Abstract
The gut microbiota are regarded as a functional organ that plays a substantial role in human health and disease. Proton pump inhibitors (PPIs) are widely used in medicine but can induce changes in the overall gut microbiome and cause disease-associated dysbiosis. The microbiome of the duodenum has not been sufficiently studied, and the effects of PPIs on the duodenal microbiome are poorly understood. In this study, we investigated the effect of PPI administration on duodenum microbiota in patients with a gastric ulcer. A total of 12 gastric ulcer patients were included, and PPI (Ilaprazole, Noltec®, 10 mg) was prescribed in all patients for 4 weeks. A total of 17 samples from the second portion of the duodenum were analyzed. Microbiome compositions were assessed by sequencing the V3–V4 region of the 16s rRNA gene (Miseq). Changes in microbiota compositions after 4 weeks of PPI treatment were analyzed. a-Diversity was higher after PPI treatment (p = 0.02, at Chao1 index), and β-diversity was significantly different after treatment (p = 0.007). Welch’s t-test was used to investigate changes in phyla, genus, and species level, and the abundance of Akkermansia muciniphila, belonging to the phylum Verrucomicrobia, and Porphyromonas endodontalis, belonging to the phylum Bacteroidetes, was significantly increased after treatment (p = 0.044 and 0.05). PPI administration appears to induce duodenal microbiome dysbiosis while healing gastric ulcers. Further large-scale studies on the effects of PPIs on the duodenal microbiome are required.
Keywords: microbiota, duodenum, biopsy, PPI
1. Introduction
PPIs are prescribed to treat several gastric acid-related diseases such as upper gastrointestinal bleeding, peptic ulcers, erosive esophagitis, gastroesophageal reflux, and certain functional dyspepsia subtypes [1,2,3,4]. A PPI is a drug that inhibits gastric acid secretion by covalently binding with activating H+K+-ATPase on the surface of parietal cells in the mucous membrane of the stomach [5,6]. When a PPI is administered, stomach pH rises rapidly to achieve the therapeutic effect. PPIs are recommended as a treatment of choice for peptic ulcers and have a strong therapeutic effect by suppressing gastric acid secretion and preventing gastrointestinal bleeding and perforation [7,8,9,10].
Gastric acid works to kill potential pathogens that might otherwise enter the digestive system from the oral cavity [11]. Nevertheless, the stomach is colonized by bacteria such as Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria (includes H. pylori), and Fusobacteria [12]. Recent studies on the gastric microbiome have shown that the diversity of these microbiomes is correlated with gastric cancer, peptic ulcer, and gastric inflammation [13,14,15,16]. PPIs can affect gastric microbiome diversity by directly targeting bacterial and fungal proton pumps or disrupting the normal gastric microenvironment by increasing gastric pH [17]. Bacteria living in the oropharynx and feces are more abundant in the gastric microbiome after PPI use [18]. Furthermore, recent studies have shown that PPIs can induce changes in the overall gut microbiome and cause dysbiosis, which is associated with disease states, such as hepatic encephalopathy or spontaneous bacterial peritonitis in cirrhotic patients, and small intestinal bacterial overgrowth [19,20,21,22,23]. According to these studies, PPI-induced bacterial overgrowth occurs due to reduced gastric acid exposure in the intestine and can increase mortality in cirrhotic patients [19,20,21,22,23].
The duodenum is the first part of the small intestine, where chyme from the stomach, bile acid, and pancreatic enzymes are mixed and processed to improve absorption in the jejunum, and it also secretes various neurotransmitters and hormones through interaction with the duodenal microbiome [24]. The gut microbiome plays an important role in maintaining physiologic homeostasis and is also considered to be an independent factor that can cause diseases [25,26]. According to a recent study, the anti-inflammatory effects of PPIs in the duodenum are due to reduced acid exposure and eosinophilia [27]. However, few studies have investigated the association between PPI use and changes in the duodenal microbiome.
Therefore, we performed next-generation sequencing (NGS)-based analysis by 16S sequencing on mucosal samples of the second portion of the duodenum to identify changes in the duodenal microbiome induced by 4 weeks of PPI treatment in patients with a gastric ulcer.
