Summary
Ischemic stroke, caused by vessel blockage, results in cerebral infarction, the death of brain tissue. Previously, quantitative trait locus (QTL) mapping of cerebral infarct volume and collateral vessel number identified a single, strong genetic locus regulating both phenotypes. Additional studies identified RAB GTPase-binding effector protein 2 (Rabep2) as the casual gene. However, there is yet no evidence that variation in the human ortholog of this gene plays any role in ischemic stroke outcomes. We established an in vivo evaluation platform in mice by using adeno-associated virus (AAV) gene replacement and verified that both mouse and human RABEP2 rescue the mouse Rabep2 knockout ischemic stroke volume and collateral vessel phenotypes. Importantly, this cross-species complementation enabled us to experimentally investigate the functional effects of coding sequence variation in human RABEP2. We chose four coding variants from the human population that are predicted by multiple in silico algorithms to be damaging to RABEP2 function. In vitro and in vivo analyses verify that all four led to decreased collateral vessel connections and increased infarct volume. Thus, there are naturally occurring loss-of-function alleles. This cross-species approach will expand the number of targets for therapeutics development for ischemic stroke.
Keywords: human RABEP2, ischemic stroke, human sequence variation, in vivo evaluation platform, cross-species complementation, coding SNPs, infarct volume, collateral vessels
Using a newly established in vivo gene rescue platform in mice, we validated Rabep2, a candidate gene modulating infarct volume after ischemic stroke. We then determined the functional consequences of human RABEP2-coding variants. This cross-species approach will expand the number of physiologically relevant therapeutic targets for human ischemic stroke.
Introduction
Stroke is the second-leading cause of death in the world, and fifteen million new cases are reported annually. In the United States, stroke is the fourth leading cause of death and the primary cause of severe, long-term disability, and almost 800,000 new or recurrent cases occur each year.1, 2, 3, 4, 5 Over 87% of strokes are ischemic, caused by a disruption of the blood supply to the brain leading to neuronal tissue damage (including infarction) in the ischemic region.6, 7, 8 Once a stroke has occurred, the only FDA-approved drug for stroke therapy is intravenous tissue plasminogen activator (IV tPA), currently given to only 2%–3% of stroke patients because of its limited time window for administration and the major risk for adverse effects.9, 10, 11, 12, 13 Therefore, there is an urgent need to identify and develop new drug targets to effectively treat stroke patients.
Decades of studies have identified individual risk factors for stroke (e.g., hypertension, smoking, diabetes, obesity, etc.).14, 15, 16, 17 Genetic risk factors for stroke susceptibility have been identified by both family-based linkage18,19 and population-based genome-wide association studies (GWASs).20, 21, 22 However, there have been no new targets identified by GWASs for stroke treatment.
In principle, new therapeutic targets could be identified by genetic studies of stroke outcomes and infarct volume (size). However, genetic mapping in the human population for stroke infarct volume is intrinsically problematic because of uncontrollable variation in the extent and location of the occluded vessel and, especially, variation in the critical time between the first recognized symptoms of stroke and medical intervention.
By contrast, the surgical occlusion mouse model of cerebral ischemia enables complete control over the variables that cannot be controlled in humans. Thus, to discover genetic variation that modulates ischemic stroke infarction, we and others have taken a quantitative trait locus (QTL) mapping (i.e., forward genetic mapping) analysis approach by using the surgical occlusion mouse model of cerebral ischemia. Through permanent occlusion of the distal middle cerebral artery (pMCAO) in 32 commonly used inbred mouse strains, we have shown that these strains demonstrate robust differences in infarct volume. The differences in infarct volume are highly reproducible within each strain and differ more than 50-fold across all strains.23,24 Importantly, the differences are caused by natural allelic variation in the mouse genome. We have exploited these differences to map the natural genetic determinants of infarct volume for several genetic loci that modulate ischemic stroke and have identified several causative genes.23, 24, 25, 26, 27, 28, 29 The strongest and most significant locus modulating infarct volume is found in multiple pairwise crosses of inbred mouse strains. This locus, cerebral infarct volume QTL 1 (Civq1), overlaps another locus on chromosome 7 that modulates cerebral collateral vessel number (collateral artery number QTL 1; Canq1).30,31 Rabep2 (GenBank: NM_030556.2), which encodes an endosomal recycling protein for vascular endothelial growth factor receptor-2 (VEGFR2 [MIM: 191306]), was subsequently identified as the causal gene at the Civq1/Canq1 locus.32,33
However, the role of human RABEP2 (MIM: 611869; GenBank: NM_024816.2) in ischemic stroke remains uncertain. To date, no GWAS of ischemic stroke phenotypes have identified SNPs within or near this gene. This could be due to the aforementioned difficulty of controlling all of the non-genetic variables in stroke outcomes. Coding variations do exist in RABEP2 and some of these variants, while rare overall in the human population, are predicted to be damaging to its function by in silico gene variant prediction algorithms. However, their effects have not been evaluated for ischemic stroke outcomes.
In the present study, we developed an in vivo evaluation platform to investigate the role of Rabep2 on ischemic stroke volume. This platform also enabled us to examine the function of RABEP2 in Rabep2 knockout (KO) mice. We then evaluated RABEP2-coding variants, focusing on four of the most common variants that are also computationally predicted to abrogate its function.
Material and methods
Animals
Rabep2 KO mice were generated by the Duke Transgenic Core with CRISPR-Cas9 technology to remove sequence from exon 3 of the gene, which was chosen to alter the reading frame downstream of the deletion. Briefly, for genome editing, the CRISPR ribonucleoprotein (RNP) including a single-guide RNA (sgRNA; 5′-GCTGCTGCCGCTCCTGTTTCAGG-3′ in exon 3) and the Cas9 protein were pre-assembled in vitro. Then the CRISPR-Cas9 RNP complex was delivered by electroporation into pronuclear-stage embryos (harvested 0.5 day post coitum [dpc] from C57BL/6J females). The electroporated embryos were transferred to the oviduct of 0.5 dpc, pseudopregnant female ICR (Hsd:ICR (CD-1)) mice to generate genetically engineered mice. Once founders were born, the targeted locus was sequenced to screen for Rabep2 KO mice. One founder acquired a 55-nucleotide deletion in exon 3 of the Rabep2 (Figure S1). This founder was maintained and expanded on a C57BL6/J background for use in this study. Mice (both males and females) were age matched (P21 and 10 weeks for collateral vessel perfusion and 12 ± 1 week for pMCAO) for all experiments.
Study approval
All animal study procedures were conducted under a protocol approved by the Duke University IACUC in accordance with NIH guidelines.
Viruses
All the AAVs used for this study were generated by the Duke Viral Vector Core. We generated multiple recombinant AAV constructs: a control virus with the CMV promoter driving the EGFP reporter gene (pAAV9-CMV-EGFP-WPRE) and six different experimental viruses with the CMV promoter driving either mouse Rabep2 (pAAV9-CMV-Rabep2-P2A-EGFP-WPRE) or human RABEP2 including wild-type RABEP2 (pAAV9-CMV-RABEP2-P2A-EGFP-WPRE) and four coding variants (pAAV9-CMV-RABEP2 (Variants)-P2A-EGFP-WPRE). The variants included rs200118396 (p.Arg508Ser), rs769480150 (p.Ser204Leu), rs184144701 (p.Arg490Trp), and rs527458355 (p.Arg543His). We validated the sequence of each variant by Sanger sequencing (Figures S4A–S4D). Briefly, 1 μL of AAV construct (minimum titer, 2 × 1013 vg/mL) was directly injected into the right hemisphere of the brain of P1 Rabep2 KO animals.
