Abstract
The strikingly rapidly mutating nature of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) genome has been a constant challenge during the coronavirus disease 2019 (COVID-19) pandemic. In this study, various techniques, including reverse transcription-quantitative polymerase chain reaction, antigen-detection rapid diagnostic tests, and high-throughput sequencing were analyzed under different scenarios and spectra for the etiological diagnosis of COVID-19 at the population scale. This study aimed to summarize the latest research progress and provide up-to-date understanding of the methodology used for the evaluation of the immunoprotection conditions against future variants of SARS-CoV-2. Our novel work reviewed the current methods for the evaluation of the immunoprotection status of a specific population (endogenous antibodies) before and after vaccine inoculation (administered with biopharmaceutical antibody products). The present knowledge of the immunoprotection status regarding the COVID-19 complications was also discussed. Knowledge on the immunoprotection status of specific populations can help guide the design of pharmaceutical antibody products, inform practice guidelines, and develop national regulations with respect to the timing of and need for extra rounds of vaccine boosters.
Keywords: COVID-19, Etiological diagnosis, Immunoprotection, SARS-CoV-2 variants, Vaccine, Complications
Graphical abstract
Highlights
-
•
Evaluating immunoprotection status may facilitate the design of pharmaceutical vaccines.
-
•
Variations in SARS-CoV-2, vaccines, and complications can affect immunoprotection.
-
•
Techniques for etiological diagnosis of COVID-19 at the population scale were discussed.
1. Introduction
According to Johns Hopkins University statistics [1], up to September 6, 2022, the number of global cases of coronavirus disease 2019 (COVID-19) had reached 605,577,075, and caused more than 6.5 million patients' death. Owing to increasingly refined medical treatments, such as extracorporeal membrane oxygenation, patients’ lives could be better saved under the condition that medical resources are fortunately sufficient. Although the risk of widespread COVID-19 remains, multiple rounds of vaccination rather than lockdowns are still the most recognized solution for epidemic prevention; thus, the protective inoculation is becoming the worldwide choice. However, new challenges in the COVID-19 pandemic, including the rapidly mutating virus genome, compromise the confidence in immunoprotection measures. This work reviewed the present research progress in variations in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), vaccine-induced protection, and complications associated with COVID-19, and provides an up-to-date understanding of how to improve the etiological diagnosis of COVID-19 and evaluate the immunoprotection status.
2. Novel coronavirus variants
SARS-CoV-2, the causative agent of COVID-19, is a positive-sense, single-stranded RNA virus belonging to the beta-coronavirus genus [2]. This virus has four essential components: the membrane (M), nucleocapsid (N), envelope (E), and spike (S) proteins, which play vital roles in host entry, viral replication, and immunoregulation of the host's antiviral immunity, thereby possibly affecting the etiological diagnosis, severity, and transmissibility of COVID-19 [3].
2.1. General structural information of several variants during the latest outbreak
Mutations within the virus's genomic regions of the virus can result in major changes in virulence; therefore, updating the global information on viral variants is crucial. Among them, the immunodominant S glycoprotein is the target of the host's anti-S neutralizing antibodies (nAbs), and the E protein is related to virus infectivity [4]. In our previous study, published in June 2021, we concluded that the variations in SARS-CoV-2 did not significantly affect the infection ability, host immune regulation, or disease severity [5]. However, during the past year, our knowledge has increasingly expanded.
According to phylodynamic analysis of SARS-CoV-2 performed using the Nextstrain platform [[6], [7]], in 2021, SARS-CoV-2 has evolved from 20H (Beta/B.1.351.3, V2) to 22C (Omicron/B.1.1.529). As we can see from Table S1 [7], within each variant, multiple changes occur in the genomic region for coding open reading frames (ORF) for the 1a/b, 3a, 6, 7a, 7b, 8, 9b, S, E, M, and N proteins, among others. Among these coding changes, those within the genomic region of the S protein are extremely frequent. Taking the latest variant of concern (VOC) 22C as an example (Table 1) [7], genomic mutations could cause thirty amino acid changes in the S protein, one in the E protein, two in the M protein, and four in the N protein.
Table 1.
Mutations in the genomic regions of the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) variants reported in 2021 (data derived from the Nextstrain platform) [7].
| Variants | S | E | M | N |
|---|---|---|---|---|
| 20H (Beta, V2) | A27S, D80A, D215G, K417N, E484K, N501Y, D614G, and A701V | P71L | G200C and T205I | |
| 20I (Alpha, V1) | N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H | D3L, R203K, G204R, and S235F | ||
| 20J (Gamma,V3) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, G1085E, and V1176F | P80R, R203K, and G204R | ||
| 21A (Delta) | T19R, R158G, L452R, T478K, D614G, P681R, and D950N | I82T | D63G and R203M | |
| 21I (Delta) | T19R, G142D, R158G, A222V, L452R, T478K, D614G, P681R, and D950N | I82T | D63G and D377Y | |
| 21J (Delta) | T19R, T95I, G142D, R158G, T208M, L452R, T478K, D614G, P681R, and D950N | I82T | D63G, R203M, G215C, and D377Y | |
| 21C (Epsilon) | S13I, W152C, L452R, and D614G | T205I, and M234I | ||
| 21D (Eta) | Q52R, A67V, D215Y, E484K, D614G, Q677H, and F888L | L21F | I82T | S2M, D3Y, A12G, and T205I |
| 21F (Iota) | L5F, T95I, D253G, E484K, D614G, and A701V | P199L, and M234I | ||
| 21G (Lambda) | G75V, T76I, D253N, L452Q, F490S, D614G, and T859N | P13L, R203K, G204R, G214C, and T325I | ||
| 21H (Mu) | T95I, Y144S, Y145N, R346K, E484K, N501Y, D614G, P681H, and D950N | R158L | T205I | |
| 21K (Omicron) | A67V, T95I, Y145D, L212I, G339D, R346K, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F | T9I | D3G, Q19E, and A63T | P13L, R203K, and G204R |
| 21L (Omicron) | T19I, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K | T9I | Q19E and A63T | P13L, R203K, G204R, and S413R |
| 21M (Omicron) | T19I, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, K417N, N440K, S477N, T478K, E484A, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K | T9I | Q19E and A63T | |
| 22A (Omicron) | T19I, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N658S, N679K, P681H, N764K, D796Y, Q954H, and N969K | T9I | Q19E and A63T | P13L, R203K, G204R, and S413R |
| 22B (Omicron) | T19I, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452R, S477N, T478K, E484A, F486V, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, and N969K | T9I | D3N, Q19E, and A63T | P13L, R203K, G204R, A397V, and S413R |
| 22C (Omicron) | T19I, A27S, G142D, V213G, G339D, S371F, S373P, S375F, T376A, D405N, R408S, K417N, N440K, L452Q, S477N, T478K, E484A, Q493R, Q498R, N501Y, Y505H, D614G, H655Y, N679K, P681H, S704L, N764K, D796Y, Q954H, and N969K | T9I | Q19E and A63T | P13L, R203K, G204R, and S413R |
2.2. SARS-CoV-2 variants and COVID-19 disease severity
During the spread of the B.1.1.529 variant in South Africa from November 2021 to January 2022, survey data from 7010 participants demonstrated that the mortality/morbidity ratio significantly decreased within these 2.5 months, indicating that the B.1.1.529 variant might cause less disease severity [8]. According to another study conducted by the US Centers for Disease Control and Prevention (CDC), patients infected with the B.1.1.529 variant had a 74% lower rate of intensive care unit admission, a 91% lower rate of mortality, and a 70% shorter hospital stay than patients infected with the Delta/B.1.617.2 variant [9].
