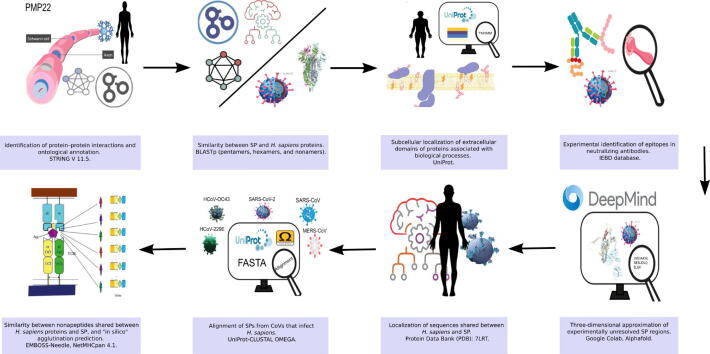
Graphical abstract
Keywords: Spike protein, SARS-CoV-2, Protein–protein interaction, In silico, Neutralizing antibodies, Nervous system, Adverse reactions
Abstract
Introduction
The development of vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in record time to cope with the ongoing coronavirus disease 2019 (COVID-19) pandemic has led to uncertainty about their use and the appearance of adverse neurological reactions. The SARS-CoV-2 spike protein (SP) is used to produce neutralizing antibodies and stimulate innate immunity. However, considering the alterations in the nervous system (NS) caused by COVID- 19, cross-reactions are plausible.
Objective
To identify peptides in Homo sapiens SP-like proteins involved in myelin and axon homeostasis that may be affected due to molecular mimicry by antibodies and T cells induced by interaction with SP.
Materials and methods
A bioinformatics approach was used. To select the H. sapiens proteins to be studied, related biological processes categorized based on gene ontology were extracted through the construction of a protein–protein interaction network. Peripheral myelin protein 22, a major component of myelin in the peripheral nervous system, was used as the query protein. The extracellular domains and regions susceptible to recognition by antibodies were extracted from UniProt. In the study of T cells, linear sequence similarity between H. sapiens proteins and SP was assessed using BLASTp. This study considered the similarity in terms of biochemical groups per residue and affinity to the human major histocompatibility complex (human leukocyte antigen I), which were evaluated using Needle and NetMHCpan 4.1, respectively.
Results
A large number of shared pentapeptides between SP and H. sapiens proteins were identified. However, only a small group of 39 proteins was linked to axon and myelin homeostasis. In particular, some proteins, such as phosphacan, attractin, and teneurin-4, were susceptible targets of B and T cells. Other proteins closely related to myelin components in the NS, such as myelin-associated glycoprotein, were found to share at least one pentamer with SP in extracellular domains.
Conclusion
Proteins involved in the maintenance of nerve conduction in the central and peripheral NS were identified in H. sapiens. Based on these findings, re-evaluation of the vaccine composition is recommended to prevent possible neurological side effects.
1. Introduction
The coronaviruses (CoVs) that infect humans are RNA viruses that show neurotropic characteristics and can be linked to severe nervous system (NS) involvement [1]. The Beta variant of SARS-CoV-2 raises particular concern because, given its current global reach, it could be associated with increased neurological involvement in the future [2].
SARS-CoV-2 infection, which causes coronavirus disease 2019 (COVID-19), has been associated with several complications in the central nervous system (CNS), such as ischemic or hemorrhagic strokes, smell and taste disorders, meningoencephalitis, and encephalitis. Peripheral nervous system (PNS) complications are mainly associated with Guillain–Barré syndrome (GBS) and its variant, Miller-Fisher syndrome [3]. Although COVID-19 can cause NS complications in cases of systemic involvement, some co-occurrences, such as acute disseminated encephalomyelitis (ADEM), GBS, and anti-NMDAR encephalitis, are better explained as parainfectious/postinfectious immune-mediated events induced by COVID-19, which result in serious implications on the quality of life and increased mortality [4], [5].
Recently, population studies have reported that GBS and other autoimmune disorders of the NS are associated with COVID-19 vaccination [6]. In addition, cases around the world have reported an association of adverse neurological events with the use of vaccines that use the SARS-CoV-2 spike protein (SP) as the main antigen for the generation of immunological memory [6], [7], [8].
The neurological complications described in the acute stage of infection and after vaccination could be the result of molecular mimicry (MM) by SP and, essentially, by SARS-CoV-2 to unknown proteins that may be related to neuronal and/or glial involvement.
This work primarily attempts to analyze the plausibility of MM by SP with the aim of establishing safe limits to minimize NS involvement by avoiding the triggering of axonal damage and demyelination due to parainfectious/postinfectious immune-mediated events when using SP.
2. Methodology
A bioinformatics approach was used to determine the plausibility of MM occurring due to the use of SP. In this regard, we assessed the potential interaction of SP to stimulate target cells of the immune system, such as B and T cells, and its affinity to major histocompatibility complex (MHC), i.e., human leukocyte antigen (HLA), supertypes. In addition, we investigated whether the SPs of CoVs affecting humans share epitopes that may interact with proteins involved in the normal functioning of neurons and glia. Fig. 1 shows the analysis as a pipeline. The tools used to perform the assessments are explained below.
Fig. 1.
Pipeline.
2.1. Identification of protein–protein interactions and ontological annotation
The STRING V 11.5 database (https://string-db.org; August 2021) [9] was used to build a two-layer protein–protein (PP) interaction network of no more than 20 interactions with a (combined) confidence level of 0.4, with default parameters and Homo sapiens (H. sapiens) as the query species.
Peripheral myelin protein 22 (PMP22) was used as the main query protein because it is one of the main components of compact myelin in the PNS. Variants of this protein have been linked to demyelinating, motor, and sensory neuropathies and described as the inflammatory immune origin of GBS (OMIN: 601097).
In addition, the relevant biological process of axon myelination were investigated by identifying gene ontology (GO) terms obtained directly from STRING or UniProt [10]. The GO annotation represents the link between a gene product and the cellular process in which it participates. In particular, the GO of a biological process defines the different molecular functions involved in the occurrence of a biological objective [11].
In this study, the said objective was defined on the basis of biological processes related to myelin and axons (GoMYA). To present an evidence-based overview of these interactions, we used Adobe Illustrator and BioRender to illustrate the findings of the P—P network.
2.2. Similarity between SARS-CoV-2 SP fragments and H. sapiens proteins
A conservative approach was employed by comparing from five nonredundant residues (pentapeptides) in SP sequences with proteins in the PMP22 network and the proteome of nonredundant sequences of H. sapiens (TAX ID: 9606) using BLASTp (https://blast.ncbi.nlm.nih.gov/). The e-value was set to e-12, and sequences with 100 % identity and 0 gaps were filtered to obtain the minimum overlap length.
2.3. Localization of proteins exposed to the extracellular fluid
The TMHMM-2.0 software (https://services.healthtech.dtu.dk/service.php?TMHMM-2.0) was used to identify the subcellular localization and the extracellular fluid–accessible domains of proteins, which were identified using BLASTp, in the networks constructed using STRING. Proteins located in the cell membrane and, especially, the specific extracellular fluid–accessible amino acids were included to identify residue sequences similar to the SARS-CoV-2 SP (SLSP) (ID-P0DTC2) for investigating potential antibody cross-reactions. Proteins that were not found through the identification of topological domains were excluded from further analyses.
2.4. Identification of experimentally validated B cell epitopes
Pentapeptide sequences of GoMYA proteins present in extracellular fluid–accessible topological regions were used to search for sequences in the linear epitopes of B cells with experimental evidence of eliciting neutralizing antibodies (NAs) against SP. For this, we used an experimental epitope database—The Immune Epitope Database (https://www.iedb.org/)—with a “substring”.
2.5. Illustrations of the SP regions sharing sequences with H. Sapiens proteins
The resolved trimeric structure of SP (ID: 7LRT, https://www.rcsb.org) was used to show the localization of shared amino acid sequences in the linear epitopes with experimental evidence of eliciting NAs. UCSF Chimera 1.15 (https://www.cgl.ucsf.edu/index.html) was used to create the illustrations.
In case the region was not experimentally resolved, an approximation of its three-dimensional structure was generated with ColabFold using the default values. ColabFold is a Google Colaboratory tool that integrates AlphaFold2 and RoseTTAFold combined with an optimized algorithm for multiple alignment, which saves computational time [12].
2.6. Alignment of SP protein orthologs from other CoVs reported to exhibit neurotropism or neuroinvasion in H. sapiens
In addition to the recently emerged SARS-CoV-2, alphacoronavirus 229E, betacoronaviruses OC-43, Middle East respiratory syndrome–related coronavirus, and SARS-CoV have been reported to potentially affect the NS. Some proteins of these viruses are orthologs of SARS-CoV-2 SP. Considering that SP is the protein under study and efforts are being made to produce a pan-vaccine,” it should be noted that SP may contain epitopes that mimic proteins related to myelination and axonal packing in H. sapiens. This may result in cross-reactivity, which should be avoided. Hence, we performed an alignment of the complete sequences of SPs of these coronaviruses in the FASTA format using the Clustal Omega program available in UniProt with default value [13]. The included proteins were as follows: P36334 (HCoV-OC43), P15423 (HCoV-229E), K9N5Q8 (MERS), P59594 (SARS), and P0DTC2 (SARS-CoV-2).
