Abstract
In response to improper kinetochore-microtubule attachments in mitosis, the spindle assembly checkpoint (SAC) assembles the mitotic checkpoint complex (MCC) to inhibit the anaphase-promoting complex/cyclosome, thereby delaying entry into anaphase. The MCC comprises Mad2:Cdc20:BubR1:Bub3. Its assembly is catalysed by unattached kinetochores on a Mad1:Mad2 platform. Mad1-bound closed-Mad2 (C-Mad2) recruits open-Mad2 (O-Mad2) through self-dimerization. This interaction, combined with Mps1 kinase-mediated phosphorylation of Bub1 and Mad1, accelerates MCC assembly, in a process that requires O-Mad2 to C-Mad2 conversion and concomitant binding of Cdc20. How Mad1 phosphorylation catalyses MCC assembly is poorly understood. Here, we characterized Mps1 phosphorylation of Mad1 and obtained structural insights into a phosphorylation-specific Mad1:Cdc20 interaction. This interaction, together with the Mps1-phosphorylation dependent association of Bub1 and Mad1, generates a tripartite assembly of Bub1 and Cdc20 onto the C-terminal domain of Mad1 (Mad1CTD). We additionally identify flexibility of Mad1:Mad2 that suggests how the Cdc20:Mad1CTD interaction brings the Mad2-interacting motif (MIM) of Cdc20 near O-Mad2. Thus, Mps1-dependent formation of the MCC-assembly scaffold functions to position and orient Cdc20 MIM near O-Mad2, thereby catalysing formation of C-Mad2:Cdc20.
Subject terms: NMR spectroscopy, Biophysics, Cell-cycle proteins, Cryoelectron microscopy
Formation of the mitotic checkpoint complex (MCC) is catalysed by a phosphorylation-dependent scaffold. This work provides structural details of how a tripartite Mad1:Bub1:Cdc20 complex presents Cdc20 to Mad2, triggering open-to-closed conversion of Mad2 to assemble the MCC.
Introduction
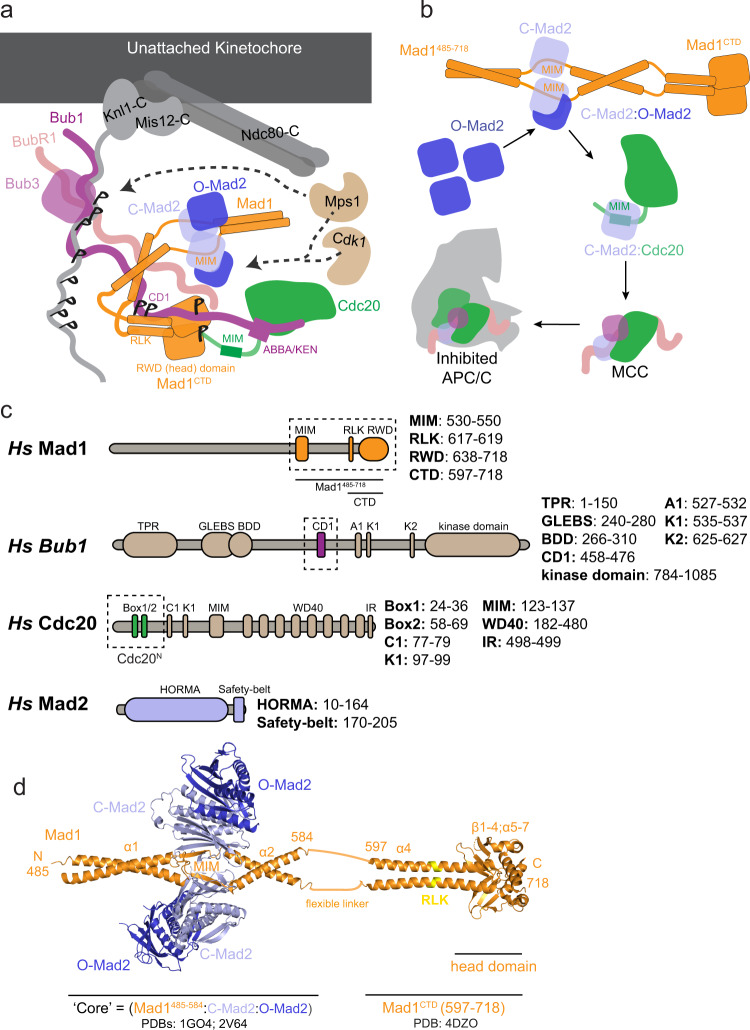
During mitosis, kinetochores assembled on centromeric chromatin act as the interface for microtubule attachment to sister chromatids1,2. In response to improper attachments, anaphase onset is delayed by kinetochore-catalysed assembly of the mitotic checkpoint complex (MCC)3–5. The MCC binds and inhibits the anaphase-promoting complex/cyclosome (APC/C) whose activity triggers mitotic exit6–9. The MCC is composed of Mad2, Cdc20 and BubR1:Bub310,11. The rate of Cdc20:Mad2 association poses a kinetic barrier to MCC formation12,13. This is overcome through a sequential assembly of checkpoint proteins onto the unattached outer kinetochore by means of an Mps1 kinase-dependent phosphorylation cascade that creates a catalytic scaffold for MCC formation14,15 (Fig. 1a). Mps1 phosphorylates multiple MELT (Met-Glu-Leu-Thr) motifs of the outer kinetochore subunit Knl1 to control recruitment of Bub1:Bub3 and BubR1:Bub316–18. Cdk1 and Mps1 then sequentially phosphorylate the conserved domain 1 (CD1) of Bub1 (Bub1CD1), enabling it to bind the Mad1:Mad2 complex by interacting with the RLK (arginine-leucine-lysine) motif within the C-terminus of Mad1 (Mad1CTD) (Fig. 1a, c, d)14,19–22. Mad1 self-dimerises through a series of coiled-coil α-helices. The coiled-coil formed from residues 485-584 is interrupted by the Mad2-interacting motif (MIM) that entraps one molecule of Mad2 per subunit to generate the Mad1:C-Mad2 tetramer (Fig. 1d)23. This segment is followed by a flexible linker (residues 585–597), after which the coiled-coil resumes, ending in a globular head domain presenting an RWD-fold (residues 597–718) (Fig. 1d)24. During MCC assembly, O-Mad2 is targeted to the outer kinetochore through self-dimerisation to the Mad1-bound C-Mad2 (Fig. 1b, d)23,25,26. A substantial remodelling of dimerised O-Mad2 into C-Mad2, comprising the C-terminal ‘safety-belt’, is then required for Mad2 to entrap the MIM of Cdc2027–30 (Fig. 1b, c). The C-Mad2:Cdc20 complex has a high affinity for BubR1:Bub3, allowing spontaneous MCC assembly (Fig. 1b)11,12,31.
Fig. 1. Assembly of the MCC at unattached kinetochores.
a The catalytic scaffold for MCC assembly onto the outer kinetochore. Phosphorylated Knl1 MELT motifs recruit the Bub3:Bub1 complex, which is sequentially phosphorylated by Cdk1 and Mps1 on the Bub1 CD1 domain. Phosphorylated Bub1 recruits the Mad1:C‐Mad2 complex via its CD1 domain and O‐Mad2 is recruited through self-dimerisation with C-Mad2. Mad1 is further phosphorylated by Mps1, which promotes interaction with Cdc20. b The catalytic scaffold shown in (a) functions to bring Cdc20 near O-Mad2. O-Mad2 undergoes conversion to C-Mad2, releasing it from Mad1:C-Mad2 and simultaneously binding Cdc20MIM. C-Mad2:Cdc20 binds BubR1:Bub3 to generate the MCC. Once formed, soluble MCC binds and inhibits the APC/C, preventing anaphase progression. c Schematics of full-length Mad1, Bub1, Cdc20 and Mad2. The truncations used in this study are highlighted by the dashed boxes. MIM Mad2-interacting motif. RLK Arg‐Leu‐Lys motif. RWD RING, WD40, DEAD domain. TPR tetratricopeptide repeat, GLEBS Gle2‐binding‐sequence, BDD Bub dimerisation domain, CD1 conserved domain 1, A1 ABBA motif, C1 C-box, K1/2 KEN-box motifs, IR IR-tail, HORMA Hop1p, Rev7p, Mad2 proteins. d A side view of the Mad1485–718:C-Mad2:O-Mad2 complex. The Mad1 dimer encompassing residues 485-584 is depicted in orange, bound to two C-Mad2 molecules in light blue (PDB 1GO4)23. Two O-Mad2 molecules (dark blue) dimerised to C-Mad2 are fitted using the structure of the O-C Mad2 dimer (PDB 2V64)26. The coiled-coil is then interrupted by a 16-residue segment after which the coiled-coil is extended to the C-terminal domain of Mad1 (residue 597–718), which is depicted in orange (PDB 4DZO)24. The RLK motif with Mad1CTD is highlighted in yellow.
In vitro, conversion of Mad2 takes several hours, whereas, in cells, the SAC response is established within minutes of kinetochore-microtubule detachment12,13,29,32. However, in the presence of components of the catalytic scaffold (Fig. 1a), nearly spontaneous conversion of Mad2 occurs in vitro12,33. Understanding how the identified catalysts (Mad1:C-Mad2, Bub1 and Mps1) promote Mad2 conversion and concomitant Cdc20 association is of key interest and remains largely unanswered. A critical requirement of a functional checkpoint in vivo, and catalytic MCC formation in vitro, is Mps1 phosphorylation of the Mad1 C-terminus14,33–35 (Fig. 1c, d). Phosphorylated Mad1 interacts with a conserved basic motif at the N-terminus of Cdc20 (Box1) (Fig. 1c)14,33. Disrupting either phosphorylation of the Mad1 C-terminus, or abolishing the Cdc20:Mad1 interaction, does not prevent Cdc20 kinetochore localisation, but causes a defective checkpoint, and impairs catalytic Cdc20:C-Mad2 formation in vitro14,33,35. These findings led to the suggestion that the Cdc20:Mad1 interaction allows Cdc20 to be incorporated into the checkpoint by promoting both the accessibility of the MIM, and its presentation to Mad2 for safety-belt entrapment33. This mechanism is likely dependent on both relieving Cdc20 autoinhibition and optimally positioning its MIM close to Mad2 as it undergoes conversion33,35. However, the molecular mechanisms for how Mad1 phosphorylation mediates interaction with Cdc20, and how this interaction promotes MIM accessibility and catalytic MCC formation are unknown.
In this study, we applied complementary approaches to investigate how Mad1 phosphorylation regulates its interaction with Cdc20, and how this interaction likely promotes Cdc20:Mad2 binding. We determined that the Cdc20:Mad1 interaction is modulated by the phosphorylation of Mad1 Thr716 (pThr716). Using NMR spectroscopy, we demonstrated that an N-terminal segment of Cdc20 binds across Mad1CTD through two regions with residual α-helicity (α1 and Box1). Biophysical characterisation and fluorescence anisotropy measurements identified that a single copy of both Bub1CD1 and Cdc20N concurrently bind to a phosphorylated Mad1CTD, and AlphaFold2 was used to predict an experimentally supported model of this tripartite assembly. Using a combination of cryo-EM and cross-linking mass spectrometry, we also identified a folded state of the Mad1:Mad2 complex. Together with the Cdc20:Mad1CTD interaction, this suggests a model for how the MIM of Cdc20 is presented to Mad2. Lastly, we used NMR to show that Cdc20MIM alone is sufficient to induce Mad2 conversion, highlighting the importance of the Mad1-Bub1 scaffold in orchestrating the arrangement of Cdc20 and Mad2 during catalytic MCC assembly.
Results
Thr716 is the predominant Mps1 phosphorylation site of Mad1CTD
Several Mps1-phosphorylation sites within Mad1 have been proposed to enhance MCC formation14,33–35. However, both the specific function of each of these sites, and the molecular mechanism of how Mad1 phosphorylation promotes interaction with Cdc20 are unknown. We phosphorylated Mad1CTD with full-length Mps1 in vitro to identify phosphorylation sites on Mad1CTD. The majority of phosphorylated peptides identified by mass spectrometry contain a phosphorylated Thr716 (pThr716) site, with a few being phosphorylated at Thr644 (Supplementary Table 1). Intact protein-centric mass spectrometry showed that each monomer of phosphorylated Mad1CTD and phosphorylated Mad1485–718 contained only a single phosphorylated residue (Supplementary Fig. 1a, b). We further analysed phosphorylated Mad1CTD using NMR to monitor the previously assigned backbone resonances of unphosphorylated Mad1CTD in 1H, 15N 2D HSQC spectra21 (Supplementary Fig. 1c). Marked chemical shift perturbations (CSP) were only observed for the backbone resonances corresponding to Thr716 and its neighbouring residues (713–718), consistent with preferred Thr716 phosphorylation (Supplementary Fig. 1d). Our data indicate that in vitro Thr716 is the only highly occupied Mps1 phosphorylation site of Mad1CTD. This agrees with previous reports that phosphorylation of Thr716 is critical for both maintaining a viable checkpoint in vivo14,34, and the ability of the MCC to assemble and inhibit the APC/C in vitro14.