2.3. SARS-CoV-2 variants and COVID-19 etiological diagnosis at the population scale
Several techniques are widely used for the etiological diagnosis of COVID-19 (Fig. 1). High-throughput sequencing (HTS) produces full-length genome sequences [10], allowing identification of viral variants. Although HTS is a powerful technique for identifying the increasingly variating SARS-CoV-2 genome [11], its high cost prevents its popularization.
Fig. 1.
Techniques used for the etiological diagnosis of coronavirus disease 2019 (COVID-19) at the population scale. SARS-CoV-2: severe acute respiratory syndrome coronavirus 2. Figure is created with BioRender.com.
In our most recent retrospective study, we evaluated the real-world effectiveness of and protection from SARS-CoV-2 inactivated vaccines in 231 patients with COVID-19 hospitalized in Xi'an, China during the latest outbreak that occurred from December 2021 to January 2022 [12]. After whole-genome sequencing of 36 of these cases was carried out by the Chinese Center for Disease Control and Prevention (CDC), the Delta strain was identified as the VOC. Because sample pooling of 1 in 10, 1 in 20, or 1 in 50 was applied at various alert levels in the reverse transcription-quantitative polymerase chain reaction (RT-qPCR) detection, we have been rallying for future practical research to determine whether and how the pooling sample method should be optimized for HTS during each outbreak.
The gold standard and most frequently used RT-qPCR method relies on specific primer binding [13] within various genomic regions. For example, the Chinese CDC uses two sets of primers that specifically bind to the ORF and N protein regions of the SARS-CoV-2 genome [12]. One year ago, with much less information about the SARS-CoV-2 genome variation, we were concerned about the possibility that the potential viral variants could affect the RT-qPCR results [13]. To date, the evidence-based analysis is relatively optimistic, given the low rate of false-negative RT-qPCR results caused by SARS-CoV-2 variants [14]. However, a handful of reports occasionally indicate that certain SARS-CoV-2 variants can affect the RT-qPCR results and lead to false-negative data, such as the C26340U point mutation detected using the E gene binding primer [15]; the G29195T point mutation detected using the N gene binding primer used by US CDC [16]; and four mutations including Del28877-28894, GGG to AAC (28881–28883), Del28877-28878 [17], and Del28896-28898, as well as three single mutations including G28881A, G28882A, and G28883C [18] detected using the N gene binding primer. To minimize the mutations caused by primer binding failure in RT-qPCR tests, the evidence-based knowledge seems to be very helpful. Potential primer candidates could be developed from 286 genomic regions longer than 20 base pairs with low variability [19], and they should be validated before use. According to Section 2.1, which is summarized in Table 1, special attention should be paid when the intended PCR template is approaching or spanning the existing genomic mutations that cause thirty amino acid changes in the S protein, one in the E protein, two in the M protein, and four in the N protein.
Over the last two years, equipment independent antigen-detection rapid diagnostic tests (Ag-RDTs), especially in the form of immunochromatographic lateral-flow test cards, have been widely used, allowing the patients to conduct a self-test using readily available nasopharyngeal or throat swabs instead of traditional blood samples. Despite its lower sensitivity than that of HTS or the RT-qPCR method, the Ag-RDT method has high practical value and is highly welcomed for its scale-up testing capacity in low prevalence and at-home settings owing to its user-friendliness and cost-efficiency [20]. Despite its specificity, the sensitivity can highly vary. The World Health Organization recommends using products with a sensitivity of >80% for practical use [21]. Most reported Ag-RDTs use the membrane component N protein as the target antigen [22], whereas some use the S protein [23]. Others use the pooled antigens such as the N protein combined with the receptor-binding domain (RBD) antigen [24]. Correspondingly, it is not surprising that N sequence variants such as the D399N and T205I are found to affect the performance of Ag-RDT kits [25]; in particular, their performance against the B.1.617.2 strain is not satisfactory, and is especially limited in the etiological diagnosis of B.1.1.529 strains, which is far more concerning.
For regular regional management of COVID-19, we recommend adjustments for the specific local outbreaks. At checkpoints in airports, railway stations, and expressways, efficient crowd dispersal can be as important as accurate diagnostic testing. We recommend the use of duplicate sampling, where one sample is used for a “free to go” decision made upon obtaining negative results from the speedy Ag-RDT, whereas the other sample is reserved for RT-qPCR inspection, which requires a longer period for detection and sample transportation.
3. Immunoprotection against COVID-19
Immunoprotection, including the innate and adaptive immune response, against SARS-CoV-2 is a powerful protection against COVID-19 and can be obtained via a previous COVID-19 infection, exposure to other deactivated CoV-RBDs, or via proactive vaccine inoculation.
3.1. Immune response to COVID-19 attack during infection
After suffering one round of COVID-19 (with or without detectable symptoms), one can obtain automatic immunity through the generation of circulating antibodies against the specific SARS-CoV-2 variant. Following innate immunity, adaptive immunity emerges, involving T and B cells as well as antibodies, which play important roles in viral infection and vaccine action and are considered critical for COVID-19 management. After recovery from SARS-CoV-2 infection, CoV-specific immune memory includes CD4+ T helper cells, CD8+ T killer cells, antibodies, and memory B cells [26]. In one study comprising 69 participants, the longevity of detectable CD4+ and CD8+ T cells ranged from 26 to 266 days (8.9 months) [27]; such long-term effect is considered strikingly profound.
Post-infection, immunoprotection against COVID-19 can be established via the use of nAbs. In 65 patients diagnosed with COVID-19 in the UK, nAbs were detected 8 days after onset of symptoms [28], which indicates the potential immunoprotection from re-infection. Although post-infection, IgG and nAb levels were decreased with the alleviation of the disease course [29], individuals were still protected for a relatively long time. Particularly in the short term, such as over a 2-month follow-up period, none out of 804 recovered Italian participants died or had recurrence [30]. In a study involving 3,276 UK workers, the levels of anti-S IgG antibody were found to be stable even after 6 months post infection [31]. Moreover, the presence of anti-S antibodies had been found related to a lower risk of COVID-19 infection (validated using RT-qPCR) in a 6-month longitudinal study involving 12,541 participants from the UK by the end of 2020. The infection incidence was found negatively correlated with baseline anti-S and anti-N antibody titers [32].
Such evidence seems to suggest a strong active adaptive immune response to COVID-19; however, the innate immunity should not be underestimated. Overaggressively surging neutrophils, neutrophil extracellular trap activation and related cytokines [33], as well as inflammasome activation, contribute to COVID-19 pathophysiology [34]. When an uncontrolled immune response to SARS-CoV-2 results in hyperinflammation, the clinical outcomes can be pathological and cause serious complications. Immunoprotection-associated complications are discussed further in Section 5.
