Abstract
The spike glycoprotein is a major neutralizing antigen of bovine coronavirus (BCV). Conformational neutralizing epitopes of group A and group B monoclonal antibodies (MAbs) have previously been mapped to two domains at amino acids 351 to 403 (domain I) and amino acids 517 to 621 (domain II). To further map antigenic sites, neutralization escape mutants of BCV were selected with a group A MAb which has both in vitro and in vivo virus-neutralizing ability. The escape mutants were demonstrated to be neutralization resistant to the selecting group A MAb and remained sensitive to neutralization by a group B MAb. In radioimmunoprecipitation assays, the spike proteins of neutralization escape mutants were shown to have lost their reactivities with the selecting group A MAb. Sequence analysis of the spike protein genes of the escape mutants identified a single nucleotide substitution of C to T at position 1583, resulting in the change of alanine to valine at amino acid position 528 (A528V). The mutation occurs in domain II and in a location which corresponds to the hypervariable region of the spike protein of the coronavirus mouse hepatitis virus. Experimental introduction of the A528V mutation into the wild-type spike protein resulted in the loss of MAb binding of the mutant protein, confirming that the single point mutation was responsible for the escape of BCV from immunological selective pressure.
Bovine coronavirus (BCV) is a member of the family Coronaviridae of the order Nidovirales (3) and is closely related to the coronavirus mouse hepatitis virus (MHV). An enteropathogenic virus, BCV causes severe diarrhea in neonatal calves and winter dysentery in adult cattle (13, 29, 31, 33). BCV has also been associated with bovine respiratory disease, which is observed with the most severity in feedlot cattle (18, 29, 34).
An enveloped virus, BCV is composed of five structural proteins and contains a large positive-stranded RNA genome of 31,043 nucleotides (D. Yoo and Y. Pei, VIIIth Int. Symp. Nidoviruses (Coronaviruses and Arteriviruses 2000). The five structural proteins are the nucleocapsid protein (N; molecular weight, 52,000 [52K]), the membrane associated protein (M; molecular weight, 25K), the small membrane protein (E; molecular weight, 8K), the spike protein (S; molecular weight, 180K), and the hemagglutinin-esterase protein (HE; molecular weight, 65K) (23, 32, 44).
The BCV S protein is a very large membrane glycoprotein of 1,363 amino acids that contains two hydrophobic regions characteristic of type 1 glycoproteins: one at the N terminus of the protein that functions as a signal sequence and the other at the C terminus that functions as a membrane anchor (25, 32). Electron microscopic studies indicate that the S protein forms the club-shaped structures on the surface of the coronavirus virion (31). For BCV, the S protein is cleaved at amino acid positions 768 and 769 to form two subunits (1): S1 represents the N-terminal half of the S protein and S2 represents the C-terminal half of the protein. The S protein has several important functions including binding of the virus to susceptible cells (4, 6, 22, 28), mediation of membrane fusion (both virus-cell and cell-cell fusion) (6, 35, 36, 42), and induction of neutralizing antibody responses in the host species (10, 17, 22, 24, 37).
For BCV, virus-neutralizing anti-S monoclonal antibodies (MAbs) recognize conformational epitopes in two distinct antigenic sites, A and B, as defined in competitive binding assays (10). While both group A and group B MAbs neutralize BCV in vitro (in cell culture), only group A MAbs demonstrate in vivo virus-neutralizing protective responses in bovine intestinal-loop studies (9). Thus, antigenic site A of the BCV S protein appears to have an important function in the host species.
Previously, mapping studies by proteolysis of antigen-antibody complexes with group A and group B MAbs have demonstrated that the epitopes recognized by both groups of antibodies are located on a 37K-molecular-weight trypsin fragment of the S protein (11). It was proposed that this fragment spans amino acid positions 351 to 621 on the S1 subunit on the basis of an analysis of the fragments generated with three proteolytic enzymes (11, 40). Deletion mapping studies have identified that both group A and group B conformational epitopes consist of two domains located within amino acid residues 324 to 403 and 517 to 720 (40). Since this is in general agreement with the proposed location of the 37K-molecular-weight trypsin fragment, amino acids residues 351 to 403 (domain I) and 517 to 621 (domain II) are thought to contain the critical amino acids of these epitopes (40). In the present study, to further map the antigenic sites of the S1 protein, we have generated BCV MAb escape mutants, and using these mutant viruses, we have identified an epitope-critical amino acid that occurs in domain II.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The Quebec strain of BCV (8) was propagated in Mardin-Darby bovine kidney (MDBK) cells. MDBK cells were maintained in minimal essential medium supplemented with 10% fetal bovine serum (Cansera, Rexdale, Ontario, Canada). HeLa cells, maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, were used for vaccinia virus propagation. Vaccinia virus expressing T7 RNA polymerase (vTF7.3) (14) was used for protein expression. Preparation of MAbs was described previously (10), and in the current study group A MAb HB10-4 and group B MAb BB7-14 were used as mouse ascitic fluids.
