of these two pathways as neither is expressed under either of the growth conditions investigated (Table 2).
To compare the expression levels of each paralog in the presence and absence of VRC, we examined their FPKM values (Table 2) in each treatment group. We found that in the absence of VRC, expression levels of both erg6A (3) and cyp51A (2) were ~70 times lower than their ‘B’ paralogs (207 and 104, respectively). Conversely, when exposed to VRC, expression levels of erg6A (4186) and cyp51A (2982) were ~3 times higher than their ‘B’ paralogs (1694 and 899, respectively). This trend was not observed in the other seven (erg10, erg13, erg7, erg24, erg25, erg3 and erg5) gene duplicates where their relative expression levels (i.e., expression level of ‘B’ paralog was higher than the ‘A’ paralog, except for erg3 which was the opposite) were similar in both the presence and absence of VRC.
3.6. Differential Expression of ABC Transporters and Transcription Factors in Response to VRC
VRC induced the expression of five ABC transporters and repressed the expression of one (Table 2). Four (DN16847, DN36936, DN21770, DN29261) were PDR transporters of the ABCG subfamily and two (DN3265 and DN33072) were ABCB-type ABC transporters (Table 2). PDR transporters abc1 (DN16847), abc3 (DN36936), orthologs 1 and 3 of 9 F. vanettenii cluster B PDR transporters [46], and DN21770, ortholog 3 of 3 F. vanettenii cluster H1 PDR transporters [46], were 4.9-, 2.8- and 2.4-fold induced and the PDR transporter DN29261, ortholog 2 of 2 F. vanettenii cluster F PDR transporters [46], was 3.6-fold downregulated in response to VRC. DN16847 encodes the previously characterized F. keratoplasticum multidrug efflux pump, abc1 [46], orthologs of which are involved in virulence and/or azole drug resistance in the important fungal pathogens F. vanettenii (also known as Nectria haematococca) [70], F. graminearum [71] and F. culmorum [72]. Fk-abc3 is closely related to Fk-abc1 [46]. The F. keratoplasticum abc1 orthologs in F. graminearum (FGSG_08312; 81% protein identity) [36] and A. fumigatus (abcG1, also known as abcC or cdr1B; 60% protein identity) [34,35,45] are also involved in azole resistance. Deletion of cdr1B in A. fumigatus [73] caused increased ITC, but not VRC, susceptibility. The function of cluster F PDR transporters, ancestors of all fungal PDR transporters [69], and the possible involvement of the ABCB-type ABC transporter homologs DN3265 and DN33072 in azole resistance remains to be explored.
Five transcription factors were differentially expressed in F. keratoplasticum when exposed to VRC of which three (DN25544, DN12705, DN14162) were up-regulated and two (DN16099, DN13837) were down-regulated (Table 2). Interestingly, the highest (4.4-fold) upregulated transcription factor DN25544 is clearly the ortholog of atrR, an autoregulatory Zn2-Cys6 zinc cluster transcription factor and master regulator of cyp51 orthologs and/or ABC transporters involved in azole resistance in F. graminearum [36] and A. fumigatus [34,35]. The transcription factor DN13837, which was downregulated 2.2-fold, is a sterol regulatory element binding protein 1 (SREBP1) homolog; these proteins are the main regulators of sterol biosynthesis in many fungal species (e.g., SrbA in A. fumigatus [74]) and in higher eukaryotes including humans [75]). DN16099 (2.1-fold downregulated) is an AP-1 like transcription factor (yap5), homologs of which are involved in stress response and yeast iron regulatory mechanisms [76].
3.7. Correlation between Gene Expression Levels Measured by qPCR and RNA-Seq
The expression levels for key genes identified from the RNA-seq analysis were measured by qPCR. The Cq values were normalized with GAPDH (gpd1, Figure 6a), and their fold up- or down-regulation in response to VRC (Figure 6b) was determined using the 2−ΔΔCq method [77]. Clear differences in expression levels (Figure 6a) were observed between the cells grown in the presence and absence of VRC. The qPCR analysis revealed a 2100-fold upregulation of erg6A, a 9-fold upregulation of erg6B and a 2400-, 10- and 4-fold upregulation of cyp51A, cyp51B and cyp51C, respectively, and a 120-fold upregulation of abc1 in response to VRC (Figure 6b). These VRC responses showed similar trends to the expression levels determined by RNA-seq (R2 = 0.712; Figure 6c). In particular, erg6A and cyp51A were both expressed far less (i.e., 2142-fold and 2431-fold less), at almost undetectable levels, than their ‘B’ paralogs under normal growth conditions, but they were induced much more than their ‘B’ paralogs in response to VRC so that they reached comparable (Figure 6) or even 2- to 3-fold higher (Table 2) expression levels than their ‘B’ paralogs in the qPCR and RNA-seq experiments, respectively.
Figure 6.
Quantification of F. keratoplasticum gene expression levels. (a) qPCR quantification of F. keratoplasticum erg6A, erg6B, cyp51A, cyp51B, cyp51C and abc1 mRNA expression levels (2−ΔCq) normalised with gpd1 and (b) Log2 fold increased expression (2−ΔΔCq) of logarithmic cells in response to 80 min exposure to VRC (16 mg/L), numbers on top of the bars indicate log2 fold changes. (c) Correlation between the log2 fold change values obtained using qPCR (Y-axis) and RNA-seq (X-axis).