3.2. Protective immune response from pre-existing immunity
In contrast to the post-infection immunity reviewed above, it is theoretically possible to generate pre-existing nAbs without pathological infection after natural exposure to other deactivated CoV-RBDs. T cells reactive to the N protein of SARS-CoV-2 were detected in patients who had been previously infected with SARS-CoV-1 and had recovered [35]. Such potential immune cross-reactivity might be protective. A SARS-CoV-2-specific T cell immune response could also be found in approximately 50% of the COVID-19 free volunteers, indicating the cross-reactivity between common cold-related coronavirus and SARS-CoV-2 [35]. Such heterogenous cross-reactivity might substantially contribute to the mechanism underlying the population's diverse susceptibility to COVID-19.
In brief, our immune systems make immense efforts to establish a protective immune response during our battle with COVID-19. This is why the anti-SARS-CoV-2 vaccine, as a proactive immunoprotection measure, is highly anticipated even before its official release, and it has indeed proven to be capable of inducing critical protective immunity (SARS-CoV-2-specific IgG) [36].
3.3. Heterogeneity of immunoprotection against COVID-19 among populations
Disease severity under infection with each SARS-CoV-2 variant highly varies across the population. For instance, some infections are asymptomatic, whereas others are symptomatic and even fatal. Many factors contribute to this overall diverse phenotype. From the perspective of immune protection, the T cell-derived adaptive immune surveillance recognizes the different combinations of SARS-CoV-2 epitopes in a human leukocyte antigen-restricted pattern, resulting in different immune responses among individuals [37]. Once infected, the antibody levels of those infected also vary. In a previous study, the levels of SARS-CoV-2-specific IgG and pro- and anti-inflammatory cytokines in asymptomatic cases were much lower than those in symptomatic cases, which suggests a lower immune response in asymptomatic infections [29,31]. Although free from the critical health conditions, it is possible that such an asymptomatic population with a shorter immune memory is not very well protected from a second round of infection.
Over the past few years, we have discovered some determinants of this heterogeneity of immunoprotection [38]. First, COVID-19 displays a significant age tropism. The present data support the assumption that SARS-CoV-2 preferentially targets older adults [39]. One explanation could be the less coordinated adaptive immune system of older people [40]. Even if children are not spared from COVID-19 as previously anticipated [41], they are still spared from the severity of the disease observed in older patients, possibly due to the presence of more rapidly responding CD4+ T cells and lower hyperinflammatory neutrophil infiltration [42]. Male patients are reported to have a higher risk of mortality than female patients, possibly due to hormonal regulation of the hyperinflammatory immune state [43]. On the other hand, pregnant women have long been considered as a susceptible population for various viral infections. A previous study demonstrated that COVID-19-infected pregnant women had more severe lymphopenia and higher levels of inflammatory markers, including interleukin-6 (IL-6) and C-reactive protein (CRP) level than nonpregnant women [44]. Furthermore, according to data from the US CDC and the COVID-19 surveillance system, pregnant women are more vulnerable to severe disease than nonpregnant women [[39], [45]].
Host-derived immune dysregulation is another risk factor for COVID-19; thus, the potential association between the immune state and COVID-19 severity has been thoroughly investigated in both immunocompromised and autoimmune individuals. For some immunocompromised patients, such as HIV-infected patients, host-derived CD4 lymphocytopenia may result in poor prognosis [46]. This is probably related to SARS-CoV-2's inhibition of immune defenses, which has been related to critical clinical outcomes [47]. Patients suffering from autoimmune rheumatic disorders could benefit from their anti-tumor necrosis factor-alpha (TNF-α) therapy [46]. Comorbidities, particularly various chronic inflammatory-related disorders, such as obesity and cardiovascular disease, were found to result in higher severity of COVID-19 during early winter in 2020 [48]. Overall, the individual balanced immunoprotection state could determine the final level of susceptibility and disease severity in COVID-19.
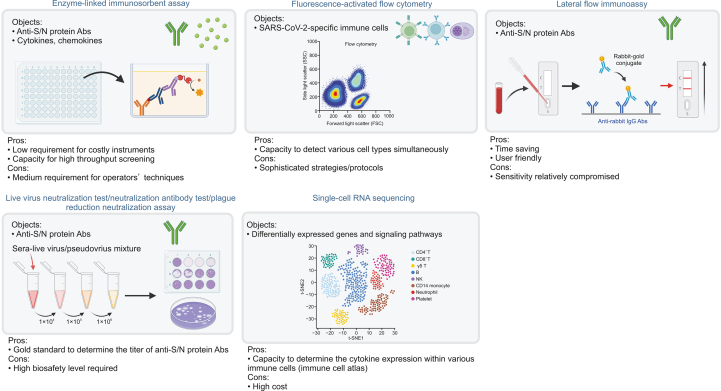
3.4. Techniques for the evaluation of immunoprotection status
Multiple techniques can be used to evaluate immunoprotection status (Fig. 2). During daily clinical practice, optional serologic investigations enable the determination of anti-S or anti-N IgG concentration. The enzyme-linked immunosorbent assay (ELISA) is a frequently used technique based on antigen-antibody specific binding for both the capture and detection followed by signal cascade amplification. ELISA for anti-S and anti-N IgG exhibits highly satisfactory sensitivity (98%) and specificity (98%) in a 96- or 384-well design for high-throughput screening [49]. Additionally, lateral flow immunoassay has an overwhelming time-saving advantage; however, its sensitivity is compromised at 65%–85% compared with that of ELISA [50].
Fig. 2.
Techniques used to evaluate immunoprotection status against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). S: spike; N: nucleocapsid; Abs: antibodies; NK: natural killer; t-SNE: t-distributed stochastic neighbour embedding. Figure is created with BioRender.com.
In addition to the detection of SARS-CoV-2-specific antibodies, ELISA is widely used for the frequent determination of related classic markers such as cytokines and chemokines (e.g. IL-1β, IL-6, IL-17, IL-18, IL-22, TNF-α, interferon-γ, and monocyte chemoattractant protein-1), which is recommended in clinical practice for monitoring the incoming cytokine storm (CS) caused by hyperinflammation [51].
Analyzing the recruitment of various immune cell populations in both innate immunity and adaptive immunity is another vital aspect for evaluating the immunoprotection status. Biased counts for neutrophils versus lymphocytes [52] could be easily accessed through automatic differential blood cell counting in clinical practice. Meanwhile, fluorescence-activated flow cytometry can detect SARS-CoV-2 (S, RBD, and N)-specific adaptive immune cells such as CD4+, CD8+ T cells, and memory B cells. However, sophisticated strategies and protocols [40,53] are needed to discriminate these cells from nonspecific cells.
Neutralization assays are the gold standard for focusing on the titer determination of SARS-CoV-2-specific nAbs. Such technique requires mixing nAb-containing serum together with live virus (live virus neutralization test) [54] or pseudovirus (surrogate neutralization antibody test) [40] and transduction into the angiotensin-converting enzyme 2-expressing engineered cell lines. These techniques are usually performed in research laboratories. The common plaque reduction neutralization assay [55] is also based on a similar principle with a different presentation of the neutralizing outcome.
In patients with COVID-19, single-cell RNA sequencing can be used to combine an intensive dataset of differentially expressed genes and signaling pathways in major immune cell types together with their cytokine expression profiles. The resulting primary finding is well recognized as a comprehensive immune cell atlas [56].
