Abstract
Background
Infants are frequently exposed to antibiotics (AB) in the first week of life for suspected bacterial infections. Little is known about the effect of AB on the developing intestinal microbiota. Therefore, we studied intestinal microbiota development with and without AB exposure in the first week of life in term born infants.
Methods
We analysed the faecal microbiota from birth until 2.5 years of age by 16S rRNA gene amplicon sequencing in a cohort with 56 term born infants, exposed to AB in the first week of life (AB+) (AB for 2–3 days (AB2, n=20), AB for 7 days (AB7, n=36)), compared with 126 healthy controls (AB-). The effects of AB and duration were examined in relation to delivery and feeding mode.
Results
AB+ was associated with significantly increased relative abundance of Enterobacteriaceae at 3 weeks and 1 year and a decrease of Bifidobacteriaceae, from 1 week until 3 months of age only in vaginally delivered, but not in C-section born infants. Similar deviations were noted in AB7, but not in AB2. After AB, breastfed infants had lower relative abundance of potentially pathogenic Enterobacteriaceae compared with formula fed infants and recovered 2 weeks faster towards controls.
Conclusions
AB exposure in the first week of life alters faecal microbiota development with deviations in the relative abundance of individual taxa until 1 year of age. These alterations can have long-term health consequences, which emphasises the need for future studies aiming at restoring intestinal microbiota after AB administration.
Keywords: Microbiology, Infant Development, Gastroenterology, Epidemiology, Neonatology
Study of the effect of antibiotics in the first week of life on fecal microbiota up to 2.5 years of age. Differences identified up to one year of age in babies who received antibiotics when compared with controls.
What is already known on this topic
Up to 20% of neonates receive antibiotics because of (suspected) early-onset neonatal sepsis. In older infants, antibiotics have been shown to disturb the intestinal microbiota, but studies in newborns with a developing microbiota are limited.
Feeding type and delivery mode also affect the microbiota, but their effect in relation to antibiotic exposure in the first week of life has not yet been studied.
What this study adds
Antibiotic exposure in the first week of life affects microbiota development throughout the first year of life, with more profound deviations in infants born at term exposed for 7 days compared with only 2–3 days.
Antibiotic exposure in the first week of life resulted in deviations in the faecal microbiota development of vaginally delivered infants but not of C-section delivered infants compared with their respective controls. This could be attributed to the difference caused by delivery mode itself, with C-section delivered infants deviating from vaginal controls up to 1 month of age regardless of antibiotic exposure.
How this study might affect research, practice and/or policy
This study underlines the importance of early cessation of antibiotics started at birth because of the prolonged effect on the intestinal microbiota and possible impact on health.
Introduction
During the first 1000 days of life, the intestinal microbiota impacts health in later life through the interdependent development of microbiota, immune system, growth and cognitive function.1 2 After birth, the intestinal microbiota develops rapidly, driven by exposure to microbes from maternal, environmental and dietary sources.3 During this early development, the intestinal microbiota is unstable and susceptible to perturbations such as those caused by antibiotic (AB) exposure. These perturbations may have long-term consequences on the developing microbiota and also on the developing immune system,4 growth5 6 and have already been associated with increased prevalence of asthma, allergies, coeliac disease, eczema, eosinophilic esophagitis, infantile colic, inflammatory bowel disease and obesity.7–11
In mice-based studies, AB exposure in the first week of life altered microbiota composition and immune function, but gavage with mature untreated microbiota, restored the perturbation and reduced the negative health effects.12 To understand the full impact in humans and develop restoration strategies, more knowledge is needed.
Worldwide, up to 20% of all neonates are prescribed ABs because of (suspected) early-onset neonatal sepsis, although in most cases, sepsis is unconfirmed and ABs could be discontinued after 48–72 hours.13–15 More prolonged AB exposure can gradually reduce overall diversity16 and richness17 of the neonate’s intestinal ecosystem. Additionally, AB type also determines the microbial perturbation through specific mechanisms of action and host interactions. Studies in infants have been inconclusive,17–19 but indicated types with faster recovery towards the microbiota composition of controls.20 Therefore, more comparative studies are needed between durations and types in AB regimes to optimise AB administration.21
In this study, we investigated the microbiota development in a subset of the Intestinal Microbiota Composition after AB treatment in early life (INCA) cohort.22 The primary aim was to investigate the impact of AB exposure in the first days of life on microbiota development during the first 2.5 years of life. Secondary aims were to examine (1) short (2–3 days) versus long (7 days) AB administration, (2) different AB types and (3) the impact of feeding and delivery mode on AB perturbation.
Materials and methods
Study design
This prospective, observational study has been described previously.10 22 To study the impact of feeding mode on AB-induced deviations, we selected a subset of 182 infants with 1128 samples who were exclusively breast-fed (BF) or formula-fed (FF) in the first 3 months of life. An overview of collected samples and their selection for analyses reported here is provided in online supplemental figure 1). All AB+ infants received gentamicin, which was combined with penicillin (ABPen), amoxicillin (ABAMX) or amoxicillin with clavulanic acid (ABAMC).
fetalneonatal-2021-322861supp001.pdf (1MB, pdf)
Data and faeces collection
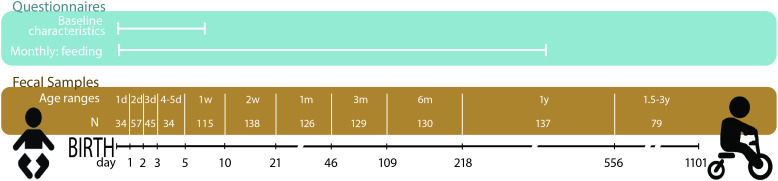
Baseline characteristics such as birth mode were assessed through hospital records. Feeding mode was reported monthly during the first year of life. Nine faecal samples were collected from infants (figure 1). Until discharge from the hospital, faeces were sampled from diapers by hospital staff and immediately frozen at −20°C. Sampling continued at home by the parents, using sampling instructions and freezer storage. After 1 year, parents delivered the samples to the clinic after transport on ice. A final sample was taken around 2 years, stored in the home freezer and collected by the study nurse. At the hospital, all samples were stored at −20°C.