Mad1CTD Thr716 phosphorylation promotes the binding of a single copy of Cdc20
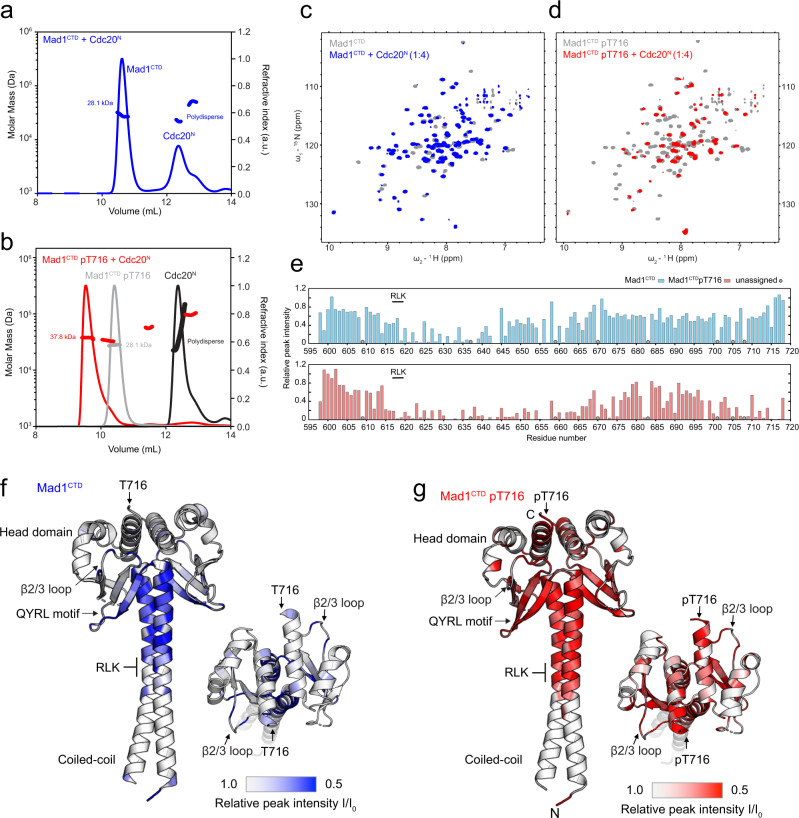
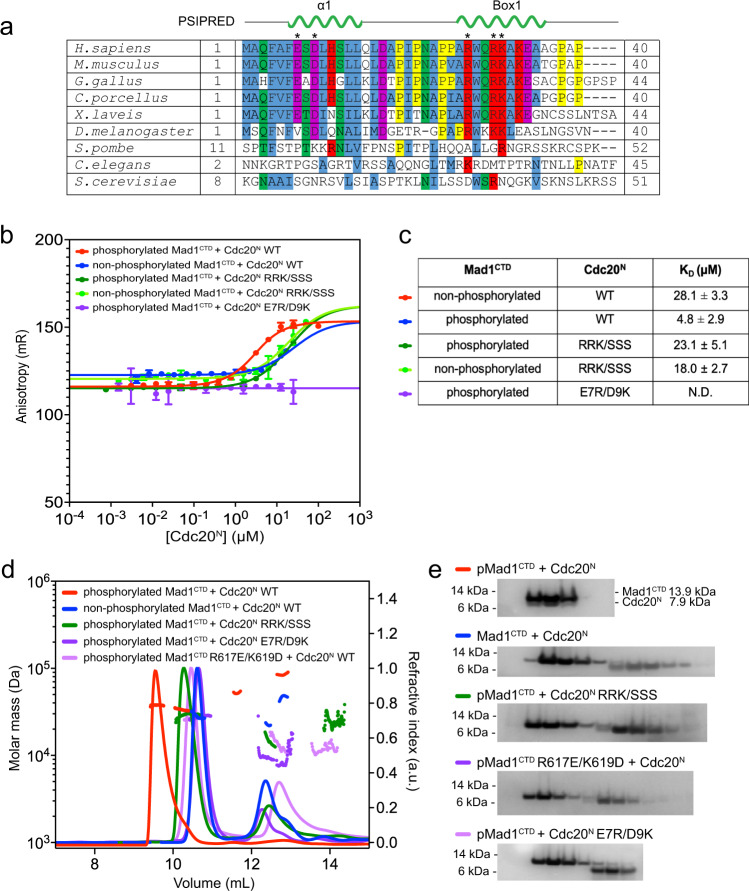
The N-terminus of Cdc20, specifically its Box1 motif, was proposed to interact with Mad1CTD in a phosphorylation-dependent manner14,33. We sought to investigate this interaction using Mad1CTD and an N-terminal truncation of Cdc20 (residues 1–73; Cdc20N) (Fig. 1c). The interaction of Cdc20N and Mad1CTD was analysed by SEC-MALS. Although Cdc20N did not bind to unphosphorylated Mad1CTD (Fig. 2a), it formed a stable complex with Mad1CTD phosphorylated at Thr716 (pMad1CTD) (Fig. 2b). This indicated a strong interaction between pMad1CTD and Cdc20N, dependent on phosphorylation of Mad1 at Thr716. The elution peak corresponding to pMad1CTD:Cdc20N was monodispersed with an observed mass of 38 kDa (Fig. 2b), close to the expected mass of a pMad1CTD dimer with one Cdc20N bound (36 kDa). AUC-SE was used to further investigate the stoichiometry of the Cdc20N:pMad1CTD complex. These data also indicated that only a single copy of Cdc20N bound to the pMad1CTD homodimer (Supplementary Fig. 2), even with a four-fold molar excess of Cdc20N at concentrations well above the assumed KD, judged by the strong interaction of Mad1CTD and Cdc20N in SEC, and later confirmed by fluorescent anisotropy experiments discussed below.
Fig. 2. A direct Cdc20-Mad1 interaction mediated by phosphorylated Mad1CTD pThr716.
a SEC-MALS analysis of unphosphorylated Mad1CTD + Cdc20N (blue). The first elution peak consists of free Mad1CTD (expected mass 28 kDa), while Cdc20N (expected mass 7.8 kDa) alone elutes in the second polydispersed peak (later confirmed to be homogenous monomer by AUC-SE in Supplementary Fig. 2), suggesting there is no binding between unphosphorylated Mad1CTD and Cdc20N. b SEC-MALS analysis of free Mad1CTD pT716 (grey), free Cdc20N (black) and Mad1CTD pT716 + Cdc20N (red). Mad1CTD pT716 alone (grey) eluted as one monodispersed species with an apparent mass of 28.1 kDa. Cdc20N alone (black) elutes as a polydispersed peak with an undefined mass. Mad1CTD pT716 + Cdc20N (red) gives a distinct monodispersed species of 37.8 kDa, indicative of a stable complex with one Mad1CTD pT716 dimer and a single molecule of Cdc20N (expected 36 kDa). c 1H,15N 2D HSQC showing 15N-labelled unphosphorylated Mad1CTD alone (grey) or in the presence of four molar excess Cdc20N (blue). d 1H,15N 2D HSQC showing 15N-labelled Mad1CTD pT716 alone (grey) or in the presence of four molar excess Cdc20N (red). e Relative peak intensities from the 1H,15N 2D HSQC of Mad1CTD (blue) or Mad1CTD pT716 (red) in the presence of Cdc20N at 1:1 molar ratio were mapped onto the Mad1CTD sequence. Peak intensities were normalised to the C-terminal residue Ala718 for the spectra of unphosphorylated Mad1CTD, where the C-terminus was clearly not involved in binding. For phosphorylated Mad1CTD the peak intensities were adjusted to match the spectra collected for unphosphorylated Mad1CTD for comparison, as the experimental conditions were nearly identical. Unassigned peaks are denoted as grey circles. f Line broadening induced by Cdc20N binding to unphosphorylated Mad1CTD was mapped onto the structure of Mad1CTD (PDB 4DZO)24. Blue in the colour scale indicates regions with significant line broadening. The RLK motif (617-619) and QYRL motif (648-651) are labelled. g Line broadening induced by Cdc20N binding to Mad1CTD pT716 was mapped onto the structure of Mad1CTD. Red in the colour scale indicates regions with significant line broadening.
Characterisation of the Cdc20N:Mad1CTD interaction by NMR
NMR spectroscopy was used to obtain structural insights into the Cdc20N:Mad1CTD interaction. Unlabelled Cdc20N was titrated into either 15N-labelled Mad1CTD or pMad1CTD (Fig. 2c, d and Supplementary Fig. 3a, b). Significant signal attenuation was observed in 1H, 15N 2D HSQC spectra for resonances corresponding to residues 616–660 of both Mad1CTD and pMad1CTD (Fig. 2e). Mapping these line broadened residues onto the Mad1CTD crystal structure (PDB 4DZO)24 indicated that regions of Mad1CTD comprising the RLK motif, the coiled-coil, and first β-strand of the head domain, were perturbed upon Cdc20N binding (Fig. 2f). Line broadening was more pronounced and extensive for pMad1CTD, with signal attenuation observed in the entire β-sheet, the C-terminal α-helix, and the pThr716 site (Fig. 2g). Our results are consistent with a previous study which showed that mutation of the conserved QYRL motif (Q648A and R650A) within the Mad1CTD β-sheet, and an RLK/AAA (residues 617-619) mutation, impaired binding of Cdc20N to Mad1 (Fig. 2e–g)33. Overall, the extent of line broadening observed upon Cdc20N binding suggests that Cdc20N forms an extensive binding interface with Mad1CTD, and/or it induces a global conformational change within Mad1CTD.
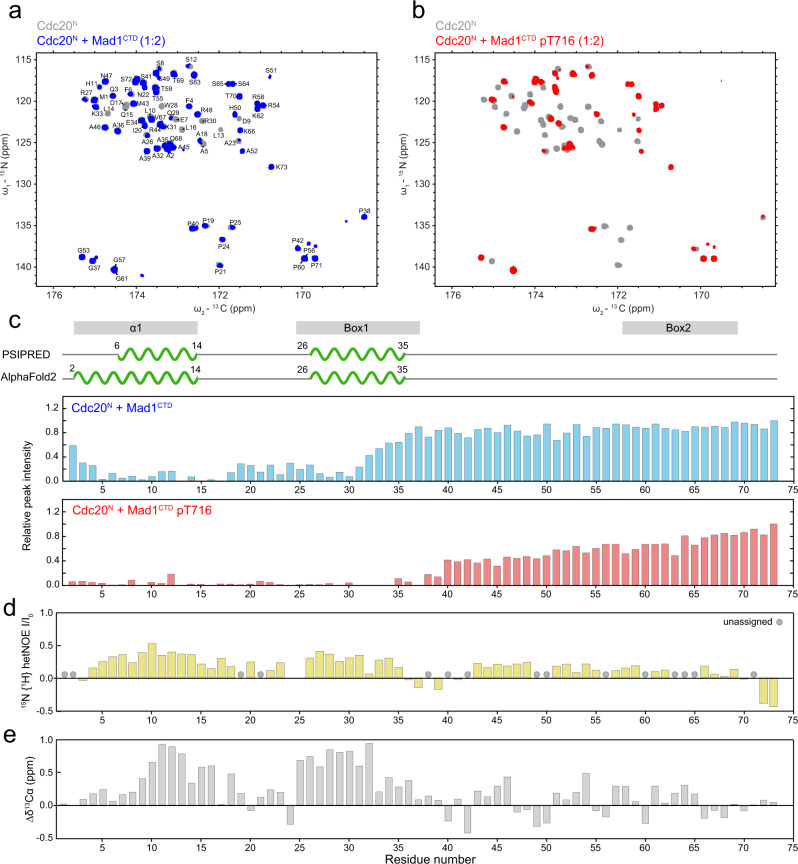
The N-terminus of Cdc20 contains two conserved basic motifs, Box1 (27-RWQRKAKE-34), which is predicted to be α-helical, and Box2 (58-RTPGKSSSKVQT-69) which is predicted to be unstructured (Supplementary Fig. 4). Box1 has been implicated in binding pMad1CTD and found to be functionally important for checkpoint formation14,33. N-terminal to Box1 and Box2 is another predicted α-helix (Cdc20 α1) with an unknown function (Supplementary Fig. 4). To obtain structural information on Cdc20N, and to reciprocally define Mad1CTD-binding sites on Cdc20N, we characterised isotopically labelled Cdc20N using NMR. The narrow dispersion of 1H chemical shifts indicated that Cdc20N was largely disordered (Supplementary Fig. 5a, b). Several backbone amide signals were missing in the 1H, 15N 2D HSQC spectrum comprising prolines and residues that underwent rapid exchange with the solvent (Supplementary Fig. 5a, b). To obtain a complete assignment of the backbone resonances of Cdc20N we employed experiments that directly detect the 13C nuclei (13C-detected) to define residues based on their 15N and 13C carbonyl frequencies (Fig. 3a). For the same reason, we used 13C-detected 2D CON spectra that report the correlation between backbone carbonyl 13C and amide 15N, to provide a complete picture of the effect of Mad1CTD binding on Cdc20N (Fig. 3a, b). The relative peak intensity changes were mapped onto the Cdc20N sequence (Fig. 3c). When either phosphorylated or unphosphorylated Mad1CTD were titrated, substantial signal attenuation in every residue of the N-terminal half of Cdc20N was observed, whereas the C-terminal half including Box2 was largely unaffected. This indicated that both Cdc20 α1 and Box1, but not Box2, are involved in Mad1CTD binding. Notably, four prolines located in the loop connecting Cdc20 α1 and Box1, are also involved in the interaction, suggesting these residues might provide some rigidity to orientate these α-helical segments. More significant signal attenuation occurred with pMad1CTD (Fig. 3 and Supplementary Fig. 5), confirming the importance of Mad1 phosphorylation in promoting this interaction.
Fig. 3. Characterisation of Cdc20N and its interaction with Mad1CTD.
a 13C-detected 2D CON spectra of Cdc20N alone (grey) and in the presence of Mad1CTD (blue) at a four molar excess, shown with the assignment of Cdc20N backbone resonances. b 13C-detected 2D CON spectra of Cdc20N alone (grey) and in the presence of Mad1CTDpT716 (red) at a twofold molar excess. c Relative peak intensities from the CON spectra of Cdc20N in the presence of Mad1CTD (blue) and Mad1CTD pT716 (red) were mapped onto the Cdc20N sequence. Peak intensities were normalised to the C-terminal residue Gly73. Regions showing significant line broadening coincide with regions that were predicted to be α-helical by both PSIPRED and AlphaFold2, as shown in the schematics above. d 15N{1H}-heteronuclear NOE measuring the backbone dynamics in Cdc20N and reported as mean values in technical duplicates. A higher (more positive) value indicates higher rigidity of the backbone amide bond, while a lower value indicates higher flexibility. e Secondary chemical shifts (∆δ13Cɑ) reveal residual secondary structure in Cdc20N. Positive values suggest α-helical propensity and negative values suggest a propensity for β-strand conformation.