The sole or combined application of the above-mentioned techniques can be very informative. Molecular diagnosis-assisted immunological testing is highly beneficial for evaluation. For instance, a combined SARS-CoV-2 nucleic acid-negative, anti-SARS-CoV-2 IgM-negative, and anti-SARS-CoV-2 IgG-positive result could better estimate the immunoprotection status. However, the mechanisms underlying achieving this status remain unclear. There are several explanations for a standing immunoprotection against COVID-19, some of which include immune memory protection through voluntary vaccination, COVID-19 recovery, and acquired cross-reactive immunity from a previous common cold.
4. Immunoprotection and vaccine inoculation
Vaccines are designed to prime the protective immune memory. Therefore, since the beginning of the COVID-19 pandemic, tremendous efforts have been made in vaccine development and inoculation at the population level.
4.1. Recent knowledge on currently available vaccines applied at a national/regional scale
According to Johns Hopkins University statistics [1], a total of 12.1 billion doses of vaccine had been administered worldwide up to September 6, 2022. Increasing efforts have been directed toward the development of an anti-SARS-CoV-2 vaccine, with encouraging results. To date, multiple vaccine types have emerged, including inactivated virus vaccines, live attenuated vaccines, protein subunit vaccines, DNA/RNA vaccines, vector-based vaccines, and virus-like particle vaccines [57]. The immunodominant S protein, a major glycoprotein on the surface of the virus envelope, is the main target of nAbs formed due to natural infection [58], and thus becomes the target of interest for vaccine design.
According to the updated International Vaccine Access Center [59], among 156 candidate vaccines which entered clinical trials, 57 candidate vaccines targeting the S protein have been developed and tested in various phases of clinical trials (as summarized in Table S2). Among them, five products have reached the market, entering phase IV clinical trials and being used for worldwide vaccination; these include mRNA-1273 [60] and BNT162b2 [61]. Inactivated virus vaccines, including BBIBP-CorV [62] and CoronaVac [63], were developed according to the inactivated primary SARS-CoV-2 strain.
Vaccine inoculation is a potent strategy to establish our adaptive immunity against SARS-CoV-2; however, the very first principle of vaccine inoculation is that the benefits should always outweigh the risks. Hence, an optimized vaccination program should be individualized. The individual's disease status, disease history, and medicine treatment should be considered before vaccination. For example, for patients with multiple sclerosis, all types of COVID-19 vaccines, except live attenuated vaccines, are recommended and considered safe [64].
4.2. Vaccines protect the population against disease onset and severity
Studies of fully vaccinated populations can provide valuable information about whether the vaccine has any positive effect on disease onset incidence or severity. A previous study described 39 mild-to-asymptomatic infective cases among 1,497 Israeli workers with high occupational exposure risk after completion of the mRNA vaccine program. The nAb titers within the infected population were only approximately one third of those of matched controls during the peri-infection period [65]. This indicates that both vaccine and personal immune protection conditions (the actively established vaccine-derived immune memory) are vital in achieving favorable clinical outcomes.
4.3. Immunoprotection against viral variants through vaccination
As described in Table 1, the S protein is the vital component of SARS-CoV-2, and various related mutations occurred in this protein in the VOCs that emerged in the recent pandemic. In addition to various random natural mutations, the virus sometimes mutates to better evade the host antiviral immunity by affecting epitopes for nAb recognition. Under immune selective pressure, SARS-CoV-2 variants can rapidly evolve to better evade our antiviral immune barrier; therefore, the vaccine effects can eventually be compromised. Moreover, because of the high heterogeneity of instinct immunoprotection barriers among the population, vaccines work differently in different individuals. For most newly emerged variants, there is yet no solid evidence from longitudinal studies, despite the value of primary findings in clinical trials with relatively shorter time windows.
After full vaccination, breakthrough infection was still detected among workers with high occupational exposure risk. Such infections mainly (85%) occurred with the B.1.1.7 variant compared with the primary virus [65]. Because of cross-reactive immunoprotection, it is very likely that vaccines based on antiquated virus information will still be able to identify many variants. Moreover, for the common variant D614G (B.1), despite its high shedding [66,67], it could still be identified and neutralized by plasma from donors infected with the primary virus [67]. Further, in a pseudovirus-neutralization assay, despite the immune escape against 50% of all detected monoclonal antibodies resulting from mutations including N501Y, N439K, and S477N, inactivated virus vaccine-elicited sera had favorable efficacy and responded to the D614G + L18F + A222V and D614G + A222V variants [68]. Theoretically speaking, based on the solid evidence of human adaptive immune response, such as mediated by CD4+ and CD8+ T cells, to diverse SARS-CoV-2 epitopes within components including S, M, and N proteins and ORFs [35,37], nAbs in our immune system recognize the S protein as the signature SARS-CoV-2 antigen, while the key epitope of the large RBD of the S antigen hinders escaping from our polyclonal serum neutralization [69,70].
In addition, the effectiveness of seven representative, widely used vaccines against the pandemic strains B.1.617.2 and B.1.1.529 is summarized in Table 2 [[71], [72], [73], [74], [75], [76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102]]. Vaccine effectiveness appears to be much lower against these recent variants than against the original D614G (B.1) strain. Despite the possibility of a booster immunization, vaccine effectiveness against B.1.1.529 is significantly reduced.
Table 2.
Vaccine effectiveness against the pandemic Delta and Omicron strains of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).
| Strains | CoronaVac | Ad26.COV 2.S | AZD1222 (ChAdOx1) | BBIBP-CorV (Sinopharm COVID-19 Vaccine) | mRNA-1273 | Covaxin | BNT162b (Pfizer-BioNTech) |
|---|---|---|---|---|---|---|---|
| Effectiveness of non-mutant strain | 80.50% (2 doses) [71] | 72% (1 dose) [72] | 70.4% (2 doses) [73] | 79.34% (2 doses) [74] | 94.1% (2 doses) [75] | 93.4% (2 doses) [76] | 95% (2 doses) [77] |
| B.1.617.2 (Delta) mutant | −2.1 (2 doses) [78] | −1.6 (1 dose) [79] | 67.0% (2 doses) [80] | −1.7 (2 doses) [81] | −3.3 (2 doses) [82] | 85% (2 doses) [83] | −5.8 (2 doses) [84] |
| 74.6% (2 doses) [85] | −5.4 (1 dose) [86] | −4.01 (2 doses) [84] | 95% (2 doses) [87] | −2.9 (2 doses) [88] | −2.06 (2 doses) [89] | −2.5 (2 doses) [90] | |
| 82.25% (2 doses) [91] | −4.3 (2 doses) [90] | −3.3 (2 doses) [86] | −8.4 (2 doses) [92] | ||||
| 76.08% (2 doses) [91] | 88.0% (2 doses) [80] | ||||||
| 98% (2 doses) [87] | |||||||
| −11.3 (2 doses) [84] | |||||||
| −2.6 (2 doses) [93] | |||||||
| B.1.1.529 (Omicron) mutant | −7.1 (2 doses) [94] a | −2.5 (1 dose) [95] | −1.6 (2 doses) [95] | −5.1 (2 doses) [81] | −8.6 (2 doses) and −6.5 (booster) [95] | −12.49 (2 doses) [89] | −21 (2 doses) and −6.5 (booster) [95] |
| −17 (1 dose) and −13 (booster) [96] | −21 (2 doses) [97] | −21.3 (2 doses) and −5.1 (booster) [98] | −12 (2 doses) and −4 (booster) [96] | ||||
| −12.7 (2 doses) [99] | −43 (2 doses) and −6 (booster) [96] | −37 (2 doses) [97] | |||||
| −39 (2 doses) [97] | −23 (2 doses) and −7.5 (booster) [100] | ||||||
| −42 (2 doses) and −16.7 (booster) [100] | −22 (2 doses) [101] | ||||||
| −34 (2 doses) and −8 (booster) [102] | |||||||
| −14.2 (2 doses) [99] |
Vaccine effectiveness against Delta or Omicron variants is represented as minus fold change.