Figure 1.
Overview of the collected questionnaires, faecal sampling points and the age categories based on the age (days) at sampling. N, number of infants for which samples were available for a given time point. d(ays), m(onths) and y(ears) indicate the age category ranging from birth until around 2 years of age.
16S rRNA gene amplicon sequencing
DNA was extracted from a maximum of 200 mg of the homogenised faecal sample at GenProbio srl (Parma, Italy) with the QIAamp DNA Stool Mini kit according to manufacturer’s instructions (Qiagen Ltd, Strasse, Germany). Sequencing libraries were prepared according to the 16S Metagenomic Sequencing Library Preparation Protocol (Part No. 15 044 223 Rev. B—Illumina) at GenProbio srl (Parma, Italy). Minor adaptations to the published protocol23 are noted in online supplemental file 1). Sequencing resulted in ~44 934 (SD 17205) reads per sample. Data were processed using NG-Tax 2.0 on demultiplexed fastq files, using default settings.24 Taxonomy was assigned using SILVA reference database V.128.25 Amplicon sequence variants (ASVs) were defined as unique sequence variants. Two synthetic mock communities were sequenced as positive controls.26
Statistical analysis of the baseline characteristics
Baseline characteristics were calculated using R 3.6.127 and tableone28 (table 1). Differences between groups were examined by using the Fisher exact test for the categorical variables and analysis of variance (ANOVA) for continuous variables. Not normally distributed variables were tested with the Kruskal-Wallis test and indicated by their median and IQR. Bonferroni correction was performed to correct for multiple testing.
Table 1.
Baseline characteristics of the INCA cohort subset included in this study
| N | AB-
|
AB+
|
AB2
|
AB7
|
| 126 | 56 | 20 | 36 | |
| GA, weeks (IQR)* | 39.4 (38.5, 40.4) |
40.4 (39.4, 41.1) |
40.05 (39.3, 41.0) |
40.55 (40.1, 41.2) |
| Birth weight, mean grams (SD) * | 3478 (515) | 3711 (484) | 3734 (428) | 3699 (518) |
| Birth weight for GA z-score (SD) | 0.15 (1.2%) | 0.39 (1%) | 0.30 (1%) | 0.57 (0.9%) |
| Sex (Female %) | 58 (46%) | 28 (50%) | 13 (65%) | 15 (42%) |
| Delivery mode (Vaginal %) | 83 (66%) | 41 (73%) | 15 (75%) | 26 (72%) |
| Exclusive breast feeding at 3 m (Yes %) | 47 (37%) | 23 (41%) | 17 (47%) | 6 (30%) |
| Additional AB 1–6 m (Yes %)*,** | 10 (8.2%) | 13 (24%) | 1 (5%) | 12 (34%) |
| Additional AB 7–12 m (Yes %) | 30 (27%) | 9 (19%) | 2 (10%) | 7 (24%) |
Statistically significant differences are indicated with *p<0.05 compared with AB-, **p<0.05 compared with AB2.
AB+, infants who received AB during their first week of life. Birth weight for GA z-score is calculated according to the z-score formula.77 AB2: AB exposure for 2 to 3 days in the first week of life, AB7: AB exposure for 7 days.
AB, antibiotics; AB-, control infants; GA, gestational age; m, months.
Bioinformatic and statistical analysis of the sequence data
All analyses were performed in R 3.6.1.27 Samples were stratified into 11 age-based and right-closed intervals for statistical analysis (figure 1). The Jenks Natural Breaks Classification (classInt package) was used to calculate the optimal ranges.29 Because of increasing increments between sampling, the x-axis (age) was log2 transformed for visualisation.
Alpha diversity (within sample diversity) was calculated at ASV level using Picante30 and Microbiome31 packages and following metrics: Faith’s phylogenetic diversity,32 ASV richness, Shannon and Inverse Simpson. All except Shannon diversity needed logarithmic transformation to obtain normal distribution for one-way analysis of covariance. Consecutive analyses were corrected for the baseline characteristic that differed significantly between AB+ and AB- and between AB2 and AB7.
Temporal trends in relative abundance were visualised using local regression with locally estimated scatterplot smoothing using ggplot2. These relative abundances did not meet normality requirements and were therefore compared using beta regression with BetaReg33 per age interval. The effects of AB on the relative abundance of phyla were modelled including and excluding delivery mode, feeding mode and additional AB exposure between one and 6 months. The optimal beta regression models, based on Akaike and Bayesian information criteria, only included AB exposure in the first week of life without additional terms.
Beta diversity (between sample diversity) was calculated using pairwise Weighted (WU)34 and Unweighted UniFrac (UU) distances.35 WU takes the relative abundance of each ASV into account, whereas UU uses presence or absence of ASVs. Pairwise UU and UW distance matrices were used to plot principle response curves (PRCs).36 PRC analysis is a redundancy analysis for interpreting longitudinal data. It visualises multivariate responses in a repeated observation design.36 37 The method was designed to analyse the treatment effect over time compared with controls, as it can disentangle the time-dependent effects from other possible determinants.38 39 In this study, time was displayed on the x-axis and the intestinal microbiota development was shown compared with AB- infants as the baseline reference group. Differences between AB groups were assessed per age interval using ANOVA in the vegan package.40
Results
Baseline characteristics of INCA cohort subset
The baseline characteristics differed between AB- and AB+ for gestational age, birth weight and additional AB exposure between one to 6 months (table 1). Birth weight z-score (birth weight corrected for gestational age), however, was comparable. AB2 differed from AB7 with regard to additional AB exposure between 1 and 6 months (5% vs 34%, respectively, p=0.019). Baseline characteristics were comparable between the AB type groups (online supplemental table S1).