We further delineated the binding sites of α1 and Box1, modelled as short peptides, on Mad1CTD by monitoring the chemical shift perturbations (CSPs) upon peptide titrations (Supplementary Figs. 3, 6). As expected, these CSPs were mapped to more localised regions within Mad1CTD. Cdc20 Box1 binds pMad1CTD but not Mad1CTD and CSPs were observed around pThr716, the β2 strand, and β2/β3 loop adjacent to pThr716 (residues 652-661, Fig. 2g and Supplementary Fig. 6b, c). Meanwhile, very small CSPs were observed when Cdc20 α1 was titrated to pMad1CTD, even at high concentrations of peptide, suggesting a weak interaction between Cdc20 α1 and pMad1CTD. These CSPs were mapped to the β-sheet and proximal coiled-coil of pMad1CTD (Supplementary Fig. 6a). The low affinity of these interactions suggests that cooperative binding involving α1 and Box1 is required to create a stable interaction between Cdc20N and Mad1CTD. The interaction of Box1 close to pThr716 and Cdc20 α1 near the Mad1 RLK motif would suggest that Cdc20N binds Mad1CTD in a parallel orientation, and is in agreement with cross-linking mass spectrometry data that Cdc20Box1 interacts with the head domain of Mad1CTD [ref. 33].
Although AlphaFold2 and PSIPRED predictions modelled Cdc20 as having two α-helical segments at its N-terminus (Supplementary Fig. 4), Cdc20N lacks a stable secondary structure in solution as indicated by its 1H chemical shift dispersion (Supplementary Fig. 5a, b). The absence of a stable secondary structure is supported by 15N{1H}-heteronuclear NOE experiments that sample 15N backbone dynamics on a fast picosecond timescale. These showed only a slight decrease in flexibility for the backbone amides in the predicted α-helical regions compared with predicted disordered regions (Fig. 3d). To identify possible residual α-helicity in these regions, we examined the secondary chemical shifts of Cdc20N by comparing the differences in Cα chemical shifts between native and denatured states36. Secondary chemical shifts are a reliable measurement of secondary structure propensity, especially in disordered proteins37. The secondary chemical shifts of Cdc20N indicated a clear α-helical propensity for both α1 and Box1 (Fig. 3e). Substantial line broadening occurred when Cdc20N interacted with pMad1CTD, preventing us from acquiring similar backbone dynamics and secondary chemical shift data on the pMad1CTD:Cdc20N complex. However, the residual α-helical propensity would be consistent with the idea that Cdc20 α1 and Box1 adopt an α-helical conformation upon binding to pMad1CTD. Altogether, our data suggest an extensive interaction between Mad1CTD and the N-terminal half of Cdc20N.
A tripartite assembly of Cdc20N and Bub1CD1 on phosphorylated Mad1CTD
We previously showed that only a single Bub1CD1 binds to the Mad1CTD homodimer, most likely a result of the inherent asymmetry within Mad1CTD in which the coiled-coil is bent with respect to the head domain, observed in both the Bub1CD1 bound and apo-Mad1CTD X-ray structures in multiple different crystal lattices21,24. Interestingly, the Mad1CTD homodimer also binds only one Cdc20N (Fig. 2b and Supplementary Fig. 2). We reasoned that Mad1CTD asymmetry might also play a role in defining the Cdc20N:Mad1CTD stoichiometry. As shown by NMR, residues of Mad1CTD that bind Cdc20N almost entirely overlap in sequence with those that bind Bub1CD1 (Fig. 2c–g)21.
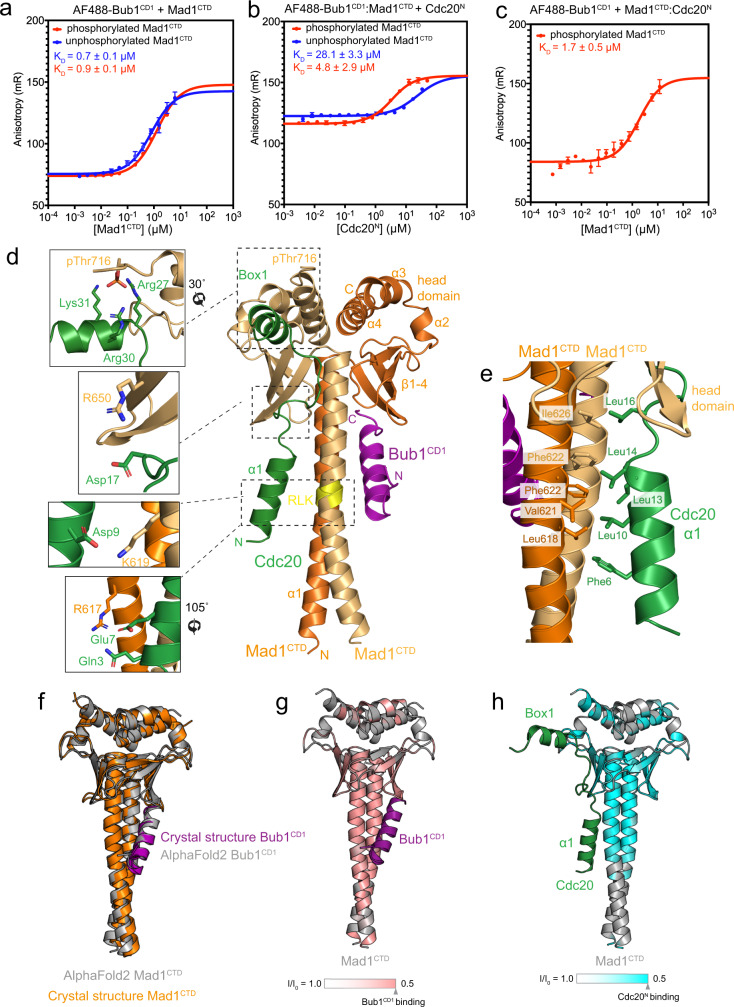
To investigate whether Cdc20N and Bub1CD1 either compete for binding to pMad1CTD, or bind concurrently, we measured the change in fluorescence anisotropy of a Bub1 peptide bound to Mad1CTD as a function of Cdc20N concentration. For this, we conjugated Aurora Fluor 488 (AF488) to the flexible N-terminus of a phosphorylated Bub1CD1 peptide. AF488-Bub1CD1 and Mad1CTD formed a complex with a binding affinity of 0.7 µM for Mad1CTD and 0.9 µM for pMad1CTD (Fig. 4a). This agrees with the 2.7 µM affinity previously determined by ITC21, indicating that the fluorescent tag does not interfere with Mad1CTD binding. The equivalent affinities of AF488-Bub1CD1 for phosphorylated and unphosphorylated Mad1CTD are in agreement with our previous crystal structure, showing that pThr716 would not contact Bub1CD1 [ref. 21]. Next, we titrated Cdc20N into the preformed AF488-Bub1CD1:pMad1CTD complex. In the case of competition, the introduction of unlabelled Cdc20N would displace AF488-Bub1CD1, resulting in a decrease in fluorescence anisotropy as its tumbling motion increases in the unbound state. Instead, titration of Cdc20N into the AF488-Bub1CD1:Mad1CTD or AF488-Bub1CD1:pMad1CTD complex increased the fluorescence anisotropy (Fig. 4b). This indicated that Cdc20N and Bub1CD1 bind to Mad1CTD at two distinct sites simultaneously. Cdc20N binds to the preformed Mad1CTD:AF488-Bub1CD1 complex with a KD of 4.8 µM for pMad1CTD and 28 µM for Mad1CTD (Fig. 4b). We next determined the affinity of AF488-Bub1CD1 for a preformed pMad1CTD:Cdc20N complex. This showed a similar affinity (1.7 µM) to the binding of AF488-Bub1CD1 to pMad1CTD alone (0.9 µM), indicating that Cdc20 and Bub1 binding to pMad1CTD is unlikely to be cooperative (Fig. 4c). A low micromolar affinity of the Cdc20N interaction agrees with the timescales observed in NMR where line broadening of Mad1CTD peaks occurred upon Cdc20N binding (Fig. 2c, d). These affinities are also consistent with the stability of the complex observed in SEC when Mad1CTD is phosphorylated but not when Mad1CTD is unphosphorylated (Fig. 2a, b). Thus, our data indicate that Bub1CD1 and Cdc20N can bind simultaneously to pMad1CTD to form a stable tripartite complex.
Fig. 4. A tripartite assembly of Cdc20 and Bub1 on phosphorylated Mad1CTD.
a Fluorescent anisotropy measurements with 20 nM AF488-Bub1CD1 and titrating Mad1CTD. The binding of Mad1CTD and Mad1CTD pThr716 to AF488-Bub1CD1 gave calculated KD of 0.9 and 0.7 µM, respectively. b Fluorescent anisotropy measurements with preformed AF488-Bub1CD1:Mad1CTD (20 nM: 2.1 µM) where Mad1 was phosphorylated or unphosphorylated and Cdc20N was titrated. The binding of Cdc20N to the Mad1CTD:AF488-Bub1CD1 complex where Mad1CTD were unphosphorylated and phosphorylated gave calculated KD of 28.1 and 4.8 µM, respectively. c Fluorescence anisotropy measurements for the titration of AF488-Bub1CD1 to preformed pMad1CTD:Cdc20N (20 nM: 15 µM), giving a KD of 1.7 µM. For data in a–c, error bars are the derived SD from three independent measurements. d AlphaFold2 prediction of the Mad1CTD homodimer (dark/light orange) bound to Bub1CD1 (purple) and Cdc20N (green). The model was generated using Cdc201–73:Bub1448–550:Mad1597–718 with a defined stoichiometry of 1:1:2. Only the first 40 residues of Cdc20 are shown, including the α1 and Box1 α-helices. Cdc20N:Mad1CTD interactions discussed in the text are highlighted in the left panels, where key residues are displayed as sticks. pThr716 is shown to visualise how the phosphate is positioned to contact the positively charged residues of Box1 (top panel). e The hydrophobic interface between Cdc20N and Mad1CTD in the AlphaFold2 prediction. f Superimposition of the Mad1CTD:Bub1CD1 crystal structure21 and the Mad1CTD:Bub1CD1 AlphaFold2 model shows their similarity, despite the Bub1CD1 input sequence not containing phosphorylated Ser459 and Thr461 on Bub1CD1. g Line broadening induced by Bub1CD1 binding to 15N-labelled Mad1CTD pT716, as previously reported21, was mapped onto the Mad1CTD AlphaFold2 model. Pink in the colour scale indicates regions with a significant line broadening and are likely involved in binding Bub1CD1. The AlphaFold2 model of Bub1CD1 is independently coloured in purple. h Line broadening induced by Cdc20N binding to Mad1CTD pT716, as reported in this study (Fig. 2d, e, g), was mapped onto the Mad1CTD AlphaFold2 model. Cyan in the colour scale indicates regions with a significant line broadening and are likely involved in binding Cdc20N. The AlphaFold2 model of Cdc20N is independently coloured in green.
The pMad1CTD-Cdc20N complex failed to crystallise using a variety of Cdc20N peptides and supplementing Bub1CD1. We, therefore, used AlphaFold238 to predict the structure of the Cdc20N:Bub1CD1:Mad1CTD tripartite assembly. On imposing a 1:1:2 stoichiometry of Cdc20N:Bub1448–550:Mad1CTD, AlphaFold2 predicted a tripartite complex with high confidence (>90 predicted local distance difference test (pLDDT) for Bub1CD1 and 70-90 pLDDT for Cdc20N) (Fig. 4d and Supplementary Fig. 7a–c). The predicted aligned error (PAE) plot also indicates high confidence for the orientation of Bub1CD1 and Cdc201–35 with respect to Mad1CTD (Supplementary Fig. 7d). A comparable tripartite complex was also predicted when full-length Bub1 and Cdc20 were used (data not shown). The predicted model of the Bub1CD1:Mad1CTD interaction was nearly identical to our Bub1CD1:Mad1CTD crystal structure (PDB: 7B1F)21 and agreed with the binding sites mapped by NMR (Fig. 4f, g)21, providing confidence in the AlphaFold2 model. The crystal structure of Mad1-Bub121 was used to build pSer459 and pThr461 of Bub1, and to model pThr716 of Mad1. For the Cdc20N:Mad1CTD interaction, AlphaFold2 predicts that the first 35 residues of Cdc20N are involved in the Cdc20:Mad1CTD interaction and α1 of Cdc20 runs parallel to the Mad1CTD coiled-coil, consistent with our NMR data (Figs. 3, 4h and Supplementary Fig. 6).
In this predicted model, the Cdc20N α1 helix lies diagonally across the Mad1CTD coiled-coil with its N-terminus close to the Mad1 RLK motif of one monomer, and the C-terminus contacting the head domain β-sheet of the adjacent monomer, in a manner that strikingly mimics the Bub1CD1:Mad1CTD interaction (Fig. 4d). Similar to Bub1CD1, the interaction of Cdc20 α1 with Mad1CTD would largely be driven by hydrophobic interactions (Fig. 4e). This model predicts that Gln3 and Glu7, and Asp9 of Cdc20 α1 interact with Arg617 and Lys619, respectively of the Mad1CTD RLK motif (Fig. 4d). This is consistent with a previous report that a Mad1CTD RLK/AAA mutant abolished Cdc20 binding33, and our NMR data where the RLK residues of Mad1CTD were perturbed upon Cdc20N binding (Fig. 2e). An interaction of Cdc20 Asp9 with Mad1 Lys619 might also explain how a Mad1CTD K619A mutant disrupted the checkpoint, even though the Mad1CTD K619A mutant did not disrupt Bub1CD1 binding significantly21,24. To further test the interaction of Cdc20 α1 with the RLK motif of Mad1, we assessed the binding of a Cdc20N E7R/D9K mutant to pMad1CTD and found that their interaction could no longer be observed at the concentrations used in our fluorescent anisotropy assay or by SEC-MALS (Fig. 5). Additionally, as judged by SEC-MALS, wild-type Cdc20N did not bind a Mad1CTD R617E/K619D mutant (Fig. 5d, e). Because the Mad1CTD R617E/K619D mutant disrupted the binding of Bub1CD1, fluorescent anisotropy could not be used to further test the interaction of Mad1CTD R617E/K619D with wild-type Cdc20N. SEC-MALS did not show an interaction between Mad1CTD R617E/K619D and Cdc20N E7R/D9K, suggesting that this interaction could not simply be restored by swapping the charges. As mentioned earlier, mutation of the conserved QYRL motif (Q648A/R650A) within Mad1CTD impaired binding to Cdc2033. This is explained in our model where Arg650 of Mad1CTD is predicted to contact Asp17 of Cdc20N (Fig. 4d).