4. Discussion
A recent landmark study by Liu et al. [33] highlighted a novel transcription factor in F. graminearum, FgSR, which binds to a 16 bp cis-element present in the promoter of 119 genes, 20 of which were involved in ergosterol biosynthesis including cyp51A, cyp51B, cyp51C, erg6A, and erg6B. Fourteen of these 20 genes were also significantly downregulated in the ∆FgSR strain [32]. Fg-erg5A and Fg-erg4 were two notable exceptions. Although FgSR did not bind to either promoter, both genes were also significantly downregulated in the ∆FgSR strain [32]. Deletion of FgSR in F. graminearum had an inverse effect to that of VRC exposure on all these orthologs in F. keratoplasticum (Figure 5a). Again, the only exceptions were erg5A and erg4 that were either not expressed under either growth condition (erg5A) or fell just below (0.98; erg4) the log2-fold change threshold of 1.0 (Supplementary File S1). These striking similarities in the regulation of each of these ergosterol biosynthesis orthologs indicates that FkSR is also likely the key transcription factor that regulates ergosterol biosynthesis in F. keratoplasticum, and it too possibly functions via phosphorylation mediated by the kinases of the high osmolarity glycerol response (HOG) pathway [33]. A similar 16 bp cis-element binding site is present in the cyp51A and cyp51B but not the cyp51C promoters of F. keratoplasticum [25]. We also found this motif in all cyp51A and cyp51B promoters of two other FSSC species [25] as well as in F. graminearum, F. verticillioides, and F. oxysporum [33]. As expected, FkSR, the ortholog of FgSR (Table 2), was not differentially expressed by VRC exposure which is consistent with the findings in Liu et al. [33] where F. graminearum cells exposed to tebuconazole or depleted in ergosterol in a mutant deleted of erg4 did not alter FgSR protein levels. This confirms that the novel sterol regulator SR is indeed likely to be a characteristic feature of all Leotiomycetes and Sordariomycetes species [33].
Fk-atrR was the transcription factor most significantly upregulated (4.4-fold; Table 2) in response to VRC. FkAtrR is clearly the ortholog (63% protein identity) of AfAtrR (ABC transporter regulator; their phylogenetic relationship is shown in Figure 1 of [36]) that mediates resistance to azoles in A. fumigatus by possibly autoregulating its own expression [33] and co-regulating srbA, cyp51A and abcG1/cdr1B expression [34,35,45]. AtrR orthologs act similarly in fusaria species. Deletion of F. graminearum atrR (FGSG_06810; 84% protein identity) [36] caused the downregulation of cyp51A (FGSG_04092; 19-fold), erg5A (FGSG_03686; 29-fold) and erg6A (FGSG_05740; 46-fold) which are clearly the orthologs of Fk-cyp51A (81% protein identity), Fk-erg5A (80%) and Fk-erg6A (86%), respectively (Supplementary Figure S1a,b). Unfortunately, FGSG_03686 and FGSG_05740 were named Fg-erg5B and Fg-erg6B and not Fg-erg5A and Fg-erg6A, respectively, in that publication, even though, like for the Fg-cyp51A and Fg-cyp51B orthologs, they are less well conserved than Fg-erg5B (FGSG_01959) and Fg-erg6B (FGSG_02783) and expressed at much lower levels than Fg-erg5B and Fg-erg6B under normal growth conditions and two of the most downregulated genes in the ∆FgSR strain (Figure 5a and ‘Fgra (gene)’ sheet in Supplementary File S1). Thus, for consistency and to avoid confusion, we modified the nomenclature for all F. graminearum ergosterol biosynthesis gene duplicates so that all fusaria orthologs are labelled with the same ‘A’ (salvage pathway) or ‘B’ (default pathway) extensions (Figure 4 and Figure 5b). Aspergillus species have only one erg6 ortholog which appears to be the common ancestor of all fusaria erg6B orthologs (Supplementary Figure S1b). As expected for the FgAtrR ortholog of AfAtrR which possibly autoregulated its own expression and co-regulated srbA expression in A. fumigatus [34], Fg-atrR expression was not affected in the ∆FgSR knock-out strain [33], but it was upregulated (3.3-fold) in response to tebuconazole [68], which is very similar to our findings with Fk-atrR being 4.4-fold upregulated in response to VRC.
Previous studies found the F. graminearum Fk-abc3 ortholog FGSG_08312 was down-regulated 4.5-fold in the ∆AtrR strain [36] and upregulated 6.4-fold in response to tebuconazole treatment [68]. Fk-abc3 was the second highest upregulated (2.8-fold) transporter in this study, after Fk-abc1 (4.9-fold). A. fumigatus abcG1 [34,35,45] is the closest homolog of both Fk-abc1 and Fk-abc3 (60% and 63% protein identity, respectively; Table 1). Interestingly, Fg-abc3 (FGSG_04580) [71], the ortholog (81% identical) of Fk-abc1 [46], was not significantly down-regulated in F. graminearum strain ΔAtrR [36] and not upregulated when F. graminearum was treated with tebuconazole [68]. Thus, F. keratopasticum AtrR possibly co-regulates the expression of abc3, but not abc1, with cyp51A as was reported for their respective orthologs or closest homologs in F. graminearum [36] and A. fumigatus [34,35,45], respectively. Fk-abc1 orthologs in a number of fungal plant pathogens have been implicated in azole resistance and virulence by protecting cells against azoles and phytoalexins [46,71]. Thus, both the abc1 and abc3 orthologs are likely to be involved in azole antifungal drug resistance in F. keratoplasticum and related fungal species.