2. Materials and Methods
2.1. Study Subjects
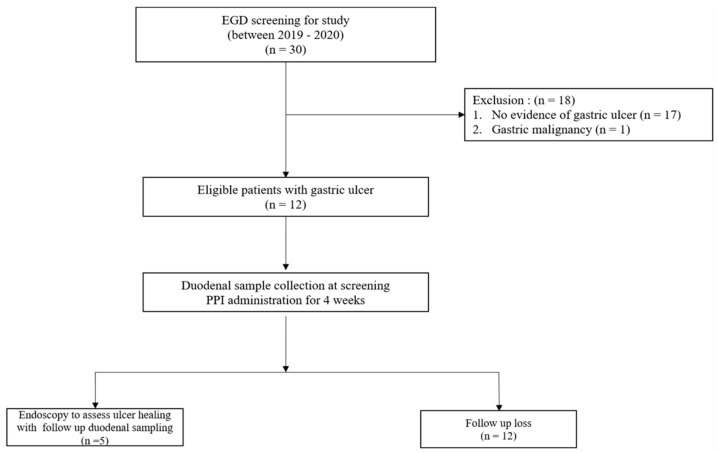
Patients scheduled to undergo screening gastroscopy for the evaluation of epigastric pain at Inha University Hospital from March 2019 to February 2020 were recruited. Informed consent was obtained when a patient met the inclusion criteria. The inclusion criteria were as follows: (1) age > 18 years at the time of treatment, (2) gastric ulcer confirmed by endoscopy, (3) those who can take PPI treatment for more than 4 weeks, and (4) no evidence of dementia or cognitive impairment. The exclusion criteria were as follows: (1) allergic symptoms or hypersensitivity to PPIs such as ilaprazole, (2) those who have undergone treatment or surgery that may affect gastric acid secretion within 3 months of registration, (3) those who have taken antibiotics within 3 months of registration, (4) those who have taken probiotics within 1 month of registration, and (5) a pregnant status or breastfeeding. Thirty patients were screened, and of these, we excluded seventeen without evidence of gastric ulcer and one patient diagnosed with malignant gastric ulcer. Finally, 12 patients were enrolled (Figure 1). However, 7 of the 12 were lost to follow-up, and only 5 underwent a follow-up biopsy. The study was approved by the Institutional Review Board of Inha University Hospital, Incheon, South Korea (Approval number: 2020-01-029-009).
Figure 1.
Flow diagram of the study.
2.2. Study Procedure and Collection of Duodenal Samples
Duodenoscopy (GIF-HQ290, Olympus, Tokyo, Japan) was performed using scrupulously cleaned equipment according to standard endoscopic cleaning guidelines [28]. Sterile working channel plugs were used in all cases. Mucosal biopsies were all performed on the second portion of the duodenum.
Biopsies were performed using a standard endoscopic forceps. Each specimen was placed in a sterile tube and stored at −80 °C until required for analysis. After duodenoscopy the patients took ilaprazole (Noltec®, 10 mg) once daily for 4 weeks. After that, five patients underwent a follow-up biopsy on the second duodenal portion during endoscopy to confirm the healing of their gastric ulcer.
2.3. DNA Extraction from Duodenal Biopsy Samples
Whole bacterial genomic DNA extraction from biopsy tissue was performed using a Maxwell RSC PureFood GMO and Authentication Kit (Promega, Madison, WI, USA) following the manufacturer’s instruction. DNA concentrations were calculated using a UV-vis spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific, Waltham, MA, USA) and quantified using a QuantiFluor ONE dsDNA System (Promega). DNA samples were stored at −20 °C until required for experiments.
2.4. PCR Amplification of the V3–V4 Region of the Bacterial 16S Ribosomal RNA (rRNA) Gene
The V3–V4 region of the bacterial 16S rRNA gene was amplified from extracted DNA using the F319 (5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACG-GGNGGCWGCAG) and R806 (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACA-GGACTACHVGGGTATC-TAATCC-3′) primer sets. DNA templates (12.5 ng/uL) were amplified using a KAPA HiFi Hotstart PCR kit (Kapa Biosystems, Wilmington, NC, USA) with 5 uM primers. PCR products were subjected to 2% agarose gel electrophoresis and purified using AMPure XP magnetic beads (Beckman Coulter, Wycombe, UK). The qualities of purified amplicons were measured using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Secondary amplification was then performed over 8 cycles to attach Illumina Nextera barcodes (Illumina, Inc., San Diego, CA, USA) using i5 forward and i7 reverse primer. Amplified products were purified using AMpure XP magnetic beads (Beckman Coulter, Brea, CA, USA) according to the manufacturer’s protocol. Purified amplicons were quantified using a QuantiFluor ONE dsDNA System (Promega, Madison, MI, USA). Amplicon sizes and qualities were evaluated using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Pooled libraries were sequenced using an illumina MiSeq instrument and the MiSeq v3 Reagent Kit (Illumina, Inc., San Diego, CA, USA).
2.5. Data Analysis
An analysis of the raw data of the 16S rRNA gene sequences was processed using the QIIME (v1.9.1) bioinformatics pipeline. Using qualified sequences (paired-end, Phred ≥ Q20), the operational taxonomic units (OTUs) were assigned based on an open-reference picking method using 97% identity to entries in the Greengenes database (V13_8) using UCLUST. Alpha diversity was calculated using phylogenetic distances and numbers of observed OTUs. For beta diversity comparison between groups, UniFrac distances were evaluated. The comparisons between groups were analyzed using Welch’s t-test; a p value was assessed as significant when <0.05.