Collateral vessel density measurement
As we have shown that collateral vessel traits are established by 3 weeks of age and remain constant for many months,25 the collateral vessel phenotype was measured at P21 and 10 weeks as previously described.26, 27, 28, 29 Mice were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg), and the ascending thoracic aorta was cannulated. The animals were perfused with freshly made buffer (1 mg/mL adenosine, 40 g/mL papaverins, and 25 mg/mL heparin in PBS) to remove the blood. The pial circulation was then exposed after removal of the dorsal calvarium and adherent dura mater. The cardiac left ventricle was cannulated and a polyurethane solution with a viscosity sufficient to minimize capillary transit (1:1 resin to 2-butanone, PU4ii, VasQtec) was slowly infused; cerebral circulation was visualized under a stereomicroscope during infusion. The brain surface was topically rinsed with 10% PBS-buffered formalin and the dye solidified within 20 min. After post-fixation with 10% PBS-buffered formalin, pial circulation was imaged. All collateral interconnecting the anterior cerebral artery (ACA) and middle cerebral artery (MCA) trees of both hemispheres were counted.
pMCAO
Focal cerebral ischemia was induced by direct permanent occlusion of the distal MCA as previously described.26, 27, 28, 29 Briefly, adult mice were anesthetized with ketamine (100 mg/kg) and xylazine (5 mg/kg), and then 0.5% bupivacaine (5 mg/mL) was also administrated by injection at the incision site. The right MCA was exposed by a 0.5 cm vertical skin incision midway between the right eye and ear under a dissecting microscope. After the temporalis muscle was split, a 2 mm burr hole was made with a high-speed micro drill at the junction of the zygomatic arch and the squamous born through the outer surface of the semi-translucent skull. The MCA was clearly visible at the level of the inferior cerebral vein. The inner layer of the skull was removed with fine forceps, and the dura was opened with 32-gauge needle. While visualizing under an operating microscope, the right MCA was electrocauterized. The cauterized MCA segment was then transected with microscissors to verify permanent occlusion. The surgical site was closed with 6-0 sterile nylon sutures, and 0.5% bupivacaine was applied. The temperature of each mouse was maintained at 37°C with a heating pad during the surgery and then the mouse was placed in a recovery chamber (set temperature 37°C) until the animal was fully recovered from the anesthetic. Mice were then returned to their cages and allowed free access to food and water in an air-ventilated room with the ambient temperature set to 25°C.
Infarct volume measurement
Cerebral infarct volumes were measured 24 h after distal permanent MCA occlusion, the time point when the size of the cortical infarct is largest and is stable. 24 h after pMCAO surgery, the animals were euthanized, and the brains were carefully removed. The brains were placed in a brain matrix, chilled at −80°C for 4 min to slightly harden the tissue, and then sliced into 1 mm coronal sections. Each brain slice was placed in one well of a 24-well plate and incubated for 20 min in a solution of 2% 2,3,5-triphenyltetrazolium chloride (TTC) in PBS at 37°C in the dark. The sections were then washed once with PBS and fixed with 10% PBS-buffered formalin at 4°C. Then, 24 h after fixation, the caudal face of each section was scanned with a flatbed color scanner. The scanned images were used to determine infarct volume. Image-Pro software (Media Cybernetics, MD) was used to calculate the infarcted area of the hemisphere to minimize error introduced by edema. The total infarct volume was calculated by summing the individual slices from each animal.
Immunohistochemistry
Immunohistochemistry (IHC) was performed by Servicebio (Servicebio, MA). Rabep2 KO mice either 3 weeks or 10 weeks after CMV-Rabep2 injection were fixed with 4% PFA and then mouse brains were shipped for sample preparations. Briefly, brain samples were processed, embedded in paraffin, and sagittally sectioned at 4 μm. Paraffin sections were then deparaffinized with ethanol and rehydrated gradually with distilled water at room temperature. The sections were submerged in 0.01 M sodium citrate buffer and boiled for 10 min for retrieval of antigen. The sections were washed with PBS three times and treated 3% H2O2 for 15 min and blocked with blocking solution (5% BSA) for 1 h at room temperature before application of primary antibody. The sections were incubated with rabbit anti-GFP (1:1,000, Cell Signaling Technology, MA) overnight at 4°C. Subsequently, the sections were immunohistochemically stained with Alexa Fluor 488 (1:1,000, Molecular Probes, OR) for 1 h at room temperature. Whole slide scanning was performed on a Panorama Midi II scanner (3DHISTECH, Hungary) and images were captured with CaseViewer software (CaseViewer 2.3, 3DHISTECH, Hungary).
In silico prediction algorithms
Coding polymorphisms were examined for potential functional consequences with two independent in silico prediction algorithms, Polymorphism Phenotyping v2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/index.shtml) and Sorting Intolerant From Tolerant (SIFT, http://sift.jcvi.org).
Molecular dynamics simulation of human RABEP2-RAB5 complex structure
The RABEP1-RAB5 complex structure, which was previously reported, was directly downloaded from the Protein Data Bank (PDB: 1TU3).34 To predict three-dimensional complex structure of the RAB5-RABEP2, the human RABEP2 protein structure was obtained by AlphaFold2,35,36 and then, with the HDOCK homology software (HDOCK server; http://hdock.phys.hust.edu.cn)37 based on the RABEP1-RAB5 complex structure, the RABEP2-RAB5 complex structure was modeled. For structure modeling of RABEP2 containing histidine at amino acid position 543, the backbone-dependent rotamer library, which is embedded in the Pymol software (https://pymol.org/2/), was used to generate four possible rotamer confirmations of histidine residue. Within the four possible histidine rotamers, the most structurally dominant rotamer was chosen for study.
Cell culture
Human microvascular endothelial cells (HMEC-1, CRL-3243, ATCC, VA) derived from human dermal endothelium were cultured at 37°C in a 5% CO2 humidified incubator with MCDB-131 medium containing microvascular growth supplement (MVGS), 10 mM L-glutamine, and 5% FBS.
In vitro scratch assay
Prior to the cell culture, vertical and horizontal reference lines were made on the bottom of a 0.1% gelatin coated 6-well plate to obtain the same field for each image acquisition. Then, the HMECs were seeded in the plate at a concentration of 8 × 104 cells/well and incubated 24 h. To reduce endogenous human RABEP2 mRNA expression by RNA interference, 10 μM of non-specific small interfering RNA (siRNA) (D-001910-10-05, Dharmacon, CO) or the human RABEP2-specific siRNA (M-009001-01-0005, Dharmacon, CO) was treated as recommended and the cells were maintained at 37°C in a 5% CO2 humidified incubator. 48 h after siRNA treatment, different AAV constructs were infected for an additional 48 h. When the infected HMECs reached over 95% confluence in the 6-well plate, the wells were scratched with a 200 μL sterile pipette tip. The cells were then washed with pre-warmed PBS to remove detached cells and pre-warmed regular culture media was added. Using the reference lines as guides, the plate was placed under a phase-contrast microscope (EVOS FL, Thermofisher Scientific, MA) and the wound scratch was photographed at 0, 4, 8, and 10 h after wounding. Wound area was calculated by manually tracing the cell-free area in captured images with the Fiji (ImageJ). The wound closure rate was expressed as the percentage of area reduction.
Cell membrane biotinylation assay
To determine the amount of VEGFR2 present on the cell membrane, HMECs were seeded on a 100 mm dish coated with 0.1% gelatin. Subsequently, siRNA and different AAV rescue constructs were added to the cultured cells as described in the in vitro scratch assay section. When the HMECs reached 90% confluence, cell membrane proteins were labeled with membrane-impermeable EZ-Link Sulfo-NHS-SS-Biotin (A44390, Thermofisher Scientific, MA). The assay was performed according to the manufacturer’s instruction. The biotinylated membrane proteins were incubated with 10 mM DTT, collected and boiled in a SDS sample buffer, and then loaded on an SDS-PAGE gel for immunoblot analysis.
Quantitative PCR
mRNA levels were measured by quantitative PCR (qPCR) as previously described.26,27 To quantitate mRNA levels of RABEP2, HMECs treated with either control siRNA or RABEP2-specific siRNA (si-RABEP2) were used. All samples were run in triplicate, and an additional assay for endogenous GAPDH was performed to control for input cDNA template quantity.
Immunoblot analysis
To determine siRNA efficiency, protein samples were collected from HMECs treated with either control siRNA or si-RABEP2. In addition, to measure RABEP2, protein samples were collected from HMECs infected with either CMV-control (control) or CMV-RABEP2 (rescued) after si-RABEP2 treatment. Briefly, HMECs were incubated in cold lysis buffer (50 mM Tris-HCl [pH 7.8], 150 mM NaCl, 0.2% Triton X-100) containing protease and phosphatase inhibitor cocktail (Thermofisher Scientific, MA). Protein samples (30–50 μg) were electrophoresed on either 10% or 12% polyacrylamide gel and then transferred on PVDF membrane with Trans-Blot Turbo Transfer System (Bio-Rad Laboratories, CA). Membranes were incubated with primary and secondary antibodies, and the level of protein was visualized via chemiluminescence (ECL Detection Kit, Thermofisher Scientific, MA) The following primary antibodies were used for immunoblot analysis; rabbit anti-RABEP2 (14625-1-AP, Proteintech Group, IL), rabbit anti-VEGFR2 (#2479, Cell Signaling Technology, MA), and mouse anti-GAPDH (sc-32233, Santa Cruz Biotechnology, TX). HRP-conjugated anti-mouse and anti-rabbit (Santa Cruz Biotechnology, Inc., TX) secondary antibodies were used to detect proteins.
Statistics
Statistical analyses were performed with GraphPad Prism (GraphPad Software, La Jolla, CA) and/or R (https://www.R-project.org). Significant differences between datasets were determined with either a two-tailed Student’s t test when comparing two groups or one-way or two-way ANOVA followed by Tukey’s test for multiple comparisons. Data are represented as the mean ± SD. p < 0.05 was considered significant. Table S2 provides statistical analyses for all data shown in figures.