Fig. 5. Binding of Cdc20N and Mad1CTD mutants by fluorescent anisotropy and SEC-MALS.
a A multiple sequence alignment of Cdc201–40. Secondary structure prediction by PSIPRED is shown above for the α1 and Box1 predicted helices. The asterisks denote residues which were mutated and tested by fluorescent anisotropy or SEC-MALS in b–d. b Fluorescent anisotropy with preformed AF488-Bub1CD1:Mad1CTD complex (20 nM: 2.1 µM) using pMad1CTD or Mad1CTD and where various Cdc20N mutants were titrated. The data were presented as mean values ± SD derived from three independent measurements. c Summary of the calculated KD of the interactions analysed in (b). d SEC-MALS analysis of Cdc20N interaction with pMad1CTD or Mad1CTD using various mutants. Theoretical masses for Mad1CTD dimer, Cdc20N, and Mad1CTD dimer with a single Cdc20N bound are 28, 7.8 and 36 kDa respectively. e SDS-PAGE analysis of the SEC-MALS experiments shown in (d), using a 4–12% Bis-Tris Glycine gel with the SeeBlue™ Plus2 Protein Ladder (Thermo Fisher Scientific). Three independent measurements of each sample were completed with similar results.
In agreement with our NMR data (Fig. 3 and Supplementary Figs. 3, 5, 6), the Box1 motif α-helix, is predicted to bind the head domain near the C-terminus of Mad1CTD. This positions three positively charged residues (Arg27, Arg30, Lys31) of Box1 near the negatively charged pThr716 of Mad1CTD (Fig. 4d), and this is consistent with our observation that Box1 interacts with pThr716 and the β2/β3 loop adjacent to pThr716 (Fig. 2g and Supplementary Fig. 6b, c). The AlphaFold2 prediction was generated without including a phosphorylated Thr716 in the input sequence. We assume that Thr716 of Mad1 co-evolves with the basic residues of Box1, and the multisequence alignment (MSA) algorithm of AlphaFold2 likely creates a close distance constraint between these residues38. This is supported by the accurate prediction of the Bub1CD1:Mad1CTD interaction despite the sequence of Bub1CD1 not containing a phosphorylated Thr461 which is essential for the Bub1CD1:Mad1CTD interaction14,20,21. Mutation of Arg27, Arg30, Lys31 within Cdc20N to Ser (Cdc20N RRK/SSS), abolished its interaction with pMad1CTD as measured by SEC-MALS (Fig. 5d, e), and reduced the KD of their interaction as measured by fluorescent anisotropy to 23 μM (Fig. 5b, c). The comparable affinities of Cdc20N RRK/SSS binding for pMad1CTD and wild-type Cdc20N for unphosphorylated Mad1CTD (28 μM) is consistent with the interaction of these residues at the pThr716 site. This is supported by the similar affinity of Cdc20N RRK/SSS for unphosphorylated Mad1CTD (18 μM) (Fig. 5b, c). Altogether, the Cdc20N:Mad1CTD AlphaFold2 prediction is in good agreement with our NMR data, illustrated by mapping the line broadening observed in the Cdc20N:Mad1CTD titrations onto the model (Fig. 4h). It also explains how Cdc20 and Bub1 bind to Mad1CTD in a non-competitive manner to form a tripartite complex, as indicated by our fluorescence anisotropy assays (Fig. 4b), and the role of pThr716 in enhancing Cdc20N binding.
Fold-over of Mad1CTD within the Mad1-Mad2 complex
One factor likely contributing to reducing the kinetic barrier for the formation of the C-Mad2:Cdc20 complex is a mechanism to promote the accessibility and proximity of Cdc20MIM relative to its binding site on Mad233,35. Because Mad1 phosphorylation is required for catalysing MCC formation but not Cdc20 kinetochore recruitment14,33,35, we next sought to further investigate how the tripartite assembly of Bub1CD1, Cdc20N and Mad1CTD might contribute to catalysing the formation of the C-Mad2:Cdc20 complex. Mad1:C-Mad2, by means of O-Mad2 dimerisation to C-Mad2 and the interaction of Cdc20 to Mad1:Bub1, is the platform by which Mad2 and Cdc20 are brought together (Fig. 1b). Similar to other reports, we were unable to reconstitute a stable complex comprising the entire ‘pre-MCC assembly’ (pBub1:Bub3:pMad1:C-Mad2:O-Mad2:Cdc20)12,14,33. We, therefore, employed a combination of cryo-EM, cross-linking mass spectrometry and SAXS to investigate the architecture of the Mad1:C-Mad2 complex (Fig. 1d).
We prepared Mad1:C-Mad2 tetramer and Mad1:C-Mad2:O-Mad2 hexamer, using Mad1 residues 485–718 (Mad1485), in the phosphorylated and unphosphorylated states, and confirmed their homogeneity by SEC-MALS (Supplementary Fig. 8a, b). This truncated form of Mad1 is sufficient for catalytic MCC assembly in vitro12. The majority of phospho-peptides identified by mass spectrometry for Mps1-phosphorylated Mad1485 contained a phosphorylated Thr716 (pThr716) (Supplementary Table 1). Lower abundant phospho-peptides with phosphorylation at Thr500, Ser538, Thr540, Thr550 and Ser551 were also detected (Supplementary Table 1). Intact mass spectrometry showed that each monomer of phosphorylated Mad1485 contained predominantly only a single phosphorylated residue (Supplementary Fig. 1b), suggesting, and consistent with our previously presented data for Mad1CTD, that Thr716 is the predominant Mps1 phosphorylation site within Mad1485 (Supplementary Fig. 1a, c, d).
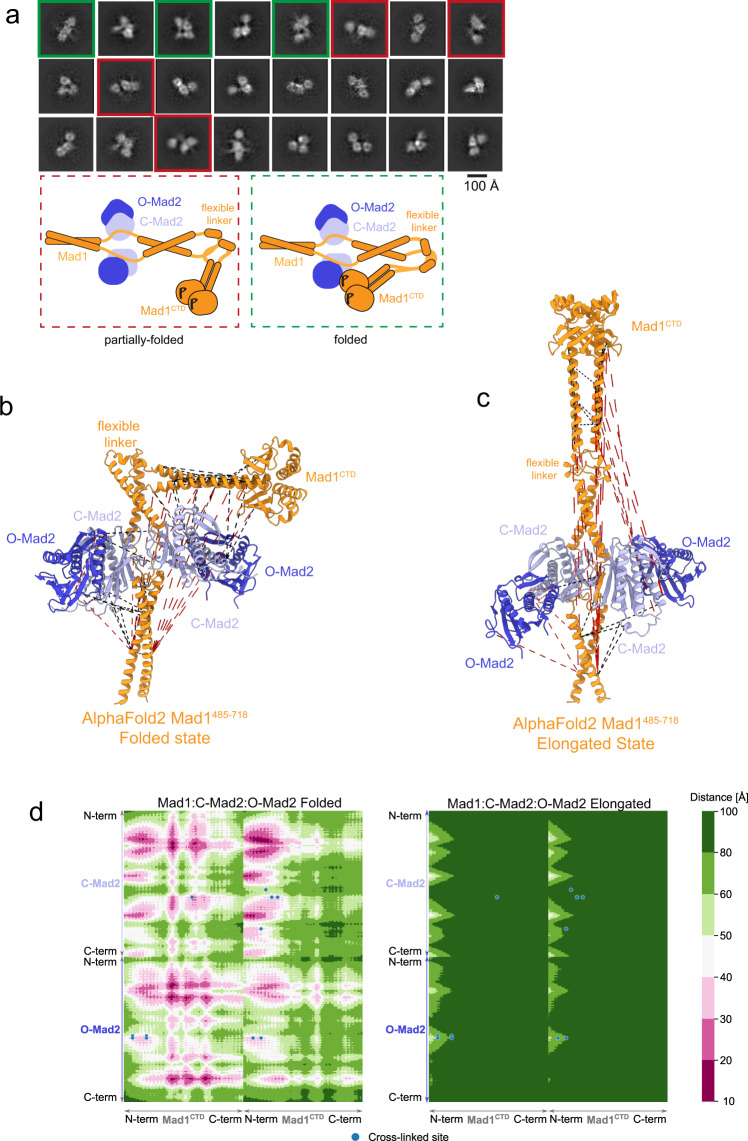
Two-dimensional class averages calculated from a cryo-EM data set of non-phosphorylated Mad1:C-Mad2:O-Mad2 revealed features corresponding to the Mad1:C-Mad2:O-Mad2 core and adjoining coiled-coil segments. The coiled-coil segment C-terminal to the Mad1:C-Mad2:O-Mad2 core is connected to Mad1CTD through a flexible linker region (Fig. 1d and Supplementary Fig. 9a–d) and matches the expected architecture of this complex (Supplementary Fig. 9e). These cryo-EM data allowed a 3D reconstruction of a medium resolution (10 Å) structure of the complex, although EM volumes for the flexible Mad1CTD were not recovered (Supplementary Fig. 9f, h). In contrast, 2D class averages of the phosphorylated Mad1:C-Mad2:O-Mad2 complex showed that conformational variability within the flexible linker region allowed this region to act as a hinge, with Mad1CTD able to adopt multiple conformations, including positions close to, and contacting the Mad1:C-Mad2:O-Mad2 core, as well as more extended states (Fig. 6a). This suggested that phosphorylated Mad1:C-Mad2:O-Mad2 exists in variable conformations ranging from a closed folded state (Mad1CTD proximal to, or in contact with the core), to an elongated state with Mad1CTD being distal to the core. Folded states of Mad1:Mad2 would be consistent with low-angle metal shadowing electron micrographs of Mad1:Mad233. More recently, fold-over of Mad1 has also been detected in vivo using fluorescence-lifetime imaging (FLIM)39. The small size and conformational variability of this complex, despite been cross-linked, precluded a high-resolution cryo-EM reconstruction and suggests there is no stable folded state of Mad1CTD but that Mad1CTD remains highly dynamic (Supplementary Fig. 10a, b). A medium-resolution 3D reconstruction (11 Å at FSC = 0.143 or 16 Å at FSC = 0.5) of a folded state of Mad1 allowed the fitting of crystal structures21,23,24,26 and visualisation of Mad1CTD positioned next to the core (Supplementary Fig. 10d, e, g). The presence of ‘empty’ density around Mad1CTD in this reconstruction is likely from this subset of particles containing Mad1CTD in various orientations. Density for the N-terminal helices was not recovered, which may be due to how the folded particles adhere to the EM grid, or due to Mad1CTD fold-over increasing their flexibility. Interestingly, AlphaFold2 predicted multiple conformations of Mad1485 that varied between elongated and folded states depending on the degree of bending at the flexible linker region connecting the Mad1:C-Mad2:O-Mad2 core with Mad1CTD (Supplementary Fig. 8c–e). Modelling Mad1:C-Mad2:O-Mad2, based on structures of Mad1:C-Mad2 (PDB: 1GO4)23 and the C-Mad2:O-Mad2 dimer (PDB: 2V64)26, onto Mad1MIM in a folded state predicted by AlphaFold2 to have the highest confidence, generated a structure that resembles our cryo-EM reconstruction of a folded Mad1:C-Mad2:O-Mad2 state (Supplementary Fig. 10e and Fig. 6b).
Fig. 6. Fold-over within the Mad1:Mad2 complex.
a 2D averages of BS3 cross-linked pMad1:C-Mad2:O-Mad2 complex by cryo-EM, representative of 125 2D classes. Classes showing complexes that appear only partially folded are boxed in red, and classes showing complexes that appear fully folded are boxed in green. A schematic of the suggested partially folded and folded states seen in the 2D averages is shown below. b Identified cross-links for the phosphorylated Mad1:C-Mad2:O-Mad2 complex were mapped onto a folded Mad1CTD state as predicted by AlphaFold2 (Supplementary Fig. 8d) where alignment of the folded AlphaFold2 Mad1485–718 structure onto the crystal structure of Mad1485–584:C-Mad2 (PDB 1GO4)23 and C-Mad2:O-Mad2 dimer (PDB 2V64)26 was used to generate a model of the hexameric complex. Mad1 subunits (residues 485–718) are shown in orange, whereas C-Mad2 and O-Mad2 are coloured in light and dark blue, respectively. Mapped cross-links with a distance below 40 Å are indicated as black dashed lines and all others in red. For all obtained cross-links, only the shortest option is depicted. c The same as in (b) but identified cross-links are mapped onto the structure of Mad1:C-Mad2:O-Mad2 in the elongated state as predicted by AlphaFold2 (Supplementary Fig. 8c). d Cross-link contact map for the folded and elongated Mad1:C-Mad2:O-Mad2 complexes. Cross-link sites reflecting inter Mad1CTD:Mad2 cross-links are shown as blue circles. For simplicity, only contacts between dimeric Mad1CTD and the closest Mad2 heterodimer (C-Mad2 and O-Mad2) were calculated. Intra- and intermolecular distances are shown in accordance with the coloured scale bar.