2.7. Search for potential T cell epitopes based on similarity to SP
H. sapiens proteins sharing at least one pentapeptide with the SARS-CoV-2 SP and GO terms previously retrieved from UniProt were transformed into nonredundant nonapeptides. BLASTp was used with the same settings as previously described. All possible nonredundant nonapeptides of SP were used as the base query. Fragments showing identity greater than 60 % were transformed into FASTA format and used as the input in a neural network–based algorithm to predict T cell epitopes showing strong binding capacity to HLA subtypes using NetMHCpan − 4.1 (https://services.healthtech.dtu.dk/service.php?NetMHCpan-4.1 [14]. As HLAs exhibit high polymorphism, the following HLA supertypes representatives were used: HLA-A01:01(A1), HLA-A02:01(A2), HLA-A03:01(A3), HLA-A24:02(A24), HLA-A26:01(A26), HLA-B07:02(B7), HLA-B08:01(B8), HLA-B27:05(B27), HLA-B39:01(B39), HLAB40:01(B44), HLA-B58:01(B58), and HLA-B15:01(B62). This approach enabled the distinction of T cell epitopes indiscriminately recognized by HLA from viral and self-antigens [15] Further, similarity was calculated by aligning sequence pairs using “Needle” (https://www.ebi.ac.uk/Tools/psa/emboss_needle/).
3. Results
3.1. PMP22 is involved in several P—P interactions occurring in different cellular compartments related to myelin and axons
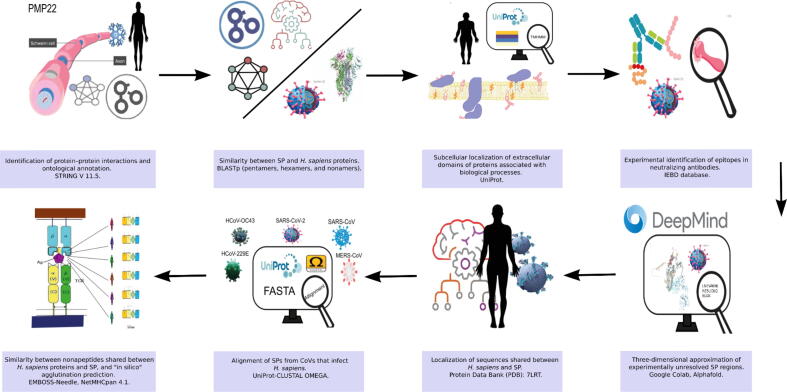
As an overview, we present the main findings of a two-layer P—P network that was constructed using PMP22 as the only query protein in STRING, revealing at least 40 expected interactions. This network showed at least four well-defined groups, indicating related biological processes. Group 4 was the most heterogeneous (Fig. 2A). Subsequently, GO related to GoMYA processes was extracted, avoiding other overlapping processes in the network (Fig. 2B).
Fig. 2.
Network of P—P interactions involving PMP22 established using STRING.
PMP22 is implicated in P—P interactions involved in myelin-related biological processes. It interacts with proteins that enhance myelin formation (GO: 0031643), promote its maintenance in the CNS and PNS (GO: 0042552), and modulate the frequency, rate, or extent of sheath formation around axons (GO: 0031641), thereby promoting axon insulation (GO: 0008366) as well as myelin formation and regeneration (GO: 0061564). Moreover, these processes can be performed in different cellular compartments (Fig. 2C).
When the proteins in the different groups were filtered using the GoMYA terms, the highest number of related P—P interactions was identified in Group 4 (light green) (Fig. 2B and C). Although this group showed the greatest heterogeneity, it contained proteins with more than one GoMYA, including myelin-associated glycoprotein (MAG) and transcription factors such as the transcription factor SOX-10 (SOX10) and the E3 SUMO-protein ligase EGR2 (EGR2), highlighting its biological importance.
Some proteins, such as integrin beta-1 (ITGB1), catenin beta-1 (CTNNB1), and reticulon-4 (RTN4) receptor, involved in GoMYA were not directly linked to PMP22. These were part of the intragroup nodes, indicating common biological processes that may overlap with PMP22. ITGB1 was a part of Group 1 together with laminin subunit gamma-2 (LAMC2) and laminin subunit alpha-1 (LAMA1) within GO: 0061564. CTNNB1 was a part of Group 3, which contained regulatory factors, mostly representing GO: 0031641, establishing P—P interactions with SOX-10 in Group 4 and ITGB1 in Group 1. Although RTN4 was a part of Group 4, like ITGB1, it was related to GO: 0061564 and formed P—P interactions with MAG and myelin basic protein (MBP), which are important myelin components.
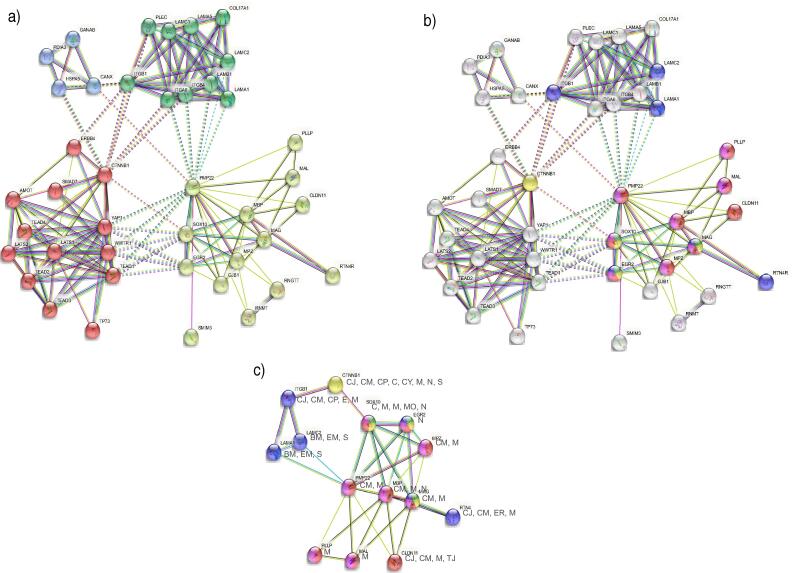
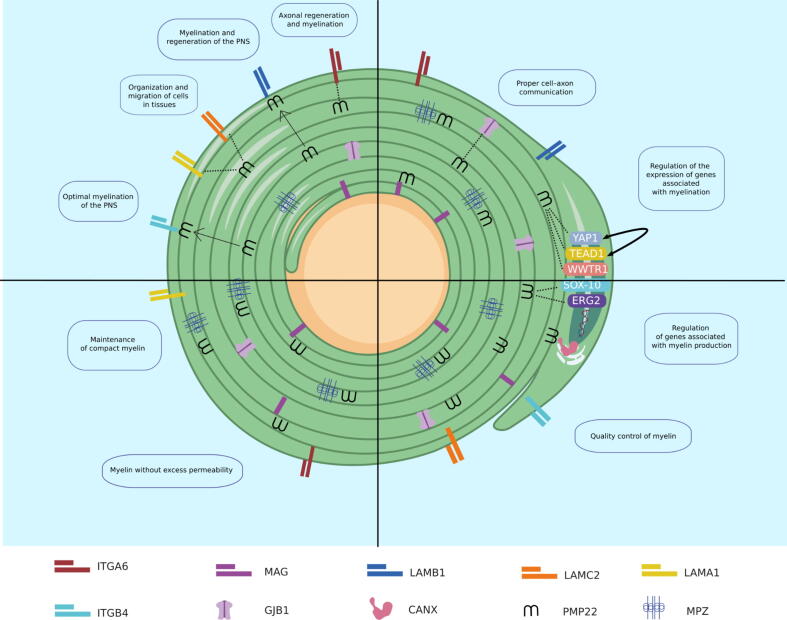
In the network that included only proteins involved in GoMYA (Fig. 2C), a majority of interconnected nodes revealed particular interaction with regulatory proteins (SOX10 and EGR2) and major components of myelin (PMP22, MBP, MAG, and myelin protein P0 [MPZ]). Fig. 3 shows the related biological processes based on the evidence supporting the P—P interactions directly associated with PMP22.
Fig. 3.
Biological processes related to PMP22 in the PNS.
The P—P interactions involving PMP22 are presented with a confidence level of 0.4, with two layers and no more than 20 interactions.
-
a)
Lines connecting proteins indicate functional and physical or computer-predicted interactions from curated databases (turquoise), experimentally determined interactions (purple), as well as interactions predicted based on gene proximity (green), fusion (red), and co-occurrence (blue). Interactions based on text mining (yellow), co-expression (black), and homology at the protein level (purple) are also presented. The groups are depicted by colors within the circles: Group 1 (green), Group 2 (blue), Group 3 (red), and Group 4 (lime).
-
b)
The proteins determined by GoMYA and their corresponding definitions are as follows: Green: Positive regulation of myelination (GO: 0031643), “any process that activates or increases the frequency, rate, or extent of the formation of myelin sheath around nerve axons.” Yellow: Regulation of myelination (GO: 0031641), “any process that modulates the frequency, rate, or extent of the formation of myelin sheath around nerve axons.” Red: Axon ensheathment (GO: 0008366), “any process in which the axon of a neuron is insulated, and that insulation maintained, thereby preventing the dispersion of the electrical signal.” Fuchsia: Myelination (GO: 0042552), “the process in which myelin sheaths are formed and maintained around neurons. Oligodendrocytes in the brain and spinal cord and Schwann cells in the PNS wrap axons with compact layers of their plasma membrane. Adjacent myelin segments are separated by a nonmyelinated stretch of axon called a node of Ranvier.” Purple: Axon development (GO: 0061564), “the progression of an axon over time. It includes axonogenesis (de novo generation of an axon) and axon regeneration (regrowth) as well as processes pertaining to the progression of the axon over time (fasciculation and defasciculation).” Proteins related to other biological processes are shown in gray.
-
c)
Cell topology of the group of proteins that directly or indirectly interact with PMP22 and are involved in GoMYA. Cell junction: CJ, Cell membrane: CM, Cell projection: CP, Endosome: E, Membrane: M, Basement membrane: BM, Extracellular matrix: EM, Secreted: S, Cytoplasm: C, Cytoskeleton: CY, Nucleus: N, Synapse: S, Mitochondrion: M, Mitochondrion outer membrane: MO, Endoplasmic reticulum: ER, Tight junction: TJ.