3. Results
3.1. Baseline Clinical Characteristics
Twelve patients were enrolled in the study group, and a follow-up biopsy was performed on five of them. The age range of patients was 43–85 years (median, 67.5 years), and six were men. Body mass indices ranged from 17.4–27.6 (median, 21 kg/m²). Ulcer locations and stages are listed in Table 1. Ulcers were located mainly in the antrum and lower stomach body. Ulcer stages were evaluated using the Sakita-Miwa classification [29]. All gastric ulcers were confirmed to be in the healing process during follow-up endoscopy.
Table 1.
The patients’ information.
| Groups | Sex | Age | BMI (kg/m2) | Ulcer Location | Ulcer Stage | Follow-Up Biopsy |
|---|---|---|---|---|---|---|
| A1 | M | 68 | 27.6 | Antrum | A2 | YES |
| A2 | M | 70 | 19.7 | Low body | A2 | NO |
| A3 | F | 68 | 19.7 | Mid body | A2 | NO |
| A4 | F | 72 | 24.4 | Antrum | H1 | YES |
| A5 | M | 63 | 27.3 | Antrum | A2 | YES |
| A6 | M | 43 | 17.4 | Antrum | H1 | NO |
| A7 | F | 66 | 25.8 | Antrum and Body | A2 | YES |
| A8 | M | 61 | 21.5 | Antrum | H1 | NO |
| A9 | F | 67 | 20.5 | Antrum | A2 | YES |
| A10 | F | 77 | 18.3 | Antrum | H1 | NO |
| A11 | M | 67 | 19.3 | Antrum | A1 | NO |
| A12 | M | 85 | 27.6 | Low body | H1 | NO |
3.2. Bacterial Composition in Duodenum
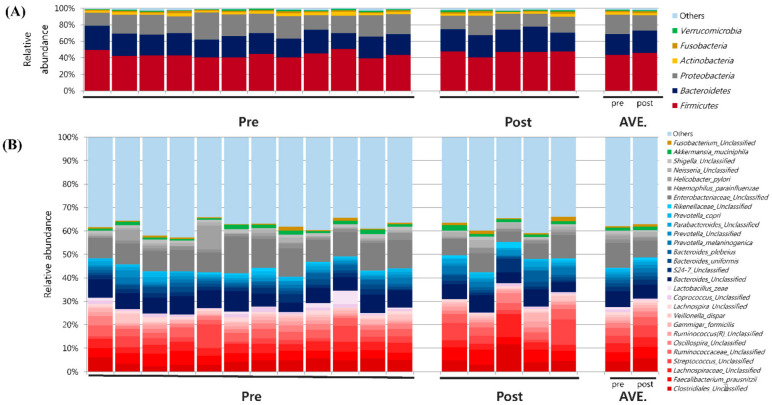
At the phylum level, the predominant bacteria groups were five in the pre- and post-PPI group at an average percentage composition of ≥1% of total microbiota. Firmicutes predominated in pre- and post-treatment samples (average percentage composition, pre-PPI: 43.5% vs. post PPI: 45.9%). Percentages of Bacteroidetes (25.4% vs. 26.9%) and Proteobacteria (23.4% vs. 18.8 %) were also high, followed by Actinobacteria (3.0% vs. 2.5%) and Fusobacteria (1.8% vs. 2.1%) (Figure 2A). At the genus level, the family Clostridiaceae (phylum Firmicutes) was present at the highest percentage (13.4% vs. 16.2%), followed by Faecalibacterium (11.9% vs. 13.7%) and Lachnospiraceae (11.3% vs. 11.8%). In the phylum Bacteroidetes, the percentage of Bacteroides was highest (55.4% vs. 51.3%), followed by Prevotella (17.9% vs. 21.3%) (Figure 2B).
Figure 2.
Microbial analysis of second duodenal portion mucosal samples obtained from gastric ulcer patients. (A) Microbiome composition at phylum level, and (B) at genus level.