To further investigate the conformation of Mad1CTD in hexameric Mad1:C-Mad2:O-Mad2 (phosphorylated and unphosphorylated) and tetrameric Mad1:C-Mad2 (unphosphorylated) states, we performed in-gel cross-linking mass spectrometry (IGX-MS)40 (Fig. 6b, c and Supplementary Fig. 11). We observed multiple cross-links between Mad1CTD and both Mad2 and the Mad1 N-terminal coiled-coils in all samples (Fig. 6b, c and Supplementary Fig. 11e–g). Mapping these cross-links onto the AlphaFold2-predicted Mad1:Mad2 structures indicated that the majority of cross-links are compatible with a folded conformation, but not with the elongated state (Fig. 6b, c). To further determine whether the cross-linking results distinguish between the elongated and folded states, a contact map for the respective Mad1CTD and Mad2 interfaces was generated (Fig. 6d). For simplicity, we calculated the distances only between Mad1CTD and the closest C-Mad2:O-Mad2 dimer. This analysis confirmed that the identified cross-links are consistent with a folded model of Mad1:C-Mad2:O-Mad2, and incompatible with the elongated conformation. Because the cross-linking pattern was similar for all three Mad1:Mad2 samples, we conclude that both the phosphorylated Mad1:Mad2 hexamer and the unphosphorylated Mad1:Mad2 hexamer and tetramer are capable of adopting a folded state. However, we note that these data do not indicate the relative proportions of folded and elongated conformations at equilibrium, but rather highlight the high flexibility of Mad1CTD. In agreement with a compact model, cross-linking captures Mad1CTD orientations in close proximity (up to 30 Å) to C-Mad2:O-Mad2. Additionally, cross-linking also likely traps a more compacted state than is presented in the AlphaFold2 and cryo-EM models, explaining why even in our folded model, several of the cross-links were still identified as outliers (Fig. 6b and Supplementary Figs 8d, 10e).
Our IGX-MS experiments revealed that a folded state of the Mad1:Mad2 hexamer does not require Mad1CTD phosphorylation, contrasting with our cryo-EM results where a folded state was observed only for phosphorylated Mad1:Mad2. To assess the conformation and flexibility of the Mad1:Mad2 complexes in solution, we used size-exclusion-coupled small angle X-ray scattering (SEC-SAXS) (Supplementary Fig. 12). Our SEC-SAXS data showed that all Mad1:Mad2 complexes eluted as a single peak having a high degree of compositional homogeneity, with an average Rg across the peak of 46 Å. This Rg value compares to a calculated Rg of 43 Å for the folded states of the hexameric and tetrameric Mad1:Mad2 species, and 57 Å for the elongated states41,42. Plotting the pairwise distances between scattering points within the molecule, the P(r) distribution (Supplementary Fig. 12f), indicates that the maximum distance extends to approximately 176 Å (Dmax) in all samples, suggesting there are no significant differences in the extent of Mad1CTD fold-over between the complexes. The fully extended Mad1:Mad2 model (Fig. 6c) has a maximum distance of ~202 Å, whereas the folded model (Fig. 5b) has a maximum distance of approximately 151 Å. In addition, the hydrodynamic radius (Rh) was measured from the quasi-elastic light scattering (QELS) data collected during the SEC-MALS experiments (Supplementary Fig. 8a) giving a value of 53 Å (Supplementary Fig. 12g). The ratio of Rg/Rh is indicative of the overall shape of the complex43. The ratio of Rg collected from SEC-SAXS (46 Å) to the Rh (53 Å) from QELS of 0.87 compared to a value of 0.78 for a spherical mass, indicates a compact conformation. Thus, SEC-SAXS and SEC-MALS-QELS aligns with our IGX-MS experiments, allowing us to conclude that in solution, Mad1:Mad2 adopts variable conformations with a tendency to adopt a folded state, regardless of Mad1CTD Thr716 phosphorylation.
A possible explanation for why a folded state of non-phosphorylated Mad1:Mad2 was absent in our cryo-EM micrographs, is the marked preferred orientation of this molecule on cryo-EM grids (Supplementary Fig. 9g), in contrast to the random orientations of phosphorylated Mad1:Mad2 (Supplementary Fig. 10f). This indicates interactions of non-phosphorylated Mad1:Mad2 with the air-water interface that are likely to differentially affect the stability of the folded and elongated conformations.
Cdc20MIM induces O-Mad2 to C-Mad2 conversion
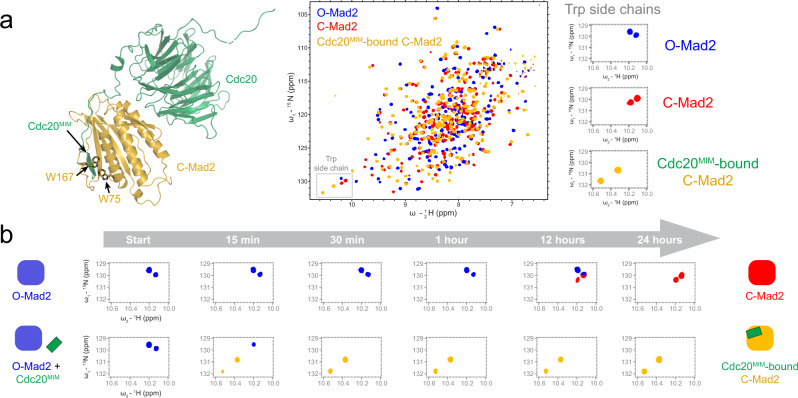
We also used NMR to analyse the rate of O-Mad2 to C-Mad2 conversion (Fig. 7a, b and Supplementary Fig. 13). The substantial remodelling of O-Mad2 into C-Mad2 results in distinctly different 1H, 15N HSQC spectra for Mad2 in different conformations (Fig. 7a). In particular insertion of Cdc20MIM near Trp75 and Trp167 in Cdc20MIM-bound C-Mad2 results in a marked chemical shift perturbation of their corresponding resonances (Fig. 7a)27,44. These resonances are useful reporters of Mad2 conformation in solution. We found that at 25 °C, O-Mad2 (100 μM) converted to C-Mad2 in 24 h (in line with previous reports)12,29,32 (Fig. 7b and Supplementary Fig. 13b). In the presence of only a two-molar excess of a Cdc20MIM peptide (200 μM), complete Mad2 conversion and entrapment of Cdc20MIM occurred in less than 30 min (Fig. 7b and Supplementary Fig. 13a). We also showed that such spontaneous conversion cannot be achieved using a substoichiometric concentration of Cdc20MIM (1:50 to Mad2, Supplementary Fig. 13c). This suggests that Cdc20MIM functions trigger a conversion and stabilises Cdc20MIM-bound C-Mad2. The capacity of Cdc20MIM to induce the O-Mad2 to C-Mad2 conversion is reminiscent of the O-Mad2 to C-Mad2 conversion induced by Mad1MIM [ref. 29].
Fig. 7. NMR-monitor conversion of O-Mad2 to C-Mad2 shows that Cdc20MIM greatly enhances the rate of Mad2 conversion.
a C-Mad2 (orange) bound to Cdc20 (green) from PDB 6TLJ90, with Mad2 Trp167 and Trp75 side chains depicted as sticks. The middle panel shows a 1H, 15N 2D HSQC of 15N-labelled O-Mad2 (blue), C-Mad2 (red), and Cdc20MIM-bound C-Mad2 (orange). All Mad2 samples contain an R133A mutation to prevent Mad2 dimerisation23. The C-Mad2 sample additionally contains an L13A mutation which favours the C-Mad2 state57. Cdc20MIM-bound C-Mad2 sample was prepared by adding a 1:2 excess of Cdc20MIM peptide to O-Mad2 with an R133A mutation. The indole N-H resonances for Mad2 tryptophan side chains were used as reporters for Mad2 conformation and are boxed in the spectra. The right panel shows the close-up views of the two tryptophan indole N-H resonances of O-Mad2 (blue), empty C-Mad2 (red), and Cdc20MIM-bound C-Mad2 (orange). As Cdc20MIM is entrapped by the safety-belt, the changes in the chemical environment of Trp167 and Trp75 result in distinct chemical shifts of their side chain resonances. b The MIM motif of Cdc20 induces the conversion of O-Mad2 into C-Mad2. Using the side chain resonances of Trp167 and Trp75 as reporters, the O-to-C conversion of Mad2 was traced in a 24-h time course at 25 °C. The schematics show the conformation of Mad2 as indicated by the tryptophan side chain resonances and the full spectra are shown in Supplementary Fig. 13a, b. O-Mad2 in the presence of Cdc20-MIM converts to Cdc20MIM-bound C-Mad2 (orange) within 30 min, while O-Mad2 alone takes up to 24 h to fully convert to C-Mad2 (red).
Discussion
Phosphorylation of Mad1CTD by Mps1, that functions to promote interaction with Cdc20N, together with Mps1-mediated phosphorylation of Bub1, have been identified as crucial steps in catalysing MCC assembly14,33,35. Additionally, O-Mad2 dimerisation with C-Mad2 contributes to catalysing the O-Mad2 to C-Mad2 conversion, although at a more modest rate that does not account for the rate of MCC assembly in cells during an active SAC13,32. Here, we found that the Mps1 phosphorylation of Mad1 Thr716 directly promotes an extensive interaction between Cdc20N and Mad1CTD. We find that, similar to Bub1CD1, only one Cdc20N binds to the Mad1CTD homodimer. Cdc20 and Bub1 share a comparable binding interface on Mad1CTD, but instead of competing, the two proteins bind simultaneously to Mad1CTD, forming a tripartite complex. We employed AlphaFold2 to predict a model of Bub1CD1:Cdc20N:Mad1CTD. The resultant model was consistent with our NMR and biophysical characterisation of the Cdc20N:Mad1CTD interaction, and correctly predicted our previously reported Bub1CD1-Mad1CTD crystal structure21. It also illustrated how dimeric Mad1CTD can accommodate both Bub1CD1 and Cdc20N. Using cryo-EM and IGX-MS, we additionally identified a mechanism by which Mad1CTD folds over towards Mad1-bound C-Mad2.
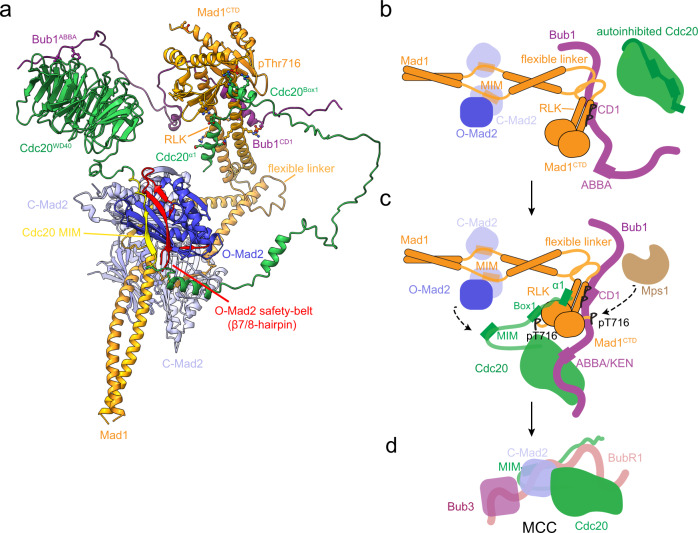
Utilising our discovery of a tripartite Bub1CD1:Cdc20N:Mad1CTD complex, together with Mad1CTD fold-over, we can present a plausible model for the structure of the catalytic scaffold posed for MCC formation (Fig. 8a and Supplementary Movie 1). Importantly, this model outlines how this assembly would position Cdc20MIM in close proximity to O-Mad2 for their interaction (Fig. 8a–d). Detachment of the safety-belt (β7/8 hairpin) concomitant with the O-Mad2 to C-Mad2 conversion would trap Cdc20MIM within C-Mad2. In our model (Fig. 8a), the configuration of Cdc20 in the context of the MCC-assembly scaffold orientates Cdc20MIM antiparallel to the β6 strand of O-Mad2, with which it pairs as an antiparallel β-sheet in the C-Mad2:Cdc20 complex. Thus, both the close proximity and optimal orientation of Cdc20MIM relative to O-Mad2, facilitated by the MCC-assembly scaffold, would contribute to catalysing C-Mad2:Cdc20 formation.
Fig. 8. A model for how tripartite assembly of pBub1:Cdc20:pMad1 in conjunction with Mad1CTD fold-over orchestrates MCC formation.
a Model of pBub1:Cdc20:pMad1:C-Mad2:O-Mad2 complex (MCC-assembly scaffold) posed for MCC formation (Supplementary Movie 1). The model (Supplementary Data 1) was generated using PDBs 1GO4, 2V64, 6TLJ, 7B1F21,23,26,90 and the AlphaFold2 models of Bub1448–534:Mad1CTD:Cdc20 and the folded model of Mad1485–718, and displayed using ChimeraX. Residues for the Mad1 pThr716 and Cdc20Box1, Cdc20α1 Asp9 and Mad1 Lys619, Mad1 R617 and Bub1 pThr461 interactions are depicted as sticks, as well as the Bub1 ABBA interaction with the Cdc20 WD40 domain. b–d A schematic of catalytic MCC formation. b The doubly phosphorylated Bub1 CD1 domain targets the Mad1:C-Mad2 complex to kinetochores which then acts as a platform for O-Mad2 binding and O-to-C Mad2 conversion. Cdc20, on its own, likely exists in an autoinhibited state, which would impair the Cdc20:C-Mad2 interaction and MCC formation. c Phosphorylation of the C-terminus of Mad1 at Thr716 promotes its interaction with the N-terminus of Cdc20. Interaction between the WD40 domain of Cdc20 and the ABBA/KEN1 motif of Bub1 also occurs and this likely promotes Cdc20 kinetochore targeting and positions Cdc20 close to Mad1:C-Mad2. The Cdc20:pMad1CTD interaction together with Mad1CTD fold-over then promotes Cdc20-MIM accessibility and leads to Cdc20:C-Mad2 formation. d The Cdc20:C-Mad2 complex rapidly binds BubR1:Bub3 to form the MCC.