Antibiotic-induced alterations to intestinal microbiota development
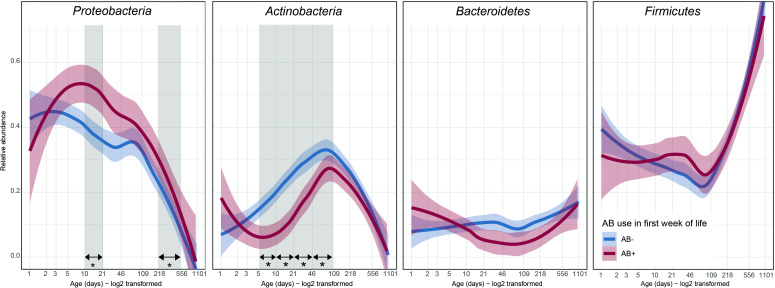
AB exposure during the first week of life did not alter microbial alpha diversity between birth and 2.5 years (online supplemental figure 1). The temporal patterns of the four major phyla (95% of the average relative abundance) were compared with univariate analyses (figure 2). Relative abundance of Proteobacteria was high overall, during the first months of life. AB exposure further increased this relative abundance at 1 month (mean 43.6% AB+, 31.5% AB-) and 1 year (mean 17.6% AB+, 6.5% AB-). Actinobacteria peaked around 3 months, but AB exposure decreased their relative abundance at 1 week (mean 6.3% AB+, 18.2% AB-), 2 weeks (mean 8.4% AB+, 24.4% AB-), 1 month (mean 17.5% AB+, 27.2% AB-) and 3 months (mean 26.5% AB+, 34.4% AB-). The average relative abundance of Bacteroidetes was stable at approximately 10% during the first year of life. Firmicutes drastically increased in relative abundance towards 2.5 years. Both were unaffected by AB.
Figure 2.
Temporal trajectories of the relative abundance of the four main phyla in the developing intestinal microbiota from birth to 2.5 years of age. Thick lines represent the average with shading showing the 95% CI. Differences between AB+ and AB- were calculated using beta regression for each age category. * and vertical grey shading: difference in relative abundance (p<0.05), AB+, infants who received AB during their first week of life, AB-, infants who did not receive AB during their first week of life, LOESS, locally estimated scatterplot smoothing.
Effect of antibiotic duration on long-term microbiota development
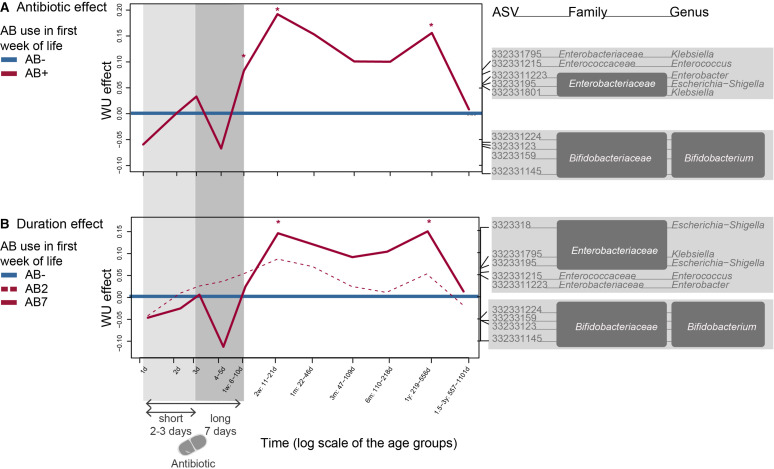
Based on univariate statistics, the temporal trajectories of Actinobacteria and Proteobacteria were most affected by AB. Based on UU, ABs increased several members of the Enterobacteriaceae and decreased Bifidobacteriaceae at one and 2 weeks (online supplemental figure 2A). For WU, ABs impacted similar taxa at 1 year (p<0.05) (figure 3A). These AB deviations were similar in AB2 and AB7, but only the microbiota composition of AB7 differed from AB- baseline (figure 3B and online supplemental figure 2B).
Figure 3.
WU-based PRC analysis is a special case of RDA for multivariate responses in a repeated observation design, which is applied here on the longitudinal infant intestinal microbiota dataset from birth till 2.5 years of age. (A) The infants who did not receive antibiotics during the first week of life (AB-), were compared as a baseline to the antibiotic exposed infants (AB+). Bacterial genera shown are the main drivers of the differences between AB+ and AB-: taxa on the same side of baseline as the curve are linked to an increased relative abundance at that time point, opposite sides indicate a decrease. (B) AB- was also compared as a baseline with the different antibiotic durations of 2–3 (AB2) or 7 days (AB7). Significance was tested at the different time points using an ANOVA like permutation test (*p<0.05 compared with baseline AB-). Covariates that were controlled for included additional AB exposure between the age of one and 6 months. AB+, infants who received AB during their first week of life; AB-, infants who did not receive AB during their first week of life; AB2, AB exposure for 2–3 days in the first week of life indicated with light grey shading; AB7, AB exposure for 7 days indicated with grey shading; ANOVA, analysis of variance; ASV, amplicon sequence variants; PRC, principal response curve; RDA, redundancy analysis; WU, weighted UniFrac.
AB types had a different impact on the microbiota (online supplemental figure 3A, B) with ABAMX not deviating from AB- baseline and ABPEN deviating at 1 week and 1 year with increased relative abundance of Enterobacteriaceae members in WU and UU. UU-based deviations between ABAMC and AB- baseline were limited to week one and involved different ASVs compared with ABPEN. ABAMC affected WU in the long-term with increased Enterobacteriaceae and Enterococcaceae at 2 weeks and 1 month and also Bifidobacterium at 1 week and 2.5 years.