Results
Absence of Rabep2 modulates both cerebral collateral vessel anatomy and ischemic infarct volume
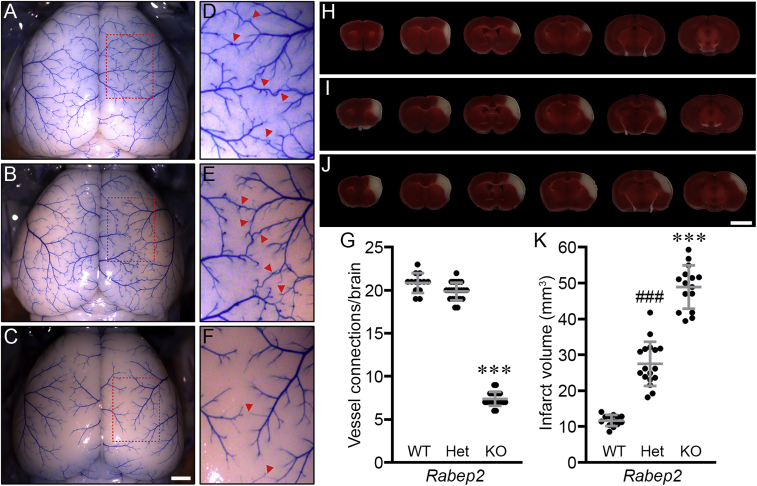
To investigate whether Rabep2 is the major modulator of both pial collateral vessel density and infarct volume, we generated Rabep2 KO mice by using CRISPR-Cas9 technology (Figure S1). We first examined the effect of the loss of Rabep2 on collateral vessel connections between the anterior cerebral artery (ACA) and the middle cerebral artery (MCA) and infarct volume after pMCAO. In concordance with a previous report,32 collateral vessel connections were dramatically reduced with loss of Rabep2 (Figures 1A–1G), leading to a concomitant increase in infarct size (Figures 1H–1K) compared to both Rabep2 wild-type (WT) and heterozygous KO (Het) animals. This is consistent with a collateral vessel-dependent effect for Rabep2 in modulating ischemic stroke infarction. Our data corroborate recent studies showing that Rabep2 modulates infarct volume via the collateral circulation (Figures 1G and 1K).
Figure 1.
Collateral vessel density and infarct volume for Rabep2 KO mice confirm a role for the gene in both phenotypes
(A–C) Representative mouse brain images are shown for Rabep2 wild-type (WT) (A), heterozygous KO (Het) (B), and homozygous KO (KO) (C) strains. Scale bar: 1 mm.
(D–F) (D), (E), and (F) are three times magnified from (A), (B), and (C), respectively, and red arrowheads indicated collateral vessel connections between the ACA and MCA.
(G) The scatter plots show the number of collateral vessel connections between the ACA and MCA in the brain for each animal. The total number of animals for Rabep2 WT, Het, and KO were 13, 23, and 21 mice, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗∗p < 0.001 versus Rabep2 WT and Het).
(H–J) Serial brain sections (1 mm) for each genotype of Rabep2 WT (H), Het (I), and KO (J) are shown. The infarct appears as white tissue after 2% TTC staining. Scale bar: 5 mm.
(K) The scatter plots show the infarct volume 24 h after pMCAO for each animal. The total number of animals for Rabep2 WT, Het, and KO were 13, 17, and 15 mice, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗∗p < 0.001 versus Rabep2 WT and Het; ###p < 0.001 versus Rabep2 WT and KO).
Rabep2 expression rescues cerebral collateral vessel connections as well as infarct volume phenotypes
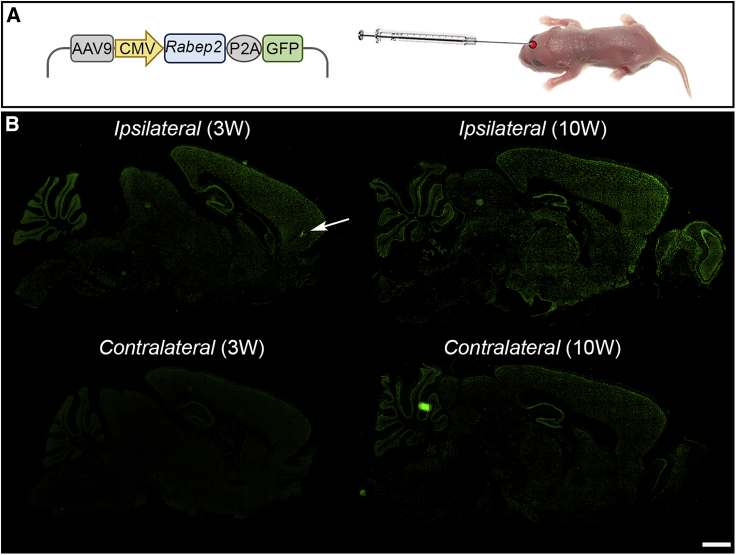
To further validate the role of Rabep2 in cerebral infarction, we sought an approach that could restore the functional effects of the gene. We chose an “add-back” gene rescue strategy, where, in the knockout mice, we could express exogenous Rabep2 delivered via recombinant AAV. We drove production of mouse RABEP2 and GFP via a CMV promoter that yields ubiquitous expression. To determine the efficiency of the AAV delivery system, we first evaluated Rabep2 expression in vivo by using AAV9-CMV-mouse Rabep2 (CMV-Rabep2) that was injected directly into the right hemisphere of the brain of P1 Rabep2 KO mice (Figure 2A). Given the nature of the AAV construct, GFP serves as a proxy for RABEP2 production. Within days of injection, GFP is stably and efficiently produced across the entire mouse brain (Figure 2B). Stronger GFP signal was observed in the right hemisphere (Ipsilateral) compared to the left hemisphere (Contralateral), providing an internal control. This expression pattern demonstrates that recombinant AAV provides an ideal platform for gene rescue in our mouse model of ischemic stroke.
Figure 2.
Genetic cargo of a single in vivo injection of recombinant AAV is widely expressed for many weeks in the mouse brain
(A) Schematic map of AAV containing mouse Rabep2 and GFP driven by a CMV (ubiquitous) promoter. The recombinant AAV was injected into a P1 mouse brain (right hemisphere).
(B) Immunostaining for recombinant GFP expressed upon AAV injection 3 and 10 weeks after AAV9-CMV-Rabep2 injections. The white arrow indicates where the virus was injected into P1 mouse brain. Scale bar: 2 mm.
We first determined whether either the collateral vessel or infarct volume phenotypes were affected by injection of this AAV vector platform. There was no difference in the collateral vessel number of Rabep2 KO mice injected with AAV empty vector containing GFP (CMV-control) compared to the Contralateral hemisphere (3 and 10 weeks) (Figure S2A). Furthermore, CMV-control-injected Rabep2 WT, Het, and KO mice showed a similar number of collateral vessel connections as Rabep2 WT, Het, and KO mice with no virus (Table S2). We then examined the effect of the AAV injection on ischemic infarct volume in the permanent occlusion model. 12 weeks after injection with the CMV-control, the infarct volumes in Rabep2 WT, Het, and KO animals were consistent with the infarct volumes in Rabep2 WT, Het, and KO mice with no virus (Figures 1K and S2B). Thus, the injection of AAV and production of GFP have no effect on these phenotypes.