To date, detailed mechanisms of ergosterol biosynthesis remain underexplored in mould pathogens like fusaria. The discovery of the human ∆8,∆7-sterol isomerase ortholog ebp1 in F. keratoplasticum and all related FSSC species, and even two paralogs ebp1A and ebp1B in A. fumigatus, was rather surprising given that fungi have evolved their own characteristic ∆8,∆7-sterol isomerase erg2. Plants and animals, on the other hand, use EBP orthologs for the ∆8,∆7-sterol isomerase reaction [78,79]. Even though murine EBP could complement a yeast mutant deleted in ERG2 which exhibited a very similar sterol composition to wildtype S. cerevisiae, murine EBP and ScErg2 share no similarities and are phylogenetically completely different proteins [66]. erg2 encodes a typical bitopic membrane protein with an N-terminal transmembrane span while Fk-ebp1 and all related fungal ebp1 homologs encode proteins with five predicted transmembrane spans, respectively (Supplementary File S1). Perhaps this is why these ebp1 homologs have been overlooked for so long in the model moulds A. fumigatus and F. graminearum.
Because Fk-ebp1 was one of the sterol biosynthesis genes most highly upregulated in response to VRC and because its presence forced us to ascertain how it might possibly fit into the commonly assumed ergosterol biosynthesis pathway in F. keratoplasticum, we were interested to find out whether this enzyme had simply been missed in previous investigations, by searching their data. That was indeed the case. The F. graminearum ebp1 ortholog FGSG_13888 was actually the most dramatically down-regulated sterol biosynthesis gene (919-fold) in the ∆FgSR strain (we found it among the list of DEGs in the Supplementary Data 3 file of [33]) and also one of the most down-regulated (8.6-fold) sterol biosynthesis genes in the ∆FgAtrR strain (we found it among the list of DEGs in the Supplementary Table S2 of [36]). The cyp51A and ebp1 orthologs were also the highest upregulated genes (163-fold each) in response to exposing F. graminearum cells to tebuconazole [68]. In addition, one of the two A. fumigatus orthologs, ebp1B, was one of only four sterol biosynthesis genes (i.e., erg24A, erg25A, erg3B and ebp1B) that was dramatically downregulated in a ∆SrbA strain when the cells were grown under hypoxic growth conditions [74]. Despite its obvious significance, ebp1 was not recognized as the ‘missing link’ that it appears to be for an alternative ergosterol biosynthesis salvage pathway that is induced by azole antifungal treatment of fusaria and possibly also A. fumigatus. However, the rather different set of erg paralogs in A. fumigatus suggests a different salvage pathway is induced by azole exposure in this Eurotiomycetes model species [42,80].
We propose two major changes to the ergosterol biosynthesis pathway in response to the inhibition of cyp51B by VRC. Erg6A acts after Ebp1 because it possibly prefers non-C24 methylated substrates, like its mammalian homologs (C24 methylation is a typical feature of fungal and plant sterols), and Ebp1 acts after the Erg25/26/27/28 complex right where it is placed in the cholesterol biosynthesis pathway. Cyp51A possibly prefers lanosterol as a substrate. erg7A, erg25A and erg5A (in orange fonts in Figure 5b) possibly play no part in either pathway because their expression levels are simply too low under either growth condition (i.e., with and without VRC). Erg24A and Erg24B are both upregulated to metabolise both types of sterol intermediates, those that are C24 methylated by Erg6B and processed by the only partially inhibited Cyp51B (F. keratoplasticum 2781 had MICVRC > 32 mg/L [25]) and those that are not methylated. These two intermediates are further processed by the Erg25B/26/27/28 complex which can process both types of intermediates to generate fecosterol and zymosterol, respectively. Fecosterol and zymosterol are then processed by the two ∆8,∆7-sterol isomerases Erg2 and Ebp1, respectively. Further progression in the two pathways is not clear. However, because there are two erg3 paralogs, both of which are expressed at similar levels under normal growth conditions (Table 2) and erg3A is significantly more induced by VRC (5.9-fold) than erg3B (2.1-fold), there are possibly two separate pathways leading all the way to the production of ergosterol. The final four enzymatic reactions necessary for ergosterol biosynthesis may involve a different series of intermediate steps (hence the question marks for the placements of erg6A and erg3A and an X for different intermediates in the proposed salvage pathway in Figure 5b). Three alternative pathways were proposed for the conversion of fecosterol to ergosterol in A. fumigatus. Each pathway involved a different series of enzymatic reactions carried out by Erg2, three Erg3, Erg5 and two Erg4 paralogs, each preferring different intermediates as their respective substrates (see Figure 3 in [42]). In all three of these proposed pathways the first step involved the conversion of fecosterol to episterol by Erg2. However, our discovery of two ebp1 paralogs, one of which, ebp1B, and erg3B were required for adaption to hypoxic growth [74], suggests one or both of these ebp1 paralogs may play a part in these alternative pathways.
