Summary
As single- and mixed-species biofilms, Staphylococcus aureus and Pseudomonas aeruginosa cause difficult-to-eradicate chronic infections. In P. aeruginosa, pseudomonas quinolone (PQS)-dependent quorum sensing regulates virulence and biofilm development that can be attenuated via antagonists targeting the transcriptional regulator PqsR (MvfR). Here, we exploited a quinazolinone (QZN) library including PqsR agonists and antagonists for their activity against S. aureus alone, when co-cultured with P. aeruginosa, and in combination with the aminoglycoside tobramycin. The PqsR inhibitor, QZN 34 killed planktonic Gram-positives but not Gram-negatives. QZN 34 prevented S. aureus biofilm formation, severely damaged established S. aureus biofilms, and perturbed P. aeruginosa biofilm development. Although P. aeruginosa protected S. aureus from tobramycin in mixed biofilms, the combination of aminoglycoside antibiotic with QZN 34 eradicated the mixed-species biofilm. The mechanism of action of QZN 34 toward Gram-positive bacteria is shown to involve membrane perturbation and dissipation of transmembrane potential.
Keywords: Staphylococcus aureus, Pseudomonas aeruginosa, biofilms, quinazolinone, antimicrobials, quorum sensing, PqsR, MvfR
Graphical abstract
Highlights
-
•
QZNs inhibit PQS-dependent quorum sensing and reduce biofilm formation in Pseudomonas aeruginosa
-
•
A subset of QZNs is bactericidal for planktonic Staphylococcus aureus and severely damages biofilms
-
•
In mixed species biofilms, P. aeruginosa protects S. aureus from tobramycin
-
•
Tobramycin plus QZN eradicates mixed P. aeruginosa and S. aureus biofilms
P. aeruginosa and S. aureus cause biofilm-associated infections. Murray et al. demonstrate that quinazolinone inhibitors of P. aeruginosa quorum-sensing can be chemically modified to kill S. aureus biofilms and prevent P. aeruginosa-mediated protection of S. aureus against tobramycin. Tobramycin plus quinazolinone eradicates mixed P. aeruginosa and S. aureus biofilms.
Introduction
Pseudomonas aeruginosa and Staphylococcus aureus are opportunistic human pathogens that are major causes of hospital- and community-acquired infections (Peleg and Hooper, 2010; Tong et al., 2015). They are members of the ESKAPE group of multi- and pan-antibiotic-resistant bacteria that cause acute and chronic infections in skin, blood, bone, and soft tissues as well as in the respiratory and urinary tracts (Peleg and Hooper, 2010; Tong et al., 2015; De Oliveira et al., 2020). They are also associated with difficult-to-eradicate biofilm-centered infections involving implanted medical devices and can be considered as shared ecological niche competitors, since they often co-exist at an infection site (Percival et al., 2015; Orazi and O’Toole, 2019). For example, non-healing diabetic foot ulcer infections, a serious complication of diabetes, are usually polymicrobial and biofilm-centered and often harbor both P. aeruginosa and S. aureus (Fazli et al., 2009; DeLeon et al., 2014). Similarly, their co-infection of the lungs of individuals with cystic fibrosis is indicative of poorer clinical outcomes than in those infected with either species alone (Orazi and O'Toole, 2019).
To facilitate co-existence within the same environmental niche, bacteria have evolved intricate regulatory networks that enable them to evade, counter, inhibit, or suppress other bacterial species and gain mutual benefit through increased biofilm production, high levels of tolerance to antimicrobial agents, and enhanced virulence (Hotterbeekx et al., 2017; Orazi and O'Toole, 2019). In this context and in contrast with the clinical situation (DeLeon et al., 2014), planktonic co-culture of P. aeruginosa with S. aureus usually results in the inhibition of staphylococcal growth and virulence factor production. This is mainly as a consequence of exposure to small-molecule exoproducts produced by P. aeruginosa, including phenazines, hydrogen cyanide, N-acylhomoserine lactones (AHLs), and 2-alkyl-4(1H)-quinolones (AQs) (Hotterbeekx et al., 2017; Orazi and O'Toole, 2019). In P. aeruginosa AHLs and AQs co-ordinately control secondary metabolite production, virulence gene expression and biofilm development through co-ordinated cell-cell communication or quorum sensing (QS) (Williams and Camara, 2009). However, P. aeruginosa QS signal molecules can also impact on other microbes including S. aureus sharing the same ecological niche. For example, N-(3-oxododecanoyl)-L-homoserine lactone and its base-catalyzed rearrangement product 5-hydroxyethyl-3-decanoyltetramic acid inhibit S. aureus growth (Kaufmann et al., 2005) and, at sub-inhibitory concentrations, virulence factor production through blockade of agr-dependent QS (Qazi et al., 2006; Murray et al., 2014).
P. aeruginosa also produces a diverse range of AQs and AQ N-oxides of various alkyl chain lengths and saturation states (Heeb et al., 2010). Apart from their role in AQ-dependent QS, AQs exhibit a range of biological activities from immune modulation and cytochrome inhibition to antimicrobial and iron-chelating activities (Hooi et al., 2004; Diggle et al., 2007; Nguyen and Oglesby-Sherrouse, 2016). For example, the AQ N-oxide 2-heptyl-4-hydroxyquinoline N-oxide (HQNO) (Figure 1) and the unsaturated analogue (E)-4-hydroxy-2-(non-1-en-1-yl)quinoline 1-oxide are effective inhibitors of S. aureus growth. Paradoxically, these N-oxides increase S. aureus persistence since, at sub-inhibitory concentrations and prolonged growth, they generate small-colony variants (SCVs) that show increased tolerance to aminoglycoside antibiotics (Orazi and O'Toole, 2019). When treated with HQNO, S. aureus biofilms become more sensitive to fluoroquinolones and membrane-targeting antibacterial agents but exhibit reduced susceptibility to vancomycin (Orazi and O'Toole, 2019; Orazi et al., 2019).
Figure 1.
Structures of the AQs, PQS, HHQ, and HQNO and a generalized structure for the QZN series
Given the global health threat posed by multi- and pan-antibiotic-resistant bacterial pathogens in single or polymicrobial infections, novel antibacterial agents are required that have unique modes of action and potent efficacy against biofilm-centered infections. In this context, the attenuation of bacterial virulence and biofilm formation have been viewed as attractive targets for the development of “anti-virulence” or “anti-pathogenic” agents that could be used individually or as antibiotic “adjuvants” in combination therapy (Williams, 2017). QS systems have therefore been considered promising targets for drug development (Rampioni et al., 2014; Williams, 2017).
In P. aeruginosa, AQ-dependent QS regulates virulence, biofilm development, and secondary metabolite production. P. aeruginosa mutants defective in AQ biosynthesis or perception are attenuated in experimental animal infection models (Déziel et al., 2005; Rampioni et al., 2010; Starkey et al., 2014; Dubern et al., 2015). Furthermore, the presence of AQs in sputum, blood, and urine correlates with clinical status cystic fibrosis patients suffering with chronic P. aeruginosa lung infections (Barr et al., 2015, 2017). Consequently, there has been considerable interest in AQ-dependent QS as a P. aeruginosa drug target and in particular the transcriptional regulator PqsR (also called MvfR) (Ilangovan et al., 2013; Starkey et al., 2014; Williams, 2017). 2-Heptyl-3-hydroxy-4(1H)-quinolone (PQS) and its biosynthetic precursor HHQ (2-heptyl-4-hydroxyquinoline) (Figure 1) function as the primary QS signal molecules in this system (Xiao et al., 2006; Heeb et al., 2010; Ilangovan et al., 2013). PQS and HHQ and their C9 congeners bind to and activate PqsR to trigger a positive feedback loop increasing expression of the pqsABCDE operon and, hence, AQ biosynthesis. PQS is also a ferric iron chelator and controls virulence, biofilm development, and the iron-starvation response via PqsR-dependent and PqsR-independent pathways (Diggle et al., 2007; Rampioni et al., 2016).
Potent inhibition of PqsR-mediated pqsABCDE expression and, hence, AQ biosynthesis has been achieved through ligand-based design, which has yielded PqsR antagonists ranging from quinolinones and quinazolinones (QZNs; Figure 1) to benzamide-benzimidazole and hydroxybenzamide derived molecules (Soukarieh et al., 2018). In addition to attenuating virulence, PqsR inhibitors have the added advantage of reducing the formation of antibiotic-tolerant persister cells (Starkey et al., 2014). From a series of (3H)-quinazolinones (3aza-AQs) screened as potential PqsR antagonists (Ilangovan et al., 2013), 12 QZN PqsR antagonists were identified, including 3-NH2-7-Cl-C9-QZN (IC50 5 μM; Table 1 compound 19), which was shown to inhibit virulence factor production and biofilm development in P. aeruginosa (Ilangovan et al., 2013). Confirmation that 19 acted as a competitive PqsR antagonist was obtained by determination of the complex crystal structure of 19 bound within the PqsR ligand-binding domain. This revealed a similar orientation within the hydrophobic ligand-binding pocket as native AQ agonists (Ilangovan et al., 2013).
Table 1.
QZN and BTD chemical structures and S. aureus growth-inhibitory properties
 | ||||||
|---|---|---|---|---|---|---|
| No. | R2 | R3 | R6 | R7 | Abbreviation | MIC (μM) |
| 1 | n-C7H15 | H | H | H | C7-QZN | >100 |
| 2 | n-C9H19 | H | H | H | C9-QZN | >100 |
| 3 | n-C9H19 | H | H | F | 7F-C9-QZN | >100 |
| 4 | n-C9H19 | H | H | Cl | 7Cl-C9-QZN | >100 |
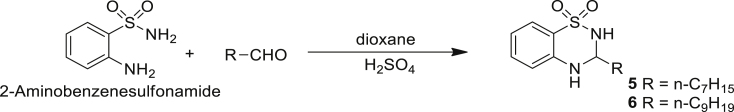
| 5 | n-C7H15 | H | H | H | C7-BTD | >100 |
| 6 | n-C9H19 | H | H | H | C9-BTD | >100 |
| 7 | n-C5H11 | NH2 | H | H | 3-NH2-C5-QZN | >100 |
| 8 | n-C7H15 | NH2 | H | H | 3-NH2-C7-QZNa | >100 |
| 9 | n-C9H19 | NH2 | H | H | 3-NH2-C9-QZNa | >100 |
| 10 | n-C11H23 | NH2 | H | H | 3-NH2-C11-QZNa | >100 |
| 11 | NH2 | H | H | 3-NH2-C9:3-QZN | >100 | |
| 12 | Ph(CH2)3 | NH2 | H | H | 3-NH2-PhC3-QZN | >100 |
| 13 | Cy(CH2)3 | NH2 | H | H | 3-NH2-CyC3-QZN | >100 |
| 14 | NH2 | H | H | 3-NH2-(2,5-dioxa-C9)-QZN | >100 | |
| 15 | NH2 | H | H | 3-NH2-(3-Me-C8)-QZN | >100 | |
| 16 | NH2 | H | H | 3-NH2-(7-Me-C8)-QZN | >100 | |
| 17 | n-C1H3 | NH2 | H | H | 3-NH2-C1-QZN | >100 |
| 18 | n-C9H19 | NH2 | H | F | 3-NH2-7F-C9-QZNa | >100 |
| 19 | n-C9H19 | NH2 | H | Cl | 3-NH2-7Cl-C9-QZNa | >100 |
| 20 | n-C9H19 | NH2 | Cl | H | 3-NH2-6Cl-C9-QZN | >100 |
| 21 | n-C9H19 | NH2 | F | H | 3-NH2-6F-C9-QZN | >100 |
| 22 | n-C9H19 | NH2 | F | F | 3-NH2-6F,7F-C9-QZNa | >100 |
| 23 | n-C9H19 | NH2 | OMe | OMe | 3-NH2-6OMe,7OMe-C9-QZN | >100 |
| 24 | n-C9H19 | NH2 | H | CF3 | 3-NH2-7CF3-C9-QZN | >100 |
| 25 | Ph(CH2)3 | NH2 | H | Cl | 3-NH2-7Cl-PhC3-QZNa | >100 |
| 26 |  |
NH2 | H | Cl | 3-NH2-7Cl-(Tri-OMeC6H2)C2-QZN | >100 |
| 27 |  |
NH2 | H | Cl | 3-NH2-7Cl-(3-Ind) C2-QZN | >100 |
| 28 | NH2 | H | Cl | 3-NH2-7Cl-BiPhC1-QZN | >100 | |
| 29 | NH2 | H | Cl | 3-NH2-7Cl-PhOBn-QZN | >100 | |
| 30 | n-C9H19 | (CH2)2NH2 | H | H | 3-C2NH2-C9-QZNa | 50 |
| 31 | n-C9H19 | (CH2)2NH2 | Cl | H | 3-C2NH2-6Cl-C9-QZNa | 25–50 |
| 32 | n-C9H19 | (CH2)2NH2 | H | Cl | 3-C2NH2-7Cl-C9-QZNa | 12.5 |
| 33 | Ph(CH2)3 | (CH2)2NH2 | H | Cl | 3-C2NH2-7Cl-PhC3-QZNa | 100 |
| 34 | n-C9H19 | (CH2)3NH2 | H | Cl | 3-C3NH2-7Cl-C9-QZNa | 6.25c |
| 35 | n-C9H19 | (CH2)4NH2 | H | Cl | 3-C4NH2-7Cl-C9-QZN | 12.5 |
| 36 | n-C7H15 | OMe | H | H | 3-OMe-C7-QZN | >100 |
| 37 | n-C9H19 | OMe | H | H | 3-OMe-C9-QZN | >100 |
| 38 | n-C9H19 | OMe | H | Cl | 3-OMe-7Cl-C9-QZNb | >100 |
| 39 | n-C9H19 | OMe | H | F | 3-OMe-7F-C9-QZNb | >100 |
| 40 | n-C9H19 | OMe | F | F | 3-OMe-6F,7F-C9-QZNb | >100 |
| 41 | n-C7H15 | NMe2 | H | H | 3-NMe2-C9-QZN | >100 |
| 42 | n-C9H19 | OH | H | H | 3-OH-C9-QZNb | 100 |
| 43 | n-C9H19 | OH | H | F | 3-OH-7F-C9-QZNb | 100 |
| 44 | n-C9H19 | OH | H | Cl | 3-OH-7Cl-C9-QZNb | 50 |
| 45 | n-C9H19 | CH2OH | H | Cl | 3-C2OH-7Cl-C9-QZN | >100 |
indicates known PqsR antagonist.