3.3. Bacterial Community Diversity Pre- vs. Post-PPI Treatment
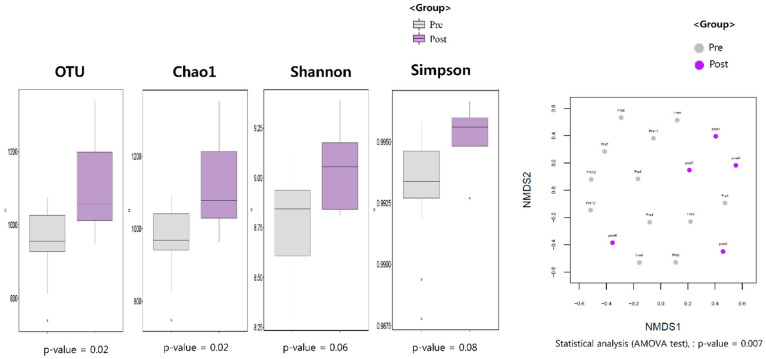
The number of OTUs observed in the samples was significantly higher after PPI treatment (p = 0.02). Alpha diversity metrics, the Chao1 estimator, and Shannon diversity indices were used to evaluate species abundances, and the Simpson index was used to evaluate species evenness. Obviously, there were significant differences in the Chao1 index pre- and post-PPI treatment (p = 0.02, Figure 3A). This shows that bacterial community diversity increased after PPI treatment with statistical significance. Significance was determined by analysis-of-variance (ANOVA). The Shannon and Simpson indices for the pre-PPI group were lower than for the post-PPI group, indicating lower bacterial community diversity. However, there was no statistically significant value. (Figure 3A, p = 0.06 and 0.08, respectively). We also assessed the significance of bacterial community composition (i.e., beta diversity) between individuals using NMDS plots on rank-order Bray–Curtis distances. As shown in Figure 3B, clustering by sample showed significant differences in beta diversity between the pre- and post-treatment group (p = 0.007).
Figure 3.
(A) Microbial richness assessed by number of OTUs, Chao1, and biodiversity index, including Shannon, Simpson indices (B) Beta diversity visualized using non-metric multidimensional scaling (NMDS) plot with Bray–Curtis dissimilarity distances.
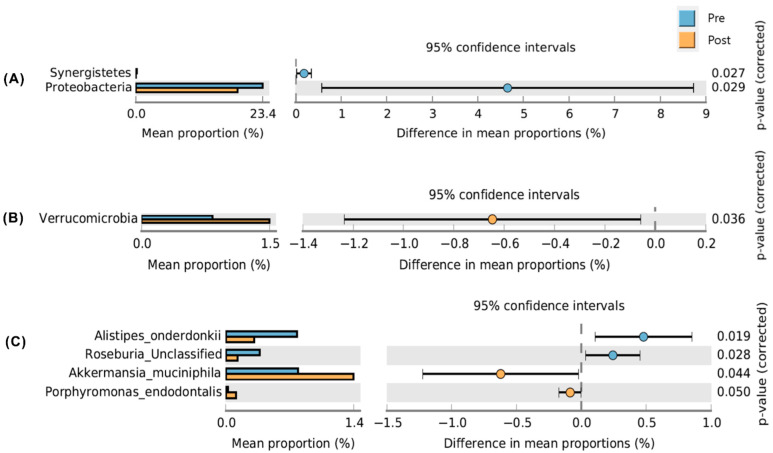
Welch’s t-test was used to compare profiles at the phylum level and showed that the abundance of Proteobacteria was significantly decreased (p = 0.029), and that of Verrucomicrobia was increased in the post-PPI group (p = 0.036) (Figure 4A,B). At the genus level (Figure 4B), Welch’s t-test showed that the abundances of Enterococcaceae, Coprococcus, Enterobacteriaceae, and Synergistes were significantly decreased, and those of Porphyromonas and Akkermansia were significantly increased after treatment (Figure 4C).
Figure 4.
Welch’s t-test to compare the profiles at the phylum level (A) genus level (B) and species level (C).
4. Discussion
In this prospective study, we compared and analyzed the duodenal microbiomes of gastric ulcer patients treated with a PPI. The study provides initial data on how the diversity of the duodenal microbiome in gastric ulcer patients is altered by PPI treatment. A duodenal biopsy was performed during gastroduodenoscopy in the second portion of the duodenum before PPI treatment and 4 weeks after treatment. Significant differences in duodenal microbiomes were observed after treatment. In particular, Chao1 index values were significantly increased by treatment, which means that duodenal microbiome richness was significantly increased by PPI treatment. Beta diversity was also significantly altered by PPI treatment.
The small intestine, including the duodenum, unlike the stomach and colon, has only one type of surface mucus, and this mucus is unattached and easily removed [30]. In addition, the duodenum contains a high concentration of bile acid, which has antimicrobial effects, so it is not a good environment for bacteria. Accordingly, the upper two-thirds of the small intestine have a much lower bacterial concentration than the colon (103–4 vs. 1011–12 microorganisms per mL) [31], which in part explains the lack of study of the duodenal microbiome. In previous studies, Lactobacillus sp., Escherichia coli, and Enterococci were found to be the predominant species in the duodenum and jejunum [31,32]. However, because these analyses were performed on the microbiome of the small intestine, including the jejunum, the results obtained cannot explain the characteristics of the duodenum microbiome. Therefore, the present analysis of the characteristics of the microbiome of duodenal mucosa contributes to current knowledge. In this study, Firmicutes was most abundant (43.5 %), followed by Bacteroidetes (25.4%), Proteobacteria (23.4%), Acinetobacteria (3.0%), and Fusobacteria (1.8%), which concurs with the results of recent studies on duodenal fluid [28,33,34].
