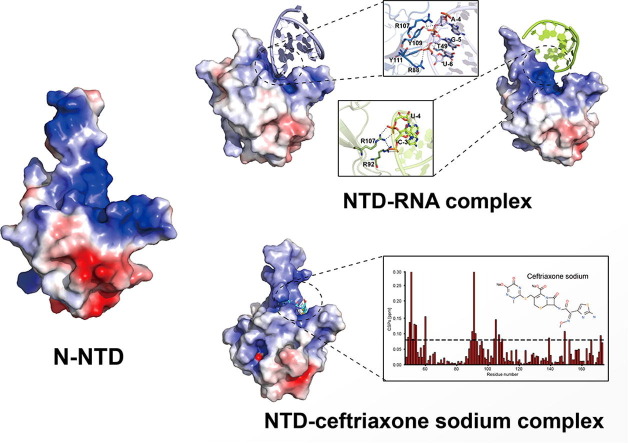
Graphical abstract
Keywords: SARS-CoV-2, Nucleocapsid protein, N-terminal domain, Ceftriaxone sodium
Abstract
Nucleocapsid (N) protein plays crucial roles in the life cycle of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), including the formation of ribonucleoprotein (RNP) complex with the viral RNA. Here we reported the crystal structures of the N-terminal domain (NTD) and C-terminal domain (CTD) of the N protein and an NTD-RNA complex. Our structures reveal a unique tetramer organization of NTD and identify a distinct RNA binding mode in the NTD-RNA complex, which could contribute to the formation of the RNP complex. We also screened small molecule inhibitors of N-NTD and N-CTD and discovered that ceftriaxone sodium, an antibiotic, can block the binding of RNA to NTD and inhibit the formation of the RNP complex. These results together could facilitate the further research of antiviral drug design targeting N protein.
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive single-stranded RNA virus (+ssRNA) with a genome of ∼30 kb [1]. According to sequence alignment and bioinformatics analysis results, SARS-CoV-2 is identified as a member of the genus Betacoronavirus, which also contains SARS-CoV and MERS-CoV with sequence homology to SARS-CoV-2 as high as 79% and 50%, respectively [2], [3]. The genome of SARS-CoV-2 mainly encodes four structural proteins, sixteen nonstructural proteins, and other accessory proteins. The four structural proteins are spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins [1], [4]. Spike protein mediates the recognition and membrane fusion with host cells by angiotensin-converting enzyme 2 (ACE2), which is the main target of antibody and vaccine development [5], [6], [7]. Sixteen nonstructural proteins, including nsp1–16, mainly mediate viral synthesis and processing, and play important roles in virus proliferation [8], [9].
Nucleocapsid protein is one of the main structural proteins of coronavirus. N protein binds viral RNA to form ribonucleoprotein (RNP) complex, which can be arranged in “beads on a string” to protect the viral genome and package virion [10], [11]. N protein also regulates viral replication, transcription, and interaction with membrane protein [12], [13], [14]. In addition, many immunological studies have shown that N protein interacts with host proteins to induce immune responses or regulate the cell cycle of host cells after viral infection [15], [16], [17], [18]. Many antibodies to the N protein can be detected in patients with COVID-19 to be used as a basis for diagnosis and treatment [19], [20]. The nucleocapsid protein of coronavirus contains two main domains: the N-terminal domain (NTD) and the C-terminal domain (CTD) [21]. They are sandwiched between three intrinsically disordered regions (IDRs). The middle IDR, which connects NTD and CTD, is called the central linker region (LKR) including a Ser/Arg-rich region [22]. Previous studies have reported that NTD is responsible for RNA binding, and CTD is responsible for dimerization, both of which contribute to the formation of RNP [21]. Given the importance of the N protein, its RNA binding and dimerization domains may be crucial targets for drug development.
The structure of the full-length N protein is still unknown due to its intrinsically disordered regions, but the structures of NTD and CTD of the N protein (designated as N-NTD and N-CTD) of viruses from genus Betacoronavirus, such as SARS-CoV-2, SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), and human coronavirus (HCoV)-OC43, have been reported [23], [24], [25], [26], [27], [28], [29], [30], [31]. Several key residues involved in RNA binding and interaction have also been identified. However, the crystal structure of N-NTD bound to RNA and the molecular mechanism of RNP formation remain unclear.
Here, we report the crystal structure of NTD of the N protein of SARS-CoV-2 at a resolution of 1.90 Å, the crystal structure of NTD bound RNA at a resolution of 2.25 Å, and the crystal structure of CTD at a resolution of 3.0 Å. Through the Nuclear Magnetic Resonance (NMR) titration experiment, we identified ceftriaxone sodium, which is the third-generation cephalosporin and a commonly used antibiotic in the clinic, as an inhibitor of RNA binding to NTD. The new crystal structural information from this paper provides a basis for the study of the molecular mechanism of RNP formation in SARS-CoV-2, and could contribute to the development of other antiviral drugs targeting NTD of the N protein and its RNA binding sites.
2. Materials and methods
2.1. Cloning, expression, and purification of N-NTD and N-CTD
The genes coding SARS-CoV-2 N-NTD (residues 48–173) and N-CTD (residues 250–365) were chemically synthesized with codon optimization (GenScript, Nanjing, China). Two genes were cloned into the pET-28a vector (Vazyme Biotech Co., Ltd., Nanjing, China) with a 5′ ATG starting sequence and C-terminal tobacco etch virus (TEV) protease site followed by a hexa-histidine (6 × His) tag separately. N-NTD and N-CTD constructs were extracted from E. coli DH5α (Tiangen Biotech Co., Ltd., Beijing, China) and then expressed into E. coli BL21(DE3) (Mei5 Biotechnology Co., Ltd., Beijing, China) grown in LB medium. When bacterial cultures were grown to an A 600 nm of 0.6–0.8 at 37 °C, isopropyl β-D-1-thiogalactopyranoside (IPTG) with a final concentration of 1 mmol/L was added to induce the expression at 16 °C for 18–20 h.
