Abstract
The mycobiome is the fungal component of the human microbial ecosystem that represents only a small part of this environment but plays an essential role in maintaining homeostasis. Colonization by fungi begins immediately after birth. The initial mycobiome is influenced by the gestational age of a newborn, birth weight, delivery method and feeding method. During a human’s life, the composition of the mycobiome is further influenced by a large number of endogenous and exogenous factors. The most important factors are diet, body weight, age, sex and antibiotic and antifungal therapy. The human mycobiome inhabits the oral cavity, gastrointestinal tract, respiratory tract, urogenital tract and skin. Its composition can influence the gut–brain axis through immune and non-immune mediated crosstalk systems. It also interacts with other commensals of the ecosystem through synergistic and antagonistic relationships. Moreover, colonization of the gut by opportunistic fungal pathogens in immunocompromised individuals can lead to clinically relevant disease states. Thus, the mycobiome represents an essential part of the microbiome associated with a variety of physiological and pathological processes. This review summarizes the current knowledge on the composition of the mycobiome in specific sites of the human body and its role in health and disease.
Keywords: fungi, gut mycobiome, oral mycobiome, skin mycobiome, genitourinary tract mycobiome, respiratory tract mycobiome, colonization, composition, dysbiosis
1. Introduction
The ecosystem represents a complex interconnected ecological community of living matter that forms the Earth’s biosphere, where organisms do not live separately, but rather in mutual relationships with each other. Likewise, a healthy human body is colonized by diverse microorganisms taxonomically belonging to all three cellular domains of life (Bacteria, Eukarya and Archaea), as well as non-cellular entities [1].
Fungi are the most abundant eukaryotes, with an estimated number of 2.2 to 3.8 million species, but it is supposed that only up to 8% are completely described [2]. In the past, they were considered as human pathogens and studied as individual species, not as a proportion of the human body’s ecosystem forming mutual relationships. Moreover, traditional cultivation methods could only provide information on morphology and viability but did not reflect an authentic composition of the fungal community due to the high percentage of non-culturable species [3,4]. However, the introduction of new high-throughput amplicon sequencing, shotgun metagenomic technologies and increasing knowledge about the importance of microorganisms as commensals in human health has turned the attention to all microbial members and their intra- and interdomain interactions.
Analogous to the terms bacteriome, archaeome or virome, the human mycobiome refers to the collective genome of all fungi (mycobiota when speaking of living matter itself), specifically unicellular yeasts and filamentous micromycetes, inhabiting our bodies [5,6]. The origin of the scientific phrase human mycobiome dates to 2010, when it was initially used in the article by Ghannoum et al. (2010). To our knowledge, this was the first study that described the oral mycobiome of healthy individuals and showed a significantly higher diversity of oral fungal commensals in comparison with standard culture-based methods [7]. Since then, the number of publications containing the keyword mycobiome has been increasing every year, and the published articles described the mycobiome not only in the oral cavity but also at the other body sites [8,9,10,11].
Although culture-based methods have been largely replaced by molecular-based high-throughput amplicon sequencing methods, the studying of the mycobiome still faces some technological limitations [12]. Different extraction methods can alter the bacterial community composition [12,13]. Among frequently used methods is the targeted sequencing of specific genes encoding ITS1, ITS2 and 18S rRNA regions [12,14]. Unfortunately, the primers designed to amplify these fungal rRNA genes show not only variable results, but also amplify genes of other eukaryotes. At the same time, none of the currently known primers can amplify the genes of all fungi. Moreover, eukaryotic rRNA operons have a variable ITS region length and number of tandem repetitions in different fungal species modifying the results of relative abundances [14,15]. In addition, the choice of reference database can play a role since some species do not have a united taxonomy of teleomorph and anamorph [16]. Thus, these limitations may lead to inconsistent results among studies performed by different research groups. Most of them agree at the phylum level; however, the major differences can be observed at the genus level. An extensive explanation of mycobiome methodology and its challenges are described in the review articles by Tiew and colleagues (2020) [17] and Cui and colleagues (2013) [18].
Up to now, the presence of the human mycobiome has been reported at five sites of the healthy human body, including the oral cavity, intestinal tract, respiratory tract, genitourinary tract and skin. However, it is not excluded that other mucosal surfaces are inhabited as well [7,8,11,19,20]. Naturally, the main focus is on the intestinal (gut) mycobiome, where the fungi play the important role in human health, immune system development and maintaining homeostasis [21,22,23,24]. Despite a high fungal diversity, the mycobiome takes only a minor part of the human microbial community. Its abundance varies across human body sites; for example, in the gastrointestinal tract, it covers less than 0.1%, whereas, on the skin, it takes up to 10% [24,25].
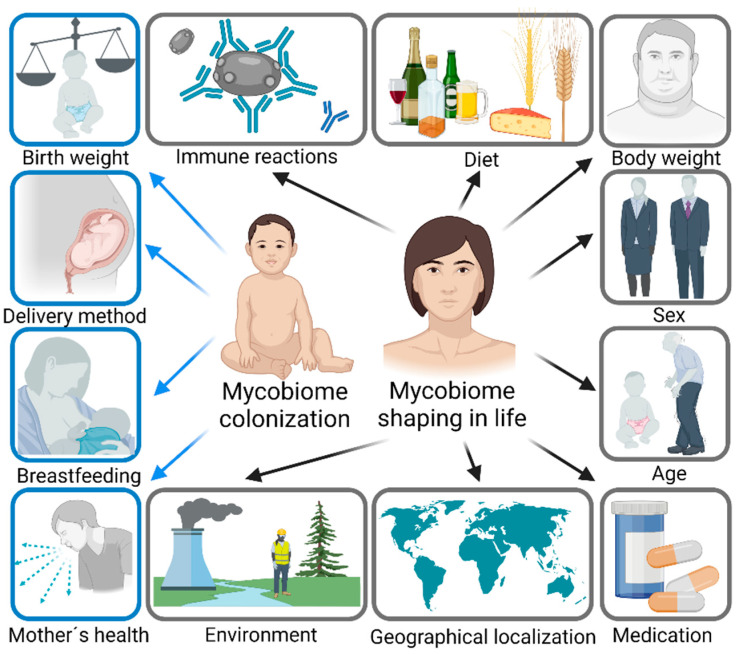
Contrary to the bacterial microbiome, which is relatively stable over time, the composition of the mycobiome varies substantially. Moreover, the human mycobiome is defined by a high interindividual and intraindividual variability [11]. The main factors influencing the development and composition of the mycobiome are described in Figure 1. The human mycobiome is shaped from birth and the source of fungal microorganisms can be either the mother (vertical transmission) or the environment (horizontal transmission) [26,27]. The gestational age, birth weight and delivery method can influence the development of the mycobiome [27,28]. Later on, the composition and the diversity of the mycobiome are largely modulated by the feeding method [26,27]. During adult life, the main factors influencing mycobiome composition are diet [29,30] and body weight [31,32,33]. Fungal microorganisms metabolize and simplify nutrient extraction, and help with digestion through enzyme and vitamin production [31,33,34]. Additional factors include the sex [31,35,36], age [36], antibiotic and antifungal use [37,38], lifestyle, hygiene and geography [19,39,40].
Figure 1.
The main factors influencing the colonization and shaping the composition of the mycobiome in life. The colonization starts immediately after birth. Birth weight, related to gestational age, delivery method and breastfeeding vs. formula milk, as well as health and microbial colonization of relatives, can play a role. During life, the mycobiome is primarily shaped by diet, body weight, sex, age, medication and immune responses, but also the environment and geographical localization. Created with BioRender.com (accessed on 28 September 2022).
As part of the human body’s interconnected ecological community, they form interdomain and intradomain interactions with other colonizers. These interactions were described and summarized in a comprehensive review by Mishra et al. (2021) [41]. It seems that the cooperation of microorganisms could play a crucial role in health and disease. Forming mixed biofilms and the microbial co-habitation of bacteria and fungi could be implicated in, e.g., Crohn´s disease, cystic fibrosis and wound infections, as shown in vitro and in vivo [41].
The Mycobiome as a source of antigens is essential to the training of the immune system and its responses. Fungi activate fungus-specific pattern recognition receptors and immune mechanisms mediating defense against pathogens and tolerance towards helpful commensals [18]. However, there is limited knowledge about these host–fungal interactions in comparison with bacteriome studies [17].
Since the latest studies have described the human mycobiome as a minor but inherent component of the human microbial ecosystem, more efforts should be put into its analysis, with a focus on its role in health and disease. As the colonization of the human body starts along with birth, microorganisms accompany humans and shape their immunity from the very beginning. Modulating the immune system could be only one of many possible ways in which microorganisms influence us. Understanding the compositional changes in the gut mycobiome as well as its communication with other microbes can help us to understand its role in the pathogenesis of various disorders at body sites colonized by these microorganisms.
In the following sections of the article, we describe the mycobial colonization of five human body sites and the composition of their core mycobiome, as well as endogenous and exogenous factors known to influence the composition over life, with the focus on the diseases associated with disturbed mycobiome.
For this complex overview, databases Web of Science and PubMed were searched, using the keywords gut mycobiome, oral mycobiome, skin mycobiome, genitourinary tract mycobiome, respiratory tract mycobiome, colonization, composition and dysbiosis. No exclusion criteria on the date and type of article were applied; however, up-to-date articles were highly preferred. From the total number of searched results, we chose 167 clinical studies, experimental studies and reviews.
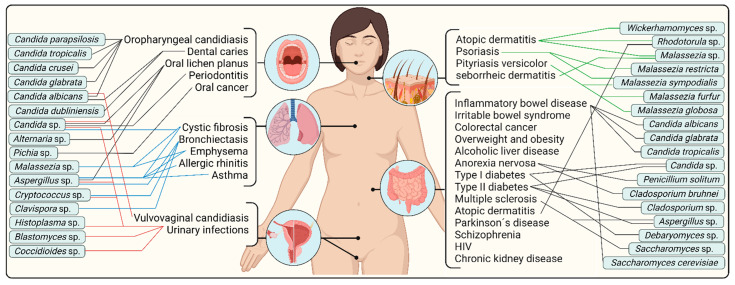
Figure 2 shows a summary scheme of diseases in which the mycobiome could be implicated, as well as fungal taxa potentially involved in the pathogenesis. However, numerous previous studies have already discussed the perturbances in pathophysiologic states [17,18,41]. Therefore, herein, we also focus on the complex overview of a healthy human mycobiome, including less explored body parts to date. The summaries of the most common colonizers of the core mycobiome at various body sites are described extensively in tabular overviews. These provide a comprehensive overview of the most common fungal genera, the relative abundance of the genera or their frequency in clinical samples, the most common species and the list of all species that have been recently described at a particular body site. We believe that knowledge of these, as well as the initial colonization and dynamics of the mycobiome in a healthy person, are key to understanding compositional changes and mechanisms in the disease state.
Figure 2.
The summary of diseases in which gut mycobiome dysbiosis at various body sites could play a role. Fungal taxa potentially involved in the pathogenesis are included, specifically oral cavity mycobiome dysbiosis (black lines on the left), respiratory tract mycobiome dysbiosis (blue lines on the left), genitourinary tract mycobiome dysbiosis (red lines on the left), skin mycobiome dysbiosis (green lines on the right) and gastrointestinal mycobiome dysbiosis (black lines on the right). Created with BioRender.com (accessed on 28 September 2022).
2. Gut Mycobiome
The most studied human body site is the gut, colonized by both morphologic stages of microscopic fungi, yeasts and filamentous micromycetes. Gut colonizers facilitate the digestion of human-indigestible micronutrients and macronutrients, e.g., fiber and oligosaccharides. They produce a wide range of metabolites, such as short and medium chain fatty acids or gases arising from their fermentative metabolisms that could modulate the transition from yeast to a filamentous form (hyphae), oftentimes considered pathogenic, as reviewed in the publication by Sam and colleagues (2017) [42,43]. Moreover, the transport of these molecules could mediate an intercellular communication and be involved in the gut–brain axis, similarly to the bacterial microbiome [22]. In addition, the gut mycobiome plays an important role in the protection of the epithelial barrier against pathogens and stimulates immunity, e.g., through T-cell 17 immune mechanisms [44]. Thus, the gut mycobiome could be implicated in healthy mucosal homeostasis.
2.1. Composition of Gut Mycobiome
It is controversial to define a “core” gut mycobiome of the healthy individual due to high interindividual and intraindividual variability [11]. The presence of the mycobiome has been detected in the gut of at least 70% of healthy adults [45,46]. The most prevalent phylum is Ascomycota, covering 48% to 99% of all present species. Less abundant is the phylum Basidiomycota, with the relative abundance usually ranging from 0.5% to 14% of identified microscopic fungi, followed by the phylum Mucoromycota. It is assumed that these cover approximately 400 species of gut fungi [31,45,46]. A detailed list of chosen fungal genera is described in Table 1.
Table 1.