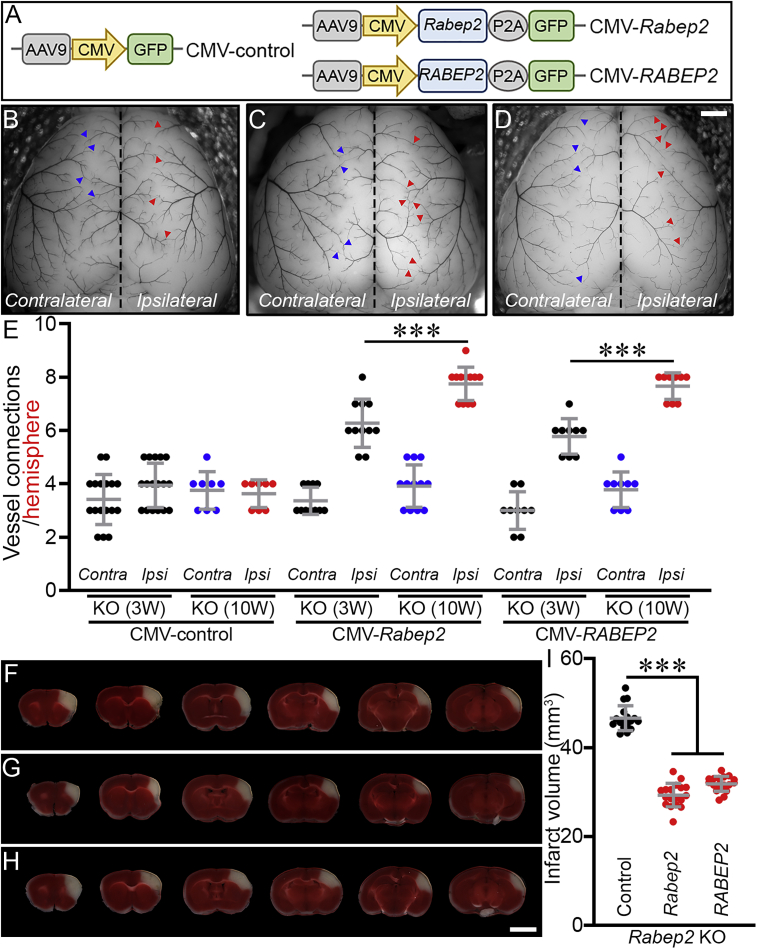
Using this platform, we attempted to rescue the phenotype in Rabep2 KO mice with AAV vectors containing mouse Rabep2 under the control of a CMV promoter (Figure 3A). The brains of Rabep2 KO mice injected with CMV-Rabep2 showed a time-dependent increase in collateral vessel connections compared to the Contralateral hemisphere as well as when compared to Rabep2 KO mice injected with CMV-control (Figures 3B, 3C, and 3E). Rabep2 KO mice injected with CMV-Rabep2 exhibited a dramatic reduction of infarct volume after pMCAO compared to Rabep2 KO mice injected with CMV-control (Figures 3F, 3G, and 3I), consistent with the role of collateral vasculature in promoting reperfusion of the ischemic territory thereby reducing infarct volume.
Figure 3.
Exogenous mouse RABEP2 rescues the collateral vessel connection phenotype as well as the infarct volume phenotype of Rabep2 KO mice
(A) Schematic map of AAV constructs.
(B–D) Representative mouse brain images of Rabep2 KO mice injected with either CMV-control (B), CMV-Rabep2 (C), or CMV-RABEP2 (D). Blue and red arrowheads indicate collateral vessel connections either on the Contralateral (no virus) or the Ipsilateral (with viruses containing only GFP [B], mouse Rabep2 [C], and human RABEP2 [D], respectively) hemisphere. Scale bar: 1 mm.
(E) The scatter plots show the number of collateral vessel connections between the ACA and MCA of each hemisphere 3 and 10 weeks after AAV injections. The total number of animals for Rabep2 KO either 3 or 10 weeks after CMV-control, Rabep2 KO either 3 or 10 weeks after CMV-Rabep2, and Rabep2 KO either 3 or 10 weeks after CMV-RABEP2 are 17, 8, 11, 12, 9, and 9 animals, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗∗p < 0.001).
(F–H) Serial brain sections (1 mm) for Rabep2 KO mice injected with either CMV-control (F), CMV-Rabep2 (G), or CMV-RABEP2 (H). Scale bar: 5 mm.
(I) The scatter plots show the infarct volume for Rabep2 KO mice injected with CMV-control, CMV-Rabep2, and CMV-RABEP2; 17, 18, and 18 animals, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗∗p < 0.001).
Although the collateral vessel network is thought to be set during embryonic collaterogenesis,32 these data show that new collateral vessels also develop in early postnatal life (Figure 3E). We note however, that there was no increase in collateral vessel connections on the Contralateral hemisphere even 10 weeks after virus injection (Figures 3C and 3E), despite what appears to be relatively strong expression of Rabep2 through the entire brain (Figure 2B). This suggests a narrow threshold for either gene dose or timing, or both, for collateral vessel formation after birth.
Human RABEP2 rescues the collateral vessel and infarct volume phenotypes of Rabep2 KO mice
We next determined whether human RABEP2 could rescue the Rabep2 KO phenotypes (Figures 3E and 3I). We expressed the human RABEP2 by using the same AAV platform (CMV-RABEP2) (Figure 3A), and we determined the effect of injection of RABEP2 in Rabep2 KO mice. Expression of human RABEP2 increased collateral vessel number (Figures 3D and 3E) and reduced infarct volume of Rabep2 KO mice upon ischemic stroke (Figures 3H and 3I). Within 3 weeks, the rescued KO mice had established new collateral vessel connections, and by 10 weeks the number of these connections increased further. At 12 weeks of age, upon stroke induction, the resulting infarct volume was also reduced in the rescued Rabep2 KO mice. Of necessity, we injected recombinant AAV expressing RABEP2 into the brains of newborn (P1) mice, the late stages of this natural process of collateral vessel growth. Thus, in this specific model and experimental approach, 100% rescue may not be feasible. Nonetheless, the human RABEP2 rescues Rabep2 KO mouse phenotypes. Furthermore, both RABEP2 and Rabep2 rescue collateral vessel and infarct volume phenotypes to similar levels (Figure 3). This strongly supports the hypothesis that Rabep2 shares the same function in both humans and mice.
Prediction of functional consequences of human RABEP2-coding variants
Since we confirmed that human RABEP2 is fully functional in the mouse, we investigated whether coding sequence variants of RABEP2 impact ischemic infarction. If coding variants in this gene alter infarct volume in vivo, this gene gains further “human genetics” support as a therapeutic target. Using the full list of RABEP2-coding SNPs found in the Genome Aggregation Database (gnomAD [v2.1.1]; https://gnomad.broadinstitute.org/), we first ordered all coding SNP alleles on the basis of overall population allele frequency. Then, we examined the predicted functional consequences of these SNPs by two independent in silico algorithms, SIFT and PolyPhen-2. Using these algorithms, we prioritized our initial functional analysis to three coding SNP variants that are strongly predicted to be damaging (Table S1; represented in bold font).
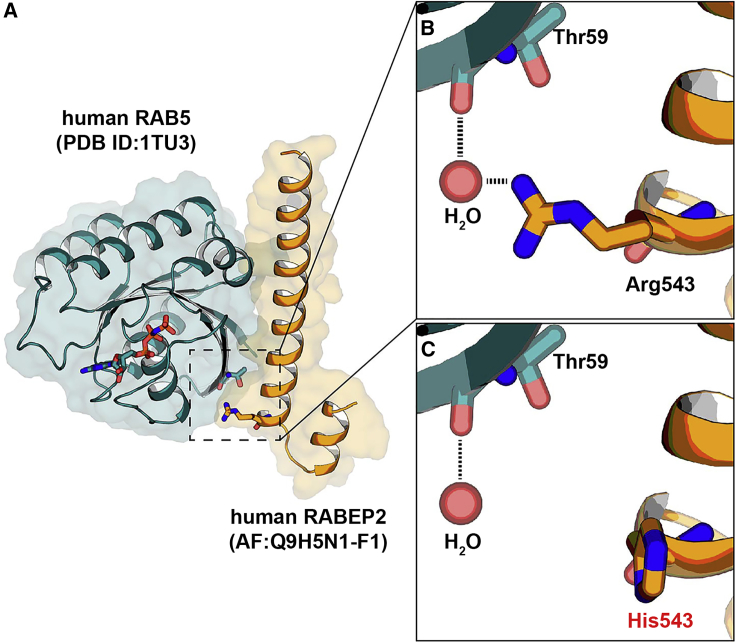
We sought to identify additional potential candidate coding variants by focusing on the cellular role of RABEP2, which regulates membrane trafficking in the early endocytic pathway with RAB5.38 A previous study reported that human RABEP1 (MIM: 603616) interacts with RAB5 (MIM: 611714) and this interaction complex is disrupted by the mutations affecting any one of the interaction residues (Asp820, Gln826, and Gln829) in RABEP134 (Figure S3A). To further identify potential interaction residue(s) between RABEP1 and RAB5, we performed structural analysis by using the RABEP1-RAB5 complex structure and then identified four amino acid residues (Val817, Leu824, Glu833, and Gln837) in RABEP1 (Figure S3A). These four amino acid residues have also been suggested as interaction residues with RAB5.34 Because human RABEP1 and RABEP2 share a high protein sequence identity and both interact with RAB5,38 we aligned the C-terminal RAB5-binding motifs of both RABEP1 and 2 that share over 80% amino acid similarity (Figure S3B). We found that all seven interaction residues (experimentally confirmed [red] and structurally predicted [blue]) are conserved between RABEP1 and RABEP2 (Figures S3A and S3B). To analyze the RABEP2-RAB5 complex structure, we performed molecular dynamics simulation. We first obtained human RABEP2 protein structure from Alpha-Fold 2, an artificial intelligence-based protein structure prediction program.35,36 Then, using HDOCK software to predict the binding complexes between two molecules,37 we modeled the RABEP2-RAB5 complex structure. On the basis of these structural and in silico analyses, we identified additional coding SNP variants from the human population database that lie at or near these known or predicted contact points. A coding SNP variant at amino acid position 543, substituting Histidine (His) for Arginine (Arg) (Table S1; represented in bold font), is predicted to be damaging due to disruption of critical ionic interactions (Figures 4A–4C). This variant is also predicted to be damaging by the in silico prediction algorithms. This variant became our fourth variant for study.