The possible contribution of the third Cyp51C paralog to ergosterol biosynthesis remains to be investigated even though it was claimed to have no sterol 14-α demethylase activity [38]. This assumption was based on the fact that Fg-cyp51A and Fg-cyp51B, but not Fg-cyp51C, could complement a S. cerevisiae mutant deleted in ERG11. However, the authors did not explore whether Fg-cyp51C was actually expressed nor did they consider the possibility that FgCyp51C may not accept lanosterol as its preferred substrate. The same authors also created null-mutants for all three Fg-cyp51 paralogs and the double deletion mutants for the A,C and B,C paralog combinations. However, the authors did not create a double deletion mutant for the A,B paralog combination because they “considered it to be lethal” [38]. Yet, all indications are that cyp51C is part of the ergosterol biosynthesis pathway. It too is upregulated in response to azole stress, deletion of Fg-cyp51C also affected the sterol composition of F. graminearum cells and it was required for virulence on host wheat ears [38]. It is therefore conceivable that a Fg-cyp51A/Fg-cyp51B double deletion mutant is viable and possibly uses another alternative ergosterol biosynthesis pathway to produce enough ergosterol or sterol intermediates for cell survival. F. keratoplasticum erg7A, erg25A and erg5A were not expressed under either growth condition tested, yet they are conserved in all FSSC species investigated. This suggests the possibility of an additional ergosterol biosynthesis salvage pathway that may be induced by hypoxia or other growth conditions not tested in the current study.
Dhcr7 and Dhcr24 catalyse the final two steps of cholesterol biosynthesis by reducing the double bonds at C7 and C24, respectively [67]. The presence of one dhcr7 and three dhcr24 orthologs in fusaria (Figure 4) indicates that fusaria may also be able to produce cholesterol or cholesterol-like compounds under certain stress conditions. A viable ‘cholesterol yeast’ was successfully engineered by deletion of S. cerevisiae ERG5 and ERG6 and replacing them with fish DHCR7 and DHCR24, respectively [81]. Chytridiomycota are examples of fungi that naturally synthesise cholesterol and other Δ5 sterols as their predominant membrane sterols [82]. In addition, an entire cholesterol biosynthesis pathway has also recently been reported for tomato plants that can produce significant quantities of cholesterol [79]. Fk-dhcr7 and Fk-dhcr24_3 were not expressed to any significant amount under either of the two growth conditions investigated. However, Fk-dhcr24_2, and to a lesser extent Fk-dchr24_1, were expressed at reasonable levels (Table 2) indicating they are functionally relevant under normal growth conditions. Perhaps F. keratoplasticum produces cholesterol or cholesterol-like compounds at higher temperatures or during host invasion by upregulating Fk-dhcr7 and Fk-dhcr24_3 and downregulating Fk-erg5B and Fk-erg6B, respectively. Cholesterol may provide a more rigid membrane environment enabling cells to grow at higher temperatures. No further possible plant, fungal or human sterol biosynthesis homologs were found in any of the fungal genomes of F. graminearum, F. solani FSSC5 or F. vanettenii.
5. Conclusions
The highlight of this study is the discovery of mould ebp homologs that are possibly an integral part of an alternative ergosterol biosynthesis salvage pathway that is upregulated in fusaria species upon azole antifungal exposure. VRC exposure of F. keratoplasticum also caused the upregulation of a number of ABC transporters including the major multidrug efflux pump genes abc1 and abc3. The antifungal drug resistance response of F. keratoplasticum appears rather well conserved in all fusaria species with orthologs of the sterol biosynthesis master regulator, SR, and the pleiotropic drug resistance regulator, AtrR, working together to protect fusaria from the effects of azole antifungals. We also discovered the elements of a conserved cholesterol biosynthesis pathway that may be expressed at higher temperature or under different stress conditions. Future biochemical studies are required to confirm these discoveries.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/jof8101070/s1, Figure S1: Phylogeny (midpoint rooted) of (a) erg7, cyp51, erg24 and erg25/26/27 complex genes, (b) ebp, erg6, erg2, erg3, erg5 and erg4, (c) dhcr7 and dhcr24 and of (d) mevalonate pathway genes erg10, erg13 and hmg with duplicated genes in some or all investigated species. Supplementary File S1: Inventory for all F. keratoplasticum (ergo)sterol biosynthesis, ABC transporter and transcription factor gene transcripts and their orthologs in F. solani, F. vanettenii, F. graminearum, A. fumigatus and S. cerevisiae. Supplementary File S2: A list of significant DEGs in F. keratoplasticum 2781 cells grown in the presence of VRC (16 mg/L) for 80 min.