indicates known PqsR agonist (llangovan et al., 2013).
MIC increased to 25 μM in the presence of 10% FBS.
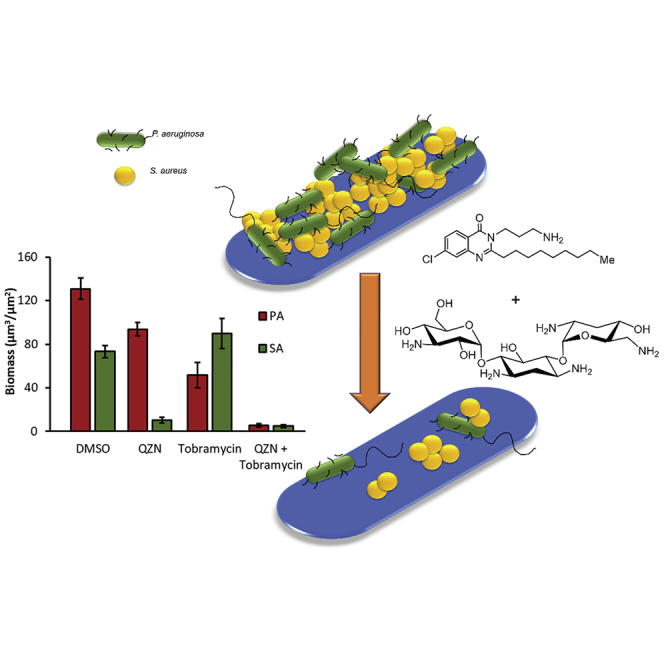
Given that 19 inhibits AQ-dependent QS in P. aeruginosa and that structurally related AQNOs display anti-staphyloccocal activity, we screened our library of 45 QZNs and related benzo[1,2,4]thiadiazines for activity against S. aureus and a range of other clinically relevant bacteria. Here, we present data that indicate that a subset of QZNs structurally related to 19, and most notably 3-C3NH2-7Cl-C9-QZN 34 (PqsR IC50 15 μM), kill planktonic Gram-positive pathogens including S. aureus, Staphylococcus epidermidis, Streptococcus pyogenes, and Clostridioides difficile but not Gram-negative bacteria such as P. aeruginosa and Escherichia. coli. Moreover, 34 inhibited S. aureus biofilm formation, damaged established S. aureus biofilms, and perturbed P. aeruginosa biofilm development. In mixed S. aureus and P. aeruginosa biofilms, 34 provided partial protection for P. aeruginosa. Although P. aeruginosa protected S. aureus from tobramycin in mixed biofilms, the combination of aminoglycoside and 34 eradicated the mixed-species biofilm. We also show that the mechanism of action of 34 involves binding and perturbation of the bacterial cytoplasmic membrane and disruption of transmembrane membrane potential.
Results
Structure-activity relationship determination reveals the QZN structural features required for S. aureus growth inhibition
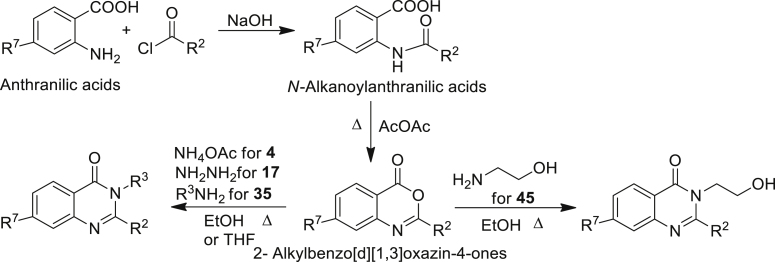
To evaluate the antibacterial activity of the QZN series, S. aureus strain RN6390B was first grown in the presence of 100 μM of each compound (Table 1). These included 43 QZNs and two related benzo[1,2,4]thiadiazines (BTDs) that were synthesized (see Schemes 1 and 2 for examples) with mono-, di-, tri-, and tetra-ring substitutions. For each QZN that abolished growth, a minimum inhibitory concentration (MIC) was generated (Table 1). QZNs with varying 2-alkyl chains and especially with the 3-position unsubstituted (1–4) or substituted with NH2 (7–29), OMe (36–40), NMe2 (41), and CH2OH (45) had MICs in excess of 100 μM. These results strongly suggest that endocyclic hydrazide and O-methyl hydroxamate functionalities would result in inactive analogs. Two related BTD derivatives (5 and 6) incorporating cyclic sulfonamide (-SO2NH) (sultam) motif were also inactive. Compound 42 (3OH-C9-QZN), however, gave a MIC of 100 μM that improved with chlorine substitution at the 7-position (44). Growth-inhibitory activity improved markedly when the 3-position was substituted with an aminoalkyl [(CH2)nNH2, where n = 2, 3, or 4] group (30–35)]. The MICs for this class of QZN suggested that, besides an aminoalkyl substituent at position-3, the presence of a 9-carbon alkyl chain at position-2 and a chlorine at position-7 were required for optimal activity. These systematic modifications resulted in the lead compounds 32 (3-C2NH2-7Cl-C9-QZN) and 34 (3-C3NH2-7Cl-C9-QZN), which gave the lowest MICs (12.5 μM [4.4 μg/mL] and 6.25 μM [2.3 μg/mL], respectively). We next sought to improve the potency of 34 by extending the aminoalkyl chain from C3NH2 to C4NH2 (35); however, this increased the MIC 2-fold to 12.5 μM.
Scheme 1.
Synthesis of 2-alkyl-4(3H)-quinazolinones (4, 17, 35, and 45)
Scheme 2.
Synthesis of 3-alkyl-3,4-dihydro-2H-benzo[e][1,2,4]thiadiazine 1,1-dioxide (5 and 6)
Growth-inhibitory properties of 3-C2NH2-7Cl-C9-QZN 32 and 3-C3NH2-7Cl-C9-QZN 34
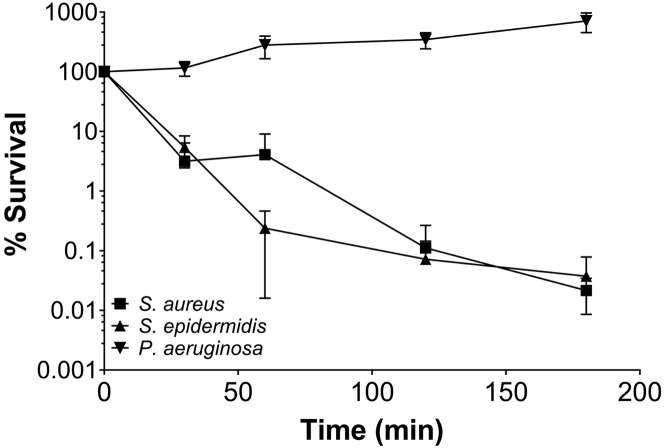
Compound 32 was assessed for the ability to inhibit the growth of clinically relevant bacterial pathogens. The MIC data shown in Table S1 demonstrate that 32 inhibits the growth of Gram-positives, including methicillin-sensitive S. aureus (MSSA) strains RN6390B and SH1000 and S. aureus hospital- (BH1CC) and community-acquired (USA300) methicillin-resistant S. aureus (MRSA) strains, as well as S. epidermidis, Streptococcus pyogenes, Streptococcus agalactiae and C. difficile. Nine recent MRSA isolates resistant to diverse conventional antibiotics were also tested and found to be similarly susceptible to 32, with a MIC of 12.5 μM; 32 was, however, inactive as a growth inhibitor of the Gram negatives, E. coli, and P. aeruginosa. To determine whether the active QZNs were bacteriostatic or bactericidal, S. aureus, S. epidermidis, and P. aeruginosa were assessed in a bactericidal assay using a dose of 50 μM of 34 (8× MIC for S. aureus). Figure 2 shows that the number of viable planktonic S. aureus and S. epidermidis cells was reduced by three logs within 3 h, indicating that 34 is indeed bactericidal against both S. aureus and S. epidermidis, whereas the viable count for P. aeruginosa increased over the equivalent time frame.
Figure 2.
3-C3NH2-7Cl-C9-QZN (QZN 34) is bactericidal for S. aureus and S. epidermidis but not for P. aeruginosa
Planktonic S. aureus USA300 JE2, S. epidermidis 1457 or P. aeruginosa PAO1 were treated with 50 μM (8× the S. aureus MIC) of compound 34. The data presented are the mean viable counts (CFU) per mL from three independent experiments ±SD.