The list of chosen gut mycobiome colonizers. The table summarizes the relative abundance (or frequency) of representative taxa, number and summary of all species described in the studies.
| Genus | The Most Common Species |
Abundance/ Frequency |
Number of Species | List of all Species | References |
|---|---|---|---|---|---|
| Saccharomyces | S. cerevisiae | ~100% [47]; 96.8%[11]; 23% [48]; 12.80% [49]; 6.94% [31]; 5.1% [50]; 1.14% [35] | 5 | S. cerevisiae, S. paradoxus, S. caariocanus, S. bayanus, S. castellii | [11,31,35,47,48,49,50,51,52,53,54,55,56,57] |
| Candida | Candida sp. | 59.72% [31]; 2.48% [49] | 21 | C. albicans, C. tropicalis, C. dubliniensis, C. glabrata, C. krissi, C. rugosa, C. austromarina, C. parapsilosis, C. lusitaniae, C. pararugosa, C. deformans, C. fermentati, C. intermedia, C. metapsilosis, C. zeylanoides, C. sake, C. krusei, C. kefyr, C. guilliermondii, C. lambica, C. humilis | [11,29,31,32,35,47,48,49,50,51,52,53,54,55,56,58,59] |
| C. albicans | 80.8%[11]; 62.7% [50]; 39.8% [35]; ~30% [47]; 28% [48]; 16.67% [31]; 11.16% [49]; 5.81% [56] | ||||
| Malassezia | M. restricta | 88.30% [11] | 5 | M. slooffiae, M. globosa, M. pachydermatis, M. restricta, M. sympodialis | [29,32,47,49,51,54,58] |
| Penicillium | Penicillium sp. | 65.28% [31]; 7% [11] | 14 | P. turbatum, P. chrysogenum, P. freii, P. camemberti, P. brevicompactum, P. allii, P. dipodomyicola, P. sacculum, P. italicum, P. glabrum, P. verrucosum, P. crutosum, P. paneum, P. roqueforti | [11,29,31,32,35,49,52,54,55,58] |
| P. brevicompactum | 3.72% [35] | ||||
| Aspergillus | Aspergillus sp. | 58.33% [31] | 16 | A. cristasus, A. flavipes, A. glaucus, A. pseudoglaucus, A. oryzeae, A. flavus, A. fumigatus, A. cf. niger, A. cf. carbonarius, A. cf. foetidus, A. cf. tubingensis, A. restrictus, A. versicolor, A. clavatus, A. ochraceoroseus, A. rambelli | [11,29,31,32,35,47,49,54,55,57,58] |
| A. versicolor | 4.95% [49] | ||||
| Galactomyces | G. candidum | 46.1%[11]; 6.98% [56]; 0.83% [49] | 2 | G. geotrichum, G. candidum | [11,32,49,51,56,58] |
| Trichosporon | Trichosporon sp. | 30.56% [31] | 4 | T. asahii, T. faecale, T. caseorum, T. cutaneum | [31,32,35,51,52,59] |
| T. asahii | 0.57% [35] | ||||
| Rhodotorula | Rhodotorula sp. | 8.50% [50] | 4 | R. mucilaginosa, R. rubra, R. graminis, R. glutinis | [31,32,35,50,54,55,56] |
| R. mucilaginosa | 15.28% [31]; 12.6% [35]; 6.98% [56] | ||||
| Geotrichum | G. silvicola | 9.72% [31] | 1 | G. silvicola | [31,32,50,51,59] |
| Cryptococcus | C. carnescens | 4.65% [56] | 6 | C. amylolyticus, C. carnescens, C. neoformans, C. saitoi, C. albidus, C. luteolus | [29,35,47,50,56] |
| Pichia | P. manshurica | 3.43% [35]; 1.39% [31] | 5 | P. kudriavzevii, P. fermentans, P. manshurica, P. kluyveri, P. caribica | [11,29,31,35,54] |
| Exophiala | E. dermatitidis | 1.39% [31] | 4 | E. mesophila, E. heteromorpha, E. equina, E. dermatitidis | [29,31,32,57] |
| Cladosporium | Cladosporium sp. | 1.16% [56] | 1 | C. cladosporioides | [11,54,55,56,58] |
| C. cladosporioides | not found | ||||
| Mucor | M. circinelloides | 0.57% [35] | 2 | M. circinelloides, M. racemosus | [31,35,53,54] |
| Alternaria | A. alternata | not found | 7 | A. alternata, A. cf. arborescens, A cf. brassicola, A. cf. citri, A.cf. mali, A. cf. tenuissima, A. metachromatica | [4,11,29,32,48,55] |
| Debaryomyces | D.fabryi | not found | 3 | D. hansenii, D. fabryi, D. prosopidis | [11,32,52,54,55,57] |
Across the phylum Ascomycota, numerous classes have been identified, namely Saccharomycetes, Dothideomycetes, Sordariomycetes and Eurotiomycetes. At the family level, Saccharomycetaceae, Aspergillaceae, Cladosporiaceae, Debaryomycetaceae, Dipodascaceae and Pichiaceae dominate, followed by less abundant families Ceratocystidaceae, Hypocreaceae, Metschnikowiaceae, Nectriaceae, Thermoascaceae or Microascaceae [30,33,45,60]. The most frequently described yeasts are genera Candida and Saccharomyces [11,31,49,51,52,53,54,55]. Various studies have described tens of other genera, e.g., yeasts Debaryomyces, Meyerozyma, Toluraspora, Pichia, Clavispora, Cyberlidnera, Hanseniaspora, Geotrichum, Galactomyces and Zygosaccharomyces [30,33,45,60]. When analyzing filamentous genera, the most common are Paecilomyces, Cladosporium, Aspergillus and Penicillium [11,31,52,54,55,58]. Some studies have also mentioned Claviceps, Fonsecaea, Exophiala, Eurotium, Phialophora and Scopulariopsis [30,31,33,45,61,62].
From the phylum Basidiomycota, most of the yeasts identified in the gut mycobiome belong to the classes Malasseziomycetes, Tremellomycetes, Agaricomycetes, Microbotryomycetes and Cystobasidiomycetes [31,33,45]. These are represented by families Malasseziaceae, Cryptococcaceae, Corticiaceae, Sporidiobolaceae and Erythrobasidiaceae [31,33,45]. The most commonly described genera are Malassezia, Cryptococcus and Rhodotorula, but Filobasidium and Trichosporon have also been reported [31,33,45].
The least abundant phylum Mucoromycota is represented by the class Mucoromycetes, family Mucoraceae and filamentous genera Mucor and Rhizopus [31,45].
Two mycotypes can be distinguished in the gut. Mycotype 1 is characterized by a high abundance of the genus Saccharomyces, but also other unclassified genera. On the contrary, Mycotype 2 preferably consists of the genera Penicillium, Malassezia and Mucor [55].
2.2. Gut Colonization and Shaping of Gut Mycobiome
The intestinal tract colonization of a newborn can take place both vertically (from mother to child) shortly after its birth or horizontally (from the environment to child) for a long period of time [27,63]. A common example of colonization impact could be a delivery mode. According to the study of European newborns, the earliest colonizers of C-section-born infants are the genus Saccharomyces and the class Exobasidiomycetes, whereas meconium samples of vaginally delivered children are colonized predominantly by the Dothideomycetes and Pezizomycotina [27]. Another method of colonization could be through drinking breast milk. The transition from maternal milk to a more diverse diet shifts its composition [26].
Over life, the diet is considered a crucial factor in modulating the gut mycobiome [34,64,65]. Food-borne fungi could be found in numerous food products: vegetables, fruit, fermented dairy products, meat and fermented beverages as shown in Table 2.
Table 2.
The list of food products colonized or contaminated by food-borne microscopic fungi.
| Food Product | Microscopic Fungi | References |
|---|---|---|
| Fruit and vegetables | ||
| Fresh citrus and grape | Candida prunicola, Pichia fermentans | [66] |
| Fresh apple, plum and pear | Saccharomyces cerevisiae, Pichia kluyveri, Pichia kudriavzevii, Galactomyces candidus, Hanseniaspora uvarum, Hanseniaspora guilliermondii | [67] |
| Peeled fruit salads | Candida sp., Debaryomyces sp., Rhodotorula sp., Penicillium sp., Cladosporium sp. | [68] |
| Dried fruit | Cladosporium sp., Aspergillus niger, Aspergillus tubingiensis, Penicillium palitans | [68,69] |
| Various fresh vegetables (salad, tomato, cucumber, green inion, lettuce, spinach, etc.) | Geotrichum sp., Alternaria sp., Cladosporium sp., Penicillium sp. | [68,70] |
| Dairy | ||
| Various cheeses (Blue cheese, Camembert, Cheddar) | Penicillium sp., Candida sp., Scopulariopsis sp. | [64] |
| Acidophilus milk | Saccharomyces fragilis, Candida pseudotropicalis | [71,72] |
| Meat | ||
| Various meats (fermented sausage, dried meat, salami, ham) |
Debaryomyces sp., Penicillium sp. | [64] |
| Beverages | ||
| Wine | Hanseniaspora sp., Saccharomyces sp. | [73,74] |
| Beer | Brettanomyces sp., Saccharomyces sp. | [73,74] |
| Sake | Aspergillus sp., Saccharomyces sp. | [73] |
| Other | ||
| Various nuts (pecan, almond, walnut, pine nut) | Aspergillus sp. Penicillium sp., Alternaria sp., Cladosporium sp., Rhizopus sp., Fusarium sp. | [69] |
| Koji | Aspergillus sp., Rhizopus sp. | [75] |
| Soy sauce | Aspergillus sp., Hansenula sp., Zygosaccharomyces sp. | [73] |
| Steamed pastry | Wickerhamomyces anomalus | [76,77] |
Some dietary components show a correlation even with the abundance of the specific genus or multiple species [64,65]. Genera Saccharomyces and Hannaella showed positive correlations with butter and animal fats, whereas the genus Aspergillus had a positive correlation with eggs and refined grain. On the contrary, Saccharomyces and Aspergillus correlated negatively with whole grain, but Hannaella correlated negatively with fish and shellfish [78]. Moreover, the genus Fusarium was identified in 88% of vegetarians, but only in 3% of omnivores [29,30]. A short-term switch to a strictly animal-based diet increased the relative abundance of the genus Penicillium, but decreased the genera Debaryomyces and Candida [64].Only approximately 20% of microscopic fungi colonizing the gut permanently inhabit this environment, e.g., the genus Candida and species Geotrichum candidum and Rhodotorula mucilaginosa. The rest, 80%, are considered allochthonous environmental and food-borne fungi, e.g., Aspergillus and Penicillium [30,45].
The gut mycobiome is related to lipid and carbohydrate metabolism through positive and negative correlations with various fungal taxa as shown in Table 3 and Table 4.
Table 3.
The list of negative and positive correlations between lipid metabolism indices and gut mycobiome taxa.
| Lipid Metabolism | |||||
|---|---|---|---|---|---|
| BMI Index and Fat Mass | Serum Total Cholesterol Fasting Triglycerides |
LDL 1 Cholesterol |
HDL 2 Cholesterol |
References | |
|
Negative
correlations |
Agaricomycetes | Eurotiomycetes | Mucoraceae | Sacharomycetes | [33,79] |
| Nectriaceae | Hypocraceae | Tremellomycetes | |||
| Mucoraceae | Mucoraceae | Mucor sp. | Cystobasidiomycetes | ||
| Mucor sp. | Mucor sp. | Candida sp. | Erythrobasidiaceae | ||
| Penicillium sp. | Candida sp. | ||||
|
Positive
correlations |
Sacharomycetes | Dipodascaceae | _ | Aspergillaceae | [33,65,78]. |
| Tremellomycetes | Penicillium sp. | ||||
| Cystobasidiomycetes | Eurotiomycetes | ||||
| Erythrobasidiaceae | Candida dubliniensis | ||||
| Dipodascaceae | |||||
| Aspergillus sp. | |||||
| Hannaella sp. | |||||
1 Low-density lipoprotein cholesterol. 2 High-density lipoprotein cholesterol.
Table 4.
The list of negative and positive correlations between carbohydrate metabolism indices and gut mycobiome taxa.
| Carbohydrate Metabolism | |||||
|---|---|---|---|---|---|
| Glucose | Insulin | Glycated Haemoglobin | References | ||
|
Negative
correlations |
Candida sp. C. dubliniensis |
_ | Agaricomycetes | Mucor sp. | [33,80] |
| Ceratocystidaceae | Monilliela sp. | ||||
| Debariomycetaceae | Eupenicillium sp. | ||||
| Mucoraceae | |||||
| Corticiaceae | Ceratocystis sp. | ||||
|
Positive
correlations |
Eurotium sp. | Ascomycota | Eurotium sp. | - | [33] |
Lipid metabolic factors altering the gut mycobiome are the body mass index, body fat mass, fasting triglycerides, serum total cholesterol, low-density lipoprotein cholesterol and high-density lipoprotein cholesterol [33]. Carbohydrate metabolic parameters that could shape the gut mycobiome composition are fasting glycated hemoglobin, insulin and fasting glucose [33,43]. The gut mycobiomes of overweight and obese individuals differ from healthy eutrophic controls and have their specific composition [31,33]. Predominant genera of overweight patients are yeast Candida and Pichia and filamentous Bipolaris, Beauveria, Exophiala, Syncephalastrum and Helminthosporium [31]. On the contrary, in obese patients, genera Candida, Nakaseomyces and Penicillium, Chaetomium and Emmonsia dominate [31,33].
2.3. Gut Mycobiome Dysbiosis in Disease
It is important to distinguish what is and is not a healthy body state anymore and influence the onset of pathogenic processes. Intestinal dysbiosis leads to altered interactions among members of the microbiome and mycobiome, respectively, and these influence microbial penetration across the gut barrier [24].
A modified gut mycobiome composition was shown in numerous conditions: inflammatory bowel disease [81], irritable bowel syndrome [82], colorectal cancer [57], obesity [31,33], alcoholic liver disease [83], diabetes type I and II [79,84,85,86], multiple sclerosis [78,87], chronic kidney disease [88], atopic dermatitis [89], Parkinson’s disease [90], schizophrenia [91], etc. Moreover, correlations of fungal taxa with innate and adaptive immunity, e.g., effector CD4+ T cells and CD8+ T cells, memory CD4+ T cells, NK cells, regulatory B cells and other proinflammatory cytokines, were observed [78,92].
The gut mycobiome of patients diagnosed with inflammatory bowel disease displayed compositional differences compared to the control group; however, there were no disturbances between mycobiomes of Crohn´s disease and ulcerative colitis [56,81]. When investigating the most common gut colonizers, an increased abundance of C. albicans [81], C. glabrata and C. tropicalis was observed in the patients with IBD [93,94,95]. Additionally, the relative abundance of C. albicans significantly correlated with the remission and relapse stages of inflammatory bowel disease, while another species, C. tropicalis, displayed interactions with anti-Saccharomyces cerevisiae antibodies that are known as biomarkers associated with Crohn´s disease [81,94]. However, the proportion of the species Saccharomyces cerevisiae appeared controversial [81,96]. Whereas Sokol et al. (2017) described a decrease in the relative abundance of this species, Lewis et al. (2015) reported its increase in Crohn´s disease [96].
A considerable biodiversity decrease was illustrated in overweight and obese individuals in contrast to healthy eutrophic controls, respectively. A comprehensive analysis of young anorexic woman exposed unique mycobiome species Penicillium solitum and Cladosporium bruhnei [32]. Nevertheless, a study of 59 anorexia nervosa patients mentioned no changes in alpha diversity [97].