The figure shows a Schwann cell and all the interactions between PMP22 and different proteins related to GoMYA. The upper left quadrant contains membrane proteins, more specifically a group of laminins (LAMB1, LAMC2, and LAMA1) and a group of integrins (ITGA6 and ITGB4) that interact with PMP22. The dotted lines represent interactions with factors without binding, and the solid lines represent binding to form a heterocomplex. The upper right quadrant shows several transcription factors (YAP1, TEAD1, WWTR1, SOX-10, and ERG2) related to PMP22 and GoMYA processes as well as a transmembrane protein (GJB1) that, upon interaction with PMP22, is responsible for proper communication between the cell and axon. The lower left quadrant shows the primary myelin components that interact with PMP22 (MAG and MPZ) and are responsible for the maintenance of compact myelin, thereby preventing excessive permeability. In the lower right quadrant, in addition to the abovementioned transcription factors, an endoplasmic reticulum protein (CANX) is presented, which forms a heterocomplex with PMP22 and is related to the control of myelin quality.
3.2. At least a quarter of H. sapiens proteins share pentapeptides with SP, but only some are found in extracellular regions and are related to myelin and axons
Due to the limitations of the established PMP22 network (40proteins) in comparison with H. sapiens proteins (>20,000 proteins), a scheme was proposed to provide a complete picture that included the CNS and PNS, with particular focus on the biological processes involved in the PMP22 network. Herein, GoMYA was used as a filter (Fig. 4).
Fig. 4.
Nonredundant proteins of H. sapiens in extracellular regions sharing linear sequences with SP.
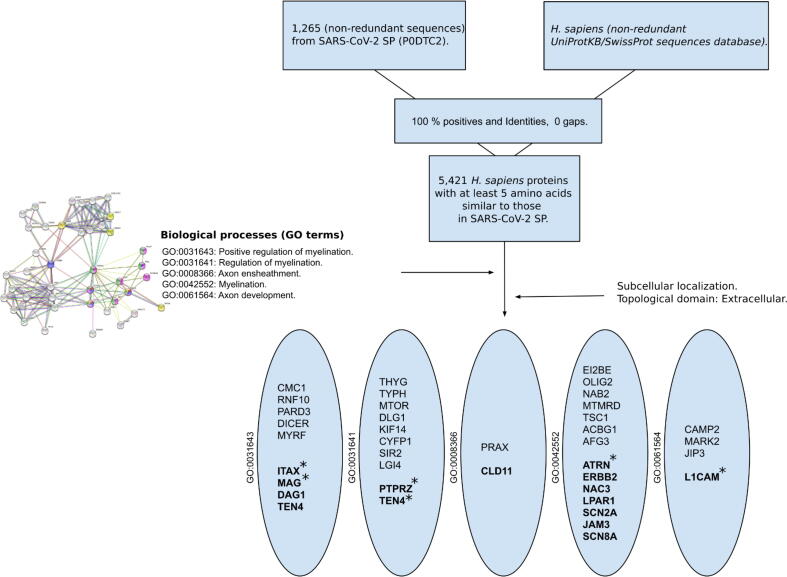
Overall, 5,421 H. sapiens proteins were found to share at least one pentapeptide with SP. After filtering by GoMYA previously obtained in the PMP22 network in UniProt and the localization of extracellular fluid–accessible sequences shared with SP (SCSP), we found 14 proteins with up to a maximum of 6 similar residues (hexapeptides). These hexapeptides were specifically found in a neural cell adhesion molecule (L1CAM) and receptor-type tyrosine-protein phosphatase zeta (PTPRZ1/phosphacan). When this same approach was used for the PMP22 network, only ITGB1 and MAG showed SCPS in extracellular domains. MAG was the only protein in common in the two independent searches. A total of eight H. sapiens proteins involved in GoMYA processes were found to share extracellular regions with at least one pentapeptide in common with SP (Table 1).
Table 1.
Similar peptides between SARS-CoV-2 SP and H. sapiens proteins with extracellular domains.
| UniProt ID | Protein-Gene | Position | Range of extracellular amino acids | Sequences similar to SARS-CoV-2 SP (pentapeptides and hexapeptides) |
Localization in SP subunit |
Pentapeptides/hexapeptides in other CoVs | B cell neutralizing antibody epitopes (Fig. 9) | Experimental evidence, ID in IEBD |
|---|---|---|---|---|---|---|---|---|
| P32004 (*) | Neural cell adhesion molecule L1- L1CAM |
1 | 20–1120 | DLQELG | S2 | (SARS-CoV) | LNEVAKNLNESLIDLQELGK | 1,309,518 (in vivo admin) |
| P23471 (*) | Receptor-type tyrosine-protein phosphatase zeta-PTPRZ1 | 2 | 25–1636 | SASFST/Iso3 | S1 and RBD | |||
| 3 | 25–1636 | AVLYQ | S1 | (SARS-CoV) | ||||
| 4 | 25–1636 | SQILP | S2 | (SARS-CoV) | PIKDFGGFNFSQILPDPSKP | 1334475;133 4476 | ||
| 5 | 25–1636 | SASFS/iso3 | S1 and RBD | |||||
| 6 | 25–1636 | ASFST/iso3 | S1 and RBD | |||||
| Q6N022 (*) | Teneurin-4-TENM4 | 7 | 367–2769 | GTTLD | S1 | |||
| 8 | 367–2769 | LLAGT | S2 | |||||
| 9 | 367–2769 | QIITT | S2 | (SARS-CoV) | QRNFYEPQIITTDNT | 1,310,750 | ||
| 10 | 367–2769 | VAIHA | S1 | |||||
| 11 | 367–2769 | RLFRK | S1 and RBD | NLDSKVGGNYNYLYRLFRK SN |
1,330,538 | |||
| P04626 (*) | Receptor tyrosine-protein kinase erbB-2- ERBB2 |
12 | 23–652 | PLQPE/iso2,3 | S2 | (SARS-CoV) | ||
| P20702 (*) | Integrin alpha-X-ITGAX | 13 | 20–1107 | DVVIG | S2 | (SARS-CoV) | ||
| O75882 (*) | Attractin-ATRN | 14 | 84–1279 | FGGFN | S2 | (SARS-CoV) | PIKDFGGFNFSQILPDPSKP | 1334475; 1,334,476 |
| P05556 (**) | Integrin beta-1-ITGB1 | 15 | 21–728 | GEVFN | S1 and RBD | (SARS-CoV) | ||
| P20916 (**) | Myelin-associated glycoprotein - MAG |
16 | 20–516 | PPLLT | S2 | (SARS-CoV) | ||
| *Global search result for H. Sapiens ** Network results obtained for PMP22 in STRING /pentapeptide or hexapeptide that is not present in any isoform | ||||||||
Proteins that share pentapeptide sequences with SP and are located in extracellular regions are shown in bold. * indicates proteins that share extracellular residues with SP. The distribution of proteins based on GO terms is shown in the ovals. Teneurin-4 (entry name in UNIPROT as “TEN4”) was the only protein for which two GO terms were obtained in UniProt: “positive regulation of myelination” and “regulation of myelination.”
3.3. Several proteins with SCSP in extracellular domains interact with major myelin components
PMP22, MAG and ITGB1 were found to be associated with GoMYA and share extracellular fluid–accessible sequences with SP (Fig. 2C). Using a more general approach that considered the H. sapiens proteome, at least six other proteins with similar characteristics were identified.
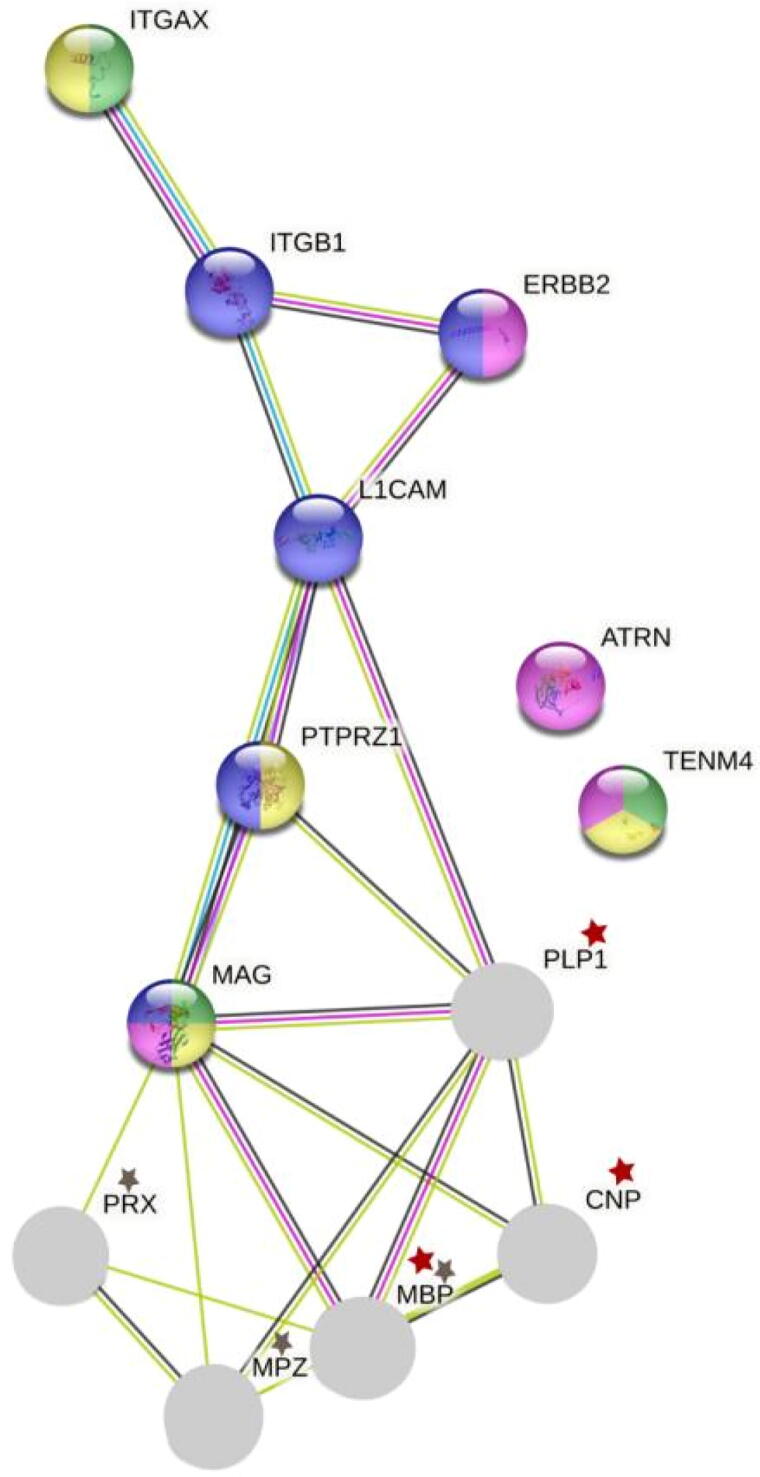
A single-layer network was constructed in STRING to determine potential interactions with key myelin proteins in the CNS and PNS. The CNS proteins included myelin proteolipid protein (PLP1), MBP, and 2′, 3′-cyclic nucleotide 3′-phosphodiesterase (CNP), corresponding up to 72.62 % [16] the PNS proteins included MPZ, MBP, and periaxin (PRX), corresponding up to 78.63 % [17] (Fig. 5). This network shows direct P—P interactions between most H. sapiens proteins bearing extracellular domains with SCSP and the major myelin components in the NS, only with the exception of TEMN4 and attractin (ATRN). In the CNS, PLP1 was found to interact with PTRZ1, L1CAM, and MAG; the latter was also associated with CNP. PRX and MPZ showed direct P—P interaction only with MAG, which was directly related to the major myelin components of both CNS and PNS. An interaction with PTRZ1 and L1CAM was observed, similar to that observed for PLP1. Other distant P—P interactions were observed among L1CAM, ErbB-2 receptor tyrosine-protein kinase (ERBB2), and ITGB1.The latter also interacted with integrin alpha-X (ITGAX).