Impact of delivery and feeding mode on AB-associated deviations in the faecal microbiota development
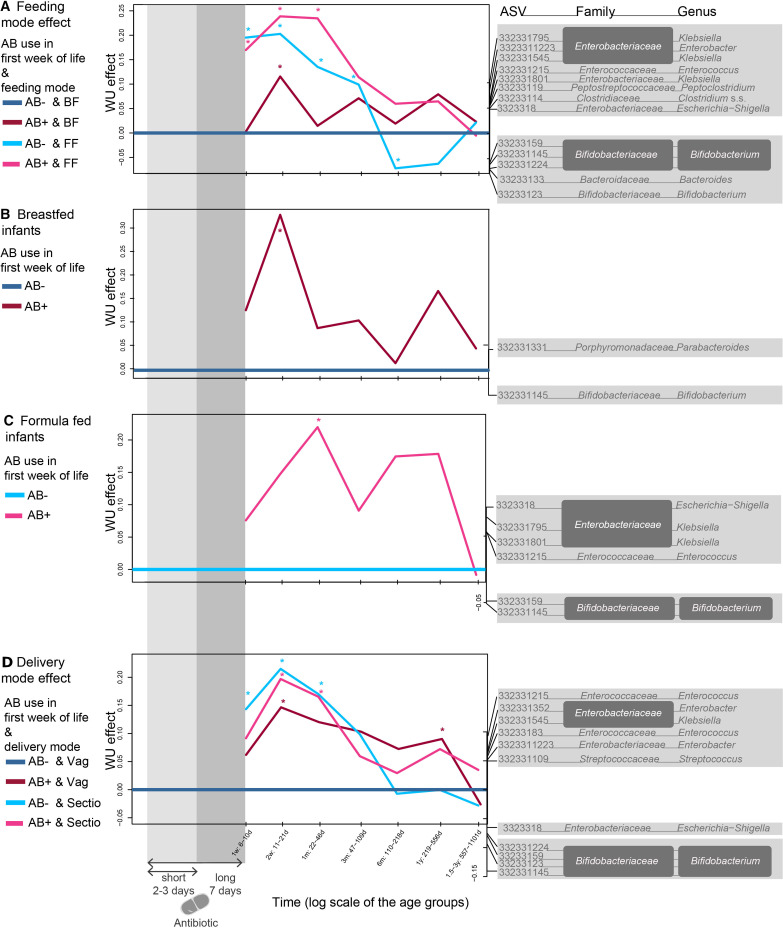
Due to the relatively low number of faecal samples in the first week per feeding and delivery type (figure 1), effects were only reported in samples collected between 1 week and 2.5 years. Within AB-, microbiota deviated based on delivery mode from 1 week until 1 month (figure 4D). In vaginally delivered infants, the AB effect on the microbiota was still significant at 1 year with an increase of several Enterobacteriaceae, Enterococcaceae and Streptococcaceae and decrease in Bifidobacterium and Escherichia-Shigella. In contrast, no AB-mediated deviations were noted between C-section born infants.
Figure 4.
WU-based PRC analyses in the different delivery and feeding mode groups. Bacterial genera shown are the main drivers for differences between the groups and the baseline: positive effect on the curve is linked to increased ASVs in the positive spectrum and decrease of those in the negative spectrum. (A) Breastfed controls (AB-BF) were compared as a baseline to antibiotic exposed (AB+) and FF infants. (B) Specifically featuring the AB effect in breastfed children, with breastfed control children as a baseline (AB-BF) and (C) formula-fed children, with formula-fed control children as a baseline (AB-). (D) Vaginally delivered control infants (AB-Vag) were compared as a baseline to antibiotic exposed (AB+) and C-section delivered (Sectio) infants. Significance was tested at the different time points using an ANOVA like permutation test (*p<0.05). Delivery mode analyses were controlled for feeding mode and vice versa. AB-, infants who did not receive AB during their first week of life; AB+, infants who received AB during their first week of life also indicated within grey shading; ASV, amplicon sequence variants; BF, infants exclusively breast-fed in the first 3 months of life; FF, infants exclusively formula-fed in the first 3 months of life; PRC, principal response curve; Sectio, infants delivered through C-section, Vag, vaginally delivered infants; WU, weighted UniFrac.
Compared with AB- BF baseline, AB+BF infants only deviated at 2 weeks, whereas AB+FF infants showed longer deviations from the first week up to 1 month (figure 4A). Because there was also a feeding effect during the first 6 months, the AB effect was also analysed within the separate feeding groups. AB+ was associated with decreased Bifidobacterium relative abundance in FF (1 month) (figure 4C), which occurred later than in BF infants (2 weeks) (figure 4B). In turn, AB+ was associated with increased relative abundance of Parabacteroides in BF, and with increased relative abundance of Enterobacteriaceae and Enterococcus in FF infants.
