Summary
GABAB receptors are obligatory heterodimers responsible for prolonged neuronal inhibition in the central nervous system. The two receptor subunits are encoded by GABBR1 and GABBR2. Variants in GABBR2 have been associated with a Rett-like phenotype (MIM: 617903), epileptic encephalopathy (MIM: 617904), and milder forms of developmental delay with absence epilepsy. To date, however, no phenotypes associated with pathogenic variants of GABBR1 have been established. Through GeneMatcher, we have ascertained four individuals who each have a monoallelic GABBR1 de novo non-synonymous variant; these individuals exhibit motor and/or language delay, ranging from mild to severe, and in one case, epilepsy. Further phenotypic features include varying degrees of intellectual disability, learning difficulties, autism, ADHD, ODD, sleep disorders, and muscular hypotonia. We functionally characterized the four de novo GABBR1 variants, p.Glu368Asp, p.Ala397Val, p.Ala535Thr, and p.Gly673Asp, in transfected HEK293 cells. GABA fails to efficiently activate the variant receptors, most likely leading to an increase in the excitation/inhibition balance in the central nervous system. Variant p.Gly673Asp in transmembrane domain 3 (TMD3) renders the receptor completely inactive, consistent with failure of the receptor to reach the cell surface. p.Glu368Asp is located near the orthosteric binding site and reduces GABA potency and efficacy at the receptor. GABA exhibits normal potency but decreased efficacy at the p.Ala397Val and p.Ala535Thr variants. Functional characterization of GABBR1-related variants provides a rationale for understanding the severity of disease phenotypes and points to possible therapeutic strategies.
Keywords: gene, mendelian disease, GABBR1
Pathogenic mutations in the human gene GABBR1 (encoding a protein that forms a receptor in neuronal cells) cause a rare autosomal dominant disorder with neurodevelopmental delay and epilepsy. These findings are important for diagnosis and genetic counseling; in addition, they provide the opportunity for development of new therapies.
Introduction
The major inhibitory neurotransmitter in the central nervous system is γ-aminobutyric acid (GABA). GABA acts at ionotropic GABAA/GABAC receptors and metabotropic GABAB receptors.1 GABAB receptors are G protein-coupled receptors that reduce cAMP levels and regulate Kir3-type potassium and voltage-sensitive calcium channels.1,2 GABAB receptors are expressed throughout the brain in glial cells and neurons. In neurons, they act at pre- and postsynaptic sites, where they inhibit neurotransmitter release and generate the late phase of inhibitory postsynaptic potentials.2 GABAB receptors in microglia have been implicated in the sculpting of inhibitory circuits.3 In astrocytes, GABAB receptors mediate calcium elevations and influence hippocampal theta and gamma oscillations.4,5
GABAB receptors are obligatory heterodimers made up of a GB1a or GB1b with a GB2 subunit, which are encoded by the GABBR1 (MIM: 603540) and GABBR2 genes (MIM: 607340).2 GB1a and GB1b are subunit variants generated by alternative promoter usage. GB1a and GB1b have similar functional and pharmacological properties when expressed in heterologous cells.2 GB1 and GB2 subunits share 54% amino acid sequence and 35% nucleotide sequence similarity. Heterodimerization of the GB1 and GB2 subunits is necessary for receptor trafficking from the endoplasmic reticulum to the cell surface.6, 7, 8, 9 GB1 and GB2 subunits consist of three domains: (1) an extracellular Venus-fly-trap domain (VFTD) composed of an N-terminal domain (LB1) and a C-terminal domain (LB2), (2) a heptahelical transmembrane domain (7TMD), and (3) a C-terminal intracellular domain.1,10 The VFTD of the GB1a and GB1b subunits contains the orthosteric binding site while the VTFD on the GB2 subunit is coupling to the G protein. The LB1 and LB2 domains of the GB1 and GB2 VFTD interact with each other to form the extracellular heterodimer interface, which is essential for receptor activation.1,10
GABAB receptor dysfunctions have been implicated in neurological and neurodevelopmental disorders including anxiety, autism, depression, addiction, epilepsy, and cognition11, 12, 13, 14. GABBR2 variants are implicated in Rett-like symptoms, epileptic encephalopathy, and severe intellectual disability.11, 12, 13, 14 As GABAB receptors are obligatory heterodimers, one could expect a similar phenotypic spectrum for pathological variants in GABBR1. Indeed, mice lacking the GB1 or GB2 subunits develop similar neurological disorders, including clonic type seizures, but also absence and tonic-clonic seizures as well as hyperactivity, hyperalgesia, and poor memory performance.15,16 In humans, one study suggested that a GABBR1 non-coding polymorphic variant is associated with temporal lobe epilepsy, but this finding is controversial.17,18
In this study, we report four individuals with heterozygous GABBR1 de novo missense variants, all presenting neurodevelopmental motor and/or language delays. Two of the four individuals have varying degrees of intellectual disability, learning difficulties, and a sleep disorder. Two individuals have attention-deficit/hyperactivity disorder (ADHD), and one individual additionally presents with other concomitant psychiatric disorders (autism, borderline personality disorder, oppositional defiant disorder [ODD], social anxiety, and one episode of major depressive disorder [MDD]). One individual is treated for epileptic seizures. Functional analyses reveal that all four variants produce dysfunctional receptors, supporting that these de novo variants are pathogenic.
Material and methods
Individuals
Four individuals were evaluated at their respective clinical institutions and the GABBR1 variants were respectively added to the web-based platform GeneMatcher19 and collected. Each clinic obtained a written consent form to participate in the publication from the legal guardians. The study was performed in accordance with the principles of Swiss Society of Medical Genetics.
Whole-exome sequencing
Each center performed a whole-exome sequencing (WES) in trio (proband and parents). The methodology of each center is provided in the supplemental information.