Cells were harvested by centrifugation (4000 r/min, 15 min, 4 °C) (Beckman Coulter, Bria, USA) and then resuspended in lysis buffer (50 mmol/L Tris-HCl pH 8.0, 500 mmol/L NaCl, 5% glycerol). Cells were lysed with a high-pressure homogenizer operating (ATS Engineering Limited, Suzhou, China) at 700–900 bar. Lysates were cleared by centrifugation (18,000 r/min, 30 min, 4 °C) (Beckman Coulter). The supernatant was loaded on Ni-NTA beads (Qianchun Bio, Yancheng, China) and then washed with buffer containing 40 and 60 mmol/L imidazole. Finally, the protein was eluted with lysis buffer containing 250 mmol/L imidazole. The protein was incubated with TEV protease at 4 °C for overnight and passed through Ni-NTA beads (Qianchun Bio) again to remove the 6 × His tag (protein: TEV protease = 100:1). Protein without 6 × His tag was concentrated by Centrifugal Filter Unit (Merk Millipore Ltd., Tullagreen, Ireland), and then size-separated by a Superdex 75 Increase 10/300 GL column (GE Healthcare, Fairfield, USA) in the SEC buffer (20 mmol/L Tris-HCl pH 8.0, 150 mmol/L NaCl). The purity of protein was checked by 15 % sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (Yeasen Biotechnology Co., Ltd., Shanghai, China). Purified protein was concentrated for future use.
Purification of protein samples for NMR is similar to the above protocol. E. coli BL21(DE3) (Mei5 Biotechnology Co., Ltd.) should be grown in M9 medium containing U-[13C, 15N] (Cambridge Isotope Laboratories, Inc., Turksbury, USA). The SEC buffer for purified protein should contain 20 mmol/L Na2HPO4 pH 5.5, 50 mmol/L NaCl, 0.01% NaN3.
2.2. Preparation of RNA
To examine the binding of RNA and N-NTD, dsRNA was prepared by two RNA oligonucleotides with sequence of 5′-CACUGAC-3′ and 5′-GUCAGUG-3′. To anneal the RNA duplex, two RNA oligonucleotides were mixed at an equal molar ratio in annealing buffer (10 mmol/L Tris-HCl pH 8.0, 25 mmol/L NaCl, 2.5 mmol/L ethylenediamine tetraacetic acid (EDTA)), denatured by heating to 94 °C for 5 min and then slowly cooled to room temperature.
2.3. Crystallization and data collection
SARS-CoV-2 N-NTD, N-CTD, and N-NTD and dsRNA complex were crystallized by the sitting drop method. Crystals of N-NTD were grown with crystal buffer (10 mmol/L ZnCl2, 100 mmol/L 2-morpholinoethanesulfonic acid (MES) pH 6.0, 20% polyethylene glycol (PEG) 6000 at 20 °C. Crystals of N-CTD were grown with crystal buffer (27 mmol/L NaNO3, 27 mmol/L Na2HPO4, 27 mmol/L (NH4)2SO4, 100 mmol/L Tris, 1 mol/L bicine pH 8.5, 20% polyethylene glycol monomethyl ethers (PEGMME) 500, 10% PEG 20,000, 100 μmol/L 1,8-anilinonaphthalene sulfonate) at 20 °C. Crystals of N-NTD and dsRNA complex were grown with crystal buffer (100 mmol/L MIB buffer pH 6.0, 25% PEG 1500) at 20 °C. The MIB buffer is prepared by mixing sodium malonate, imidazole, and boric acid according to 2:3:3. Crystals are frozen directly in liquid nitrogen.
2.4. Structure determination and refinement
X-ray diffraction data were collected at beamline using a wavelength of 0.979361 Å (NTD), 1.18057 Å (NTD-RNA), or 0.979150 Å (CTD). The structure was determined by molecular replacement (MR) with Phaser (STFC Daresbury Lab, Warrington, UK), using the X-ray crystal structure of SARS-CoV N-NTD (PDB ID: 2OFZ) or N-CTD (PDB ID: 2CJR) as the search model [26], [28], [32]. All initial models were further refined with Refmac5 (STFC Daresbury Lab, Warrington, UK) as part of CCP4 program (STFC Daresbury Lab) and interactively adjusted by COOT (STFC Daresbury Lab) [33]. The X-ray diffraction and structure refinement statistics are summarized in Table S1 (online).
2.5. Surface plasmon resonance (SPR) assay
The binding affinity of ceftriaxone sodium (MedChemExpress, Monmouth Junction, USA) and RNA to SARS-CoV-2 N-NTD were verified with an SPR assay by a Biacore S200 instrument (GE Healthcare). SARS-CoV-2 N-NTD was immobilized on a CM5 sensor chip (GE Healthcare). The channel of the blank group flowed through the chip with PBS buffer, and the channels of the experimental group were diluted to different concentrations of ceftriaxone sodium (MedChemExpress) or RNA with PBS buffer and flowed through the chip. Equilibrium dissociation constant (K D) values were calculated and analyzed by Biacore S200 analysis software (GE Healthcare). In the competition experiment between ceftriaxone sodium (MedChemExpress) and RNA, 100 μmol/L ceftriaxone sodium (MedChemExpress) was added to the PBS buffer of blank groups and experimental groups.
2.6. NMR spectroscopy
The NMR experiments were carried out at 25 °C on a Bruker Avance 600 MHz spectrometer (Bruker, Billerica, USA) equipped with a 5 mm triple-resonance TCI cryogenic probe. The U-[13C, 15N] NMR samples (Cambridge Isotope Laboratories, Inc.) were prepared in the SEC buffer, 5% D2O/90%–95% H2O. A series of double- and triple-resonance spectra were recorded to obtain sequence-specific resonance assignments, including 2D 1H-15N HSQC, 3D HNCACB, and 3D CBCA(CO)NH. For NMR titrations with candidate small molecule compounds (TargetMol Chemicals Inc., Boston, USA), 2D 1H-15N HSQC spectra were collected on the 15N-labelled N-NTD/N-CTD in the absence and in the presence of small molecules at different ratios. Small molecular compounds are usually dissolved into DMSO, which may induce shifts in the NMR peaks. To eliminate the effect of DMSO, we dissolved small molecular compounds into the SEC buffer of N-NTD/N-CTD and repeated NMR titration experiments. All NMR spectra were processed and analyzed with Sparky (University of California, San Francisco, USA) and NMRPipe (National Institute of Standards and Technology (NIST), Rockville, USA). The chemical shift perturbations (CSPs) were calculated using the equation:
| (1) |
here, the significantly perturbed residues are defined as those with the CSP values > average + STD.