Fig. 5.
Single-layer P—P network of proteins with GoMYA containing SCSP in the extracellular domains and major myelin components in the CNS and PNS.
Overall, these results revealed P—P interactions between myelin components of the NS and proteins with SCSP susceptible to recognition by antibodies against SP, particularly MAG, PTRZ1, and L1CAM. L1CAM, in addition to interacting directly with MAG, shares related biological processes such as axonal regeneration in both CNS and PNS, whereas other proteins with SCSP, such as ITGAX, ITGB1, and PTRZ1, are involved in brain development (Fig. 5, Fig. 6, Fig. 7).
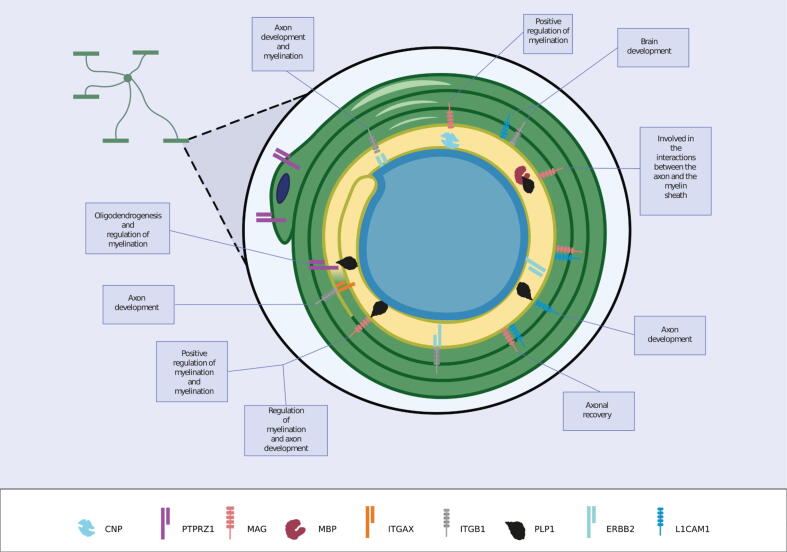
Fig. 6.
Interactions between proteins with SCSP and GoMYA terms in an oligodendrocyte.
Fig. 7.
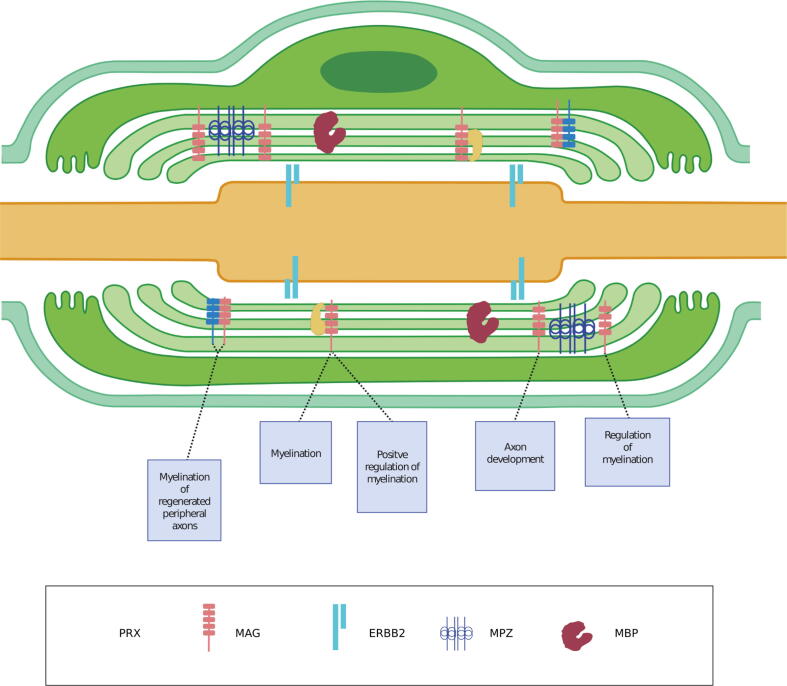
Interactions between proteins with SCSP and GoMYA terms in a Schwann cell.
The protein components of myelin in the gray circles are depicted by a red (CNS) or gray (PNS) star [18]. From the network obtained, GOs were most frequently linked to processes of myelin maintenance and formation as well as axonal morphogenesis and regeneration. Green: Positive regulation of myelination (GO: 0031643), Yellow: Regulation of myelination (GO: 0031641). Fuchsia: Myelination (GO: 0042552), and Purple: Axon development (GO: 0061564).
The image shows the interactions between proteins with SCSP and GoMYA terms in an oligodendrocyte. MAG (protein with SCSP) forms a heterocomplex with PLP1 and MBP (primary myelin components), which results in interactions between the axon and myelin sheath. The interaction between MAG and CNP leads to the positive regulation of myelination. The interaction between MAG and L1CAM facilitates axonal regeneration. The interaction between L1CAM and ITGB1 is implicated in brain development, whereas that of L1CAM with ITGB1 and ERBB2 is important for increasing sensitization to neuroregulins. The interaction between ITGB1 and ITGAX facilitates axonal development, whereas that between PTPRZ1 and PLP1 is implicated in oligodendrogenesis and myelin regulation.
The image shows the interactions between proteins with SCSP and GoMYA terms in a Schwann cell. MAG (protein with SCSP) interacts with primary myelin components, such as PRX and MPZ, thereby leading to four biological processes—myelination, positive regulation of myelination, axon development, and regulation of myelination. The interaction between MAG and L1CAM (protein with SCSP) is implicated in the myelination of regenerated peripheral axons.
3.4. SP epitopes related to the production of NAs contain sequences similar to those in H. sapiens proteins with GoMYA
The aim of a vaccine is to elicit a specific immune response, in part by producing NAs. The use of SP could cause secondary cross-reactions because it contains pentapeptides/hexapeptides similar to those in the extracellular regions of H. sapiens proteins.
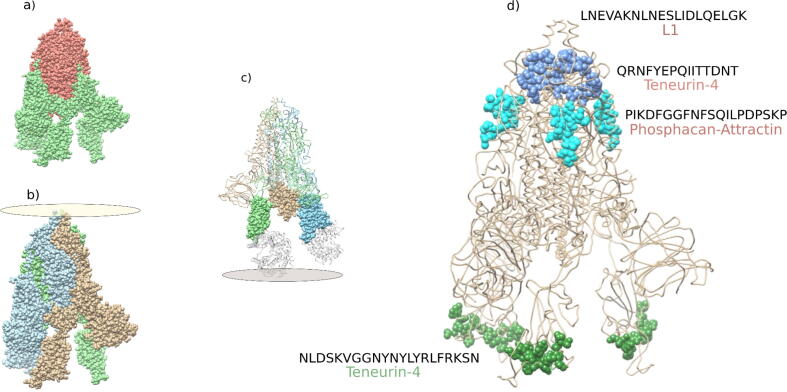
From the experimental information available in the IEBD, we explored the linear epitopes of SP that shared pentapeptides/hexapeptides with H. sapiens proteins and were experimentally demonstrated to elicit NAs (Table 1). In general, L1CAM, PTPRZ1, TENM4, and ATRN were susceptible to NAs against SP. Most epitopes were identified in subunit 2 of SP (SP2) (Fig. 8, Fig. 9). The only linear epitope in subunit 1 of SP (S1) with the ability to elicit NAs was “NLDSKVGGNYNYLYRLFRKSN,” which is present in the extracellular domain of TENM4 and the receptor-binding domain (RBD) of SP that binds with angiotensin-converting enzyme 2 (ACE2) (Fig. 8, Fig. 9).