Author Contributions
Conceptualization, methodology, validation and data curation J.E.J. and E.L.; software, J.E.J. and E.L.; writing—original draft preparation, J.E.J.; supervision and writing—review and editing, E.L., R.D.C. and J.S.; project administration and funding acquisition, J.S. and R.D.C.; All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The RNA-seq raw data can be accessed at the NCBI Short Read Archive with accession number SRS14719455. GenBank accession numbers for the manually curated F. keratoplasticum 2781 mRNA transcript sequences are provided in Supplementary File S1.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Ministry of Education Malaysia under Fundamental Research Grant Scheme (FRGS/1/2018/SKK11/UKM/02/1). J.E.J. was the recipient of a Sir Thomas Kay Sidey Postgraduate Visiting Fellowship from the University of Otago Faculty of Dentistry.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Coleman J.J. The Fusarium solani species complex: Ubiquitous pathogens of agricultural importance. Mol. Plant Pathol. 2016;17:146–158. doi: 10.1111/mpp.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kazan K., Gardiner D.M. Transcriptomics of cereal–Fusarium graminearum interactions: What we have learned so far. Mol. Plant Pathol. 2018;19:764–778. doi: 10.1111/mpp.12561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Urbaniak C., Massa G., Hummerick M., Khodadad C., Schuerger A., Venkateswaran K. Draft genome sequences of two Fusarium oxysporum isolates cultured from infected Zinnia hybrida plants grown on the International Space Station. Genome Announc. 2018;6:e00326-18. doi: 10.1128/genomeA.00326-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nucci M., Anaissie E. Fusarium infections in immunocompromised patients. Clin. Microbiol. Rev. 2007;20:695–704. doi: 10.1128/CMR.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horn D.L., Freifeld A.G., Schuster M.G., Azie N.E., Franks B., Kauffman C.A. Treatment and outcomes of invasive fusariosis: Review of 65 cases from the PATH Alliance registry. Mycoses. 2014;57:652–658. doi: 10.1111/myc.12212. [DOI] [PubMed] [Google Scholar]
- 6.Nucci M., Anaissie E. Cutaneous infection by Fusarium species in healthy and immunocompromised hosts: Implications for diagnosis and management. Clin. Infect. Dis. 2002;35:909–920. doi: 10.1086/342328. [DOI] [PubMed] [Google Scholar]
- 7.Espinel-Ingroff A., Colombo A.L., Cordoba S., Dufresne P.J., Fuller J., Ghannoum M., Gonzalez G.M., Guarro J., Kidd S.E., Meis J.F., et al. International evaluation of MIC distributions and epidemiological cutoff value (ECV) definitions for Fusarium species identified by molecular methods for the CLSI broth microdilution method. Antimicrob. Agents Chemother. 2016;60:1079–1084. doi: 10.1128/AAC.02456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaize M., Normand A.C., Imbert S., Al-Hatmi A.M.S., Chryssanthou E., Cassaing S., Schuttler C., Hasseine L., Mahinc C., Costa D., et al. Antifungal susceptibility of 182 Fusarium species isolates from 20 European centers: Comparison between EUCAST and gradient concentration strip methods. Antimicrob. Agents Chemother. 2021;65:e0149521. doi: 10.1128/AAC.01495-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stempel J.M., Hammond S.P., Sutton D.A., Weiser L.M., Marty F.M. Invasive fusariosis in the voriconazole era: Single-center 13-year experience. Open Forum Infect. Dis. 2015;2:ofv099. doi: 10.1093/ofid/ofv099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lortholary O., Obenga G., Biswas P., Caillot D., Chachaty E., Bienvenu A.L., Cornet M., Greene J., Herbrecht R., Lacroix C., et al. International retrospective analysis of 73 cases of invasive fusariosis treated with voriconazole. Antimicrob. Agents Chemother. 2010;54:4446–4450. doi: 10.1128/AAC.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoenigl M., Salmanton-García J., Walsh T.J., Nucci M., Neoh C.F., Jenks J.D., Lackner M., Sprute R., Al-Hatmi A.M.S., Bassetti M., et al. Global guideline for the diagnosis and management of rare mould infections: An initiative of the European Confederation of Medical Mycology in cooperation with the International Society for Human and Animal Mycology and the American Society for Microbiology. Lancet Infect. Dis. 2021;21:e246–e257. doi: 10.1016/S1473-3099(20)30784-2. [DOI] [PubMed] [Google Scholar]
- 12.Okada K., Endo T., Hashimoto D., Saga T., Ara T., Ogasawara R., Yasumoto A., Ibata M., Takahata M., Shigematsu A., et al. Disseminated fusariosis emerged from prolonged local genital infection after cord blood transplantation. J. Infect. Chemother. 2018;24:660–663. doi: 10.1016/j.jiac.2017.12.018. [DOI] [PubMed] [Google Scholar]
- 13.Silva G.M., Silveira A.R., Betania C.A., Macedo D.P., Neves R.P. Disseminated fusariosis secondary to neuroblastoma with fatal outcome. Mycopathologia. 2013;176:233–236. doi: 10.1007/s11046-013-9674-8. [DOI] [PubMed] [Google Scholar]
- 14.Esnakula A.K., Summers I., Naab T.J. Fatal disseminated Fusarium infection in a human immunodeficiency virus positive patient. Case Rep. Infect. Dis. 2013;2013:379320. doi: 10.1155/2013/379320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nucci M., Anaissie E.J., Queiroz-Telles F., Martins C.A., Trabasso P., Solza C., Mangini C., Simoes B.P., Colombo A.L., Vaz J. Outcome predictors of 84 patients with hematologic malignancies and Fusarium infection. Cancer. 2003;98:315–319. doi: 10.1002/cncr.11510. [DOI] [PubMed] [Google Scholar]