We identified significant differences after PPI treatment at the genus and species levels. The diversity and richness of the mucosa-associated microbiome were greater after treatment. The dominant difference in the bacterial composition in the pre-PPI treatment group was observed in Alistipes_onderdonkii and Roseburia_unclassified. Alistipes_onderdonkii has been suggested to have protective effects against some diseases, including liver fibrosis, colitis, and cardiovascular disease, and against cancer immunotherapy [35]. Roseburia spp. are known to produce short-chain fatty acids, specifically butyrate, which affects colon motility, the maintenance of immunity, and anti-inflammatory properties [36]. In contrast, the dominant difference in bacterial composition in the PPI-treated group was the presence of Akkermansia mucinophila and Porphyromonas endodontalis. Previous studies have shown that Porphyromonas endodontalis is associated with periodontitis [37,38]. A. mucinophilia is considered a gut symbiont associated with numerous health-enhancing effects [39]. These observed changes indicate that PPI treatment induces duodenal dysbiosis.
In this study, the average percentage and proportion of Akkermansia at the genus level increased significantly after PPI treatment, and the average proportion of Akkermansia muciniphila also increased significantly. A. muciniphila has been reported to have the ability to obtain nutrients by breaking down mucin and modulating the inflammatory response in a mouse model [40,41]. Also, the abundance of A. muciniphila has been found to be inversely proportional to the severity of appendicitis, as well as being present in lower amounts in patients with inflammatory bowel disease, suggesting an association with human health problems [42,43]. A. muciniphila is a viable strain due to its decomposing mucin in the outer layer of mucus secreted by goblet cells in gastric mucosa [44]. In patients with gastric ulcer, the healing process begins within 2–3 days after ulceration. After re-epithelialization begins at the ulcer margin, granulated tissue of the ulcer base restores epithelial continuity [45]. Furthermore, the mucus produced during re-epithelialization can become the medium for A. muciniphila growth, and the host may benefit due to the anti-inflammatory effects of the metabolites produced. This was interpreted as a result opposite to the detrimental effect of PPI-induced bacterial overgrowth in gastric ulcer patients. Conversely, the increase in A. muciniphila may have effects such as increased permeability and duodenal dysbiosis, as in small intestinal bacterial overgrowth, so further, subsequent studies are needed [46,47].
In general, dysbiosis means that the richness and abundance of the gut microbiome are decreased, and the diversity also tends to decrease. However, all three increased after PPI treatment. The colon contains more than 10⁷ times the number of microorganisms found in the duodenum, and the bacteria present produce metabolites and interact with host immune cells. Thus, dysbiosis in the colon is defined as a decrease in bacterial diversity. On the other hand, it is difficult to apply the same definition to the duodenum because the number of microorganisms is relatively small, as food materials pass through the duodenum quickly, which means the duodenal environment militates against bacterial survival. Further studies are needed to define duodenal dysbiosis accurately.
The peculiarity of our study is that it is the result of analyzing a biopsy specimen and considering the characteristics of the duodenal mucosa. In addition, the results of this study are consistent with the results of recent studies. PPIs induce duodenal bacterial overgrowth and have been shown to be associated with the development of severe acute pancreatitis and recurrent cholelithiasis, and our study suggests important microbiome changes in these developments after PPI use [48,49].
Despite its prospective design, this study has some limitations. First, the number of patients included in the evaluation was small. Second, healthy controls with no upper gastrointestinal abnormalities such as gastric ulcer were not included in this study. Despite the above two limitations, we consider the study meaningful, as it provides statistically derived information on the effects of a PPI on the duodenal microbiome in a background of gastric ulcer. Third, there was no analysis of H. pylori infection and eradication, which can change the microbiome of the stomach and duodenum. Because this study was designed to analyze changes in the microbiome of the duodenum according to the use of PPIs, it was not handled. Nevertheless, it is necessary to evaluate the effect of H. pylori infection and eradication, so a large-scale subsequent study should be conducted in the future. Fourth, this study showed changes in duodenal bacterial compositions but did not explain how the composition of the bacteria affects host health, including the gut–brain axis. A well-designed large-scale study is needed to explore this issue.
In conclusion, this study showed that species abundance increased, and microbiome compositions changed significantly after PPI treatment in the duodenum of gastric ulcer patients. PPI treatment decreased the abundances of the beneficial bacteria Alisipes onderodonkii and Roseburia, and it increased the abundance of the known pathogen Porphyromonas endodontalis. Furthermore, the abundance of Akkermansia mucinophilia increased after PPI treatment, which we suggest was related to increased duodenal mucus and gastric ulcer healing with an increase in gastric pH. These findings lead us to speculate that PPI use is associated with the development of dysbiosis in the duodenal microbiome.