Functional analysis
A functional characterization of the four de novo GB1 subunit variants (p.Glu368Asp, p.Ala397Val, p.Ala535Thr, and p.Gly673Asp)]was carried out in transfected HEK293 cells.
Plasmids and reagents
Human GB1b and GB2 plasmids were used earlier;20 SRE-FLuc (28) was a gift from M. Choi, Seoul. Mutant GB1b subunits and N-terminally HA-tagged subunits were generated with the Q5 Site-Directed Mutagenesis kit (New England Biolabs, Ipswich, USA). The following primers were used: p.Glu368Asp_For ACTTTTCTATgacACTGAAGCCCGG and p.Glu368Asp_Rev CCCACGATGATTCGGGCA; p.(Ala397Val) was generated with the following mutagenic primers: p.(Ala397Val)_For TGGGTGGTATgttGACAATTGGT and p.(Ala397Val)_Rev ATGAGGAACCAGACGTAC; p.(Ala535Thr)_For CTCTCGGATGacaTGGACGCTTATC and p.(Ala535Thr)_Rev CCCACGATGATTCGGGCA; p.(Gly673Asp) _For CCTGGGCCTGgacTTTAGTCTGGGC and p.(Gly673Asp) _Rev AGCCAGAGGCGGGCCTGG (small caps indicate the mutation). The N-terminal HA-tag was inserted with GB1b_HA_For ttatgcgggcggcggcagcTCCCACTCCCCCCATCTCC and GB1b_Rev tccggcacatcatacggataGGCCCACACCGGAGCCAC (small caps indicate HA-tag). GABA and CGP54626 were from Tocris Bioscience (Bristol, England). Anti-HA antibodies (clone 3F10) were from Roche (Basel, Switzerland). Anti-GB1 antibodies (#55051) were from Abcam (Cambridge, UK). Anti-βactin antibodies (#4970) were from Cell Signaling Technology (Danvers, USA). AF568-conjugated anti-rat and AF488-conjugated anti-mouse secondary antibodies were from Invitrogen. HRP-conjugated anti-mouse and anti-rabbit antibodies were from GE Healthcare (Chicago, USA). Poly-L-Lysine was from Sigma (Burlington, USA). The following transfection reagents were used: Lipofectamine 2000 from Invitrogen (Waltham, USA) and PEI from Sigma. The Plasma Membrane Protein Extraction Kit was from Abcam.
Cell surface expression
HEK293 cells were transiently transfected (Lipofectamine 2000, 2:1 ratio) with HA-GB1b and GB2 constructs. 6 h later, cells were detached and plated on PLL-coated glass coverslips in a 24-well plate at a density of 45,000 cells/well. After 2 days, the growth medium was replaced with Opti-MEM-GlutaMAX supplemented with anti-HA antibody (1:1,000) for 1 h. Subsequently, cells were fixed (4% PFA + 4% sucrose, 10 min, RT), permeabilized (PBS + 0.2% Triton X-100), and incubated overnight at 4°C with the anti-GB1 antibody (1:1,000). The following day, cells were washed with 1×PBS and incubated for 90 min at room temperature with AF568 (HA-tag) and AF488-conjugated (anti-GB1) secondary antibodies (1:500). Imaging was performed on Zeiss LSM880 confocal microscope with a PLAN APO 63× objective. Samples were excited at 488 and 555 nm. Image processing and analysis was done in Fiji (ImageJ). Maximum projection images were generated from Z stacks, and average fluorescence intensity was measured in regions of interest (ROIs) encompassing individual cells. We used ROIs of the same shape to obtain background values for the same imaging channel. HA-to-GB1 fluorescence intensity ratios were calculated in Microsoft Excel and exported to GraphPad Prism for statistical analysis. Data were tested for normality with the Shapiro-Wilk normality test and analyzed via Kruskal-Wallis with Dunn’s multiple comparison tests.
Protein fractionation
HEK293 cells (single 15 cm diameter Petri dish per GB1 variant) were transiently transfected (PEI, 3:1 ratio, 40 μg total DNA) with plasmids encoding GB1b and GB2. After 24 h, cells were harvested and fractionated with the Plasma Membrane Protein Extraction Kit according to the manufacturer’s protocol. Cellular fractions were probed with anti-GB1 (1:300) and anti-βactin (1:1,000) antibodies. Immunoblots were scanned with Vilber Fusion FX camera system. Densitometric analysis was done in Fiji.
SRE-luciferase accumulation assay
HEK293 cells stably expressing Gαqi were transiently transfected with WT or mutant GB1b, GB2, and SRE-FLuc (Lipofectamine 2000, 2:1 ratio, 4 μg total DNA). In variant and WT receptor co-expression experiments, equal amounts DNA encoding WT and mutant GB1b were used. Transfected cells were seeded into 96-well microplates (Greiner Bio-One) at a density of 70,000 cells/well. After 24 h, the culture medium was replaced with Opti-MEM-GlutaMAX. GB1b/2 receptors were activated with various concentrations of GABA (10, 50, 100, 500 nM; 1, 5, 10, 50, 100, 500 μM; 1, 5, 10 mM) for 6 h. FLuc activity in lysed cells was measured via the Dual-Luciferase Assay Kit (Promega) with a Spark microplate reader. Luminescence signals were adjusted by subtracting the luminescence obtained when expressing SRE-FLuc fusion proteins alone. For CGP54626 dose response assays, the culture medium was replaced with Opti-MEM-GlutaMAX + 10 μM or 100 μM GABA. GB1b/2 receptors were blocked with various concentrations of CGP54626 (1, 5, 10, 50, 100, 500 nM; 1, 5, 10, 50, 100 μM) or treated with the same amount of vehicle (DMSO) for 6 h. Data were analyzed and plotted in GraphPad Prism 8. Dose response curves were fitted to the raw data. Curve parameters (tonic activity, Emax, EC50, IC50) were tested for normal distribution and analyzed with Kruskal-Wallis or ANOVA. Appropriate corrections were made in cases where SD were significantly different (Brown-Forsythe test for ANOVA and F-test for t test).