Generation of MAb-resistant (mar) mutants.
Neutralizing MAb escape mutants were generated by incubating an equal volume of neat wild-type BCV (∼106 PFU) and a 1:100 dilution of MAb HB10-4 for 60 min at 37°C. Cells propagated in a 100-mm dish were inoculated with the mixture for 1 h at 37°C. The inoculum was removed and the cells were overlaid with 0.7% agarose containing a 1:1,000 dilution of MAb HB10-4. At 3 days of incubation, the cells were stained with neutral red to visualize plaques. Plaques were picked with a Pasteur pipette and were resuspended in 1 ml of medium. The plaque-picked virus was propagated in MDBK cells in the presence of a 1:1,000 dilution of MAb HB10-4 for three passages until a cytopathic effect was evident. Tenfold dilutions of the passaged virus were then incubated with a 1:100 dilution of MAb HB10-4 or without antibody and were propagated in the plaque assay to confirm an MAb resistance phenotype and to generate plaque-purified (subcloned) mutant viruses. Subclones of the escape virus mutant were propagated as described above, retested for the mar phenotype, aliquoted, and stored at −70°C.
cDNA cloning.
Cells were infected with mar viruses and incubated for 2 days at 37°C in the presence of MAb HB10-4. Total RNA was extracted from the cells by using Trizol (Gibco BRL, Burlington, Ontario, Canada) according to the manufacturer's instructions. cDNA was synthesized from virus-infected total cellular RNA equivalent to that from approximately 105 cells by using Superscript II RNase H− reverse transcriptase (Gibco BRL) and a primer specific for the S gene of BCV representing nucleotide positions 2256 to 2282 (downstream primer SMr1 [5′-CACACAGTAACCACTACCTACTGTGAGATCA-3′]). The reverse transcription reaction was carried out for 1 h at 39°C in the presence of 1 mM each dCTP, dGTP, dATP, and dTTP; 10 mM dithiothreitol; 50 mM Tris-HCl (pH 8.3); 75 mM KCl; and 3 mM MgCl2 in a reaction volume of 20 μl. The second-strand DNA was synthesized by PCR amplification with the upstream primer (SR954 [5′-CAGCCAATTGCAGATGTTTACCGAC-3′]) representing nucleotide positions 946 to 970 of the BCV S gene and the downstream primer (SMr1) that was used to make the first-strand cDNA. For PCR, 4 μl of the first-strand cDNA reaction mixture was added to the PCR mixture containing a final concentration of 0.15 μg of the upstream and downstream primers, 20 mM Tris-HCl (pH 8.4), 5 mM MgCl2, 50 mM KCl, each deoxynucleoside triphosphate at a concentration of 1 mM, and 0.5 U of Vent DNA polymerase (New England Biolabs, Beverly, Mass). The PCR was performed in a thermocycler (Tyler Instrument, Edmonton, Alberta, Canada) for 30 cycles with the following parameters: 94°C for 30 s for denaturation, 62°C for 30 s for annealing, and 72°C for 2.5 min for extension, followed by a 10-min elongation at 72°C after the final cycle. The PCR product was cloned into the SmaI site of plasmid pGEM3Zf(+) (Promega, Madison, Wis.).
Determination of nucleotide sequence.
Sequences were determined in both directions by the dideoxynucleotide chain termination method with a T7 DNA sequencing kit (Pharmacia, Baie d'Urfe, Quebec, Canada) according to the manufacturer's instruction. Approximately 1 to 2 μg of double-stranded DNA denatured in 0.2 N NaOH was incubated with 20 ng of either forward or reverse sequencing primers for 20 min at 37°C, and sequencing reactions for each nucleotide were carried out with T7 DNA polymerase and [α-35S]dATP (specific activity, 500 Ci/mmol; New England Nuclear, Boston, Mass). The reactions were resolved on an 8 M urea–5% polyacrylamide gel until bromophenol blue dye runoff with an IBI STS-45 sequencing gel apparatus (Kodak). The gel was air dried at 37°C, and the images were visualized by exposure to X-ray film at −70°C.
Site-directed mutagenesis.
