Abstract
Ubiquitous eukaryotic non-coding circular RNAs regulate transcription and translation. We have reported full-length intronic circular RNAs (flicRNAs) in Entamoeba histolytica with esterified 3′ss and 5′ss. Their 5′ss GU-rich elements are essential for their biogenesis and their suggested role in transcription regulation. Here, we explored whether exonic, exonic-intronic, and intergenic circular RNAs are also part of the E. histolytica and E. invadens ncRNA RNAome and investigated their possible functions. Available RNA-Seq libraries were analyzed with the CIRI-full software in search of circular exonic RNAs (circRNAs). The robustness of the analyses was validated using synthetic decoy sequences with bona fide back splice junctions. Differentially expressed (DE) circRNAs, between the virulent HM1:IMSS and the nonvirulent Rahman E. histolytica strains, were identified, and their miRNA sponging potential was analyzed using the intaRNA software. Respectively, 188 and 605 reverse overlapped circRNAs from E. invadens and E. histolytica were identified. The sequence composition of the circRNAs was mostly exonic although different to human circRNAs in other attributes. 416 circRNAs from E. histolytica were virulent-specific and 267 were nonvirulent-specific. Out of the common circRNAs, 32 were DE between strains. Finally, we predicted that 8 of the DE circRNAs could function as sponges of the bioinformatically reported miRNAs in E. histolytica, whose functions are still unknown. Our results extend the E. histolytica RNAome and allow us to devise a hypothesis to test circRNAs/miRNAs/siRNAs interactions in determining the virulent/nonvirulent phenotypes and to explore other regulatory mechanisms during amoebic encystment.
Keywords: CIRIfull, back splice junction, reverse overlap, differential expression, EIcirRNA, flicRNA, parasite, virulence
1. Introduction
From their discovery 45 years ago [1,2], the widely conserved circular transcripts or circRNAs have been extensively studied, displaying important features such as exonuclease-resistance and ability to interact with proteins and other RNAs [3]. CircRNA molecules can be monoexonic, multi-exonic, exon-intronic, or intronic generated by co-transcriptional backsplicing, in which an upstream 3′ss acceptor is linked to a downstream 5´ss donor producing exonic circular molecules. Skipped exons within a lariat produced from linear splicing resulting may undergo post-transcriptional backsplicing originating exon-intron circRNAs (EIciRNAs) [4,5,6].
Backsplicing is also activated by splicing proteins and cis-elements. Alu repeats in flanking introns facilitate circularization by looping out of one or more exons [7,8], and CRISPR/Cas9-mediated Alu repeats gene editing prevents circularization [9]. In addition, the RNA binding proteins (RBP) FUS, and the splicing factors QKI and MBL have been associated with the circRNAs biogenesis [10,11,12]. Furthermore, intronic circRNAs are covalently closed lariat products resulting from processed intron lariats [6,13].
In keeping with their localization, circRNAs have different functions. Those with exonic sequences localize mainly to the cytoplasm and perform regulatory functions such as microRNAs and protein sponging, translation regulation, mRNA stability, and 5′ cap-independent translation [10,14,15,16,17,18]. Intronic circRNAs localize heterogeneously within the cell. In the nucleus, they are linked to gene expression regulation, while the function of cytoplasmatic intron circles remains unknown [19,20]. Some nuclear EIciRNAs are part of the RNA polymerase II (Pol II) binding complex able to downregulate their parental genes [4,21]. This is the case of the human 2′–5′ intronic circRNAs (ciRNAs), ci-ankrd52, and ci-sirt7, which directly bind to the serine 2-phosphorylated C-terminus domain (CTD) of Pol II, indicating that ciRNA-CTD interaction occurs during transcription elongation [21].
Shortly following its release from the spliceosome, a lariat becomes a circular intronic molecule after trimming off the 3´-end tail and before it is hydrolyzed by the debranching enzyme (Dbr1). However, numerous introns show cis-elements that confer them stability and accumulate inside the cell, like the stable sequence introns (sisRNAs) [19]. SisRNAs include 2′–5′ covalently linked intronic circular RNAs, which are basically processed lariats, and 3′–5′ covalently linked species with untrimmed 3´-end tails.
Entamoeba histolytica is the causative agent of amebiasis, a neglected tropical disease that still accounts for 55,500 deaths and nearly 2.4 million disability-adjusted life years [22]. Much effort has been devoted to understanding the mechanisms leading to infection establishment and development of disease and to treating such infections as well. In search of alternative theragnostic target molecules, we recently identified full-length intronic circular RNAs (flicRNAs) in the human protozoan parasite E. histolytica [23]. In agreement with previous reports [24], the 5′ss and 3′ss ends of these molecules are covalently linked with an average of 10 nucleotides between the branch point (BP) and the 3´ss in their lariat precursors [23]. We observed that flicRNAs are post-splicing products originating from the lariat by mechanisms not fully understood. In vivo splicing assays showed that the conserved GU-rich 5′ss is required for flicRNAs biogenesis. Furthermore, mutation of such elements decreased flicRNAs production and increased expression of their parental genes, indicating their role in the gene expression regulation [23]. Strikingly, although circular RNAs are expressed throughout the biota, they have been reported in a small number of protozoans [25]. Notwithstanding, other non-coding (nc) RNAs are expressed in Amoebozoa. For example, although not fully characterized, Mar-Aguilar et al. [26] confirmed the presence of microRNA-like molecules that had been predicted bioinformatically in E. histolytica [27]. miRNAs have been identified in Dictyostelium discoideum also [28]. Other ncRNAs in Entamoeba are the stress-induced self-circularized 5′-external transcribed spacer rRNAs in E. histolytica [29] and the encystation-related long ncRNAs in E. invadens [30].
To cope with protist and bacterial infections, recent works proposed that hosts’ immune response trigger circRNAs–miRNAs–mRNA regulatory networks [31,32,33]. However, little is known about whether these regulatory networks exist in E. histolytica and if they enable the onset of parasitic infections and invasive diseases. To address this question, we mined available RNA-seq libraries in search of circular exonic transcripts with miRNA sponging potential differentially expressed (DE) in HM1:IMSS (virulent) and Rahman (avirulent) E. histolytica strains. We also searched for circular transcripts in the reptile parasite E. invadens. After validation of the software using synthetic decoy sequences with bona fide back splice junctions (BSJ), we identified 188 and 605 reverse overlapped (RO) circRNAs from the E. invadens and E. histolytica libraries, respectively. The sequence composition of the identified circRNAs was mostly exonic. A total of 416 circRNAs from E. histolytica were virulent-specific and 267 were avirulent-specific. In addition, out of the common circRNAs, 32 were DE between strains, showing no correlation between intron length or the number of introns per locus with the number of circRNAs produced in a given locus. Finally, we predicted that eight of the DE circRNAs could function as sponges of the reported miRNAs in E. histolytica, whose functions are still unknown. Our results allow us to devise a working hypothesis to test the relationships between circRNAs and miRNAs in determining virulent/nonvirulent phenotypes and to explore additional regulatory mechanisms during amoebic trophozoite to cyst differentiation.
2. Results
2.1. Computational Identification of Entamoeba circRNAs
CircRNAs were identified based on backspliced RO reads as described previously [34] (Figure S1 depicts the workflow used here). To identify the circRNAs in E. histolytica and E. invadens, available RNAseq libraries [35,36] were mined using the CIRI-full package. Respectively, six E. histolytica (run access codes ERR058005–ERR058007 for the virulent HM1:IMSS strain and ERR058008–ERR058010 for the avirulent Rahman strain) and four E. invadens (run access codes GSM4108195–GSM4108198) libraries were used, and the access codes for the reference genomes were GCF_000208925 and GCA_000330505.
Despite the robustness of the CIRI-full package [34] to ensure that it could identify circRNAs derived from an AT-rich genome such as E. histolytica, we performed a prospective run in a fraction of the libraries and found a putative circRNA from locus EHI_110790. Next, we used the exon end sequences as back splice junctions and manually added random sequences from the same locus to obtain 10 circular RNA decoys with displaced backsplice junctions. These sequences were incorporated into the partial libraries, and circRNAs were re-identified. In addition to the previous results, all decoy sequences were rescued in the analysis (Figure S2).
2.2. E. histolytica circRNAs
In E. histolytica, 958 circRNAs were identified. After filtering for duplicates, 605 circRNAs remained. Of these, 433 were found in the HM1:IMSS virulent strain and 276 in the Rahman avirulent strain; 104 of these circRNAs were shared in both strains; therefore, 329 circRNAs were virulent-specific, and 172 were avirulent-specific. A total of 20 out of 605 circRNAs originated from intergenic regions (17 were from the virulent strain, 9 were from the avirulent strain, and they shared 6 circRNAs), and 585 circRNAs were associated with a particular locus (Table 1). Notably, only 35 of the 585 circRNAs were formed by two exons.
Table 1.
Summary of Entamoeba histolytica and Entamoeba invadens circRNAs.
| Entamoeba histolytica | Entamoeba invadens | |
|---|---|---|
| Total circRNAs | 958 | 188 |
| Without duplicates | 605 | 143 |
| mono elements | 570 | 143 |
| 2+ elements | 35 | 0 |
| Exonic | 585 | 131 |
| Intronic | 0 | 2 |
| Intergenic | 20 | 10 |
| Virulent-specific | 329 | nd 1 |
| Shared between virulent and avirulent |
104 | nd |
| Avirulent-specific | 172 | nd |
| Cyst-specific | nd | 40 |
1 not determined.
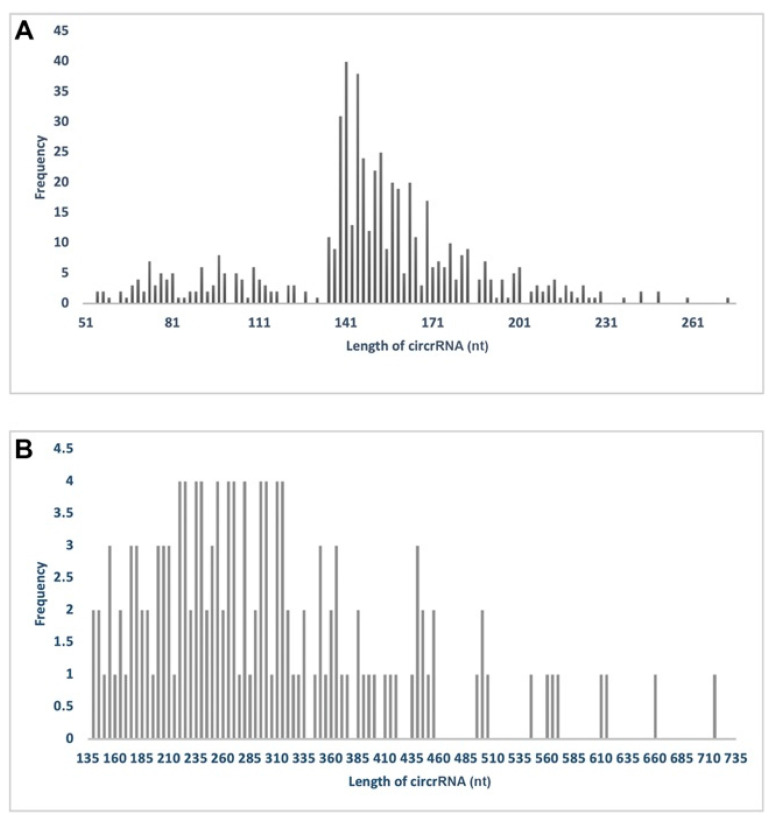
The length distribution of E. histolytica circRNAs was limited, ranging from nearly 50 to 250 nucleotides (nt), peaking at 140 nt (Figure 1A), and the expression of circRNAs was practically proportional to the expression of their respective linear RNAs (Figure S3).
Figure 1.
Length frequency distribution of Entamoeba circRNAs. The histograms show the E. histolytica (A) and E. invadens (B) circRNA length-frequency distribution.
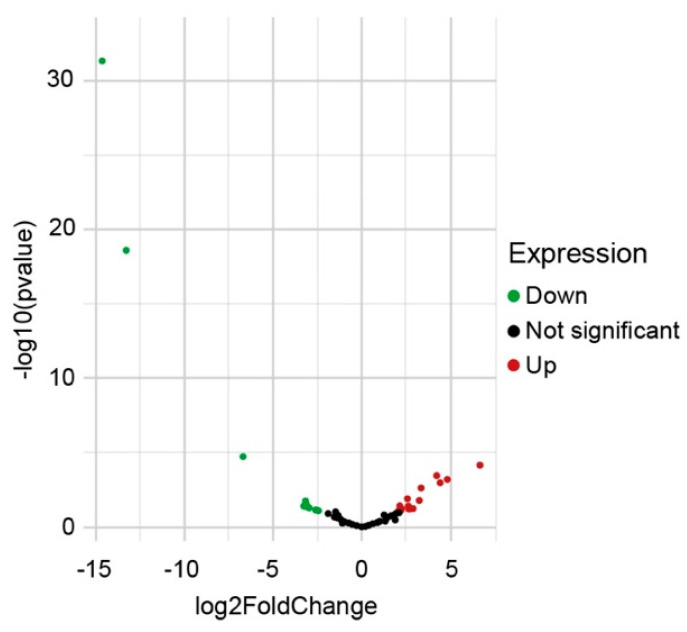
Despite the E. histolytica libraries were previously analyzed, we wanted to ensure the detection of differential expression (DE) of circRNAs between the virulent and avirulent strains. To this end, the normalized expression of circRNAs between samples was analyzed. Box plot analysis and principal components analysis (PCA) both show that the circRNAs from the three avirulent replicas have similar distribution and variation, whereas the circRNAs from one of the virulent samples are distributed slightly different and appear to have more variation than the other two samples, while separated from the avirulent circRNAs (Figure S4). The normalized expression counts were analyzed using DEseq2 library and the statistic p value and a fold change of 2 were determined. A volcano plot was constructed to evidence that the circRNAs were underexpressed (green dots) and overexpressed (red dots) in the E. histolytica virulent strain with respect to the avirulent strain (Figure 2); thus, an overexpressed circRNA in the virulent strain is underexpressed in the avirulent strain and vice-versa. Usually, circular RNA expression is lower than their linear counterparts, and for this reason, some authors prefer lower p values to assess DE [37]. However, in E. histolytica, the abundance of circRNAs expression is heterogeneous; therefore, we used two p values. Using p < 0.05, twenty DE circRNAs were identified, and twelve more were detected when a p < 0.1 was used. The plot shows the 32 DE circRNAs identified using both p values. Interestingly, three circRNAs are particularly underexpressed, all of them derived from locus EHI_169670 (a gene with three exons and two introns); one circRNA has one exon element, and the other two have two exonic elements.
Figure 2.
Differentially expressed E. histolytica circRNAs in HM1:IMSS strain with respect to Rahman strain. The volcano plot shows the DE circRNAs (p < 0.05 and p < 0.1 data are combined). 32 circRNAs were identified: 20 overexpressed (red dots) and 12 underexpressed (green dots).
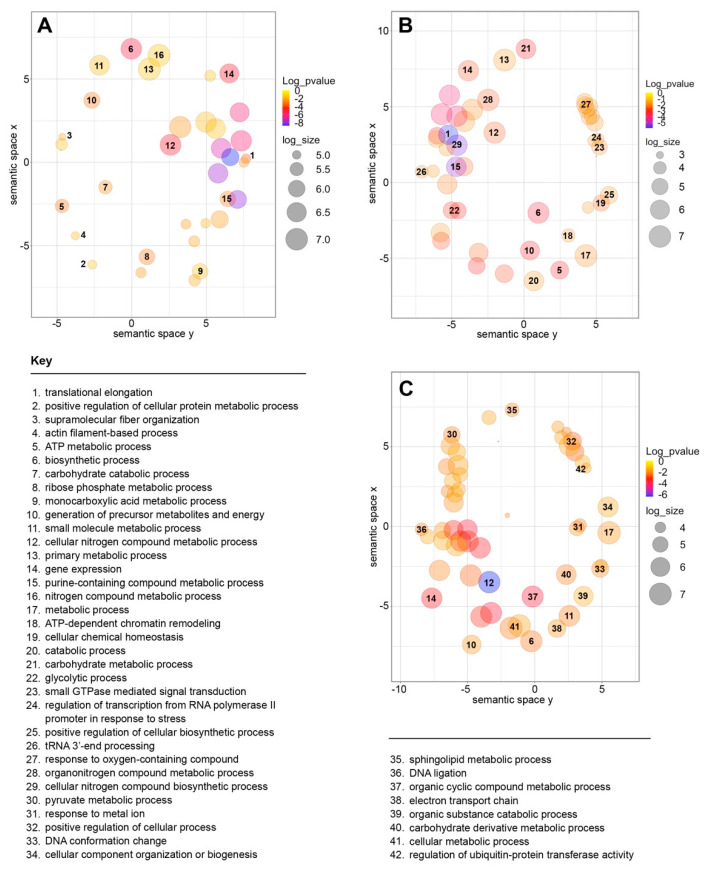
Next, we used the virulent-specific and the avirulent-specific circRNAs to perform a gene ontology (GO) analysis focusing on metabolic processes. Clearly, the metabolic processes potentially regulated by the circRNAs from virulent and avirulent E. histolytica strains substantially differ (respectively, Figure 3A,B and Tables S1 and S2). For example, 35 GO terms were best represented in the HM1:IMSS strain, but in the Rahman strain were 47. Notably, 26 of these terms were represented in both strains, but their abundance and significance were differentially distributed. For example, taking into account the dispensability of the metabolic processes only, translational elongation, ATP metabolic processes, cellular nitrogen compound metabolic process, and gene expression appear in different X coordinates between strains. Additionally, biosynthetic processes, the generation of precursor metabolites and energy, as well as purine-containing compound metabolic processes were distributed diametrically opposed. However, as expected from metabolic processes of the same species with different virulent attributes, the term primary metabolic process showed little change between strains. This set of data suggests that the DE circRNAs might be involved in gene expression regulation in the virulent and avirulent phenotypes.
Figure 3.
Metabolic process GO enrichment terms were identified in E. histolytica and E. invadens circRNAs. Semantic distribution for HM1:IMSS strain (A), Rahman strain (B), and 20 h encysting E. invadens circRNAs (C) are shown.
When we searched for circRNAs sponging potential, remarkably, eight of the twenty DE circRNAs were predicted to have one or more Ehi-miRNA-like target sites (Table 2), suggesting the possibility of circRNA-miRNA-mRNA regulatory mechanisms existing in Entamoeba. For instance, according to their prediction score, the GRIP domain RUD3 circRNAs 01_169670 and 18_169670 could potentially sponge Ehi-miR-18, Ehi-miR-136, Ehi-miR-160, Ehi-miR-50, Ehi-miR-88, and Ehi-miR-82, and Ehi-miR-17, Ehi-miR-18, Ehi-miR-160, and Ehi-miR-50, respectively.
Table 2.
Differentially expressed Entamoeba histolytica circRNAs 1 with predicted anti-Ehi-miRNA-like target sites.
| circRNA | Element | Product | C/L 2 | DE 3 | Ehi-miR-like Sites In 4 |
|---|---|---|---|---|---|
| 01_169670 | exon | GRIP domain RUD3 | ↓/↓ | ↓ | Ehi-miR-18, Ehi-miR-136, Ehi-miR-160, Ehi-miR-50, Ehi-miR-88, Ehi-miR-82 |
| 02_169670 | exon | GRIP domain RUD3 | ↓/↓ | ↓ | |
| 03_169670 | exon | GRIP domain RUD3 | ↓/↓ | ↓ | |
| 04_130700 | exon | Enolase | ↑/⎯ | ↑ | Ehi-miR-56 |
| 05_146110 | exon | hp 5 | ↑/↑ | ↑ | |
| 06_DS571557 | intergenic | nd 6 | na 6 | ↑ | Ehi-miR-193, Ehi-miR-39 |
| 07_036530 | exon | Ribosomal S27a | ↑/⎯ | ↑ | Ehi-miR-69 |
| 08_130700 | exon | Enolase | ↑/⎯ | ↑ | |
| 09_146110 | exon | hp | ↑/↑ | ↑ | |
| 10_146370 | exon | Ribosomal L27 | ↑/↑ | ↑ | |
| 11_014170 | exon | GRIP domain RUD3 | ↓/↑ | ↓ | |
| 12_026410 | intergenic | Ribosomal S20 | ↑/↓ | ↓ | |
| 13_086110 | exon | HMG domain | ↑/⎯ | ↑ | |
| 14_116360 | exon | Serine-rich | ↑/↑ | ↓ | Ehi-miR-23, Ehi-miR-51 |
| 15_193440 | exon | hp | ↑/↑ | ↑ | |
| 16_183480 | exon | Ribosomal L27 | ↓/↓ | ↓ | |
| 17_027380 | exon | hp | ↑/↑ | ↑ | |
| 18_169670 | exon | GRIP domain RUD3 | ↓/↓ | ↓ | Ehi-miR-17, Ehi-miR-18, Ehi-miR-160, Ehi-miR-50 |
| 19_125950 | exon | ADH | ↑/⎯ | ↑ | Ehi-miR-177 |
| 20_DS571344 | intergenic | nd | na | ↑ | Ehi-miR-150, Ehi-miR-7 |
1 The list includes those detected with p-value < 0.05 only; 2 C (circRNA)/L (linear RNA composed of spliced and unspliced variants) overexpressed (↑), underexpressed (↓) or unchanged (⎯); 3 DE, differential expression of the virulent HM1:IMSS strain with respect to the avirulent Rahman strain; 4 The numbers represent the Ehi-miRNA-like molecules as described by Mar-Aguilar et al. [26], possibly sponged by the corresponding circRNA, ordered according to the highest target probability; 5 Hypothetical protein; 6 Not determined/available.
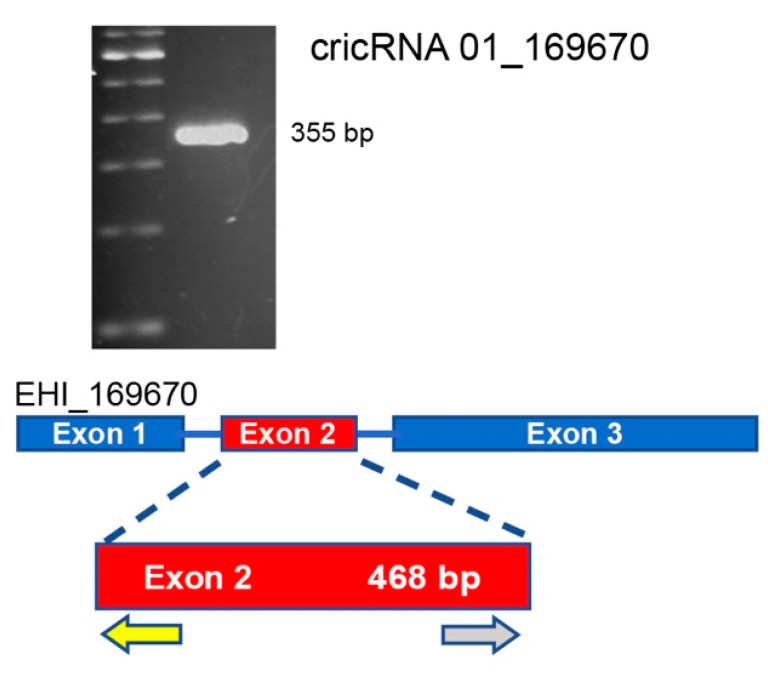
Lastly, to verify that circRNAs are expressed in E. histolytica, we used RT-PCR to amplify circRNA 01_169670 (Figure 4). Sequence analysis confirmed the identity of the circRNA.
Figure 4.
RT-PCR amplification of circRNA 01_169670. The 355 bp fragment amplified corresponds to exon 2. Arrows underneath correspond to the divergent primers used in the reaction.
2.3. E. invadens circRNAs
In 20 h, encystment induced E. invadens, 188 circRNAs were identified, and after duplicates elimination 143 remained. All of these circRNAs were formed of single elements, where 10 of them were intergenic, 101 were exonic, and 2 were intronic (Table 1). The length distribution of E. invadens circRNAs was broad, ranging from nearly 130 to 710 nt, and peaked at 250 nt (Figure 1B).
Since mature cyst formation is reached after 72 h incubation in an encystment medium, the circRNAs found in trophozoites transcriptomes were also filtered, including the intergenic circRNAs whose cell-stage expression profile cannot be univocally assigned, thus 40 cyst-specific circRNAs were identified (Table 1 and Table 3). Interestingly, three circRNAs are E. invadens-specific (the Jacob coding EIN_023210, the cadmium metallothionein coding EIN_381500, and the high mobility protein B3 locus EIN_284560), and the rest of the circRNA-producing loci have corresponding orthologs with other amoebozoans. Three circRNAs have orthologs with the free-living Mastigamoeba balamuthi (EIN_061000, EIN_369710, and EIN_475930), and fifteen have more than one E. histolytica orthologs. Remarkably, locus EIN_461630 which codes for the splicing factor U1-70 kDa, produced three different circRNAs and has two E. histolytica orthologs.
Table 3.
Entamoeba invadens encystment-specific circRNAs.
| 20 h Cysts (#) 1 | Gene Annotation in E. invadens/Gene Annotation in Orthologs 2 | Orthologs 2 |
|---|---|---|
| EIN_023210 | Jacob/na 3 | |
| EIN_031580 | hp/hp 4 | EHI_030890 |
| EIN_044390 | hp/hp | EHI_054800 |
| EIN_057320 | hp/C2-DCP 5 | EHI_069320 + 6 |
| EIN_061000 (3) | hp/Neuroendocrine convertase 1-like | MBAL_005770 |
| EIN_065890 | hp/hp | EHI_166920 + |
| EIN_065940 | Sphingomyelinase C precursor/Endo- exonuclease, phosphatase-DCP | EHI_125790 + |
| EIN_082550 (3) | hp/Zinc finger protein | EHI_055700 |
| EIN_092220 | hp/Actin | EHI_198930 |
| EIN_093520 | ATP-dependent RNA helicase DBP6/DEAD/DEAH box helicase | EHI_052790 |
| EIN_118080 | hp/hp | EHI_016340 + |
| EIN_129500 | 60S ribosomal protein L21/60S ribosomal protein L21 | EHI_069110 + |
| EIN_145900 | DNA topoisomerase/DNA topoisomerase | EHI_120640 |
| EIN_153430 | Pyruvate phosphate dikinase/Pyruvate phosphate dikinase | EHI_009530 + |
| EIN_162170 (3) | hp/Deoxycytidine triphosphate deaminase | EHI_140240 |
| EIN_176430 | Vesicle-fusing ATPase/Vesicle-fusing ATPase | EHI_004640 |
| EIN_230150 (3) | hp/WD-DCP | EHI_135040 |
| EIN_231040 | hp/hp | EHI_160980 + |
| EIN_274360 | hp/hp | EHI_002780 |
| EIN_281330 (3) | hp/hp | EHI_012130 + |
| EIN_284560 | High mobility group protein B3/na | |
| EIN_284970 | GAPDH/GAPDH | EHI_187020 + |
| EIN_327660 | Caldesmon/Glutamic acid-rich protein precursor | EHI_182620 |
| EIN_335410 | Phospholipase D/Phospholipase D | EHI_082560 + |
| EIN_369450 | hp/hp | EHI_152970 |
| EIN_369520 | hp/hp | EHI_091120 |
| EIN_369710 | hp/hp | MBAL_010850 |
| EIN_377620 | hp/hp | EHI_142970 |
| EIN_381500 | Cadmium metallothionein precursor/na | |
| EIN_398110 | hp/hp | EHI_030480 |
| EIN_403300 | Myosin-2 heavy chain/Myosin heavy chain | EHI_110180 |
| EIN_409300 | 60S ribosomal protein L4/60S ribosomal protein L4 | EHI_000510 + |
| EIN_409670 | hp/hp | EHI_024550 |
| EIN_416970 | Actin binding protein/Actin binding protein | EHI_094030 + |
| EIN_424210 | dTDP-glucose 4,6-dehydratase/dTDP-glucose 4,6-dehydratase | EHI_125700 + |
| EIN_461630 (3) | U1-70 kDa/U1 small nuclear ribonucleoprotein subunit | EHI_153670 + |
| EIN_468500 | Pyruvate dehydrogenase/Pyruvate:ferredoxin oxidoreductase | EHI_051060 |
| EIN_475930 | Pyrroline-5-carboxylate reductase/Pyrroline-5-carboxylate reductase | MBAL_004253 |
| EIN_486020 | hp/hp | EHI_196570 |
| EIN_487250 (5) | Rho GTPase/Rho family GTPase | EHI_178680 + |
1 The additional number of circRNAs in their respective loci appear between parenthesis; 2 As registered in the AmoebaDB for E. invadens/E. histolytica or Mastigamoeba balamuthi; 3 not available; 4 Hypothetical protein; 5 DCP (domain-containing protein); 6 + denotes at least one additional ortholog.
Finally, we analyzed the metabolic processes GO terms of 20 h encysting E. invadens and their corresponding E. histolytica orthologs (Figure 3C). Fifty-four metabolic processes are represented in encysting amoebas indicating that distinct metabolic processes are involved during cell differentiation in Entamoeba. When we compared the GO terms of 20 h cysts with the virulent strain (Table S3), again, we observed that they differed both in dispensability and distribution. For example, the metabolic process was in opposite X-axis coordinates, and the small molecule metabolic process was diametrically opposed.
3. Discussion
Here we bioinformatically identified circular RNA molecules from exons and intergenic regions of E. histolytica and E. invadens. Given the available suboptimal libraries used here, the number of circRNAs detected is more likely a sub-estimation. In agreement, we interpret that the lower number of circRNAs identified in E. invadens might not reflect the actual repertoire. For example, Mar-Aguilar and coworkers identified 199 Ehi-miRNA-like molecules [26]; however, when small RNA-specific libraries were analyzed, nearly half a million reads of 27nt small RNAs were identified in E. histolytica and, depending on the differentiation stage, from over 1.2 to nearly 1.9 million reads in E. invadens [38]. However, because of their structure, biogenesis, and half-life, we do not expect such high numbers for the circRNA repertoire.
Ninety-seven percent of the E. histolytica circRNAs without duplicates are exonic and constituted of a single element, and only 3% are intergenic (Table 1). We suspect that the intron mean size in the amoebic genome was the main cause for the null detection of intron-derived circRNAs. In E. invadens, in keeping with its gene structure, 6.99% intergenic and 1.3% intronic circRNAs were identified (Table 1). The same holds true for the distribution lengths of their respective circRNAs; E. invadens circRNAs double in length E. histolytica (Figure 1).
Mostly one circRNA per locus was identified. However, 44 loci from the virulent and 28 loci from the avirulent E. histolytica strains produce two or more circRNAs and up to nine circRNAs for locus EHI_169670 (Table 2 and repository). Likewise, seven loci of encystation-specific E. invadens also produce additional (up to five) circRNAs (Table 3). Additionally, in agreement with apparent stochastic alternative 5′ss and 3′ss selection reported in the forward-splicing of E. histolytica introns [35], we observed different BSJ of some circRNAs derived from a given locus. This suggests that certain circRNAs are required in larger molar quantities to achieve their roles or that they might be involved in additional metabolic tasks.
Significantly, different from the human circRNAs [39,40], the expression of Entamoeba circRNAs positively correlates with the expression of their linear counterparts (Supplementary Figure S2). Furthermore, whereas human circRNAs originate from longer-than-average exons flanked by large introns [41,42], in both Entamoeba species, we observed no correlation between intron length or the number of introns per locus with the number of circRNAs produced in a given locus (Table 2 and Table 3, repository). Altogether, these data support their suggested role in gene or protein expression regulation, as has been demonstrated in the intronic flicRNAs [23,43].
As expected, we observed the DE of circRNAs between E. histolytica virulent and avirulent strains and detected circRNAs during E. invadens cell differentiation as well (Figure 3). Our data are in agreement with previous observations for amoebic small ncRNAs [38,44,45] and lncRNAs [30,46]. Interestingly, circRNA 14_116360, underexpressed in the virulent strain (i.e., overexpressed in the avirulent strain), originated from a locus that codes for a Serine-rich protein implicated in abiotic stress, whose phosphorylation might be controlled by ncRNAs [47]. Other circRNAs overexpressed in the Rahman strain correspond to loci coding for GRIP domain RUD3 proteins involved in the Golgi Apparatus structuring and trafficking (01_169670, 02_069670, 03_069670, 11_ 014170, and 18_069670) and to loci coding for the S20 and L27 ribosomal proteins (12_026410 and 16_183480, respectively). Therefore, one-quarter of the DE circRNAs (i.e., five out of seven of those overexpressed in the avirulent strain) originate from two loci coding for GRIP domain RUD3, suggesting that significant cell trafficking might be implicated in the control of core metabolic processes to maintain the avirulent phenotype, and probably in the virulent phenotype as well. This general feature reflects the differences in gene expression regulation required in distinct virulence and encystment phenotypes, encompassing all levels of control, from transcription to translation and protein modification.
Furthermore, the differences in GO terms for biological processes such as gene expression, translation elongation, ATP-metabolic process, cellular nitrogen and generation of metabolite precursors, and energy generation processes between E. histolytica strains and encysting E. invadens support this notion. The fact that some primary metabolic processes do not change between strains strengthens this view, indicating that not all biological processes are differentially regulated between the virulent and avirulent phenotypes.
Noteworthy is the encystment-specific circRNAs detected here (Table 3). As expected, and casting insights of the value of E. invadens as an encystment model, three E. invadens-specific circRNAs were identified. Locus EIN_023210 codes for protein Jacob, locus EIN_284560 codes for a putative high mobility group B3 protein/SWI/SNF related chromatin-binding protein, and locus EIN_381500 codes for a putative cadmium metallothionein precursor. Jacob is the most abundant glycoprotein that has chitin-binding domains and is thus an integral part of the cyst wall [48]. The fact that three loci have Mastigamoeba balamuthi orthologs only (Table 3) reflects the deep-branching pre-parasitic origins of encystment-related circRNAs, which may be possibly involved in encystment regulation.
When we searched for anti-Eh-miRNA target sites within the identified circRNAs, we found that eight circRNAs contain at least one and as many as six target sites for Eh-miRNAs. Probably, this number is an underestimation since we used very stringent base complementary parameters. This finding suggests a sponging function of these circRNAs, a possibility that we are currently investigating. Nevertheless, because the putative sponging circRNAs are differentially expressed in virulent amoebas (circRNAs 01_169670, 14_116360, and 18_169670 are underexpressed, and circRNAs 04_130700, 06_ DS571557, 07_036530, 19_125950, and 20_ DS571344 are overexpressed) we can envisage a circRNA-Eh-miRNA-mRNA virulence/avirulence regulatory network in which, for example, the translation of a virulence-related mRNA could be repressed by an avirulent regulatory Eh-miRNA-like molecule, and such translation repression could be relieved by the overexpression of a virulent regulatory circRNA, which will titter the avirulent regulatory Eh-miRNA (or silencing RNA molecules), and vice-versa. Recently, additional small RNA and antisense Entamoeba databases have been reported [38,49]. However, a statistically significant miRNA/small RNA DE comparison between strains has proved unmanageable, masking any DE relationship between circRNA and miRNA-like molecules in these strains.
So far, regulatory networks have been predicted at the transcriptional level using the Bayesian inference [50]. Here, we propose additional regulatory networks impacting post-transcriptional expression events. Central to these regulatory networks include circRNAs that, in addition to sponging miRNA/siRNA molecules, could also interact with other ncRNAs or transcription or translation regulatory proteins and even may be translated into proteins.
In conclusion, we identified 143 and 605 reverse overlapped circRNAs from the E. invadens and E. histolytica libraries, respectively. The identified circRNAs were mostly exonic. 416 circRNAs from E. histolytica were virulent-specific, 267 were avirulent-specific, and 32 were shared between strains. From the latter, 32 were DE between strains, showing no correlation between intron length or the number of introns per locus with the number of circRNAs produced in a given locus. Finally, we predicted that eight of the DE circRNAs could function as sponges of the reported miRNAs in E. histolytica, whose functions are still unknown, although other epigenetic regulatory roles must be also explored. Our results allowed us to devise a working hypothesis to test the relationships between circRNAs and miRNA-like molecules in determining virulent/nonvirulent phenotypes and to explore additional regulatory mechanisms during amoebic encystment. Furthermore, the in silico prediction of circRNAs in this study would facilitate a deeper comprehension of regulatory epigenetic processes in such pathogenic amoebic protozoa. However, E. histolytica virulence is widely variable worldwide across several strains, and the relevant mechanisms controlling the differential virulent expression need to be investigated more prospectively.
4. Materials and Methods
4.1. Libraries
Total RNA E. histolytica RNA-Seq libraries were downloaded from the European Bioinformatics Institute, project PRJEB2766 (Available online: https://www.ebi.ac.uk/ena/browser/view/PRJEB2766?show=reads (21 September 2022)). Three libraries were from the HM1:IMSS virulent strain (113,997,619; 91,291,028; and 81,271,700 pair-end reads, respectively) and three from the Rahman avirulent strain (70,821,013; 68,278,102; and 108,168,110 pair-end reads) were used. Total RNA E. invadens RNA-Seq libraries were downloaded from the GenBank GEO database (Available online: htpps://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138449 (21 September 2022)). Four libraries from 24 h encystment induced E. invadens (34,897,385; 46,698,915; 33,531,808; and 36,965,892 pair-end reads) were used.
4.2. Identification of Linear and Circular RNAs
The quality control of the RNA libraries was made using the FASTQC (v0.11.8) and Trimmomatic (v0.39) software packages. To detect and quantify the linear RNAs, we improved the STAR-HTSeq pipeline. STAR software (v2.7.10a) [51] was used to make the indexed, mapping, and annotation of linear RNAs. To quantify the expression of the linear RNAs was made by the HTSeq software with the htseq-count module. circRNAs were detected using the software CIRI-full (v2.1.1) [34,52]. For the indexing and mapping of the RNA libraries, we used the program BWA (v0.7.17), as recommended by the authors of CIRI2. The quantification and annotation were made with the RO1 and RO2 modules of CIRI-full as recommended using the author’s pipeline.
4.3. Differential Expression of RNAs
To make the differential expression of linear RNAs and circRNAs, we used the R-base (v4.1), Rstudio IDE (v1.1.4), and the R libraries DEseq2 and ggplot2 to build the normalization of the expression counts, differential expression, statistical analysis, and plots.
4.4. Gene Ontology Analysis
GO analyses were carried out using the AmoebaDB data analysis suite. The gene ID of all detected circRNAs in both E. histolytica strains and E. invadens were uploaded and analyzed by the amoebaDB web (https://amoebadb.org) (accessed on 16 August 2022) to obtain the annotation, GO terms and GO enrichment analysis for each locus. Finally, with this data, we used the Revigo web tool (http://revigo.irb.hr) (accessed on 16 August 2022) to build the GO enrichment graph, and, with Rstudio IDE (v1.1.4), the graph was visualized.
4.5. Identification of circRNAs as miRNAs Sponges
To assess the prediction of circRNAs as sponges of miRNAs, we used the software intaRNA [53], which is capable of detecting RNA-RNA interactions through an alignment. The parameters used to make the predictions were a seed sequence of at least 7 nucleotides, canonical interactions between miRNA and circRNA in nucleotides 1 to 5, the possibility of gaps in the alignment, and favorable thermodynamics interaction ΔG < −7 Kcal/mol.
4.6. CIRI-Full Control
We designed in silico RNA decoy reads from exon end sequences of the locus EHI_110740 of E. histolytica. These reads were constructed by fragments of this locus to make 10 RNA library decoy reads with a length of 100 nucleotides which includes a BSJ. Additionally, we prepared the RNA libraries extracting 10% of all RNA reads, and we identified the circRNAs presented in this modified library with the described method. Next, we included the 10 artificial reads to this modified library and reidentified the circRNAs in the library.
4.7. RNA Isolation, Retrotranscription, and Polymerase Chain Reactions
Total RNA was isolated using the TRIzol Reagent as specified by the manufacturer (Invitrogen). For circular RNA isolation, preparations were treated with RNase R (Epicentre). Outward-facing primers targeted the second exon of E. histolytica locus EHI_169670 (E2as 5′CTTCTTTTTCTTTTTCTAATTCTTCACCC3´ and E2s 5′AAGAAAGTTAATGATTCTGAGAAAGAG3′) was designed as described [54] for the detection of circular RNA molecules. Retro-transcription reactions were carried out in the presence of 5 µM Actinomycin D (SigmaAldrich). The drug was added immediately after the denaturing step [55]. PCR products were amplified as described [43] and were resolved in 2.5% agarose gels.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ncrna8050065/s1, Figure S1: Workflow for identification and partial characterization of circRNAs from Entamoeba histolytica and Entamoeba invadens, Figure S2: Design and rescue of synthetic circRNA decoy sequence from limited Entamoeba histolytica total RNA-Seq libraries, Figure S3: Near-linear correlation between circRNA and mRNA expression in Entamoeba histolytica, Figure S4: Principal Components Analysis of the six total RNA-Seq libraries used in this work, Table S1: Metabolic process GO terms of Entamoeba histolytica HM1:IMSS strain circRNAs, Table S2: Metabolic process GO terms of Entamoeba histolytica Rahman strain circRNAs, Table S3: Metabolic process GO terms of Table S3. Metabolic process GO terms of 20 h encysting Entamoeba invadens circRNAs.
Author Contributions
Conceptualization, J.V., A.M.-T., and M.Á.L.-L.; methodology, M.Á.L.-L., C.J.C.P.-M., J.A.G.-L., D.L.-A., R.H.-R., and O.S.-C.; writing—original draft preparation, M.Á.L.-L., A.M.-T. and J.V.; writing—review and editing, project administration, and funding acquisition, J.V. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data found in this work is currently being uploaded in the AmoebaDB.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This research was funded by SEP-CINVESATAV, grant number 56 and by CONACYT, grant number 194163 and the postgraduate scholarship 988721 to M.Á.L.-L.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 3.Lasda E., Parker R. Circular RNAs: Diversity of form and function. RNA. 2014;20:1829–1842. doi: 10.1261/rna.047126.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 5.Chen L.L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 2016;17:205–211. doi: 10.1038/nrm.2015.32. [DOI] [PubMed] [Google Scholar]
- 6.Holdt L.M., Kohlmaier A., Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol. Life Sci. 2018;75:1071–1098. doi: 10.1007/s00018-017-2688-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 8.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 10.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 11.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Ebbesen K.K., Hansen T.B., Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14:1035–1045. doi: 10.1080/15476286.2016.1271524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talhouarne G.J., Gall J.G. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA. 2014;20:1476–1487. doi: 10.1261/rna.045781.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e29. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B., Feng C., Zhu S., Zhang J., Irwin D.M., Zhang X., Wang Z., Zhang S. Identification of candidate circular RNAs underlying intramuscular fat content in the donkey. Front. Genet. 2020;11:587559. doi: 10.3389/fgene.2020.587559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X.X., Xiao L., Chung H.K., Ma X.X., Liu X., Song J.L., Jin C.Z., Rao J.N., Gorospe M., Wang J.Y. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol. Cell. Biol. 2020;40:e00492-19. doi: 10.1128/MCB.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talhouarne G.J.S., Gall J.G. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc. Natl. Acad. Sci. USA. 2018;115:E7970–E7977. doi: 10.1073/pnas.1808816115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panda A.C., De S., Grammatikakis I., Munk R., Yang X., Piao Y., Dudekula D.B., Abdelmohsen K., Gorospe M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 2017;45:e116. doi: 10.1093/nar/gkx297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 22.Turkeltaub J.A., McCarty T.R., 3rd, Hotez P.J. The intestinal protozoa: Emerging impact on global health and development. Curr. Opin. Gastroenterol. 2015;31:38–44. doi: 10.1097/MOG.0000000000000135. [DOI] [PubMed] [Google Scholar]
- 23.Mendoza-Figueroa M.S., Alfonso-Maqueira E.E., Velez C., Azuara-Liceaga E.I., Zarate S., Villegas-Sepulveda N., Saucedo-Cardenas O., Valdes J. Postsplicing-derived full-length intron circles in the protozoan parasite Entamoeba histolytica. Front. Cell. Infect. Microbiol. 2018;8:255. doi: 10.3389/fcimb.2018.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taggart A.J., Lin C.L., Shrestha B., Heintzelman C., Kim S., Fairbrother W.G. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017;27:639–649. doi: 10.1101/gr.202820.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mar-Aguilar F., Trevino V., Salinas-Hernandez J.E., Tamez-Guerrero M.M., Barron-Gonzalez M.P., Morales-Rubio E., Trevino-Neavez J., Verduzco-Martinez J.A., Morales-Vallarta M.R., Resendez-Perez D. Identification and characterization of microRNAs from Entamoeba histolytica HM1-IMSS. PLoS ONE. 2013;8:e68202. doi: 10.1371/journal.pone.0068202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De S., Pal D., Ghosh S.K. Entamoeba histolytica: Computational identification of putative microRNA candidates. Exp. Parasitol. 2006;113:239–243. doi: 10.1016/j.exppara.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 28.Avesson L., Reimegard J., Wagner E.G., Soderbom F. MicroRNAs in Amoebozoa: Deep sequencing of the small RNA population in the social amoeba Dictyostelium discoideum reveals developmentally regulated microRNAs. RNA. 2012;18:1771–1782. doi: 10.1261/rna.033175.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta A.K., Panigrahi S.K., Bhattacharya A., Bhattacharya S. Self-circularizing 5′-ETS RNAs accumulate along with unprocessed pre-ribosomal RNAs in growth-stressed Entamoeba histolytica. Sci. Rep. 2012;2:303. doi: 10.1038/srep00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ojha S., Singh N., Bhattacharya A., Bhattacharya S. The ribosomal RNA transcription unit of Entamoeba invadens: Accumulation of unprocessed pre-rRNA and a long noncoding RNA during encystation. Mol. Biochem. Parasitol. 2013;192:30–38. doi: 10.1016/j.molbiopara.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Broadbent K.M., Broadbent J.C., Ribacke U., Wirth D., Rinn J.L., Sabeti P.C. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genom. 2015;16:454. doi: 10.1186/s12864-015-1603-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin Y.L., Liu T.L., Yao Q., Wang Y.X., Wu X.M., Wang X.T., Yang X., Song J.K., Zhao G.H. Circular RNA ciRS-7 affects the propagation of Cryptosporidium parvum in HCT-8 cells by sponging miR-1270 to activate the NF-kappaB signaling pathway. Parasit. Vectors. 2021;14:238. doi: 10.1186/s13071-021-04739-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiu Y., Jiang G., Zhou S., Diao J., Liu H., Su B., Li C. Identification of potential immune-related circRNA-miRNA-mRNA regulatory network in intestine of Paralichthys olivaceus during Edwardsiella tarda infection. Front. Genet. 2019;10:731. doi: 10.3389/fgene.2019.00731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng Y., Ji P., Chen S., Hou L., Zhao F. Reconstruction of full-length circular RNAs enables isoform-level quantification. Genome Med. 2019;11:2. doi: 10.1186/s13073-019-0614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hon C.C., Weber C., Sismeiro O., Proux C., Koutero M., Deloger M., Das S., Agrahari M., Dillies M.A., Jagla B., et al. Quantification of stochastic noise of splicing and polyadenylation in Entamoeba histolytica. Nucleic Acids Res. 2013;41:1936–1952. doi: 10.1093/nar/gks1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozano-Amado D., Avila-Lopez P.A., Hernandez-Montes G., Briseno-Diaz P., Vargas M., Lopez-Rubio J.J., Carrero J.C., Hernandez-Rivas R. A class I histone deacetylase is implicated in the encystation of Entamoeba invadens. Int. J. Parasitol. 2020;50:1011–1022. doi: 10.1016/j.ijpara.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Wang J., Yin J., Wang X., Liu H., Hu Y., Yan X., Zhuang B., Yu Z., Han S. Changing expression profiles of mRNA, lncRNA, circRNA, and miRNA in lung tissue reveal the pathophysiological of bronchopulmonary dysplasia (BPD) in mouse model. J. Cell. Biochem. 2019;120:9369–9380. doi: 10.1002/jcb.28212. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H., Ehrenkaufer G.M., Manna D., Hall N., Singh U. High throughput sequencing of Entamoeba 27nt small RNA population reveals role in permanent gene silencing but no effect on regulating gene expression changes during stage conversion, oxidative, or heat shock stress. PLoS ONE. 2015;10:e0134481. doi: 10.1371/journal.pone.0134481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-S. [DOI] [PubMed] [Google Scholar]
- 40.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Lerena J.A., Gonzalez-Blanco G., Saucedo-Cardenas O., Valdes J. Promoter-bound full-length intronic circular RNAs-RNA polymerase II complexes regulate gene expression in the human parasite Entamoeba histolytica. Noncoding RNA. 2022;8:12. doi: 10.3390/ncrna8010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang H., Alramini H., Tran V., Singh U. Nucleus-localized antisense small RNAs with 5′-polyphosphate termini regulate long-term transcriptional gene silencing in Entamoeba histolytica G3 strain. J. Biol. Chem. 2011;286:44467–44479. doi: 10.1074/jbc.M111.278184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., Ehrenkaufer G.M., Hall N., Singh U. Small RNA pyrosequencing in the protozoan parasite Entamoeba histolytica reveals strain-specific small RNAs that target virulence genes. BMC Genomics. 2013;14:53. doi: 10.1186/1471-2164-14-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saha A., Bhattacharya S., Bhattacharya A. Serum stress-responsive gene EhslncRNA of Entamoeba histolytica is a novel long noncoding RNA. Sci. Rep. 2016;6:27476. doi: 10.1038/srep27476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kishor P.B.K., Suravajhala R., Rajasheker G., Marka N., Shridhar K.K., Dhulala D., Scinthia K.P., Divya K., Doma M., Edupuganti S., et al. Lysine, lysine-rich, serine, and serine-rich proteins: Link between metabolism, development, and abiotic stress tolerance and the role of ncrnas in their regulation. Front. Plant Sci. 2020;11:546213. doi: 10.3389/fpls.2020.546213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frisardi M., Ghosh S.K., Field J., Van Dellen K., Rogers R., Robbins P., Samuelson J. The most abundant glycoprotein of amebic cyst walls (Jacob) is a lectin with five Cys-rich, chitin-binding domains. Infect. Immun. 2000;68:4217–4224. doi: 10.1128/IAI.68.7.4217-4224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mornico D., Hon C.C., Koutero M., Weber C., Coppee J.Y., Dillies M.A., Guillen N. Genomic determinants for initiation and length of natural antisense transcripts in Entamoeba histolytica. Sci. Rep. 2020;10:20190. doi: 10.1038/s41598-020-77010-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hackney J.A., Ehrenkaufer G.M., Singh U. Identification of putative transcriptional regulatory networks in Entamoeba histolytica using Bayesian inference. Nucleic Acids Res. 2007;35:2141–2152. doi: 10.1093/nar/gkm028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao Y., Wang J., Zhao F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mann M., Wright P.R., Backofen R. IntaRNA 2.0: Enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel J., Hess W.R., Borner T. Precise branch point mapping and quantification of splicing intermediates. Nucleic Acids Res. 1997;25:2030–2031. doi: 10.1093/nar/25.10.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Houseley J., Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data found in this work is currently being uploaded in the AmoebaDB.