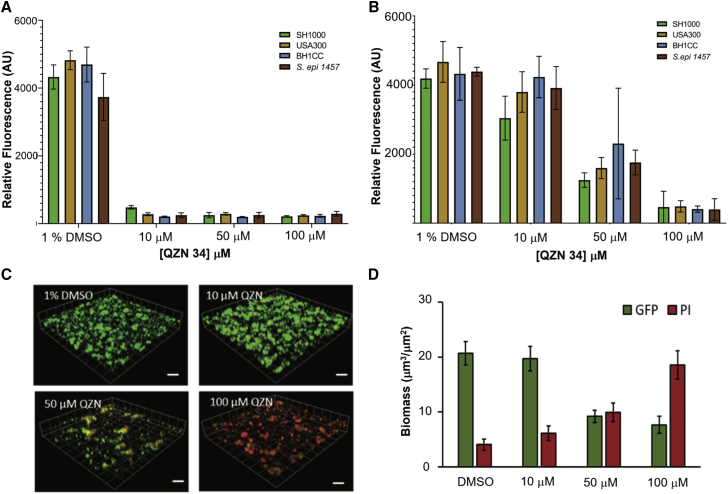
Anti-staphylococcal biofilm properties of 3-C3NH2-7Cl-C9-QZN 34
The impact of 34 on biofilms formed by S. aureus USA300, the hospital-acquired MRSA strain BH1CC, as well as the laboratory prototype S. aureus and S. epidermidis strains SH1000 and 1457 was investigated. Biofilms were treated with 34 either immediately after cell attachment or once the biofilm had established (24 h post-attachment). Conversion of the redox-sensitive dye resazurin to the fluorescent product resorufin was used as an indicator of biofilm metabolic activity. Figure 3A indicates high fluorescence for all four staphylococcal biofilms exposed to the DMSO vehicle control, suggesting that a metabolically active biofilm had established after 24-h growth. Exposure to as little as 10 μM of 34 immediately after attachment (Figure 3A) resulted in a substantial loss of metabolic activity. Addition of 34 to a mature staphylococcal biofilm (Figure 3B), at a concentration of 50 μM (8× MIC), resulted in a 2-fold reduction in fluorescence compared with the vehicle control, whereas treatment with 100 μM 34 (16× MIC) reduced fluorescence to background for all four staphylococcal strains tested. Although resazurin provides useful information on biofilm viability, alternative methods are required to determine the impact of 34 on biofilm architecture. To further investigate our initial findings, biofilms were established over 24 h with S. aureus cells (SH1000) constitutively expressing green fluorescent protein (GFP) and counterstained with propidium iodide (PI) for live/dead quantification after challenge with increasing concentrations of 34 for 2 h. Confocal microscopy was used to generate the representative images shown in Figure 3C and quantified with respect to biofilm biomass and dead cells (Figure 3D). The data clearly show that S. aureus biofilms treated with the vehicle control had a biomass composed of cells expressing GFP with no visible red fluorescence (PI), indicative of viable, undamaged S. aureus cells and biofilms. Treatment with increasing concentrations of 34 revealed a concentration-dependent increase in red fluorescence (PI) and a reciprocal reduction in green fluorescence, confirming that the bacterial cells were either severely damaged or dead. In addition, viable count experiments demonstrated that whereas 34 completely inhibited biofilm formation, mature biofilms treated with 34 were more refractory, exhibiting an ∼1.5 log reduction in viable counts (Figure S1). Taken together, the data shown in Figures 2, 3, and S1 show that 34 can damage and kill both planktonic and biofilm S. aureus.
Figure 3.
3-C3NH2-7Cl-C9-QZN 34 inhibits staphylococcal biofilm formation
(A and B) Resazurin was used to assess the metabolic activity of S. aureus MRSA (USA300 and BH1CC), MSSA (SH1000) strains, and S. epidermidis staphylococcal biofilms treated with increasing concentrations of 34 directly after attachment (A) or after formation of mature biofilms (B). Data are means from 3 independent experiments ± SD.
(C) 3D confocal microscope images of mature GFP-labeled S. aureus biofilms treated with 34 for 4 h.
(D) Quantification of biofilm biomasses shown in (C). Dead cells and extracellular DNA (red) were stained with propidium iodide (PI; 2μM). Scale bars, 50 μm. Data are means from 3 independent experiments ± SD.
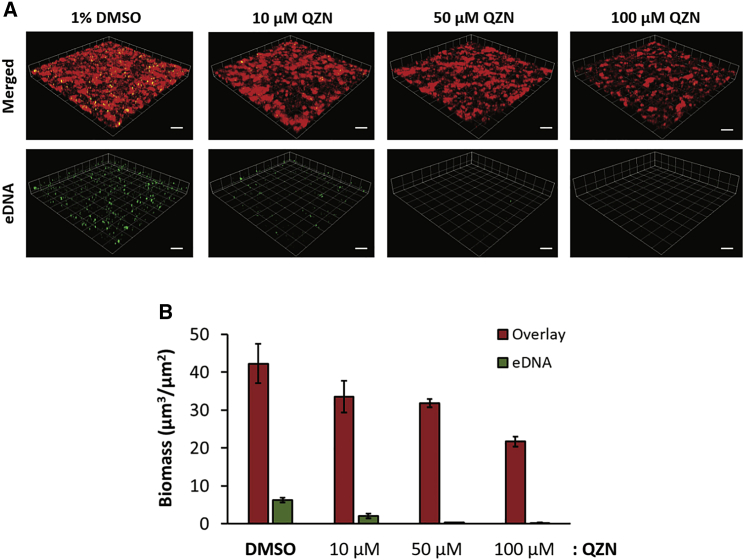
3-C3NH2-7Cl-C9-QZN 34 perturbs P. aeruginosa biofilm development
Since PqsR inhibitors including 19 (3NH2-7Cl-C9-QZN) inhibit P. aeruginosa biofilm development (Ilangovan et al., 2013), we examined the effect of 34, which has an IC50 for PqsR of 15.5 μM Figure 4 shows the impact of 34 on P. aeruginosa biofilm formation. Biofilm biomass and extracellular DNA (eDNA) content were reduced by as little as 10 μM 34, with a substantial reduction in biomass (∼50%) clearly apparent at 100 μM 34 and with very little eDNA detectable (Figure 4).
Figure 4.
Effect of 3-C3NH2-7Cl-C9-QZN 34 on P. aeruginosa PAO1 biofilm formation
(A) 3D confocal microscope images showing mCherry-tagged P. aeruginosa biofilms grown in RPMI 1640 for 24 h in the presence of either DMSO (vehicle control) or QZN 34 (10, 50, and 100 μM). Scale bars, 50 μm.
(B) Biomass quantification of P. aeruginosa biofilms. Extracellular DNA was stained with Yoyo-1 (2 μM). Data are means from 3 independent experiments ± SD.
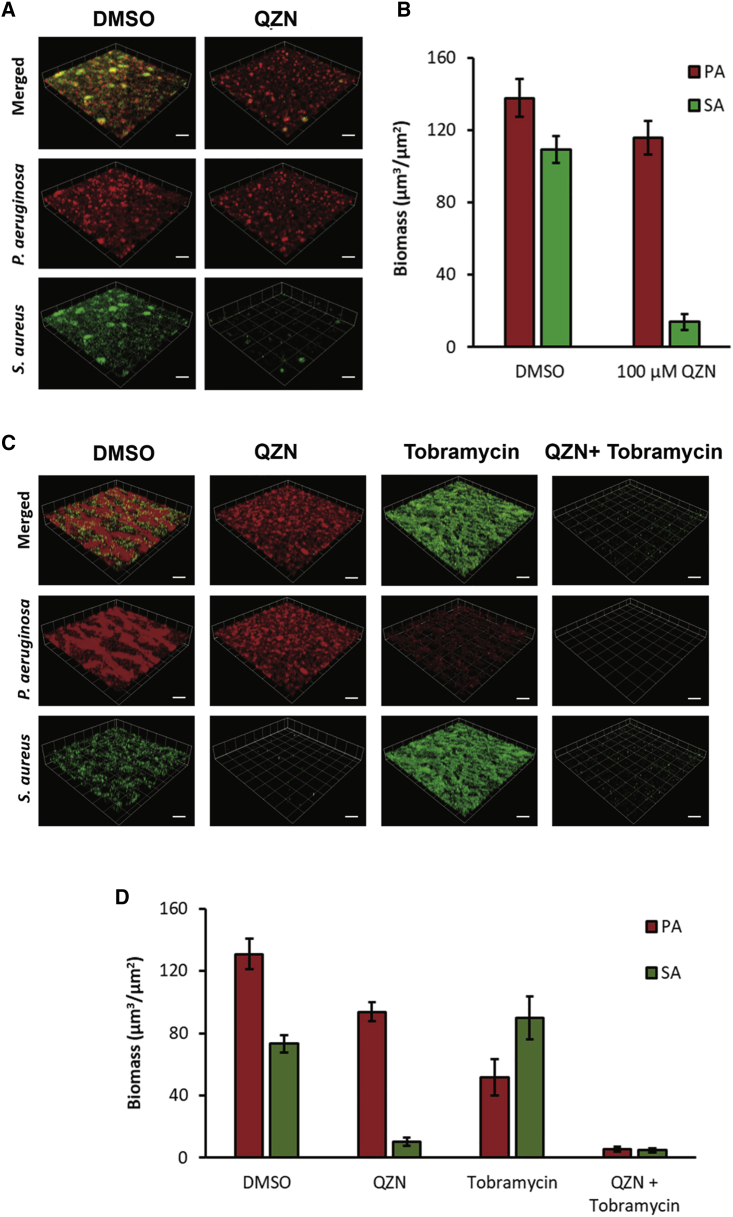
S. aureus protects P. aeruginosa in a mixed-species biofilm from the effects of 3-C3NH2-7Cl-C9-QZN 34
Fluorescently labeled S. aureus (GFP, green) and P. aeruginosa (mCherry, red) were co-cultured to form mixed biofilm. Figure 5A shows that the biomass of the mixed biofilm is significantly greater (∼5×) than that formed by each individual organism (compare Figures 3D and 4B with 5B). When incubated with 34 at 100 μM, GFP fluorescence is reduced by ∼90%, suggesting that S. aureus, as with the single-species biofilm (Figure 3), succumbs to treatment with 34 (Figures 5A and 5B). However, despite a prolonged treatment time (4 h instead of 2 h) the red fluorescence of P. aeruginosa is only marginally reduced (∼15%), suggesting that in the presence of S. aureus P. aeruginosa is protected to some extent from the effects of 34.
Figure 5.
Impact of 3-C3NH2-7Cl-C9-QZN 34 and tobramycin on P. aeruginosa and S. aureus mixed biofilms
(A and B) 3-C3NH2-7Cl-C9-QZN 34 confers partial protection of P. aeruginosa (PA) in a mixed-species biofilm with S. aureus (SA). Biofilms were allowed to form on glass after sequential inoculation with GFP-labeled S. aureus SH1000 (green) and mCherry labeled P. aeruginosa PAO1 (red) in a 10:1 ratio in RPMI 1640 containing either DMSO (vehicle control) or QZN 34 (100 μM) and incubated for 48 h.
(C and D) Combined effect of 3-C3NH2-7Cl-C9-QZN 34 and tobramycin on a mixed-species biofilm. The biofilm was allowed to form on glass after sequential inoculation with GFP-labeled S. aureus SH1000 (green) and mCherry labeled P. aeruginosa PAO1 (red) in a 10:1 ratio in RPMI 1640 containing either DMSO (vehicle control) or QZN 34 (100 μM) and incubated for 48 h. For some experiments, tobramycin (100 μg/mL) was also added, and the biofilms were cultured for a further 4 h.
(A and C) show 3D confocal microscope images. (B and D) show biomass quantification of the mixed-species biofilms after treatment. Scale bars, 50 μm. Data are means from 3 independent experiments ± SD.
3-C3NH2-7Cl-C9-QZN and tobramycin in combination are highly active against mixed biofilms
The aminoglycoside tobramycin is commonly used to treat chronic biofilm-associated P. aeruginosa infections in cystic fibrosis patients (Beaudoin et al., 2017). To determine whether 3-C3NH2-7Cl-C9-QZN 34 in combination with tobramycin could eradicate both P. aeruginosa and S. aureus in mixed biofilms, we first examined the susceptibility of S. aureus and P. aeruginosa monoculture biofilms to 34 in the presence and absence of tobramycin (Figure S2). For both pathogens, the combination of tobramycin and QZN was more effective than either compound alone. When we co-cultured with P. aeruginosa in the presence of tobramycin alone, we observed that the S. aureus biomass was protected, whereas the P. aeruginosa biomass was reduced by ∼60% (Figures 5C and 5D). However, when 34 and tobramycin were used in combination, they reduced biofilm biomass by >95% for both bacterial species in the mixed biofilm (Figures 5C and 5D).