Fig. 8.
Localization of pentapeptides/hexapeptide in SARS-CoV-2 SP that are similar to extracellular fluid–accessible H. sapiens proteins.
Fig. 9.
Localization of pentapeptides/hexapeptide shared by SP and GoMYA proteins in linear epitopes with experimental evidence of eliciting NAs.
Interestingly, the linear epitope “PIKDFGGFNFSQILPDPSKP” of SP contains pentapeptides that are present in the extracellular domains of PTPRZ1 and ATRN, demonstrating a high possibility that a single NA could affect more than one protein (Fig. 8, Fig. 9).
For some pentapeptides/hexapeptides, no experimental evidence was found in the IEBD regarding their presence in the linear epitopes of SP associated with NAs. However, the possibility that they are part of a longer epitope should not be rejected. TENM4, PTPRZ1, and ITGB1 showed the highest number of SCSPs, present in both S1 and S2 of SP as well as RBD. As the current primary approach for NAs aims to avoid the interaction between RBD and ACE2r; these proteins could be candidates for analyzing whether they are affected not only by vaccination but also during the acute phase of natural infection.
The figure shows the three-dimensional structure of SP (PDB ID: 7LRT). The extracellular sequences of H. sapiens proteins that overlap with GoMYA proteins are presented in different colors. * represents localization in the RBD. SCSP in L1CAM was not found in the experimental model; therefore, it was resolved using Colab Fold. Structural alignment was performed in Chimera. The monomer extracted from PDB: 7LRT is shown in cyan; the structure resolved using Colab Fold is shown in tan.
In Fig. 9, (a) shows subunits S1 (green) and S2 (red) of SP. (b) shows a homotrimer formed by SP; each monomer is shown in a different color and the oval represents its position with respect to the virion. (c) Shows each homotrimer, which presents a domain in S1 that interacts with ACE2r (gray) at the distal end with respect to the virion. The gray oval represents the host cell membrane. (d) Shows linear epitopes in SP with experimental evidence of eliciting NAs (cyan, green, and blue) and SCPS of GoMYA-related H. sapiens proteins exposed in extracellular domains.
3.5. Pentapeptides/hexapeptides present in NS-related proteins of H. Sapiens that are also present in the SPs of SARS-CoV and SARS-CoV-2
The presence of pentapeptides/hexapeptide common between SP and GoMYA-related H. sapiens proteins was assessed through alignment with other CoVs that affect H. sapiens and contain SP orthologs (Supplement Table 1). More than half of the pentapeptides/hexapeptide were found to be shared between the extracellular domains of H. sapiens GoMYA proteins and SP, particularly from SARS-CoV and SARS-CoV-2. In conjunction with the experimental evidence retrieved from the IEBD, it could be argued that there are potential immune regions shared between SARS-CoV SP and SARS-CoV-2 SP that contain pentapeptides/hexapeptide similar to those in H. sapiens NS proteins and could be associated with the neurological pathophysiology of CoV infection. These could preferably be prevented or optimized in the development of a pan-vaccine.
3.6. Cross-reactivity of T cells elicited by SP could increase susceptibility of myelin- and axon-related proteins
Immune cross-reactions have been associated with viral infections, in part due to the similarity of self-epitopes, survival of T cells weakly reactive to self-epitopes, nonrigid TCR–peptide–HLA interactions, and HLA-I susceptibility, which may be linked to demyelinating entities or axonal damage [19], [20], [21], [22]. In this research, two approaches were used to evaluate possible cross-reactivity with GoMYA proteins having at least one pentapeptide in common with SP (Fig. 4). We used the minimum number of residues required to elicit a cellular or humoral immune response (including non-extracellular domains) as a filter [23] as well as the similarity of side chain residues present in SP nonapeptides/octapeptides and H. sapiens GoMYA proteins [23], [24] supported by an in silico prediction of binding with HLA supertypes (Table 1). The presence of nonapeptides/octapeptides common between SP and GoMYA-related H. sapiens proteins was assessed through alignment with other CoVs that affect H. sapiens and contain SP orthologs (Supplement Table 1). Using these approaches, we identified 58 nonredundant pentapeptides and 3 nonredundant hexapeptides in common with SP, which were present in 39 GoMYA proteins, in addition to 13 nonapeptides/octapeptides contained in GoMYA proteins biochemically similar among some nonapeptides/octapeptides also conserved in SARS-CoV (Supplement Table 1).
The similarity analysis of the side chain residues of nonapeptides in H. sapiens GoMYA proteins and SP yielded 35 nonredundant nonapeptides with up to 100 % similarity, which were present in 16 proteins. Some of these also showed binding with HLA supertypes (Table 2). The SP nonapeptide “VLKGVKLHY” with 88.9 % similarity to “VLKGSKLHF” in the kinesin-like protein (KIF14-Q15058) was an experimentally validated epitope [25] activated in 25 % of the assays performed with HLA-A*29:02-restricted CD8+ T cells recovered from the serum of convalescent patients with COVID-19.
Table 2.
Similarity between the nonapeptides of SP and GoMYA proteins and predictions of binding with HLA supertypes.
| Protein-UniProt ID | Position | SP nonapeptide | H. sapiens nonapeptide | Localization in SP subunit | Similarity per side chain | Percent identity (Needle) | Agglutination - SP nonapeptide |
Agglutination - H. sapiens nonapeptide |
|---|---|---|---|---|---|---|---|---|
| Attractin-O75882 | 1 | DFCGKGYH | NICGIGWH | S2 | :.||.|:| | 75.0 % | ||
| 2 | FCGKGYHLM | ICGIGWHLV | S2 | .||.|:||: | 77.8 % | |||
| 3 | CGKGYHLM | CGIGWHLV | S2 | ||.|:||: | 87.5 % | |||
| Thymidine phosphorylase-P19971 | 4 | CVLGQSKRV | CIVGQSEQL | S2 | |::|||::: | 100.0 % | ||
| Receptor-type tyrosine-protein phosphatase zeta-P23471 | 5 | SNFRVQPT | NNFSVQPT | S1 | :||.|||| | 87.5 % | ||
| 6 | SNFRVQPTE | NNFSVQPTH | S1 | :||.||||. | 77.8 % | |||
| 7 | NFRVQPTES | NFSVQPTHT | S1 | ||.||||.: | 77.8 % | |||
| Disks large homolog 1-Q12959 | 8 | ESLIDLQEL | QALIDIQEF | S2 | ::|||:||. | 88.9 % | HLA-B*58:01, HLA-A*26:01 | |
| Kinesin-like protein KIF14-Q15058 | 9 | KRISNCVAD | RRISGCLHD | S1 and RBD | :|||.|:.| | 77.8 % | ||
| 10 | PVLKGVKLH | AVLKGSKLH | S2 | .||||.||| | 77.8 % | HLA-A*03:01 | ||
| 11 | VLKGVKLHY | VLKGSKLHF | S2 | ||||.|||: | 88.9 % | HLA-A*03:01, HLA-B*15:01, HLA-A*26:01 | HLA-B*15:01, HLA-B*08:01, HLA-A*24:02 | |
| 12 | LKGVKLHY | LKGSKLHF | S2 | |||.|||: | 87.5 % | |||
| Teneurin-4-Q6N022 | 13 | FGTTLDSK | YGTTLDEE | S1 | :|||||.: | 87.5 % | ||
| Serine/threonine-protein kinase MARK2-Q7KZI7 | 14 | EKSNIIRG | SKSNMIRG | S1 | .|||:||| | 87.5 % | ||
| 15 | TEKSNIIRG | SSKSNMIRG | S1 | :.|||:||| | 88.9 % | |||
| 16 | STECSNLL | STDCENLL | S2 | ||:|.||| | 87.5 % | |||
| Cytoplasmic FMR1-interacting protein 1-Q7L576 | 17 | HRSYLTPGD | NRMYLTPSE | S1 | :|.||||.: | 77.8 % | ||
| 18 | RSYLTPGD | RMYLTPSE | S1 | |.||||.: | 75.0 % | |||
| 19 | EELDKYFKN | SKIDKYFKQ | S2 | .::|||||. | 77.8 % | |||
| 20 | ELDKYFKN | KIDKYFKQ | S2 | ::|||||. | 87.5 % | |||
| Myotubularin-related protein | 21 | YQDVNCTE | YDDVSCTQ | S1 | |.||:||: | 87.5 % | ||
| 13-Q86WG5 | 22 | VLYNSASF | VLFHSASF | S1 and RBD | ||::|||| | 100.0 % | HLA-B*15:01 | HLA-B*15:01 |
| Leucine-rich repeat LGI family member 4-Q8N135 | 23 | TQTNSPRR | TQTLAPRR | S1 | |||.:||| | 87.5 % | ||
| RING finger protein 10- Q8N5U6 |
24 | PAYTNSFTR | PSFQNSFSQ | S1 | |::.|||:: | 88.9 % | ||
| Partitioning defective 3 homolog-Q8TEW0 | 25 | DGYFKIYS | DGHMKVFS | S1 | ||:.|::| | 87.5 % | ||
| 26 | VLPPLLTD | VLPPHLSD | S2 | ||||.|:| | 87.5 % | |||
| 27 | ITDAVDCAL | ISDSADCSL | S1 | |:|:.||:| | 88.9 % | HLA-A*01:01 | HLA-A*01:01 | |
| 28 | DAVDCALDP | DSADCSLSP | S1 | |:.||:|.| | 77.8 % | |||
| Sodium channel protein type 2 subunit alpha-Q99250 | 29 | FGAISSVLN | LGAIPSIMN | S2 | .|||.|::| | 77.8 % | ||
| Junctional adhesion molecule C-Q9BX67 | 30 | GCLIGAEH | GCLIGAVN | S1 | ||||||.: | 87.5 % | ||
| 31 | GCLIGAEHV | GCLIGAVNL | S1 | ||||||.:: | 88.9 % | |||
| Sodium channel protein type 8 subunit alpha-Q9UQD0 | 32 | DLPQGFSAL | DIPQGLVAV | S1 | |:|||..|: | 77.8 % | HLA-B*39:01, HLA-A*26:01, HLA-B*08:01, HLA-B*39:01 | HLA-A*26:01 |
| 33 | PDKVFRSS | PDKVFRSS | S1 | |:|||:|. | 87.5 % | |||
| 34 | EHVNNSYEC | EDVNNKTEC | S1 | |.|||..|| | 66.7 % | |||
| Myelin regulatory factor- Q9Y2G1 |
35 | NDLCFTNV | SDLCFPDI | S1 | :||||.:: | 87.5 % |
SP nonapeptide: Nonredundant nonapeptides in SP that are similar to nonapeptides in H. sapiens GoMYA proteins. H. sapiens nonapeptide: Nonredundant nonapeptides in GoMYA proteins that are similar to nonapeptides in SP. Localization in SP subunit: Localization of the nonapeptide in the SP subunit. Similarity per side chain based on the alignment of nonapeptides in SP and H. sapiens proteins: (:) Similarity by biochemical group,(|) identical residue and (.) different residue. Percent identity (Needle): Identity (expressed as a percentage) obtained from the alignment in Needle. Agglutination - SP and H. sapiens nonapeptides: In silico prediction of the agglutination of nonapeptides in SP or H. sapiens proteins; weak agglutinations are presented in italics.