Site-directed mutagenesis was carried out on the basis of the QuickChange Site-Directed Mutagenesis Protocol (Strategene, La Jolla, Calif.). Approximately 15 ng of pTZ19-S1 plasmid DNA was mixed with 300 ng of paired primers (forward primer, 5′-CATAATGCTGTCCAATGTGAT-3′; reverse primer, 5′-ATCACATTGGACAGCATTATG-3′) in the presence of all four deoxynucleoside triphosphates at a concentration of 0.1 mM, 10 mM KC1, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.75), 2 mM MgSO4, 0.1% Triton X-100, and 100 μg of bovine serum albumin per ml. DNA polymerase Pfu (2.5 U; Stratagene) was added to the mixture, and the final volume was adjusted to 100 μl by addition of water. The reaction was subjected to PCR for 30 cycles at 95°C for 30 s, 55°C for 1 min, and 68°C for 8 min with a thermocycler (model PE2400; Perkin-Elmer). Five microliters of the PCR product was retained for gel electrophoresis and for use as a control for transformation. The remaining sample was digested with 10 U of DpnI by incubation for 1 h at 37°C in order to remove the unmethylated template DNA strands. Competent XL-1 Blue cells (Stratagene) were transformed with approximately 1 μg of the PCR product. Transformants were picked randomly, and plasmid DNA was prepared from each transformant. Mutant plasmids were screened by DNA sequencing as described above.
Protein expression and radiolabeling.
Plasmid DNA was prepared with a plasmid purification column kit (Qiagen Inc., Santa Clarita, Calif.) according to the manufacturer's instructions. HeLa cells, grown to 90% confluence in 100-mm dishes, were infected with vTF7-3 vaccinia virus expressing T7 RNA polymerase at a multiplicity of infection of 5 to 10. A mixture of 10 μg of plasmid DNA and 40 μl of LipofectACE (Gibco BRL) was incubated in Opti-MEM serum reduced medium (Gibco BRL) for 45 min at room temperature prior to transfection. The culture medium was removed from cells at 2 h postinfection, and the cells were transfected with the mixture for 8 h in 5 ml of Opti-MEM. At 10 h postinfection, the supernatant was removed and the cells were radiolabeled for 6 h with 50 μCi of [35S]methionine (specific activity, 407 MBq/ml; New England Nuclear) per ml in methionine-free Eagle's medium (Sigma) supplemented with 2% dialyzed fetal bovine serum. The cells were harvested by scraping with a rubber policemen and were resuspended in lysis buffer (0.1% Triton X-100, 10 mM Tris-HCl [pH 7.4]). After incubation of the cells for 20 min on ice, the cell lysates were centrifuged in a microcentrifuge for 10 min. The supernatant containing the cytoplasmic fraction was collected for immune precipitation studies.
RIPA and SDS-polyacrylamide gel electrophoresis.
Aliquots of radiolabeled cell lysates were adjusted with radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 0.1% sodium dodecyl sulfate [SDS]) to a final volume of 100 μl and were incubated for 2 h at room temperature with 1 μl of antibody. The immune complexes were adsorbed to 10 mg of protein A-Sepharose CL-4B beads (Pharmacia) for 16 h at 4°C in 800 μl of RIPA buffer containing a final concentration of 0.3% SDS. The Sepharose beads, collected by centrifugation at 6,000 rpm (Micromax; International Equipment Co., Needham Heights, Mass.) for 2 min, were washed twice with RIPA buffer and once with washing buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl). Twenty microliters of sample buffer (10 mM Tris-HCl [pH 6.8], 5% glycerol, 10% SDS, 10% β-mercaptoethanol, 0.12% [wt/vol] bromophenol blue) was added to the beads, followed by heating for 5 min at 95°C. The beads were pelleted at 10,000 rpm for 5 min, and the supernatant was analyzed by SDS-polyacrylamide gel electrophoresis on 7.5% polyacrylamide gels, followed by autoradiography. Alternatively, RIPA was performed with rabbit anti-mouse immunoglobulin G immunobeads (Bio-Rad Laboratories, Richmond, Calif.) as described previously (10).
Nucleotide sequence accession number.
The sequence reported in this work has been deposited in the GenBank database under accession number AF313395.
RESULTS
Generation and characterization of mar mutants.
Two domains, domains I and II, associated with BCV neutralizing epitopes have previously been mapped to amino acid positions 351 to 403 and 517 to 621 of the S protein, respectively (40). Both domains lie within the S1 subunit of the S protein, and domain II overlaps sequences of the corresponding hypervariable region of the MHV S protein (amino acid positions 456 to 592 in BCV). To dissect the neutralizing epitope recognized by group A MAb HB10-4, mutant BCVs resistant to the neutralizing MAb were generated. From a total of 30 plaques picked from residual virus after incubation of the wild-type BCV with MAb HB10-4, 13 viable viruses were obtained after three passages in cell culture. All of these viable viruses were of the MAb-resistant (mar) phenotype and after incubation with a 1:100 dilution of MAb HB10-4 had titers which were within 10-fold of the titers obtained without MAb. In contrast, the titer of wild-type BCV was reduced by greater than 1,000-fold after incubation with MAb HB10-4. Initial titers of the mar mutants ranged from 1 × 104 to 5 × 106/ml. Four viruses (HBm1, HBm5, HBm9, and HBm13) which gave a range of low to high titers were selected for further study. One of these (HBm9) with the lowest titer also initially showed plaques with a smaller morphology. However, after propagation of the subcloned virus, HBm9 demonstrated plaques whose sizes were similar to those of other mar mutants. The reason for the apparent change was not ascertained.