Author Contributions
J.-H.L. and J.S. were responsible for the study concept and design, the acquisition, analysis, and interpretation of data, and the drafting of the manuscript. J.-H.L. and J.S. contributed equally to this work and share first authorship. Study concept and design: J.-H.L., J.S. and J.-S.P.; collection and assembly of data, J.-H.L., J.S. and J.-S.P.; analysis and interpretation of data, J.-H.L., J.S. and J.-S.P.; drafting of the article, J.-H.L. and J.S.; critical revision of the manuscript, J.-S.P. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of Inha University Hospital, Incheon, South Korea (Approval number: 2020-01-029-009, date of approval: 23 March 2020).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by an INHA UNIVERSITY Research Grant and by the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (Grant no. NRF-202R1F1A1067621).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sharma V.K., Leontiadis G.I., Howden C.W. Meta-analysis of randomized controlled trials comparing standard clinical doses of omeprazole and lansoprazole in erosive oesophagitis. Aliment. Pharmacol. Ther. 2001;15:227–231. doi: 10.1046/j.1365-2036.2001.00904.x. [DOI] [PubMed] [Google Scholar]
- 2.Bardou M., Toubouti Y., Benhaberou-Brun D., Rahme E., Barkun A.N. Meta-analysis: Proton-pump inhibition in high-risk patients with acute peptic ulcer bleeding. Aliment. Pharmacol. Ther. 2005;21:677–686. doi: 10.1111/j.1365-2036.2005.02391.x. [DOI] [PubMed] [Google Scholar]
- 3.DeVault K.R., Castell D.O. Updated guidelines for the diagnosis and treatment of gastroesophageal reflux disease. The Practice Parameters Committee of the American College of Gastroenterology. Am. J. Gastroenterol. 1999;94:1434–1442. doi: 10.1111/j.1572-0241.1999.1123_a.x. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki H., Okada S., Hibi T. Proton-pump inhibitors for the treatment of functional dyspepsia. Ther. Adv. Gastroenterol. 2011;4:219–226. doi: 10.1177/1756283X11398735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fellenius E., Berglindh T., Sachs G., Olbe L., Elander B., Sjöstrand S.E., Wallmark B. Substituted benzimidazoles inhibit gastric acid secretion by blocking (H+ + K+)ATPase. Nature. 1981;290:159–161. doi: 10.1038/290159a0. [DOI] [PubMed] [Google Scholar]
- 6.Shin J.M., Kim N. Pharmacokinetics and pharmacodynamics of the proton pump inhibitors. J. Neurogastroenterol. Motil. 2013;19:25–35. doi: 10.5056/jnm.2013.19.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamada T., Satoh K., Itoh T., Ito M., Iwamoto J., Okimoto T., Kanno T., Sugimoto M., Chiba T., Nomura S., et al. Evidence-based clinical practice guidelines for peptic ulcer disease 2020. J. Gastroenterol. 2021;56:303–322. doi: 10.1007/s00535-021-01769-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laine L., Jensen D.M. Management of patients with ulcer bleeding. Am. J. Gastroenterol. 2012;107:345–360; quiz 361. doi: 10.1038/ajg.2011.480. [DOI] [PubMed] [Google Scholar]
- 9.Søreide K., Thorsen K., Harrison E.M., Bingener J., Møller M.H., Ohene-Yeboah M., Søreide J.A. Perforated peptic ulcer. Lancet. 2015;386:1288–1298. doi: 10.1016/S0140-6736(15)00276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matuz M., Benkő R., Engi Z., Schváb K., Doró P., Viola R., Szabó M., Soós G. Use of Proton Pump Inhibitors in Hungary: Mixed-Method Study to Reveal Scale and Characteristics. Front. Pharmacol. 2020;11:552102. doi: 10.3389/fphar.2020.552102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassis C.M., Erb-Downward J.R., Dickson R.P., Freeman C.M., Schmidt T.M., Young V.B., Beck J.M., Curtis J.L., Huffnagle G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio. 2015;6:e00037. doi: 10.1128/mBio.00037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bik E.M., Eckburg P.B., Gill S.R., Nelson K.E., Purdom E.A., Francois F., Perez-Perez G., Blaser M.J., Relman D.A. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aviles-Jimenez F., Vazquez-Jimenez F., Medrano-Guzman R., Mantilla A., Torres J. Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci. Rep. 2014;4:4202. doi: 10.1038/srep04202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khosravi Y., Dieye Y., Poh B.H., Ng C.G., Loke M.F., Goh K.L., Vadivelu J. Culturable bacterial microbiota of the stomach of Helicobacter pylori positive and negative gastric disease patients. Sci. World J. 2014;2014:610421. doi: 10.1155/2014/610421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X.X., Wong G.L., To K.F., Wong V.W., Lai L.H., Chow D.K., Lau J.Y., Sung J.J., Ding C. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS ONE. 2009;4:e7985. doi: 10.1371/journal.pone.0007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrahami D., McDonald E.G., Schnitzer M.E., Barkun A.N., Suissa S., Azoulay L. Proton pump inhibitors and risk of gastric cancer: Population-based cohort study. Gut. 2022;71:16–24. doi: 10.1136/gutjnl-2021-325097. [DOI] [PubMed] [Google Scholar]