In general, the shared proteins found in B and T cells using the above mentioned approaches suggest that ATRN, PTPRZ1, and TEN4 are potentially affected, the latter two being mainly present in the CNS.
UniProt ID: Protein identifier in the UniProt database. Protein-Gene: Protein name and acronym used in the study. Range of extracellular amino acids: Range of residues in proteins (presented in the first column) exposed to the extracellular fluid. Sequences similar to SARS-CoV-2 SP: Linear sequences of pentapeptides and hexapeptides that are shared between H. sapiens proteins and SARS-CoV-2 SP. Localization in SP subunit: Localization of the pentapeptide/hexapeptide shared with H. sapiens in the SP subunit; RBD indicates that it is located in the binding domain that interacts with ACE2. Pentapeptides/hexapeptides in other CoVs: Pentapeptides/hexapeptides identified primarily in SARS-CoV-2 SP that are also found in other CoVs. B cell neutralizing antibody epitopes: Epitopes with the ability to elicit NAs against SP. Experimental evidence, ID in IEBD: IEBD identifier of the experimentally validated linear epitopes with experimental evidence of eliciting NAs (available as of September 1, 2021).
4. Discussion
A bioinformatics approach was used to study the plausibility of cross-reactions due to MM by SP with respect to GoMYA proteins. This is especially useful considering the complexity of studying potential myelin and axonal self-antigens using experimental methods [26].
We used a group of proteins in a network of P—P interactions with PMP22, grouping and identifying related proteins localized in different cellular compartments that act as scaffolds in a coordinated way in the cell membrane [27]. Biological processes related to myelin and axons were identified from this network. These included positive regulation of myelination (GO: 0031643), regulation of myelination (GO: 0031641), axon ensheathment (GO: 0008366), myelination (GO: 0042552), and axon development (GO: 0061564).
Using a proteomics approach based on shared GoMYA processes in H. sapiens, a group of proteins involved in the formation of diverse complex membrane structures that facilitate nerve impulse conduction and enable bidirectional communication between the surrounding cells and the extracellular medium were identified [27], [28]. Most of these proteins were localized in the cytoplasmic projections of Schwann cells and oligodendrocytes, which interact with axons in the CNS and PNS, respectively. In addition, some of the proteins were localized in the nodes of Ranvier and some shared related biological processes. If affected, these proteins could lead to serious disorders in both the CNS and PN [26].
In summary, we studied the plausibility that GoMYA proteins with SLSP are affected by cross-reactions due to MM and consequently, they act as triggers for severe NS disorders as part of immune events mediated by T and B cells. On the part of B cell. Regarding the maximum length in which they share SCSP in the GoMYA proteins characterized, it was possible to identify hexapeptides accessible to the cell surface and which could be part of linear B cells epitopes. No octapeptides or nonapeptides were found, being the last, the length of amino acids used as the exploration window.
On the other hand, in the study of T cells, we used some bioinformatic tools to analyze possible reactions due to MM by using a non-stochastic approach to contemplate for this purpose biochemically similar octapeptides and nonapeptides between SP in GoMYA proteins [24]. The length of these peptides are appropriate for complementary experimental validation in vivo, and having greater precision in predicting peptide binding in the HLA-1 anchor as well [29]. Nonapeptides, especially, are most often complexed with HLA-1 in nature. Biochemically, similar nonapeptides plausibly in synergy with an adjuvant or proinflammatory state could be recognized by clonotypes of T-cells, in the generation of an acute immune response [30], [31].
Similarities of up to 100 % were observed with nonapeptides in H. sapiens proteins expressed in the NS and ubiquitously present in several tissues. These were predicted to be the binding sources of HLA supertypes molecules. We further explored whether this ability was present in other CoVs that affect H. sapiens and its potential implications for vaccine development.
In the proinflammatory phase of COVID-19, peptides derived from the immune-mediated degradation of SP may activate T cells against self-antigens, causing the clonal expansion of pathological T cells in H. sapiens as well as cross-reactivity of antibodies produced by B cells, particularly in the acute phase [22], [32]. These cells could be activated by a lower load of self-antigens, eventually leading to the onset of autoimmune diseases [33].
Possible links between viral infections and autoimmune CNS diseases have been previously described in H. sapiens [34], [35]. These are supported by molecular structural approaches that have identified self-reactive T cells towards human myelin proteins and recognize viral immunodominant peptides. T cell cross-reactivity to myelin components (MBP and PLP1) has been observed in patients with previous CoV infection.
Of the epitopes found and described in Table 2, only the SP nonapeptide “VLKGVKLHY” with 88.9 % similarity to “VLKGSKLHF” in kinesin-like protein (KIF14-Q15058) was an experimentally validated epitope [25]. It was activated in 25 % of the assays performed with HLA-A*29:02-restricted CD8+ T cells recovered from the serum of convalescent patients with COVID-19. Moreover, in accordance with the in silico analyses, this peptide exhibited in vitro binding to HLA-B*15:01 of peripheral blood mononuclear cells of individuals without any evidence of prior SARS-CoV-2 infection, suggesting heterologous immunity [36]. In this study, the nonapeptide was validated as a potential epitope of SP that could elicit the cross-reactivity of T cells to KIF14. This reaction as well as reactions for the other 34 nonapeptides present in 16 proteins with similar characteristics should be further validated to identify their pathogenic potential.
HLA has been described as a risk factor for autoimmune diseases of the NS [26], [37]. In particular, HLA-B*15:01, HLA-A*01:01, and HLA-A*26:01 (Table 1) were found to agglutinate nonapeptides with high similarity to H. sapiens proteins and SP in silico. This allows the grouping of individuals more likely to present cross-reactivity due to the recognition of a greater number of diverse immunodominant peptides, which paradoxically has also been associated with a better clinical outcome in COVID-19 [36]. For example, asymptomatic individuals with COVID-19 are eight times more likely to carry the HLA-B*15:01 allele. Further, homozygous carriers are likely to be asymptomatic. In the case of synergy with the HLA-DRB1*04:01 allele, there is a greater possibility of asymptomatic or mild disease. However, the latter allele has been associated with increased susceptibility to multiple sclerosis in fine-mapping studies [38], [39], [40].
Due to the rapid involvement of antibodies and the effects not correlated with viral load, it has been suggested that cross-reactions may be part of the clinical manifestations of COVID-19. A few months after the COVID-19 outbreak, the plausible involvement of autoantibodies against pulmonary surfactant was reported based on SP pentapeptides similar to amino acid sequences in human surfactant proteins [32]. In addition, strong reactions of IgG and IgM antibodies directed against the S1 subunit that also recognized several tissue proteins, including MBP, were detected by ELISA in patients with COVID-19 [41]. Hamada et al. [42] indirectly identified the strong reaction of IgG antibodies directed against SP with neurofilament light polypeptide (NF-L), which is linked to variants of Charcot–Marie–Tooth disease [43].
In support of the theoretical possibility of cross-reactions resulting from the degradation of SARS-CoV-2 SP, before focusing on GoMYA proteins, we showed that at least a quarter of the H. sapiens proteome shares pentapeptides with SP (Fig. 4). This finding is in line with a previous report showing that unlike nonhuman primates or other species that do not develop severe COVID-19 [44], the H. sapiens proteome shares several hexapeptides and pentapeptides with SP [44].
Only eight GoMYA proteins with high differential expression in the brain were found to have SP-like pentapeptides in their extracellular domains. Some of these proteins have been proposed as potential antigens associated with acute and chronic polyneuropathies owing to their involvement in antibody-mediated electrophysiological blockade [45]. In particular, these include proteins located in the node of Ranvier, where disruption affects the axon and leads to motor and sensory disabilities [40], [46], [47].
An approach to study the potential effect of antibodies on these proteins is based on the manifestations of IgM monoclonal gammopathy (IgMMG), in which IgM antibodies are directed against the post-translational modification mediated by GlcUAT-P (B3GA1_HUMAN) (present in phosphacan, MAG, and L1) [48], [49], [50]. This study focused on one hexapeptide and four pentapeptides in phosphacan and one pentapeptide in MAG, which are similar to those in SP and not adjacent to this epitope. They are exposed in the antibody-accessible extracellular domains, yielding neoepitopes of [48], [51]. MAG is particularly important as it is associated with a large number of GoMYA biological processes involved in myelination and is also related to axons through key myelin components in the PNS and CNS.