This model also demonstrates how the tripartite assembly of Bub1CD1 and Cdc20N on Mad1CTD is compatible with Bub1ABBA-mediated Cdc20 kinetochore recruitment45–48. The parallel orientation of Bub1CD1 and Cdc20N with respect to Mad1CTD, optimally positions Bub1ABBA, located C-terminal to Bub1CD1, and the C-terminal WD40 domain of Cdc20 (Cdc20WD40) for their interaction (Fig. 8a, c). Self-interaction of the N- and C-termini of Cdc20 has been reported35,49,50, leading to the suggestion that the catalytic scaffold also acts to relieve Cdc20 autoinhibition33,35. Our model suggests dual anchorage of Cdc20N to pMad1CTD and Cdc20WD40 to Bub1ABBA as a plausible mechanism for both relieving Cdc20 autoinhibition and promoting the accessibility of Cdc20MIM for Mad2 safety-belt entrapment. This also likely explains why both Bub1CD1 and Bub1ABBA contribute to catalytic MCC assembly in vitro33, and how a viable checkpoint requires Bub1CD1 even when Mad1 is tethered to kinetochores20,51,52.
Our model for the catalytic scaffold also provides insight into the relevance of Mad1CTD fold-over, mediated through the flexible linker. In the elongated state of Mad1 (Supplementary Fig. 14), the Mad1CTD is separated from the Mad1:Mad2 dimerisation platform compared to the Mad1 folded state (Fig. 8a). Modelling suggests that the segments connecting both Bub1CD1 with Bub1ABBA and Cdc20N with Cdc20MIM are of insufficient length to allow Cdc20MIM to be positioned proximal to the O-Mad2 safety-belt. This is supported by a study which reported that catalytic MCC formation is impaired by decreasing the distance between the Cdc20 Box1 and MIM motifs33. In a more recent complementary study, Chen and colleagues demonstrate that the flexible hinge within Mad1 which allows Mad1CTD fold-over is essential for catalytic MCC assembly in vitro and SAC signalling in vivo39. They also identified that catalysis induced by the flexible linker was dependent upon an intact Mad1:Mad2 interaction with Bub1CD1 and Cdc20Box1, thereby coupling Mad1 flexibility and tripartite Bub1CD1:Cdc20N:Mad1CTD assembly with catalytic Mad2:Cdc20 formation.
Our observation that Cdc20MIM induces the O-Mad to C-Mad2 conversion is significant because it further confirms that the key function of the catalytic scaffold is to promote the accessibility and repositioning of Cdc20MIM for Mad2 binding. Ultimately, this likely explains how Mad1 phosphorylation and its interaction with Cdc20 accelerates MCC formation and provides significance to our model of Cdc20MIM poised to bind Mad2 and subsequently induce Mad2 conversion and, therefore its own incorporation into the MCC (Fig. 8a).
In summary, our study provides mechanistic insights into how Mps1 phosphorylation of Mad1 modulates interaction with the N-terminus of Cdc20, completing a catalytic scaffold which acts to promote the Cdc20:C-Mad2 interaction, thereby overcoming the key kinetic barrier to MCC formation. Understanding how Cdc20MIM directly induces the O-Mad2 to C-Mad2 conversion is an outstanding future challenge.
Methods
Expression and purification
A gallery of all purified proteins in this study by SDS-PAGE is shown in Supplementary Fig. 15. Mad1CTD was expressed and purified as previously described21. The coding region of human Cdc20N was cloned by USER® (NEB) into a modified pRSFDuet-1 vector (71341-3, Sigma-Aldrich) with an N-terminal His6-MBP-tag, followed by a tobacco etch virus (TEV) protease cleavage site53,54. Cdc20 was transformed into RosettaTM 2 (DE3) SinglesTM Competent Cells (71400, Novagen) for expression. Expression was induced with 0.3 mM IPTG and grown overnight at 20 °C. Cells were lysed in 25 mM HEPES pH 8.2, 300 mM NaCl, 10% glycerol, supplemented with lysozyme, and Complete™ EDTA-free protease inhibitors (Roche). His6-MBP-Cdc20N was then purified over a HiTrap TALON® Crude column (Cytiva), and the tag was cleaved with TEV protease overnight at 4 °C. Cleaved Cdc20N was then passed over a HisTrap column (GE Healthcare) and two rounds of cation-exchange Resource S (GE Healthcare) and Superdex 75 (GE Healthcare) purification. The size-exclusion and storage buffer consisted of 20 mM HEPES pH 7.0, 100 mM NaCl, 1 mM TCEP. Mad1CTD and Cdc20N mutants were generated using the QuikChangeTM Lightning Site-Directed Mutagenesis Kit (Agilent), developed by Stratagene Inc. (La Jolla, CA)55.
The Mad1:C-Mad2 tetrameric complexes (His6-DoubleStrep-TEV-Mad1485–718:Mad2 and His6-DoubleStrep-TEV-Mad1485–718:Mad2R133A), as well as GST-Mps1, were cloned into a modified MultiBac system as previously described56. The Mad1:Mad2 complexes were purified as previously described14. Mad2 R133A is a dimerisation-deficient Mad2 mutant and was used when forming a tetrameric-only control complex to prevent super-stoichiometric amounts of Mad2 being present due to Mad2 self-dimerisation25. We found that during baculovirus expression, the amount of Mad2 in the tetrameric complex was substoichiometric, and therefore we individually purified Mad2 WT or Mad2 R133A with an N-terminal double strep-tag followed by a TEV protease site using the same protocols as for Mad1CTD [ref. 21]. Excess Mad2 was added to the Mad1:C-Mad2 complex and re-purified by SEC to produce a homogenous 2:2 complex of Mad1:C-Mad2. Mad1:C-Mad2:O-Mad2 hexameric complex was formed by adding an excess of Mad2LL (Mad2 kinetically locked in the open state), using the same construct and purification strategy as previously described26. The homogeneity of the various Mad1:Mad2 tetrameric and hexameric complexes was confirmed by SEC-MALS (Supplementary Fig. 8a, b). Expression and purification of Mad2 in the open state for use in the Mad2 conversion NMR experiments was carried out as previously described in ref. 57.
GST-Mps1 was purified using a lysis buffer of 25 mM HEPES, pH 7.0, 500 mM NaCl, 10% glycerol and 2 mM DTT supplemented with 25 mg/ml lysozyme, and 0.05% NP-40 and purified over 10 mL of glutathione sepharose™ 4B resin. GST-Mps1 was eluted overnight at 4 °C without removal of the GST-tag to keep full-length Mps1 soluble, using an elution buffer containing 25 mM HEPES pH 8.0, 500 mM NaCl, 2 mM DTT, 10% glycerol and 20 mM reduced glutathione. The eluate was concentrated and then run over a Superdex 200 16/600 column in 20 mM HEPES pH 8.0, 100 mM NaCl and 1 mM TCEP.
Peptide synthesis
Cdc20MIM: NLNGFDVEEAKILRLSGKPQNAPEGYQNRLKVLY, was synthesised by Designer Bioscience, UK. All other peptides were synthesised by Cambridge Research Biochemicals, UK. Cdc20 α1: MAQFAFESDLHSLL. Cdc20 Box1: PPARWQRKAKEAA. AF488-Bub1CD1: [Cys(AF(488)]-W-KVQP-[pSer]-P-[pThr]-VHTKEALGFIMNMFQAPTS, where AF488 is Aurora Fluor 488 and is structurally identical to Alexa Fluor®488 C5 maleimide.
NMR spectroscopy
Uniformly-labelled proteins were expressed in M9 minimal media (6 g/L Na2HPO4, 3 g/L KH2PO4, 0.5 g/L NaCl) supplemented with 1.7 g/L yeast nitrogen base without NH4Cl and amino acids (Sigma Y1251). 1 g/L 15NH4Cl and 4 g/L unlabelled glucose were supplemented for 15N labelling. For 13C/ 15N double-labelled samples, unlabelled glucose was replaced with 3 g/L 13C-glucose. Prior to all NMR experiments, proteins were dialysed into 20 mM HEPES, pH 7.0, 100 mM NaCl and 1 mM TCEP.
The following protein concentrations were used in NMR studies: 1H,15N HSQC of Mad1CTD were collected with a 100 µM sample, whereas a 50 µM sample was used for titrations with Cdc20N and Cdc20α1. Cdc20Box1 peptide was added at mM concentrations to minimise dilution during titration. Limited by solubility, Cdc20N or Cdc20α1 peptides were pre-mixed with Mad1CTD to achieve a final concentration of 50 µM Mad1CTD. 1H,15N HSQC and 13C,15N CON of Cdc20N were collected at 50 and 200 µM respectively. Mad1CTD was concentrated to 3 mM for titrations into Cdc20N. 1H,15N HSQC of Mad2 were collected at 100 µM. All Mad2 NMR samples contain an R133A mutation to prevent Mad2 dimerisation23. The C-Mad2 sample additionally contains an L13A mutation which favours the C-Mad2 state58. Cdc20MIM-bound C-Mad2 sample was prepared by adding a 1:2 excess of Cdc20MIM peptide to an O-Mad2 sample with R133A mutation. To follow the O-to-C conversion of Mad2, the samples were prepared on ice and monitored using BEST 1H,15N-TROSY HSQC. The spectra were acquired with 16 scans and a recycle delay of 400 msec, giving an experimental time of 12 min per spectrum.
Mad1CTD and Mad2 experiments were performed using an in-house 800 MHz Avance III spectrometer and Cdc20N experiments were performed using an in-house 700 MHz Avance II + spectrometer, both equipped with triple resonance TCI CryoProbes. To retrieve signals that suffer from exchange broadening, we also utilised the Bruker 950 MHz Avance III spectrometer located at MRC Biomedical NMR Centre (Francis Crick Institute). Mad1CTD and Mad2 spectra were collected at 298 K. Cdc20N spectra were collected at 278 K to minimise exchange broadening for the largely disordered Cdc20N.
Backbone resonances of Cdc20N were obtained with BEST TROSY versions of triple resonance experiments: HNCO, HN(CA)CO, HNCACB and HN(CO)CACB (Bruker). All 3D datasets were collected with non-uniform sampling at 10–20% and processed in MddNMR59 using compressed sensing reconstruction. Additional non-amide resonances were assigned using 1H start versions of 13C-detected CON, CBCACON and CBCANCO60. Backbone resonances were assigned using in-house scripts and Mars61. Topspin 4.1.1 (Bruker) was used for processing and NMRFAM-Sparky 1.47 for data analysis62.
15N{1H}-heteronuclear NOE values were measured from technical duplicates using standard Bruker pulse sequences and expressed as I/I0 ratio, with interleaved on- (I) and off-resonance (I0) saturation, using a recycle delay of 5 seconds. Secondary chemical shifts were calculated using the equation δCɑobs - δCɑrc where δCɑobs are the observed Cɑ chemical shifts and δCɑrc are the Cɑ chemical shifts for random coils36,37. Random coil chemical shifts were obtained by assigning the backbone resonances of Cdc20N in the presence of 2 M urea using the experiments described above.
For binding studies, the relative peak intensities were normalised to the C-terminal residue Ala718 of Mad1CTD or Gly73 of Cdc20N and expressed as PIbound/PIfree, with PIfree and PIbound being the peak heights of the free and bound forms, respectively. Weighted chemical shift perturbations were calculated using the equation63: Δδ = [(ΔδHNWHN)2 + (ΔδNWN)2)2]1/2 where ΔδHN and ΔδN are the chemical shift perturbations in 1H and 15N dimensions respectively64. The weight factors were determined from the average variances of chemical shifts in the BMRB database65, where WHN = 1 and WN = 0.16.
In vitro protein phosphorylation
Mad1CTD or Mad1:Mad2 complexes and GST-Mps1 were buffer exchanged into 25 mM HEPES pH 8.0, 5% glycerol, 0.1 μM okadaic acid, 15 mM BGP, 2 mM ATP, 10 mM MgCl2, 0.5 mM TCEP, using a PD-10 column (GE Healthcare). Mad1 and GST-Mps1 were mixed at a 5:1 ratio and incubated at 28 °C for 3.5 h before removing the phosphorylation buffer and GST-Mps1 from Mad1 by size-exclusion chromatography.
Mass spectrometry
Identification of phosphorylation sites on Mad1CTD and Mad1:C-Mad2 was performed at the MRC-LMB mass spectrometry facility. Intact proteins were subjected to LC-MS analysis. A modified NanoAcquity (Waters, UK) delivered a flow of approximately 50 µl/min, and proteins were injected directly on a C4 BEH 1.7 µm, 1.0 × 100 mm UPLC column (Waters, UK). Proteins were eluted with a 20-min gradient of acetonitrile (2 to 80%). The analytical column outlet was directly interfaced via an electrospray ionisation source, with a hybrid quadrupole time-of-flight (Q-TOF) mass spectrometer (Xevo G2, Waters, UK). Data were acquired over an m/z range of 350–2000, in positive ion mode with a cone voltage of 30 v. Scans were summed together manually and deconvoluted using MaxEnt1 (Masslynx, Waters, UK).
Peptide mass spectrometry was completed after reduction with 10 mM DTT and alkylation with 55 mM iodoacetamide and protein digestion overnight at 37 °C at a 1:50 ratio of trypsin (Promega, UK). Tryptic peptides were analysed by nano-scale capillary LC-MS/MS with an Ultimate U3000 HPLC (ThermoFisher Scientific Dionex, San Jose, USA) set to a flow rate of 300 nL/min. Peptides were trapped on a C18 Acclaim PepMap100 5 μm, 100 μm × 20 mm nanoViper (Thermo Fisher Scientific Dionex, San Jose, USA) prior to separation on a C18 T3 1.8 μm, 75 μm × 250 mm nanoEase column (Waters, Manchester, UK). A gradient of acetonitrile eluted the peptides, and the analytical column outlet was directly interfaced using a nano-flow electrospray ionisation source, with a quadrupole Orbitrap mass spectrometer (Q-Exactive HFX, ThermoFisher Scientific, USA). For data-dependent analysis a resolution of 60,000 for the full MS spectrum was used, followed by twelve MS/MS. The data were then searched against an LMB in-house database using a Mascot search engine (Matrix Science)66, and the peptide identifications were validated using the Scaffold programme (Proteome Software Inc.)67.