Results
Identification of GABBR1 variants
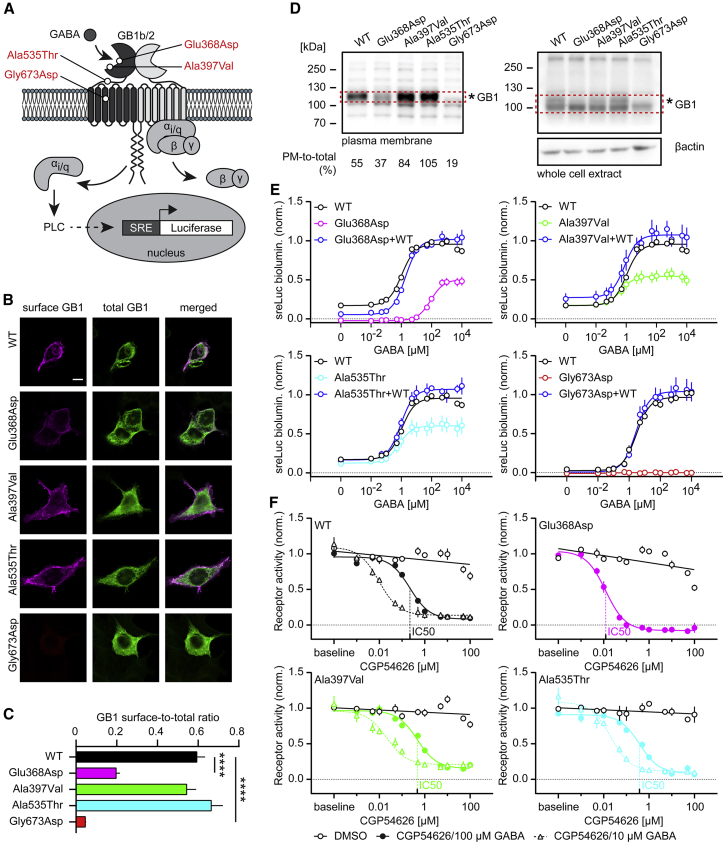
Individual 1 carries a heterozygous de novo missense p.Ala535Thr variant in GABBR1 (RefSeq accession number GenBank: NM_001470.4; hg19). Sanger sequencing confirmed its absence in the proband’s parents. The variant is absent from gnomAD v.2.1.1 and from BRAVO databases.21,22 The codon is conserved and the variant is predicted as deleterious by SIFT, Mutation Taster, and Polyphen-2 (Table 1). The variant is localized in the linker between the VFTD and TMD1 of the GB1 subunit (Figure 1A). Using Genematcher, we identified three other individuals carrying heterozygous de novo missense GABBR1 variants: p.Gly673Asp, p.Glu368Asp, and p.Ala397Val. All three variants are absent from gnomAD and predicted as deleterious by the SIFT, Mutation Taster, and Polyphen-2 (Table 1). The variant p.Gly673Asp is located in TMD3 of the GB1 subunit. The p.Glu368Asp and p.Ala397Val variants are located in the VFTD containing the orthosteric GABA-binding site (Figure 1A). All of these codons were well conserved in evolution (Figure S1).
Table 1.
In silico prediction and conservation of the four GABBR1 variants
| Variant in hg19 | Individual 1, c.1603G>A (p.Ala535Thr) (GenBank: NM_001470.4) |
Individual 2, c.1104G>C (p.Glu368Asp) (GenBank: NM_001470.4) |
Individual 3, c.1190C>T (p.Ala397Val) (GenBank: NM_001470.4) |
Individual 4, c.2018G>A (p.Gly673Asp) (GenBank: NM_001470.4) |
|---|---|---|---|---|
| chr6: 29580355C>T | chr6: 29589556C>G | chr6: 29589011G>A | chr6: 29574970C>T | |
| Prediction Tools | ||||
| gnomAD (non-TOPMed) | absent in 244,660 alleles | absent in 244,844 alleles | absent in 240,972 alleles | absent in 244,834 alleles |
| BRAVO | absent in 264,690 alleles | absent in 264,690 alleles | absent in 264,690 alleles | absent in 264,690 alleles |
| CADD | 29 | 23.3 | 29.4 | 26.4 |
| SIFT | Deleterious | deleterious | deleterious | deleterious |
| Mutation Taster | Deleterious | deleterious | deleterious | deleterious |
| Polyphen-2 | Deleterious | deleterious | deleterious | deleterious |
| Conservation | ||||
| GERP NR | 5.86 | 4.55 | 5.45 | 4.57 |
| GERP RS | 5.86 | 2.23 | 5.45 | 4.57 |
| Codon conservation | Conserved | poor conservation at the nucleotide level, conserved at amino acid level | conserved | conserved |
Figure 1.
Functional characterization of de novo GB1 variants in HEK293 cells
(A) Location of GB1 variants in the heterodimeric GBR. The scheme additionally depicts the assay used to monitor PLC-dependent RLuc expression under control of the serum response element (SRE). GB1b/2 receptors were artificially coupled to PLC by stable expression of the chimeric G protein subunit Gαqi in HEK293T cells. GB1b, GB2, and SRE-RLuc were transiently transfected.
(B) Surface and total GB1b expression levels of WT and mutant HA-tagged GB1b subunits co-expressed with GB2 in transfected HEK293 cells. Surface GB1b expression levels were determined with anti-HA antibodies in non-permeabilized cells. Total GB1b expression levels were determined with anti-GB1 antibodies in permeabilized cells. Scale bar: 10 μm.
(C) Bar graphs of the GB1b surface-to-total expression ratio. Kruskal-Wallis (p < 0.0001) and Dunn’s multiple comparison tests were used because of failed Shapiro-Wilk normality test. ∗∗∗∗p < 0.0001; WT, n = 28; p.Ala397Val, n = 38; p.Ala535Thr, n = 32; p.Glu368Asp, n = 32; p.Gly673Asp, n = 26.