Modulating the gut microbial colonizers through dietary components could have an anti-diabetic activity [98]. The gut mycobiome in type II diabetes displayed compositional differences at the phylum level, as well as at the genus level, compared to healthy individuals [84,85]. An increase in the relative abundance of Candida, Meyerozyma, Aspergillus and Cladosporium was shown. In contrast, 21 genera were significantly reduced in patients, e.g., Saccharomyces, Trichoderma, Clavispora, Rhizopus and Wickerhamomyces [84,85]. The bacterial and fungal dysbiosis in type I diabetic children with beta-cell autoimmunity was in association with intestinal inflammation [86]. An increased relative abundance of the genus Debaryomyces and decreased abundance of Malassezia were disclosed [86]. An increased abundance of the genus Candida was reported in diabetic patients [79,99]. This increased abundance of saccharide-metabolizing yeast could probably be a consequence of elevated blood glucose levels or modified immunological responses.
The gut mycobiome of multiple sclerosis patients had a higher alpha diversity, interindividual variability and over-representation of genera Saccharomyces and Aspergillus compared to a healthy control group [78]. Despite that, it preserved its stability for a 6-month period, suggesting that common species are permanent colonizers over time. Gut mycobiome enrichment and numerous correlations with bacterial taxa were reported in people with HIV [92]. The overgrowth of the genus Rhodotorula was related to atopic dermatitis manifestation in infants [89].
Provided data suggest that fungal dysbiosis may contribute to the pathogenesis of numerous intestinal and extraintestinal disorders. Knowledge of the intradomain and interdomain interaction of fungal and bacterial colonizers in gut mycobiome dysbiosis could lead to the development of novel therapeutic strategies in the future.
3. Oral Mycobiome
The oral microbiome is one of the most complex and diverse microbial ecosystems in the human body [100]. While eating, foodborne microscopic fungi (or their conidia) pass through our digestive system. Starting from the oral cavity, the oral mycobiome has been relatively well described [7]. The inter-kingdom interactions between bacteria and fungi are essential in the oral cavity and contribute to oral health and the establishment of distal gastrointestinal tract functions [100,101,102]. These interactions are crucial for maintaining the balance between health and disease, not only locally but also systematically [103]. Highly abundant genera, such as Candida, are associated with a distinct oral ecology of a decreased pH. Some of these fungal species can serve as the main etiological agent of oral mucosal mycosis. However, the role of other metabolically active colonizers, such as the lipid-dependent Malassezia, is still unclear. Less abundant oral mycobiome species could potentially serve as opportunistic pathogens in immunocompromised hosts [104].
3.1. Composition of Oral Mycobiome
The composition of the oral mycobiome is stable, yet it shows a high interindividual variability [100]. The majority of the oral adult mycobiome composition represents the phylum Ascomycota, Basidiomycota, Glomeromycota and Mucoromycota. These four groups include approximately 81 genera and 101 species of fungi [7,105]. A detailed list of chosen fungal genera is described in Table 5.
Table 5.
The list of chosen oral mycobiome colonizers. The table summarizes the relative abundance (or frequency) of representative taxa, number and summary of all species described in the studies.
| Genus | The Most Common Species |
Abundance/ Frequency |
Number of Species | List of All Species | References |
|---|---|---|---|---|---|
| Candida | Candida sp. | 100% [106]; 100% [105]; 85% [101]; 75% [7]; 67% [100] | 6 | C. albicans, C. khmerensis, C. metapsilosis, C. parapsilosis, C. tropicalis, C. glabrata | [7,61,100,101,105,106,107,108] |
| Aspergillus | Aspergillus sp. | 100% [106]; 100% [105]; 75% [100]; 45% [101]; 35% [7] | 7 | A. amstelodami, A. caesiellus, A. flavus, A. oryzae, A. penicillioides, A. ruber, A. niger | [7,61,100,101,105,106,107,108] |
| Penicillium | Penicillium sp. | 97% [105]; 85% [100]; 70% [101]; 50% [106] | 4 | P. brevicompactum, P. glabrum, P. spinulosum, P. citrinum | [7,61,100,101,105,107] |
| Cladosporium | Cladosporium sp. | 100% [106]; 72.5% [100]; 65% [7] | 4 | C. cladosporioides, C. herbarum, C. sphaerospermu, C. teniussimum | [7,61,100,106,107,108] |
| Phoma | Phoma sp. | 100% [106]; 65% [101] | 3 | P. foveata, P. plurivora, P. herbarum | [7,61,101,106] |
| Malassezia | Malassezia sp. | 100% [106]; 40% [101] | 3 | M. restricta, M. globosa, M. sympodialis | [106,107,108] |
| Cryptococcus | Cryptococcus sp. | 100% [106]; 20% [7] | 2 | C. cellulolyticus, C. diffluens | [7,106] |
| Alternaria | Alternaria sp. | 100% [106]; 5% [100] | 4 | A. tenuissima, A. triticina, A. alternata, A. armoraciae | [7,100,101,106,107,108] |
| Fusarium | Fusarium sp. | 83% [106]; 30% [7] | 3 | F. oxysporum, F. culmorum, F. poae | [7,107,108] |
| Rhodotorula | Rhodotorula sp. | 75% [100] | 1 | R. mucilaginosa | [100,107,108] |
| Aureobasidium | Aureobasidium sp. | 67% [106]; 50% [7] | 0 | not classified | [7,100,108] |
| Saccharomyces | Saccharomyces sp. | 50% [7]; 50% [106]; 45% [101] | 3 | S. bayanus, S. cerevisiae, S. ellipsoideus | [7,61,101,107] |
| Trichoderma | Trichoderma sp. | 50% [106]; 45% [101]; 10% [100] | 1 | T. aureoviride | [100,101,107] |
The most common representatives of the basal oral mycobiome originate from the phylum Ascomycota, namely the yeast-like genus Candida [7,61,101,109,110]. Less prevalent yeasts that have been reported are Aureobasidium, Saccharomyces and Pichia [7,61,100,107,108]. Some of the identified filamentous micromycetes are genera Penicillium, Cladosporium, Aspergillus and Alternaria [7,61,100,101,105,106,107,108]. Some studies reported airborne fungi, genera Phoma and Epicoccum, whose spores could be inhaled by the study participants without the subsequent colonization of the oral cavity [7,61,106].
Basidiomycota representatives include Rhodotorula, Malassezia and Cryptococcus. Minor relative abundances of other representatives are rarely reported [7,61,105,106].
3.2. Oral Cavity Colonization and Shaping of Oral Mycobiome
The colonization of the oral cavity by fungi starts during childbirth [103] and progresses with age [102]. The abundance of Candida, one of the most abundant genera of the oral cavity, is age-dependent. Its relative abundance is lower in infants compared to one-year-old children [111]. The mycobiome composition is similar in children born by either vaginal or C-section delivery, but the relative abundance of certain species, such as C. orthopsilosis, is significantly higher in section-born infants [112,113]. The fungal colonization is higher in neonates born vaginally than in those born by C-section. Late colonization may also occur due to longer stays in the neonatal intensive care unit [103]. The very low birth weight neonates were more frequently colonized by non-albicans Candida sp. than neonates with a birth weight above 1500 g, who were mostly colonized by C. albicans [103]. Children’s oral cavity contains over 40 fungal species with the dominant species C. albicans [40]. Most studies suggest that the type of feeding does not influence the oral mycobiome composition [102,103]. Candida species abundance is the same for infants who are breastfed, bottle-fed or those who are on both patterns of feeding [114]. Candida species can also contribute to dental caries development [40].
The significant associations of the oral mycobiome composition with an increased age, teeth loss [105], low salivary flow rates and presence of dental caries [40,112], but also diet, personal oral hygiene, stress and drug exposure [115], were observed. Geographical differences were also described, e.g., a lower oral Candida abundance in healthy people in Asia compared with Western countries [116]. In addition, C. glabrata and C. dubliniensis were detected in the oral mycobiome of people with a low BMI [116].
3.3. Oral Mycobiome Dysbiosis in Disease
The dysbiosis of the oral mycobiome influences interactions between fungi and bacteria and might be associated with numerous conditions, e.g., dental caries [117], oropharyngeal candidiasis [118], oral lichen planus [101], periodontitis [105] and others.
Numerous studies have suggested a possible role of the genus Candida, the common isolate, in oral dysbiosis. This genus can participate in biofilm structure and function. It is able to form dentinal tubules and bind to denatured collagen and secrete aspartyl proteases, resulting in the demineralization and dissolution of the dental hard tissues [40]. The abundance of species C. albicans and C. dubliniensis correlated with dental caries in early childhood, indicating that C. dubliniensis could be implicated in disease pathogenesis [117]. A higher abundance of C. glabrata resulted in tongue infections and the interactions between C. glabrata and hyphae of C. albicans established oropharyngeal candidiasis [118]. Other etiological agents of oropharyngeal candidiasis were C. parapsilosis, C. tropicalis and C. crusei [118]. Fungal dysbiosis was also associated with the oral lichen planus, the chronic oral mucosa disease. Significantly higher abundances of genera Candida and Aspergillus in patients with reticular oral lichen planus and of Alternaria and Sclerotiniaceae in patients with erosive oral lichen planus were observed [101]. On the contrary, no compositional changes in the overall mycobiome diversity were shown in patients with periodontal disease compared to healthy study individuals [105]. Peters et al. (2017) manifested that the abundance of the genus Candida was increased in patients, but not significantly [105].
A recent study proposed that compositional alterations of the oral mycobiome caused by smoking tobacco could be implicated in oral cancer by increasing the abundance of pathogenic fungi that promote opportunistic infections [119]. A decreased oral fungal diversity and increase in the genus Pichia correlated with the severity of the oral lesions.
4. Skin Mycobiome
The skin is the first barrier between the environment and the human body, protecting against potential pathogens [120]. However, it is colonized by numerous commensal microorganisms, maintaining homeostasis through the production of antimicrobial peptides and protection against external harmful microorganisms, and mediating lipid and urea degradation by lipase and urease enzymes secretion [121,122]. These commensals contribute to immune response development and are involved in potential microbial communication with bacteria, modulating their physiology and virulence [123].
4.1. Composition of Skin Mycobiome
The human skin fungal ecosystem represents less than 10% of all skin microbes [9]. The skin mycobiome consists of at least 168 genera, taxonomically originating from the phyla Basidiomycota and Ascomycota [19,39,124], and these are body-site-specific [39]. Parts of the body in pairs, such as forearms and palms, are colonized by the same dominant taxa [39]. A detailed list of chosen fungal genera is described in Table 6.
Table 6.
The list of chosen skin mycobiome colonizers. The table summarizes the relative abundance (or frequency) of representative taxa, number and summary of all species described in the studies.
| Site | Genus | The Most Common Species | Abundance/ Frequency |
Number of Species | List of All Species | References |
|---|---|---|---|---|---|---|
| Scalp | Malassezia | M sympodialis | 23.8–57.1% [125] | 7 | M. sympodialis, M. furfur, M. globosa, M. restricta, M. sympodialis, M. dermatis, M. slooffiae | [125,126] |
| Candida | C. albicans | 50–65% [126] | 2 | C. albicans, C. metapsilosis | [125,126] | |
| Aspergillus | Aspergillus sp. | not found | 0 | not classified | [125] | |
| Cryptococcus | Cryptococcus sp. | not found | 0 | not classified | [126] | |
| Aureobasidium | Aureobasidium sp. | not found | 0 | not classified | [125] | |
| Acremonium | Acremonium sp. | not found | 0 | not classified | [126] | |
| Cladosporium | Cladosporium sp. | not found | 0 | not classified | [125] | |
| Hortaea | Hortaea sp. | not found | 0 | not classified | [125] | |
| Trichosporon | Trichosporon sp. | not found | 0 | not classified | [125] | |
| Pallidocercospora | Pallidocercospora sp. | not found | 0 | not classified | [125] | |
| Didymella | Didymella sp. | not found | 0 | not classified | [126] | |
| Areas with a high density of sebaceous glands |
Malassezia | M. globosa | 91% [127] 13.5–56.8% [125] | 6 | M. globosa, M. furfur, M. restricta, M. sympodialis, M. dermatis, M. slooffiae | [125,127] |
| Candida | C. parapsilosis | 6.25–35.1% [125] | 2 |
C. parapsilosis, C. orthopsilosis |
[125] | |
| Alternaria | Alternaria sp. | not found | 0 | not classified | [120] | |
| Cladosporium | Cladosporium sp. | not found | 0 | not classified | [120] | |
| Cryptococcus | Cryptococcus sp. | not found | 0 | not classified | [120] | |
| Schizophillum | Schizophillum sp. | not found | 0 | not classified | [120] | |
| Rhodotorula | R. mucilaginosa | 13.5% [120]; 0.62% [125] | 1 | R. mucilaginosa | [120,125] | |
| Penicillium | Penicillium sp. | not found | 0 | not classified | [125] | |
| Lodderomyces | L. elongisporus | not found | 1 | L. elongisporus | [125] | |
| Rhodosporium | R. toloroides | not found | 1 | R. toloroides | [125] | |
| Naganisha | N. albidosimilis | not found | 1 | N. albidosimilis | [125] | |
| Areas with a low density of sebaceous glands |
Malassezia | Malassezia sp. | 24.48–43.43% [120] | 6 | M. globosa M. restricta, M. furfur, M. obtuse, M. sympodialis, M. japonica | [120] |
| Mucor | Mucor sp. | not found | 0 | not classified | [127] | |
| Neurospora | Neurospora sp. | not found | 0 | not classified | [127] | |
| Aspergillus | Aspergillus sp. | not found | 0 | not classified | [127] | |
| Preussia | Preussia sp. | not found | 0 | not classified | [127] | |
| Saccharomyces | Saccharomyces sp. | not found | 0 | not classified | [127] | |
| Rasamsonia | Rasamsonia sp. | not found | 0 | not classified | [120] | |
| Alternaria | Alternaria sp. | not found | 0 | not classified | [120] | |
| Cladosporium | Cladosporium sp. | 5.71–9.85% [120] | 0 | not classified | [120] | |
| Candida | Candida sp. | 4.42–7.05% [120] | 0 | not classified | [120] | |
| Cryptococcus | Cryptococcus sp. | not found | 0 | not classified | [120] | |
| Rhodotorula | Rhodotorula sp. | not found | 0 | not classified | [120] | |
| Penicillium | Penicillium sp. | not found | 0 | not classified | [120] |
The most common colonizer of human skin is the yeast of the genus Malassezia. It dominates several parts of the body, such as the inner side of the elbows, back, outer auditory canal, nostril and various facial areas. Within each observed sample, it represents more than 57% of the microscopic community on average [19,124].