SEC-MALS
Size-exclusion chromatography coupled with multi-angle static light scattering (SEC-MALS), was performed using an Agilent 1200 series LC system with an online Dawn Helios ii system (Wyatt) equipped with a QELS + module (Wyatt) and an Optilab rEX differential refractive index detector (Wyatt). About 100 μl purified protein at ~3.0 mg/ml was auto-injected onto Superdex 75 or a Superdex 200 Increase 10/300 GL column (GE Healthcare) and run at 0.5 ml/min. The molecular masses were analysed with ASTRA 7.3.0.11 (Wyatt). Data were plotted using Prism 8.4.3 (GraphPad Software, Inc). Purified Cdc20N and Cdc20N mutants were incubated with Mad1CTD wild-type or mutants at a 1:1.2 ratio of Mad1CTD dimer to Cdc20N.
AUC-SE
Sedimentation equilibrium experiments were carried out in a buffer of 20 mM HEPES pH 7.5, 100 mM NaCl and 1 mM TCEP. Samples were loaded into 12 mm six-sector cells, placed in an An-50Ti rotor and centrifugated at 10,200, 12,200 and 21,000 rpm at 20 °C until equilibrium had been reached using an Optima XL-I analytical ultracentrifuge (Beckman). The data were analysed in SEDPHAT 15.2b68. The partial-specific volumes (v-bar) were calculated using Sednterp69. The density and viscosity of the buffer were determined with a DMA 4500 M density metre (Anton Parr) and an AMVn viscometer (Anton Paar). Data were plotted with the program GUSSI70.
Fluorescent anisotropy
All reactions were completed in a buffer of 25 mM HEPES pH 7.75, 1 mM TCEP, 100 mM NaCl, 0.05% (v/v) Tween-20. To analyse the binding of Bub1CD1 to Mad1CTD, a twofold dilution series of Mad1CTD from 100 μM to 6 nM was prepared and mixed 1:1 with fluorescently-labelled AF488-Bub1CD1 at 40 nM. To analyse the binding of Cdc20N to the Mad1CTD:AF488-Bub1CD1 complex, a mixture of 40 nM AF488-Bub1CD1 and 4.2 μM Mad1CTD was added 1:1 with a twofold dilution series of Cdc20N from 100 μM to 6 nM. To analyse the binding of Bub1CD1 to the Mad1CTD:Cdc20N complex and test for cooperativity, a twofold dilution series of Mad1CTD from 96 μM to 2.9 nM was mixed 1:1 with 92 μM of Cdc20N, incubated on ice for 30 min and then mixed 1:1 with 40 nM of AF488-Bub1CD1. Measurements were performed with a PheraStar plate reader FSX plate reader (BMG Labtech) using an optic module for λex = 485 nm, λem = 520 nm. Reactions were carried out in a total volume of 40 μl at 25 °C in a black, flat-bottom, non-binding surface 384-well plate (Corning). All experiments were performed as technical triplicates and data were analysed in PRISM 9.3.1 (GraphPad Software). For titration of wild-type Cdc20N and Cdc20N the fit for the top value of the anisotropy was constrained to be a shared value between the phosphorylated and non-phosphorylated Mad1CTD-Bub1CD1 samples, as further titration of Cdc20N to saturation was not possible due to large increases in the total fluorescence intensity indicating aggregation or non-specific binding to the plate.
BS3 cross-linking for cryo-EM
Purified homogenous Mad1:C-Mad2:O-Mad2 complex in the unphosphorylated or phosphorylated state were cross-linked with BS3 (bis[sulfosuccinimidyl] suberate) (Thermo Fisher Scientific) before analysis by cryo-electron microscopy. About 2 mM BS3 was incubated with Mad1:Mad2 complexes at 0.3 mg/ml for 1 h at 4 °C. BS3 was then quenched by the addition of Tris-HCl pH 8.0 to 100 mM and incubated on ice for 30 min. The cross-linked complex was then concentrated to 3.5 mg/ml and re-purified by size-exclusion chromatography, first on a Superdex 200 15/300 column (GE Healthcare), and then again on a Superdex 200 5/150 (GE Healthcare).
Cryo-EM grid preparation and data acquisition
Copper Quantifoil 1.2/1.3, 300 mesh, grids were used. Grids were treated with a 9:1 argon: oxygen plasma for 45 s, after which 3.5 μl of the purified complex at 1.5–2.5 mg/ml supplemented with 0.05% NP-40 (1 h before grid application) was pipetted onto the grid at 4 °C and 100% humidity before immediately blotting for 2 s at blot force −10 and plunging into liquid ethane using an FEI Vitrobot Mark III (Thermo Fisher Scientific). Data were collected at 300 kV on a Titan Krios, equipped with a K3 detector with a pixel size of 1.1 Å (Gatan). A volta-phase plate (VPP) was additionally used with the phosphorylated Mad1:C-Mad2:O-Mad2, as the VPP was found to improve the data quality and allowed seeing the coiled-coil connection between the flexible Mad1CTD and core. Data acquisition statistics are outlined in Supplementary Table 2.
Cryo-EM data processing
Data were pre-processed using Warp v1.09, including training and then implementing a model for particle picking71. All further processing was carried out in CryoSparc v2.15.072. Particles were cleaned up using repetitive 2D classifications. Initial 3D reconstructions were created using an SGD algorithm72. For the unphosphorylated Mad1:C-Mad2:O-Mad2 complex, heterogenous refinement was used to further clean up the data, after which homogenous refinement was used to obtain a final resolution of 8.9 Å (Supplementary Table 2). For the phosphorylated Mad1:C-Mad2:O-Mad2 complex, 2D classification was used to remove open-state particles (7.6% of the total particles) and particles where the head domain was not closely folded towards the core (12.3% of total particles). Further repetitive rounds of 2D classification were performed until classes which clearly presented the head domain folded towards the core with the coiled-coil being visible. These classes were then used to make an initial model (4,714 particles). Attempts at 3D classification and heterogenous refinement did not help to produce a higher resolution map, due to the high flexibility of this complex. Many different models with the head domain in slightly different degrees of folding could be obtained, and no stable conformation could be identified. Ultimately non-uniform refinement in cryoSPARC was used to produce a model for a folded state of Mad1:Mad2 with a final resolution of 11.1 Å at 0.143 FSC and 16.3 Å at 0.5 FSC73. The reported estimated final map resolutions were calculated based on the gold standard Fourier shell correlation (GS-FSC) 0.143 or 0.5 criterion74–77. Crystal structures of Mad1CTD (PDB ID: 4DZO24), Mad1:C-Mad2 (PDB ID: 1GO423) and C-Mad2:O-Mad2 (PDB ID: 2V6426) were manually docked into the EM density maps using in Chimera78.
In-gel cross-linking mass spectrometry (IGX-MS)
Purified complexes of the tetrameric Mad1485–718:C-Mad2 (using Mad2R133A) complex, as well as the unphosphorylated and phosphorylated hexameric Mad1485–718:C-Mad2:O-Mad2 complexes (with O-Mad2 being the Mad2LL mutant described earlier) were cross-linked following the published IGX-MS workflow40. Respective complexes were mixed with NativePAGE sample buffer, and subsequently, 15 µg of each sample was run onto a Bis-Tris gel (3–12%). For cross-linking, bands corresponding to tetrameric and hexameric Mad1:Mad2 complexes were excised, rinsed with distilled H2O and subsequently cross-linked (in triplicates) with 1.5 mM DSS for 30 min at 25 °C. The reaction was quenched by the addition of 1 M Tris pH 8.5 to a final concentration of 50 mM. Following classical in-gel digestion79, cross-linked proteins were washed, reduced and alkylated prior to trypsin digestion. For MS analysis of cross-linked peptides, the samples were re-suspended in 2% formic acid and analysed using an UltiMate™ 3000 RSLCnano System (Thermo Fisher Scientific) coupled online to an Orbitrap Exploris 480 (Thermo Fischer Scientific). Peptides were trapped for 5 min in 0.1% formic acid (FA) in water, using a 100-µm inner diameter 2-cm trap column (packed in-house with ReproSil-Pur C18-AQ, 3 µm) prior to separation on an analytical column (50 cm of length, 75 µM inner diameter; packed in-house with Poroshell 120 EC-C18, 2.7 µm). Trapped peptides were eluted following a 60 min gradient from 9–40% of 80% ACN, 0.1% FA. Full scan MS spectra from 350–1600 m/z were acquired in the Orbitrap at a resolution of 60,000 with a normalised AGC target of 300% and maximum injection time of 120 ms. Only peptides with charged states 3–8 were fragmented, and dynamic exclusion properties were set to n = 1, for a duration of 30 s. Fragmentation was performed using a stepped HCD collision energy mode (27, 30, 33%) in the ion trap and acquired in the Orbitrap at a resolution of 30,000 with an AGC target set to 500%, an isolation window of 1.4 m/z and a maximum injection time of 54 ms. Raw files obtained for respective IGX-MS experiments were subsequently analysed with pLink280. FDR (controlled at PSM level for cross-linked spectrum matches and separately computed for intra and inter cross-links) rate was set to 5%. Acetylation (protein N-terminus), oxidation (Met) and phosphorylation (Ser/Thr/Tyr) were set as dynamic modifications. Carbamidomethylation (Cys) was set as a static modification.
Crosslinks observed in two out of three technical replicates for each sample were used for further analysis. A total of 41 cross-links were detected in unphosphorylated Mad1:C-Mad2 tetramer, and 68 and 62 cross-links for unphosphorylated and phosphorylated Mad1:C-Mad2:O-Mad2 hexamer, respectively. Crosslinks were plotted onto the Alphafold2 structures of tetrameric and hexameric Mad1:Mad2 complexes using the XMAS bundle in ChimeraX81. Venn diagrams showing the overlap between replicates and Boxen plots to visualise cross-link distances were generated using seaborn and matplotlib in python. The Boxen plots were created using the letter-value plot (Boxen plot) method82. Boxen plots are similar to a box plot, but they additionally plot the quartile values, starting with the median (Q2, 50th percentile) as the centreline, and each successive level outwards contains half of the remaining data (thereby describing the box bounds), continuing until the outlier level (5–8 outliers per tail) is reached. The box for each level out is depicted in lighter colour shades. A boxen plot was used to improve visualisation of the distribution of cross-link distances for Mad1:Mad2 complexes, which are highly flexible and thus have more outliers. The contact maps for highlighting the cross-links with respect to elongated and folded Mad1:C-Mad2:O-Mad2 were generated using the Bio.PDB package83 in combination with seaborn and matplotlib in python.
SEC-SAXS
The Mad1:Mad2 complexes (tetrameric Mad1485–718:C-Mad2R133A and hexameric Mad1485–718:C-Mad2:O-Mad2 in the unphosphorylated and phosphorylated states, where O-Mad2 is the Mad2LL [ref. 26] were analysed by size-exclusion chromatography coupled to small angle X-ray scattering (SEC-SAXS) at the BL21 beamline at Diamond Lightsource (DLS) (Harwell Campus, Didcot, United Kingdom), using a Shodex KW-403 column (Shodex) pre-equilibrated in 25 mM HEPES pH 7.5, 100 mM NaCl, 1 mM TCEP84. To confirm column performance and instrument calibration, 40 μl of BSA at 10 mg/mL was analysed prior to experiments. In total, 40 μl of each sample was injected at 9, 6 or 3 mg/mL. No significant difference was seen between samples at different dilutions, and therefore only the highest concentration (9 mg/ml) of each sample is presented. Raw SAXS images were processed with the DAWN85 processing pipeline at the DLS beamline to produced normalised and integrated 1-d un-subtracted SAXS curves. SEC-SAXS buffer subtraction and analyses were performed in Scatter IV (BioISIS). The expected Rg values of the modelled folded and elongated states of Mad1:Mad2 were calculated using the FoXS server41.
Computational methods
Molecular analyses were performed with the UCSF Chimera package78. Chimera is developed by the Resource for Biocomputing, Visualisation, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). Molecular graphics were produced in PyMOL Molecular Graphics System, Version 2.3.3 Schrödinger, LLC. Protein structure predictions were generated using AlphaFold238 using the notebook-based ColabFold environment and the MMseqs2 MSA option86. The majority of protein secondary structure predictions were performed with PHYRE2 or PSIPRED proteins sequence analysis workbench87–89. Multiple sequence alignments were completed in BLAST90.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
E.S.F. was funded by a Gates Cambridge Scholarship. This work was funded by an MRC grant (MC_UP_1201/6) and a CRUK grant (C576/A14109) to D.B. The XL-MS experiments were supported by the Horizon 2020 INFRAIA project Epic-XS (Project 823839 to A.J.R.H.). We thank K. Yan and the MRC-LMB EM facility for helping with EM data collection. J. Grimmett and T. Darling for computing and J. Shi for help with insect cell expression. We acknowledge S. Tamara (University of Utrecht) for additional cross-linking mass spectrometry experiments and T. Dendooven for advice in SEC-SAXS analyses. We thank Ajit Jogelkar and Andrea Musacchio for discussing unpublished work and for useful discussions regarding Mad1 fold-over and Mad2 conversion, and Andrea Musacchio for helpful comments on this manuscript. Some of the NMR studies were supported by the Francis Crick Institute through access to the MRC Biomedical NMR Centre. The Francis Crick Institute receives its core funding from Cancer Research UK (FC001029), the UK Medical Research Council (FC001029), and the Wellcome Trust (FC001029). The authors also thank the B21 SAXS beamline at the Diamond Light Source, Oxfordshire, for the measurements of mail-in SAXS samples.
Source data
Author contributions
E.S.F. cloned, purified proteins, performed the biochemical assays, analysed SAXS data, collected and processed the cryo-EM data. C.W.H.Y. and S.M.V.F. acquired and analysed the NMR data. J.F.H. performed IGX-MS with supervision from A.J.R.H. S.H.M. completed AUC-SE and SEC-MALS. S.L.M. performed in-house mass spectrometry. E.S.F., C.W.H.Y., and D.B. wrote the manuscript with input from all authors.