Cytotoxicity of 3-C2NH2-7Cl-C9-QZN 32 and 3-C3NH2-7Cl-C9-QZN 34 for eukaryotic cells
Since 34 showed excellent antibacterial activity, cytotoxicity for eukaryotic cells was assessed. Hemolysis assays (Figure S3A) indicated that 34 has an IC50 of 173 μM, which is ∼28 times higher than the MIC (6.25 μM) for S. aureus and ∼69 times the MIC generated for C. difficile (Table S1). Similar results were also observed with 32 (see Figure S3A). The non-growth-inhibitory P. aeruginosa PqsR-inhibitory QZN compound 19 was not hemolytic (Figure S3A). The release of lactate dehydrogenase from Jurkat cells after 24-h incubation with 34 is shown in Figure S3B. Data indicate that exposure to 34 resulted in an ∼30% loss of cytoplasmic lactate dehydrogenase at concentrations above 6.3 μM; however, the total amount of lactate dehydrogenase released did not increase above 50% in the presence of up to 100 μM of 34; therefore, an IC50 could not be generated. However, since the Jurkat cell assay medium includes 10% fetal bovine serum (FBS), we repeated the S. aureus MIC for 34 with FBS. This increased the MIC to 25 μM (Table 1), suggesting that the QZN binds to serum proteins, which would also reduce mammalian cytotoxicity. These data collectively suggest that 32 and 34 possess some limited selectivity for bacterial membranes.
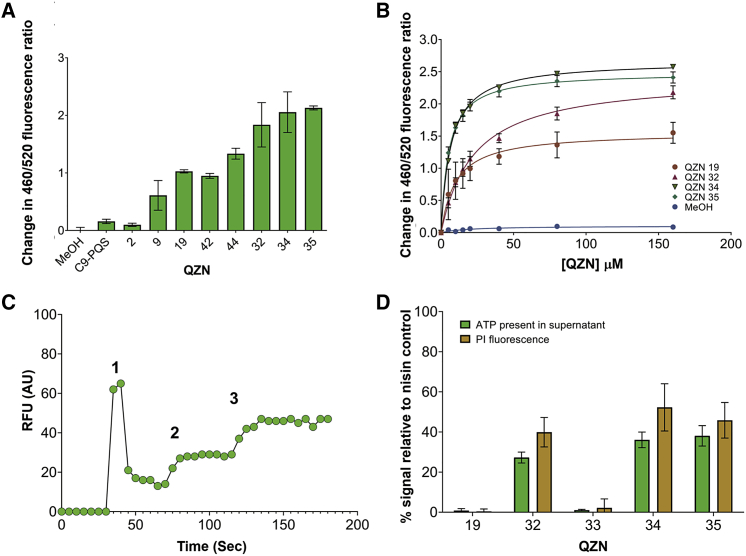
Mechanism of action of 3-C2NH2-7Cl-C9-QZN and 3-C3NH2-7Cl-C9-QZN for S. aureus
A potential explanation for QZN-mediated staphylococcal growth-inhibitory activities given the structural similarity to the AQs was reduced availability of ferric iron, since some AQs are ferric iron chelators. Although 3OH-C9-QZN 42 in common with PQS sequestered iron, neither 32 nor 34 possessed iron-chelating properties (Figure S4). The potency of 34 increased when the assay medium osmolality was reduced by replacing LB with sterile water. Under these conditions, 34 at 10 μM (∼1.5× MIC for S. aureus) resulted in a rapid 2.5 log reduction in viable S. aureus cells after only 1 h of exposure (Figure S5). The provision of ionic solutes (NaCl, KCl, or CaCl2) protected S. aureus from the bactericidal activity of 34. These data suggested that 3-C3NH2-7Cl-C9-QZN 34 was likely to bind to and perturb the staphylococcal cell membrane.
Antibacterial agents that target the cytoplasmic membrane often disrupt essential electrical potentials, such as the transmembrane potential, that that are required to maintain essential cellular processes. Consequently, we first determined whether the QZNs interact with staphylococcal membranes, by utilizing the fluorescent dye di-8-ANEPPS, to monitor changes in dipole potential that permit quantification of membrane binding affinities and information on binding specificity (Murray et al., 2014). Figure 6A shows the changes in fluorescence ratio after the addition of a series QZNs based on their structures and SAR for S. aureus growth inhibition initially at a fixed concentration (40 μM). The PqsR agonist C9-PQS and closely related QZN, C9-QZN 2 had little impact on membrane dipole potential, which increased following the introduction of 3NH2, 3OH, and 7Cl substituents as for 9, 19, 42, and 44, although these compounds exhibited either no (9, 19, and 42) or very weak (44) growth-inhibitory activity. The greatest changes in dipole potential were noted for the growth-inhibitory QZNs 32, 34, and 35. Subsequently, binding curves were constructed for selected QZNs by plotting changes in dipole potential against concentration (Figure 6B). These show that 32, 34, and 35 bound in a saturable-specific manner with similar high affinities (Kds 22 ± 1.7, 6.6 ± 0.2, and 5.2 ± 0.3 μM, respectively) and that the data obtained fit a hyperbolic function consistent with a non-cooperative, single-site receptor-binding model (Figure 6B).
Figure 6.
Affinity for and perturbation of the S. aureus cytoplasmic membrane by selected AQs and QZNs
(A) QZNs disturb the membrane dipole potential. Changes in dipole potential were determined using di-8-ANEPPS to measure the variation in the fluorescence ratio R(460/520) at a fixed concentration of 40 μM for each compound. Data are means from 3 independent experiments±SD
(B) Binding curves as a function of QZN concentration. The binding profiles for MeOH, 2, 19, 32, 34, and 35. The data plotted are the mean values of three independent experiments; error bars represent standard deviations.
(C) Depolarization of transmembrane potential by 34. (1) The cationic fluorescent dye DiSC3(5) was added to cells, followed (2) by 10 μM of test compound. Complete depolarization of the membrane potential was achieved by the addition of 5 μM valinomycin (3). Experiments were repeated on three independent occasions, with representative data shown.
(D) Membrane permeabilization. Cytoplasmic ATP release and PI fluorescence relative to nisin (0.6 μM; positive control) after treatment of S. aureus with 10 μM of compounds 19, 32, 33, 34, and 35. The data plotted are the mean values of three independent experiments; error bars represent standard deviations.
The bacterial membrane potential (ΔΨ) is a key component of the proton motive force (PMF) and is generated by selective ionic inclusion/expulsion. The cationic fluorescent dye Disc(3)5 can be used to assess the membrane-perturbing effects of the QZNs on ΔΨ. Figures 6C and S6 show that, after initial uptake and quenching of the fluorescent dye, addition of the growth-inhibitory QZNs 32 or 35 results in an increase in fluorescence as the cationic dye is released down a concentration gradient. In contrast, the non-growth-inhibitory QZN 19, despite its ability to bind to the membrane, does not increase fluorescence (Figure S6A). Addition of the ionophore valinomycin as a positive control (Figures 6C and S6) but not antibiotics such as novobiocin and chloramphenicol (which do not target the cytoplasmic membrane) confirmed dye release as dependent on ΔΨ depolarization. Although significant, the loss of ΔΨ after addition of either 32 or 35 is unlikely to fully account for rapid cell death (Figure 2A). Consequently, the impact of the QZNs on membrane integrity was also investigated by evaluating uptake of the membrane-impermeable dye PI and the loss of ATP. The data presented in Figure 6D show that incubation with 32, 34, or 35, but not 19 or 33, resulted in significant cellular ATP loss alongside the reciprocal uptake of PI.
Discussion
To discover compounds capable of attenuating biofilm formation and rendering biofilms more susceptible to conventional antibiotics for S. aureus individually and in co-culture with P. aeruginosa, we screened our library of QZNs for staphylococcal growth inhibition. Whereas 36 out of 45 compounds screened had MICs >100 μM, 3-OH-7Cl-C9-QZN 44 had a MIC of 50 μM for S. aureus. However, for P. aeruginosa, this compound is a PqsR agonist (Ilangovan et al., 2013) and therefore likely to enhance rather than inhibit P. aeruginosa virulence and biofilm formation. Replacement of the 3-hydroxy with 3-amino converted 3-OH-7Cl-C9-QZN 44 from a PqsR agonist to a potent antagonist (3-NH2-7Cl-C9-QZN 19) (Ilangovan et al., 2013). Although the amino substitution at the 3-position failed to improve the MIC for S. aureus, anti-staphylococcal activity was markedly enhanced by an aminoalkyl group (30–32; 34, 35), with 34 being the most potent compound (MIC 6.25 μM; 2.3 μg/mL). The MICs for this class of QZN suggested that, besides an aminoalkyl substituent at position-3, the presence of a 9-carbon alkyl chain at position-2 and a chlorine at position-7 were required for optimal activity. Whereas these compounds are PqsR antagonists with 30 being the most potent (IC50 9.3 μM), compounds 32 and 34 were more active toward staphylococci and other Gram-positive pathogens while retaining good PqsR-inhibitory activity (IC50s 19.3 and 15.5 μM, respectively; Ilangovan et al., 2013). These compounds were therefore selected for further evaluation. Although bactericidal for S. aureus, 32 and 34 do not inhibit P. aeruginosa planktonic growth. However, PQS-dependent QS regulates the release of eDNA, which is an important component of the P. aeruginosa biofilm extracellular matrix and is involved in the attachment, aggregation, and stabilization of microcolonies during P. aeruginosa biofilm formation (Allesen-Holm et al., 2006). eDNA also contributes to the tolerance of biofilms toward antibiotics such as tobramycin (Chiang et al., 2013; Wilton et al., 2015). 3-C3NH2-7Cl-C9-QZN 34 substantially inhibited P. aeruginosa biofilm formation and reduced eDNA to almost undetectable levels, consistent with its activity as a PqsR inhibitor.
Apart from its bactericidal activity against planktonic S. aureus strains as well as S. epidermidis, 34 substantially inhibited the metabolic activity of staphylococcal biofilms at concentrations as low 10 μM, while at 100 μM it effectively damaged mature biofilms. In mixed biofilms with P. aeruginosa, 34 eradicated S. aureus, although in this context it showed reduced efficacy against P. aeruginosa. Interspecies interactions have long been known to impact on the sensitivity profiles of bacteria to antimicrobial agents within multispecies biofilms via different mechanisms (Orazi and O’Toole, 2019). However, here, it is unclear whether the increased resistance of P. aeruginosa is simply due to the much greater biomass of the mixed- compared with the single-species biofilms so reducing the dose of 34 or whether it is a consequence of sequestration into staphylococcal membranes, given the mode of action of the QZN. Alternatively, the presence of S. aureus within the biofilm may alter the physiology of P. aeruginosa, resulting in a more QZN-resistant phenotype. In this context, cell aggregation caused by the binding of S. aureus protein A adhesin to the P. aeruginosa exopolysaccharide Psl has been reported to increase tobramycin resistance in P. aeruginosa (Beaudoin et al., 2017).
Following exposure of P. aeruginosa cell-free culture supernatants, S. aureus biofilms have been reported to become more sensitive to membrane-permeabilizing antibiotics and biocides (Orazi et al., 2019). This increase was associated with HQNO as well as the P. aeruginosa siderophores pyochelin and pyoverdine. HQNO biosynthesis depends on the PqsR-dependent expression of the pqsABCDE operon. Consequently, PqsR antagonists such as 34 inhibit the production of all AQs including HQNO as well as reducing siderophore production, since PQS also regulates the latter (Rampioni et al., 2016). Therefore, in mixed biofilms with P. aeruginosa, the susceptibility of S. aureus to the membrane-active 34 cannot be enhanced by HQNO, given that the QZN at 100 μM (over six times the IC50 for PqsR) is likely to have abolished HQNO production when provided following the initial seeding inoculation of both species in the biofilm assay. However, in mixed biofilms under these experimental conditions, 34 appears to be more active in reducing the S. aureus biofilm biomass than in the single-species biofilm (∼7-fold compared with ∼4-fold). Further work will therefore be required to determine whether the dual properties of 34 as both an inhibitor of HQNO production and a membrane-permeabilizing agent are counter-productive in a single antimicrobial agent in mixed biofilms at lower concentrations.