- 16.Zubrod J.P., Bundschuh M., Arts G., Bruhl C.A., Imfeld G., Knabel A., Payraudeau S., Rasmussen J.J., Rohr J., Scharmuller A., et al. Fungicides: An overlooked pesticide class? Environ. Sci. Technol. 2019;53:3347–3365. doi: 10.1021/acs.est.8b04392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meis J.F., Chowdhary A., Rhodes J.L., Fisher M.C., Verweij P.E. Clinical implications of globally emerging azole resistance in Aspergillus fumigatus. Philos. Trans. R Soc. Lond. B Biol. Sci. 2016;371:20150460. doi: 10.1098/rstb.2015.0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chowdhary A., Kathuria S., Xu J., Meis J.F. Emergence of azole-resistant Aspergillus fumigatus strains due to agricultural azole use creates an increasing threat to human health. PLoS Pathog. 2013;9:e1003633. doi: 10.1371/annotation/4ffcf1da-b180-4149-834c-9c723c5dbf9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao D., Wang F., Yu S., Dong S., Wu R., Cui N., Ren J., Xu T., Wang S., Wang M., et al. Prevalence of azole-resistant Aspergillus fumigatus is highly associated with azole fungicide residues in the fields. Environ. Sci. Technol. 2021;55:3041–3049. doi: 10.1021/acs.est.0c03958. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J., van den Heuvel J., Debets A.J.M., Verweij P.E., Melchers W.J.G., Zwaan B.J., Schoustra S.E. Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proc. Biol. Sci. 2017;284:20170635. doi: 10.1098/rspb.2017.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang S.E., Sumabat L.G., Melie T., Mangum B., Momany M., Brewer M.T. Evidence for the agricultural origin of resistance to multiple antimicrobials in Aspergillus fumigatus, a fungal pathogen of humans. G3. 2022;12:jkab427. doi: 10.1093/g3journal/jkab427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snelders E., Huis In ’t Veld R.A., Rijs A.J., Kema G.H., Melchers W.J., Verweij P.E. Possible environmental origin of resistance of Aspergillus fumigatus to medical triazoles. Appl. Environ. Microbiol. 2009;75:4053–4057. doi: 10.1128/AEM.00231-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhary A., Sharma C., Hagen F., Meis J.F. Exploring azole antifungal drug resistance in Aspergillus fumigatus with special reference to resistance mechanisms. Future Microbiol. 2014;9:697–711. doi: 10.2217/fmb.14.27. [DOI] [PubMed] [Google Scholar]
- 24.Otu A., Osaigbovo I., Orefuwa E., Ebenso B., Ojumu T. Collaborative One Health approaches can mitigate increasing azole-resistant Aspergillus fumigatus in Africa. Lancet Microbe. 2021;2:e490–e491. doi: 10.1016/S2666-5247(21)00218-4. [DOI] [PubMed] [Google Scholar]
- 25.James J.E., Lamping E., Santhanam J., Milne T.J., Abd Razak M.F., Zakaria L., Cannon R.D. A 23 bp cyp51A promoter deletion associated with voriconazole resistance in clinical and environmental isolates of Neocosmospora keratoplastica. Front. Microbiol. 2020;11:272. doi: 10.3389/fmicb.2020.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tava V., Prigitano A., Cortesi P., Esposto M.C., Pasquali M. Fusarium musae from diseased bananas and human patients: Susceptibility to fungicides used in clinical and agricultural settings. J. Fungi. 2021;7:784. doi: 10.3390/jof7090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bard M., Lees N.D., Turi T., Craft D., Cofrin L., Barbuch R., Koegel C., Loper J.C. Sterol synthesis and viability of Erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 28.Ghannoum M.A., Rice L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999;12:501–517. doi: 10.1128/CMR.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bien C.M., Espenshade P.J. Sterol regulatory element binding proteins in fungi: Hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell. 2010;9:352–359. doi: 10.1128/EC.00358-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown M.S., Goldstein J.L. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/S0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 31.Yang H., Tong J., Lee C.W., Ha S., Eom S.H., Im Y.J. Structural mechanism of ergosterol regulation by fungal sterol transcription factor Upc2. Nat. Commun. 2015;6:6129. doi: 10.1038/ncomms7129. [DOI] [PubMed] [Google Scholar]
- 32.MacPherson S., Akache B., Weber S., De Deken X., Raymond M., Turcotte B. Candida albicans zinc cluster protein Upc2p confers resistance to antifungal drugs and is an activator of ergosterol biosynthetic genes. Antimicrob. Agents Chemother. 2005;49:1745–1752. doi: 10.1128/AAC.49.5.1745-1752.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Z., Jian Y., Chen Y., Kistler H.C., He P., Ma Z., Yin Y. A phosphorylated transcription factor regulates sterol biosynthesis in Fusarium graminearum. Nat. Commun. 2019;10:1228. doi: 10.1038/s41467-019-09145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul S., Stamnes M., Thomas G.H., Liu H., Hagiwara D., Gomi K., Filler S.G., Moye-Rowley W.S. AtrR is an essential determinant of azole resistance in Aspergillus fumigatus. mBio. 2019;10:e02563-18. doi: 10.1128/mBio.02563-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagiwara D., Miura D., Shimizu K., Paul S., Ohba A., Gonoi T., Watanabe A., Kamei K., Shintani T., Moye-Rowley W.S., et al. A novel Zn2-Cys6 transcription factor AtrR plays a key role in an azole resistance mechanism of Aspergillus fumigatus by co-regulating cyp51A and cdr1B expressions. PLoS Pathog. 2017;13:e1006096. doi: 10.1371/journal.ppat.1006096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., Sun H., Li J., Ju C., Huang J. The transcription factor FgAtrR regulates asexual and sexual development, virulence, and DON production and contributes to intrinsic resistance to azole fungicides in Fusarium graminearum. Biology. 