From the Basidiomycota, the skin of healthy people is colonized by the genus Cryptococcus [19,39]. Rarely identified are other genera, e.g., Aspergillus, Penicillium and Epicoccum [19,123].
4.2. Skin Colonization and Shaping of Skin Mycobiome
The colonization of the skin by fungi starts during childbirth and is characterized by a high inter-individual variability [27,113,120]. Children born vaginally most likely acquire microscopic fungi from the mother´s vagina, whereas children born via C-section acquire fungi primarily found on the mother´s skin [27,113]. The relative abundance of specific taxa, including Saccharomyces cerevisiae, Candida tropicalis, C. parapsilosis, C. albicans and C. orthopsilosis, was influenced by the delivery mode in the study of American infants [113]. No alpha diversity differences were recorded during the first month of life between vaginal and C-section delivery. However, only vaginally derived infants showed an alpha diversity increase over the first month of life [113]. On the contrary, a study of Chinese infants proposed no significant change in the mycobiome composition within the first six months of life [120].
The skin mycobiome changes and develops compositionally from childhood to adulthood, with a profound shift in community composition during puberty. The most common species of pre-puberty children were Aspergillus, Epicoccum and Phoma. The dominant species of Malassezia before puberty was M. globosa. On the contrary, individuals in the post-pubertal age were predominantly colonized by M. restricta. Gender also affected the composition of the skin mycobiome. Epicoccum and Cryptococcus were more abundant in adolescent boys, whereas, in girls, it was the genus Malassezia [36].
During life, the skin mycobiome could vary across body site localization or hygiene as shown by culture-dependent and independent methods [19,128,129,130]. Sebaceous body sites (e.g., face, chest and back) had a higher fungal diversity of lipophilic species compared to moist or dry skin sites [120,128]. Site specification is typical for Malassezia: the higher abundance of M. globosa on the scalp and forehead and lower abundance on the back were observed, while the relative abundance of M. sympodialis showed an inverse pattern [123]. Malassezia arunalokei was more abundant on the forehead and cheek compared to the scalp [129]. Seasonal and gender-related changes in the skin mycobiome were observed [131,132].
4.3. Skin Mycobiome Dysbiosis in Disease
The skin mycobiome comprises harmless commensals; however, it also comprises opportunistic pathogens, whose overgrowth leads to morphological changes from budding yeasts to a pseudohyphal form invading the stratum corneum layer and promoting skin infections [133]. Inter-kingdom and inter-species microbial interactions can worsen the disease or change relationships from opportunistic to pathogenic [131,134,135]. Fungal dysbiosis could be associated with numerous skin diseases often present in a site-specific manner, including atopic dermatitis in the arm and leg creases [136], psoriasis on the elbows and knees [132], pityriasis versicolor [137], seborrheic dermatitis [138], etc. The prevalence of fungal infections on the skin, hair and nails varies from 20 to 25% worldwide [139].
The dysbiosis of the major commensal skin fungi, Malassezia sp., is associated with numerous skin disorders [123,137]. The species Malassezia globosa colonized antecubital flexures at the neck of healthy individuals but was absent in patients with atopic dermatitis [136]. Similarly, a higher relative abundance of M. globosa in controls was reported by another study [140]. This growth restriction could be a consequence of dry skin and insufficient growth conditions usually provided by the bacterium Cutibacterium acnes, which were decreased in patients with atopic dermatitis as well [136]. Furthermore, Malassezia restricta and Malassezia sympodialis were significantly associated with psoriasis at the back and elbow body sites as shown by LEfSe analysis [135]. In addition, Malassezia globosa has been considered a common etiological agent of Pityriasis versicolor (also known as tinea versicolor), present as hyperpigmented and hypopigmented skin lesions [133,141,142]. Nevertheless, species Malassezia furfur and Malassezia sympodialis have been isolated from the clinical isolates as well [137]. Lastly, an increased relative abundance of the genus Malassezia and decreased fungal diversity were reported in the study of patients with facial seborrheic dermatitis [38]. These were modified by ketoconazole treatment.
Besides the genus Malassezia, the perturbances of the skin mycobiome have been shown; for instance, in the study of nine 12-month-old infants with atopic dermatitis. Specifically, genera Rhodotorula, Wickerhamomyces, Kodamaea, Acremonium and Rhizopus showed the highest rates of perturbance [89].
5. Genitourinary Tract Mycobiome
Fungi are important members of the vaginal ecosystem in healthy women and the vaginal mycobiome was first described by Castellani in 1929 [8,143]. Candida albicans, an opportunistic fungal pathogen, colonizes 20% of women without causing any overt symptoms, yet is one of the leading causes of infectious vaginitis. Understanding its mechanisms of commensalism and pathogenesis is essential to developing more effective therapies [144]. In contrast, before the advent of culture-independent analyses, the uninfected urinary tract had been assumed to be a sterile environment [145].
5.1. Composition of Genitourinary Mycobiome
The vaginal mycobiome consists primarily of yeast-like microorganisms originating from Ascomycota and Basidiomycota [8,143]. The research of the team Guo et al. (2012) [146] mentions peronosporomycota (oomycota) as well, but these have not been reported in other studies so far. At least 22 genera of microscopic fungi naturally inhabit the vaginal microenvironment of women [144]. A detailed list of chosen fungal genera is described in Table 7.
Table 7.
The list of chosen genitourinary tract mycobiome colonizers. The table summarizes the relative abundance (or frequency) of representative taxa, number and summary of all species described in the studies.
| Site | Genus | The Most Common Species | Abundance/ Frequency |
Number of Species | List of all Species | References |
|---|---|---|---|---|---|---|
| Urinary tract | Candida | Candida sp. | not found | 1 | not classified | [145,147] |
| Saccharomyces | Saccharomyces sp. | not found | 1 | not classified | [145] | |
| Malassezia | Malassezia sp. | not found | 1 | not classified | [145] | |
| Vagina | Candida | Candida sp. | 1–69% [148]; 36.9% [8] | 9 | C. albicans, C. glabrata, C. krusei, C. tropicalis, C. parapsilosis, C. dubliniensis, C. guillermondi, C. kefyr, C. humicolus, | [8,148] |
| C. albicans | 34.1 [8] | |||||
| Saccharomyces | S. cerevisiae | 5% [148] | 1 | S. cerevisiae | [148,149] | |
| Cladosporium | C. perangustum | >1% [8] | 1 | C. perangustum | [8] | |
| Alternaria | A. alternata | >1% [8] | 1 | A. alternata | [8] | |
| Rhodotorula | Rhodotorula sp. | >1% [8] | 0 | not classified | [8] |
The vast majority of genera belong to Ascomycota and order Saccharomycetales, similarly to the oral cavity and gut mycobiome [8]. The most ubiquitous natural colonizer, which is a part of the vaginal mycobiome of almost every healthy woman, is the genus Candida and, namely, the species C. albicans [8,148]. The second most common species is C. glabrata [148,149], but numerous other species have been identified in the vaginal mycobiome so far, e.g., C. krusei, C. tropicalis, C. parapsilosis, C. dubliniensis and C. guillermondi [8,146,149]. Another representative commonly occurring in the vaginal mycobiome is the species S. cerevisiae [149].
Other taxa from the Ascomycota of yeasts and filamentous fungi that have been identified are from orders Capnodiales, Eurotiales, Pleosporales and Helotiales, namely the species Cladosporium perangustum, Eurotium amslodami and Alternaria alternata [8]. From the Basidiomycota, the yeast-like genus Rhodotorula has been reported [8].
The mycobiome of the urinary tract has not been clarified at present. However, one study described genera Candida, Saccharomyces and Malassezia as potential colonizers [145].
5.2. Genitourinary Tract Colonization and Shaping of Genitourinary Mycobiome
Within weeks after birth, fungi colonize various body sites [150]. A chronic colonization of the urogenital tract with fungi is common, especially for Candida spp. The carriage rate of Candida among healthy adults ranges from 30 to 70% [126].
The source of the vaginal mycobiome is a dietary intake, environmental contacts and living habits [146]. Similarly to other body sites, the composition of the initial vaginal mycobiome is influenced by the type of delivery, as vaginally delivered children possess a mycobiome originating from their mother´s vagina, whereas the children delivered by C-section obtain mycobiome from the mother’s skin and environment [150,151]. The vaginal mycobiome is different in women with a menstruation cycle, and in pre-adolescent, post-climacteric or pregnant women. The composition changes with the vaginal epithelium structure and depends on the level of sex hormones [150].
The composition of the urinary tract mycobiome is influenced by a multitude of factors, including age, gender, environmental variations and hygiene [145].
5.3. Genitourinary Mycobiome Dysbiosis in Disease
The vaginal microbiome and mycobiome are closely associated with the development of reproductive disorders. The dysbiosis of the vaginal microenvironment, together with immune defects and an infraction of epithelial integrity, leads to vulvovaginal candidiasis, one of the most common forms of infectious vaginitis caused by C. albicans [144]. The dysbiosis of Lactobacillus spp. and other lactic-acid bacteria, producing short chain fatty acids and lactate, probably allows for the switch of this dimorphic fungus from budding yeast to a virulent pseudohyphal form [144,152,153].
The study of intrauterine adhesion patients described Candida-related differences in the gut mycobiome composition and numerous fungal–bacterial correlations in cervical canal and middle vagina sites [154]. There are multiple studies describing the role of Candida strains in the context of vaginal pathophysiology, but there are no studies describing the comprehensive impact of vaginal mycobiome dysbiosis on women´s health. However, some studies hypothesized that vaginal microbial dysbiosis could contribute to human papillomavirus infection and carcinogenesis [155].
Fungal urinary infections are usually caused by the dysbiosis of Candida spp., but also the common invasive fungal species, including Cryptococcus, Aspergillus, Mucoraceae, Histoplasma, Blastomyces and Coccidioides. Fungal urinary tract infections are life-threatening due to the limitations in culture methodologies, frequent low suspicion of fungal involvement and the lack of preventative and therapeutic options [145]. As the urinary tract was considered sterile until recently and only a very limited number of articles have been devoted to the urinary tract mycobiome, it is not possible to deduce conclusions regarding the dysbiosis of this body site.
6. Respiratory Tract Mycobiome
Until recently, it was thought that the lungs of healthy people are sterile. However, it was found that the respiratory tract, including the lungs, have diverse microbiota influencing the pathogenesis of different diseases [156,157]. The lung mycobiome has its composition and evolution, which is unique and specific to each individual [158]. In healthy people, fungal spores, which are parts of the environment, are effectively removed from the respiratory tract through mucociliary clearance and phagocytosis [159,160]. In contrast, in immunosuppressed patients and in patients with a defect such as neutropenia, asthma or cavitating lung disease, fungal spores can persist and colonize, leading to the development of disease [160,161].
6.1. Composition of Respiratory Mycobiome
The number of studies describing respiratory mycobiome is limited. Despite that, already published literature describes mostly Ascomycota and Basidiomycota phyla at various sections of the respiratory tract, as well as those obtained by various sampling methods [20,110,159]. A detailed list of chosen fungal genera is described in Table 8.
Table 8.
The list of chosen respiratory tract mycobiome colonizers. The table summarizes representative taxa, number and summary of all species described in the studies.
| Site | Genus | The Most Common Species | Number of Species | List of Species | References |
|---|---|---|---|---|---|
| Lungs | Cladosporium | Cladosporium sp. | 0 | not classified | [156] |
| Penicillium | Penicillium sp. | 0 | not classified | [156] | |
| Aspergillus | Aspergillus sp. | 0 | not classified | [156] | |
| Candida | Candida sp. | 0 | not classified | [156] | |
| Malassezia | Malassezia sp. | 0 | not classified | [156] | |
| Kluyveromyces | Kluyveromyces sp. | 0 | not classified | [156] | |
| Pneumocystis | Pneumocystis sp. | 0 | not classified | [126,156] | |
| Bronchoalveolar lavage | Cladosporium | Cladosporium sp. | 0 | not classified | [145] |
| Candida | C. guilliermondii | 2 | C. guilliermondii, C. albicans | [20,156] | |
| Aspergillus | Aspergillus sp. | 2 | A. fumigatus, A. flavus | [145,156] | |
| Penicillium | Penicillium sp. | 0 | not classified | [145] | |
| Saccharomyces | S. cerevisiae | 0 | not classified | [20] | |
| Pichia | Pichia sp. | 2 | P. Jadini, P. brevicompactum | [20] | |
| Exhaled breath Condensate (EBC) |
Cladosporium | Cladosporium sp. | 1 | C. herbarum | [159] |
| Aspergillus | A. sydowii | 1 | A. sydowii | [159] | |
| Penicillium | P. brevicompactum | 5 | P. brevicompactum, P. Expansum, P. glabrum, P. olsonii, P. bilaiae | [159] | |
| Alternaria | A. alternata | 2 | A. alternata, A. infectoria | [159] | |
| Sputum | Saccharomyces | Saccharomyces sp. | 0 | not classified | [162] |
| Candida | Candida sp. | 2 | C. albicans, C. dubliniensis | [162] | |
| Ganoderma | Ganoderma sp. | 0 | not classified | [20] | |
| Malassezia | Malassezia sp. | 0 | not classified | [162] | |
| Aspergillus | A. fumigatus | 1 | A. fumigatus | [20] | |
| Cryptococcus | C. magnus | 1 | C. magnus | [20] | |
| Peniophora | P. incarnata | 2 | P. Incarnata, P. Cinerea | [20] | |
| Daedaleopsis | D. confragosa | 1 | D. confragosa | [20] | |
| Sistotrema | S. brinkmannii | 1 | S. brinkmannii | [20] | |
| Stereum | S. hirsutum | 1 | S. hirsutum | [20] |
Through the sequencing of the exhaled breath condensate samples, genera Cladosporium, Aspergillus and Alternaria and numerous Penicillium species were described [159]. On the contrary, analysis of the bronchoalveolar lavage samples revealed yeast-like fungi Saccharomyces cerevisiae, Candida, Meyerozyma guilliermondii (Candida guilliermondii), Pichia jadini and Debaryomyces and filamentous Cladosporium, Aspergillus and Penicillium [20,110,161]. However, the abundance of some of these taxa could have been a result of the lung transplantation that patients underwent [110].