Figure 4.
Protein-protein docking between human RABEP2 and human RAB5
(A) Homologous modeling of the interaction of human RABEP2 with human RAB5 showing the secondary structure with HDOCK homology software.
(B and C) Closed-up views of the interacting interfaces of human RAB5 with human RABEP2 (p.Arg543) (B) and human RAB5 with human RABEP2-coding variant (p.His543) (C). The black dotted lines indicate water-mediated hydrogen bonds. A water molecule is shown as a red sphere.
RABEP2-coding variants reduce endothelial cell migration
To investigate the role of RABEP2 on endothelial cell biology in vitro, we examined the effects of the gene on endothelial cell migration (Figures 3A and S4). We first determined the extent of protein loss by acute knockdown of RABEP2 expression in human microvascular endothelial cells (HMECs) by using RABEP2-specific siRNA (si-RABEP2). The efficiency of siRNA delivery in HMECs was evaluated for up to 72 h. RABEP2 mRNA transcript levels in HMECs showed a significant and gradual reduction after transfection of si-RABEP2 compared to that observed after transfection with non-specific siRNA (control siRNA) (Figure S5A). RABEP2 abundance concomitantly decreased (Figure S5B). We were able to restore RABEP2 protein levels for an additional 48 h after infection with CMV-RABEP2. RABEP2 abundance increased approximately 12-fold compared to control (CMV-control) (Figure S5C).
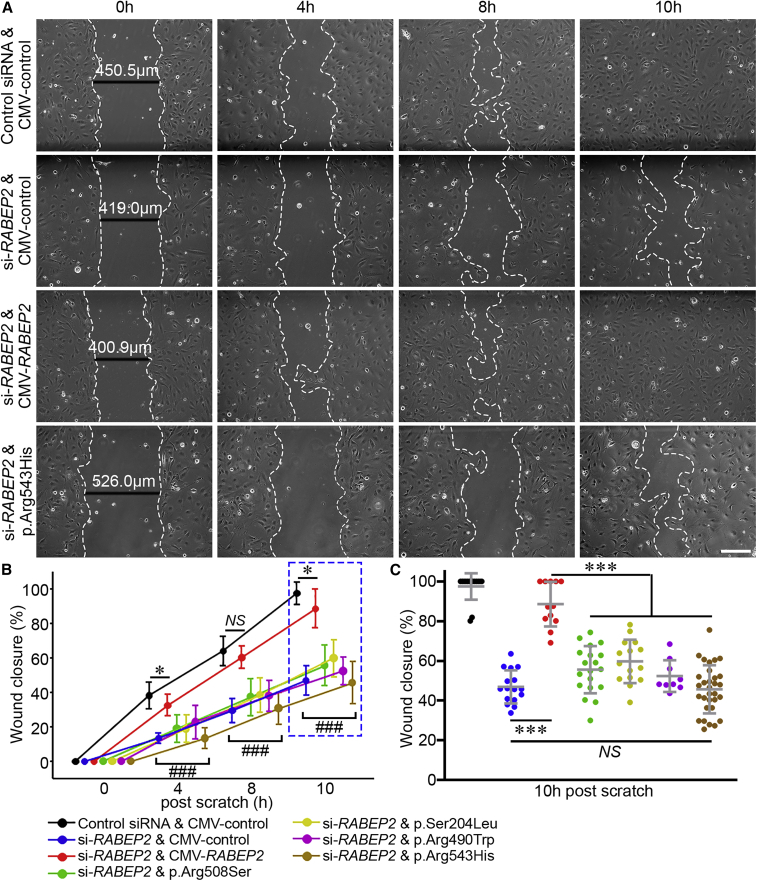
To determine whether RABEP2-coding variants would affect endothelial cell migration, we performed in vitro scratch assays (i.e., cell migration) after knockdown of RABEP2 expression by si-RABEP2 in HMECs, expressing different AAV constructs (Figure 5). As shown in Figure 5A, HMECs expressing CMV-control after treatment of si-RABEP2 (loss of function [LoF]) showed dramatically reduced cell migration as compared to HMECs treated with control siRNA and infected with the CMV-control. By contrast, the cell migration defect was rescued when exogenous RABEP2 was delivered (Figures 5A and 5B), indicating that RABEP2 is able to influence endothelial cell migration. The cell migration in HMECs expressing RABEP2 p.Arg543His was drastically reduced (Figures 5A and 5B), comparable to the migration of the LoF si-RABEP2. We also performed the cell migration assay by using the three other human RABEP2-coding variants (encoding p.Arg508Ser, p.Ser204Leu, and p.Arg490Trp). Cell migration rate was comparable across all four variants (Figures 5B and 5C), suggesting they each represent hypomorphic or loss-of-function alleles that segregate in the human population.
Figure 5.
Human RABEP2-coding variants impair cell migration in an in vitro scratch wound assay
(A) Representative bright-field images display four different groups: control infected with CMV-control after control siRNA treatment, loss of function (LoF) infected with CMV-control after si-RABEP2 treatment, rescued infected with CMV-RABEP2 after si-RABEP2 treatment, and p.Arg543His infected with CMV-RABEP2-p.Arg543His after si-RABEP2 treatment. A fully confluent (100%) HMEC monolayer for each treatment was scratched with the tip of a 200 μL pipet. At different time intervals (0, 4, 8, and 10 h), the degree of migration of HMECs was imaged.
(B) The line graph shows wound closure ratio. The wound closure rate was quantified as the percentage of area reduction at each time point. The total number of scratch experiments for control, LoF, rescued, p.Arg508Ser, p.Ser204Leu, p.Arg490Trp, and p.Arg543His were 15, 16, 12, 19, 15, 9, and 31, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗p < 0.05; ### (individual group) p < 0.001 versus control and rescued groups; NS, not significant).
(C) The scatterplots show the percentage of wound closure 10 h after scratch. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗∗p < 0.001).
A recent study provided mechanistic insight into the role of RABEP2 in arteriogenesis, where RABEP2 regulates endothelial cell surface localization of a receptor for vascular endothelial growth factor, VEGFR2.33 In agreement with their observation, cell-surface-localized VEGFR2 was reduced in HMECs after transfection of si-RABEP2 (Figure S6). We sought to determine whether the previously investigated coding SNPs in RABEP2 show a defect in VEGFR2 cell surface localization. After knockdown of RABEP2 in HMECs, we introduced human RABEP2 either with CMV-RABEP2 WT or with p.Arg543His. Whereas WT RABEP2 rescued the VEGFR2 cell surface protein to near normal levels, the p.Arg543His variant did not (Figure S6). These data show that at least one of the variants is defective in VEGFR2 localization and also validate an important cellular function of RABEP2 in endothelial cells.
RABEP2-coding variants abolish the ability of RABEP2 to rescue the collateral vessel and infarct volume phenotypes in Rabep2 KO mice
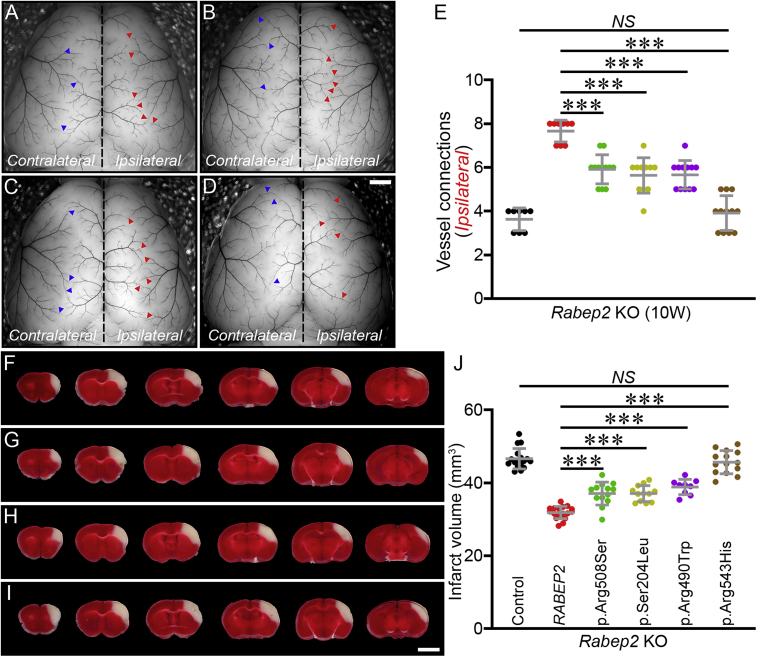
To investigate whether human RABEP2-coding variants could modulate clinically relevant phenotypes of collateral vessel number and ischemic stroke infarct volume, we examined these phenotypes in Rabep2 KO mice expressing the different RABEP2-coding variants. We first measured the number of collateral vessel connections between the ACA and MCA in Rabep2 KO mice. The number of collateral vessel connections in Rabep2 KO mice injected with either CMV-RABEP2-p.Arg508Ser, CMV-RABEP2-p.Ser204Leu, or CMV-RABEP2-p.Arg490Trp were slightly increased compared to Rabep2 KO mice injected with CMV-control (Figures 6A–6E); however, these collateral vessel connections were not as fully developed as those we observed in Rabep2 KO mice injected with CMV-RABEP2 (Figure 6E). Interestingly, CMV-RABEP2-p.Arg543His, which is predicted to disrupt the interaction between RABEP2 and RAB5, has no ability to rescue the collateral vessel defect (Figure 6E). The number of collateral vessel connections in Rabep2 KO mice injected with CMV-RABEP2-p.Arg543His is similar to Rabep2 KO mice injected with CMV-control (Figure 6E). Rabep2 KO mice injected with either CMV-RABEP2-p.Arg508Ser, CMV-RABEP2-p.Ser204Leu, or CMV-RABEP2-p.Arg490Trp did show increased collateral vessel connections between 3 and 10 weeks after AAV injection (Figure S7). However, Rabep2 KO mice injected with CMV-RABEP2-p.Arg543His showed no difference in collateral vessel connections between 3 and 10 weeks after AAV injection (Figure S7). Thus, this coding SNP, p.Arg543His, of RABEP2 is effectively a null allele.