2022;11:326. doi: 10.3390/biology11020326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellado E., Diaz-Guerra T.M., Cuenca-Estrella M., Rodriguez-Tudela J.L. Identification of two different 14-alpha sterol demethylase-related genes (CYP51A and CYP51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 2001;39:2431–2438. doi: 10.1128/JCM.39.7.2431-2438.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fan J., Urban M., Parker J.E., Brewer H.C., Kelly S.L., Hammond-Kosack K.E., Fraaije B.A., Liu X., Cools H.J. Characterization of the sterol 14alpha-demethylases of Fusarium graminearum identifies a novel genus-specific CYP51 function. New Phytol. 2013;198:821–835. doi: 10.1111/nph.12193. [DOI] [PubMed] [Google Scholar]
- 39.Lucio J., Gonzalez-Jimenez I., Rivero-Menendez O., Alastruey-Izquierdo A., Pelaez T., Alcazar-Fuoli L., Mellado E. Point mutations in the 14-alpha sterol demethylase Cyp51A or Cyp51C could contribute to azole resistance in Aspergillus flavus. Genes. 2020;11:1217. doi: 10.3390/genes11101217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warrilow A.G., Melo N., Martel C.M., Parker J.E., Nes W.D., Kelly S.L., Kelly D.E. Expression, purification, and characterization of Aspergillus fumigatus sterol 14-alpha demethylase (Cyp51) isoenzymes A and B. Antimicrob. Agents Chemother. 2010;54:4225–4234. doi: 10.1128/AAC.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bean T.P., Cools H.J., Lucas J.A., Hawkins N.D., Ward J.L., Shaw M.W., Fraaije B.A. Sterol content analysis suggests altered eburicol 14alpha-demethylase (Cyp51) activity in isolates of Mycosphaerella graminicola adapted to azole fungicides. FEMS Microbiol. Lett. 2009;296:266–273. doi: 10.1111/j.1574-6968.2009.01645.x. [DOI] [PubMed] [Google Scholar]
- 42.Alcazar-Fuoli L., Mellado E., Garcia-Effron G., Lopez J.F., Grimalt J.O., Cuenca-Estrella J.M., Rodriguez-Tudela J.L. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids. 2008;73:339–347. doi: 10.1016/j.steroids.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Alcazar-Fuoli L., Mellado E. Ergosterol biosynthesis in Aspergillus fumigatus: Its relevance as an antifungal target and role in antifungal drug resistance. Front. Microbiol. 2012;3:439. doi: 10.3389/fmicb.2012.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhattacharya S., Esquivel B.D., White T.C. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio. 2018;9:e01291-18. doi: 10.1128/mBio.01291-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul S., Diekema D., Moye-Rowley W.S. Contributions of both ATP-binding cassette transporter and Cyp51A proteins are essential for azole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 2017;61:e02748-16. doi: 10.1128/AAC.02748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James J.E., Lamping E., Santhanam J., Cannon R.D. PDR transporter ABC1 is involved in the innate azole resistance of the human fungal pathogen Fusarium keratoplasticum. Front. Microbiol. 2021;12:673206. doi: 10.3389/fmicb.2021.673206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James J.E., Santhanam J., Zakaria L., Mamat Rusli N., Abu Bakar M., Suetrong S., Sakayaroj J., Abdul Razak M.F., Lamping E., Cannon R.D. Morphology, phenotype, and molecular identification of clinical and environmental Fusarium solani species complex isolates from Malaysia. J. Fungi. 2022;8:845. doi: 10.3390/jof8080845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Andrew S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Babraham Institute; Cambridge, UK: 2015. 0.11.4. [Google Scholar]
- 49.Martin M. Cutadapt: Removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 50.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haas B.J. TransDecoder: Find Coding Regions Within Transcripts. 2016. 3.0.0.
- 52.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li B., Dewey C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Warnes G.R., Bolker B., Bonebakker L., Gentleman R., Huber W., Liaw A., Lumley T., Maechler M., Magnusson A., Moeller S., et al. gplots: Various R Programming Tools for Plotting Data. R package version; Vienna, Austria: 2022. 3.1.3. [Google Scholar]
- 57.R-core-team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2022. 4.2.0. [Google Scholar]
- 58.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mesny F., Miyauchi S., Thiergart T., Pickel B., Atanasova L., Karlsson M., Huttel B., Barry K.W., Haridas S., Chen C., et al. Genetic determinants of endophytism in the Arabidopsis root mycobiome. Nat. Commun. 2021;12:7227. doi: 10.1038/s41467-021-27479-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coleman J.J., Rounsley S.D., Rodriguez-Carres M., Kuo A., Wasmann C.C., Grimwood J., Schmutz J., Taga M., White G.J., Zhou S., et al. The genome of Nectria haematococca: Contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009;5:e1000618. doi: 10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuomo C.A., Guldener U., Xu J.R., Trail F., Turgeon B.G., Di Pietro A., Walton J.D., Ma L.J., Baker S.E., Rep M., et al. The Fusarium graminearum genome reveals a link between localized polymorphism and pathogen specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 62.Nierman W.C., Pain A., Anderson M.J., Wortman J.R., Kim H.S., Arroyo J., Berriman M., Abe K., Archer D.B., Bermejo C., et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 63.Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., et al. Life with 6000 genes. Science. 1996;274:546–547. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 64.Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., et al. MycoCosm portal: Gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang T., Yuan D., Xie J., Lei Y., Li J., Fang G., Tian L., Liu J., Cui Y., Zhang M., et al. Evolution of the cholesterol biosynthesis pathway in animals. Mol. Biol. Evol. 2019;36:2548–2556. doi: 10.1093/molbev/msz167. [DOI] [PubMed] [Google Scholar]