The induced sputum examination showed C. albicans, C. dubliniensis and Aspergillus. From the Basidiomycota, Cryptococcus magnus was identified, but also numerous other species, such as Peniophora incarnata, Peniophora cinerea, Daedaleopsis confragosa, Sistotrema brinkmannii and Stereum hirsutum, that occur in nature in connection with plants [20]. We assume that these species could get into the airways with microaspiration and are not permanent colonizers of the respiratory tract.
6.2. Respiratory Tract Colonization and Shaping of Respiratory Mycobiome
In the past, studies on respiratory tract colonization focused mostly on microorganisms in the context of pathogens that could potentially trigger diseases in humans [163,164]. Due to this, respiratory tract mycobiome studies have been lacking. Up to 2015, it was not clear whether there is a respiratory tract mycobiome in immunocompetent individuals without the pathogenic condition or not [20]. Some studies indicated that the presence of eukaryotic microorganisms in the respiratory tract is transient since the abundance of these species fluctuates over time [164]. The entry of these species into the airways can be a result of micro-aspiration from the oral cavity [100]. Other studies proposed that, although the composition of the mycobiome from individual airway sections overlaps, these sections also contain genera specific to only one of them. In addition, the respiratory mycobiome also shows differences compared to oral cavity mycobiome [20]. From these results, we suppose that the respiratory tract can be colonized by fungi permanently.
The respiratory tract mycobiome varies throughout parts of the respiratory system as shown above in the composition chapter [157]. It is also influenced by the composition of other mycobiomes. The intestinal microbiome influences the oropharyngeal mycobiome, which may influence the respiratory mycobiome and host immune response through micro-aspiration [100,158]. Other factors influencing the respiratory tract mycobiome are geographic variability (climate, humidity and air quality), genetics, lifestyle and dietary differences [17].
6.3. Respiratory Tract Mycobiome Dysbiosis in Disease
Mycobiome dysbiosis contributes to respiratory disease pathogenesis. Characteristic mycobiome profiles exist in chronic respiratory diseases [17]. The increased abundance of Malassezia, Candida and Aspergillus in patients with cystic fibrosis was observed [17,158,165]. In addition, a shift in the Basidiomycota:Ascomycota ratio was observed [161,165]. A higher abundance of Aspergillus, Cryptococcus and Clavispora was observed in patients with bronchiectasis [17]. Aspergillus and Malassezia are associated with combined pulmonary fibrosis and emphysema.
A human invariably inhales fungal spores, which are potential allergens [166]. Aspergillus spores can be implicated in the airway’s allergic disease production, such as allergic rhinitis and asthma [24]. An increased chance of asthma and allergies is also influenced by antibiotics use, which allows for the overgrowth of commensal fungi [24,167]. The most frequent species in the airways of asthma patients is Malassezia pachydermatis, the species associated with atopic dermatitis [166]. In addition to this, an increased abundance of Psathyrella, Malassezia, Termitomyces and Grifola was observed [17]. However, the effect of respiratory tract mycobiome dysbiosis on systemic health is currently not known.
7. Conclusions
Despite constituting a small proportion of the whole microbial ecosystem, the mycobiome plays an essential role in maintaining host homeostasis. Gut and oral mycobiomes have already been well-described, but studies on the skin, genitourinary and respiratory tract mycobiomes are lacking. Many already published papers have focused either on specific yeasts and cultivation methods or have attributed the presence of fungi to infection and not to the natural microflora. Therefore, we believe that the added value of this article is a complex tabular overview of a healthy mycobiome of less-explored body sites provided on the basis of recent literature. Knowledge of the natural fungal flora would help us to understand the changes in dysbiosis.
One possible explanation for why mycobiome research is still falling behind the bacterial microbiome could be that the research methodology is not fully standardized yet compared with the 16S rRNA microbiome analysis, and the fungi represent a minor part of the microbial composition, increasing the risk of contamination. Moreover, only limited information is known about its interactions with other commensals at the moment. Therefore, more efforts should be put into analyzing not only its variable composition but also the intercommunication with other microorganisms and released microbial metabolites, interactions with the host organism and their complex impact on human health. We believe that these could be potential studies in future research. Knowledge of these could provide a comprehensive point of view, helping to enlighten the pathogenesis of various disorders at body sites colonized by these microorganisms.
Acknowledgments
Artwork was created in BioRender.com (accessed on 9 September 2022).
Author Contributions
Conceptualization, P.B., P.S. and P.V.; writing—original draft preparation, P.B.; writing—review and editing, R.G.; visualization, P.B.; supervision, P.S., P.V. and R.G.; funding acquisition, R.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Slovak Research and Development Agency under the contract no. APVV-17-0505 and APVV-21-0370 and by the Ministry of Education, Science, Research and Sport of the Slovak Republic under the contract no. VEGA 1/0649/21. Authors thank the Research Infrastructure RECETOX (No. LM2018121) financed by the Ministry of Education, Youth and Sports and the OP RDE-project CETOCOEN EXCELLENCE (No CZ.02.1.01/0.0/0.0/17_043/0009632) and CETOCOEN Plus (CZ.02.1.01/0.0/0.0/15_003/0000469) for supportive background. This work was supported from the European Union’s Horizon 2020 research and innovation programme under grant agreement 857560. This publication reflects only the author’s view and the European Commission is not responsible for any use that may be made of the information it contains.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Matijasic M., Mestrovic T., Paljetak H.C., Peric M., Baresic A., Verbanac D. Gut Microbiota beyond Bacteria-Mycobiome, Virome, Archaeome, and Eukaryotic Parasites in IBD. Int. J. Mol. Sci. 2020;21:2668. doi: 10.3390/ijms21082668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hawksworth D.L., Lücking R. Fungal Diversity Revisited: 2.2 to 3.8 Million Species. Microbiol. Spectr. 2017;5:4. doi: 10.1128/microbiolspec.FUNK-0052-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seed P.C. The Human Mycobiome. Cold Spring Harb. Perspect. Med. 2015;5:a019810. doi: 10.1101/cshperspect.a019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gouba N., Drancourt M. Digestive Tract Mycobiota: A Source of Infection. Médecine Et Mal. Infect. 2015;45:9–16. doi: 10.1016/j.medmal.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Underhill D.M., Iliev I.D. The Mycobiota: Interactions between Commensal Fungi and the Host Immune System. Nat. Rev. Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santus W., Devlin J.R., Behnsen J. Crossing Kingdoms: How the Mycobiota and Fungal-Bacterial Interactions Impact Host Health and Disease. Infect. Immun. 2021;89:e00648–20. doi: 10.1128/IAI.00648-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghannoum M.A., Jurevic R.J., Mukherjee P.K., Cui F., Sikaroodi M., Naqvi A., Gillevet P.M. Characterization of the Oral Fungal Microbiome (Mycobiome) in Healthy Individuals. PLoS Pathog. 2010;6:e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drell T., Lillsaar T., Tummeleht L., Simm J., Aaspõllu A., Väin E., Saarma I., Salumets A., Donders G.G.G., Metsis M. Characterization of the Vaginal Micro- and Mycobiome in Asymptomatic Reproductive-Age Estonian Women. PLoS ONE. 2013;8:e54379. doi: 10.1371/journal.pone.0054379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh J., Freeman A.F., NISC Comparative Sequencing Program. Park M., Sokolic R., Candotti F., Holland S.M., Segre J.A., Kong H.H. The Altered Landscape of the Human Skin Microbiome in Patients with Primary Immunodeficiencies. Genome Res. 2013;23:2103–2114. doi: 10.1101/gr.159467.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Woerden H.C., Gregory C., Brown R., Marchesi J.R., Hoogendoorn B., Matthews I.P. Differences in Fungi Present in Induced Sputum Samples from Asthma Patients and Non-Atopic Controls: A Community Based Case Control Study. BMC Infect. Dis. 2013;13:69. doi: 10.1186/1471-2334-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash A.K., Auchtung T.A., Wong M.C., Smith D.P., Gesell J.R., Ross M.C., Stewart C.J., Metcalf G.A., Muzny D.M., Gibbs R.A., et al. The gut mycobiome of the Human Microbiome Project healthy cohort. Microbiome. 2017;5:153. doi: 10.1186/s40168-017-0373-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frau A., Kenny J.G., Lenzi L., Campbell B.J., Ijaz U.Z., Duckworth C.A., Burkitt M.D., Hall N., Anson J., Darby A.C., et al. DNA Extraction and Amplicon Production Strategies Deeply Inf Luence the Outcome of Gut Mycobiome Studies. Sci. Rep. 2019;9:9328. doi: 10.1038/s41598-019-44974-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huseyin C.E., Rubio R.C., O’Sullivan O., Cotter P.D., Scanlan P.D. The Fungal Frontier: A Comparative Analysis of Methods Used in the Study of the Human Gut Mycobiome. Front. Microbiol. 2017;8:1432. doi: 10.3389/fmicb.2017.01432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Filippis F., Laiola M., Blaiotta G., Ercolini D. Different Amplicon Targets for Sequencing-Based Studies of Fungal Diversity. Appl. Environ. Microbiol. 2017;83:e00905-17. doi: 10.1128/AEM.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schoch C.L., Seifert K.A., Huhndorf S., Robert V., Spouge J.L., Levesque C.A., Chen W., Bolchacova E., Voigt K., Crous P.W., et al. Nuclear Ribosomal Internal Transcribed Spacer (ITS) Region as a Universal DNA Barcode Marker for Fungi. Proc. Natl. Acad. Sci. USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huseyin C.E., O’Toole P.W., Cotter P.D., Scanlan P.D. Forgotten Fungi—the Gut Mycobiome in Human Health and Disease. Fems Microbiol. Rev. 2017;41:479–511. doi: 10.1093/femsre/fuw047. [DOI] [PubMed] [Google Scholar]
- 17.Tiew P.Y., Mac Aogain M., Ali N.A.B.M., Thng K.X., Goh K., Lau K.J.X., Chotirmall S.H. The Mycobiome in Health and Disease: Emerging Concepts, Methodologies and Challenges. Mycopathologia. 2020;185:207–231. doi: 10.1007/s11046-019-00413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui L., Morris A., Ghedin E. The Human Mycobiome in Health and Disease. Genome Med. 2013;5:63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J.A., Schoenfeld D., Nomicos E., Park M., NIH Intramural Sequencing Center Comparative Sequencing Program et al. Topographic Diversity of Fungal and Bacterial Communities in Human Skin. Nature. 2013;498:367. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cui L., Lucht L., Tipton L., Rogers M.B., Fitch A., Kessinger C., Camp D., Kingsley L., Leo N., Greenblatt R.M., et al. Topographic Diversity of the Respiratory Tract Mycobiome and Alteration in HIV and Lung Disease. Am. J. Respir. Crit. Care Med. 2015;191:932–942. doi: 10.1164/rccm.201409-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin V.K., Yong V.C., Chong P.P., Amin Nordin S., Basir R., Abdullah M. Mycobiome in the Gut: A Multiperspective Review. Mediat. Inflamm. 2020;2020:9560684. doi: 10.1155/2020/9560684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enaud R., Vandenborght L.-E., Coron N., Bazin T., Prevel R., Schaeverbeke T., Berger P., Fayon M., Lamireau T., Delhaes L. The Mycobiome: A Neglected Component in the Microbiota-Gut-Brain Axis. Microorganisms. 2018;6:UNSP 22. doi: 10.3390/microorganisms6010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallen-Adams H.E., Suhr M.J. Fungi in the Healthy Human Gastrointestinal Tract. Virulence. 2016;8:352–358. doi: 10.1080/21505594.2016.1247140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai G.C., Tan T.G., Pavelka N. The Mammalian Mycobiome: A Complex System in a Dynamic Relationship with the Host. Wiley Interdiscip. Rev. Syst. Biol. Med. 2018;11:e1438. doi: 10.1002/wsbm.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes J.D., Bernstein C.N., Tremlett H., Van Domselaar G., Knox N.C. A Fungal World: Could the Gut Mycobiome Be Involved in Neurological Disease? Front. Microbiol. 2019;9:3249. doi: 10.3389/fmicb.2018.03249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schei K., Avershina E., Øien T., Rudi K., Follestad T., Salamati S., Ødegård R.A. Early Gut Mycobiota and Mother-Offspring Transfer. Microbiome. 2017;5:107. doi: 10.1186/s40168-017-0319-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wampach L., Heintz-Buschart A., Hogan A., Muller E.E.L., Narayanasamy S., Laczny C.C., Hugerth L.W., Bindl L., Bottu J., Andersson A.F., et al. Colonization and Succession within the Human Gut Microbiome by Archaea, Bacteria, and Microeukaryotes during the First Year of Life. Front. Microbiol. 2017;8:738. doi: 10.3389/fmicb.2017.00738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward T.L., Knights D., Gale C.A. Infant Fungal Communities: Current Knowledge and Research Opportunities. BMC Med. 2017;15:30. doi: 10.1186/s12916-017-0802-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suhr M.J., Banjara N., Hallen-Adams H.E. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett. Appl. Microbiol. 2016;62:209–215. doi: 10.1111/lam.12539. [DOI] [PubMed] [Google Scholar]