(D) Fractionation of plasma membrane (PM) proteins in HEK293 cells transiently expressing GB1b/2 receptors. GB1 proteins were identified on immunoblots. WT and variant GB1 protein in the red rectangles was quantified densitometrically. The percentage of PM-to-total GB1 protein is indicated. Immunostaining for β-actin was used to control for loading. The results are from a single experiment. (∗) indicates the glycosylated form of GB1. Of note, the loss of glycosylated plasma membrane GB1 protein observed with the p.Glu368Asp and p.Gly673Asp variants is consistent with impaired maturation and surface transport of these proteins.
(E) Dose-response curves showing GABA-induced RLuc activity by WT, variant, and WT+variant (1:1 GB1b plasmid DNA ratio) GBRs in transfected cells. Three parameter log(GABA) versus response nonlinear regression curve fit was used for GABA dose response curves, with the exception of p.Gly673Asp, for which a linear regression curve fit was used. Table S1 provides information about EC50, Emax, and basal activity values derived from the GABA concentration-response curves and a statistical analysis.
(F) Dose-response curves showing inhibition of GBR activity (10 μM or 100 μM GABA) by the antagonist CGP54626. Three parameter log(inhibitor) versus response nonlinear regression curve fit was used for CGP54626 dose-response curves and linear regression curve fits for the DMSO controls. The curve fits depict DMSO controls for 100 μM GABA only. All data are mean ± SEM. The number of independent experiments is indicated. Table S2 provides IC50 values and a statistical analysis of the CGP54626 inhibition curves.
Clinical description of the GABBR1 variant carriers
Individual 1 (GABBR1 p.Ala535Thr) is a 30-month-old female with normal weight and height parameters (weight P25–50, height P50–75), known for neurodevelopmental delays of both motor and language skills. She sat unsupported at 12 months, crawled between 13 and 14 months, and walked at 17 months. She presented poor functional use of her hands and started to self-feed at 27 months. She began babbling, then regressed to no sounds. To date, she presents a significant speech delay with no words except “mama” and communicates through non-verbal language. She presents stereotypical movements of the hands, fingers, and feet and is noted for repetitive circular movements of the tongue as well as diurnal and nocturnal bruxism. Further, she presents a particular intense and “sticky” gaze. She shows axial hypotonia and peripheral hypertonia. There is no history of epilepsy. She suffers from constipation that requires medication and from periodic fever without focus since 12 months of age.
Individual 2 (GABBR1 p.Glu368Asp) is a 4-and-a-half-year-old female with normal weight and height parameters (weight at P25–50, height P10–25), presenting morphological particularities including long philtrum, hinted epicanthus, broad nasal root, clinodactyly of the fifth fingers of both hands, and hypoplasia of the teeth. She has a profound psychomotor and language delay. Opposition of the thumb was acquired around 9 months, she sat independently at 30 months, crawled at 36 months, and currently has no independent gait. She presents poor functional use of her hands (she is fed by her parents). With regard to language, she is nonverbal. She presents stereotypical movements consisting of periodic flapping of the feet. She shows combined axial and peripheral hypotonia. Further, at the neurological level, she presents five episodes of prolonged generalized seizures with conscious disorder and hypoxia during a febrile state. A brain MRI was performed at age 4 that showed a progressive cerebellar atrophy with no changes in the white matter or in the cortex. The electroencephalogram (EEG) revealed monomorphic activity with generalized slow SW (spike-wave) complexes, some with clinical correlate of absence seizures. She is treated with levetiracetam with a positive medical response. She presents with a severe intellectual disability, along with severe learning difficulties and sleep difficulties. With regard to other visceral particularities, at the ocular level she has an optic nerve hypoplasia, cortical visual disorders, nystagmus, and hypermetropia and a scoliosis at the osteo-muscular level.
Individual 3 (GABBR1 p.Ala397Val) is a 13-year-old male with neurodevelopmental delay; he walked independently at 36 months and also presented language delay. Currently, he speaks in short sentences with a limited vocabulary and problems with articulation and grammar. At a younger age, he presented with repetitive wheel-spinning, which is currently resolved. He has a mild intellectual disability (IQ at 58) and moderate learning difficulties. Further, he has an ADHD, encopresis, and sleeping difficulties. There is no notion of epilepsy, and an MRI performed at 2 years of age was normal.
Individual 4 (GABBR1 p.Gly673Asp) is a 17-year-old young woman of average weight and height (P50–75 and P10–25, respectively) with diagnoses of autism spectrum disorder, ADHD, borderline personality disorder, opposition defiant disorder, and social anxiety and one episode of major depressive disorder. She has no intellectual disability, but her autism is severe with extremely rigid thinking and profound defects in social motivation, although she is highly capable of social communication and exchange. Earlier in life, she had significant receptive language delay and moderate expressive speech delay with normal motor milestones (sitting at 7 months and walking at 17 months). At present, her language is fluent but with some pressured speech and fragmentation of flow of expressed ideas. She continues to have anger fits and crying attacks, and she has significant daytime sleepiness. She also has constipation, mood-associated headaches, as well as acne vulgaris, xerosis cutis, multiple melanocytic nevi, and nail dystrophy. She displays congenital bilateral anteversion of the femur and a bilateral tight heel chord with knee pain. There have been no known seizures.
A summary of the phenotypes can be found in Table 2.
Table 2.