The propagated subcloned viruses were retested to determine if they retained their mar phenotype upon subcloning and whether they could be neutralized by group B MAb BB7-14. All four viruses remained resistant to MAb HB10-4, and all were efficiently neutralized with BB7-14 (a 1:200 dilution of the ascitic fluid completely neutralized ≥600 PFU of each mutant). This indicated that the mutation that affected antigenic site A did not affect the integrity of antigenic site B.
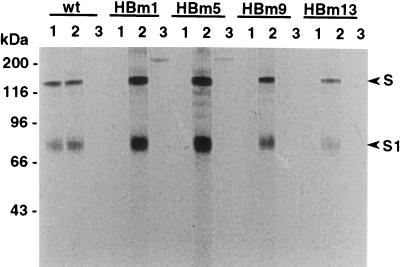
To determine if the S protein of the mutant viruses could be bound by MAb HB10-4, cells were infected with mutant viruses and radiolabeled with [35S]methionine. Cell lysates were prepared and subjected to immunoprecipitation with MAb HB10-4 (Fig. 1, lanes 1), MAb BB7-14 (Fig. 1, lanes 2), or MAb 115, a negative control antibody specific for the NS2-3 (125 kDa) and NS3 (80 kDa) proteins of bovine viral diarrhea virus (12) (Fig. 1, lanes 3). MAb HB10-4 precipitated proteins of ∼150 and ∼80kDa from cells infected with wild-type BCV (lane 1 for the wild type in Fig. 1) but did not precipitate any proteins from cells infected with mar mutants HBm1, HBm5, HBm9, and HBm13 (Fig. 1, lanes 1). The 150-kDa protein is the uncleaved form of the S protein, and the 80-kDa protein represents the N-terminal cleaved product (S1 subunit) of the S protein (20, 32, 41). Since the S1 proteins of the mar mutants, which were immune precipitated by BB7-14 (Fig. 1, lanes 2), did not show a discernible change in size from that of the wild-type BCV, it was apparent that notable deletions of the protein were not involved in changes that resulted in the loss of reactivity with MAb HB10-4. Previously, for some MHV mar mutants, large deletions in the S protein were observed (15, 26).
FIG. 1.
Immunoprecipitations of BCV mar mutants with MAb HB10-4. MDBK cells were infected with mar mutants and radiolabeled with [35S]methionine. Total cell lysates were prepared and subjected to immunoprecipitation with BCV-specific neutralizing MAbs. Lanes 1, selecting MAb HB10-4; lanes 2, nonselecting MAb BB7-14; lanes 3, bovine viral diarrhea virus-specific MAb 115. wt, wild-type BCV; HBm1 through HBm13, BCV mar mutants 1, 5, 9, and 13, respectively; S, uncleaved form of the BCV spike protein; S1, N-terminal half cleavage product of the BCV spike protein.
Identification of substituted amino acid in mar mutants.
To identify the change in sequence responsible for the loss of immune reactivity with MAb HB10-4, a portion of the S1 gene of the mar mutants was cloned and sequenced. To minimize misincorporation rates which might occur during the PCR cloning step, a DNA polymerase containing a proofreading activity was used throughout the PCR experiments. Since the domains recognized by MAb HB10-4 were identified within amino acid positions 324 to 720, the nucleotide sequences representing this region were specifically examined in all four mutants. Plaque-purified virus from the original stock of parental wild-type BCV was also cloned and sequenced in parallel with the mar mutants in order to compare the mar sequence directly to the wild-type BCV sequence.
When the S1 sequences of the mar mutants were compared to that of the wild-type BCV, only a single nucleotide substitution was observed, and the same substitution occurred in all four mutant viruses. This change (C to T) occurred at position 1583 of the S1 gene (Fig. 2). The mutation of C to T resulted in the change of the codon for alanine to valine at amino acid position 528 of the S1 protein (Fig. 3A). It is interesting that this mutation occurs within the region of domain II which overlaps the hypervariable region of the MHV S1 protein (Fig. 3A, and B). Furthermore, comparison of the sequence with those of different strains of MHV shows that the mutated codon occurs in a region of sequence that is deleted in MHV JHM and MHV A59. This is consistent with our finding that MAb HB10-4 does not recognize the S proteins of these strains of MHV (unpublished data), although they are classified in the same serogroup with BCV (19).