- 30.Hallen-Adams H.E., Kachman S.D., Kim J., Legge R.M., Martínez I. Fungi Inhabiting the Healthy Human Gastrointestinal Tract: A Diverse and Dynamic Community. Fungal Ecol. 2015;15:9–17. doi: 10.1016/j.funeco.2015.01.006. [DOI] [Google Scholar]
- 31.Borges F.M., de Paula T.O., Sarmiento M.R.A., de Oliveira M.G., Pereira M.L.M., Toledo I.V., Nascimento T.C., Ferreira-Machado A.B., Silva V.L., Diniz C.G. Fungal Diversity of Human Gut Microbiota Among Eutrophic, Overweight, and Obese Individuals Based on Aerobic Culture-Dependent Approach. Curr. Microbiol. 2018;75:726–735. doi: 10.1007/s00284-018-1438-8. [DOI] [PubMed] [Google Scholar]
- 32.Gouba N., Raoult D., Drancourt M. Gut Microeukaryotes during Anorexia Nervosa: A Case Report. BMC Res. Notes. 2014;7:33. doi: 10.1186/1756-0500-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mar Rodriguez M., Perez D., Javier Chaves F., Esteve E., Marin-Garcia P., Xifra G., Vendrell J., Jove M., Pamplona R., Ricart W., et al. Obesity Changes the Human Gut Mycobiome. Sci. Rep. 2015;5:14600. doi: 10.1038/srep14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G.D., Lewis J.D., Bushman F.D. Archaea and Fungi of the Human Gut Microbiome: Correlations with Diet and Bacterial Residents. PLoS ONE. 2013;8:e66019. doi: 10.1371/journal.pone.0066019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strati F., Di Paola M., Stefanini I., Albanese D., Rizzetto L., Lionetti P., Calabrò A., Jousson O., Donati C., Cavalieri D., et al. Age and Gender Affect the Composition of Fungal Population of the Human Gastrointestinal Tract. Front. Microbiol. 2016;7:1227. doi: 10.3389/fmicb.2016.01227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jo J.-H., Deming C., Kennedy E.A., Conlan S., Polley E.C., Ng W.-I., Segre J.A., Kong H.H. Diverse Human Skin Fungal Communities in Children Converge in Adulthood. J. Invest. Dermatol. 2016;136:2356–2363. doi: 10.1016/j.jid.2016.05.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezeonu I.M., Ntun N.W., Ugwu K.O. Intestinal Candidiasis and Antibiotic Usage in Children: Case Study of Nsukka, South Eastern Nigeria. Afr. Health Sci. 2017;17:1178–1184. doi: 10.4314/ahs.v17i4.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tao R., Wang R., Wan Z., Song Y., Wu Y., Li R. Ketoconazole 2% Cream Alters the Skin Fungal Microbiome in Seborrhoeic Dermatitis: A Cohort Study. Clin. Exp. Dermatol. 2022;47:1088–1096. doi: 10.1111/ced.15115. [DOI] [PubMed] [Google Scholar]
- 39.Leung M.H.Y., Chan K.C.K., Lee P.K.H. Skin Fungal Community and Its Correlation with Bacterial Community of Urban Chinese Individuals. Microbiome. 2016;4:46. doi: 10.1186/s40168-016-0192-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fechney J.M., Browne G.V., Prabhu N., Irinyi L., Meyer W., Hughes T., Bockmann M., Townsend G., Salehi H., Adler C.J. Preliminary Study of the Oral Mycobiome of Children with and without Dental Caries. J. Oral Microbiology. 2019;11:1536182. doi: 10.1080/20002297.2018.1536182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra K., Bukavina L., Ghannoum M. Symbiosis and Dysbiosis of the Human Mycobiome. Front. Microbiol. 2021;12:636131. doi: 10.3389/fmicb.2021.636131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sam Q.H., Chang M.W., Chai L.Y.A. The Fungal Mycobiome and Its Interaction with Gut Bacteria in the Host. Int. J. Mol. Sci. 2017;18:330. doi: 10.3390/ijms18020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mims T.S., Abdallah Q.A., Stewart J.D., Watts S.P., White C.T., Rousselle T.V., Gosain A., Bajwa A., Han J.C., Willis K.A., et al. The Gut Mycobiome of Healthy Mice Is Shaped by the Environment and Correlates with Metabolic Outcomes in Response to Diet. Commun. Biol. 2021;4:281. doi: 10.1038/s42003-021-01820-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leonardi I., Gao I.H., Lin W.-Y., Allen M., Li X.V., Fiers W.D., De Celie M.B., Putzel G.G., Yantiss R.K., Johncilla M., et al. Mucosal Fungi Promote Gut Barrier Function and Social Behavior via Type 17 Immunity. Cell. 2022;185:831–846. doi: 10.1016/j.cell.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raimondi S., Amaretti A., Gozzoli C., Simone M., Righini L., Candeliere F., Brun P., Ardizzoni A., Colombari B., Paulone S., et al. Longitudinal Survey of Fungi in the Human Gut: ITS Profiling, Phenotyping, and Colonization. Front. Microbiol. 2019;10:1575. doi: 10.3389/fmicb.2019.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schulze J., Sonnenborn U. Yeasts in the Gut: From Commensals to Infectious Agents. Dtsch. Arztebl. Int. 2009;106:837–842. doi: 10.3238/arztebl.2009.0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Q., Wang C., Tang C., He Q., Li N., Li J. Dysbiosis of Gut Fungal Microbiota Is Associated with Mucosal Inflammation in Crohn’s Disease. J. Clin. Gastroenterol. 2014;48:513–523. doi: 10.1097/MCG.0000000000000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huët M.A.L., Wong L.W., Goh C.B.S., Hussain M.H., Muzahid N.H., Dwiyanto J., Lee S.W.H., Ayub Q., Reidpath D., Lee S.M., et al. Investigation of Culturable Human Gut Mycobiota from the Segamat Community in Johor, Malaysia. World. J. Microbiol. Biotechnol. 2021;37:113. doi: 10.1007/s11274-021-03083-6. [DOI] [PubMed] [Google Scholar]
- 49.Chen Y., Chen Z., Guo R., Chen N., Lu H., Huang S., Wang J., Li L. Correlation between Gastrointestinal Fungi and Varying Degrees of Chronic Hepatitis B Virus Infection. Diagn. Microbiol. Infect. Dis. 2011;70:492–498. doi: 10.1016/j.diagmicrobio.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Khatib R., Riederer K.M., Ramanathan J., Baran J. Faecal Fungal Flora in Healthy Volunteers and Inpatients. Mycoses. 2001;44:151–156. doi: 10.1046/j.1439-0507.2001.00639.x. [DOI] [PubMed] [Google Scholar]
- 51.Hamad I., Sokhna C., Raoult D., Bittar F. Molecular Detection of Eukaryotes in a Single Human Stool Sample from Senegal. PLoS ONE. 2012;7:e40888. doi: 10.1371/journal.pone.0040888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Motooka D., Fujimoto K., Tanaka R., Yaguchi T., Gotoh K., Maeda Y., Furuta Y., Kurakawa T., Goto N., Yasunaga T., et al. Fungal ITS1 Deep-Sequencing Strategies to Reconstruct the Composition of a 26-Species Community and Evaluation of the Gut Mycobiota of Healthy Japanese Individuals. Front. Microbiol. 2017;8:238. doi: 10.3389/fmicb.2017.00238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pandey P.K., Siddharth J., Verma P., Bavdekar A., Patole M.S., Shouche Y.S. Molecular Typing of Fecal Eukaryotic Microbiota of Human Infants and Their Respective Mothers. J. Biosci. 2012;37:221–226. doi: 10.1007/s12038-012-9197-3. [DOI] [PubMed] [Google Scholar]
- 54.Kabwe M.H., Vikram S., Mulaudzi K., Jansson J.K., Makhalanyane T.P. The Gut Mycobiota of Rural and Urban Individuals Is Shaped by Geography. BMC Microbiol. 2020;20:257. doi: 10.1186/s12866-020-01907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Botschuijver S., Roeselers G., Levin E., Jonkers D.M., Welting O., Heinsbroek S.E.M., de Weerd H.H., Boekhout T., Fornai M., Masclee A.A., et al. Intestinal Fungal Dysbiosis Is Associated with Visceral Hypersensitivity in Patients with Irritable Bowel Syndrome and Rats. Gastroenterology. 2017;153:1026–1039. doi: 10.1053/j.gastro.2017.06.004. [DOI] [PubMed] [Google Scholar]
- 56.Ott S.J., Kühbacher T., Musfeldt M., Rosenstiel P., Hellmig S., Rehman A., Drews O., Weichert W., Timmis K.N., Schreiber S. Fungi and Inflammatory Bowel Diseases: Alterations of Composition and Diversity. Scand. J. Gastroenterol. 2008;43:831–841. doi: 10.1080/00365520801935434. [DOI] [PubMed] [Google Scholar]
- 57.Coker O.O., Nakatsu G., Dai R.Z., Wu W.K.K., Wong S.H., Ng S.C., Chan F.K.L., Sung J.J.Y., Yu J. Enteric Fungal Microbiota Dysbiosis and Ecological Alterations in Colorectal Cancer. Gut. 2019;68:654–662. doi: 10.1136/gutjnl-2018-317178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gouba N., Raoult D., Drancourt M. Plant and Fungal Diversity in Gut Microbiota as Revealed by Molecular and Culture Investigations. PLoS ONE. 2013;8:e59474. doi: 10.1371/journal.pone.0059474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Agırbaslı H., Özcan S.A.K., Gedikoğlu G. Fecal Fungal Flora of Pediatric Healthy Volunteers and Immunosuppressed Patients. Mycopathologia. 2005;159:515–520. doi: 10.1007/s11046-005-3451-2. [DOI] [PubMed] [Google Scholar]
- 60.Robert V., Stegehuis G., Stalpers J. The MycoBank Engine and Related Databases. 2005. [(accessed on 15 August 2022)]. Available online: www.mycobank.org.
- 61.Auchtung T.A., Fofanova T.Y., Stewart C.J., Nash A.K., Wong M.C., Gesell J.R., Auchtung J.M., Ajami N.J., Petrosino J.F. Investigating Colonization of the Healthy Adult Gastrointestinal Tract by Fungi. mSphere. 2018;3:e00092. doi: 10.1128/mSphere.00092-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richard M.L., Lamas B., Liguori G., Hoffmann T.W., Sokol H. Gut Fungal Microbiota: The Yin and Yang of Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015;21:656–665. doi: 10.1097/MIB.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 63.Nagata R., Nagano H., Ogishima D., Nakamura Y., Hiruma M., Sugita T. Transmission of the Major Skin Microbiota, Malassezia, from Mother to Neonate. Pediatr. Int. 2012;54:350–355. doi: 10.1111/j.1442-200X.2012.03563.x. [DOI] [PubMed] [Google Scholar]
- 64.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sun Y., Zuo T., Cheung C.P., Gu W., Wan Y., Zhang F., Chen N., Zhan H., Yeoh Y.K., Niu J., et al. Population-Level Configurations of Gut Mycobiome Across 6 Ethnicities in Urban and Rural China. Gastroenterology. 2021;160:272–286. doi: 10.1053/j.gastro.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 66.Moubasher A.-A.H., Abdel–Sater M.A., Soliman Z. Biodiversity of Filamentous and Yeast Fungi in Citrus and Grape Fruits and Juices in Assiut Area, Egypt. JMBFS. 2018;7:353–365. doi: 10.15414/jmbfs.2018.7.4.353-365. [DOI] [Google Scholar]
- 67.Vadkertiová R., Molnárová J., Vránová D., Sláviková E. Yeasts and Yeast-like Organisms Associated with Fruits and Blossoms of Different Fruit Trees. Can. J. Microbiol. 2012;58:1344–1352. doi: 10.1139/cjm-2012-0468. [DOI] [PubMed] [Google Scholar]
- 68.Tournas V.H., Heeres J., Burgess L. Moulds and Yeasts in Fruit Salads and Fruit Juices. Food Microbiol. 2006;23:684–688. doi: 10.1016/j.fm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 69.Tournas V., Niazi N., Kohn J. Fungal Presence in Selected Tree Nuts and Dried Fruits. Microbiol. Insights. 2015;8:1–6. doi: 10.4137/MBI.S24308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tournas V.H. Moulds and Yeasts in Fresh and Minimally Processed Vegetables, and Sprouts. Int. J. Food Microbiol. 2005;99:71–77. doi: 10.1016/j.ijfoodmicro.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 71.Reed G., Nagodawithana T.W. Yeast Technology. Springer; Dordrecht, The Netherlands: 1990. Use of Yeasts in the Dairy Industry; pp. 441–445. [Google Scholar]
- 72.Griffin S., Falzon O., Camilleri K., Valdramidis V.P. Bacterial and Fungal Contaminants in Caprine and Ovine Cheese: A Meta-Analysis Assessment. Food Res. Int. 2020;137:109445. doi: 10.1016/j.foodres.2020.109445. [DOI] [PubMed] [Google Scholar]
- 73.Venturini Copetti M. Yeasts and Molds in Fermented Food Production: An Ancient Bioprocess. Curr. Opin. Food Sci. 2019;25:57–61. doi: 10.1016/j.cofs.2019.02.014. [DOI] [Google Scholar]
- 74.Jolly N.P., Varela C., Pretorius I.S. Not Your Ordinary Yeast: Non-Saccharomyces Yeasts in Wine Production Uncovered. FEMS Yeast Res. 2014;14:215–237. doi: 10.1111/1567-1364.12111. [DOI] [PubMed] [Google Scholar]
- 75.Endo A., Irisawa T., Dicks L., Tanasupawat S. FERMENTED FOODS|Fermentations of East and Southeast Asia. In: Batt C.A., Tortorello M.L., editors. Encyclopedia of Food Microbiology. 2nd ed. Academic Press; Oxford, UK: 2014. pp. 846–851. [Google Scholar]