List of phenotypic features of the four probands
| Individual 1 | Individual 2 | Individual 3 | Individual 4 | |
|---|---|---|---|---|
| GABBR1 variant hg19 | c.1603G>A (p.Ala535Thr) (GenBank: NM_001470.4) | c.1104G>C (p.Glu368Asp) (GenBank: NM_001470.4) | c.1190C>T (p.Ala397Val) (GenBank: NM_001470.3) | c.2018G>A (p.Gly673Asp) (GenBank: NM_001470.4) |
| Gender | F | F | M | F |
| Age | 2 years 6 months | 4 years 6 months | 13 years | 17 years |
| Growth parameters (percentile) | ||||
| Head circumference (cm) | P25 | P25 | P95 | P97 |
| Height (cm) | P50–P75 | P10–P25 | P79 | P10–P25 |
| Weight (kg) | P25–P50 | P25–P50 | P85 | P50–P75 |
| Development | ||||
| Motor delay | + | + | + | – |
| Speech delay/abnormalities | + | + | + | + |
| Hypotonia | + | + | – | – |
| Repetitive/stereotypical movements | + | – | + (but resolved with age) | – |
| Epilepsy | – | + | – | – |
| Neuropsychiatric problems | ||||
| Intellectual disability | not documented | + | + | – |
| Learning difficulties | not documented | + | + | – |
| Autism spectrum disorder | – | – | – | + |
| ADHD | – | – | + | + |
| Encopresis | – | – | + | – |
| Sleeping difficulties | – | + | + (but resolved with age) |
+ |
| Ocular | – (note sticky and intense gaze) | + (optic nerve hypoplasia, cortical visual disorders, intermittent nystagmus, hypermetropia) | – | – |
| Musculo-skeletal | – | scoliosis | – | congenital bilateral anteversion of femur; bilateral tight heel chord; knee pain |
| Dermatological | – | – | – | acne vulgaris; xerosis cutis; multiple melanocytic nevi; nail dystrophy |
Functional analysis of GABBR1 variants
We functionally characterized four de novo GABBR1 variants, p.Glu368Asp, p.Ala397Val, p.Ala535Thr, and p.Gly673Asp, in transfected HEK293 cells. These variants localize to (1) the VFTD containing the orthosteric GABA-binding site (p.Glu368Asp, p.Ala397Val), (2) the linker between VFTD and TMD1 (p.Ala535Thr), and (3) TMD3 (p.Gly673Asp) (Figure 1A). We first addressed whether HA-tagged wild-type (WT) and mutant GB1b subunits traffic to the cell surface when co-expressed with GB2. We determined cell surface levels GB1b protein with an HA antibody and total GB1b protein after cell permeabilization with a GB1 antibody (Figure 1B). The percentage of GB1b cell surface expression was calculated as the ratio of surface/total immunofluorescence (Figure 1C). As the ratio distribution for some of the receptor variants was not normal (Shapiro-Wilk: HA-p.WT: p = 0.9752, HA-p.Glu368Asp: p = 0.0535, HA-p.Ala397Val: p = 0.0003, HA-p.Ala535Thr: p = 0.0024, and HA-p.Gly673Asp: p = 0.0119), non-parametric Kruskal-Wallis and Dunn’s multiple comparison tests were used. The data show that receptors assembled with p.Ala397Val and p.Ala535Thr traffic normally to the cell surface while p.Glu368Asp and p.Gly673Asp surface expression is significantly reduced or absent, respectively (Kruskal-Wallis: p < 0.0001; Dunn’s multiple comparison: WT versus p.Glu368Asp and WT versus p.Gly673Asp p < 0.0001; remaining pairwise comparisons are not significantly different). We additionally used cellular fractionation of HEK293 cells transiently expressing WT and variant GBRs to study plasma membrane expression of GB1. The GB1 cell surface/total ratios from the fractionation experiments (Figure 1D) are in good agreement with the ratios determined in immunohistochemistry experiments (Figure 1C). Importantly, the fractionation experiments yield the same rank order of variant GB1 surface expression as the immunocytochemical analysis (Ala535Thr > Ala397Val > Glu368Asp > Gly673Asp). The data confirm that transport of the variant receptors assembled with p.Glu368Asp and p.Gly673Asp to the plasma membrane is impaired, while receptors assembled with p.Ala397Val and p.Ala535Thr have close to normal surface expression (Figure 1D). We further determined GABA dose-response curves for WT and mutant GBRs by using an assay based on artificially coupling GBRs via chimeric Gαqi proteins to phospholipase C (PLC)23 (Figure 1A). We monitored PLC activity monitored with a serum responsive element-luciferase (SRE-Luciferase) reporter. Increasing concentrations of GABA yielded sigmoidal dose-response curves for WT and mutant receptors, with the exception of p.Gly673Asp that does not traffic to the cell surface (Figure 1E). p.Glu368Asp exhibited decreased constitutive activity, a reduced Emax, and a nearly 100-fold increased EC50. p.Ala397Val and p.Ala535Thr exhibited a decrease in the Emax but similar constitutive activity and GABA potency as WT receptor. Because the affected individuals are heterozygous for the variations, we also addressed whether co-expression of a variant receptor affects activity of WT receptors (Figure 1E). The data show that the potency of GABA at a mixed population of p.Glu368Asp and WT receptors is significantly decreased versus WTs receptor alone, consistent with the reduced potency of GABA at p.Glu368Asp receptors. In general, the concentration-response curves of mixed populations of variant and WT receptors are similar to those of WT receptors alone, showing that the variant receptors exert no overt dominant-negative effects in heterologous cells. Table S1 provides information about EC50, Emax, and basal activity values derived from the GABA concentration-response curves and the statistical analysis. Inhibition of 10 or 100 μM GABA by increasing concentrations of the GBR antagonist CGP54626 yielded similar IC50 values for WT, p.Ala397Val, and p.Ala535Thr (Figure 1F). The IC50 value of CGP54626 at p.Glu368Asp was decreased, consistent with the lower potency of GABA at this mutant. Table S2 provides IC50 values and the statistical analysis of the CGP54626 inhibition curves.