Phosphacan, located primarily in the brain (cerebral cortex, cerebellum, hippocampus, and basal ganglia) [50], has a pentapeptide in an epitope linked to the production of NAs against S2 of SP. However, its role in IgMMG neuropathy remains unclear. anti-phosphacan antibodies, unlike anti-MAG antibodies, have been associated with poor prognosis and/or treatment resistance in IgMMG [46], which may be of special interest for further studies on the involvement of this protein in cross-reactions with SP. Most of the proteins with pentapeptides and hexapeptides in their extracellular domains showed P—P interactions with axonal or glial components, with the exception of TEMN4 and ATRN even though they were linked to biological processes involved in myelination. This could be indicative of distant processes with respect to the myelin components analyzed. TEMN4, a protein expressed predominantly in the brain (cerebral cortex, hippocampus, and basal ganglia) [50] facilitates myelination by promoting the maturation and adhesion of oligodendrocytes [52] to axons through homophilic and heterophilic interactions with other teneurins (teneurins 1–3) through its ectodomain [52]. It contains two pentapeptides located in two epitopes that bind to NAs directed against S2 and S1, specifically against the RBD of SP (Fig. 8, Fig. 9). Considering their high homology, it is likely that some pentapeptides are also present in other teneurins (teneurins 1–3), which should be taken into account in future studies [52].
ATRN is a glycoprotein that acts as a receptor, shows low abundance in the brain and medium abundance in multiple organs [50], and has been reported to be altered in hypomyelinating leukodystrophies in humans [53]. It contains one pentapeptide similar to that in S2 of SP, which together with the pentapeptide in phosphacan, is part of a linear epitope related to the production of NAs against SP. As it is a receptor, in addition to affecting adhesion processes, it can alter the signaling of glial products or neurons [54], these findings should be further studied.
On the basis of prior evidence and our findings on B and T cells, it can be argued that during the acute phase of COVID-19, SP, the protein with the highest level of translation in this phase and the main antigen used as a target for vaccination, might result in cross-reactivity against ubiquitous proteins and NS-specific proteins as part of the pathogenesis [5], [55], [56]. In addition, COVID-19 and other factors, including HLA and cytokines such as IL-6 and IL-17, could lead to increased effects on the blood–brain barrier, allowing the migration of T or B cells and viruses into the CNS, consequently causing localized clinical manifestations.
Other CoVs that affect humans have been linked to neurological manifestations due to brain tissue tropism [1]. However, the possibility of their involvement via MM and alteration of the NS have not been studied in depth. In this study, SARS-CoV SP was shown to contain more than half of the pentapeptides and hexapeptides identified.
The similarity among pentapeptides in GoMYA proteins, SARS-CoV-2 SP, and SARS-CoV SP may be associated with neurological conditions, including stroke, encephalopathy, autoimmune disorders such as GBS, meningoencephalitis, and seizure [4]. In addition, encephalopathy and immune-mediated disorders may contribute to autoimmune complications over time. It is interesting to explore the heterologous immunity in populations previously affected by SARS-CoV and now by SARS-CoV-2 as this could provide important insights on whether prior immunity to SP is beneficial or pathogenic [57].
Efforts are currently underway to develop a pan-vaccine against SARS-CoV, SARS-CoV-2, and other CoVs with pandemic potential. As evinced in this work, in addition to antigens conserved between subspecies, antigens with low similarity to H. sapiens proteins should be used. This poses additional challenges for vaccine development as researchers delve deeper into the response and characteristics of cellular immunity against SARS-CoV-2 [58].
With the emergence of SARS-CoV-2 variants capable of evading the NA response to SP as well as the recognition of cellular immunity as a protection mechanism against severe COVID-19 [59] an alternative approach using SARS-CoV-2 sequences with low similarity to NS proteins could be proposed. In this way, cellular immunity could be specifically stimulated with a lower antigen load.
5. Limitations
We evaluated the plausibility of MM by SP in SARS-CoV-2. Nevertheless, our theoretical and limited approach used only some proteins related to GoMYA processes, which were obtained from P—P interaction networks described for neurological conditions. We might have underestimated other proteins that may act directly and indirectly. In addition, our approach did not comprehensively study physicochemical properties such as hydrophobicity nor did it simulate the proposed mechanistic interactions. Moreover, we only evaluated linear sequences, with conformational sequences being the most frequent in nature, thereby disregarding a large number of possible interactions. We have likely underestimated the number of GoMYA proteins as well as conformational epitopes that may induce NA production during the acute phase of infection or after vaccination.
Although some proteins are associated with biological processes involved in the formation of normal neuronal tissue and myelin and regeneration of axons and may share hexapeptide and pentapeptides with SP, it does not necessarily mean that they cross-react with B or T cells. However, it suggests the potential involvement of the immune system in acute and chronic manifestations in the NS induced by COVID-19. Therefore, further experimental studies should be conducted to make better medium- and long-term decisions regarding the current pandemic.
Although we used in silico tools that yielded experimentally validated predictions, we only analyzed similarities with HLA supertypes class I and did not evaluate any theoretical responses by HLA class II. Synergy of the two could lead to interesting phenotypes. In addition, there may be class I alleles capable of recognizing peptides that do not belong to the supertypes used herein.
6. Conclusions
Current evidence on the impact of SARS-CoV-2 suggests that COVID-19 does not only cause acute conditions and supports that cross-reactivity is a part of the pathophysiology of COVID-19. Using this theoretical approach, we identified some myelin- and axon-related membrane proteins specifically involved in the biological processes of maintenance, regeneration, and axonogenesis. These proteins can be affected due to MM by SP, and their alteration can lead to severe neurological conditions.
Although this was a theoretical study, our results support the search for a safe vaccination approach, which could focus on the stimulation of long-lasting cellular immunity. A rapid experimental evaluation of MM should be performed and lower antigen loads should be used in vaccines. As in other viral diseases of global importance, vaccines should be adjusted periodically with special focus on specific protection against circulating strains. The methodology used in this study can be used to explore immune-mediated events driven by COVID-19. Finally, we support the search for safe vaccines and the avoidance of SP in the current mass vaccination campaigns because it causes long-term neurological effects.
CRediT authorship contribution statement
Andrés Felipe Cuspoca: Conceptualization, Data curation, Formal analysis, Methodology, Project administration, Writing – original draft, Writing – review & editing. Pablo Isaac Estrada: Data curation, Writing – review & editing. Alberto Velez-van-Meerbeke: Conceptualization, Methodology, Project administration, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.10.022.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Carod-Artal F.J. Neurological complications of coronavirus and COVID-19. Rev Neurol. 2020;70(9):311–322. doi: 10.33588/rn.7009.2020179. [DOI] [PubMed] [Google Scholar]
- 2.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., et al. Neurologic manifestations of hospitalized patients with Coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020;77(6):683–690. doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orrù G., Conversano C., Malloggi E., Francesconi F., Ciacchini R., Neurological G.A., et al. Neurological complications of COVID-19 and possible neuroinvasion pathways: a systematic review. Int J Environ Res Public Health. 2020;17(18) doi: 10.3390/ijerph17186688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almqvist J., Granberg T., Tzortzakakis A., Klironomos S., Kollia E., Öhberg C., et al. Neurological manifestations of coronavirus infections - a systematic review. Ann Clin Transl Neurol. 2020;7(10):2057–2071. doi: 10.1002/acn3.51166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vasilevska V., Guest P.C., Bernstein H.G., Schroeter M.L., Geis C., Steiner J. Molecular mimicry of NMDA receptors may contribute to neuropsychiatric symptoms in severe COVID-19 cases. J Neuroinflammation. 2021;18(1):245. doi: 10.1186/s12974-021-02293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patone M., Handunnetthi L., Saatci D., Pan J., Katikireddi S.V., Razvi S., et al. Neurological complications after first dose of COVID-19 vaccines and SARS-CoV-2 infection. Nat Med. 2021 Oct;25:1–10. doi: 10.1038/s41591-021-01556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao SJ, Khurana S, Murthy G, Dawson ET, Jazebi N, Haas CJ. A case of Guillain–Barre syndrome following Pfizer COVID-19 vaccine [Internet]. Vol. 11, Journal of Community Hospital Internal Medicine Perspectives. 2021. p. 597–600. Available from: 10.1080/20009666.2021.1954284. [DOI] [PMC free article] [PubMed]
- 8.McKean N., Chircop C. Guillain-Barré syndrome after COVID-19 vaccination. BMJ Case Reports CP. 2021;14(7):e244125. doi: 10.1136/bcr-2021-244125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman M.H., Peng S., Hu X., Chen C., Rahman M.R., Uddin S., et al. Network-based bioinformatics approach to identify molecular biomarkers for type 2 diabetes that are linked to the progression of neurological diseases. Int J Environ Res Public Health. 2020;17(3) doi: 10.3390/ijerph17031035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill D.P., Smith B., McAndrews-Hill M.S., Blake J.A. Gene Ontology annotations: what they mean and where they come from. BMC Bioinf. 2008 Apr 29;9(5):1–9. doi: 10.1186/1471-2105-9-S5-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mirdita M, Ovchinnikov S, Steinegger M. ColabFold - Making protein folding accessible to all [Internet]. Available from: 10.1101/2021.08.15.456425. [DOI] [PMC free article] [PubMed]