FIG. 2.
Electropherogram of sequencing gel for the S1 gene of BCV mar mutants. Arrows indicate sequencing directions. wt, wild-type BCV; HBm1 through HBm13, BCV mar mutants 1, 5, 9, and 13, respectively. Arrowheads indicate the changed nucleotides.
FIG. 3.
(A) Structural illustration of the S1 protein of BCV. Arabic numbers indicate amino acid positions. Two antigenic regions that have previously been identified are indicated as domains I and II, respectively. Vertical arrows and underlined boldface characters indicate the substitution. The darkened area at the N terminus depicts a hydrophobic signal sequence. Shaded areas indicate antigenic domains or the hypervariable region. aa, amino acid; wt, wild type. (B) Comparisons of the sequences of the hypervariable regions of various coronaviruses. Dotted lines indicate deletions. BCV, bovine coronavirus (wild-type BCV sequence, GenBank accession number D00662; mar mutant sequence, GenBank accession number AF313395); JHM, mouse hepatitis virus strain JHM (GenBank accession number D00093); A59, mouse hepatitis virus strain A59 (GenBank accession number M18379); SDAV, sialodacryoadenitis rat coronavirus (GenBank accession number AF188193); MHV4, mouse hepatitis virus strain 4 (GenBank accession number S51114).
Experimental introduction of the mutation.
Since the sequence of the mar mutants was determined for nucleotide positions 972 to 2160 of the S1 gene only, it was conceivable that mutations other than that coding for amino acid residue 528 may have occurred in other regions of the S1 gene and may be responsible or partially responsible for the loss of antibody reactivity. To exclude this possibility, we introduced the same mutation into the full-length S1 gene of wild-type BCV. By site-directed mutagenesis, the C at nucleotide position 1583 was precisely replaced by T to alter the codon of GCC for alanine to GTC for valine at amino acid position 528 to create the mutant A528V S1 gene. Thus, the mutant A528V S1 protein would be identical to the wild-type full-length S1 protein except for the single amino acid at position 528. Both the wild-type S1 gene and the A528V S1 gene were individually expressed in cells with the T7 vaccinia virus expression system, and the cell lysates were subjected to RIPA with MAb HB10-4 or MAb BB7-14 (Fig. 4). As observed, the A528V mutant S1 protein was precipitated by MAb BB7-14 (Fig. 4, lane 4). In contrast, the mutant protein was not recognized by MAb HB10-4 (Fig. 4, lane 3), although the MAb was able to precipitate the wild-type S1 protein (Fig. 4, lane 2). These results demonstrate that the amino acid change of alanine to valine at position 528 was sufficient to confer resistance to the HB10-4 mar mutants.
FIG. 4.
Immunoprecipitations of the mutant S1 protein expressed in HeLa cells. Cells were infected with recombinant vaccinia virus vTF7-3 and transfected with the S1 gene. Cells were radiolabeled with [35S]methionine and subjected to immunoprecipitation with selecting MAb HB10-4 (lanes 1, 2, and 3) or nonselecting MAb BB7-14 (lane 4). The immune complexes were resolved by SDS-polyacrylamide gel electrophoresis on 7.5% polyacrylamide gels, and the gel was autoradiographed. Lanes: 1, vTF7-3-infected but DNA-untransfected cell lysate; 2, wild-type BCV S1 gene, transfected; 3, A528V mutant S1 gene, transfected; 4, mutant S1 gene, transfected (A528V) and immunoprecipitated with MAb BB7-14.
The amino acid change of alanine to (the larger) valine is not considered a conservative substitution. According to the PAM250 matrix, a mutation probability index, an alanine-to-valine change occurs in closely related proteins at a frequency similar to that observed for alanine to asparagine, aspartic acid, glutamic acid, or glutamine (7). Changes of alanine to the small amino acids glycine, serine, threonine, and proline occur more frequently. Thus, the alanine-to-valine change may have caused a local or a more extensive disruption in the S1 structure, causing it to be no longer recognized by MAb HB10-4.
DISCUSSION
Of the two cleavage products of S, the S2 subunit is highly conserved among coronaviruses. In contrast, the S1 portion generally shows a low level of sequence homology, and in MHV an extensive heterogeneity has been shown to exist. When the amino acid sequence of the BCV S1 protein is compared to those of various strains of MHV, large deletions of 49 and 138 amino acids are identified within the region between positions 456 and 592 in MHV strains A59 and JHM, respectively (23). Similarly, MHV 2 has deletions of 150 amino acids within the same region. In contrast, MHV 4 and the rat coronavirus sialodacryoadenitis virus have only minor deletions of 9 and 12 amino acids, respectively, in this region (26, 43). Thus, the region between positions 456 and 592 in the S protein is considered hypervariable in rodent coronaviruses. The hypervariable region appears to be biologically significant in MHV, and studies have indicated that it acts as a pathogenic determinant. For MHV JHM and MHV 4, two highly neurotropic viruses which produce acute fatal encephalitis in mice, large deletions and single or multiple point mutations are observed in this region for viral mutants with reduced neurovirulence (15, 26, 38).