Figure 6.
Human RABEP2-coding variants significantly impair rescue of both collateral vessel density and infarct volume phenotypes
(A–D) Representative mouse brain images of Rabep2 KO mice injected with either p.Arg508Ser (A), p.Ser204Leu (B), p.Arg490Trp (C), or p.Arg543His (D). Blue and red arrowheads indicate collateral vessel connections on the Contralateral (no virus) and on the Ipsilateral (with virus) hemisphere, respectively. Scale bar: 1 mm.
(E) The scatter plots show the number of collateral vessel connections between ACA and MCA of the Ipsilateral hemisphere, 10 weeks after AAV injection. The total number of animals for Rabep2 KO injected with CMV-control, CMV-RABEP2, p.Arg508Ser, p.Ser204Leu, p.Arg490Trp, and p.Arg543His are 8, 9, 12, 11, 12, and 12 animals, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗∗p < 0.001; NS, not significant).
(F–I) Serial brain sections (1 mm) for Rabep2 KO mice injected with either p.Arg508Ser (F), p.Ser204Leu (G), p.Arg490Trp (H), or p.Arg543His (I). Scale bar: 5 mm.
(J) The scatter plots show the infarct volume for Rabep2 KO mice injected with CMV-control, CMV-RABEP2, p.Arg508Ser, p.Ser204Leu, p.Arg490Trp, and p.Arg543His; 17, 18, 13, 12, 9, and 14 animals, respectively. Data represent the mean ± SD and statistical significance was determined by one-way ANOVA followed by Tukey’s multiple comparison test (∗∗p < 0.01; ∗∗∗p < 0.001; NS, not significant).
We next examined the effect of the human RABEP2-coding variants on ischemic stroke in the permanent occlusion model. We introduced each human RABEP2-coding variant with recombinant AAV injection into Rabep2 KO mice and performed pMCAO to measure infarct volume for each individual coding variant (Figures 6F–6I). As observed with the collateral vessel phenotypes in Rabep2 KO mice with different coding variants, the infarct volume in Rabep2 KO mice injected with either CMV-RABEP2-p.Arg508Ser, CMV-RABEP2-p.Ser204Leu, or CMV-RABEP2-p.Arg490Trp was reduced compared to Rabep2 KO mice injected with CMV-control (Figure 6J). However, these infarct volumes were still larger than that observed in Rabep2 KO mice injected with CMV-RABEP2, suggesting that these coding variants are hypomorphic alleles (Figure 6J). The infarct volume in Rabep2 KO mice injected with CMV-RABEP2-p.Arg543His showed no difference from Rabep2 KO mice injected with CMV-control (Figure 6J). Together with the results from the rescue experiment for collateral vessel number, it appears that this variant is a null allele. These data show that certain human RABEP2-coding variants are either hypomorphs or complete loss-of-function alleles for both phenotypes of collateral vessel connections and infarct volume.
The overall population allele frequencies for these four variants are quite low. While p.Arg508Ser is found in multiple populations, the other three of these variants exclusively segregate within distinct populations. As shown in Table 1, p.Ser204Leu, p.Arg490Trp, and p.Arg543His are predominantly found in the European, East Asian, and African/African American populations, respectively. These coding variants are potentially a strong determinant of stroke outcome in these populations.
Table 1.
Population frequencies for four RABEP2-coding variants
| Population | p.Arg508Ser |
p.Ser204Leu |
p.Arg490Trp |
p.Arg543His |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele |
Allele |
allele |
allele |
|||||||||
| Count | num. | freq. | count | num. | freq. | count | num. | freq. | count | num. | freq. | |
| Ashkenazi Jewish | 29 | 10,036 | 0.00289 | 0 | 10,168 | 0 | 0 | 10,326 | 0 | 0 | 10,268 | 0 |
| European (non-Finnish) | 136 | 124,560 | 0.001092 | 7 | 124,038 | 0.0000564 | 0 | 128,000 | 0 | 0 | 125,522 | 0 |
| European (Finnish) | 1 | 23,762 | 0.0000421 | 84 | 24,870 | 0.003378 | 0 | 24,992 | 0 | 0 | 21,320 | 0 |
| African/African American | 2 | 23,348 | 0.0000857 | 0 | 23,748 | 0 | 2 | 24,130 | 0.0000829 | 15 | 23,982 | 0.0006255 |
| Latino/admixed American | 22 | 34,648 | 0.000635 | 0 | 35,250 | 0 | 0 | 35,346 | 0 | 0 | 35,236 | 0 |
| East Asian | 1 | 19,204 | 0.0000521 | 0 | 19,486 | 0 | 31 | 19,514 | 0.001589 | 0 | 19,490 | 0 |
| South Asian | 34 | 29,398 | 0.001157 | 0 | 30,506 | 0 | 1 | 30,600 | 0.0000328 | 1 | 30,534 | 0.0000327 |
| Other | 14 | 6,904 | 0.002028 | 0 | 7,058 | 0 | 0 | 7,128 | 0 | 0 | 7,052 | 0 |
| XX | 91 | 123,556 | 0.0007365 | 44 | 124,658 | 0.000353 | 17 | 127,538 | 0.0001333 | 9 | 123,862 | 0.0000727 |
| XY | 148 | 148,304 | 0.000998 | 47 | 150,466 | 0.0003124 | 17 | 152,498 | 0.0001115 | 7 | 149,542 | 0.0000468 |
| Total | 239 | 271,860 | 0.0008791 | 91 | 275,124 | 0.0003308 | 34 | 280,036 | 0.0001214 | 16 | 273,404 | 0.0000585 |
The list displays population-based allele frequencies of RABEP2-coding variants. While one (p.Arg508Ser) is widely distributed across populations, p.Ser204Leu, p.Arg490Trp, or p.Arg543His exclusively segregate in the European, East Asian, or African/African American populations, respectively.
Discussion
Although mouse models of ischemic stroke have been widely used to gain insight into pathobiological mechanisms, translation of these findings to the clinic have been largely disappointing.39, 40, 41 As an alternative strategy, identification of drug targets, has more recently focused on candidate gene identification through human genetics.42 Therapeutic targets identified as a result of their genetic variants that predict risk or disease outcome are twice as likely to progress through the phases of clinical development as those identified through other experimental approaches.43,44
In this study, we focused on investigating and validating a potential candidate gene for ischemic stroke, Rabep2, by examining the effects of human sequence variants on clinically relevant phenotypes. Rabep2 was identified via a forward genetic mapping approach in mouse inbred strains,23,24,26 and a single locus containing Rabep2 was identified as modulating infarct volume through a collateral vessel-dependent pathway.32 Yet to date, there is no evidence that human RABEP2 plays a role in either ischemic stroke or stroke outcomes. Instead, GWASs have shown that SNPs near or within RABEP2 are associated with body mass, obesity, type 2 diabetes, and neurological phenotypes (e.g., Parkinson disease [MIM: 605909], intelligence, cortical surface size).45, 46, 47, 48, 49, 50, 51
We have developed an in vivo evaluation platform to investigate the functional effects of human sequence variations in RABEP2. This evaluation platform is based on an “add-back” gene rescue strategy and an in vivo rescue platform. We express the missing gene in knockout mice by using recombinant AAV. Employing this in vivo evaluation platform, we show that we can rescue the Rabep2 KO phenotype. Significantly, we also show that RABEP2 rescues the mouse Rabep2 KO ischemic stroke phenotypes. This cross-species complementation allows us to experimentally investigate the functional effects of sequence variation in the human gene.