- 13.Söding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21(7):951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 14.Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48(W1):W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9(1):1–15. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siems S.B., Jahn O., Eichel M.A., Kannaiyan N., Wu L.M.N., Sherman D.L., et al. Proteome profile of peripheral myelin in healthy mice and in a neuropathy model. Elife [Internet] 2020;9 doi: 10.7554/eLife.51406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahn O., Siems S.B., Kusch K., Hesse D., Jung R.B., Liepold T., et al. The CNS myelin proteome: deep profile and persistence after post-mortem delay. Front Cell Neurosci. 2020;19(14):239. doi: 10.3389/fncel.2020.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quarles R.H. Myelin-associated glycoprotein (MAG): past, present and beyond. J Neurochem. 2007;100(6):1431–1448. doi: 10.1111/j.1471-4159.2006.04319.x. [DOI] [PubMed] [Google Scholar]
- 19.Shahrizaila N., Lehmann H.C., Kuwabara S. Guillain-Barré syndrome. Lancet. 2021;397(10280):1214–1228. doi: 10.1016/S0140-6736(21)00517-1. [DOI] [PubMed] [Google Scholar]
- 20.Bukowski J.F., Welsh R.M. Enhanced susceptibility to cytotoxic T lymphocytes of target cells isolated from virus-infected or interferon-treated mice. J Virol. 1986;59(3):735–739. doi: 10.1128/jvi.59.3.735-739.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welsh R.M., Che J.W., Brehm M.A., Selin L.K. Heterologous immunity between viruses. Immunol Rev. 2010;235(1):244–266. doi: 10.1111/j.0105-2896.2010.00897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sewell A.K. Why must T cells be cross-reactive? Nat Rev Immunol. 2012;12(9):669–677. doi: 10.1038/nri3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanduc D. Homology, similarity, and identity in peptide epitope immunodefinition. J Pept Sci. 2012;18(8):487–494. doi: 10.1002/psc.2419. [DOI] [PubMed] [Google Scholar]
- 24.Frankild S., de Boer R.J., Lund O., Nielsen M., Kesmir C. Amino acid similarity accounts for T cell cross-reactivity and for “holes” in the T cell repertoire. PLoS One. 2008;3(3):e1831. doi: 10.1371/journal.pone.0001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Reports Medicine. 2021;2(2):100204. [DOI] [PMC free article] [PubMed]
- 26.Seil FJ. Myelin Antigens and Antimyelin Antibodies. Antibodies (Basel) [Internet]. 2018;7(1). Available from: 10.3390/antib7010002. [DOI] [PMC free article] [PubMed]
- 27.Anitei M., Pfeiffer S.E. Myelin biogenesis: sorting out protein trafficking. Curr Biol. 2006;16(11):R418–R421. doi: 10.1016/j.cub.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Mittendorf K.F., Marinko J.T., Hampton C.M., Ke Z., Hadziselimovic A., Schlebach J.P., et al. Peripheral myelin protein 22 alters membrane architecture. Sci Adv. 2017;3(7):e1700220. doi: 10.1126/sciadv.1700220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMHCpan 4.0: Improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data [Internet]. Available from: 10.1101/149518. [DOI] [PMC free article] [PubMed]
- 30.Petrova G., Ferrante A., Gorski J. Cross-reactivity of T cells and its role in the immune system. Crit Rev Immunol. 2012;32(4):349–372. doi: 10.1615/critrevimmunol.v32.i4.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lang H.L.E., Jacobsen H., Ikemizu S., Andersson C., Harlos K., Madsen L., et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat Immunol. 2002;3(10):940–943. doi: 10.1038/ni835. [DOI] [PubMed] [Google Scholar]
- 32.Kanduc D., Shoenfeld Y. On the molecular determinants of the SARS-CoV-2 attack. Clin Immunol. 2020;215 doi: 10.1016/j.clim.2020.108426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Welsh R.M., Fujinami R.S. Pathogenic epitopes, heterologous immunity and vaccine design. Nat Rev Microbiol. 2007;5(7):555–563. doi: 10.1038/nrmicro1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boucher A., Desforges M., Duquette P., Talbot P.J. Long-term human coronavirus-myelin cross-reactive T-cell clones derived from multiple sclerosis patients. Clin Immunol. 2007;123(3):258–267. doi: 10.1016/j.clim.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Huang Y., Lue J., Quandt J.A., Martin R., Mariuzza R.A. Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule. EMBO J. 2005;24(17):2968–2979. doi: 10.1038/sj.emboj.7600771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat Immunol. 2020;22(1):74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 37.Bhagavati S. Autoimmune disorders of the nervous system: pathophysiology, clinical features, and therapy. Front Neurol. 2021 doi: 10.3389/fneur.2021.664664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Augusto D.G., Yusufali T., Peyser N.D., Butcher X., Marcus G.M., Olgin J.E., et al. HLA-B*15:01 is associated with asymptomatic SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.05.13.21257065. [DOI] [Google Scholar]
- 39.Matzaraki V., Kumar V., Wijmenga C., Zhernakova A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017;18(1):1–21. doi: 10.1186/s13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stathopoulos P., Alexopoulos H., Dalakas M.C. Autoimmune antigenic targets at the node of Ranvier in demyelinating disorders. Nat Rev Neurol. 2015;11(3):143–156. doi: 10.1038/nrneurol.2014.260. [DOI] [PubMed] [Google Scholar]
- 41.Vojdani A., Kharrazian D. Potential antigenic cross-reactivity between SARS-CoV-2 and human tissue with a possible link to an increase in autoimmune diseases. Clin Immunol. 2020;217 doi: 10.1016/j.clim.2020.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamada Y., Hirano M., Kuwahara M., Samukawa M., Takada K., Morise J., et al. Binding specificity of anti-HNK-1 IgM M-protein in anti-MAG neuropathy: possible clinical relevance. Neurosci Res. 2015;91:63–68. doi: 10.1016/j.neures.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Vojdani A., Vojdani E., Kharrazian D. Reaction of human monoclonal antibodies to SARS-CoV-2 proteins with tissue antigens: implications for autoimmune diseases. Front Immunol. 2021 doi: 10.3389/fimmu.2020.617089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanduc D., Shoenfeld Y. Molecular mimicry between SARS-CoV-2 spike glycoprotein and mammalian proteomes: implications for the vaccine. Immunol Res. 2020;68(5):310–313. doi: 10.1007/s12026-020-09152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stathopoulos P., Alexopoulos H., Dalakas M.C. Autoimmune antigenic targets at the node of Ranvier in demyelinating disorders [Internet] Nat Rev Neurol. 2015;11:143–156. doi: 10.1038/nrneurol.2014.260. [DOI] [PubMed] [Google Scholar]
- 46.Schnaar R.L., Gerardy-Schahn R., Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol Rev. 2014;94(2):461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexopoulos H., Biba A., Dalakas M.C. Anti-B-cell therapies in autoimmune neurological diseases: rationale and efficacy trials. Neurotherapeutics. 2016;13(1):20–33. doi: 10.1007/s13311-015-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matà S., Ambrosini S., Saccomanno D., Biagioli T., Carpo M., Amantini A., et al. Anti-MAG IgM: differences in antibody tests and correlation with clinical findings. Neurol Sci. 2020;41(2):365–372. doi: 10.1007/s10072-019-04089-7. [DOI] [PubMed] [Google Scholar]
- 49.Matsui T., Hamada Y., Kuwahara M., Morise J., Oka S., Kaida K., et al. Association of variability in antibody binding affinity with a clinical course of anti-MAG neuropathy. J Neuroimmunol. 2020;15(339) doi: 10.1016/j.jneuroim.2019.577127. [DOI] [PubMed] [Google Scholar]
- 50.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 51.Delmont E., Attarian S., Antoine J.C., Paul S., Camdessanché J.P., Grapperon A.M., et al. Relevance of anti-HNK1 antibodies in the management of anti-MAG neuropathies. J Neurol. 2019;266(8):1973–1979. doi: 10.1007/s00415-019-09367-0. [DOI] [PubMed] [Google Scholar]
- 52.Hayashi C., Suzuki N., Mabuchi Y., Kikura N., Hosoda Y., de Vega S., et al. The extracellular domain of teneurin-4 promotes cell adhesion for oligodendrocyte differentiation. Biochem Biophys Res Commun. 2020;523(1):171–176. doi: 10.1016/j.bbrc.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Shahrour M.A., Ashhab M., Edvardson S., Gur M., Abu-Libdeh B., Elpeleg O. Hypomyelinating leukodystrophy associated with a deleterious mutation in the ATRN gene. Neurogenetics. 2017;18(3):135–139. doi: 10.1007/s10048-017-0515-7. [DOI] [PubMed] [Google Scholar]
- 54.Paz J., Yao H., Lim H.S., Lu X.Y., Zhang W. The neuroprotective role of attractin in neurodegeneration. Neurobiol Aging. 2007;28(9):1446–1456. doi: 10.1016/j.neurobiolaging.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 55.Al Saiegh F., Ghosh R., Leibold A., Avery M.B., Schmidt R.F., Theofanis T., et al. Status of SARS-CoV-2 in cerebrospinal fluid of patients with COVID-19 and stroke. J Neurol Neurosurg Psychiatry. 2020;91(8):846–848. doi: 10.1136/jnnp-2020-323522. [DOI] [PubMed] [Google Scholar]
- 56.Duan L., Zheng Q., Zhang H., Niu Y., Lou Y., The W.H., et al. The SARS-CoV-2 spike glycoprotein biosynthesis, structure, function, and antigenicity: implications for the design of spike-based vaccine immunogens. Front Immunol. 2020 doi: 10.3389/fimmu.2020.576622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584(7821):457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 58.Khanolkar A. Elucidating T cell and B cell responses to SARS-CoV-2 in humans: gaining insights into protective immunity and immunopathology. Cells. 2021;11(1):67. doi: 10.3390/cells11010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilhelm A., Widera M., Grikscheit K., Toptan T., Schenk B., Pallas C., et al. Reduced neutralization of SARS-CoV-2 omicron variant by vaccine sera and monoclonal antibodies. medRxiv. 2021;12(7):21267432. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.