The cellular receptor binding region of the MHV S protein occurs distally from the hypervariable region in the first 330 amino acid residues of the protein (22). In contrast, for the enteropathic transmissible enteritis coronavirus (TGEV) of swine, the cellular receptor binding region occurs in the location on the S protein that corresponds to the hypervariable region of MHV (16) and enteric tropism determinants occur in the N-terminal region (2, 21). Respiratory porcine coronaviruses, which are nonenteropathic variants of TGEV, demonstrate large amino acid deletions in the N-terminal region of the S protein (39).
The S gene of BCV has been sequenced for several cell culture-adapted reference strains and some low-level cell culture-passaged clinical isolates. Although S1 gene sequences appear to be highly conserved among strains and isolates of BCV and sequence deletions or insertions have not been observed, a polymorphic region in the gene is apparent. Low-level cell culture-passaged clinical isolates recovered from diarrheic calves show sequence differences which cluster in the region representing amino acid positions 456 to 592 (27), which corresponds to the MHV hypervariable region. For low-level-passaged respiratory BCV isolates, sequence differences also cluster in this region and in the N-terminal region of the S1 protein in comparison with the locations in enteric BCV strains (5).
For the (polymorphic) region from amino acids 456 to 592, a 6 to 9% variation in amino acid sequence occurs between our reference strain and the enteric and respiratory BCV clinical isolates and a 4 to 6% variation occurs between the enteric and respiratory isolates (5, 25, 27). Amid the clustering of sequence variation in the region, the alanine at position 528 and the surrounding sequence from amino acids 511 to 530 are fully conserved among all BCV strains and isolates sequenced to date, suggesting that functional constraints may exist for this portion of the polymorphic region. The polymorphic region contains 15 conserved cysteine residues, a large number for its size, many of which are likely involved in disulfide linkages and confer a complex structure. Our analysis of the region for respiratory and enteric BCVs reveals that only three amino acid changes occur consistently between the two groups; two of these are conservative substitutions at amino acid positions 510 and 578 (serine and threonine are interchanged). The third change, a nonconservative change between groups, occurs very close to critical residue 528 identified in this study at amino acid position 531, where an aspartic acid or asparagine residue occurs in enteric BCV isolates and a glycine occurs in respiratory BCV isolates.
The BCV S protein binds to sialic acid residues (30), but the cellular receptor protein and viral receptor binding region have not yet been identified. The polymorphic region from amino acids 456 to 592 may be involved in receptor binding, as in TGEV, or may be a pathological determinant like MHV. Sequence differences between respiratory and enteric BCV isolates suggest that tropism determinants may occur in the polymorphic region, perhaps involving residue 531, or in the N-terminal region of the S protein.
It remains to be directly demonstrated if the region of the S protein from amino acids 456 to 592 plays a significant role in BCV pathogenesis in cattle. However, the finding that it harbors a critical amino acid essential for the reactivity with a MAb with demonstrated in vivo neutralizing ability strongly suggests that it has an important biological role in virus-cell interactions. Development of a system which will enable the introduction of specific modifications into the coronavirus genome, such as an infectious cDNA clone, is essential to further study the biological significance in vivo of this and other regions of the BCV S protein.
ACKNOWLEDGMENTS
This study was supported by the Medical Research Council of Canada, Ontario Cattlemen's Association, and Ontario Ministry of Agriculture Food and Rural Affairs (OMAFRA).