Discussion
In this prospective, observational INCA study, we examined the microbiota development after AB exposure during the first week of life and found perturbations in the faecal microbiota development from 1 week until 1 year of age. These perturbations included decreased relative abundance of Bifidobacteriaceae while potentially pathogenic Enterobacteriaceae increased. This study adds new insights into long-term compositional shifts after neonatal AB exposure.16 41 Our results corroborate findings in older infants with increased Enterobacteriaceae and decreased Bifidobacteriaceae after AB administration.16 20 42–44 Importantly, the severity and duration of AB-mediated microbiota perturbations increased with longer AB administration (5–7 vs 2–3 days). The results also align with a small study in preterm infants, where >5 days AB exposure intensified perturbations compared with 2–3 days.45
Bifidobacteriaceae form a cornerstone in the early development of the immune system. They were shown to promote B-cell maturation and associations with decreased inflammatory responses and T-regulatory cell acquisition.46 Enterobacteriaceae, on the other hand, produce toxins and have lipopolysaccharides on their outer membranes, which causes inflammation.47–49 Therefore, it is not surprising that reduced Bifidobacteriaceae, often combined with an increase in potentially pathogenic Enterobacteriaceae, like Shigella, Klebsiella and Enterobacter, have been associated with immune-mediated disorders like asthma.50 Similar deviations were also found in functional disorders like infantile colic51 and irritable bowel syndrome.52
The long-term microbiota effect of ABs and its associated negative health outcomes reinforce the need for implementing AB stewardship programs53–55 to avoid AB overuse.21 56 57 The microbiota perturbations were only significant after 5–7 days compared with 2–3 days AB which could explain previous findings from the INCA cohort: namely higher incidence of infantile colic, wheezing and food allergies in infants exposed for 5–7 days, but not for 2–3 days.10 58 If this observed difference between 2–3 days and 5–7 days exposure is the result of longer AB exposure or the result of a concomitant infection or inflammatory response is yet unclear. The AB7 infants were treated because of suspected early onset sepsis (EOS). EOS is rare in term infants,59–61 but it is difficult to distinguish from normal neonatal physiology after birth, and laboratory tests cannot always reliably detect or rule out EOS.62 Because the consequences of delaying treatment are significant, on average 82 newborns without EOS are treated for each case.15 63–65 In our study population, only two of the 36 AB7 infants had a positive blood culture. The others were also treated for 5–7 days because of elevated inflammatory markers or clinical symptoms. Uzan-Yulzari et al showed that the association between neonatal AB exposure and growth was independent of the neonatal infection state.6 This suggests that the differences in microbiota development after AB treatment in our study are more likely the result of the AB treatment duration itself than caused by a possible EOS. Our new findings emphasise the need for microbiota restoration to minimise aberrant immune development. Suggested strategies include prebiotics, probiotics and synbiotics66 but also faecal transfers, which partially restored the microbiota of mice exposed to AB for 7 days.12
In vaginally delivered infants, the AB effect was most pronounced with microbiota deviations in the second week of life. C-section born infants, however, showed similar perturbations regardless of AB exposure. After C-section, microbiota perturbations occurred due to reduced vertical mother-infant transmission of important intestinal microbes such as Bacteroides and Bifidobacterium, while transmission of other microbes like skin and mouth bacteria increased,67 as well as due to maternal AB administration prior to cord clamping.3 C-section delivery already showed decreases in Bifidobacterium spp and increases in opportunistic pathogens from hospital environments like Enterobacter, Enterococcus and Klebsiella.68 This resembled the AB effect, which might explain the lack of an additional AB effect in C-section infants as these infants already lack the affected microbial groups from birth.
Feeding also has a major impact on early life microbiota development.69 In our study, ABs in the first week of life perturbed the microbiota of both BF and FF infants, but potentially pathogenic Enterobacteriaceae only increased in FF infants. Moreover, AB perturbations were still notable at 1 month in FF infants but only until 2 weeks in BF infants. Breastmilk probably aids restoration through components like human milk oligosaccharides and live bacteria, which stimulate the growth of bifidobacteria and reduce (potential) pathogens.70
The strengths of this study are the quantity of samples and long-term follow-up. This enabled the investigation of the interplay of AB with delivery and feeding modes. Moreover, the quantity of sampling points allowed for detailed and long-term detection of AB-induced perturbations within individuals. This was relevant as AB impact was not uniform over time, suggesting that limited sampling points could lead to misinterpretation. Finally, the number of infants receiving additional courses of AB in the first year was low in this cohort, thereby reducing an important confounding factor.
The methodology for profiling the intestinal microbiota, which targeted the V3 hypervariable regions of the bacterial 16S rRNA gene, provides a cost-effective overview of bacterial community composition, however, resolution at species level is limited, and amplification bias cannot be unequivocally ruled out.71 72 The applied Probio_Uni and Probio_Rev primers were validated and, compared with other primers, they seemed to represent relevant members of the intestinal microbiota such as bifidobacteria more accurately, which makes them especially fit for analysing the intestinal microbiota of infants.23 For future research, whole genome shotgun sequencing could be used to increase the accuracy of species and strain detection.73 74 Another limitation may be that environmental factors and maternal-infant interactions could have been confounders, because AB+infants were admitted to neonatal wards, whereas AB- infants stayed with their mothers on the maternity ward and were discharged earlier. Last, we did not have sufficient data on perinatal AB exposure and were therefore unable to correct for it, although it is questionable to what extent this confounder is important to take into account.75 76 Additionally, the study was not primarily designed (and thus underpowered) to conclude on AB types. Nevertheless, our results suggest that ABAMX induced less perturbations as it did not result in any differences from AB- (online supplemental figure 3B). The addition of the β-lactamase inhibitor clavulanic acid (ABAMC) was associated with higher levels of bifidobacteria compared with other AB types, which supports an earlier finding in a single subject.20 Dedicated studies are, however, needed to further elucidate the optimal regime with the least microbial perturbations.
In conclusion, AB exposure in the first week of life in term born infants disturbed the microbiota up to 1 year, with more significant deviations after longer AB exposure (5–7 days). Both C-section delivery and AB administration in the first week of life are associated with deviant intestinal microbiota, but the two combined are not associated with further deviation. Breastfeeding was associated with reduced severity and duration of perturbations compared with formula feeding. Our observations may help to elucidate why AB-exposed infants have more health problems. It may also support the development of preventive and curative strategies after AB exposure to stimulate the growth of beneficial microbiota in order to prevent future health problems.
Acknowledgments
First of all, we want to thank the mothers and infants who participated in the INCA cohort for their time and effort. Second, we would like to acknowledge the help of all the hospital staff in the recruitment and collection of data, and especially Carin Bunkers and Nicole Rutten for their support. Last, we thank Marta Magnifesta at GenProbio srl (Parma, Italy) for performing the DNA extraction and sequencing of the faecal samples.
Footnotes
RMvE and JK contributed equally.
Contributors: JK and RMvE contributed equally to this paper. EVD conceptualized the research questions, wrote the manuscript, performed the analyses and data interpretation, wrote and revised the manuscript. KK provided data and revised the manuscript. AMV conceptualised the original cohort study and provided a clinical outlook for data interpretation and writing and revised the manuscript. GH conceptualized the research questions, provided support for the data analysis and revised the manuscript. CM performed data analysis and revised the manuscript. MV performed data analysis and revised the manuscript. CB acquired funding and revised the manuscript. HS supervised the research activity and revised the manuscript. RMvE conceptualised the original cohort study and provided a clinical outlook for data interpretation and writing and revised the manuscript. JK is guarantor, supervised the research activity and revised the manuscript.