We prioritized four different coding variants in RABEP2 that were predicted to be damaging by in silico prediction algorithms. One of these was also identified as potentially damaging by structural analyses. Although relatively rare, three of these are population-specific variants. Coding variant p.Arg543His, which lies near a critical interaction residue for RAB5, appears to be a complete loss-of-function variant. It segregates exclusively in the African/African American population.
We examined the effects of the prioritized variants on an endothelial cell biological phenotype (scratch wound migration) in VEGFR2 localization and, in vivo, in the rescue of the phenotypes of collateral vessel number and infarct volume after pMCAO. When injected into the Rabep2 KO mice, we showed that each of these four variants are at best hypomorphic alleles, and one variant (encoding p.Arg543His) appeared as a null allele for all four phenotypes. In silico analyses have further identified other coding variants in RABEP2 that are predicted to be damaging. Thus, RABEP2 appears to be rich in potentially damaging coding variations that may influence human stroke outcomes. By cross-species validation, we show that RABEP2 variation might play a role in human ischemic stroke by modulating infarct volume through effects of the vascular circulation. We note, however, that in the Rabep2 KO mice, the two ischemic stroke phenotypes, collateral vessel density and infarct volume, are differentially rescued by different RABEP2-coding variants, suggesting a more complex role for this gene in ischemic stroke.
This study again demonstrates the importance Rabep2 on ischemic stroke outcome. We have also identified RABEP2 as a therapeutic target candidate in human ischemic stroke, although further studies are required to determine the RABEP2 mechanism of action in ischemic stroke. Additionally, we show that an in vivo evaluation platform in mice that uses AAV gene replacement is an effective and efficient method for screening the functional effects of gene variants. Using this system, we can query the functional consequences coding variants of almost any human gene relevant to ischemic stroke in the mouse. This will expand the number of physiologically relevant target candidates with the goal of identifying therapeutics for human ischemic stroke.
Acknowledgments
The authors thank Mr. Christian R. Benavides for animal husbandry. This study was supported by grants from the NIH (grant 5R01NS100866 to D.A.M.) and the Foundation Leducq Transatlantic Network Of Excellence in Neurovascular Disease (17 CVD 03 to D.A.M.) and American Heart Association Career Development Award 938553 (to H.K.L.).
Author contributions
H.K.L. and D.A.M. designed research. H.K.L. performed research. D.H.K. performed structure modeling. H.K.L., D.L.A., and D.A.M. analyzed the data. H.K.L. and D.A.M. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: September 26, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.09.003.
Contributor Information
Han Kyu Lee, Email: hankyu.lee@duke.edu.
Douglas A. Marchuk, Email: douglas.marchuk@duke.edu.
Supplemental information
Data and code availability
This study did not generate datasets.
References
- 1.Bogousslavsky J., Aarli J., Kimura J., Board of Trustees, World Federation of Neurology Stroke: time for a global campaign? Cerebrovasc. Dis. 2003;16:111–113. doi: 10.1159/000070589. [DOI] [PubMed] [Google Scholar]
- 2.Johnson W., Onuma O., Owolabi M., Sachdev S. Stroke: a global response is needed. Bull. World Health Organ. 2016;94 doi: 10.2471/BLT.16.181636. 634-634A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krishnamurthi R.V., Feigin V.L., Forouzanfar M.H., Mensah G.A., Connor M., Bennett D.A., Moran A.E., Sacco R.L., Anderson L.M., Truelsen T., et al. Global and regional burden of first-ever ischaemic and haemorrhagic stroke during 1990-2010: findings from the Global Burden of Disease Study 2010. Lancet. Glob. Health. 2013;1:e259–e281. doi: 10.1016/S2214-109X(13)70089-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim J., Thayabaranathan T., Donnan G.A., Howard G., Howard V.J., Rothwell P.M., Feigin V., Norrving B., Owolabi M., Pandian J., et al. Global Stroke Statistics 2019. Int. J. Stroke. 2020;15:819–838. doi: 10.1177/1747493020909545. [DOI] [PubMed] [Google Scholar]
- 5.Writing Group Members. Benjamin E.J., Fullerton H.J., Arnett D.K., Jiménez M.C., Cushman M., Huffman M.D., de Ferranti S., Kissela B.M., Lisabeth L.D., et al. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association. Circulation. 2016;133 doi: 10.1161/CIR.0000000000000350. e38–360. [DOI] [PubMed] [Google Scholar]
- 6.Roger V.L., Go A.S., Lloyd-Jones D.M., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., et al. Heart disease and stroke statistics-2012 update: a report from the American Heart Association. Circulation. 2012;125 doi: 10.1161/CIR.0b013e31823ac046. e2–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamin E.J., Muntner P., Alonso A., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Das S.R., et al. Heart disease and stroke statistics-2019 update: a report from the American heart association. Circulation. 2019;139 doi: 10.1161/CIR.0000000000000659. e56–528. [DOI] [PubMed] [Google Scholar]
- 8.Virani S.S., Alonso A., Benjamin E.J., Bittencourt M.S., Callaway C.W., Carson A.P., Chamberlain A.M., Chang A.R., Cheng S., Delling F.N., et al. Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association. Circulation. 2020;141 doi: 10.1161/CIR.0000000000000757. e139–596. [DOI] [PubMed] [Google Scholar]
- 9.Goyal M., Demchuk A.M., Menon B.K., Eesa M., Rempel J.L., Thornton J., Roy D., Jovin T.G., Willinsky R.A., Sapkota B.L., et al. Randomized assessment of rapid endovascular treatment of ischemic stroke. N. Engl. J. Med. 2015;372:1019–1030. doi: 10.1056/NEJMoa1414905. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y.D., Nam H.S., Kim S.H., Kim E.Y., Song D., Kwon I., Yang S.-H., Lee K., Yoo J., Lee H.S., Heo J.H. Time-dependent Thrombus resolution after tissue-type plasminogen activator in patients with stroke and mice. Stroke. 2015;46:1877–1882. doi: 10.1161/STROKEAHA.114.008247. [DOI] [PubMed] [Google Scholar]
- 11.Muchada M., Rodriguez-Luna D., Pagola J., Flores A., Sanjuan E., Meler P., Boned S., Alvarez-Sabin J., Ribo M., Molina C.A., Rubiera M. Impact of time to treatment on tissue-type plasminogen activator-induced recanalization in acute ischemic stroke. Stroke. 2014;45:2734–2738. doi: 10.1161/STROKEAHA.114.006222. [DOI] [PubMed] [Google Scholar]
- 12.Emberson J., Lees K.R., Lyden P., Blackwell L., Albers G., Bluhmki E., Brott T., Cohen G., Davis S., Donnan G., et al. Effect of treatment delay, age, and stroke severity on the effects of intravenous thrombolysis with alteplase for acute ischemic stroke: a meta-analysis of individual patient data from randomized trials. Lancet. 2014;384:1929–1935. doi: 10.1016/S0140-6736(14)60584-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albers G.W., Goldstein L.B., Hess D.C., Wechsler L.R., Furie K.L., Gorelick P.B., Hurn P., Liebeskind D.S., Nogueira R.G., Saver J.L., STAIR VII Consortium STAIR VII Consortium. Stroke Treatment Academic Industry Roundtable (STAIR) recommendations for maximizing the use of intravenous thrombolytics and expanding treatment options with intra-arterial and neuroprotective therapies. Stroke. 2011;42:2645–2650. doi: 10.1161/STROKEAHA.111.618850. [DOI] [PubMed] [Google Scholar]
- 14.O'Donnell M.J., Xavier D., Liu L., Zhang H., Chin S.L., Rao-Melacini P., Rangarajan S., Islam S., Pais P., McQueen M.J., et al. Risk factors for ischemic and intracerebral hemorrhagic stroke in 22 countries (the interstroke study): A case-control study. Lancet. 2010;376:112–123. doi: 10.1016/S0140-6736(10)60834-3. [DOI] [PubMed] [Google Scholar]
- 15.Go A.S., Mozaffarian D., Roger V.L., Benjamin E.J., Berry J.D., Borden W.B., Bravata D.M., Dai S., Ford E.S., Fox C.S., et al. Heart disease and stroke statistics-2013 update: A report from the American heart association. Circulation. 2013;127 doi: 10.1161/CIR.0b013e31828124ad. e6–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elkind M.S.V. Why now? Moving from stroke risk factors to stroke triggers. Curr. Opin. Neurol. 2007;20:51–57. doi: 10.1097/WCO.0b013e328012da75. [DOI] [PubMed] [Google Scholar]