REFERENCES
- 1.Abraham S, Kienzle T E, Lapps W, Brian D A. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176:296–301. doi: 10.1016/0042-6822(90)90257-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballesteros M L, Sanchez C M, Enjuanes L. Two amino acid changes at the N-terminus of transmissible gastroenteritis coronavirus spike protein result in the loss of enteric tropism. Virology. 1997;227:378–388. doi: 10.1006/viro.1996.8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 4.Cavanagh D, Davis P J. Coronavirus IBV: removal of spike glycoprotein S1 by urea abolishes infectivity and hemagglutination but not attachment to cells. J Gen Virol. 1986;67:1443–1448. doi: 10.1099/0022-1317-67-7-1443. [DOI] [PubMed] [Google Scholar]
- 5.Chouljenko V N, Kousoulas K G, Lin X, Storz J. Nucleotide and predicted amino acid sequences of all genes encoded by the 3′ genomic portion (9.5kb) of respiratory coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes. 1998;17:33–42. doi: 10.1023/A:1008048916808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins A R, Knobler R L, Powell H, Buchmeier M J. Monoclonal antibodies to murine hepatitis virus-4 (strain JHM) define the viral glycoprotein responsible for attachment and cell-cell fusion. Virology. 1982;119:358–371. doi: 10.1016/0042-6822(82)90095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dayhoff M, Schwartz R M, Orcutt B C. A model of evolutionary change in proteins. In: Dayhoff M, editor. Atlas of protein sequence and structure. 5, supplement 3. Silver Spring, Md: National Biomedical Research Foundation; 1978. pp. 345–352. [Google Scholar]
- 8.Dea S, Roy R S, Begin M E. Bovine coronavirus isolation and cultivation in continuous cell lines. Am J Vet Res. 1980;41:30–38. [PubMed] [Google Scholar]
- 9.Deregt D, Gilford G A, Ijaz M K, Watts T C, Gilchrist J E, Haines D M, Babiuk L A. Monoclonal antibodies to bovine coronavirus glycoproteins E2 and E3: demonstration of in vivo virus-neutralizing activity. J Gen Virol. 1989;70:993–998. doi: 10.1099/0022-1317-70-4-993. [DOI] [PubMed] [Google Scholar]
- 10.Deregt D, Babiuk L A. Monoclonal antibodies to bovine coronavirus: characteristics and topographical mapping of neutralizing epitopes on the E2 and E3 glycoproteins. Virology. 1987;161:410–420. doi: 10.1016/0042-6822(87)90134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deregt D, Parker M D, Cox G J, Babiuk L A. Mapping of neutralizing epitopes to fragments of bovine coronavirus E2 protein by proteolysis of antigen-antibody complexes. J Gen Virol. 1989;70:647–658. doi: 10.1099/0022-1317-70-3-647. [DOI] [PubMed] [Google Scholar]
- 12.Deregt D, Masri S A, Cho H J, Bielefeldt-Ohmann H. Monoclonal antibodies to the p80/125 and gp53 proteins of bovine viral diarrhea virus: their potential use as diagnostic reagents. Can J Vet Res. 1990;54:343–348. [PMC free article] [PubMed] [Google Scholar]
- 13.Espinasse J, Viso M, Laval A, Savey M, Le Layec C, Blot J P, L'Haridon R, Cohen J. Winter dysentery: a coronavirus-like agent in the faeces of beef and dairy cattle with diarrhoea. Vet Rec. 1982;110:385. doi: 10.1136/vr.110.16.385. [DOI] [PubMed] [Google Scholar]
- 14.Furest T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallagher T M, Parker S E, Buchmeier M J. Neutralization-resistant variants of a neurotropic coronavirus are generated by deletions within the amino-terminal half of the spike glycoprotein. J Virol. 1990;64:731–741. doi: 10.1128/jvi.64.2.731-741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Godet M, Grosclaude J, Delmas B, Laude H. Major receptor-binding and neutralization determinants are located within the same domain of the transmissible gastroenteritis virus (coronavirus) spike protein. J Virol. 1994;68:8008–8016. doi: 10.1128/jvi.68.12.8008-8016.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grosse B, Siddell S G. Single amino acid changes in the S2 subunit of the MHV surface glycoprotein confer resistance to neutralization by S1 subunit-specific monoclonal antibody. Virology. 1994;202:814–824. doi: 10.1006/viro.1994.1403. [DOI] [PubMed] [Google Scholar]
- 18.Hasoksuz M, Lathrop S L, Gadfield K L, Saif L J. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am J Vet Res. 1999;60:1227–1233. [PubMed] [Google Scholar]
- 19.Hogue B G, King B, Brian D A. Antigenic relationships among proteins of bovine coronavirus, human respiratory coronavirus OC43, and mouse hepatitis coronavirus A59. J Virol. 1984;51:384–388. doi: 10.1128/jvi.51.2.384-388.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King B, Brian D A. Bovine coronavirus structural proteins. J Virol. 1982;42:700–707. doi: 10.1128/jvi.42.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krempl C, Schultz B, Laude H, Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubo H, Yamada Y K, Taguchi F. Localization of neutralizing epitopes and the receptor-binding site within the amino-terminal 330 amino acids of the murine coronavirus spike protein. J Virol. 1994;68:5403–5410. doi: 10.1128/jvi.68.9.5403-5410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luytjes W, Geerts D, Posthumus W, Meloen R, Spaan W. Amino acid sequence of a conserved neutralizing epitope of murine coronaviruses. J Virol. 1989;63:1408–1412. doi: 10.1128/jvi.63.3.1408-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker M D, Yoo D, Babiuk L A. Primary structure of the E2 peplomer gene of bovine coronavirus and surface expression in insect cells. J Gen Virol. 1990;71:263–270. doi: 10.1099/0022-1317-71-2-263. [DOI] [PubMed] [Google Scholar]
- 26.Parker S E, Gallagher T M, Buchmeier M J. Sequence analysis reveals extensive polymorphism and evidence of deletions within the E2 glycoprotein gene of several strains of murine hepatitis virus. Virology. 1989;173:664–673. doi: 10.1016/0042-6822(89)90579-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rekik M R, Dea S. Comparative sequence analysis of a polymorphic region of the spike glycoprotein S1 subunit of enteric bovine coronavirus isolates. Arch Virol. 1994;135:319–331. doi: 10.1007/BF01310017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeki K, Ohtsuka N, Taguchi F. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J Virol. 1997;71:9024–9031. doi: 10.1128/jvi.71.12.9024-9031.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saif L D, Redman R J, Brock K V, Kohler E M, Heckert R A. Winter dysentery in adult dairy cattle: detection of coronavirus in the faeces. Vet Rec. 1988;123:300–301. doi: 10.1136/vr.123.11.300. [DOI] [PubMed] [Google Scholar]
- 30.Schultz B, Gross H J, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stair E L, Rhodes M B, White R G, Mebus C A. Neonatal calf diarrhea: purification and electron microscopy of a coronavirus-like agent. Am J Vet Res. 1972;33:1147–1156. [PubMed] [Google Scholar]
- 32.St. Cyr-Coats K S, Storz J, Hussain K A, Schnorr K L. Structural proteins of bovine coronavirus strain L9: effects of the host cell and trypsin treatment. Arch Virol. 1988;103:35–45. doi: 10.1007/BF01319807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Storz J, Kaluza G, Niemann H, Rott R. On enteropathogenic bovine coronavirus. Adv Exp Med Biol. 1981;142:171–179. doi: 10.1007/978-1-4757-0456-3_14. [DOI] [PubMed] [Google Scholar]
- 34.Storz J, Stine L, Liem A, Anderson G A. Coronavirus isolation from nasal swab samples in cattle with signs of respiratory tract disease after shipping. J Am Vet Med Assoc. 1996;208:1452–1455. [PubMed] [Google Scholar]
- 35.Storz J, Rott R, Kaula G. Enhancement of plaque formation and cell fusion of an enteropathogenic coronavirus by trypsin treatment. Infect Immun. 1981;31:1214–1222. doi: 10.1128/iai.31.3.1214-1222.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sturman L S, Richard C S, Homes K V. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J Virol. 1985;56:904–911. doi: 10.1128/jvi.56.3.904-911.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takase-Yoden S, Kikuchi T, Siddell S G, Taguchi F. Localization of major neutralizing epitopes on the S1 polypeptide of the murine coronavirus peplomer glycoprotein. Virus Res. 1991;18:99–107. doi: 10.1016/0168-1702(91)90011-J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang F I, Fleming J O, Lai M M C. Sequence analysis of the spike protein gene of murine coronavirus variants: study of genetic sites affecting neuropathogenicity. Virology. 1992;186:742–749. doi: 10.1016/0042-6822(92)90041-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wesley R D, Woods R D, Cheung A K. Genetic analysis of porcine respiratory coronavirus, an attenuated variant of transmissible gastroenteritis virus. J Virol. 1991;65:3369–3373. doi: 10.1128/jvi.65.6.3369-3373.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo D, Parker M D, Song J, Cox G J, Deregt D, Babiuk L A. Structural analysis of the conformational domains involved in neutralization of bovine coronavirus using deletion mutants of the spike glycoprotein S1 subunit expressed by recombinant baculoviruses. Virology. 1991;183:91–98. doi: 10.1016/0042-6822(91)90121-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoo D, Parker M D, Babiuk L A. Analysis of the S spike (peplomer) glycoprotein of bovine coronavirus synthesized in insect cells. Virology. 1990;179:121–128. doi: 10.1016/0042-6822(90)90281-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoo D, Parker M D, Babiuk L A. The S2 subunit glycoprotein of bovine coronavirus mediates membrane fusion in insect cells. Virology. 1991;180:395–399. doi: 10.1016/0042-6822(91)90045-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo D, Pei Y, Christie N, Cooper M. Primary structure of the sialodacryoadenitis virus genome: sequence of the structural-protein region and its application for differential diagnosis. Clin Diagn Lab Immunol. 2000;7:568–573. doi: 10.1128/cdli.7.4.568-573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu X, Bi W, Weiss S R, Leibowitz J L. Mouse hepatitis virus gene 5b protein is a new virion envelope protein. Virology. 1994;202:1018–1023. doi: 10.1006/viro.1994.1430. [DOI] [PubMed] [Google Scholar]