- 17.Vesper B.J., Jawdi A., Altman K.W., Haines G.K., 3rd, Tao L., Radosevich J.A. The effect of proton pump inhibitors on the human microbiota. Curr. Drug Metab. 2009;10:84–89. doi: 10.2174/138920009787048392. [DOI] [PubMed] [Google Scholar]
- 18.Sanduleanu S., Jonkers D., De Bruine A., Hameeteman W., Stockbrügger R.W. Non-Helicobacter pylori bacterial flora during acid-suppressive therapy: Differential findings in gastric juice and gastric mucosa. Aliment. Pharmacol. Ther. 2001;15:379–388. doi: 10.1046/j.1365-2036.2001.00888.x. [DOI] [PubMed] [Google Scholar]
- 19.Nardelli S., Gioia S., Ridola L., Farcomeni A., Merli M., Riggio O. Proton Pump Inhibitors Are Associated With Minimal and Overt Hepatic Encephalopathy and Increased Mortality in Patients With Cirrhosis. Hepatology. 2019;70:640–649. doi: 10.1002/hep.30304. [DOI] [PubMed] [Google Scholar]
- 20.Zhu J., Qi X., Yu H., Yoshida E.M., Mendez-Sanchez N., Zhang X., Wang R., Deng H., Li J., Han D., et al. Association of proton pump inhibitors with the risk of hepatic encephalopathy during hospitalization for liver cirrhosis. United Eur. Gastroenterol. J. 2018;6:1179–1187. doi: 10.1177/2050640618773564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dam G., Vilstrup H., Watson H., Jepsen P. Proton pump inhibitors as a risk factor for hepatic encephalopathy and spontaneous bacterial peritonitis in patients with cirrhosis with ascites. Hepatology. 2016;64:1265–1272. doi: 10.1002/hep.28737. [DOI] [PubMed] [Google Scholar]
- 22.Fujimori S. What are the effects of proton pump inhibitors on the small intestine? World J. Gastroenterol. 2015;21:6817–6819. doi: 10.3748/wjg.v21.i22.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo W.K., Chan W.W. Proton pump inhibitor use and the risk of small intestinal bacterial overgrowth: A meta-analysis. Clin. Gastroenterol. Hepatol. 2013;11:483–490. doi: 10.1016/j.cgh.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 24.van Baar A.C.G., Nieuwdorp M., Holleman F., Soeters M.R., Groen A.K., Bergman J. The Duodenum harbors a Broad Untapped Therapeutic Potential. Gastroenterology. 2018;154:773–777. doi: 10.1053/j.gastro.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Volk N., Lacy B. Anatomy and Physiology of the Small Bowel. Gastrointest. Endosc. Clin. N. Am. 2017;27:1–13. doi: 10.1016/j.giec.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 26.Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19:55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- 27.Wauters L., Ceulemans M., Frings D., Lambaerts M., Accarie A., Toth J., Mols R., Augustijns P., De Hertogh G., Van Oudenhove L., et al. Proton Pump Inhibitors Reduce Duodenal Eosinophilia, Mast Cells, and Permeability in Patients With Functional Dyspepsia. Gastroenterology. 2021;160:1521–1531.e1529. doi: 10.1053/j.gastro.2020.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Oh H.J., Kim J.S. Clinical Practice Guidelines for Endoscope Reprocessing. Clin. Endosc. 2015;48:364–368. doi: 10.5946/ce.2015.48.5.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyake T., Suzaki T., Oishi M. Correlation of gastric ulcer healing features by endoscopy, stereoscopic microscopy, and histology, and a reclassification of the epithelial regenerative process. Dig. Dis. Sci. 1980;25:8–14. doi: 10.1007/BF01312726. [DOI] [PubMed] [Google Scholar]
- 30.Gum J.R., Jr., Hicks J.W., Toribara N.W., Siddiki B., Kim Y.S. Molecular cloning of human intestinal mucin (MUC2) cDNA. Identification of the amino terminus and overall sequence similarity to prepro-von Willebrand factor. J. Biol. Chem. 1994;269:2440–2446. doi: 10.1016/S0021-9258(17)41965-X. [DOI] [PubMed] [Google Scholar]
- 31.Aron-Wisnewsky J., Doré J., Clement K. The importance of the gut microbiota after bariatric surgery. Nat. Rev. Gastroenterol. Hepatol. 2012;9:590–598. doi: 10.1038/nrgastro.2012.161. [DOI] [PubMed] [Google Scholar]
- 32.Cotter P.D. Small intestine and microbiota. Curr. Opin. Gastroenterol. 2011;27:99–105. doi: 10.1097/MOG.0b013e328341dc67. [DOI] [PubMed] [Google Scholar]