2.7. Molecular docking
The structure of the N-NTD in complex with ceftriaxone sodium (MedChemExpress) was calculated using HADDOCK (Utrecht University Bonvin Lab, Utrecht, the Netherlands) in combination with crystallography and NMR system (CNS) [34], [35]. The actual docking was performed using the NMR structure of N-NTD (PDB ID: 6YI3) and the 3D structure of ceftriaxone sodium (PubChem CID: 5479530) (MedChemExpress) according to a standard protocol, which is described as follows. The topology and parameters files of ceftriaxone sodium (MedChemExpress) were generated with ACPYPE (Vrije Universiteit Brussel Wim F Vranken Lab, Brussels, Belgium) [36]. The N-NTD residues with CSP > 0.08 ppm and at least 20% solvent accessibility were chosen as active residues (T49, A50, W52, T54, G60, A90, T91, R92, L104, S105, R107, Y109, L139, A152, and Y172), while adjacent solvent-exposed residues were additionally selected as passive residues (N48, L55, H59, K61, R89, R93, D103, P106, A138, N140, P151, N153, F171, and A173). All atoms of ceftriaxone sodium (MedChemExpress) were defined as active for the experimentally driven docking protocol. In addition, three regions within the N-NTD were defined as fully-flexible segments for the advanced stages of the docking calculation (the N-terminal N48–A50, the central loop I94–M101, and the C-terminal G170–A173). The final set of 200 water-refined structures was clustered using a Fraction of Common Contacts approach with a default cut-off of 0.75. Finally, the structure of NTD-ceftriaxone sodium with the lowest energy score was selected for detailed analysis and display by PyMOL (The PyMOL Molecular Graphics System, version 2.1.1; Schrodinger, New York, USA).
3. Results
3.1. Crystal structure of N-NTD
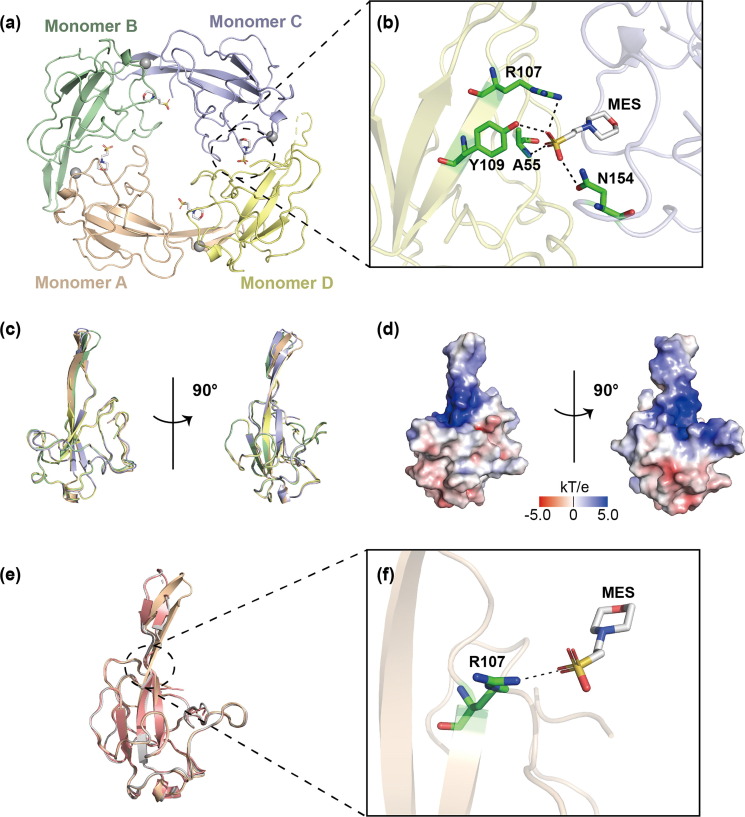
We designed a SARS-CoV-2 N-NTD construct containing 48–173 residues of the N protein, expressed and purified as the method described (Fig. S1 online). The crystal structure of SARS-CoV-2 N-NTD (PDB ID: 7XX1) was determined to a resolution of 1.90 Å by molecular replacement using the structure of SARS-CoV N-NTD (PDB ID: 2OFZ) as the search model [26]. The final structure was refined to R-factor and R-free values of 0.210 and 0.258, respectively. The additional data are given in Table S1 (online). Each asymmetric unit (ASU) contains four N-NTD molecules with overall root mean square deviation (RMSD) values of 0.43–1.06 Å between each monomer, each of which binds one zinc ion and one MES molecule (Fig. 1 a, c). In the tetramer, MES molecules, which are buffer components, are found to form hydrogen bonds with two adjacent N-NTD monomers, making the entire tetramer assume a cyclic configuration (Fig. 1b). Compared with the structures of other N-NTD tetramers, our circular tetramer structure is more stable under the action of MES molecules, which may contribute to the oligomerization of the N protein. All four monomers in one asymmetric unit of N-NTD show a right-handed shape and contain a structural core and a flexible loop similar to other coronaviruses’ N-NTDs [26], [30], [31]. The structural core composes of a four-stranded antiparallel β-sheet and a short 310 helix, while the flexible loop is a protruding β-hairpin (Fig. 1c). As depicted on the electrostatic potential surface, the junction between the protruding β-hairpin and the core structure forms a positively charged pocket, serving as a potential RNA binding site (Fig. 1d).
Fig. 1.