Alternative advantages of the 3-aminoalkyl-substituted QZNs as inhibitors of HQNO production by P. aeruginosa in biofilm co-culture is that this should avoid HQNO-mediated inhibition of the electron transport chain, which reduces the susceptibility of S. aureus to protein synthesis-inhibitory antibiotics, including the aminoglycosides, tetracyclines, and macrolides (Orazi et al., 2019).
When the mixed biofilm was treated with tobramycin (100 μg/mL), the aminoglycoside reduced the P. aeruginosa biomass by ∼60%, but the S. aureus component of the biofilm was clearly protected by the presence of the Gram negative. Although the mechanism involved has not yet been elucidated, in mixed P. aeruginosa and Streptococcus biofilms tobramycin treatment appears to enhance streptococcal biofilm formation as a consequence of a tobramycin-driven reduction in rhamnolipid production (Price et al., 2015). Furthermore, S. aureus may also be protected by P. aeruginosa biofilm matrix components including the exopolysaccharides Psl and Pel as well as eDNA (Ciofu and Tolker-Nielsen, 2019). In contrast, when the mixed biofilm was incubated with tobramycin after pre-treatment with 34, most of the biofilm was killed, with less than 5% remaining. These data suggest that, in combination, tobramycin and the QZN are capable of eradicating both bacterial species in a mixed biofilm.
The bactericidal activity of the 3-aminoalkyl-substituted QZNs toward planktonic S. aureus suggested that these QZNs may target the cytoplasmic membrane. We therefore first investigated whether they possessed any affinity for the S. aureus cytoplasmic membrane by measuring dipole potential. This membrane potential originates via the molecular dipoles associated with carbonyl, oxygen-bonded phosphate moieties of phospholipids and is orientated toward water molecules at the membrane interface (O’Shea, 2005). Previously, we successfully used this technique to investigate the interactions of AHLs and tetramic and tetronic acids with both staphylococcal and eukaryotic membranes (Davis et al., 2010; Murray et al., 2014). For example, 3-tetradecanoyltetronic acid (C14-TOA), a membrane-active agent with a Kd of 4 μM, inhibited agr-dependent quorum sensing, reduced S. aureus colonization of human nasal epithelial cells, and showed efficacy in a staphylococcal experimental mouse infection model (Murray et al., 2014). Similar affinities for staphylococcal membranes were calculated from the binding curves for QZNs such as 34 and 35, consistent with the presence of a specific saturable receptor. These data also indicated that anti-staphylococcal activity positively correlated with a low-value Kd, given the reduced membrane affinity of inactive QZNs such as compound 2. Further investigation of the interactions of 32, 34, and 35 with the cytoplasmic membrane suggested that the Gram-positive bactericidal activity of these QZNs depends on a combination of membrane depolarization and pore formation leading to ATP release and cell death. Similar results have been reported for the clinically approved antibiotic daptomycin, although this lipopeptide antibiotic has also been shown to perturb fluid microdomains and, as a consequence, to interfere with membrane-bound cell wall and lipid biosynthesis (Müller et al., 2016). However, daptomycin lacks bactericidal activity against Gram-negative bacteria, and this appears to be independent of the outer-membrane permeability barrier (Miller et al., 2016). This may also be the case for the QZNs 32 and 34, given that their antagonistic activity for PqsR requires their uptake into the P. aeruginosa cytoplasm.
Our findings of the efficacy of tobramycin in combination with the lead QZNs toward P. aeruginosa and S. aureus in mixed biofilms offers additional opportunities to exploit the QZN scaffold for the development of adjuvant drugs capable, in combination with antibiotics, of eradicating chronic wound and lung infections caused by these problematic pathogens.
Significance
Staphylococcus aureus and Pseudomonas aeruginosa commonly cause biofilm-centered chronic infections in diverse body sites that are highly refractory to host immune defences and antibiotics. Their co-existence in polymicrobial biofilms within the same infection site often results in poor clinical outcomes. To survive in the same environmental niches, bacteria have evolved intricate regulatory networks that facilitate co-operative behavior through quorum sensing (QS) that facilitates evasion or suppression of other bacterial species as well as providing mutual benefit by forming antibiotic-tolerant biofilms. Given the global health threat posed by multi-antibiotic-resistant bacterial pathogens, novel antibacterial agents are required that have potent efficacy against biofilm-centered infections. In this context, QS systems have been viewed as attractive targets for drug molecules that inhibit biofilm formation and virulence and could be used alone or as adjuvants in combination with antibiotics. In P. aeruginosa, pseudomonas quinolone (PQS)-dependent QS can be attenuated via small-molecule antagonists targeting the transcriptional regulator PqsR. Here, we explored our library of quinazolinones (QZNs), which includes PqsR agonists and antagonists, for their activity against S. aureus alone, when co-cultured with P. aeruginosa, and in combination with tobramycin. We discovered that a subset of PqsR QZN antagonists not only perturbed P. aeruginosa biofilm development but also killed planktonic cells and effectively damaged S. aureus biofilms. Although P. aeruginosa protected S. aureus from tobramycin in mixed biofilms, the combination of the aminoglycoside antibiotic with a QZN eradicated mixed-species biofilms. The mechanism of action of the active QZNs toward S. aureus is shown to involve perturbation of the cytoplasmic membrane and dissipation of essential membrane electrical potentials. Our findings of the combined activity of tobramycin and the lead QZN toward P. aeruginosa and S. aureus in mixed biofilms offers additional opportunities to exploit the QZN scaffold for anti-infective drug development.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| S. aureus | (Novick et al., 1993)_ | RN6390 |
| S. aureus | (Horsburgh et al., 2002) | SH1000 |
| S. aureus | (Fey et al., 2013) | USA300 JE2 |
| S. aureus | (O’Neil et al., 2007) | BH1CC |
| S. aureus | (Holden, 2010) | TW20 |
| S. aureus | (Castillo-Ramírez et al., 2012) | HU6 |
| S. aureus | (Castillo-Ramírez et al., 2012) | HU24 |
| S. aureus | (Castillo-Ramírez et al., 2012) | IU4 |
| S. aureus | (Castillo-Ramírez et al., 2012) | IU9 |
| S. aureus | (Castillo-Ramírez et al., 2012) | IU12 |
| S. aureus | (Castillo-Ramírez et al., 2012) | DEU9 |
| S. aureus | (Castillo-Ramírez et al., 2012) | DEU19 |
| S. aureus | (Castillo-Ramírez et al., 2012) | DEU20 |
| S. epidermidis | This laboratory | 1455 |
| E. coli | This laboratory | MG1655 |
| P. aeruginosa | This Laboratory | PA01-N |
| S. pyogenes | (Scott et al., 1986) | JRS4 |
| S. agalactiae | (Valenti-Weigand et al., 1996) | 6313 |
| C. difficile | Ed Kuijper Collection | 8085054 |
| C. difficile | Ed Kuijper Collection | 8079089 |
| B. subtilis | Bacillus Genetic Stock Centre | 168 |
| Chemicals, peptides, and recombinant proteins | ||
| Müller–Hinton broth | Oxoid | Cat# CM0405 |
| L-cysteine | Oxoid | Cat# CM1135 |
| Brain Heart Infusion broth | Oxoid | Cat# LP0021 |
| Yeast Extract | Sigma-Aldrich | Cat# C7352 |
| C. difficile supplement | Oxoid | Cat# SR0096 |
| Luria Broth | Oxoid | Cat# CM0996B |
| Defibrinated Rabbit Blood | TCS biosciences | Cat# RB052 |
| Phosphate Buffered Saline | Sigma-Aldrich | Cat# P4417 |
| Triton X-100 | Sigma-Aldrich | Cat# T8787 |
| Resazurin | Sigma-Aldrich | Cat# R7017 |
| RPMI-1640 (Biofilm Studies) | Lonza | Cat# 12-918F |
| RPMI-1640 (Cell Culture) | Sigma-Aldrich | Cat# R7509 |
| Penicillin Streptomycin mix | Sigma-Aldrich | Cat# P0781 |
| Glutamine | Sigma-Aldrich | Cat# G7513 |
| Fetal Bovine Serum | Sigma-Aldrich | Cat# F9665 |
| Tobramycin | Sigma-Aldrich | Cat# T04014 |
| Propidium Iodide | Sigma-Aldrich | Cat# P4170 |
| Nisin | Sigma-Aldrich | Cat# N5764 |
| di-8-ANEPPS | Thermo-Fisher | Cat# D3167 |
| Disc3(5) | Thermo-Fisher | Cat# D306 |
| Valinomycin | Sigma-Aldrich | Cat# V0627 |
| Critical commercial assays | ||
| LDH Cytotoxicity Assay Kit | Biochain | Cat# K6330400 |
| Cell Titre-Glo kit | Promega | Cat# G7570 |
| Experimental models: Cell lines | ||
| Jurkat T cells | Stratech | PC-006-SIG |
| Recombinant DNA | ||
| pMMR | (Popat et al., 2012) | Constitutive mCherry fluorescent reporter |
| pBK-miniTn7-egfp | (Popat et al., 2012) | Constitutive eGFP fluorescent reporter |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| Zen | Zeiss, Germany | www.zeiss.com |
| Graphpad Prism 8.0 | GraphPad Software, USA | https://www.graphpad.com |
Resource availability
Lead contact
Requests for further information and requests for responding to material, resources and reagents should be directed to and will be fulfilled by the Lead Contact Paul Williams (paul.williams@nottingham.ac.uk).
Materials availability
There are restrictions to the availability of the QZN compounds and bacterial strains due to the lack of an external centralized repository for their distribution and our need to maintain the stock. We are glad to share the QZNs with reasonable compensation by requestor for its processing and shipping. Requests for QZNs and bacterial strains should be directed to the lead contact, Paul Williams (paul.williams@nottingham.ac.uk).
Experimental model and subject details
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in the key resources table. S. aureus, S. epidermidis, P. aeruginosa, E. coli and B. subtilis were grown in Müller–Hinton broth (MHB) S. aureus was also grown in MHB with 10% fetal bovine serum (FBS). Strep. pyogenes and Strep. agalactiae were grown statically in brain heart infusion (BHI) with 5% CO2. C. difficile isolates were grown anaerobically in BHI supplemented with yeast extract, L-cysteine and C. difficile supplement. Bacteria were grown at 37°C.
Mammalian cells
Jurkat T cells were grown at 37°C in the presence of 5% CO2. Growth medium was RPMI-1640 supplemented with a final concentration of 10% fetal bovine serum, 100 units penicillin, 0.1 mg/ml streptomycin and 2 mM glycine.
Method details
Bacterial strain susceptibility testing
MICs were determined by broth microdilution in the appropriate medium for bacterial growth.
Bactericidal activity
S. aureus, S. epidermidis and P. aeruginosa were grown overnight in Luria Broth, diluted 250-fold and grown to an OD600 0.5. Cells were diluted to 1×106 per ml in challenge medium (LB plus QZN at 8 × MIC) or shock media (sterile water with or without 10 or 50 μM QZN and without or with 160 mOSm NaCl, KCl, or CaCl2). Samples were removed at intervals and viable counts determined.