- 33.Mailhe M., Ricaboni D., Vitton V., Gonzalez J.M., Bachar D., Dubourg G., Cadoret F., Robert C., Delerce J., Levasseur A., et al. Repertoire of the gut microbiota from stomach to colon using culturomics and next-generation sequencing. BMC Microbiol. 2018;18:157. doi: 10.1186/s12866-018-1304-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Seekatz A.M., Schnizlein M.K., Koenigsknecht M.J., Baker J.R., Hasler W.L., Bleske B.E., Young V.B., Sun D. Spatial and Temporal Analysis of the Stomach and Small-Intestinal Microbiota in Fasted Healthy Humans. mSphere. 2019;4:e00126-19. doi: 10.1128/mSphere.00126-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker B.J., Wearsch P.A., Veloo A.C.M., Rodriguez-Palacios A. The Genus Alistipes: Gut Bacteria With Emerging Implications to Inflammation, Cancer, and Mental Health. Front. Immunol. 2020;11:906. doi: 10.3389/fimmu.2020.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S.B., Bonnaure-Mallet M., Jolivet-Gougeon A. Roseburia spp.: A marker of health? Future Microbiol. 2017;12:157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 37.Lombardo Bedran T.B., Marcantonio R.A., Spin Neto R., Alves Mayer M.P., Grenier D., Spolidorio L.C., Spolidorio D.P. Porphyromonas endodontalis in chronic periodontitis: A clinical and microbiological cross-sectional study. J. Oral Microbiol. 2012;4:10123. doi: 10.3402/jom.v4i0.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanner A.C., Paster B.J., Lu S.C., Kanasi E., Kent R., Jr., Van Dyke T., Sonis S.T. Subgingival and tongue microbiota during early periodontitis. J. Dent. Res. 2006;85:318–323. doi: 10.1177/154405910608500407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cani P.D., Depommier C., Derrien M., Everard A., de Vos W.M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022;19:625–637. doi: 10.1038/s41575-022-00631-9. [DOI] [PubMed] [Google Scholar]
- 40.van Passel M.W., Kant R., Zoetendal E.G., Plugge C.M., Derrien M., Malfatti S.A., Chain P.S., Woyke T., Palva A., de Vos W.M., et al. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS ONE. 2011;6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Derrien M., Van Baarlen P., Hooiveld G., Norin E., Müller M., de Vos W.M. Modulation of Mucosal Immune Response, Tolerance, and Proliferation in Mice Colonized by the Mucin-Degrader Akkermansia muciniphila. Front. Microbiol. 2011;2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swidsinski A., Dörffel Y., Loening-Baucke V., Theissig F., Rückert J.C., Ismail M., Rau W.A., Gaschler D., Weizenegger M., Kühn S., et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut. 2011;60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 43.Png C.W., Lindén S.K., Gilshenan K.S., Zoetendal E.G., McSweeney C.S., Sly L.I., McGuckin M.A., Florin T.H. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 44.Li H., Limenitakis J.P., Fuhrer T., Geuking M.B., Lawson M.A., Wyss M., Brugiroux S., Keller I., Macpherson J.A., Rupp S., et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat. Commun. 2015;6:8292. doi: 10.1038/ncomms9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarnawski A.S., Ahluwalia A. The Critical Role of Growth Factors in Gastric Ulcer Healing: The Cellular and Molecular Mechanisms and Potential Clinical Implications. Cells. 2021;10:1964. doi: 10.3390/cells10081964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takashima S., Tanaka F., Kawaguchi Y., Usui Y., Fujimoto K., Nadatani Y., Otani K., Hosomi S., Nagami Y., Kamata N., et al. Proton pump inhibitors enhance intestinal permeability via dysbiosis of gut microbiota under stressed conditions in mice. Neurogastroenterol. Motil. 2020;32:e13841. doi: 10.1111/nmo.13841. [DOI] [PubMed] [Google Scholar]
- 47.Jangi S., Gandhi R., Cox L.M., Li N., von Glehn F., Yan R., Patel B., Mazzola M.A., Liu S., Glanz B.L., et al. Alterations of the human gut microbiome in multiple sclerosis. Nat. Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Huang L., Huang Z., Jiang J., Zhao C., Tong H., Feng Z., Gao J., Liu R., Zhang M., et al. The impacts of acid suppression on duodenal microbiota during the early phase of severe acute pancreatitis. Sci. Rep. 2020;10:20063. doi: 10.1038/s41598-020-77245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fukuba N., Ishihara S., Sonoyama H., Yamashita N., Aimi M., Mishima Y., Mishiro T., Tobita H., Shibagaki K., Oshima N., et al. Proton pump inhibitor is a risk factor for recurrence of common bile duct stones after endoscopic sphincterotomy—propensity score matching analysis. Endosc. Int. Open. 2017;5:E291–E296. doi: 10.1055/s-0043-102936. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.