Crystal structure of SARS-CoV-2 N-NTD. (a) The overall structure of SARS-CoV-2 N-NTD tetramer (PDB ID: 7XX1). Each monomer is represented by a different color. (b) Hydrogen bonds between an MES molecule and different N-NTD monomers. Each monomer is represented by a different color. Hydrogen bonds are represented by black dotted lines, and amino acids involved in hydrogen bond interactions are marked. (c) The superimposition of four monomers in an asymmetric unit shown as a cartoon representation in two orientations. (d) The electrostatic potential surface of SARS-CoV-2 N-NTD in two orientations. Blue indicates positive charge potential, while red indicates negative charge potential. (e) Superposition (based on Cα) of our SARS-CoV-2 N-NTD structure (PDB ID: 7XX1, wheat) and other published SARS-CoV-2 N-NTD structures (PDB ID: 6M3M, gray; PDB ID: 7CDZ, pink). (f) Hydrogen bonds between an MES molecule and an N-NTD monomer. Hydrogen bonds are represented by black dotted lines, and amino acids involved in hydrogen bond interactions are marked.
At present, there are several reports on the structures of SARS-CoV-2 N-NTD [23], [24]. To further explore the structural information, we compared our structure with the reported structures (PDB ID: 6M3M, 7CDZ) (Fig. 1e) [23], [24]. The RMSD values between our N-NTD structure and the other two N-NTD structures are 0.630–1.149 Å (PDB ID: 6M3M) and 0.635–1.190 Å (PDB ID: 7CDZ), respectively. In addition to the differences in the flexible N- and C-termini, there are still two differences in the β-hairpin region. First, compared with the other two N-NTD structures, our N-NTD structure shows an extended β-hairpin, similar to MERS-CoV N-NTD (PDB ID: 4UD1) [31]. Second, the β-hairpin shows a movement towards the RNA binding site as described later. These differences may be caused by the flexibility of β-hairpin and the influence of MES molecules. The MES molecule binds to R107 at the bottom of the β-hairpin region via hydrogen bond interactions (Fig. 1f).
3.2. Crystal structure of N-NTD and RNA complex
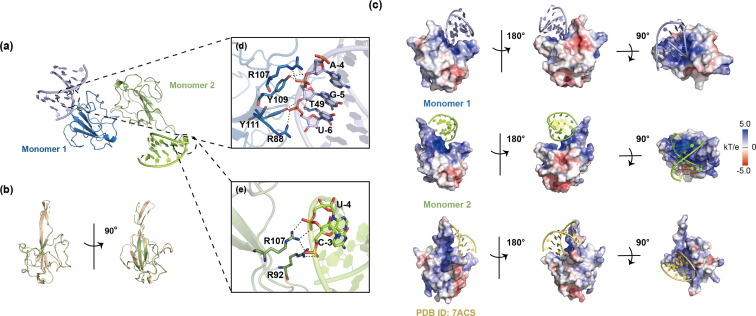
The electrostatic potential surface of N-NTD reveals a potential RNA binding site [26], [30], [37], [38]. To obtain the structure of the N-NTD and RNA complex, we selected an RNA sequence (5′-CACUGAC-3′ and 5′-GUCAGUG-3′) as a duplex of RNA (dsRNA), which binds to N-NTD [38]. The K D value of N-NTD and dsRNA detected by SPR analysis was 8 nmol/L (Fig. S2 a online). The crystal structure of the SARS-CoV-2 NTD-RNA complex (PDB ID: 7XWZ) was solved at a resolution of 2.25 Å. Each ASU contains two N-NTD molecules and two dsRNA fragments (Fig. 2a). Structural superimposition of N-NTD in the apo and RNA-bound forms reveals significant conformational changes within the β-hairpin region (Fig. 2b), highlighting the intrinsic flexibility of these RNA-interacting regions, which would allow each N-NTD monomer binds to dsRNA in different ways. Of our two N-NTD monomers, N-NTD binds predominantly to one of the strands in the dsRNA. In monomer 1, the hydrogen bond interaction between Y109, Y111, and RNA involves OP1′ atoms and O3′ atoms in the RNA phosphate backbone, the hydrogen bond between R88 and RNA includes only an OP1′ atom, while the hydrogen bond between T49 and RNA includes only an O2′ atom (Fig. 2c, d). However, hydrogen bond interactions between R92, R107, and RNA involve OP1′ atoms and OP2′ atoms in RNA phosphate backbones in monomer 2 (Fig. 2c, e). These binding modes are also different from that of the docking model of the NTD-RNA complex (PDB ID: 7ACS) by NMR (Fig. 2b) [38]. In the NMR-based model, three arginine residues (R92, R107, and R149) directly bind to RNA. Although the amino acid residues affected by the three binding modes are distinct, the binding site is obviously at the same position, the positively charged junction between the protruding β-hairpin and the core structure. Distinct binding modes may indicate that N-NTD binds to RNA as much as possible in various ways to promote the formation of RNP.
Fig. 2.
Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex. (a) The overall structure of SARS-CoV-2 N-NTD and dsRNA complex (PDB ID: 7XWZ). Each monomer is represented by a different color. (b) Superimposition of SARS-CoV-2 N-NTD between the apo form (wheat) and dsRNA-bound form (green). (c) The electrostatic potential surface of SARS-CoV-2 N-NTD monomer and its bound dsRNA in three orientations. dsRNA is shown as a cartoon representation (Monomer 1: purple; Monomer 2: green; PDB ID: 7ACS, yellow). Blue indicates positive charge potential, while red indicates negative charge potential. (d) Hydrogen bonds between N-NTD monomer 1 and dsRNA. Hydrogen bonds are represented by black dotted lines, and amino acids and nucleotides involved in hydrogen bond interactions are marked. (e) Hydrogen bonds between N-NTD monomer 2 and dsRNA. Hydrogen bonds are represented by black dotted lines, and amino acids and nucleotides involved in hydrogen bond interactions are marked.