Meanwhile, a previous study discovered a VOC (B1.1.7) for which the neutralization activity of the ChAdOx1 nCoV-19 vaccine-elicited antibody was reduced by 9 folds compared with the primary non-B.1.1.7 strain [73]. Moreover, the spike RBD N439K variant (belonging to B.1.1.529) was found to decrease around 50% of the RBD-binding serum IgG or monoclonal antibody levels compared with the wild type [70].
In summary, there is little possibility that SARS-CoV-2 variants could completely escape our humoral and cellular immune surveillance in humans [26]. However, these continually emerging variants will remain a global challenge. With the advances in vaccine boosters, we believe that our immunoprotection strategy is appropriate. With breakthroughs in the development of effective pharmacological interventions, it is possible to defeat COVID-19. Although we hope for the best, we must be prepared for the worst; thus, timely surveillance of variants using deep-sequencing technologies is necessary.
5. Immunoprotection and COVID-19 complications
COVID-19 is mostly characterized by common symptoms, including fever, dry cough, shortness of breath, and various other clinical signs [103]. For some patients, however, once infected with SARS-CoV-2, the symptoms do not quickly disappear. There are several complications that may affect the long-term health of such patients. Clinicians are diagnosing not only pneumonia following COVID-19, but also other disorders occurring within a couple of days after hospital admission.
5.1. Over immunoprotection from the host immune system accompanies many severe COVID-19 complications
Aside from the unfortunate patients who suffer respiratory failure during the acute infection phase of COVID-19 [104], survivors can also encounter extensive production and release of inflammatory cytokines, known as a “cytokine storm” [105], which can result in the multiorgan failure and critical illness [106]. Apart from immunological pneumonia during infection, SARS-CoV-2 could trigger various autoimmune disorders, including inflammatory arthritis, systemic lupus erythematosus, renal disease, rhabdomyolysis, myositis, and vasculitis (particularly large-vessel vasculitis) [107]. More importantly, many post-infectious patients are further diagnosed with the multisystem inflammatory syndrome (MIS), known as MIS-A in adults [108] and MIS-C in children [109]. Even after complete resolution of SARS-CoV-2 infection, patients could still experience various symptoms, such as fatigue, persistent cough, pain, dyspnea, headache, cognitive dysfunction, and even various cardiovascular sequelae [110], which are known as post-COVID conditions or “long COVID” [111,112]. The SARS-CoV-2-triggered overactive neuroinflammation may play a crucial role in these sequelae [113]. An overprotective antiviral response is one of the most accepted hypothesis, but the fact that SARS-CoV-2 would act as a molecular mimic is also plausible, suggesting that the dozens of SARS-CoV-2 heptapeptides that share high similarity with human heptapeptides may have high pathologically autoimmune potential [52]. Therefore, post-COVID-19 symptoms could be caused by a secondary autoimmune attack.
5.2. Real-time evaluation and monitoring of cytokine-driven hyperinflammatory responses for correctly addressing COVID-19 complications
Real-time evaluation of the hyperinflammatory state of patients can provide better etiological diagnosis and treatment. A research group from Temple University in the US has proposed novel criteria for COVID-CS. These criteria include a combination of multiple indeces as follows: ferritin > 250 ng/mL and CRP > 4.5 mg/dL (indicating hyperinflammation and tissue damage, respectively) as well as prerenal electrolyte imbalance [114]. Early diagnosis of COVID-CS would help physicians prescribe personalized medication in time, resulting in better prognoses. Hence, in addition to clinical signs including breathlessness, ultrastructure changes such as deteriorating chest radiograph [111], potential biomarker combinations available in daily clinical practice, especially in developing countries, would be desirable. For instance, the hyperinflammatory state could be profiled using immunological indicators, including abnormal immune cell populations such as excessive neutrophil recruitment [52] in peripheral blood, oversecretion of proinflammatory cytokines and chemokines including circulating IL-6, TNF-α, IL-8, IL-10 [105,115], as well as other hyperinflammatory indeces such as CRP and D-dimers [111] and autoantibodies [52].
6. Conclusion and future perspectives
Here, we reviewed the current progress in research on emerging coronavirus variants, vaccination progression, and COVID-19 complications from the perspective of immunoprotection. For better etiological diagnosis, various techniques, including RT-qPCR, Ag-RDTs, and HTS, were analyzed for use in different scenarios and spectra at the population scale. This work discusses how to evaluate the immunoprotection status in a specific population (endogenous antibodies) before and after vaccine inoculation. Our findings can help guide the design of antibody drugs, facilitate the development of practice guidelines, and help policy makers to introduce national regulations, such as the timing of and requirements for extra rounds of vaccine boosters.
According to the current knowledge, we recommend RT-qPCR for population-scale screening as normal practice, supplemented with the use of Ag-RDTs in low-prevalence settings such as schools or workplaces. For each new pandemic wave, HTS should always be applied to representative specimens for final variant validation.
With the global spread of the B.1.1.529 variant, determination of the immunoprotection status of some populations can help determine practice guidelines. A service package that evaluates an individual's immunoprotection status against the most recent pandemic SARS-CoV-2 variant should be developed. We expect these techniques to become more financially available so that the practice of anti-COVID-19 immune surveillance can become as easy as our regular programme of anti-hepatitis B virus immune surveillance.
After evaluating the performance of different vaccines against various variants, expectant management with the present technological reserves of vaccine is a matter of expediency, so far well-run. If and when the situation deteriorates, we recommend the development of an inactivated virus vaccine (with advanced and ready-to-use technologies) based on the most concerning variant (B.1.1.529, for example).
CRediT author statement
Congshan Jiang: Conceptualization, Funding acquisition, Validation, Visualization, Writing - Original draft preparation, Reviewing and Editing; Kaichong Jiang: Methodology, Writing - Original draft preparation, Reviewing and Editing; Xiaowei Li: Methodology, Investigation; Ning Zhang: Validation; Wenhua Zhu: Visualization; Liesu Meng: Visualization; Yanmin Zhang: Project administration, Supervision, Conceptualization, Funding acquisition, Writing - Reviewing and Editing; Shemin Lu: Supervision, Conceptualization, Funding acquisition, Writing - Reviewing and Editing.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Acknowledgments
We appreciate the critical reading of our work by Professor Xu Li from Center for Translational Medicine, The First Affiliated Hospital of Xi'an Jiaotong University. We are very grateful for the financial support from the National Natural Science Foundation of China (Grant Nos.: 81970029, 81974014, 82211530115, and 81470452), China Postdoctoral Science Foundation (Project No.: 2021M702591), the Natural Science Foundation of Shaanxi Province (Project No.: 2021JQ-024), Fundamental Research Funds for the Central Universities (Project No.: xjh012020026), Xi'an Health Commission (COVID-19 special project), Xi'an Talent Program (Project No.: XAYC200023), and research funds of Xi'an Children's Hospital (Project No.: 2020A03).