- 76.Suo B., Nie W., Wang Y., Ma J., Xing X., Huang Z., Xu C., Li Z., Ai Z. Microbial Diversity of Fermented Dough and Volatile Compounds in Steamed Bread Prepared with Traditional Chinese Starters. LWT. 2020;126:109350. doi: 10.1016/j.lwt.2020.109350. [DOI] [Google Scholar]
- 77.Li Z., Li H., Song K., Cui M. Performance of Non-Saccharomyces Yeasts Isolated from Jiaozi in Dough Fermentation and Steamed Bread Making. LWT. 2019;111:46–54. doi: 10.1016/j.lwt.2019.05.019. [DOI] [Google Scholar]
- 78.Shah S., Locca A., Dorsett Y., Cantoni C., Ghezzi L., Lin Q., Bokoliya S., Panier H., Suther C., Gormley M., et al. Alterations of the Gut Mycobiome in Patients with MS. EBioMedicine. 2021;71:103557. doi: 10.1016/j.ebiom.2021.103557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gosiewski T., Salamon D., Szopa M., Sroka A., Malecki M.T., Bulanda M. Quantitative Evaluation of Fungi of the Genus Candida in the Feces of Adult Patients with Type 1 and 2 Diabetes—a Pilot Study. Gut. Pathog. 2014;6:43. doi: 10.1186/s13099-014-0043-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chiaro T.R., Soto R., Stephens W.Z., Kubinak J.L., Petersen C., Gogokhia L., Bell R., Delgado J.C., Cox J., Voth W., et al. A Member of the Gut Mycobiota Modulates Host Purine Metabolism Exacerbating Colitis in Mice. Sci. Transl. Med. 2017;9:eaaf9044. doi: 10.1126/scitranslmed.aaf9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sokol H., Leducq V., Aschard H., Pham H.-P., Jegou S., Landman C., Cohen D., Liguori G., Bourrier A., Nion-Larmurier I., et al. Fungal Microbiota Dysbiosis in IBD. Gut. 2017;66:1039–1048. doi: 10.1136/gutjnl-2015-310746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hong G., Li Y., Yang M., Li G., Qian W., Xiong H., Bai T., Song J., Zhang L., Hou X. Gut Fungal Dysbiosis and Altered Bacterial-Fungal Interaction in Patients with Diarrhea-Predominant Irritable Bowel Syndrome: An Explorative Study. Neurogastroenterol. Motil. 2020;32:e13891. doi: 10.1111/nmo.13891. [DOI] [PubMed] [Google Scholar]
- 83.Lang S., Duan Y., Liu J., Torralba M.G., Kuelbs C., Ventura-Cots M., Abraldes J.G., Bosques-Padilla F., Verna E.C., Brown R.S., et al. Intestinal Fungal Dysbiosis and Systemic Immune Response to Fungi in Patients With Alcoholic Hepatitis. Hepatology. 2020;71:522–538. doi: 10.1002/hep.30832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jayasudha R., Das T., Kalyana Chakravarthy S., Sai Prashanthi G., Bhargava A., Tyagi M., Rani P.K., Pappuru R.R., Shivaji S. Gut Mycobiomes Are Altered in People with Type 2 Diabetes Mellitus and Diabetic Retinopathy. PLoS ONE. 2020;15:e0243077. doi: 10.1371/journal.pone.0243077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bhute S.S., Suryavanshi M.V., Joshi S.M., Yajnik C.S., Shouche Y.S., Ghaskadbi S.S. Gut Microbial Diversity Assessment of Indian Type-2-Diabetics Reveals Alterations in Eubacteria, Archaea, and Eukaryotes. Front. Microbiol. 2017;8:214. doi: 10.3389/fmicb.2017.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Honkanen J., Vuorela A., Muthas D., Orivuori L., Luopajärvi K., Tejesvi M.V.G., Lavrinienko A., Pirttilä A.M., Fogarty C.L., Härkönen T., et al. Fungal Dysbiosis and Intestinal Inflammation in Children with Beta-Cell Autoimmunity. Front. Immunol. 2020;11:468. doi: 10.3389/fimmu.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yadav M., Ali S., Shrode R.L., Shahi S.K., Jensen S.N., Hoang J., Cassidy S., Olalde H., Guseva N., Paullus M., et al. Multiple Sclerosis Patients Have an Altered Gut Mycobiome and Increased Fungal to Bacterial Richness. PLoS ONE. 2022;17:e0264556. doi: 10.1371/journal.pone.0264556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hu J., Wei S., Gu Y., Wang Y., Feng Y., Sheng J., Hu L., Gu C., Jiang P., Tian Y., et al. Gut Mycobiome in Patients with Chronic Kidney Disease Was Altered and Associated with Immunological Profiles. Front. Immunol. 2022;13:843695. doi: 10.3389/fimmu.2022.843695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mok K., Suratanon N., Roytrakul S., Charoenlappanit S., Patumcharoenpol P., Chatchatee P., Vongsangnak W., Nakphaichit M. ITS2 Sequencing and Targeted Meta-Proteomics of Infant Gut Mycobiome Reveal the Functional Role of Rhodotorula Sp. during Atopic Dermatitis Manifestation. J. Fungi. 2021;7:748. doi: 10.3390/jof7090748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pablo-Fernandez E.D., Gebeyehu G.G., Flain L., Slater R., Frau A., Ijaz U.Z., Warner T., Probert C. The Faecal Metabolome and Mycobiome in Parkinson’s Disease. Parkinsonism Relat. Disord. 2022;95:65–69. doi: 10.1016/j.parkreldis.2022.01.005. [DOI] [PubMed] [Google Scholar]
- 91.Zhang X., Pan L., Zhang Z., Zhou Y., Jiang H., Ruan B. Analysis of Gut Mycobiota in First-Episode, Drug-Naïve Chinese Patients with Schizophrenia: A Pilot Study. Behav. Brain Res. 2020;379:112374. doi: 10.1016/j.bbr.2019.112374. [DOI] [PubMed] [Google Scholar]
- 92.Gosalbes M.J., Jimenéz-Hernandéz N., Moreno E., Artacho A., Pons X., Ruíz-Pérez S., Navia B., Estrada V., Manzano M., Talavera-Rodriguez A., et al. Interactions among the Mycobiome, Bacteriome, Inflammation, and Diet in People Living with HIV. Gut Microbes. 2022;14:2089002. doi: 10.1080/19490976.2022.2089002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chehoud C., Albenberg L.G., Judge C., Hoffmann C., Grunberg S., Bittinger K., Baldassano R.N., Lewis J.D., Bushman F.D., Wu G.D. Fungal Signature in the Gut Microbiota of Pediatric Patients with Inflammatory Bowel Disease. Inflamm. Bowel Dis. 2015;21:1948–1956. doi: 10.1097/MIB.0000000000000454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hoarau G., Mukherjee P.K., Gower-Rousseau C., Hager C., Chandra J., Retuerto M.A., Neut C., Vermeire S., Clemente J., Colombel J.F., et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio. 2016;7:e01250-16. doi: 10.1128/mBio.01250-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liguori G., Lamas B., Richard M.L., Brandi G., da Costa G., Hoffmann T.W., Di Simone M.P., Calabrese C., Poggioli G., Langella P., et al. Fungal Dysbiosis in Mucosa-Associated Microbiota of Crohn’s Disease Patients. J. Crohns Colitis. 2016;10:296–305. doi: 10.1093/ecco-jcc/jjv209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lewis J.D., Chen E.Z., Baldassano R.N., Otley A.R., Griffiths A.M., Lee D., Bittinger K., Bailey A., Friedman E.S., Hoffmann C., et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe. 2015;18:489–500. doi: 10.1016/j.chom.2015.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Prochazkova P., Roubalova R., Dvorak J., Kreisinger J., Hill M., Tlaskalova-Hogenova H., Tomasova P., Pelantova H., Cermakova M., Kuzma M., et al. The Intestinal Microbiota and Metabolites in Patients with Anorexia Nervosa. Gut Microbes. 2021;13:1902771. doi: 10.1080/19490976.2021.1902771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li B.-Y., Xu X.-Y., Gan R.-Y., Sun Q.-C., Meng J.-M., Shang A., Mao Q.-Q., Li H.-B. Targeting Gut Microbiota for the Prevention and Management of Diabetes Mellitus by Dietary Natural Products. Foods. 2019;8:440. doi: 10.3390/foods8100440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Soyucen E., Gulcan A., Aktuglu-Zeybek A.C., Onal H., Kiykim E., Aydin A. Differences in the Gut Microbiota of Healthy Children and Those with Type 1 Diabetes. Pediatr. Int. 2014;56:336–343. doi: 10.1111/ped.12243. [DOI] [PubMed] [Google Scholar]
- 100.Monteiro-da-Silva F., Araujo R., Sampaio-Maia B. Interindividual Variability and Intraindividual Stability of Oral Fungal Microbiota over Time. Med. Mycol. 2014;52:498–505. doi: 10.1093/mmy/myu027. [DOI] [PubMed] [Google Scholar]
- 101.Li Y., Wang K., Zhang B., Tu Q., Yao Y., Cui B., Ren B., He J., Shen X., Van Nostrand J.D., et al. Salivary Mycobiome Dysbiosis and Its Potential Impact on Bacteriome Shifts and Host Immunity in Oral Lichen Planus. Int. J. Oral Sci. 2019;11:1–10. doi: 10.1038/s41368-019-0045-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oba P.M., Holscher H.D., Mathai R.A., Kim J., Swanson K.S. Diet Influences the Oral Microbiota of Infants during the First Six Months of Life. Nutrients. 2020;12:3400. doi: 10.3390/nu12113400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Azevedo M.J., Pereira M.d.L., Araujo R., Ramalho C., Zaura E., Sampaio-Maia B. Influence of Delivery and Feeding Mode in Oral Fungi Colonization—A Systematic Review. Microb. Cell. 2020;7:36–45. doi: 10.15698/mic2020.02.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Diaz P.I., Dongari-Bagtzoglou A. Critically Appraising the Significance of the Oral Mycobiome. J. Dent. Res. 2021;100:133–140. doi: 10.1177/0022034520956975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peters B.A., Wu J., Hayes R.B., Ahn J. The Oral Fungal Mycobiome: Characteristics and Relation to Periodontitis in a Pilot Study. BMC Microbiol. 2017;17:157. doi: 10.1186/s12866-017-1064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dupuy A.K., David M.S., Li L., Heider T.N., Peterson J.D., Montano E.A., Dongari-Bagtzoglou A., Diaz P.I., Strausbaugh L.D. Redefining the Human Oral Mycobiome with Improved Practices in Amplicon-Based Taxonomy: Discovery of Malassezia as a Prominent Commensal. PLoS ONE. 2014;9:e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Khadija B., Imran M., Faryal R. Keystone Salivary Mycobiome in Postpartum Period in Health and Disease Conditions. J. Mycol. Med. 2021;31:101101. doi: 10.1016/j.mycmed.2020.101101. [DOI] [PubMed] [Google Scholar]
- 108.Stehlikova Z., Tlaskal V., Galanova N., Roubalova R., Kreisinger J., Dvorak J., Prochazkova P., Kostovcikova K., Bartova J., Libanska M., et al. Oral Microbiota Composition and Antimicrobial Antibody Response in Patients with Recurrent Aphthous Stomatitis. Microorganisms. 2019;7:636. doi: 10.3390/microorganisms7120636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Diaz P.I., Hong B.-Y., Dupuy A.K., Strausbaugh L.D. Mining the Oral Mycobiome: Methods, Components, and Meaning. Virulence. 2017;8:313–323. doi: 10.1080/21505594.2016.1252015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Charlson E.S., Diamond J.M., Bittinger K., Fitzgerald A.S., Yadav A., Haas A.R., Bushman F.D., Collman R.G. Lung-Enriched Organisms and Aberrant Bacterial and Fungal Respiratory Microbiota after Lung Transplant. Am. J. Respir. Crit. Care Med. 2012;186:536–545. doi: 10.1164/rccm.201204-0693OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bandara H.M.H.N., Panduwawala C.P., Samaranayake L.P. Biodiversity of the Human Oral Mycobiome in Health and Disease. Oral Dis. 2019;25:363–371. doi: 10.1111/odi.12899. [DOI] [PubMed] [Google Scholar]
- 112.Ikebe K., Morii K., Matsuda K., Hata K., Nokubi T. Association of Candidal Activity with Denture Use and Salivary Flow in Symptom-Free Adults over 60 Years1. J. Oral Rehab. 2006;33:36–42. doi: 10.1111/j.1365-2842.2006.01527.x. [DOI] [PubMed] [Google Scholar]
- 113.Ward T.L., Dominguez-Bello M.G., Heisel T., Al-Ghalith G., Knights D., Gale C.A. Development of the Human Mycobiome over the First Month of Life and across Body Sites. mSystems. 2018;3:e00140–17. doi: 10.1128/mSystems.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Darwazeh A.M., al-Bashir A. Oral Candidal Flora in Healthy Infants. J. Oral Pathol. Med. 1995;24:361–364. doi: 10.1111/j.1600-0714.1995.tb01200.x. [DOI] [PubMed] [Google Scholar]
- 115.Burcham Z.M., Garneau N.L., Comstock S.S., Tucker R.M., Knight R., Metcalf J.L. Genetics of Taste Lab Citizen Scientists Patterns of Oral Microbiota Diversity in Adults and Children: A Crowdsourced Population Study. Sci. Rep. 2020;10:2133. doi: 10.1038/s41598-020-59016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zakaria M.N., Furuta M., Takeshita T., Shibata Y., Sundari R., Eshima N., Ninomiya T., Yamashita Y. Oral Mycobiome in Community-Dwelling Elderly and Its Relation to Oral and General Health Conditions. Oral Dis. 2017;23:973–982. doi: 10.1111/odi.12682. [DOI] [PubMed] [Google Scholar]
- 117.O’Connell L.M., Santos R., Springer G., Burne R.A., Nascimento M.M., Richards V.P. Site-Specific Profiling of the Dental Mycobiome Reveals Strong Taxonomic Shifts during Progression of Early-Childhood Caries. Appl. Environ. Microbiol. 2020;86:e02825–19. doi: 10.1128/AEM.02825-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tati S., Davidow P., McCall A., Hwang-Wong E., Rojas I.G., Cormack B., Edgerton M. Candida Glabrata Binding to Candida Albicans Hyphae Enables Its Development in Oropharyngeal Candidiasis. PLoS Pathog. 2016;12:e1005522. doi: 10.1371/journal.ppat.1005522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sajid M., Sharma P., Srivastava S., Hariprasad R., Singh H., Bharadwaj M. Smokeless Tobacco Consumption Induces Dysbiosis of Oral Mycobiome: A Pilot Study. Appl. Microbiol. Biotechnol. 2022;106:5643–5657. doi: 10.1007/s00253-022-12096-6. [DOI] [PubMed] [Google Scholar]