3.3. Structure of N-NTD and ceftriaxone sodium complex
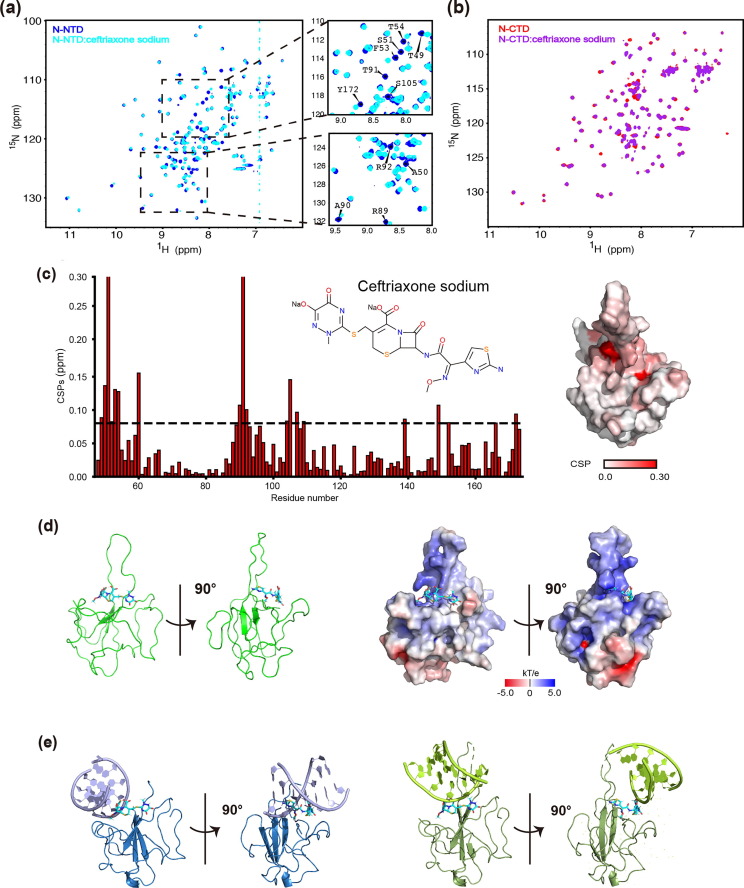
N-NTD and N-CTD contribute to the formation of RNP, so small molecule drugs that bind to them and block this process can be used as inhibitors targeting the N protein. We screened a large number of small molecule compounds from literature reports and compound libraries to identify potential drugs. Finally, we identified several candidate compounds. Because of the difficulties in growing crystals of protein and small molecular compounds, we chose to characterize the binding of these small molecule compounds to N-NTD by NMR titration experiments. Residues of N-NTD are specifically perturbed by ceftriaxone sodium dissolved in DMSO or SEC buffer on the 2D 1H-15N HSQC spectra (Fig. S3 online). Meanwhile, the SPR assay also detected the K D value of 8.3 μmol/L for N-NTD and ceftriaxone sodium (Fig. S2b online). Ceftriaxone sodium is a third-generation cephalosporin and was approved by FDA in 1984. It is mainly used for infections caused by pathogens by inhibiting the synthesis of the cell wall.
We added ceftriaxone sodium into the N-NTD protein sample at ratios from 0:1 to 25:1. Superimposition of the 2D 1H-15N HSQC spectra in the presence of ceftriaxone sodium at different ratios revealed remarkable resonance perturbations of many residues (Fig. 3 a). Surprisingly, the same small compound can also induce resonance shifts in N-CTD, although relatively few residues were found to move (Fig. 3b). These results indicate that ceftriaxone sodium specifically interacted with both N-NTD and N-CTD. To locate the binding sites of ceftriaxone sodium on the surface of N-NTD, we analyzed chemical shift perturbations (CSPs) values on a per-residue basis. According to CSPs at the max ratio (N-NTD: ceftriaxone sodium = 1:25), a total of 17 residues (T49, A50, S51, W52, F53, T54, G60, A90, T91, R92, L104, S105, R107, Y109, L139, A152, and Y172) showed significant resonance shifts, which, by mapping onto the crystal structure of N-NTD, constituted two binding sites on the surface of N-NTD (Fig. 3a, c). Subsequently, we used the HADDOCK program for the NMR-restraint driven docking simulations of ceftriaxone sodium. The 3D structure with PubChem CID of 5479530 was chosen as a starting conformation of the ceftriaxone sodium. The analysis of CSPs provided a set of “active” solvent-accessible residues on N-NTD that were expanded for surrounding “passive” residues. The selection criteria for active residues were that their CSP values were higher than 0.08 ppm and at least 20% solvent accessibility. The standard docking protocol yielded a set of 200 water-refined structures, in which the structure with the lowest energy score was selected as a representative conformation for the N-NTD in complex with ceftriaxone sodium (NTD-ceftriaxone sodium) (Fig. 3d). As shown in the resulting structure model of NTD-ceftriaxone sodium complex, two main binding sites are located within the junction region, the same location as the RNA binding site (Fig. 3e). Interestingly, significantly perturbed residues include those (R92, R107, and Y109) involved in hydrogen bond interactions under two NTD-RNA binding modes (Fig. 2d, e, Fig. 3d). In addition, we also used SPR to study ceftriaxone sodium and RNA competition for NTD binding. The experimental results revealed that in the presence of 10- to 20-fold excess of ceftriaxone sodium, no RNA binding signal to N-NTD was detected, indicating that ceftriaxone sodium can compete with RNA for N-NTD binding (Fig. S2c online). Therefore, with the N protein as the drug target, our results suggest that ceftriaxone sodium is an inhibitor by blocking the interaction between NTD and RNA.
Fig. 3.