Funding: This work was supported by JPI HDHL in conjunction with ZonMW and Danone Nutricia Research and grant number IM2015.
Competing interests: This work and the PhD research by EVD was financed by a EU Joint Programming Initiative namely A Healthy Diet for a Healthy Life (JPIHDHL, http://www.healthydietforhealthylife.eu/) inconjunction with ZonMW and Danone Nutricia Research. GH is a full full-time employee of Chr Hansen A/S since January 2020. CB received a grant financed by JPI HDHL inconjunction with ZonMW and Danone Nutricia Research. RMvE was an employee at Danone Nutricia Research till 2020. JK is a full-time employee of Danone Nutricia.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. The data are available on request with the corresponding author.
Ethics statements
Patient consent for publication
Consent obtained from parent(s)/guardian(s)
Ethics approval
The INCA study was approved by the ethical board of the St. Antonius Hospital in Nieuwegein, the Netherlands (registered as NCT02536560), and informed consent was obtained from both parents. Participants gave informed consent to participate in the study before taking part.
References
- 1. Wopereis H, Oozeer R, Knipping K, et al. The first thousand days - intestinal microbiology of early life: establishing a symbiosis. Pediatr Allergy Immunol 2014;25:428–38. 10.1111/pai.12232 [DOI] [PubMed] [Google Scholar]
- 2. Robertson RC, Manges AR, Finlay BB, et al. The Human Microbiome and Child Growth - First 1000 Days and Beyond. Trends Microbiol 2019;27:131–47. 10.1016/j.tim.2018.09.008 [DOI] [PubMed] [Google Scholar]
- 3. Van Daele E, Knol J, Belzer C. Microbial transmission from mother to child: improving infant intestinal microbiota development by identifying the obstacles. Crit Rev Microbiol 2019;45:613–48. 10.1080/1040841X.2019.1680601 [DOI] [PubMed] [Google Scholar]
- 4. Gensollen T, Iyer SS, Kasper DL, et al. How colonization by microbiota in early life shapes the immune system. Science 2016;352:539–44. 10.1126/science.aad9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kamphorst K, Oosterloo BC, Vlieger AM, et al. Antibiotic treatment in the first week of life impacts the growth trajectory in the first year of life in term infants. J Pediatr Gastroenterol Nutr 2019;69:131–6. 10.1097/MPG.0000000000002360 [DOI] [PubMed] [Google Scholar]
- 6. Uzan-Yulzari A, Turta O, Belogolovski A, et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun 2021;12:443. 10.1038/s41467-020-20495-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tamburini S, Shen N, Wu HC, et al. The microbiome in early life: implications for health outcomes. Nat Med 2016;22:713–22. 10.1038/nm.4142 [DOI] [PubMed] [Google Scholar]
- 8. Korpela K, Salonen A, Virta LJ, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 2016;7:10410. 10.1038/ncomms10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dierikx TH, Visser DH, Benninga MA, et al. The influence of prenatal and intrapartum antibiotics on intestinal microbiota colonisation in infants: a systematic review. J Infect 2020;81:190-204. 10.1016/j.jinf.2020.05.002 [DOI] [PubMed] [Google Scholar]
- 10. Oosterloo BC, van Elburg RM, Rutten NB, et al. Wheezing and infantile colic are associated with neonatal antibiotic treatment. Pediatr Allergy Immunol 2018;29:151–8. 10.1111/pai.12857 [DOI] [PubMed] [Google Scholar]
- 11. Kamphorst K, Van Daele E, Vlieger AM, et al. Early life antibiotics and childhood gastrointestinal disorders: a systematic review. BMJ Paediatr Open 2021;5:e001028. 10.1136/bmjpo-2021-001028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niu X, Daniel S, Kumar D, et al. Transient neonatal antibiotic exposure increases susceptibility to late-onset sepsis driven by microbiota-dependent suppression of type 3 innate lymphoid cells. Sci Rep 2020;10:12974. 10.1038/s41598-020-69797-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Versporten A, Sharland M, Bielicki J, et al. The antibiotic resistance and prescribing in European children project: a neonatal and pediatric antimicrobial web-based point prevalence survey in 73 hospitals worldwide. Pediatr Infect Dis J 2013;32:e242–53. 10.1097/INF.0b013e318286c612 [DOI] [PubMed] [Google Scholar]
- 14. Mukhopadhyay S, Eichenwald EC, Puopolo KM. Neonatal early-onset sepsis evaluations among well-appearing infants: projected impact of changes in CDC GBS guidelines. J Perinatol 2013;33:198–205. 10.1038/jp.2012.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fjalstad JW, Stensvold HJ, Bergseng H, et al. Early-Onset sepsis and antibiotic exposure in term infants. Pediatr Infect Dis J 2016;35:1–6. 10.1097/INF.0000000000000906 [DOI] [PubMed] [Google Scholar]
- 16. Fjalstad JW, Esaiassen E, Juvet LK, et al. Antibiotic therapy in neonates and impact on gut microbiota and antibiotic resistance development: a systematic review. J Antimicrob Chemother 2018;73:569–80. 10.1093/jac/dkx426 [DOI] [PubMed] [Google Scholar]
- 17. Rooney AM, Timberlake K, Brown KA, et al. Each additional day of antibiotics is associated with lower gut anaerobes in neonatal intensive care unit patients. Clin Infect Dis 2020;70:2553–60. 10.1093/cid/ciz698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bennet R, Eriksson M, Nord CE. The fecal microflora of 1-3-month-old infants during treatment with eight oral antibiotics. Infection 2002;30:158-60. 10.1007/s15010-002-2140-z [DOI] [PubMed] [Google Scholar]
- 19. Parm U, Metsvaht T, Sepp E, et al. Impact of empiric antibiotic regimen on bowel colonization in neonates with suspected early onset sepsis. Eur J Clin Microbiol Infect Dis 2010;29:807–16. 10.1007/s10096-010-0931-1 [DOI] [PubMed] [Google Scholar]
- 20. Korpela K, Salonen A, Saxen H. Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatr Res 2020:1–6. 10.1038/s41390-020-0761-5 [DOI] [PubMed] [Google Scholar]
- 21. van den Anker J, Allegaert K. Rational use of antibiotics in neonates: still in search of tailored tools. Healthcare 2019;7:28. 10.3390/healthcare7010028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rutten NBMM, Rijkers GT, Meijssen CB, et al. Intestinal microbiota composition after antibiotic treatment in early life: the IncA study. BMC Pediatr 2015;15:204. 10.1186/s12887-015-0519-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Milani C, Hevia A, Foroni E, et al. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 2013;8:e68739. 10.1371/journal.pone.0068739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poncheewin W, Hermes GDA, van Dam JCJ, et al. NG-Tax 2.0: a semantic framework for high-throughput amplicon analysis. Front Genet 2019;10:1366. 10.3389/fgene.2019.01366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yilmaz P, Parfrey LW, Yarza P, et al. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res 2014;42:D643–8. 10.1093/nar/gkt1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramiro-Garcia J, Hermes GDA, Giatsis C, et al. NG-Tax, a highly accurate and validated pipeline for analysis of 16S rRNA amplicons from complex biomes. F1000Res 2016;5:5. 10.12688/f1000research.9227.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. R Core Team . R: a language and environment for statistical computing, 2019. Available: https://www.r-project.org/
- 28. Yoshida K. tableone: Create ‘Table 1’ to Describe Baseline Characteristics. R package version 0.11.1, 2020. Available: https://cran.r-project.org/package=tableone
- 29. Bivand R. classInt: choose univariate class intervals, 2020. Available: https://cran.r-project.org/package=classInt [Accessed 1 Jul 2021].