Discussion
We present here monoallelic de novo GABBR1 variants that define another gene-Mendelian disease association. Probands with these de novo non-synonymous variants all presented developmental and language delays of varying severity. Furthermore, we found an association with hypotonia, intellectual disability, and sleep disorder in two probands and an association with epilepsy in one proband. We note one proband with optic nerve hypoplasia, cortical visual disorders, nystagmus, and hypermetropia.
We analyzed the consequences of the p.Glu368Asp, p.Ala397Val, p.Ala535Thr, and p.Gly673Asp changes for receptor functions in a heterologous expression system monitoring receptor-mediated G protein activation. The alteration of p.Gly673Asp in TM3 renders the receptor completely inactive as a result of lack of receptor expression at the cell surface. Our experiments indicate that p.Gly673Asp does not dimerize with GB2 and therefore does not influence the Emax of co-expressed WT GBRs. The p.Glu368Asp variant is located near the orthosteric binding site and reduces both potency and efficacy of GABA at the receptor. Previous mutagenesis experiments have shown that the neighboring variant p.Tyr367Ala similarly reduces efficacy of GABA,24 supporting that these variants directly influence the orthosteric binding site and/or trans-subunit conformational changes involved in G protein activation. Surface expression of p.Glu368Asp is impaired, which will further reduce the efficacy of signaling. GABA exhibited normal potency but decreased efficacy at p.Ala397Val and p.Ala535Thr. p.Ala397Val and p.Ala535Thr surface expression is normal, supporting that the two variants affect receptor efficacy by interfering with coupling of agonist binding to G protein activation. Previous mutagenesis experiments showed a similar decrease in agonist efficacy for the p.Trp395Ala variant.24 All affected individuals harbor monoallelic de novo variants of GABBR1. We therefore examined whether co-expression of variant and WT GBRs influences the activity of WT receptors in HEK293 cells. The potency of GABA at a mixed population of p.Glu368Asp and WT receptors is significantly reduced versus that of a WT receptor alone, consistent with the reduced potency of GABA at p.Glu368Asp receptors. In general, the concentration-response curves of mixed populations of receptors are similar to those of WT receptors alone, showing that the variant receptors exert no overt dominant-negative effects in heterologous cells. However, in neurons, WT and variant receptors may compete for limiting numbers of synaptic sites as well as trafficking, regulatory, transport, and effector proteins, which could negatively affect WT receptor activity. Therefore, one will have to analyze heterozygous mice harboring the variants to identify possible dominant-negative effects caused by these variants. Overall, our data support that all four variants are loss-of-function mutations that most likely tip the excitation/inhibition balance in the central nervous system towards more excitation, which may explain the epileptiform activity observed with the affected individual with the p.Glu368Asp variant.
Knockout Gabbr1−/− and Gabbr2−/− mice have been studied for almost 20 years.2,15,16 These mice lack electrophysiological, biochemical, and behavioral GBR responses. Moreover, they exhibit stereotypical circling, hyperlocomotor activity, delayed sleep phase, frequent seizures, memory deficits, hypothermia, hyperalgesia, hyperactivity, impaired passive avoidance learning, and increased anxiety. Heterozygous Gabbr1+/− and Gabbr2+/− mice have not been systematically analyzed. Saturation binding experiments with a high-affinity antagonist revealed only an 18% decrease in GBR-binding sites in Gabbr1+/− versus Gabbr1+/+ mice.16 Gabbr1+/− mice showed enhanced prepulse inhibition responses compared to littermate controls, indicating increased sensorimotor gating.25 This phenotype of Gabbr1+/− mice shows that the WT Gabbr1 allele cannot fully compensate for the loss of GBRs from the knockout allele.
Previous studies12,13 reported a variety of phenotypes for GABBR2 variants in affected individuals. They hypothesized that the severity of the phenotypes correlates with the loss of GABA signaling at the variant receptors. Our four GABBR1-affected individuals exhibit common phenotypes but differ in disease severity and in associated disorders. Consistent with the level of GBR impairment predicting the severity of the disease, the p.Gly673Asp variant rendering GBRs completely inactive is associated with severe phenotypes, including numerous neurodevelopmental disorders and co-morbid psychiatric disorders. Likewise, the p.Glu368Asp variant with reduced surface expression, a 100-fold loss in potency, and reduced efficacy is associated with several phenotypes, including epilepsy, severe intellectual disability, ocular problems, and dysmorphic features. The p.Ala397Val and p.Ala535Thr variants, which only exhibit reduced efficacy, produce the mildest phenotypes, mostly characterized by developmental delays. The impairment of GBR functioning caused by the GABBR1 variants is expected to tip the excitation/inhibition balance in the brain towards increased excitation.26,27 This is supported by the epileptic phenotype of the individual with the p.Glu368Asp variant. It is surprising that the individual with the p.Gly673Asp variant, which causes a complete loss of GBR function, does not suffer from seizures. We showed that the mutant allele does not interfere with the production of WT receptors from the WT allele, which apparently provides sufficient GBR activity to prevent epileptiform activity.
Baclofen, the GBR agonist marketed for the treatment of muscle pain, spasms, and stiffness in people with multiple sclerosis or spinal cord injury or disease,1 could be therapeutically used to increase GBR activity in the affected individuals. In this context, baclofen showed promising effects in animal models of GABBR2-related Rett syndrome and epileptic encephalopathy.12 Preclinical and clinical studies are also underway to test the utility of baclofen for the treatment of ASD28, 29, 30 and epilepsy.31 In addition, positive allosteric modulators of GBRs are now available.32,33 Positive allosteric modulators synergistically increase the activity of endogenous GABA at GBRs and allow increasing potency and efficacy of mutant (if still responsive to the modulator) and WT receptors in the affected individuals. Positive allosteric modulators may help to avoid the side-effects experienced with the agonist baclofen, which activates GBRs independent of the presence of endogenous GABA. In addition, treating the individuals with pathogenic GABBR1 variants with benzodiazepines, which increase GABA inhibition through GABAA receptors, would also allow for correction of the excitation/inhibition balance.