NMR characterization of N-NTD/N-CTD and ceftriaxone sodium. (a) Superimposition of HSQC spectra of N-NTD in the absence (blue) and the presence of ceftriaxone sodium at a molar ratio (N-NTD: ceftriaxone sodium) of 1:25 (cyan). The assignments of the significantly perturbed residues are labeled on the right. (b) Superimposition of HSQC spectra of N-CTD in the absence (red) and the presence of ceftriaxone sodium at a molar ratio (N-CTD: ceftriaxone sodium) of 1:100 (purple). (c) Residue-specific chemical shift perturbations (CSPs) of N-NTD in the presence of ceftriaxone sodium at a molar ratio (N-NTD: ceftriaxone sodium) of 1:25. The significantly perturbed residues are defined as those with the CSP values at 1:25 > 0.08 ppm (average + STD) (dark dashed line). The surface representation of N-NTD is colored according to the CSP values in a white-to-red scale. Color intensities correlate with CSP values, with darker colors indicating larger values. (d) The structure of the NTD-ceftriaxone sodium complex with ceftriaxone sodium in sticks and N-NTD in cartoon (left) and electrostatic potential surface (right) are shown in two orientations. (e) Superimposition (based on Cα) of ceftriaxone sodium-bound form and two dsRNA-bound forms of N-NTD reveals a similar binding interface. For the ceftriaxone sodium-bound form of N-NTD, only ceftriaxone sodium is shown.
3.4. Crystal structure of N-CTD
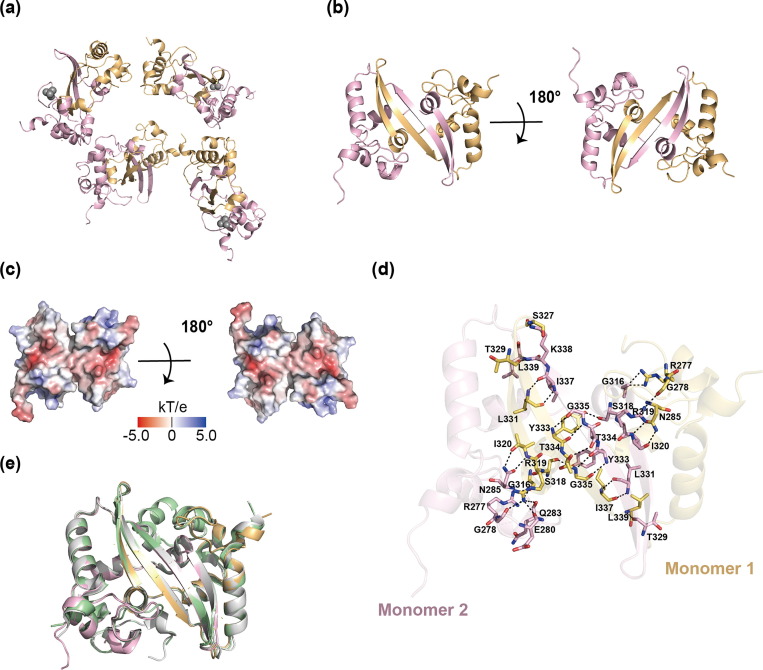
We also designed a SARS-CoV-2 N-CTD construct containing 250–365 residues of the N protein, expressed and purified as the method described (Fig. S3 online). The crystal structure of SARS-CoV-2 N-CTD (PDB ID: 7XWX) was determined by molecular replacement at a resolution of 3.0 Å. The final structure was refined to R-factor and R-free values of 0.219 and 0.268, respectively. The additional data are given in Table S1 (online). Eight N-CTD molecules are paired to form four homo-dimers in one ASU, each dimer also binding one phosphate ion (Fig. 4 a). Each monomer is composed of one 310 helix, three α-helices, and two β-strands (Fig. 4b, c). N-CTD molecules form stable dimers, consistent with the result of analytical gel filtration (Fig. S4c online). Extensive hydrogen bonds and hydrophobic interactions stabilize the dimerization of N-CTD. In the dimer, two β-strands from each monomer form an antiparallel β-sheet with eight hydrogen bonds interactions, including S318-Y333, S327-K338, T329-L339, L331-I337, Y333-G335, T334-T334, G335-Y333, I337-L331, and L339-T329 (residues written in front are from monomer 1) (Fig. 4d). There are also many hydrogen bonds in the interaction region of α-helices from two monomers, such as R277-G316, G278-R319, N285-I320, G316-R277, R319-G278, R319-E280, R319-Q283, and I320-N285. In addition, residues F286, T296, W301, A305, and Q306 from monomer 1 show hydrophobic interactions with residues A311, S312, and G316 from monomer 2 in other interaction regions (data not shown).
Fig. 4.
Crystal structure of SARS-CoV-2 N-CTD. (a) The overall structure of SARS-CoV-2 N-CTD octamer (PDB ID: 7XWX). The two monomers are shown in light orange and light pink respectively in each homo-dimer. (b) Cartoon representation of SARS-CoV-2 N-CTD dimer in two orientations. Each monomer is represented by a different color. (c) The electrostatic potential surface of SARS-CoV-2 N-CTD. Blue indicates positive charge potential, while red indicates negative charge potential. (d) Hydrogen bonds in N-CTD dimer. Hydrogen bonds are represented by black dotted lines, and amino acids involved in hydrogen bond interactions are marked. (e) Superposition (based on Cα) of our SARS-CoV-2 N-CTD structure (PDB ID: 7XWX, light orange and light pink) and other published SARS-CoV-2 N-NTD structures (PDB ID: 7CE0, gray; PDB ID: 7DE1, pale green).
We overlaid our N-CTD structure with reported structures (PDB ID: 7CE0, 7DE1) [23], [39]. Because of extensive hydrogen bonds and hydrophobic interactions, all structures show similar and stable dimer structures and characteristics with RMSD of 0.95–0.97 Å (Fig. 4e). Stable dimer structures are also essential elements in the formation of RNP.
4. Discussion and conclusion
As the pandemic of COVID-19 enters its third year, several vaccines and drugs have been approved. However, this public health crisis has not been completely controlled due to the rapid mutation of SARS-CoV-2, highlighting the urgent demand to find additional potential drug targets and new drugs. As one of the important structural proteins of SARS-CoV-2, N protein plays a crucial role in many physiological processes of the virus, especially when it combines with viral RNA to form an RNP complex [10], [11]. In this study, we presented the crystal structures of N-NTD and N-CTD and compared them with previously published structures. The comparison results indicate that these structures are very similar to each other. NTD shows a right-handed shape, while CTD exists as a stable homo-dimer.