Cytotoxicity
Haemolytic activity was assessed by incubating defibrinated rabbit blood (TCS Biosciences) in phosphate buffered saline (PBS) for 60 min in the presence of test compound or Triton X-100 (positive control). Samples were pelleted and supernatants collected. % hemolytic activity was determined as A543 relative to the Triton X-100 treated control. Cytotoxicity for mammalian cells was measured using the BioChain® LDH cytotoxicity assay kit as per the manufacturer’s instructions. Briefly, 2 x 104 Jurkat cells were incubated for 24 h in the presence of test compound or the lysis control agent. The A490 was measured and percentage cytotoxicity calculated relative to the positive lysis control.
Chrome azurol S (CAS) iron chelation assay
A 0.5 ml aliquot of phosphate buffered saline (PBS; pH 7.4) without (as a reference) or with the relevant compound (50 μM) was mixed with 0.5 ml of CAS assay solution prepared according to Schwyn and Neilands (1987). The sample (S) and reference (R) absorbances were determined at 630 nm after 15 min incubation at room temperature. The percentage of iron-chelating activity was calculated by subtracting the sample A630 from that of the reference A630 value. Siderophore units are defined as [A(r) – A(s)/A(r)] x 100.
Biofilm metabolic activity
The metabolic activity of staphylococcal biofilms was quantified using the redox sensitive dye resazurin as described previously (Zapotoczna et al., 2017). Briefly, cells were grown overnight in BHI, diluted to an OD600 of 0.1 and aliquoted into a 96 well plate. Staphylococci were incubated at 37°C for 3 h to allow for cell attachment. The growth medium was removed and the wells were washed with PBS before fresh BHI was added. Biofilms were challenged with QZN immediately after bacterial attachment or after 24 h of static growth and biofilm development at 37°C. After treatment with a QZN, biofilms were cultured for a further 24 h before Reaszurin fluorescence was quantified.
Quantification of biofilm biomass
Biofilms were cultivated on borosilicate glass coverslips in 6-well microplates (Corning). P. aeruginosa strain PAO1 (Washington subline) and S. aureus strain SH1000, were transformed respectively with plasmids pMMR or pMRE147 and pBK-miniTn7-egfp that constitutively express mCherry and a green fluorescent protein (GFP or mClover3 respectively) (Popat et al., 2012; Schlechter et al., 2018). The fluorescently tagged strains were grown at 37°C for 16 h in RPMI-1640 (Lonza, Slough, UK), diluted 1:100 (v/v) in fresh medium and allowed to grow until an OD600 of ∼0.5 was reached. These cultures were diluted to OD600 0.1 for S. aureus and 0.01 for P. aeruginosa in RPMI-1640 and inoculated into 6 well microplates containing UV sterilised borosilicate glass coverslips (22 × 22 mm, thickness no.1) (VWR, Lutterworth, UK). Bacterial cells were seeded onto the coverslip surface at 37°C under static conditions for 1 h before cultures incubated with shaking for 48 h for S. aureus or 24 h for P. aeruginosa to form mature biofilms. For the S. aureus biofilm killing assay, the medium was discarded after 48 h incubation and replaced by fresh RPMI-1640 containing 3-C3NH2-7Cl-C9-QZN 34 or tobramycin followed by a further 4h incubation. For P. aeruginosa biofilm inhibition assays, 34 was added to the inoculum and incubated for 24 h prior to tobramycin addition and a further 4 h of incubation. For the co-culture biofilm experiments, microplates were inoculated first with S. aureus and incubated statically for 1 h followed by P. aeruginosa in a 10:1 ratio. 3-C3NH2-7Cl-C9-QZN was added and the cultures incubated for 48 h at 37°C. Where required, tobramycin was then added to the 48 h-old co-cultures by a further 4h incubation. After incubation stained for extracellular DNA (eDNA) with propidium iodide (PI) or YOYO-1 (2 μM). Coverslips were directly examined under a confocal laser scanning microscope (Zeiss LSM2, Zeiss, Oberkochen, Germany) using eGFP and mCherry modes at an excitation wavelengths of 488 nm and 555 nm. Imaging was carried out using Zen 2011 imaging software (Zeiss, Oberkochen, Germany). A total of 5 Z-stacked images were collected per coverslip. Sampling was conducted at random from the central portion of each coverslip. Biomass was calculated using Image J (NIH, Bethesda, MD, USA) and Comstat 2.1. Software package (www.comstat.dk, Lyngby, Denmark). The percentage of live cells was determined as described by Ou et al. (2016).
Biofilm viability assay
Staphyloccocal cells were recovered from biofilms on coverslip surfaces by gentle sonication in an ultrasonic bath for 5 min at a frequency of 37 kHz. The removal of biofilm was confirmed using confocal microscopy. Bacterial cells were collected and OD600nm adjusted to 0.1. Cell suspensions were serial diluted down to 10-8 and 10μl spotted onto LB agar plates. These were incubated at 30°C for 16 h followed by manual colony counting. Cell numbers were expressed as log10 CFU/ml.
Bacterial membrane permeabilization assays
Membrane permeabilization was evaluated by quantifying (a) ATP release (Higgins et al., 2005) and (b) the uptake of propidium iodide (PI). S. aureus strain USA300 JE2 was grown overnight in CYGP medium (Ji et al., 1995) washed with phosphate buffer (100 mM, pH 7), re-suspended to OD600 0.1 and incubated at 37°C with a range of concentrations of each compound for 1 h. Nisin (0.6 μM) was used as a positive control. Cell-free supernatants were assayed for ATP content using the Cell Titre-Glo kit (Promega). Staphylococcal cells were re-suspended in phosphate buffer (pH 7) containing PI (10 μg/ml), incubated in the dark at room temperature for 10 min. After washing, PI fluorescence (excitation: 575 nm; emission 630 nm) was quantified.
Membrane dipole potential
S. aureus membranes were prepared and labelled with the dipole potential fluorescent sensor 1-(3-sulfonatopropyl)-4-[β-2-(di-n-octylamino)-6-naphthylvinyl] pyridinium betaine (di-8-ANEPPS) (Invitrogen) and the Kd for each compound determined using the dual wavelength ratiometric method as described by Qazi et al. (2006).
Dissipation of bacterial membrane potential
Transmembrane potential was determined as described by Breeuwer and Abee (2000). Staphylococcal cells were suspended in potassium phosphate buffer (50 mM, pH 7) containing 5 μM of the cationic fluorescent dye Disc3(5) (Invitrogen). Fluorescence output was continually monitored over 200 s (excitation 650 nm, emission 680 nM). Once the fluorescent signal had reduced and stabilised the selected QZN was added to the appropriate final concentration. To confirm that the membrane potential was responsible for the uptake of the cationic dye, 5 μM valinomycin was added to completely depolarise the remaining membrane potential. As negative controls, the antibiotics novobiocin and chloramphenicol were also included at 10 μg/ml.
Chemical synthesis
QZNs (Table 1) were synthesized and characterized as described previously (Ilangovan et al., 2013) except for 4 (7-Cl-C9-QZN), 5 (C7-BTD), 6 (C9-BTD), 17 (3-NH2-C1-QZN), 35 (3-C4NH2-7-Cl-C9-QZN) and 45 (3-C2OH-7-Cl-C9-QZN). These were synthesised by the established literature procedures (Purcell, 2007; Kirchner and Zalay, 1968, 1974; Mahmoud et al., 2007), essentially in three steps as outlined in Scheme 1.
The acylation of appropriate anthranilic acids with acid chlorides gave N-alkanoylanthranilic acids which via dehydrative intramolecular cyclisation yielded 2-alkylbenzo[d] [1,3]oxazin-4-ones. The latter when aminated in refluxing ethanol (for 4,17,45) or THF (for 35) delivered the desired quinazolinones as crystalline solids after purification.
7-Chloro-2-n-nonylquinazolin-4(3H)-one (7Cl-C9-QZN) 4
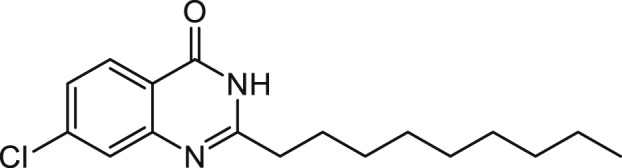
N-Decanoyl-4-chloroanthranilic acid
Decanoyl chloride (0.0275 mol) and a solution of sodium hydroxide, (0.025 mol) in water (10 mL) were simultaneously added dropwise over 15 min to a solution of 4-chloroanthranilic acid (0.025 mol) in sodium hydroxide (0.025 mol) dissolved in distilled water (10 mL) at 0-5°C. The resulting slurry was stirred at 0-5°C for 30 min followed by a further 30 min stirring at room temperature. The reaction mixture was then acidified to pH 1.0 with concentrated HCl (3.5 mL) and extracted with ethyl acetate (30 mL) which was washed with brine (10 mL), dried with magnesium sulphate and concentrated to dryness under reduced pressure. The resulting solid residue was stirred with petroleum ether bp 60-80°C (15 mL) for 1 h. The product was collected by filtration to give N-decanoyl-4-chloroanthranilic acid as a pale brown solid in 83% yield.
7-Chloro-2-n-nonyl-4H-benzo[d][1,3]oxazin-4-one
A mixture of N-decanoyl-4-chloroanthranilic acid (4.0 g) and acetic anhydride (25 mL) was stirred at reflux for 2 h. After cooling the solvent was removed under high vacuum to give the title compound.
7-Chloro-2-n-nonylquinazolin-4(3H)-one
A mixture of 7-chloro-2-n-nonyl-4H-benzo[d][1,3]-oxazin-4-one (0.3077 g, 1 mmol) and ammonium acetate (0.154 g, 2 mmol) was heated at 170°C for 2 h. The residue was cooled to room temperature and taken up in ethyl acetate (40 mL) and water (25 mL) which after 30 min of stirring the mixture was fully dissolved. The aqueous layer was removed and the organic layer was washed with saturated sodium bicarbonate, 1 M HCl, and brine. The organic layer was dried over magnesium sulphate and concentrated to dryness to give an off white solid which was purified using Flash chromatography (40% ethyl acetate in hexane) to deliver 7-chloro-2-n-nonylquinazolin-4(3H)-one as a white solid (0.096 g, 31%).
1H NMR (CDCl3) δ 0.91 (3H, t, Me), 1.25-1.5 (12H, m, (CH2)6Me), 1.87 (2H, quintet, CH2(CH2)6Me), 2.76 (2H, t, CH2(CH2)7Me), 7.44 (1H, 6-H), 7.72 (1H, 8-H), 8.22 (1H, 5-H), 10.74 (1H, s, NH), ES-MS m/z 307.1572 [M+H]+, C17H24ClN2O+ requires 307.1572).
3-Amino-2-methylquinazolin-4(3H)-one (3-NH2-C1-QZN) 17
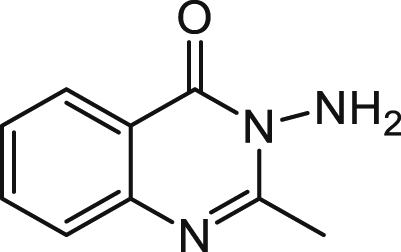
2-Methyl-4H-benzo[d][1,3]oxazin-4-one was prepared as a cream solid in quantitative yield from anthranilic acid (0.03 M) and acetic anhydride (25 ml) by the procedure described above under 5. A solution of 2-Methyl-4H-benzo[d][1,3]oxazin-4-one (1 mmol) and hydrazine monohydrate (4 mmol) in ethanol (20 mL) was stirred at reflux overnight. The solvent was removed under reduced pressure to give the title compound as a cream solid 0.640 g (73% yield) which required no further purification.
1H NMR (CDCl3) δ 2.74 (3H, t, Me), 4.92 (2H, s, NH2), 7.48 (1H, 8-H), 7.68 (1H, 6-H), 7.76 (1H, 7-H), 8.26 (1H, 5-H).
ES-MS m/z 176.0815 [M+H]+, C9H10N3O+ requires 176.0818.