Acknowledgments
We thank the families for their participation in the study. This work was supported by the Swiss National Science Foundation (31003A-152970 to B.B.) and the ChildCare Foundation to S.E.A.
Declaration of interests
S.E.A. is a cofounder and CEO of Medigenome, Swiss Institute of Genomic Medicine; he is also a member of the Scientific Advisory Board of the “Imagine Institute”, Paris. E.R. is also a cofounder and medical director of Medigenome, Swiss Institute of Genomic Medicine. M.L.C. is an intern in the federally recognized clinical training program for Genetic Medicine of Medigenome. L.N. was supported by the grant NV19-07-00136 from the Ministry of Health of the Czech Republi, and The National Center for Medical Genomics (LM2018132) for support with the WES analyses.
Published: September 13, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.08.010.
Supplemental information
Data and code availability
The datasets (FASTQ files) supporting the current study have not been deposited in a public repository because of confidentiality of medical diagnostic tests but are available from the corresponding author on request.
References
- 1.Pin J.P., Bettler B. Organization and functions of mGlu and GABAB receptor complexes. Nature. 2016;540:60–68. doi: 10.1038/nature20566. [DOI] [PubMed] [Google Scholar]
- 2.Gassmann M., Bettler B. Regulation of neuronal GABA(B) receptor functions by subunit composition. Nat. Rev. Neurosci. 2012;13:380–394. doi: 10.1038/nrn3249. [DOI] [PubMed] [Google Scholar]
- 3.Favuzzi E., Huang S., Saldi G.A., Binan L., Ibrahim L.A., Fernández-Otero M., Cao Y., Zeine A., Sefah A., Zheng K., et al. GABA-receptive microglia selectively sculpt developing inhibitory circuits. Cell. 2021;184:4048–4063.e32. doi: 10.1016/j.cell.2021.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mariotti L., Losi G., Lia A., Melone M., Chiavegato A., Gómez-Gonzalo M., Sessolo M., Bovetti S., Forli A., Zonta M., et al. Interneuron-specific signaling evokes distinctive somatostatin-mediated responses in adult cortical astrocytes. Nat. Commun. 2018;9:82. doi: 10.1038/s41467-017-02642-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perea G., Gómez R., Mederos S., Covelo A., Ballesteros J.J., Schlosser L., Hernández-Vivanco A., Martín-Fernández M., Quintana R., Rayan A., et al. Activity-dependent switch of GABAergic inhibition into glutamatergic excitation in astrocyte-neuron networks. Elife. 2016;5:e20362. doi: 10.7554/eLife.20362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White J.H., Wise A., Main M.J., Green A., Fraser N.J., Disney G.H., Barnes A.A., Emson P., Foord S.M., Marshall F.H. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- 7.Kuner R., Köhr G., Grünewald S., Eisenhardt G., Bach A., Kornau H.C. Role of heteromer formation in GABAB receptor function. Science. 1999;283:74–77. doi: 10.1126/science.283.5398.74. [DOI] [PubMed] [Google Scholar]
- 8.Kaupmann K., Malitschek B., Schuler V., Heid J., Froestl W., Beck P., Mosbacher J., Bischoff S., Kulik A., Shigemoto R., et al. GABA(B)-receptor subtypes assemble into functional heteromeric complexes. Nature. 1998;396:683–687. doi: 10.1038/25360. [DOI] [PubMed] [Google Scholar]
- 9.Jones K.A., Borowsky B., Tamm J.A., Craig D.A., Durkin M.M., Dai M., Yao W.J., Johnson M., Gunwaldsen C., Huang L.Y., et al. GABA(B) receptors function as a heteromeric assembly of the subunits GABA(B)R1 and GABA(B)R2. Nature. 1998;396:674–679. doi: 10.1038/25348. [DOI] [PubMed] [Google Scholar]
- 10.Shaye H., Stauch B., Gati C., Cherezov V. Molecular mechanisms of metabotropic GABAB receptor function. Sci. Adv. 2021;7:eabg3362. doi: 10.1126/sciadv.abg3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samanta D., Zarate Y.A. Widening phenotypic spectrum of GABBR2 mutation. Acta Neurol. Belg. 2019;119:493–496. doi: 10.1007/s13760-019-01088-5. [DOI] [PubMed] [Google Scholar]
- 12.Yoo Y., Jung J., Lee Y.N., Lee Y., Cho H., Na E., Hong J., Kim E., Lee J.S., Lee J.S., et al. GABBR2 mutations determine phenotype in rett syndrome and epileptic encephalopathy. Ann. Neurol. 2017;82:466–478. doi: 10.1002/ana.25032. [DOI] [PubMed] [Google Scholar]
- 13.Vuillaume M.L., Jeanne M., Xue L., Blesson S., Denommé-Pichon A.S., Alirol S., Brulard C., Colin E., Isidor B., Gilbert-Dussardier B., et al. A novel mutation in the transmembrane 6 domain of GABBR2 leads to a Rett-like phenotype. Ann. Neurol. 2018;83:437–439. doi: 10.1002/ana.25155. [DOI] [PubMed] [Google Scholar]
- 14.Lopes F., Barbosa M., Ameur A., Soares G., de Sá J., Dias A.I., Oliveira G., Cabral P., Temudo T., Calado E., et al. Identification of novel genetic causes of Rett syndrome-like phenotypes. J. Med. Genet. 2016;53:190–199. doi: 10.1136/jmedgenet-2015-103568. [DOI] [PubMed] [Google Scholar]
- 15.Gassmann M., Shaban H., Vigot R., Sansig G., Haller C., Barbieri S., Humeau Y., Schuler V., Müller M., Kinzel B., et al. Redistribution of GABAB(1) protein and atypical GABAB responses in GABAB(2)-deficient mice. J. Neurosci. 2004;24:6086–6097. doi: 10.1523/JNEUROSCI.5635-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuler V., Lüscher C., Blanchet C., Klix N., Sansig G., Klebs K., Schmutz M., Heid J., Gentry C., Urban L., et al. Epilepsy, hyperalgesia, impaired memory, and loss of pre- and postsynaptic GABA(B) responses in mice lacking GABA(B(1)) Neuron. 2001;31:47–58. doi: 10.1016/s0896-6273(01)00345-2. [DOI] [PubMed] [Google Scholar]