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jpha.2022.10.003.
Contributor Information
Yanmin Zhang, Email: zhangym@xjtu.edu.cn.
Shemin Lu, Email: lushemin@xjtu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Johns Hopkins University & Medicine, Coronavrius Resource Center, Global Map. https://coronavirus.jhu.edu/map.html. (Accessed 6 September 2022).
- 2.Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li X., Geng M., Peng Y., et al. Molecular immune pathogenesis and diagnosis of COVID-19. J. Pharm. Anal. 2020;10:102–108. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberfeld B., Achanta A., Carpenter K., et al. SnapShot: COVID-19. Cell. 2020;181 doi: 10.1016/j.cell.2020.04.013. 954–954.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang C., Li X., Ge C., et al. Molecular detection of SARS-CoV-2 being challenged by virus variation and asymptomatic infection. J. Pharm. Anal. 2021;11:257–264. doi: 10.1016/j.jpha.2021.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hadfield J., Megill C., Bell S.M., et al. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.The Nextstrain team, Genomic epidemiology of SARS-CoV-2 with subsampling focused globally over the past 6 months. https://nextstrain.org/ncov/gisaid/global/6m. (Accessed 16 June 2022).
- 8.Madhi S.A., Kwatra G., Myers J.E., et al. Population immunity and Covid-19 severity with Omicron variant in South Africa. N. Engl. J. Med. 2022;386:1314–1326. doi: 10.1056/NEJMoa2119658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewnard J.A., Hong V.X., Patel M.M., et al. Clinical outcomes among patients infected with Omicron (B.1.1.529) SARS-CoV-2 variant in southern California. Lancet Respir. Med. 2022;10:689–699. [Google Scholar]
- 10.Bull R.A., Adikari T.N., Ferguson J.M., et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat. Commun. 2020;11:6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chappleboim A., Joseph-Strauss D., Rahat A., et al. Early sample tagging and pooling enables simultaneous SARS-CoV-2 detection and variant sequencing. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.X. Li, Y. Xu, X. Li, et al., Real-world effectiveness and protection of SARS-CoV-2 vaccine among patients hospitalized for COVID-19 in Xi’an, China, December 8, 2021, to January 20, 2022: A retrospective study, Front. Immunol. 13 (2022), 978977. [DOI] [PMC free article] [PubMed]
- 13.Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat. Biotechnol. 2020;38:382–384. doi: 10.1038/d41587-020-00002-2. [DOI] [PubMed] [Google Scholar]
- 14.Arena F., Pollini S., Rossolini G.M., et al. Summary of the available molecular methods for detection of SARS-CoV-2 during the ongoing pandemic. Int. J. Mol. Sci. 2021;22:1298. doi: 10.3390/ijms22031298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Artesi M., Bontems S., Göbbels P., et al. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J. Clin. Microbiol. 2020;58 doi: 10.1128/JCM.01598-20. e01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ko K.K.K., Abdul Rahman N.B., Tan S.Y.L., et al. SARS-CoV-2 N gene G29195T point mutation may affect diagnostic reverse transcription-PCR detection. Microbiol. Spectr. 2022;10 doi: 10.1128/spectrum.02223-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J.C.C. Lesbon, M.D. Poleti, E.C. de Mattos Oliveira, et al., Nucleocapsid (N) gene mutations of SARS-CoV-2 can affect real-time RT-PCR diagnostic and impact false-negative results, Viruses 13 (2021), 2474. [DOI] [PMC free article] [PubMed]
- 18.Laine P., Nihtilä H., Mustanoja E., et al. SARS-CoV-2 variant with mutations in N gene affecting detection by widely used PCR primers. J. Med. Virol. 2022;94:1227–1231. doi: 10.1002/jmv.27418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain A., Rophina M., Mahajan S., et al. Analysis of the potential impact of genomic variants in global SARS-CoV-2 genomes on molecular diagnostic assays. Int. J. Infect. Dis. 2021;102:460–462. doi: 10.1016/j.ijid.2020.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peeling R.W., Olliaro P.L., Boeras D.I., et al. Scaling up COVID-19 rapid antigen tests: Promises and challenges. Lancet Infect. Dis. 2021;21:e290–e295. doi: 10.1016/S1473-3099(21)00048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antigen-detection in the Diagnosis of SARS-CoV-2 Infection Using Rapid Immunoassays: Interim Guidance. https://www.who.int/publications/i/item/antigen-detection-in-the-diagnosis-of-sars-cov-2infection-using-rapid-immunoassays. (Accessed 6 September 2022).
- 22.Xu J., Suo W., Goulev Y., et al. Handheld microfluidic filtration platform enables rapid, low-cost, and robust self-testing of SARS-CoV-2 virus. Small. 2021;17 doi: 10.1002/smll.202104009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.L. Liv, G. Çoban, N. Nakiboğlu, et al., A rapid, ultrasensitive voltammetric biosensor for determining SARS-CoV-2 spike protein in real samples, Biosens. Bioelectron. 192 (2021), 113497. [DOI] [PMC free article] [PubMed]
- 24.M. Nóra, D. Déri, D.S. Veres, et al., Evaluating the field performance of multiple SARS-Cov-2 antigen rapid tests using nasopharyngeal swab samples, PLoS One 17 (2022), e0262399. [DOI] [PMC free article] [PubMed]
- 25.J.-L. Bayart, J. Degosserie, J. Favresse, et al., Analytical sensitivity of six SARS-CoV-2 rapid antigen tests for omicron versus delta variant, Viruses 14 (2022), 654. [DOI] [PMC free article] [PubMed]
- 26.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peluso M.J., Deitchman A.N., Torres L., et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 2021;36 doi: 10.1016/j.celrep.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeffrey S., Carl G., Blair M., et al. Longitudinal evaluation and decline of antibody responses in SARS-CoV-2 infection. Nat. Biotechnol. 2020;5:1598–1607. [Google Scholar]
- 29.Long Q.-X., Tang X.-J., Shi Q.-L., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 30.Mumoli N., Vitale J., Mazzone A. Clinical immunity in discharged medical patients with COVID-19. Int. J. Infect. Dis. 2020;99:229–230. doi: 10.1016/j.ijid.2020.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lumley S.F., Wei J., O'Donnell D., et al. The duration, dynamics, and determinants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody responses in individual healthcare workers. Clin. Infect. Dis. 2021;73:e699–e709. doi: 10.1093/cid/ciab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lumley S.F., O'Donnell D., Stoesser N.E., et al. Antibody status and incidence of SARS-CoV-2 infection in health care workers. N. Engl. J. Med. 2021;384:533–540. doi: 10.1056/NEJMoa2034545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ackermann M., Anders H.-J., Bilyy R., et al. Patients with COVID-19: In the dark-NETs of neutrophils. Cell Death Differ. 2021;28:3125–3139. doi: 10.1038/s41418-021-00805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee S., Channappanavar R., Kanneganti T.-D. Coronaviruses: Innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41:1083–1099. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grifoni A., Weiskopf D., Ramirez S.I., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e5. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ewer K.J., Barrett J.R., Belij-Rammerstorfer S., et al. T cell and antibody responses induced by a single dose of ChAdOx1 nCoV-19 (AZD1222) vaccine in a phase 1/2 clinical trial. Nat. Med. 2021;27:270–278. doi: 10.1038/s41591-020-01194-5. [DOI] [PubMed] [Google Scholar]
- 37.Tarke A., Sidney J., Kidd C.K., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 39.United States COVID-19 cases and deaths by state over time. https://data.cdc.gov/Case-Surveillance/United-States-COVID-19-Cases-and-Deaths-by-State-o/9mfq-cb36. (Accessed 6 September 2022).