- 120.Zhu T., Duan Y.-Y., Kong F.-Q., Galzote C., Quan Z.-X. Dynamics of Skin Mycobiome in Infants. Front. Microbiol. 2020;11:1790. doi: 10.3389/fmicb.2020.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ratanapokasatit Y., Laisuan W., Rattananukrom T., Petchlorlian A., Thaipisuttikul I., Sompornrattanaphan M. How Microbiomes Affect Skin Aging: The Updated Evidence and Current Perspectives. Life. 2022;12:936. doi: 10.3390/life12070936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boxberger M., Cenizo V., Cassir N., La Scola B. Challenges in Exploring and Manipulating the Human Skin Microbiome. Microbiome. 2021;9:125. doi: 10.1186/s40168-021-01062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jo J.-H., Kennedy E.A., Kong H.H. Topographical and Physiological Differences of the Skin Mycobiome in Health and Disease. Virulence. 2016;8:324–333. doi: 10.1080/21505594.2016.1249093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li H., Goh B.N., Teh W.K., Jiang Z., Goh J.P.Z., Goh A., Wu G., Hoon S.S., Raida M., Camattari A., et al. Skin Commensal Malassezia Globosa Secreted Protease Attenuates Staphylococcus Aureus Biofilm Formation. J. Investig. Dermatol. 2018;138:1137–1145. doi: 10.1016/j.jid.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 125.Leong C., Schmid B., Toi M.J., Wang J., Irudayaswamy A.S., Goh J.P.Z., Bosshard P.P., Glatz M., Dawson T.L. Geographical and Ethnic Differences Influence Culturable Commensal Yeast Diversity on Healthy Skin. Front. Microbiol. 2019;10:1891. doi: 10.3389/fmicb.2019.01891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huffnagle G.B., Noverr M.C. The Emerging World of the Fungal Microbiome. Trends Microbiol. 2013;21:334–341. doi: 10.1016/j.tim.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Keum H.L., Kim H., Kim H.-J., Park T., Kim S., An S., Sul W.J. Structures of the Skin Microbiome and Mycobiome Depending on Skin Sensitivity. Microorganisms. 2020;8:E1032. doi: 10.3390/microorganisms8071032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grice E.A., Segre J.A. The Skin Microbiome. Nat. Rev. Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cho Y.-J., Kim T., Croll D., Park M., Kim D., Keum H.L., Sul W.J., Jung W.H. Genome of Malassezia Arunalokei and Its Distribution on Facial Skin. Microbiol. Spectr. 2022;10:e00506-22. doi: 10.1128/spectrum.00506-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gupta A.K., Kohli Y., Summerbell R.C., Faergemann J. Quantitative Culture of Malassezia Species from Different Body Sites of Individuals with or without Dermatoses. Med. Mycol. 2001;39:243–251. doi: 10.1080/mmy.39.3.243.251. [DOI] [PubMed] [Google Scholar]
- 131.Tong X., Leung M.H.Y., Wilkins D., Cheung H.H.L., Lee P.K.H. Neutral Processes Drive Seasonal Assembly of the Skin Mycobiome. mSystems. 2019;4:e00004-19. doi: 10.1128/mSystems.00004-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Koike Y., Kuwatsuka S., Nishimoto K., Motooka D., Murota H. Skin Mycobiome of Psoriasis Patients Is Retained during Treatment with TNF and IL-17 Inhibitors. Int. J. Mol. Sci. 2020;21:3892. doi: 10.3390/ijms21113892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Crespo-Erchiga V., Florencio V.D. Malassezia Yeasts and Pityriasis Versicolor. Curr. Opin. Infect. Dis. 2006;19:139–147. doi: 10.1097/01.qco.0000216624.21069.61. [DOI] [PubMed] [Google Scholar]
- 134.Moosbrugger-Martinz V., Hackl H., Gruber R., Pilecky M., Knabl L., Orth-Höller D., Dubrac S. Initial Evidence of Distinguishable Bacterial and Fungal Dysbiosis in the Skin of Patients with Atopic Dermatitis or Netherton Syndrome. J. Investig. Dermatol. 2021;141:114–123. doi: 10.1016/j.jid.2020.05.102. [DOI] [PubMed] [Google Scholar]
- 135.Stehlikova Z., Kostovcik M., Kostovcikova K., Kverka M., Juzlova K., Rob F., Hercogova J., Bohac P., Pinto Y., Uzan A., et al. Dysbiosis of Skin Microbiota in Psoriatic Patients: Co-Occurrence of Fungal and Bacterial Communities. Front. Microbiol. 2019;10:438. doi: 10.3389/fmicb.2019.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bjerre R.D., Holm J.B., Palleja A., Sølberg J., Skov L., Johansen J.D. Skin Dysbiosis in the Microbiome in Atopic Dermatitis Is Site-Specific and Involves Bacteria, Fungus and Virus. BMC Microbiol. 2021;21:256. doi: 10.1186/s12866-021-02302-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dylag M., Leniak E., Gnat S., Szepietowski J.C., Kozubowski L. A Case of Anti-Pityriasis Versicolortherapy That Preserves Healthy Mycobiome. BMC Dermatol. 2020;20:9. doi: 10.1186/s12895-020-00106-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Honnavar P., Prasad G.S., Ghosh A., Dogra S., Handa S., Rudramurthy S.M. Malassezia Arunalokei Sp. Nov., a Novel Yeast Species Isolated from Seborrheic Dermatitis Patients and Healthy Individuals from India. J. Clin. Microbiol. 2016;54:1826–1834. doi: 10.1128/JCM.00683-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sahni K., Singh S., Dogra S. Newer Topical Treatments in Skin and Nail Dermatophyte Infections. Indian Dermatol. Online J. 2018;9:149–158. doi: 10.4103/idoj.IDOJ_281_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chng K.R., Tay A.S.L., Li C., Ng A.H.Q., Wang J., Suri B.K., Matta S.A., McGovern N., Janela B., Wong X.F.C.C., et al. Whole Metagenome Profiling Reveals Skin Microbiome-Dependent Susceptibility to Atopic Dermatitis Flare. Nat. Microbiol. 2016;1:1–10. doi: 10.1038/nmicrobiol.2016.106. [DOI] [PubMed] [Google Scholar]
- 141.Crespo Erchiga V., Ojeda Martos A., Vera Casaño A., Crespo Erchiga A., Sanchez Fajardo F. Malassezia Globosa as the Causative Agent of Pityriasis Versicolor. Br. J. Dermatol. 2000;143:799–803. doi: 10.1046/j.1365-2133.2000.03779.x. [DOI] [PubMed] [Google Scholar]
- 142.Shah A., Koticha A., Ubale M., Wanjare S., Mehta P., Khopkar U. Identification and Speciation of Malassezia in Patients Clinically Suspected of Having Pityriasis Versicolor. Indian. J. Dermatol. 2013;58:239. doi: 10.4103/0019-5154.110841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Carter B., Jones C.P., Creadick R.N., Parker R.T., Turner V. The Vaginal Fungi. Ann. N.Y. Acad. Sci. 1959;83:265–279. doi: 10.1111/j.1749-6632.1960.tb40900.x. [DOI] [PubMed] [Google Scholar]
- 144.Bradford L.L., Ravel J. The Vaginal Mycobiome: A Contemporary Perspective on Fungi in Women’s Health and Diseases. Virulence. 2017;8:342–351. doi: 10.1080/21505594.2016.1237332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ackerman A.L., Underhill D.M. The Mycobiome of the Human Urinary Tract: Potential Roles for Fungi in Urology. Ann. Transl. Med. 2017;5:31. doi: 10.21037/atm.2016.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Guo R., Zheng N., Lu H., Yin H., Yao J., Chen Y. Increased Diversity of Fungal Flora in the Vagina of Patients with Recurrent Vaginal Candidiasis and Allergic Rhinitis. Microb. Ecol. 2012;64:918–927. doi: 10.1007/s00248-012-0084-0. [DOI] [PubMed] [Google Scholar]
- 147.Pearce M.M., Hilt E.E., Rosenfeld A.B., Zilliox M.J., Thomas-White K., Fok C., Kliethermes S., Schreckenberger P.C., Brubaker L., Gai X., et al. The Female Urinary Microbiome: A Comparison of Women with and without Urgency Urinary Incontinence. mBio. 2014;5:e01283-01214. doi: 10.1128/mBio.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Holland J., Young M., Lee O., Chen C.-A. Vulvovaginal Carriage of Yeasts Other than Candida Albicans. Sex. Transm. Infect. 2003;79:249–250. doi: 10.1136/sti.79.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Papaemmanouil V., Georgogiannis N., Plega M., Lalaki J., Lydakis D., Dimitriou M., Papadimitriou A. Prevalence and Susceptibility of Saccharomyces Cerevisiae Causing Vaginitis in Greek Women. Anaerobe. 2011;17:298–299. doi: 10.1016/j.anaerobe.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 150.Kalia N., Singh J., Kaur M. Microbiota in Vaginal Health and Pathogenesis of Recurrent Vulvovaginal Infections: A Critical Review. Ann. Clin. Microbiol. Antimicrob. 2020;19:5. doi: 10.1186/s12941-020-0347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pytka M., Kordowska-Wiater M., Jarocki P. Mikrobiome of the Women’s Genital System. Adv. Microbiol. 2019;58:227–236. [Google Scholar]
- 152.Noverr M.C., Huffnagle G.B. Does the Microbiota Regulate Immune Responses Outside the Gut? Trends Microbiol. 2004;12:562–568. doi: 10.1016/j.tim.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 153.Sabbatini S., Visconti S., Gentili M., Lusenti E., Nunzi E., Ronchetti S., Perito S., Gaziano R., Monari C. Lactobacillus Iners Cell-Free Supernatant Enhances Biofilm Formation and Hyphal/Pseudohyphal Growth by Candida Albicans Vaginal Isolates. Microorganisms. 2021;9:2577. doi: 10.3390/microorganisms9122577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu N.-N., Zhao X., Tan J.-C., Liu S., Li B.-W., Xu W.-X., Peng L., Gu P., Li W., Shapiro R., et al. Mycobiome Dysbiosis in Women with Intrauterine Adhesions. Microbiol. Spectr. 2022;10:e01324-22. doi: 10.1128/spectrum.01324-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kovachev S.M. Cervical Cancer and Vaginal Microbiota Changes. Arch. Microbiol. 2020;202:323–327. doi: 10.1007/s00203-019-01747-4. [DOI] [PubMed] [Google Scholar]
- 156.Tipton L., Ghedin E., Morris A. The Lung Mycobiome in the Next-Generation Sequencing Era. Virulence. 2017;8:334–341. doi: 10.1080/21505594.2016.1235671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Krause R., Moissl-Eichinger C., Halwachs B., Gorkiewicz G., Berg G., Valentin T., Prattes J., Högenauer C., Zollner-Schwetz I. Mycobiome in the Lower Respiratory Tract—A Clinical Perspective. Front. Microbiol. 2017;7:2169. doi: 10.3389/fmicb.2016.02169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nguyen L.D.N., Viscogliosi E., Delhaes L. The Lung Mycobiome: An Emerging Field of the Human Respiratory Microbiome. Front. Microbiol. 2015;6:89. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carpagnano G.E., Susca A., Scioscia G., Lacedonia D., Cotugno G., Soccio P., Santamaria S., Resta O., Logrieco G., Barbaro M.P.F. A Survey of Fungal Microbiota in Airways of Healthy Volunteer Subjects from Puglia (Apulia), Italy. BMC Infect. Dis. 2019;19:78. doi: 10.1186/s12879-019-3718-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gago S., Overton N.L.D., Ben-Ghazzi N., Novak-Frazer L., Read N.D., Denning D.W., Bowyer P. Lung Colonization by Aspergillus Fumigatus Is Controlled by ZNF77. Nat. Commun. 2018;9:3835. doi: 10.1038/s41467-018-06148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Martinsen E.M.H., Eagan T.M.L., Leiten E.O., Haaland I., Husebø G.R., Knudsen K.S., Drengenes C., Sanseverino W., Paytuví-Gallart A., Nielsen R. The Pulmonary Mycobiome—A Study of Subjects with and without Chronic Obstructive Pulmonary Disease. PLoS ONE. 2021;16:e0248967. doi: 10.1371/journal.pone.0248967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ali N.A.B.M., Mac Aogain M., Morales R.F., Tiew P.Y., Chotirmall S.H. Optimisation and Benchmarking of Targeted Amplicon Sequencing for Mycobiome Analysis of Respiratory Specimens. Int. J. Mol. Sci. 2019;20:4991. doi: 10.3390/ijms20204991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kim S.H., Clark S.T., Surendra A., Copeland J.K., Wang P.W., Ammar R., Collins C., Tullis D.E., Nislow C., Hwang D.M., et al. Global Analysis of the Fungal Microbiome in Cystic Fibrosis Patients Reveals Loss of Function of the Transcriptional Repressor Nrg1 as a Mechanism of Pathogen Adaptation. PLOS Pathog. 2015;11:e1005308. doi: 10.1371/journal.ppat.1005308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kramer R., Sauer-Heilborn A., Welte T., Guzman C.A., Abraham W.-R., Hoefle M.G. Cohort Study of Airway Mycobiome in Adult Cystic Fibrosis Patients: Differences in Community Structure between Fungi and Bacteria Reveal Predominance of Transient Fungal Elements. J. Clin. Microbiol. 2015;53:2900–2907. doi: 10.1128/JCM.01094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Soret P., Vandenborght L.-E., Francis F., Coron N., Enaud R., Avalos M., Schaeverbeke T., Berger P., Fayon M., Thiebaut R., et al. Respiratory Mycobiome and Suggestion of Inter-Kingdom Network during Acute Pulmonary Exacerbation in Cystic Fibrosis. Sci. Rep. 2020;10:3589. doi: 10.1038/s41598-020-60015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Weaver D., Gago S., Bromley M., Bowyer P. The Human Lung Mycobiome in Chronic Respiratory Disease: Limitations of Methods and Our Current Understanding. Curr. Fungal Infect. Rep. 2019;13:109–119. doi: 10.1007/s12281-019-00347-5. [DOI] [Google Scholar]
- 167.Huffnagle G.B. The Microbiota and Allergies/Asthma. PLOS Pathog. 2010;6:e1000549. doi: 10.1371/journal.ppat.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.