Based on the reported N-NTD structures of coronaviruses, it has been proposed that NTD binds to RNA through its positively charged pocket [26], [30], [37], [38]. Our NTD-RNA complex structure, which was not reported before, reveals that NTD and dsRNA display a specific and different RNA binding mode. The involved residues include R88, Y109, and Y111 in our monomer 1, R92, and R107 in our monomer 2, which are different from R92, R107, and R149 in the docking model as reported before [38]. Residues interacting with RNA in other coronaviruses’ N-NTDs have also been reported, such as R106, R107, and R117 in HCoV-OC43 (corresponding to R92, R93, and K102 in SARS-CoV-2) [30]. Although involved residues in different binding modes are not exactly the same, they include positively charged arginine, which may play a major role in RNA binding. Different binding modes indicate that N-NTD could bind to RNA in many different ways.
As one type of cephalosporin, ceftriaxone sodium is often used to treat bacterial infections, such as wound infections, respiratory tract infections, and intra-abdominal infections. Currently, there is an ongoing clinical trial to verify the therapeutic efficacy of ceftriaxone sodium in lung infections caused by COVID-19 [40]. Nevertheless, we first confirmed the inhibitory effect of this small molecule on the N protein of SARS-CoV-2. According to the process of RNP formation, there are two main antiviral strategies for the N protein: blocking the packaging of the viral RNA or inhibiting the oligomerization of the N protein. Drugs previously screened only inhibit the function of NTD, such as PJ34 [37], [41].
Nevertheless, ceftriaxone sodium we screened shows inhibitory effects both on NTD and CTD. According to the docking model of the NTD-ceftriaxone sodium complex, ceftriaxone sodium mainly affects 17 residues, including those binding to dsRNA (R92, R107, and Y109). Meanwhile, the SPR assay also detected that the K D value of N-NTD and ceftriaxone sodium was 8.3 μmol/L, and ceftriaxone sodium could compete with RNA for N-NTD binding within a similar concentration range. Therefore, ceftriaxone sodium blocks the interaction between NTD and RNA by occupying the RNA binding site, inhibiting the formation of RNP and further interrupting the life cycle of SARS-CoV-2. Its mechanism of inhibiting CTD remains to be further explored. Recently OMICRON (B.1.1.529) has been reported as a variant of concern. However, mutations of the N protein (P13L, R203K, and G204R) only occurred in three IDR regions, not the conserved NTD and CTD domains [42], [43]. Therefore, these sites that bind to RNA can serve as conserved drug targets, and ceftriaxone sodium may also serve as an effective inhibitor for OMICRON.
In conclusion, we reported and analyzed the crystal structures of NTD and CTD from the N protein of SARS-CoV-2 as well as the crystal structure of the NTD-RNA complex. Our structures revealed a novel tetramer configuration of NTD and identified a distinct binding mode of RNA binding by NTD. We also identified ceftriaxone sodium as an inhibitor that blocks RNA binding of NTD and inhibits the formation of RNP, which could interrupt the life cycle of SARS-CoV-2. This structural information presented in this paper could offer new insights into the follow-up studies of antiviral drug design targeting N protein.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
Acknowledgments
This work was supported by Beijing Natural Science Foundation (M21016), Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences (2021-I2M-1-003 and 2021-CAMS-JZ004), Tsinghua-Peking Center for Life Sciences (045-61020100122), and Beijing Advanced Innovation Center for Structural Biology.
Author contributions
Shuyang Zhang, H. Eric Xu, and Yi Xue designed and supervised the study. Xiaodong Luan performed the protein purification and structure analysis. Xinming Li performed NMR-related experiments and molecular docking. Yufan Li assisted in the work of Xiaodong Luan and finished the manuscript writing and figure preparation. Xinming Li, Gengchen Su, Wanchao Yin, and H. Eric Xu participated in the manuscript and figure preparation. Other authors have provided facilities and funds to assist in this study. All authors read and approved the manuscript.
Biographies

Xiaodong Luan is a Ph.D. candidate at School of Medicine, Tsinghua University. His research interest focuses on drug target structure research and new drug development.

Xinming Li received her B.S. degree from Huazhong University of Science and Technology in 2017 and Ph.D. degree from Tsinghua University in 2022. Her current research interest mainly focuses on the computational design of antibodies.

Yufan Li is a Ph.D. candidate at School of Medicine, Tsinghua University. Her main research interest is virus structural biology.

Gengchen Su received his B.S. degree from Inner Mongolia University (2021). He is currently a Ph.D. candidate at School of Medicine, Tsinghua University. His research interest focuses on the structural and biochemical research of viruses, drug discovery in potential targets of viruses, and histone acylation modification in disease generation.

H. Eric Xu obtained his bachelor’s and master’s degrees from Tsinghua University. He then went to Duke University and the University of Texas Southwestern Medical Center for his Ph.D. Following a postdoctoral fellowship at Massachusetts Institute of Technology, he moved to Institute of Glaxo Smith Kline. In 2002, he joined Van Andel Research Institute and became the founding Director of the Key Laboratory of Receptor Research of Chinese Academy of Sciences at Shanghai Institute of Materia Medica in 2013. His main research direction is protein structure study and related drug development.

Yi Xue is an assistant professor at School of Life Sciences, Tsinghua University. After receiving his Ph.D. degree from Purdue University, he did postdoctoral research at the University of Michigan and Duke University. His lab has a longstanding interest in studying the dynamics of RNAs and disordered proteins by means of NMR spectroscopy and computational structural biology.

Shuyang Zhang is the president of Peking Union Medical College Hospital. She received her M.D. degree from Peking Union Medical College in 1991. Her main research interest includes cardiovascular diseases, rare diseases, and emergent infectious diseases.