- 17.Boehme A.K., Esenwa C., Elkind M.S.V. Stroke Risk Factors, Genetics, and Prevention. Circ. Res. 2017;120:472–495. doi: 10.1161/CIRCRESAHA.116.308398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gretarsdottir S., Thorleifsson G., Reynisdottir S.T., Manolescu A., Jonsdottir S., Jonsdottir T., Gudmundsdottir T., Bjarnadottir S.M., Einarsson O.B., Gudjonsdottir H.M., et al. The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat. Genet. 2003;35:131–138. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 19.Helgadottir A., Manolescu A., Thorleifsson G., Gretarsdottir S., Jonsdottir H., Thorsteinsdottir U., Samani N.J., Gudmundsson G., Grant S.F.A., Thorgeirsson G., et al. The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat. Genet. 2004;36:233–239. doi: 10.1038/ng1311. [DOI] [PubMed] [Google Scholar]
- 20.Matarín M., Brown W.M., Scholz S., Simón-Sánchez J., Fung H.-C., Hernandez D., Gibbs J.R., De Vrieze F.W., Crews C., Britton A., et al. A genome-wide genotyping study in patients with ischemic stroke: initial analysis and data release. Lancet Neurol. 2007;6:414–420. doi: 10.1016/S1474-4422(07)70081-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matarin M., Brown W.M., Singleton A., Hardy J.A., Meschia J.F., ISGS investigators Whole genome analyses suggest ischemic stroke and heart disease share an association with polymorphisms on chromosome 9p21. Stroke. 2008;39:1586–1589. doi: 10.1161/STROKEAHA.107.502963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matarin M., Simon-Sanchez J., Fung H.-C., Scholz S., Gibbs J.R., Hernandez D.G., Crews C., Britton A., De Vrieze F.W., Brott T.G., et al. Structural genomic variation in ischemic stroke. Neurogenetics. 2008;9:101–108. doi: 10.1007/s10048-008-0119-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keum S., Marchuk D.A. A locus mapping to mouse chromosome 7 determines infarct volume in a mouse model of ischemic stroke. Circ. Cardiovasc. Genet. 2009;2:591–598. doi: 10.1161/CIRCGENETICS.109.883231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keum S., Lee H.K., Chu P.-L., Kan M.J., Huang M.-N., Gallione C.J., Gunn M.D., Lo D.C., Marchuk D.A. Natural genetic variation of integrin alpha L (Itgal) modulates ischemic brain injury in stroke. PLoS Genet. 2013;9:e1003807. doi: 10.1371/journal.pgen.1003807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chu P.-L., Keum S., Marchuk D.A. A novel genetic locus modulates infarct volume independently of the extent of collateral circulation. Physiol. Genomics. 2013;45:751–763. doi: 10.1152/physiolgenomics.00063.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H.K., Keum S., Sheng H., Warner D.S., Lo D.C., Marchuk D.A. Natural allelic variation of the IL-21 receptor modulates ischemic stroke infarct volume. J. Clin. Invest. 2016;126:2827–2838. doi: 10.1172/JCI84491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee H.K., Koh S., Lo D.C., Marchuk D.A. Neuronal IL-4Rα modulates neuronal apoptosis and cell viability during the acute phases of cerebral ischemia. FEBS J. 2018;285:2785–2798. doi: 10.1111/febs.14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee H.K., Widmayer S.J., Huang M.-N., Aylor D.L., Marchuk D.A. Novel neuroprotective loci modulating ischemic stroke volume in wild-derived inbred mouse strains. Genetics. 2019;213:1079–1092. doi: 10.1534/genetics.119.302555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee H.K., Wetzel-Strong S.E., Aylor D.L., Marchuk D.A. A neuroprotective locus modulates ischemic stroke infarction independent of collateral vessel anatomy. Front. Neurosci. 2021;15:705160. doi: 10.3389/fnins.2021.705160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., Zhang H., Wiltshire T., Sealock R., Faber J.E. Genetic Dissection of the Canq1 Locus Governing Variation in Extent of the Collateral Circulation. PLoS One. 2012;7:e31910. doi: 10.1371/journal.pone.0031910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., Prabhakar P., Sealock R., Faber J.E. Wide genetic variation in the native pial collateral circulation is a major determinant of variation in severity of stroke. J. Cereb. Blood Flow Metab. 2010;30:923–934. doi: 10.1038/jcbfm.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucitti J.L., Sealock R., Buckley B.K., Zhang H., Xiao L., Dudley A.C., Faber J.E. Variants of Rab GTPase-Effector Binding Protein-2 Cause Variation in the Collateral Circulation and Severity of Stroke. Stroke. 2016;47:3022–3031. doi: 10.1161/STROKEAHA.116.014160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kofler N., Corti F., Rivera-Molina F., Deng Y., Toomre D., Simons M. The Rab-effector protein RABEP2 regulates endosomal trafficking to mediate vascular endothelial growth factor receptor-2 (VEGFR2)-dependent signaling. J. Biol. Chem. 2018;293:4805–4817. doi: 10.1074/jbc.M117.812172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu G., Zhai P., Liu J., Terzyan S., Li G., Zhang X.C. Structural basis of Rab5-Rabaptin5 interaction in endocytosis. Nat. Struct. Mol. Biol. 2004;11:975–983. doi: 10.1038/nsmb832. [DOI] [PubMed] [Google Scholar]
- 35.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Žídek A., Potapenko A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Varadi M., Anyango S., Deshpande M., Nair S., Natassia C., Yordanova G., Yuan D., Stroe O., Wood G., Laydon A., et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2021;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yan Y., Tao H., He J., Huang S.-Y. The HDOCK server for integrated protein–protein docking. Nat. Protoc. 2020;15:1829–1852. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 38.Gournier H., Stenmark H., Rybin V., Lippé R., Zerial M. Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J. 1998;17:1930–1940. doi: 10.1093/emboj/17.7.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodge R.D., Bakken T.E., Miller J.A., Smith K.A., Barkan E.R., Graybuck L.T., Close J.L., Long B., Johansen N., Penn O., et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O'Collins V.E., Macleod M.R., Donnan G.A., Horky L.L., van der Worp B.H., Howells D.W. 1, 026 experimental treatments in acute stroke. Ann. Neurol. 2006;59:467–477. doi: 10.1002/ana.20741. [DOI] [PubMed] [Google Scholar]
- 41.Ginsberg M.D. Neuroprotection for ischemic stroke: past, present and future. Neuropharmacology. 2008;55:363–389. doi: 10.1016/j.neuropharm.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plenge R.M., Scolnick E.M., Altshuler D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 43.Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J., et al. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 44.King E.A., Davis J.W., Degner J.F. Are drug targets with genetic support twice as likely to be approved? Revised estimates of the impact of genetic support for drug mechanisms on the probability of drug approval. PLoS Genet. 2019;15:e1008489. doi: 10.1371/journal.pgen.1008489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turcot V., Lu Y., Highland H.M., Schurmann C., Justice A.E., Fine R.S., Bradfield J.P., Esko T., Giri A., Graff M., et al. Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2018;50:26–41. doi: 10.1038/s41588-017-0011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Christakoudi S., Evangelou E., Riboli E., Tsilidis K.K. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci. Rep. 2021;11:10688. doi: 10.1038/s41598-021-89176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vujkovic M., Keaton J.M., Lynch J.A., Miller D.R., Zhou J., Tcheandjieu C., Huffman J.E., Assimes T.L., Lorenz K., Zhu X., et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat. Genet. 2020;52:680–691. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berndt S.I., Gustafsson S., Mägi R., Ganna A., Wheeler E., Feitosa M.F., Justice A.E., Monda K.L., Croteau-Chonka D.C., Day F.R., et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat. Genet. 2013;45:501–512. doi: 10.1038/ng.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grasby K.L., Jahanshad N., Painter J.N., Colodro-Conde L., Bralten J., Hibar D.P., Lind P.A., Pizzagalli F., Ching C.R.K., McMahon M.A.B., et al. The genetic architecture of the human cerebral cortex. Science. 2020;367:eaay6690. [Google Scholar]
- 50.Nalls M.A., Blauwendraat C., Vallerga C.L., Heilbron K., Bandres-Ciga S., Chang D., Tan M., Kia D.A., Noyce A.J., Xue A., et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–1102. doi: 10.1016/S1474-4422(19)30320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill W.D., Marioni R.E., Maghzian O., Ritchie S.J., Hagenaars S.P., McIntosh A.M., Gale C.R., Davies G., Deary I.J. A combined analysis of genetically correlated traits identifies 187 loci and a role for neurogenesis and myelination in intelligence. Mol. Psychiatry. 2019;24:169–181. doi: 10.1038/s41380-017-0001-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets.