Abstract
A new, inexpensive method is described that enables lymphocytes to be enumerated very precisely. Normal leukocytes were simultaneously stained and fixed with a propidium iodide-paraformaldehyde solution. The preparation obtained (CellBeads) was used as an internal standard for cell enumeration by flow cytometry and was stable at 4°C for at least 60 days. Unlike synthetic beads, the CellBeads behaved similarly to normal cells during red blood cell lysis and cell washing procedures. When known numbers of CellBeads were added to whole-blood samples and the numbers of CellBeads and lymphocytes were determined, highly reproducible and accurate enumerations were obtained—far more so than when synthetic beads were used. This inexpensive method is suitable for routine use.
There is often a requirement for cells of a specific type to be accurately enumerated in absolute numbers per unit volume. For example, enumeration of CD4+ T lymphocytes is essential in the evaluation of prognosis and therapy in patients infected with human immunodeficiency virus (HIV) (9), and enumeration of CD34+ stem cells is important in the assessment of cancer patients receiving stem cell transplantation (3).
There are currently two ways of enumerating such cells. The dual-platform method calculates absolute cell numbers from the relative frequency of phenotypes (derived from a flow cytometer) and the total white blood cell (WBC) count (derived from a hematology analyzer). Unfortunately, this approach leads to great interlaboratory variation in estimation of CD4+ lymphocyte counts (3) because of errors inherent in WBC enumeration by hematology analyzers.
The single-platform method does not involve a hematology analyzer, using only a flow cytometer. It is more precise as it relies on either concomitant precise measurement of the fluid volume in which such suspended cells are enumerated or the precise addition of fluorescent calibration particles to the sample. While a new generation of flow cytometers with precision fluidics may represent the long-term solution to problems of interlaboratory variation, most laboratories still rely on flow cytometers that lack the ability to measure fluid volumes precisely.
Manufacturers of flow cytometers market systems of beads, protocols, and software that purport to provide precise enumeration, e.g., the PROCOUNT system from Becton Dickinson (2) or Flow-Count Fluorospheres from Beckman Coulter. However, assays using such commercially available beads are expensive (around an additional $6/test for the TruCount system or an additional $1.6/test for Fluorospheres), and this may be beyond the means of some laboratories. Other commercially available fluorescent beads are designed primarily for calibration of flow cytometers rather than enumeration of cells in individual samples. These beads, usually plastic, are not suitable for the latter purpose. Many experiments performed by us (results not shown) daily over the course of 3 months, produced data with very poor reproducibility, large variation, and major inaccuracy, with calculated lymphocyte counts much lower than those obtained by conventional methods. This inaccuracy is attributed to the beads segregating differentially from cells during the various staining and washing procedures involved in preparing the cells for analysis. Counting particles or beads requires addition at the start of such an analysis in order to be able to compensate for cell losses from the sample during sample handling.
The present study demonstrates that leukocytes from healthy volunteers stained with the fluorescent dye propidium iodide (PI) are stable after fixation and suitable for use as a calibrant as described above for enumeration of absolute numbers of cells by flow cytometry.
MATERIALS AND METHODS
PBS solution.
Twenty phosphate-buffered saline (PBS) tablets (Oxoid Ltd., Basingstoke, United Kingdom) were dissolved in water (2 liters). This gives sodium chloride (0.16 M), potassium chloride (0.003 M), sodium dihydrogen phosphate (0.008 M), and potassium dihydrogen phosphate (0.001 M).
Lysing solution (stock solution).
Ammonium chloride (40.1 g), sodium bicarbonate (4.2 g), and EDTA disodium salt (1.85 g) were dissolved in water (500 ml). The stock solution was stored at 4°C for not more than 6 months. Working solution was prepared daily by a 10-fold dilution in water.
One-percent PFA solution in PBS.
Paraformaldehyde (PFA; 1 g) was added to distilled water (90 ml) and heated in a water bath in a fume cupboard at 75°C for 3 h, with occasional stirring. When cool, 10 ml of concentrated PBS solution (one PBS tablet dissolved in 10 ml of water) was added.
Dye solution.
PI stock solution was prepared by dissolving PI (20 mg) in PBS (20 ml) and storing at 4°C, protected from light.
Staining-fixation solution.
Addition of 1% Tween 20 solution (0.2 ml) to 1% paraformaldehyde solution (20 ml) gave a 0.01% final concentration of Tween. The addition of PI stock solution (2 ml) gave a final PI concentration of 100 μg/ml.
Preparation of CellBeads.
Whole blood (16 ml) was collected from a healthy volunteer into four 5-ml Vacutainers containing EDTA anticoagulant. Aliquots (1 ml) were added to four sterile 25-ml plastic screw-cap tubes (Sarstedt Ltd., Leicester, United Kingdom) each containing 20 ml of lysing solution. The contents were mixed and then left for 10 min at room temperature. Following centrifugation at 300 × g for 5 min, the supernatants were discarded. The peripheral blood (PB) mononuclear cell pellets were then resuspended and combined into one tube, to which further PBS (15 ml) was added. This process was repeated a further three times. All the PB mononuclear cell aliquots were then pooled before being redivided into four tubes. These were again centrifuged at 300 × g for 5 min, and the supernatants were discarded. The cells in each tube were resuspended in freshly diluted PI dye-fixation solution (5 ml) and left at 4°C overnight. They were then centrifuged at 300 × g for 5 min, the supernatant was discarded, and the cells were resuspended in PBS (5 ml).
All cells were then combined, and an aliquot was removed after vortexing for counting in duplicate in a Neubauer-ruled, dual-chamber hemocytometer. The cells were then separated by centrifugation at 300 × g for 5 min and then resuspended in the calculated volume of 1% paraformaldehyde in PBS to give a count of approximately 106 particles/ml. The CellBeads were stored at 4°C, and daily counts of 10 aliquots were made. The mean count and coefficient of variation (CV) of each set of 10 aliquot values were found to be stable from 72 h onwards.
Stability of CellBeads.
Using five dual-chamber hemocytometers, counts of 10 aliquots of CellBeads were made on each day that patient samples were enumerated.
Enumeration of cells.
For all samples in which cells were enumerated, the numbers of CD3+, CD3+ CD4+, and CD3+ CD8+ cells were calculated using both the dual-platform method (using the flow cytometric differential and the hematology analyzer's WBC count) and the single-platform method (involving the addition of known numbers of CellBeads to the samples).
The total WBC count was obtained using an Advia (Bayer) hematology analyzer. The flow cytometer used was the FACScan (Becton Dickinson) equipped with a 15-mW argon ion laser tuned to 488 nm. The FACScan has three fluorescence detection pathways whose photomultiplier tubes detect FL1 (530 ± 30 nm, optimized for fluorescein isothiocyanate [FITC]), FL2 (585 ± 42 nm, optimized for phycoerythrin [PE]), and FL3 (>650 nm, optimized for PE–carbocyanine 5 [PE-Cy5]). Lysys II software (Becton Dickinson) was used for data acquisition, and analysis and enumeration were performed using FlowMate (Dako, Ely, United Kingdom) and ExCel (Microsoft) programs.
Dual-platform enumeration.
CD3+ T cells and the CD3+ CD4+ and CD3+ CD8+ subsets were enumerated. For each blood sample, 200-μl aliquots of whole blood were stained with the appropriate dual monoclonal antibody combinations (DUAL-TAG; Sigma Aldrich, Poole, United Kingdom)—CD45-FITC and CD14-PE, CD3-FITC and CD4-PE, and CD3-FITC and CD8-PE—by incubating in the dark for 15 min at room temperature. Then, 2 ml of FACS lysing solution (Becton Dickinson) was added, and the sample was vortexed and incubated for 10 min at room temperature. The sample was then centrifuged at 300 × g for 5 min, washed in 2 ml of PBS, centrifuged again at 300 × g for 5 min, and resuspended in 0.5 ml of 1% paraformaldehyde in PBS. Samples were then stored at 2 to 8°C in the dark for not more than 24 h before analysis. Lymphocytes are identified by low forward and low side scatter (SSC) with positivity to CD45 and negativity to CD14 as described by Nicholson (6).
Single-platform enumeration using CellBeads.
Two 100-μl aliquots of PB were stained with either CD3-FITC–CD4-PE–CD45-PE-Cy5 or CD3-FITC–CD8-PE–CD45-PE-Cy5 triple-color monoclonal antibody combinations (Dako) and incubated in the dark for 15 min at room temperature. Then, 2 ml of FACS lysing solution (Becton Dickinson) was added, the sample was vortexed, 100 μl of CellBead suspension was added, the contents were mixed well with the pipette tip and revortexed, and the sample was incubated for 10 min at room temperature. The sample was then centrifuged at 300 × g for 5 min, washed in 2 ml of PBS, centrifuged at 300 × g for 5 min, and resuspended in 0.5 ml of 1% paraformaldehyde in PBS. Samples were then stored at 2 to 8°C in the dark for not more than 24 h before enumeration by flow cytometry. Lymphocytes were identified by their CD45 and SSC characteristics on the SSC-FL3 dot plot. Separate gates were set around lymphocytes and CellBeads on this dot plot, and the ratio of lymphocytes to CellBeads was determined from the number of events in each gate. This allowed the absolute number of lymphocytes to be calculated, based on the volume and concentration of the CellBead suspension added initially. Selection of the gated lymphocyte population and display of CD3+ CD4+ or CD3+ CD8+ (FL1-FL2) allowed analogous calculation of the absolute CD4+ or CD8+ counts.
The precision of this procedure was assessed by taking separate duplicate aliquots from 20 patient samples throughout the entire procedure.
Samples enumerated.
The enumerated samples comprised samples from healthy volunteers, predominantly laboratory staff, and aliquots taken from anonymous samples submitted for CD4+ T-cell enumeration from patients with HIV.
RESULTS
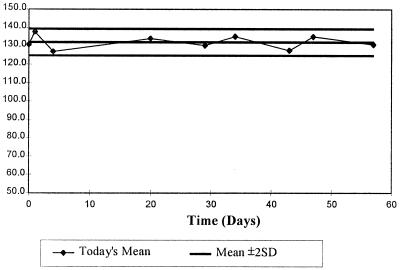
It was our experience that after 72 h, CellBead counts, performed on 10 aliquots on each day that the CellBeads were used, gave constant mean cell counts and low CVs for each lot of 10 aliquots. The quality control data for one batch of CellBeads are shown in Fig. 1.
FIG. 1.
Stability of CellBeads (PI–Tween 20 treated and fixed in 1% PFA) evaluated by replicate counts using five dual-chamber hemocytometers. The y axis shows the number of CellBeads (104/milliliter).
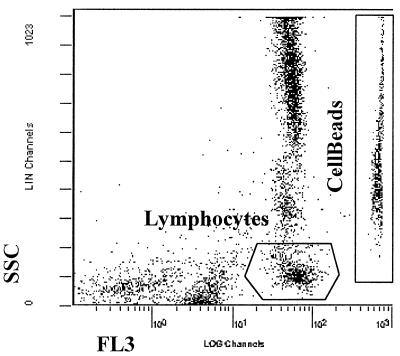
The emission spectrum of PI strongly overlaps that of PE and that of Cy5. However, the CellBeads can be clearly distinguished from the cells to be enumerated by their forward-scatter and SSC characteristics. Figure 2 shows the clear distinction achieved between patient cells stained with an anti-CD45- PE-Cy5 conjugate and CellBeads stained with PI. In spite of the wash step, some cellular debris is still apparent, but this does not interfere in enumeration of cells since it has low SSC and FL3 values.
FIG. 2.
Bivariant dot plot showing SSC and FL3 profiles for CellBeads stained with PI and patient leukocytes stained with anti-CD45-PE-Cy5.
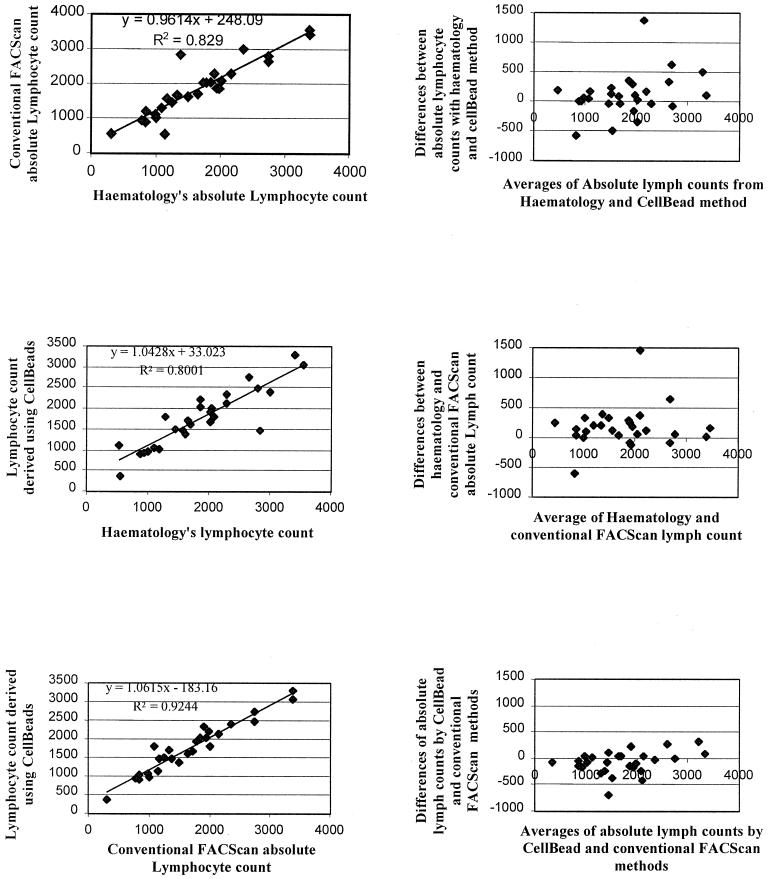
The left-hand panels of Fig. 3 show the correlations obtained between individual lymphocyte counts determined by three methods. The methods used were (i) the single-platform method using CellBeads to enumerate lymphocytes characterized by CD45-SSC characteristics, (ii) the dual-platform method (the first using the total WBC count from a hematology analyzer and the WBC differential from the flow cytometer), and (iii) the total lymphocyte count obtained from the hematology analyzer alone. It is apparent that the best agreement is between the CellBead procedure and the FACS differential procedure (coefficient of correlation, 0.9615; slope of best linear fit, 1.062).
FIG. 3.
Comparison of enumeration of absolute lymphocyte counts by three methods: lymphocyte count derived using CellBeads (single-platform flow cytometer calibrated by added CellBeads), hematology's lymphocyte count (hematology analyzer absolute lymphocyte count), and conventional FACScan absolute lymphocyte count (hematology analyzer total WBC and FACScan differential lymphocyte count: dual-platform method). Each pair of panels is compared by correlation and Altman-Bland plot.
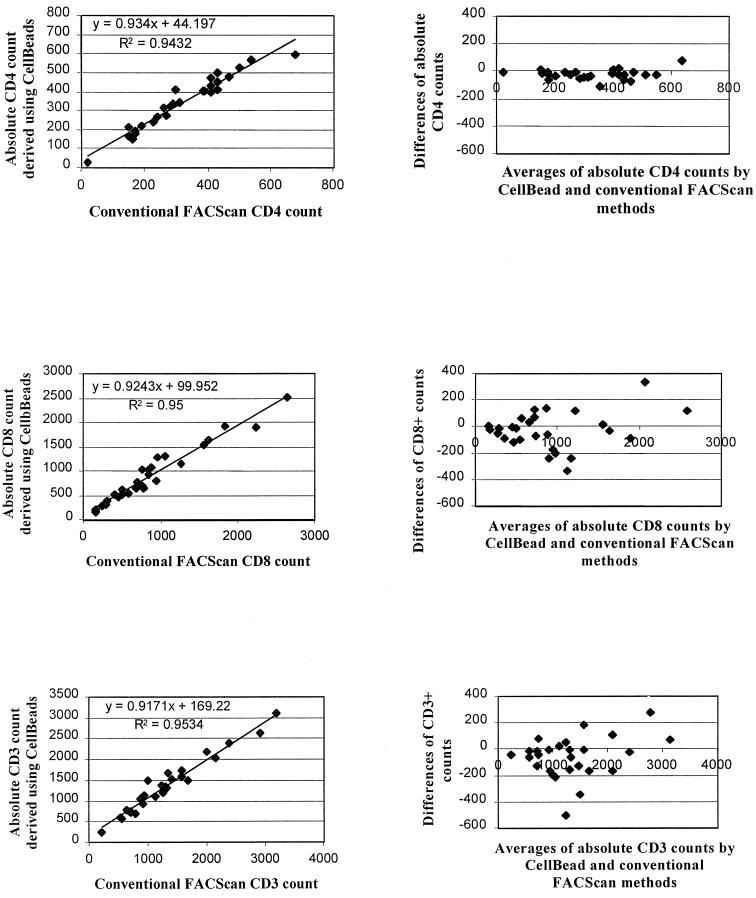
The associated Altman and Bland plots, shown in the corresponding right-hand three panels of Fig. 3, confirm satisfactory agreement between the two sets of estimates, with little indication of a tendency to increased bias at low or high lymphocyte count. Fig. 4 presents similar data for comparison of CD3+, CD3+ CD4+ and CD3+ CD8+ numbers derived by the single-platform CellBead procedure and the dual-platform method involving the hematology analyzer's total WBC and the flow cytometer's differential WBC.
FIG. 4.
Comparison of absolute CD4+, CD8+, and CD3+ counts obtained using the dual-platform procedure with a hematology WBC and flow cytometer differential versus a single-platform procedure with CellBead calibration. Regressions and Bland-Altman plots are shown.
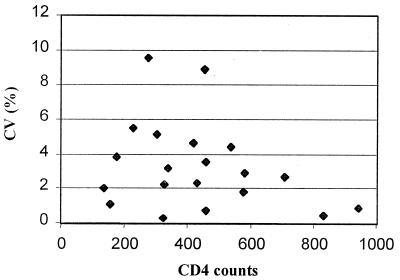
Figure 5 shows the precision profile (plot of coefficient of variance versus mean concentration) for 20 duplicate samples processed by the single-platform procedure using CellBeads. The results demonstrate that the precision averaged (root mean square) just over 4% for CD4+ determination and was significantly better than this (2.54%; n = 6) at normal CD4+ T-cell levels (>500 million/liter).
FIG. 5.
Precision profile for absolute CD3+ CD4+ lymphocyte counts evaluated by the CellBead technique.
DISCUSSION
In developing countries where HIV infection is increasing rapidly in prevalence, the reagent costs for a routine CD4 cell count are considered by some to be prohibitive (8). Major additional costs to enable a single-platform absolute CD4 cell count to be made are thus impractical. Under these circumstances, a simple economical way of obtaining accurate CD4+ cell counts might be helpful. This method describes the preparation and use of CellBeads prepared from human cells, whose behavior closely matches that of the cells in the sample from the patient.
The CellBeads were produced from human PB leucocytes that were stained with PI and fixed in paraformaldehyde solution. Our data showed that the CellBeads were stable at 4°C for at least 2 months, as indicated by the stability of the counts over time and the low CV when 10 aliquots were counted. Their very bright fluorescence clearly distinguished them from cells in the samples stained with anti-CD4 monoclonal antibody-fluorescent dye conjugates. This clear distinction lasted throughout the 3 months for which each batch was in use (data not shown). When used for enumeration, the CellBeads produced results for lymphocyte counts (based on CD45-SSC gating) that were in excellent agreement with results obtained by our standard procedure (the combination of a whole-blood count from a hemocytometer with the differential CD45-SSC proportion from the flow cytometer). There was similarly excellent agreement between absolute numbers of CD3+, CD3+ CD4+ and CD3+ CD8+ lymphocytes between the two procedures.