3-(4-Aminobutyl)-7-chloro-2-n-nonylquinazolin-4(3H)-one (3-C4NH2-7-Cl-C9-QZN) 35
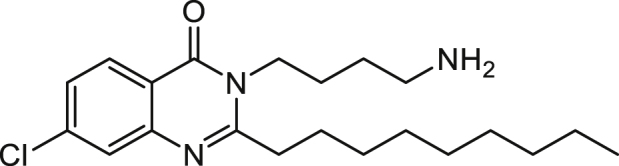
A solution of 7-chloro-2-n-nonyl-4H-benzo[d][1,3]oxazin-4-one (as described for 4) (0.615 g, 2.0 mmol) and N-Boc-1,4-butanediamine (1.506 g, 8.0 mmol) in dry THF (20 mL) was stirred at reflux overnight. The solvent was removed under reduced pressure and the residue was dissolved in ethyl acetate (40 mL) and washed with saturated NaHCO3 (2 x 15 mL), 1 M KHSO4 (2 x 15 mL) and brine (15 mL). The organic layer was dried over magnesium sulphate and concentrated to dryness to give a pale brown oily solid which was purified using flash chromatography (10% ethyl acetate in hexane followed by 15% ethyl acetate in hexane) and trituration with diethyl ether/hexane to give 3-[3-(tert-butoxycarbonylamino)butyl]-7-chloro-2-n-nonyl-4(3H)-quinazolinone as a white solid ( 0.303 g).
A solution of 3-[3-(tert-butoxycarbonylamino)butyl]-7-chloro-2-n-nonyl4(3H)-quinazolinone (0.130 g, 2.8 mmol) in DCM (7 mL) and trifluoroacetic acid (7 mL) was stirred overnight at room temperature. The solution was concentrated to dryness under reduced pressure with the aid of acetonitrile. The residue was dissolved in ethyl acetate (15 mL) and washed with saturated sodium bicarbonate (2 x 7.5 mL) and brine (7.5 mL). The organic layer was dried over magnesium sulphate and concentrated to dryness to give an oily solid which was purified by trituration with hexane and petroleum ether bp 40-60°C to give the title compound as a white solid (0.027 g, 13%).
1H NMR (CDCl3) δ 0.91 (3H, t, Me), 1.31-1.51 (12H, m, (CH2)6Me), 1.65 (2H, m, CH2CH2CH2CH2NH2), 1.83 (4H, m, CH2CH2CH2CH2NH2 and CH2(CH2)6Me), 2.11 (2H, b, NH2), 2.84 (4H, m, CH2(CH2)7Me and (CH2)3CH2NH2), 4.11 (2H, t, CH2(CH2)3NH2), 7.39 (1H, 8-H), 7.65 (1H, 6-H), 8.18 (1H, 5-H).
ES-MS m/z 378.2312 [M+H]+, C21H33ClN3O+ requires 378.2307.
7-Chloro-3-(2-hydroxyethyl)-2-n-nonylquinazolin-4(3H)-one (3-C2OH-7Cl-C9-QZN) 45
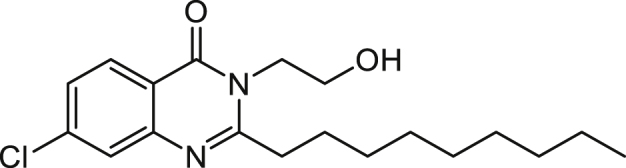
A solution of 7-chloro-2-n-nonyl-4H-benzo[d][1,3]oxazin-4-one (as described for 4) (0.461 g, 1.5 mmol) and ethanolamine (0.367 g, 6 mmol) in ethanol (20 mL) was stirred at reflux overnight. The solvent was removed under reduced pressure and the residue was dissolved in ethyl acetate (30 mL) and washed with saturated NaHCO3 (2 x 10 mL), 1 M KHSO4 (2 x 10 mL) and brine (10 mL). The organic layer was dried over magnesium sulphate and concentrated to dryness to give an off white solid which was purified using flash chromatography (10% ethyl acetate in hexane followed by 30% ethyl acetate in hexane and finally 70% ethyl acetate in hexane) to give the titled compound as a white solid (0.133 g, 25%).
1H NMR (CDCl3) δ 0.90 (3H, t, Me), 1.3-1.5 (12H, m, (CH2)6Me), 1.84 (2H, m, CH2(CH2)6Me), 2.4 (1H, b, OH) 2.91 (2H, t, CH2(CH2)7Me), 4.02 (2H, t, CH2CH2OH), 4.33 (2H, t, CH2CH2OH), 7.41 (1H, 8-H), 7.67 (1H, 6-H), 8.18 (1H, 5-H).
ES-MS m/z 351.1839 [M+H]+, C19H28ClN2O2+ requires 351.1834.
Synthesis of 3-alkyl-3,4-dihydro-2H-benzo[e][1,2,4]thiadiazine 1,1-dioxide (5 and 6)
The compounds 5 and 6 were synthesised by the acid catalysed cyclative condensation of 2-aminobenzenesulfonamide with an appropriate aldehyde in refluxing dioxane as outlined in Scheme 2.
3-n-Heptyl-3,4-dihydro-2H-benzo[e][1,2,4]thiadiazine 1,1-dioxide (C7-BTD) 5
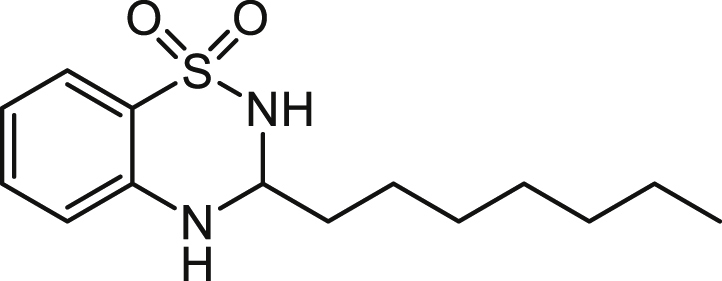
A solution of 2-aminobenzenesulfonamide (0.344 g, 2 mmol), octanal (0.282 g/0.343 ml, 2 mmol) and concentrated H2SO4 (4 drops) in dioxane (10 mL) was stirred at reflux for 3 h. The solvent was removed under reduced pressure and the orange oily solid was purified using flash chromatography (20% ethyl acetate in hexane) to give the title compound as an off white solid (0.159 g, 28%).
1H NMR (DMSO-d6) δ 0.88 (3H, t, Me), 1.29 (8H, m, (CH2)4Me), 1.46 (2H, m, CH2(CH2)4Me), 1.74 (2H, m, CH2(CH2)5Me), 4.63 (1H, m, (NH)2CHCH2(CH2)6Me), 6.69 (1H, t, 6-H), 6.81 (1H, d, 8-H), 6.97 (1H, s, 1-NH), 7.27 (1H, m, 7-H) 7.32 and 7.35 (1H, d, 3-NH), 7.45 (1H, d, 5H).
ES-MS m/z 283.1474 [M+H]+, C14H23N2O2S+ requires 283.1475.
3-n-Nonyl-3,4-dihydro-2H-benzo[e][1,2,4]thiadiazine 1,1-dioxide (C9-BTD) 6
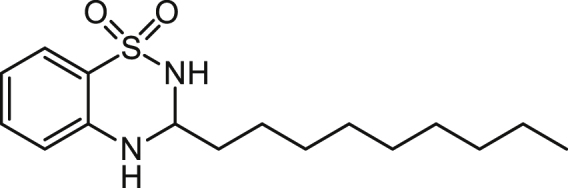
A solution of 2-aminobenzenesulfonamide (0.344 g, 2 mmol), decanal (0.343 g/0.415 ml, 2 mmol) and concentrated H2SO4 (4 drops) in dioxane (10 mL) was stirred at reflux for 3 h. The solvent was removed under reduced pressure and the brown oil was purified using flash chromatography (20% ethyl acetate in hexane) to give an impure cream solid. This was further purified by triturating with petroleum ether 60-80 (9 x 1.5 mL) to give the title compound as a white solid (0.078 g, 13%).
1H NMR (DMSO-d6) δ 0.87 (3H, t, Me), 1.27 (12H, m, (CH2)6Me), 1.45 (2H, m, CH2(CH2)6Me), 1.74 (2H, m, CH2(CH2)7Me), 4.64 (1H, m, (NH)2CHCH2(CH2)6Me), 6.69 (1H, t, 6-H), 6.81 (1H, d, 8-H), 6.99 (1H, s, 1NH), 7.27 (1H, m, 7-H) 7.33 and 7.36 (1H, d, 3NH), 7.43 (1H, d, 5H).
ES-MS m/z 311.1786 [M+H]+, C16H27N2O2S+ requires 311.1788.
Quantification and statistical analysis
Unless otherwise stated within the figure legend, each experiment was conducted on 3 independent occasions. Mean data is presented, error bars indicate the standard deviation. The biomass of each biofilm was assessed from 5 randomly selected sites and quantified using Image J software (Schneider et al., 2012).
Acknowledgments
We thank Alex Truman for QZN synthesis. This work was funded via a Medical Research Council UK program grant (MR/N010477/1) and a Wellcome Trust Senior Investigator grant (103884).
Author contributions
P.W. and S.R.C. conceived the project and supervised the work. P.W., E.M., and S.R.C. wrote the manuscript. E.M. and J.-F.D. designed and carried out the experimental work. All authors analyzed the data, contributed to and commented on the manuscript text, and approved its final version.
Declaration of interests
The authors declare no competing interests.
Published: March 7, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.chembiol.2022.02.007.
Supplemental information
Data and code availability
The published article includes all biological data generated during this study. This study did not generate code. All data reported in this paper will be shared by the lead contact upon request.