- 30. Kembel SW, Cowan PD, Helmus MR, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics 2010;26:1463–4. 10.1093/bioinformatics/btq166 [DOI] [PubMed] [Google Scholar]
- 31. Lahti L, Shetty SA. Microbiome R package, 2019. Available: http://microbiome.github.io
- 32. Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conserv 1992;61:1–10. 10.1016/0006-3207(92)91201-3 [DOI] [Google Scholar]
- 33. Cribari-Neto F, Zeileis A. Beta Regression in R. J Stat Softw 2010;34:1–24. 10.18637/jss.v034.i02 [DOI] [Google Scholar]
- 34. Lozupone CA, Hamady M, Kelley ST, et al. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol 2007;73:1576–85. 10.1128/AEM.01996-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71:8228–35. 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Van den Brink PJ, Braak CJFT. Principal response curves: analysis of time-dependent multivariate responses of biological community to stress. Environ Toxicol Chem 1999;18:138–48. 10.1002/etc.5620180207 [DOI] [Google Scholar]
- 37. Paliy O, Shankar V. Application of multivariate statistical techniques in microbial ecology. Wiley/Blackwell (10.1111), 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fuentes S, van Nood E, Tims S, et al. Reset of a critically disturbed microbial ecosystem: faecal transplant in recurrent Clostridium difficile infection. Isme J 2014;8:1621–33. 10.1038/ismej.2014.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choudhury R, Middelkoop A, Boekhorst J, et al. Early life feeding accelerates gut microbiome maturation and suppresses acute post-weaning stress in piglets. Environ Microbiol 2021;23:7201–13. 10.1111/1462-2920.15791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Oksanen J, Blanchet FG, Friendly M. Package ‘vegan’ Title Community Ecology Package Version 2.5-6, 2019. [Google Scholar]
- 41. Tapiainen T, Koivusaari P, Brinkac L, et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci Rep 2019;9:10635. 10.1038/s41598-019-46964-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Korpela K, de Vos WM. Antibiotic use in childhood alters the gut microbiota and predisposes to overweight. Microb Cell 2016;3:296–8. 10.15698/mic2016.07.514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tanaka S, Kobayashi T, Songjinda P, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol 2009;56:80–7. 10.1111/j.1574-695X.2009.00553.x [DOI] [PubMed] [Google Scholar]
- 44. Navarro-Tapia E, Sebastiani G, Sailer S, et al. Probiotic supplementation during the perinatal and infant period: effects on gut dysbiosis and disease. Nutrients 2020;12:1–42. 10.3390/nu12082243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zwittink RD, Renes IB, van Lingen RA, et al. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur J Clin Microbiol Infect Dis 2018;37:475–83. 10.1007/s10096-018-3193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lim HJ, Shin HS. Antimicrobial and Immunomodulatory Effects of Bifidobacterium Strains: A Review. J Microbiol Biotechnol 2020;30:1793–800. 10.4014/jmb.2007.07046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 2010;8:26–38. 10.1038/nrmicro2265 [DOI] [PubMed] [Google Scholar]
- 48. Vatanen T, Kostic AD, d'Hennezel E, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016;165:842–53. 10.1016/j.cell.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zeevenhooven J, Browne PD, L'Hoir MP, L’Hoir MP, et al. Infant colic: mechanisms and management. Nat Rev Gastroenterol Hepatol 2018;15:479–96. 10.1038/s41575-018-0008-7 [DOI] [PubMed] [Google Scholar]
- 50. Zimmermann P, Messina N, Mohn WW, et al. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: A systematic review. J Allergy Clin Immunol 2019;143:467–85. 10.1016/j.jaci.2018.09.025 [DOI] [PubMed] [Google Scholar]
- 51. de Weerth C, Fuentes S, Puylaert P, et al. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics 2013;131:e550–8. 10.1542/peds.2012-1449 [DOI] [PubMed] [Google Scholar]
- 52. Pittayanon R, Lau JT, Yuan Y, et al. Gut microbiota in patients with irritable bowel syndrome-a systematic review. Gastroenterology 2019;157:97–108. 10.1053/j.gastro.2019.03.049 [DOI] [PubMed] [Google Scholar]
- 53. Ramasethu J, Kawakita T. Antibiotic stewardship in perinatal and neonatal care. Semin Fetal Neonatal Med 2017;22:278-283. 10.1016/j.siny.2017.07.001 [DOI] [PubMed] [Google Scholar]
- 54. Araujo da Silva AR, Albernaz de Almeida Dias DC, Marques AF, et al. Role of antimicrobial stewardship programmes in children: a systematic review. J Hosp Infect 2018;99:117–23. 10.1016/j.jhin.2017.08.003 [DOI] [PubMed] [Google Scholar]
- 55. Ibrahim NA, Makmor Bakry M, Ishak S, et al. A review of antibiotic used in suspected early-onset neonatal sepsis from Malaysian perspective: which ones to choose and how long to give? Asian J Pharm Clin Res 2019;12:529. 10.22159/ajpcr.2019.v12i1.29489 [DOI] [Google Scholar]