It has long been known that the large variation in estimates of PB CD3+ CD4+ lymphocyte numbers is due to poor reproducibility in the lymphocyte count carried out with hematology analyzers (7). Thus, methods which avoid the use of hematology analyzers should have superior precision (3, 4) if suitably calibrated by accurate volumetric (fluidics) measurements or by the addition of precise numbers of particles as internal counting standards.
Precision in the presently described method averaged just over 4% for CD4+ cell determinations in patients with significant CD4+ T-cell lymphopenia. It was significantly better than this at normal CD4+ T-cell levels. Such precision is adequate. In summary we describe a method of counting absolute numbers of cells that is cheap, reproducible, reliable, accurate, and suitable for use in any laboratory. The CellBead preparation is one that, in our hands, behaves more like the cells that they are meant to enumerate than anything else we have tried.
REFERENCES
- 1.Barbosa I L, Sousa M E, Godinho M I, Sousa F, Carvalhais A. Single- versus dual-platform assays for human CD3+ cell enumeration. Cytometry. 1999;38:274–279. [PubMed] [Google Scholar]
- 2.Barnett D, Bird G, Hodges E, Linch D C, Matutes E, Newland A C, Reilly J T. Guidelines for the enumeration of CD4+ T lymphocytes in immunosuppressed individuals. Clin Lab Haematol. 1997;19:231–241. doi: 10.1046/j.1365-2257.1997.00091.x. [DOI] [PubMed] [Google Scholar]
- 3.Barnett D, Granger V, Whitby L, Storie I, Reilly J T. Absolute CD4+ T-lymphocyte and CD34+ stem cell counts by single-platform flow cytometry: the way forward. Br J Haematol. 1999;106:1059–1062. doi: 10.1046/j.1365-2141.1999.01632.x. [DOI] [PubMed] [Google Scholar]
- 4.Gale H B, Henry K. Measuring percent lymphocytes by flow cytometry to calculate absolute lymphocyte subset counts for HIV+ specimens. Cytometry. 1992;13:175–181. doi: 10.1002/cyto.990130211. [DOI] [PubMed] [Google Scholar]
- 5.Lopez A, Carago I, Candeias J, et al. Enumeration of CD4+ T-cells in the peripheral blood of HIV-infected patients: an interlaboratory study of the FACSCount system. Cytometry. 1999;38:231–237. doi: 10.1002/(sici)1097-0320(19991015)38:5<231::aid-cyto5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson J K A. Immunophenotyping specimens from HIV-infected persons: laboratory guidelines from the centers for disease control and prevention. Cytometry. 1994;18:55–59. doi: 10.1002/cyto.990180111. [DOI] [PubMed] [Google Scholar]
- 7.Robinson G, Morgan L, Evans M, McDermott S, Pereira S, Wansbrough-Jones M, Griffin G. Effect of type of haematology analyser on CD4 count. Lancet. 1992;340:485. doi: 10.1016/0140-6736(92)91807-k. [DOI] [PubMed] [Google Scholar]
- 8.Sherman G G, Galpin J S, Patel J M, Mendelow B V, Glencross D K. CD4+ T-cell enumeration in HIV infection with limited resources. J Immunol Methods. 1999;222:209–217. doi: 10.1016/s0022-1759(98)00172-0. [DOI] [PubMed] [Google Scholar]
- 9.Stein D S, Korvick J A, Vermund S H. CD4+ lymphocyte cell enumeration for prediction of clinical course of human immunodeficiency virus disease: a review. J Infect Dis. 1992;165:352–363. doi: 10.1093/infdis/165.2.352. [DOI] [PubMed] [Google Scholar]
- 10.Strauss K, Hannett I, Engel S, Shiba A, Ward D, Ullery S, Jinguji M G, Valinsky J, Barnett D, Orfao A, Kestens L. Performance evaluation of FACSCount: a dedicated system for clinical cellular analysis. Cytometry. 1996;26:52–59. doi: 10.1002/(SICI)1097-0320(19960315)26:1<52::AID-CYTO8>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]