References
- Allesen-Holm M., Barken K.B., Yang L., Klausen M., Webb J.S., Kjelleberg S., Molin S., Givskov M., Tolker-Nielsen T. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol. Microbiol. 2006;59:1114–1128. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- Barr H.L., Halliday N., Cámara M., Barrett D.A., Williams P., Forrester D.L., Simms R., Smyth A.R., Honeybourne D., Whitehouse J.L., et al. Pseudomonas aeruginosa quorum sensing molecules correlate with clinical status in cystic fibrosis. Eur. Respir. J. 2015;46:1046–1054. doi: 10.1183/09031936.00225214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr H.L., Halliday N., Barrett D.A., Williams P., Forrester D.L., Peckham D., Williams K., Smyth A.R., Honeybourne D., Whitehouse J.L., et al. Diagnostic and prognostic significance of systemic alkyl quinolones for P. aeruginosa in cystic fibrosis: a longitudinal study. J. Cyst. Fibros. 2017;16:230–238. doi: 10.1016/j.jcf.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin T., Yau Y., Stapleton P.J., Gong Y., Wang P.W., Guttman D.S., Waters V. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. NPJ Biofilms Microbiomes. 2017;3:25. doi: 10.1038/s41522-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breeuwer P., Abee T. Assessment of viability of microorganisms employing fluorescence techniques. Int. J. Food Microbiol. 2000;55:193–200. doi: 10.1016/s0168-1605(00)00163-x. [DOI] [PubMed] [Google Scholar]
- Castillo-Ramírez S., Corander J., Marttinen P., Aldeljawi M., Hanage W.P., Westh H., Boye K., Gulay Z., Bentley S.D., Parkhill J., et al. Phylogeographic variation in recombination rates within a global clone of methicillin-resistant Staphylococcus aureus. Genome Biol. 2012;13:R126. doi: 10.1186/gb-2012-13-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang W.C., Nilsson M., Jensen P.Ø., Høiby N., Nielsen T.E., Givskov M., Tolker-Nielsen T. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2013;57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front. Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis B.M., Jensen R., Williams P., O'Shea P. The interaction of N-acylhomoserine lactone quorum sensing signaling molecules with biological membranes: implications for inter-kingdom signaling. PLoS One. 2010;5:e13522. doi: 10.1371/journal.pone.0013522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeon S., Clinton A., Fowler H., Everett J., Horswill A.R., Rumbaugh K.P. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun. 2014;82:4718–4728. doi: 10.1128/IAI.02198-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliveira D.M.P., Forde B.M., Kidd T.J., Harris P.N.A., Schembri M.A., Beatson S.A., Paterson D.L., Walker M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020;33:e00181-19. doi: 10.1128/CMR.00181-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Déziel E., Gopalan S., Tampakaki A.P., Lépine F., Padfield K.E., Saucier M., Xiao G., Rahme L.G. The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation: multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-L-homoserine lactones. Mol. Microbiol. 2005;55:998–1014. doi: 10.1111/j.1365-2958.2004.04448.x. [DOI] [PubMed] [Google Scholar]
- Diggle S.P., Matthijs S., Wright V.J., Fletcher M.P., Chhabra S.R., Lamont I.L., Kong X., Hider R.C., Cornelis P., Cámara M., Williams P. The Pseudomonas aeruginosa 4-quinolone signal molecules HHQ and PQS play multifunctional roles in quorum sensing and iron entrapment. Chem. Biol. 2007;14:87–96. doi: 10.1016/j.chembiol.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Dubern J.F., Cigana C., De Simone M., Lazenby J., Juhas M., Schwager S., Bianconi I., Döring G., Eberl L., Williams P., et al. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ. Microbiol. 2015;17:4379–4393. doi: 10.1111/1462-2920.12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazli M., Bjarnsholt T., Kirketerp-Møller K., Jørgensen B., Andersen A.S., Krogfelt K.A., Givskov M., Tolker-Nielsen T. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol. 2009;47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P.D., Endres J.L., Yajjala V.K., Widhelm T.J., Boissy R.J., Bose J.L., Bayles K.W. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013;4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeb S., Fletcher M.P., Chhabra S.R., Diggle S.P., Williams P., Camara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev. 2010;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D.L., Chang R., Debabov D.V., Leung J., Wu T., Krause K.M., Sandvik E., Hubbard J.M., Kaniga K., Schmidt D.E., Jr., et al. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005;49:1127–1134. doi: 10.1128/AAC.49.3.1127-1134.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M.T., Lindsay J.A., Corton C., Quail M.A., Cockfield J.D., Pathak S., Batra R., Parkhill J., Bentley S.D., Edgeworth J.D. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic- and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW) J. Bacteriol. 2010;192:888–892. doi: 10.1128/JB.01255-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooi D.S., Bycroft B.W., Chhabra S.R., Williams P., Pritchard D.I. Differential immune modulatory activity of Pseudomonas aeruginosa quorum-sensing signal molecules. Infect. Immun. 2004;72:6463–6470. doi: 10.1128/IAI.72.11.6463-6470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M.J., Aish J.L., White I.J., Shaw L., Lithgow J.K., Foster S.J. Sigma B modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotterbeekx A., Kumar-Singh S., Goossens H., Maddocks S. In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell. Infect. Microbiol. 2017;7:1–13. doi: 10.3389/fcimb.2017.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilangovan A., Fletcher M., Rampioni G., Pustelny C., Rumbaugh K., Heeb S., Cámara M., Truman A., Chhabra S.R., Emsley J., Williams P. Structural basis for native agonist and synthetic inhibitor recognition by the Pseudomonas aeruginosa quorum sensing regulator PqsR (MvfR) PLoS Pathog. 2013;9:e1003508. doi: 10.1371/journal.ppat.1003508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G., Beavis R.C., Novick R.P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc. Nat. Acad. Sci. U S A. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann G.F., Sartorio R., Lee S.H., Rogers C.J., Meijler M.M., Moss J.A., Clapham B., Brogan A.P., Dickerson T.J., Janda K.D. Revisiting quorum sensing: discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc. Nat. Acad. Sci. U S A. 2005;102:309–314. doi: 10.1073/pnas.0408639102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner, F.K. and Zalay, A.W. (1968). Certain alkyl and aryl substituted 3-amino-2, 3-dihydro-4(1h)quinazolinones. US patent 3375250.
- Kirchner, F.K. and Zalay, A.W. (1974). 3-Amino-2,3-dihydro-4(1H)-quinazolinones. US patent 3843654.
- Mahmoud M.R., El-Bordany E.A.A., Hassan N.F., El-Azm F.S.M.A. New 2,3-disubstituted quinazolin-4(3H)-Ones from 2-undecyl-3,1-benzoxazin-4-one. J. Chem. Res. 2007;9:541–544. [Google Scholar]
- Miller W.R., Bayer A.S., Arias C.A. Mechanism of action and resistance to daptomycin in Staphylococcus aureus and Enterococci. Cold Spring Harb. Perspect. Med. 2016;6:a026997. doi: 10.1101/cshperspect.a026997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Wenzel M., Strahl H., Grein F., Saaki T.N.V., Kohl B., Siersma T., Bandow J.E., Sahl H.G., Schneider T., Hamoen L.W. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc. Nat. Acad. Sci. U S A. 2016;113:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray E.J., Crowley R.C., Truman A., Clarke S.R., Cottam J.A., Jadhav G.P., Steele V.R., O'Shea P., Lindholm C., Cockayne A., et al. Targeting Staphylococcus aureus quorum sensing with nonpeptidic small molecule inhibitors. J. Med. Chem. 2014;57:2813–2819. doi: 10.1021/jm500215s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R.P., Ross H.F., Projan S.J., Kornblum J., Kreiswirth B., Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.T., Oglesby-Sherrouse A.G. Interactions between Pseudomonas aeruginosa and Staphylococcus aureus during co-cultivations and polymicrobial infections. Appl. Microbiol. Biotechnol. 2016;100:6141–6148. doi: 10.1007/s00253-016-7596-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazi G., O’Toole G.A. “It takes a village”: Mechanisms underlying antimicrobial recalcitrance of polymicrobial biofilms. J. Bacteriol. 2019;202 doi: 10.1128/JB.00530-19. e00530–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazi G., Ruoff K.L., O'Toole G.A. Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio. 2019;10:e01501–e01519. doi: 10.1128/mBio.01501-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E., Pozzi C., Houston P., Smyth D., Humphreys H., Robinson D.A., O'Gara J.P. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J. Clin. Microbiol. 2007;45:1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea P. Physical landscapes in biological membranes: physico-chemical terrains for spatio-temporal control of biomolecular interactions and behaviour. Philos. Trans. A Math. Phys. Eng. Sci. 2005;363:575–588. doi: 10.1098/rsta.2004.1509. [DOI] [PubMed] [Google Scholar]
- Ou F., Mcgoverin C., Swift S., Vanholsbeeck F. Rapid evaluation of bacterial viability using the optrode – a near real time portable fluorimeter. Aust. Conf. Opt. Fiber Technol. 2016:AW3C.6. doi: 10.1364/ACOFT.2016. [DOI] [Google Scholar]
- Peleg A.Y., Hooper D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival S.L., Suleman L., Vuotto C., Donelli G. Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 2015;64:323–334. doi: 10.1099/jmm.0.000032. [DOI] [PubMed] [Google Scholar]
- Popat R., Crusz S.A., Messina M., Williams P., West S.A., Diggle S.P. Quorum-sensing and cheating in bacterial biofilms. Proc. Biol. Sci. 2012;279:4765–4771. doi: 10.1098/rspb.2012.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price K.E., Naimie A.A., Griffin E.F., Bay C., O'Toole G.A. Tobramycin-treated Pseudomonas aeruginosa PA14 Enhances Streptococcus constellatus 7155 biofilm formation in a cystic fibrosis model system. J. Bacteriol. 2015;198:237–247. doi: 10.1128/JB.00705-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell, I. C. (2007). Bacterial autoinducer derived 4-quinolones as novel immune modulators. PhD Thesis, University of Nottingham. http://eprints.nottingham.ac.uk/10319/.
- Qazi S., Middleton B., Muharram S.H., Cockayne A., Hill P., O'Shea P., Chhabra S.R., Cámara M., Williams P. N-Acylhomoserine lactones antagonize virulence gene expression and quorum sensing in Staphylococcus aureus. Infect. Immun. 2006;74:910–919. doi: 10.1128/IAI.74.2.910-919.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G., Pustelny C., Fletcher M.P., Wright V.J., Bruce M., Rumbaugh K.P., Heeb S., Cámara M., Williams P. Transcriptomic analysis reveals a global alkyl-quinolone-independent regulatory role for PqsE in facilitating the environmental adaptation of Pseudomonas aeruginosa to plant and animal hosts. Environ. Microbiol. 2010;12:1659–1673. doi: 10.1111/j.1462-2920.2010.02214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampioni G., Leoni L., Williams P. The art of antibacterial warfare: deception through interference with quorum sensing-mediated communication. Bioorg. Chem. 2014;55:60–68. doi: 10.1016/j.bioorg.2014.04.005. [DOI] [PubMed] [Google Scholar]
- Rampioni G., Falcone M., Heeb S., Frangipani E., Fletcher M.P., Dubern J.F., Visca P., Leoni L., Cámara M., Williams P. Unravelling the genome-wide contributions of specific 2-alkyl-4-quinolones and PqsE to quorum sensing in Pseudomonas aeruginosa. PLoS Pathog. 2016;12:e1006029. doi: 10.1371/journal.ppat.1006029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlechter R.O., Jun H., Bernach M., Oso S., Boyd E., Munoz-Lintz D.A., Dobson R.C.J., Remus D.M., Remus-Emsermann M.N.P. Chromatic Bacteria - a broad host-range plasmid and chromosomal insertion toolbox for fluorescent protein expression in bacteria. Front. Microbiol. 2018;9:3052. doi: 10.3389/fmicb.2018.03052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwyn B., Neilands J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Scott J.R., Guenthner P.C., Malone L.M., Fischetti V.A. Conversion of an M- group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 1986;164:1641–1651. doi: 10.1084/jem.164.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukarieh F., Williams P., Stocks M.J., Cámara M. Pseudomonas aeruginosa quorum sensing systems as drug discovery targets: current position and future perspectives. J. Med. Chem. 2018;61:10385–10402. doi: 10.1021/acs.jmedchem.8b00540. [DOI] [PubMed] [Google Scholar]
- Starkey M., Lepine F., Maura D., Bandyopadhaya A., Lesic B., He J., Kitao T., Righi V., Milot S., Tzika A., Rahme L. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014;10:e1004321. doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S.Y.C., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenti-Weigand P., Benkel P., Rohde M., Chhatwal G.S. Entry and intracellular survival of group B streptococci in J774 macrophages. Infect. Immun. 1996;64:2467–2473. doi: 10.1128/iai.64.7.2467-2473.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P., Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 2009;12:182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Williams P. Strategies for inhibiting quorum sensing. Emerg. Top. Life Sci. 2017;1:23–30. doi: 10.1042/ETLS20160021. [DOI] [PubMed] [Google Scholar]
- Wilton M., Charron-Mazenod L., Moore R., Lewenza S. Extracellular DNA acidifies biofilms and induces aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2015;60:544–553. doi: 10.1128/AAC.01650-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., Déziel E., He J., Lépine F., Lesic B., Castonguay M.H., Milot S., Tampakaki A.P., Stachel S.E., Rahme L.G. MvfR, a key Pseudomonas aeruginosa pathogenicity LTTR-class regulatory protein, has dual ligands. Mol. Microbiol. 2006;62:1689–1699. doi: 10.1111/j.1365-2958.2006.05462.x. [DOI] [PubMed] [Google Scholar]
- Zapotoczna M., Murray E.J., Hogan S., O'Gara J.P., Chhabra S.R., Chan W.C., O'Neill E., Williams P. 5-Hydroxyethyl-3-tetradecanoyltetramic acid represents a novel treatment for intravascular catheter infections due to Staphylococcus aureus. J. Antimicrob. Chemother. 2017;72:744–753. doi: 10.1093/jac/dkw482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all biological data generated during this study. This study did not generate code. All data reported in this paper will be shared by the lead contact upon request.