- 40.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lingappan K., Karmouty-Quintana H., Davies J., et al. Understanding the age divide in COVID-19: Why are children overwhelmingly spared? Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L39−L44. doi: 10.1152/ajplung.00183.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brodin P. Why is COVID-19 so mild in children? Acta Paediatr. 2020;109:1082–1083. doi: 10.1111/apa.15271. [DOI] [PubMed] [Google Scholar]
- 43.Bienvenu L.A., Noonan J., Wang X., et al. Higher mortality of COVID-19 in males: Sex differences in immune response and cardiovascular comorbidities. Cardiovasc. Res. 2020;116:2197–2206. doi: 10.1093/cvr/cvaa284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen G., Zhang Y., Zhang Y., et al. Differential immune responses in pregnant patients recovered from COVID-19. Signal Transduct. Target. Ther. 2021;6:289. doi: 10.1038/s41392-021-00703-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jamieson D.J., Rasmussen S.A. An update on COVID-19 and pregnancy. Am. J. Obstet. Gynecol. 2022;226:177–186. doi: 10.1016/j.ajog.2021.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldman J.D., Robinson P.C., Uldrick T.S., et al. COVID-19 in immunocompromised populations: Implications for prognosis and repurposing of immunotherapies. J. Immunother. Cancer. 2021;9 doi: 10.1136/jitc-2021-002630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bost P., De Sanctis F., Canè S., et al. Deciphering the state of immune silence in fatal COVID-19 patients. Nat. Commun. 2021;12:1428. doi: 10.1038/s41467-021-21702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.National SARS-CoV-2 Serology Assay Evaluation Group Performance characteristics of five immunoassays for SARS-CoV-2: A head-to-head benchmark comparison. Lancet Infect. Dis. 2020;20:1390–1400. doi: 10.1016/S1473-3099(20)30634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams E.R., Ainsworth M., Anand R., et al. Antibody testing for COVID-19: A report from the national COVID scientific advisory panel. Wellcome Open. Res. 2020;5:139. doi: 10.12688/wellcomeopenres.15927.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Merad M., Blish C.A., Sallusto F., et al. The immunology and immunopathology of COVID-19. Science. 2022;375:1122–1127. doi: 10.1126/science.abm8108. [DOI] [PubMed] [Google Scholar]
- 52.Dotan A., Muller S., Kanduc D., et al. The SARS-CoV-2 as an instrumental trigger of autoimmunity. Autoimmun. Rev. 2021;20 doi: 10.1016/j.autrev.2021.102792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dan J.M., Mateus J., Kato Y., et al. 2020. Immunological memory to SARS-CoV-2 assessed for up to eight months after infection, bioRxiv.https://pubmed.ncbi.nlm.nih.gov/33442687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Premkumar L., Segovia-Chumbez B., Jadi R., et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okba N.M.A., Müller M.A., Li W., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease Patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilk A.J., Rustagi A., Zhao N.Q., et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26:1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Callaway E. The race for coronavirus vaccines: A graphical guide. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- 58.Pallesen J., Wang N., Corbett K.S., et al. Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc. Natl. Acad. Sci. U S A. 2017;114:E7348–E7357. doi: 10.1073/pnas.1707304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.View-hub by IVAC, COVID vaccine data. https://view-hub.org. (Accessed 16 June 2022).
- 60.Corbett K.S., Edwards D.K., Leist S.R., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sahin U., Muik A., Vogler I., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 62.Wang H., Zhang Y., Huang B., et al. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721.e9. doi: 10.1016/j.cell.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y., Zeng G., Pan H., et al. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18-59 years: A randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2021;21:181–192. doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.H. Kelly, B. Sokola, H. Abboud, Safety and efficacy of COVID-19 vaccines in multiple sclerosis patients, J. Neuroimmunol. 356 (2021), 577599. [DOI] [PMC free article] [PubMed]
- 65.Bergwerk M., Gonen T., Lustig Y., et al. Covid-19 breakthrough infections in vaccinated health care workers. N. Engl. J. Med. 2021;385:1474–1484. doi: 10.1056/NEJMoa2109072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hou Y.J., Chiba S., Halfmann P., et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370:1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Korber B., Fischer W.M., Gnanakaran S., et al. Tracking changes in SARS-CoV-2 spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.J. Wu, L. Zhang, Y. Zhang, et al., The antigenicity of epidemic SARS-CoV-2 variants in the United Kingdom, Front. Immunol. 12 (2021), 687869. [DOI] [PMC free article] [PubMed]
- 69.Li Q., Wu J., Nie J., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomson E.C., Rosen L.E., Shepherd J.G., et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Borges R.P.M.C., Brango H.A., et al. 2021. Projeto S: A stepped-wedge randomized trial to assess CoronaVac effectiveness in Serrana, Brazil, Preprints with the Lancet.https://papers.ssrn.com/sol3/papers.cfm?abstract_id=3973422 [Google Scholar]
- 72.Sadoff J., Gray G., Vandebosch A., et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N. Engl. J. Med. 2021;384:2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emary K.R.W., Golubchik T., Aley P.K., et al. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): An exploratory analysis of a randomised controlled trial. Lancet. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sinopharm COVID-19 vaccine (BBIBP-CorV). https://www.precisionvaccinations.com/vaccines/sinopharm-covid-19-vaccine-bbibp-corv. (Accessed 16 June 2022).