- 56. Wagstaff JS, Durrant RJ, Newman MG, et al. Antibiotic treatment of suspected and confirmed neonatal sepsis within 28 days of birth: a retrospective analysis. Front Pharmacol 2019;10:1191. 10.3389/fphar.2019.01191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Thaulow CM, Berild D, Blix HS, et al. Can we optimize antibiotic use in Norwegian neonates? A prospective comparison between a university hospital and a district hospital. Front Pediatr 2019;7:440. 10.3389/fped.2019.00440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kamphorst K, Vlieger AM, Oosterloo BC, et al. Higher risk of allergies at 4-6 years of age after systemic antibiotics in the first week of life. Allergy 2021;76:2599–602. 10.1111/all.14829 [DOI] [PubMed] [Google Scholar]
- 59. Schrag SJ, Farley MM, Petit S, et al. Epidemiology of invasive early-onset neonatal sepsis, 2005 to 2014. Pediatrics 2016;138. 10.1542/peds.2016-2013 [DOI] [PubMed] [Google Scholar]
- 60. Braye K, Foureur M, de Waal K, et al. Epidemiology of neonatal early-onset sepsis in a geographically diverse Australian health district 2006-2016. PLoS One 2019;14:e0214298. 10.1371/journal.pone.0214298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Benitz WE, Achten NB. Finding a role for the neonatal early-onset sepsis risk calculator. EClinicalMedicine 2020;19:100255. 10.1016/j.eclinm.2019.100255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sharma D, Farahbakhsh N, Shastri S. Biomarkers for diagnosis of neonatal sepsis: a literature review;31:1646–1659, 2017. Available: https://doi.org/101080/1476705820171322060 [DOI] [PubMed]
- 63. Kerste M, Corver J, Sonnevelt MC. Application of sepsis calculator in newborns with suspected infection. 29:3860–3865, 2016. Available: https://doi.org/103109/1476705820161149563 [DOI] [PubMed]
- 64. van Herk W, Stocker M, van Rossum AMC. Recognising early onset neonatal sepsis: an essential step in appropriate antimicrobial use. J Infect 2016;72 Suppl:S77–82. 10.1016/j.jinf.2016.04.026 [DOI] [PubMed] [Google Scholar]
- 65. Goel N, Shrestha S, Smith R, et al. Screening for early onset neonatal sepsis: NICE guidance-based practice versus projected application of the Kaiser Permanente sepsis risk calculator in the UK population. Arch Dis Child Fetal Neonatal Ed 2020;105:118–22. 10.1136/archdischild-2018-316777 [DOI] [PubMed] [Google Scholar]
- 66. Sohn K, Underwood MA. Prenatal and postnatal administration of prebiotics and probiotics. Semin Fetal Neonatal Med 2017;22:284–9. 10.1016/j.siny.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015;17:690–703. 10.1016/j.chom.2015.04.004 [DOI] [PubMed] [Google Scholar]
- 68. Shao Y, Forster SC, Tsaliki E, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 2019;574:117–21. 10.1038/s41586-019-1560-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Martin R, Makino H, Cetinyurek Yavuz A, et al. Early-Life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 2016;11:e0158498. 10.1371/journal.pone.0158498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Le Doare K, Holder B, Bassett A, et al. Mother's milk: a purposeful contribution to the development of the infant microbiota and immunity. Front Immunol 2018;9:361. 10.3389/fimmu.2018.00361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rintala A, Pietilä S, Munukka E, et al. Gut microbiota analysis results are highly dependent on the 16S rRNA gene target region, whereas the impact of DNA extraction is minor. J Biomol Tech 2017;28:19. 10.7171/jbt.17-2801-003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chen Z, Hui PC, Hui M, et al. Impact of preservation method and 16S rRNA hypervariable region on gut microbiota profiling. mSystems 2019;4. 10.1128/mSystems.00271-18. [Epub ahead of print: 26 02 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ranjan R, Rani A, Metwally A, et al. Analysis of the microbiome: advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun 2016;469:967–77. 10.1016/j.bbrc.2015.12.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jovel J, Patterson J, Wang W, et al. Characterization of the gut microbiome using 16S or shotgun Metagenomics. Front Microbiol 2016;7:459. 10.3389/fmicb.2016.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dhudasia MB, Spergel JM, Puopolo KM, et al. Intrapartum group B streptococcal prophylaxis and childhood allergic disorders. Pediatrics 2021;147. 10.1542/peds.2020-012187. [Epub ahead of print: 08 04 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dierikx T, Berkhout D, Eck A, et al. Influence of timing of maternal antibiotic administration during caesarean section on infant microbial colonisation: a randomised controlled trial. Gut 2021. 10.1136/gutjnl-2021-324767. [Epub ahead of print: 21 Nov 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hoftiezer L, Hof MHP, Dijs-Elsinga J, et al. From population reference to national standard: new and improved birthweight charts. Am J Obstet Gynecol 2019;220:383.e1-383.e17. 10.1016/j.ajog.2018.12.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
fetalneonatal-2021-322861supp001.pdf (1MB, pdf)
Data Availability Statement
Data are available on reasonable request. The data are available on request with the corresponding author.