- 66.Silve S., Dupuy P.H., Labit-Lebouteiller C., Kaghad M., Chalon P., Rahier A., Taton M., Lupker J., Shire D., Loison G. Emopamil-binding protein, a mammalian protein that binds a series of structurally diverse neuroprotective agents, exhibits delta8-delta7 sterol isomerase activity in yeast. J. Biol. Chem. 1996;271:22434–22440. doi: 10.1074/jbc.271.37.22434. [DOI] [PubMed] [Google Scholar]
- 67.Bae S.H., Lee J.N., Fitzky B.U., Seong J., Paik Y.K. Cholesterol biosynthesis from lanosterol. Molecular cloning, tissue distribution, expression, chromosomal localization, and regulation of rat 7-dehydrocholesterol reductase, a Smith-Lemli-Opitz syndrome-related protein. J. Biol. Chem. 1999;274:14624–14631. doi: 10.1074/jbc.274.21.14624. [DOI] [PubMed] [Google Scholar]
- 68.Becher R., Weihmann F., Deising H.B., Wirsel S.G. Development of a novel multiplex DNA microarray for Fusarium graminearum and analysis of azole fungicide responses. BMC Genom. 2011;12:52. doi: 10.1186/1471-2164-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lamping E., Baret P.V., Holmes A.R., Monk B.C., Goffeau A., Cannon R.D. Fungal PDR transporters: Phylogeny, topology, motifs and function. Fungal Genet. Biol. 2010;47:127–142. doi: 10.1016/j.fgb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coleman J.J., White G.J., Rodriguez-Carres M., Vanetten H.D. An ABC transporter and a cytochrome P450 of Nectria haematococca MPVI are virulence factors on pea and are the major tolerance mechanisms to the phytoalexin pisatin. Mol. Plant Microbe Interact. 2011;24:368–376. doi: 10.1094/MPMI-09-10-0198. [DOI] [PubMed] [Google Scholar]
- 71.Abou Ammar G., Tryono R., Doll K., Karlovsky P., Deising H.B., Wirsel S.G. Identification of ABC transporter genes of Fusarium graminearum with roles in azole tolerance and/or virulence. PLoS ONE. 2013;8:e79042. doi: 10.1371/journal.pone.0079042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hellin P., King R., Urban M., Hammond-Kosack K.E., Legreve A. The adaptation of Fusarium culmorum to DMI fungicides is mediated by major transcriptome modifications in response to azole fungicide, including the overexpression of a PDR transporter (FcABC1) Front. Microbiol. 2018;9:1385. doi: 10.3389/fmicb.2018.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fraczek M.G., Bromley M., Buied A., Moore C.B., Rajendran R., Rautemaa R., Ramage G., Denning D.W., Bowyer P. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 2013;68:1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- 74.Willger S.D., Puttikamonkul S., Kim K.H., Burritt J.B., Grahl N., Metzler L.J., Barbuch R., Bard M., Lawrence C.B., Cramer R.A., Jr. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 2008;4:e1000200. doi: 10.1371/journal.ppat.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Todd B.L., Stewart E.V., Burg J.S., Hughes A.L., Espenshade P.J. Sterol regulatory element binding protein is a principal regulator of anaerobic gene expression in fission yeast. Mol. Cell. Biol. 2006;26:2817–2831. doi: 10.1128/MCB.26.7.2817-2831.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramos-Alonso L., Romero A.M., Martinez-Pastor M.T., Puig S. Iron regulatory mechanisms in Saccharomyces cerevisiae. Front. Microbiol. 2020;11:582830. doi: 10.3389/fmicb.2020.582830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 78.Moebius F.F., Striessnig J., Glossmann H. The mysteries of sigma receptors: New family members reveal a role in cholesterol synthesis. Trends Pharmacol. Sci. 1997;18:67–70. doi: 10.1016/S0165-6147(96)01037-1. [DOI] [PubMed] [Google Scholar]
- 79.Sonawane P.D., Pollier J., Panda S., Szymanski J., Massalha H., Yona M., Unger T., Malitsky S., Arendt P., Pauwels L., et al. Plant cholesterol biosynthetic pathway overlaps with phytosterol metabolism. Nat. Plants. 2016;3:16205. doi: 10.1038/nplants.2016.205. [DOI] [PubMed] [Google Scholar]
- 80.Long N., Xu X., Zeng Q., Sang H., Lu L. Erg4A and Erg4B are required for conidiation and azole resistance via regulation of ergosterol biosynthesis in Aspergillus fumigatus. Appl. Environ. Microbiol. 2017;83:e02924-16. doi: 10.1128/AEM.02924-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Souza C.M., Schwabe T.M., Pichler H., Ploier B., Leitner E., Guan X.L., Wenk M.R., Riezman I., Riezman H. A stable yeast strain efficiently producing cholesterol instead of ergosterol is functional for tryptophan uptake, but not weak organic acid resistance. Metab. Eng. 2011;13:555–569. doi: 10.1016/j.ymben.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 82.Weete J.D., Abril M., Blackwell M. Phylogenetic distribution of fungal sterols. PLoS ONE. 2010;5:e10899. doi: 10.1371/journal.pone.0010899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq raw data can be accessed at the NCBI Short Read Archive with accession number SRS14719455. GenBank accession numbers for the manually curated F. keratoplasticum 2781 mRNA transcript sequences are provided in Supplementary File S1.