- 17.Xi B., Chen J., Yang L., Wang W., Fu M., Wang C. GABBR1 gene polymorphism(G1465A)isassociated with temporal lobe epilepsy. Epilepsy Res. 2011;96:58–63. doi: 10.1016/j.eplepsyres.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 18.Ma S., Abou-Khalil B., Sutcliffe J.S., Haines J.L., Hedera P. The GABBR1 locus and the G1465A variant is not associated with temporal lobe epilepsy preceded by febrile seizures. BMC Med. Genet. 2005;6:13. doi: 10.1186/1471-2350-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaupmann K., Schuler V., Mosbacher J., Bischoff S., Bittiger H., Heid J., Froestl W., Leonhard S., Pfaff T., Karschin A., Bettler B. Human gamma-aminobutyric acid type B receptors are differentially expressed and regulate inwardly rectifying K+ channels. Proc. Natl. Acad. Sci. USA. 1998;95:14991–14996. doi: 10.1073/pnas.95.25.14991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141, 456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taliun D., Harris D.N., Kessler M.D., Carlson J., Szpiech Z.A., Torres R., Taliun S.A.G., Corvelo A., Gogarten S.M., Kang H.M., et al. Sequencing of 53, 831 diverse genomes from the NHLBI TOPMed Program. Nature. 2021;590:290–299. doi: 10.1038/s41586-021-03205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stefan E., Aquin S., Berger N., Landry C.R., Nyfeler B., Bouvier M., Michnick S.W. Quantification of dynamic protein complexes using Renilla luciferase fragment complementation applied to protein kinase A activities in vivo. Proc. Natl. Acad. Sci. USA. 2007;104:16916–16921. doi: 10.1073/pnas.0704257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geng Y., Bush M., Mosyak L., Wang F., Fan Q.R. Structural mechanism of ligand activation in human GABA(B) receptor. Nature. 2013;504:254–259. doi: 10.1038/nature12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prosser H.M., Gill C.H., Hirst W.D., Grau E., Robbins M., Calver A., Soffin E.M., Farmer C.E., Lanneau C., Gray J., et al. Epileptogenesis and enhanced prepulse inhibition in GABA(B1)-deficient mice. Mol. Cell. Neurosci. 2001;17:1059–1070. doi: 10.1006/mcne.2001.0995. [DOI] [PubMed] [Google Scholar]
- 26.Heaney C.F., Kinney J.W. Role of GABA(B) receptors in learning and memory and neurological disorders. Neurosci. Biobehav. Rev. 2016;63:1–28. doi: 10.1016/j.neubiorev.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Lujan R., Ciruela F. GABAB receptors-associated proteins: potential drug targets in neurological disorders? Curr. Drug Targets. 2012;13:129–144. doi: 10.2174/138945012798868425. [DOI] [PubMed] [Google Scholar]
- 28.Mahdavinasab S.M., Saghazadeh A., Motamed-Gorji N., Vaseghi S., Mohammadi M.R., Alichani R., Akhondzadeh S. Baclofen as an adjuvant therapy for autism: a randomized, double-blind, placebo-controlled trial. Eur. Child Adolesc. Psychiatry. 2019;28:1619–1628. doi: 10.1007/s00787-019-01333-5. [DOI] [PubMed] [Google Scholar]
- 29.Erickson C.A., Veenstra-Vanderweele J.M., Melmed R.D., McCracken J.T., Ginsberg L.D., Sikich L., Scahill L., Cherubini M., Zarevics P., Walton-Bowen K., et al. STX209 (arbaclofen) for autism spectrum disorders: an 8-week open-label study. J. Autism Dev. Disord. 2014;44:958–964. doi: 10.1007/s10803-013-1963-z. [DOI] [PubMed] [Google Scholar]
- 30.Veenstra-VanderWeele J., Cook E.H., King B.H., Zarevics P., Cherubini M., Walton-Bowen K., Bear M.F., Wang P.P., Carpenter R.L. Arbaclofen in Children and Adolescents with Autism Spectrum Disorder: A Randomized, Controlled, Phase 2 Trial. Neuropsychopharmacology. 2017;42:1390–1398. doi: 10.1038/npp.2016.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeman A., Hoefeijzers S., Milton F., Dewar M., Carr M., Streatfield C. The GABAB receptor agonist, baclofen, contributes to three distinct varieties of amnesia in the human brain - A detailed case report. Cortex. 2016;74:9–19. doi: 10.1016/j.cortex.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 32.Kalinichev M., Girard F., Haddouk H., Rouillier M., Riguet E., Royer-Urios I., Mutel V., Lütjens R., Poli S. The drug candidate, ADX71441, is a novel, potent and selective positive allosteric modulator of the GABAB receptor with a potential for treatment of anxiety, pain and spasticity. Neuropharmacology. 2017;114:34–47. doi: 10.1016/j.neuropharm.2016.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Urwyler S., Mosbacher J., Lingenhoehl K., Heid J., Hofstetter K., Froestl W., Bettler B., Kaupmann K. Positive allosteric modulation of native and recombinant gamma-aminobutyric acid(B) receptors by 2, 6-Di-tert-butyl-4-(3-hydroxy-2, 2-dimethyl-propyl)-phenol (CGP7930) and its aldehyde analog CGP13501. Mol. Pharmacol. 2001;60:963–971. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets (FASTQ files) supporting the current study have not been deposited in a public repository because of confidentiality of medical diagnostic tests but are available from the corresponding author on request.