Footnotes
Supplementary materials to this article can be found online at https://doi.org/10.1016/j.scib.2022.10.021.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Chan J.F., Kok K.H., Zhu Z., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu B., Guo H., Zhou P., et al. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R., Zhao X., Li J., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brian D.A., Baric R.S. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. 2005;287:1–30. doi: 10.1007/3-540-26765-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C., Park Y.J., Tortorici M.A., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snijder E.J., Decroly E., Ziebuhr J. The nonstructural proteins directing coronavirus RNA synthesis and processing. Adv Virus Res. 2016;96:59–126. doi: 10.1016/bs.aivir.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arya R., Kumari S., Pandey B., et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433 doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao H., Song Y., Chen Y., et al. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein S., Cortese M., Winter S.L., et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zúñiga S., Cruz J.L., Sola I., et al. Coronavirus nucleocapsid protein facilitates template switching and is required for efficient transcription. J Virol. 2010;84:2169–2175. doi: 10.1128/JVI.02011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narayanan K., Maeda A., Maeda J., et al. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J Virol. 2000;74:8127–8134. doi: 10.1128/jvi.74.17.8127-8134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurst K.R., Kuo L., Koetzner C.A., et al. A major determinant for membrane protein interaction localizes to the carboxy-terminal domain of the mouse coronavirus nucleocapsid protein. J Virol. 2005;79:13285–13297. doi: 10.1128/JVI.79.21.13285-13297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo L., Li Z., Zhao T., et al. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci Bull. 2021;66:1194–1204. doi: 10.1016/j.scib.2021.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Surjit M., Liu B., Chow V.T.K., et al. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. 2006;281:10669–10681. doi: 10.1074/jbc.M509233200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu X., Ja P., Tao J., et al. SARS-CoV nucleocapsid protein antagonizes IFN-β response by targeting initial step of IFN-β induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes. 2011;42:37–45. doi: 10.1007/s11262-010-0544-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon D.E., Jang G.M., Bouhaddou M., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burbelo P.D., Riedo F.X., Morishima C., et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar R., Christensen N.D., Kaddis Maldonado R.J., et al. Monoclonal antibodies to S and N SARS-CoV-2 proteins as probes to assess structural and antigenic properties of coronaviruses. Viruses. 2021;13:1899. doi: 10.3390/v13101899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang C.K., Hou M.H., Chang C.F., et al. The SARS coronavirus nucleocapsid protein – forms and functions. Antiviral Res. 2014;103:39–50. doi: 10.1016/j.antiviral.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng T.Y., Lee K.R., Tarn W.Y. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 2008;275:4152–4163. doi: 10.1111/j.1742-4658.2008.06564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peng Y., Du N., Lei Y., et al. Structures of the SARS-CoV-2 nucleocapsid and their perspectives for drug design. EMBO J. 2020;39:e105938. doi: 10.15252/embj.2020105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang S., Yang M., Hong Z., et al. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 2020;10:1228–1238. doi: 10.1016/j.apsb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jayaram H., Fan H., Bowman B.R., et al. X-ray structures of the N- and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saikatendu K.S., Joseph J.S., Subramanian V., et al. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takeda M., Chang C.K., Ikeya T., et al. Solution structure of the C-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method. J Mol Biol. 2008;380:608–622. doi: 10.1016/j.jmb.2007.11.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C.Y., Chang C.K., Chang Y.W., et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 2007;368:1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen T.H.V., Lichière J., Canard B., et al. Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein. Acta Crystallogr D Struct Biol. 2019;75:8–15. doi: 10.1107/S2059798318014948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen I.J., Yuann J.M., Chang Y.M., et al. Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochim Biophys Acta Proteins Proteom. 2013;1834:1054–1062. doi: 10.1016/j.bbapap.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papageorgiou N., Lichière J., Baklouti A., et al. Structural characterization of the N-terminal part of the MERS-CoV nucleocapsid by X-ray diffraction and small-angle X-ray scattering. Acta Crystallogr D Struct Biol. 2016;72:192–202. doi: 10.1107/S2059798315024328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P., Lohkamp B., Scott W.G., et al. Features and development of Coot. Acta Crystallogr D Struct Biol. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 35.Brünger A.T., Adams P.D., Clore G.M., et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Struct Biol. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Sousa da Silva AW, Vranken WF. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res Notes 2012;5:367. [DOI] [PMC free article] [PubMed]
- 37.Lin S.Y., Liu C.L., Chang Y.M., et al. Structural basis for the identification of the N-terminal domain of coronavirus nucleocapsid protein as an antiviral target. J Med Chem. 2014;57:2247–2257. doi: 10.1021/jm500089r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dinesh D.C., Chalupska D., Silhan J., et al. Structural basis of RNA recognition by the SARS-CoV-2 nucleocapsid phosphoprotein. PLoS Pathog. 2020;16:e1009100. doi: 10.1371/journal.ppat.1009100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang M., He S., Chen X., et al. Structural insight into the SARS-CoV-2 nucleocapsid protein C-terminal domain reveals a novel recognition mechanism for viral transcriptional regulatory sequences. Front Chem. 2021;8 doi: 10.3389/fchem.2020.624765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lennie Derde. Randomized, embedded, multifactorial adaptive platform trial for community- acquired pneumonia (REMAP-CAP). ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/study/NCT02735707?cond=ceftriaxone&draw=3. Accessed 8 July 2022.
- 41.Mukherjee S., Dasgupta S., Adhikary T., et al. Structural insight to hydroxychloroquine-3C-like proteinase complexation from SARS-CoV-2: inhibitor modelling study through molecular docking and MD-simulation study. J Biomol Struct Dyn. 2021;39:7322–7334. doi: 10.1080/07391102.2020.1804458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thakur V., Ratho R.K. OMICRON (B.1.1.529): a new SARS-CoV-2 variant of concern mounting worldwide fear. J Med Virol. 2022;94:1821–1824. doi: 10.1002/jmv.27541. [DOI] [PubMed] [Google Scholar]
- 43.Bansal K., Kumar S. Mutational cascade of SARS-CoV-2 leading to evolution and emergence of omicron variant. Virus Res. 2022;315 doi: 10.1016/j.virusres.2022.198765. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.