Peer review
Peer review information
Nature Communications thanks Beat Vögeli and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The data that support this study are available from the corresponding authors upon request. The NMR assignments of Cdc20N were deposited to the BMRB database (http://www.bmrb.wisc.edu/) with the accession number 51304. The in-gel cross-linking mass spectrometry (IGX-MS) data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/) partner repository with the dataset identifier PXD031872. PDBs used in this study: 4DZO, 1GO4, 2V64, 7B1F. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Elyse S. Fischer, Conny W. H. Yu.
Contributor Information
Elyse S. Fischer, Email: efischer@mrc-lmb.cam.ac.uk
David Barford, Email: dbarford@mrc-lmb.cam.ac.uk.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-34058-2.
References
- 1.Monda, J. K. & Cheeseman, I. M. The kinetochore-microtubule interface at a glance. J. Cell Sci. 131, jcs214577 (2018). [DOI] [PMC free article] [PubMed]
- 2.Pesenti ME, Weir JR, Musacchio A. Progress in the structural and functional characterization of kinetochores. Curr. Opin. Struct. Biol. 2016;37:152–163. doi: 10.1016/j.sbi.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Foley EA, Kapoor TM. Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat. Rev. Mol. Cell Biol. 2013;14:25–37. doi: 10.1038/nrm3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musacchio A. The molecular biology of spindle assembly checkpoint signaling dynamics. Curr. Biol. 2015;25:R1002–R1018. doi: 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 5.Sacristan C, Kops GJPL. Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol. 2015;25:21–28. doi: 10.1016/j.tcb.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Herzog F, et al. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izawa D, Pines J. The mitotic checkpoint complex binds a second CDC20 to inhibit active APC/C. Nature. 2015;517:631–634. doi: 10.1038/nature13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alfieri C, et al. Molecular basis of APC/C regulation by the spindle assembly checkpoint. Nature. 2016;536:431–436. doi: 10.1038/nature19083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaguchi M, et al. Cryo-EM of mitotic checkpoint complex-bound APC/C reveals reciprocal and conformational regulation of ubiquitin ligation. Mol. Cell. 2016;63:593–607. doi: 10.1016/j.molcel.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudakin V, Chan GKT, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J. Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chao WCH, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 12.Faesen AC, et al. Basis of catalytic assembly of the mitotic checkpoint complex. Nature. 2017;542:498–502. doi: 10.1038/nature21384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonetta M, et al. The influence of catalysis on Mad2 activation dynamics. PLoS Biol. 2009;7:e1000010. doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji Z, Gao H, Jia L, Li B, Yu H. A sequential multi-target Mps1 phosphorylation cascade promotes spindle checkpoint signaling. Elife. 2017;6:e22513. doi: 10.7554/eLife.22513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lara-Gonzalez P, Pines J, Desai A. Spindle assembly checkpoint activation and silencing at kinetochores. Semin. Cell Dev. Biol. 2021;117:86–98. doi: 10.1016/j.semcdb.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr. Biol. 2012;22:900–906. doi: 10.1016/j.cub.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shepperd LA, et al. Phosphodependent recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 kinase maintains the spindle checkpoint. Curr. Biol. 2012;22:891–899. doi: 10.1016/j.cub.2012.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat. Cell Biol. 2012;14:746–752. doi: 10.1038/ncb2515. [DOI] [PubMed] [Google Scholar]
- 19.London N, Biggins S. Mad1 kinetochore recruitment by Mps1-mediated phosphorylation of Bub1 signals the spindle checkpoint. Genes Dev. 2014;28:140–152. doi: 10.1101/gad.233700.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, et al. Bub1 positions Mad1 close to KNL1 MELT repeats to promote checkpoint signalling. Nat. Commun. 2017;8:15822. doi: 10.1038/ncomms15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, E. S. et al. Molecular mechanism of Mad1 kinetochore targeting by phosphorylated Bub1. EMBO Rep. 22, e52242 (2021). [DOI] [PMC free article] [PubMed]
- 22.Klebig C, Korinth D, Meraldi P. Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 2009;185:841–858. doi: 10.1083/jcb.200902128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc. Natl Acad. Sci. USA. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 26.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 27.Lou X, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 28.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 29.Luo X, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat. Struct. Mol. Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 30.Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–1625. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lad L, Lichtsteiner S, Hartman JJ, Wood KW, Sakowicz R. Kinetic analysis of Mad2-Cdc20 formation: conformational changes in Mad2 are catalyzed by a C-Mad2-ligand complex. Biochemistry. 2009;48:9503–9515. doi: 10.1021/bi900718e. [DOI] [PubMed] [Google Scholar]
- 33.Piano V, et al. CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science. 2021;371:67–71. doi: 10.1126/science.abc1152. [DOI] [PubMed] [Google Scholar]
- 34.Ji W, Luo Y, Ahmad E, Liu ST. Direct interactions of mitotic arrest deficient 1 (MAD1) domains with each other and MAD2 conformers are required for mitotic checkpoint signaling. J. Biol. Chem. 2018;293:484–496. doi: 10.1074/jbc.RA117.000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lara-Gonzalez P, Kim T, Oegema K, Corbett K, Desai A. A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science. 2021;371:64–67. doi: 10.1126/science.abc1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kakeshpour T, et al. A lowly populated, transient β-sheet structure in monomeric Aβ1-42 identified by multinuclear NMR of chemical denaturation. Biophys. Chem. 2021;270:106531. doi: 10.1016/j.bpc.2020.106531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spera S, Bax A. Empirical correlation between protein backbone conformation and Cα and Cβ 13C nuclear magnetic resonance chemical shifts. J. Am. Chem. Soc. 1991;113:5490–5492. [Google Scholar]
- 38.Jumper J, et al. Highly accurate protein structure prediction with AlphaFold. Nat 2021 5967873. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, C. et al. The structural flexibility of MAD1 facilitates the assembly of the mitotic checkpoint complex. Preprint at bioRxiv10.1101/2022.06.29.498198 (2022). [DOI] [PMC free article] [PubMed]
- 40.Hevler JF, et al. Selective cross‐linking of coinciding protein assemblies by in‐gel cross‐linking mass spectrometry. EMBO J. 2021;40:e106174. doi: 10.15252/embj.2020106174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. Accurate SAXS profile computation and its assessment by contrast variation experiments. Biophys. J. 2013;105:962–974. doi: 10.1016/j.bpj.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneidman-Duhovny D, Hammel M, Tainer JA, Sali A. FoXS, FoXSDock and MultiFoXS: Single-state and multi-state structural modeling of proteins and their complexes based on SAXS profiles. Nucleic Acids Res. 2016;44:W424–W429. doi: 10.1093/nar/gkw389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burchard W, Schmidt M, Stockmayer WH. Information on polydispersity and branching from combined quasi-elastic and integrated scattering. Macromolecules. 1980;13:1265–1272. [Google Scholar]
- 44.Kim S, et al. Phosphorylation of the spindle checkpoint protein Mad2 regulates its conformational transition. Proc. Natl Acad. Sci. USA. 2010;107:19772–19777. doi: 10.1073/pnas.1009000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lischetti T, Zhang G, Sedgwick GG, Bolanos-Garcia VM, Nilsson J. The internal Cdc20 binding site in BubR1 facilitates both spindle assembly checkpoint signalling and silencing. Nat. Commun. 2014;5:1–12. doi: 10.1038/ncomms6563. [DOI] [PubMed] [Google Scholar]
- 46.Diaz-Martinez LA, et al. The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J. Biol. Chem. 2015;290:2431–2443. doi: 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiFiore B, et al. The ABBA Motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev. Cell. 2015;32:358–372. doi: 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Di Fiore B, Wurzenberger C, Davey NE, Pines J. The mitotic checkpoint complex requires an evolutionary conserved cassette to bind and inhibit active APC/C. Mol. Cell. 2016;64:1144–1153. doi: 10.1016/j.molcel.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol. Cell. Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han JS, et al. Catalytic assembly of the mitotic checkpoint inhibitor BubR1-Cdc20 by a Mad2-induced functional Switch in Cdc20. Mol. Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang, G. et al. Efficient mitotic checkpoint signaling depends on integrated activities of Bub1 and the RZZ complex. EMBO J. 38, e100977 (2019). [DOI] [PMC free article] [PubMed]
- 52.Heinrich S, et al. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 2014;15:291–298. doi: 10.1002/embr.201338114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bitinaite J, et al. USERTM friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Demple B, Linn S. On the recognition and cleavage mechanism of Escherichia coli endodeoxyribonuclease V, a possible DNA repair enzyme. J. Biol. Chem. 1982;257:2848–2855. [PubMed] [Google Scholar]
- 55.Nøhr J, Kristiansen K. Site-directed mutagenesis. Methods Mol. Biol. 2003;232:127–131. doi: 10.1385/1-59259-394-1:127. [DOI] [PubMed] [Google Scholar]
- 56.Zhang Z, Yang J, Barford D. Recombinant expression and reconstitution of multiprotein complexes by the USER cloning method in the insect cell-baculovirus expression system. Methods. 2016;95:13–25. doi: 10.1016/j.ymeth.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Luo X, Yu H. Purification and assay of Mad2: a two-state inhibitor of anaphase-promoting complex/cyclosome. Methods Enzymol. 2005;398:246–255. doi: 10.1016/S0076-6879(05)98020-8. [DOI] [PubMed] [Google Scholar]
- 58.Yang M, et al. Insights into Mad2 regulation in the spindle checkpoint revealed by the crystal structure of the symmetric Mad2 dimer. PLoS Biol. 2008;6:0643–0655. doi: 10.1371/journal.pbio.0060050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaravine VA, Zhuravleva AV, Permi P, Ibraghimov I, Orekhov VY. Hyperdimensional NMR spectroscopy with nonlinear sampling. J. Am. Chem. Soc. 2008;130:3927–3936. doi: 10.1021/ja077282o. [DOI] [PubMed] [Google Scholar]
- 60.Bermel W, et al. H-start for exclusively heteronuclear NMR spectroscopy: the case of intrinsically disordered proteins. J. Magn. Reson. 2009;198:275–281. doi: 10.1016/j.jmr.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 61.Jung YS, Zweckstetter M. Backbone assignment of proteins with known structure using residual dipolar couplings. J. Biomol. NMR. 2004;30:25–35. doi: 10.1023/B:JNMR.0000042955.14647.77. [DOI] [PubMed] [Google Scholar]
- 62.Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2015;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williamson MP. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Ayed A, et al. Latent and active p53 are identical in conformation. Nat. Struct. Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 65.Mulder FAA, Schipper D, Bott R, Boelens R. Altered flexibility in the substrate-binding site of related native and engineered high-alkaline Bacillus subtilisins. J. Mol. Biol. 1999;292:111–123. doi: 10.1006/jmbi.1999.3034. [DOI] [PubMed] [Google Scholar]
- 66.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 67.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 68.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 2003;320:104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 69.Cole JL, Lary JW, P. Moody T, Laue TM. Analytical ultracentrifugation: sedimentation velocity and sedimentation equilibrium. Methods Cell Biol. 2008;84:143–179. doi: 10.1016/S0091-679X(07)84006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brautigam, C. A. Calculations and publicationquality illustrations for analytical ultracentrifugation data. Methods Enzymol.562, 109–133 (2015). [DOI] [PubMed]
- 71.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. CryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 73.Punjani A, Zhang H, Fleet DJ. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods. 2020;17:1214–1221. doi: 10.1038/s41592-020-00990-8. [DOI] [PubMed] [Google Scholar]
- 74.Scheres SHW, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neumann P, Dickmanns A, Ficner R. Validating resolution revolution. Structure. 2018;26:785–795.e4. doi: 10.1016/j.str.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 77.Ramírez-Aportela E, et al. FSC-Q: a CryoEM map-to-atomic model quality validation based on the local Fourier shell correlation. Nat. Commun. 2021;12:1–7. doi: 10.1038/s41467-020-20295-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pettersen EF, et al. UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 79.Shevchenko A, Tomas H, Havliš J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 80.Chen ZL, et al. A high-speed search engine pLink 2 with systematic evaluation for proteome-scale identification of cross-linked peptides. Nat. Commun. 2019;10:1–12. doi: 10.1038/s41467-019-11337-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lagerwaard, I. M., Albanese, P., Jankevics, A. & Scheltema, R. A. Xlink mapping and analySis (XMAS) - smooth integrative modeling in chimeraX. Preprint at bioRxiv10.1101/2022.04.21.489026 (2022).
- 82.Hofmann H, Wickham H, Kafadar K. Letter-value plots: boxplots for large data. J. Comput. Graph. Stat. 2017;26:469–477. [Google Scholar]
- 83.Cock PJA, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cowieson NP, et al. Beamline B21: high-throughput small-angle X-ray scattering at diamond light source. J. Synchrotron Radiat. 2020;27:1438–1446. doi: 10.1107/S1600577520009960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Basham M, et al. Data analysis workbeNch (DAWN) J. Synchrotron Radiat. 2015;22:853–858. doi: 10.1107/S1600577515002283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mirdita, M., Ovchinnikov, S. & Steinegger, M. ColabFold - Making protein folding accessible to all. Nat Methods19, 679–682 (2022). [DOI] [PMC free article] [PubMed]
- 87.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 89.Buchan DWA, Jones DT. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that support this study are available from the corresponding authors upon request. The NMR assignments of Cdc20N were deposited to the BMRB database (http://www.bmrb.wisc.edu/) with the accession number 51304. The in-gel cross-linking mass spectrometry (IGX-MS) data have been deposited to the ProteomeXchange Consortium via the PRIDE (https://www.ebi.ac.uk/pride/) partner repository with the dataset identifier PXD031872. PDBs used in this study: 4DZO, 1GO4, 2V64, 7B1F. Source data are provided with this paper.