- 75.Baden L.R., El Sahly H.M., Essink B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ella R., Reddy S., Blackwelder W., et al. Efficacy, safety, and lot-to-lot immunogenicity of an inactivated SARS-CoV-2 vaccine (BBV152): Interim results of a randomised, double-blind, controlled, phase 3 trial. Lancet. 2021;398:2173–2184. doi: 10.1016/S0140-6736(21)02000-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Polack F.P., Thomas S.J., Kitchin N., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu L., Mok B.W.Y., Chen L.L., et al. Neutralization of severe acute respiratory syndrome coronavirus 2 omicron variant by sera from BNT162b2 or CoronaVac vaccine recipients. Clin. Infect. Dis. 2022;75:e822–e826. doi: 10.1093/cid/ciab1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alter G., Yu J., Liu J., et al. Immunogenicity of Ad26.COV2.S vaccine against SARS-CoV-2 variants in humans. Nature. 2021;596:268–272. doi: 10.1038/s41586-021-03681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez Bernal J., Andrews N., Gower C., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (delta) variant. N. Engl. J. Med. 2021;385:585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao X., Li D., Ruan W., et al. Effects of a prolonged booster interval on neutralization of omicron variant. N. Engl. J. Med. 2022;386:894–896. doi: 10.1056/NEJMc2119426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Choi A., Koch M., Wu K., et al. Serum neutralizing activity of mRNA-1273 against SARS-CoV-2 variants. J. Virol. 2021;95 doi: 10.1128/JVI.01313-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhatnagar T., Chaudhuri S., Ponnaiah M., et al. Effectiveness of BBV152/Covaxin and AZD1222/Covishield vaccines against severe COVID-19 and B.1.617.2/Delta variant in India, 2021: A multi-centric hospital-based case-control study. Int. J. Infect. Dis. 2022;122:693–702. doi: 10.1016/j.ijid.2022.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis C., Logan N., Tyson G., et al. Reduced neutralisation of the Delta (B.1.617.2) SARS-CoV-2 variant of concern following vaccination. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ma C., Sun W., Tang T., et al. Effectiveness of adenovirus type 5 vectored and inactivated COVID-19 vaccines against symptomatic COVID-19, COVID-19 pneumonia, and severe COVID-19 caused by the B.1.617.2 (Delta) variant: Evidence from an outbreak in Yunnan, China, 2021. Vaccine. 2022;40:2869–2874. doi: 10.1016/j.vaccine.2022.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tada T., Zhou H., Samanovic M.I., et al. 2021. Comparison of neutralizing antibody titers elicited by mRNA and adenoviral vector vaccine against SARS-CoV-2 variants, bioRxiv.https://www.biorxiv.org/content/10.1101/2021.07.19.452771v3 [Google Scholar]
- 87.Mousa M., Albreiki M., Alshehhi F., et al. Similar effectiveness of the inactivated vaccine BBIBP-CorV (Sinopharm) and the mRNA vaccine BNT162b2 (Pfizer-BioNTech) against COVID-19 related hospitalizations during the Delta outbreak in the UAE. J. Travel Med. 2022;29 doi: 10.1093/jtm/taac036. taac036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Edara V.V., Pinsky B.A., Suthar M.S., et al. Infection and vaccine-induced neutralizing-antibody responses to the SARS-CoV-2 B.1.617 variants. N. Engl. J. Med. 2021;385:664–666. doi: 10.1056/NEJMc2107799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yadav P.D., Sapkal G.N., Sahay R.R., et al. Elevated neutralization of Omicron with sera of COVID-19 recovered and breakthrough cases vaccinated with Covaxin than two dose naïve vaccinees. J. Infect. 2022;84:834–872. doi: 10.1016/j.jinf.2022.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu C., Ginn H.M., Dejnirattisai W., et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184:4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Suah J.L., Tng B.H., Tok P.S.K., et al. Real-world effectiveness of homologous and heterologous BNT162b2, CoronaVac, and AZD1222 booster vaccination against Delta and Omicron SARS-CoV-2 infection. Emerg. Microbes Infect. 2022;11:1343–1345. doi: 10.1080/22221751.2022.2072773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mlcochova P., Kemp S.A., Dhar M.S., et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021;599:114–119. doi: 10.1038/s41586-021-03944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lustig Y., Zuckerman N., Nemet I., et al. Neutralising capacity against Delta (B.1.617.2) and other variants of concern following Comirnaty (BNT162b2, BioNTech/Pfizer) vaccination in health care workers, Israel. Euro. Surveill. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.26.2100557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pérez-Then E., Lucas C., Monteiro V.S., et al. Neutralizing antibodies against the SARS-CoV-2 Delta and Omicron variants following heterologous CoronaVac plus BNT162b2 booster vaccination. Nat. Med. 2022;28:481–485. doi: 10.1038/s41591-022-01705-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu L., Iketani S., Guo Y., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602:676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 96.Garcia-Beltran W.F., Denis K.J., St., Hoelzemer A., et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185:457–466. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cameroni E., Bowen J.E., Rosen L.E., et al. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zeng C., Evans J.P., Chakravarthy K., et al. COVID-19 mRNA booster vaccines elicit strong protection against SARS-CoV-2 Omicron variant in patients with cancer. Cancer Cell. 2022;40:117–119. doi: 10.1016/j.ccell.2021.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dejnirattisai W., Huo J., Zhou D., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Carreño J.M., Alshammary H., Tcheou J., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 101.Cele S., Jackson L., Khoury D.S., et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. 2022;602:654–656. doi: 10.1038/s41586-021-04387-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoffmann M., Krüger N., Schulz S., et al. The Omicron variant is highly resistant against antibody-mediated neutralization: Implications for control of the COVID-19 pandemic. Cell. 2022;185:447–456. doi: 10.1016/j.cell.2021.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wiersinga W.J., Rhodes A., Cheng A.C., et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): A review. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 104.Wilcox S.R. Management of respiratory failure due to covid-19. BMJ. 2020;369:m1786. doi: 10.1136/bmj.m1786. [DOI] [PubMed] [Google Scholar]
- 105.Copaescu A., Smibert O., Gibson A., et al. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J. Allergy Clin. Immunol. 2020;146:518–534.e1. doi: 10.1016/j.jaci.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Berlin D.A., Gulick R.M., Martinez F.J. Severe Covid-19. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 107.Zacharias H., Dubey S., Koduri G., et al. Rheumatological complications of covid 19. Autoimmun. Rev. 2021;20 doi: 10.1016/j.autrev.2021.102883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Joshi S., Bhatia A., Tayal N., et al. Rare multisystem inflammatory syndrome in young adult after COVID-19 immunization and subsequent SARSCoV-2 infection. J. Assoc. Physicians India. 2022;69:11–12. [PubMed] [Google Scholar]
- 109.Diorio C., Henrickson S.E., Vella L.A., et al. Multisystem inflammatory syndrome in children and COVID-19 are distinct presentations of SARS-CoV-2. J. Clin. Invest. 2020;130:5967–5975. doi: 10.1172/JCI140970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raman B., Bluemke D.A., Lüscher T.F., et al. Long COVID: Post-acute sequelae of COVID-19 with a cardiovascular focus. Eur. Heart J. 2022;43:1157–1172. doi: 10.1093/eurheartj/ehac031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mandal S., Barnett J., Brill S.E., et al. 'Long-COVID': A cross-sectional study of persisting symptoms, biomarker and imaging abnormalities following hospitalisation for COVID-19. Thorax. 2021;76:396–398. doi: 10.1136/thoraxjnl-2020-215818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seeßle J., Waterboer T., Hippchen T., et al. Persistent symptoms in adult patients 1 year after coronavirus disease 2019 (COVID-19): A prospective cohort study. Clin. Infect. Dis. 2022;74:1191–1198. doi: 10.1093/cid/ciab611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Song W.-J., Hui C.K.M., Hull J.H., et al. Confronting COVID-19-associated cough and the post-COVID syndrome: Role of viral neurotropism, neuroinflammation, and neuroimmune responses. Lancet Respir. Med. 2021;9:533–544. doi: 10.1016/S2213-2600(21)00125-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Caricchio R., Gallucci M., Dass C., et al. Preliminary predictive criteria for COVID-19 cytokine storm. Ann. Rheum. Dis. 2021;80:88–95. doi: 10.1136/annrheumdis-2020-218323. [DOI] [PubMed] [Google Scholar]
- 115.Vaninov N. In the eye of the COVID-19 cytokine storm. Nat. Rev. Immunol. 2020;20:277. doi: 10.1038/s41577-020-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.