Abstract
The ability of the nervous system to detect a wide range of noxious stimuli is crucial to avoid life-threatening injury and to trigger protective behavioral and physiological responses. Pain represents a complex phenomenon, including nociception associated with cognitive and emotional processing. Animal experimental models have been developed to understand the mechanisms involved in pain response, as well as to discover novel pharmacological and non-pharmacological anti-pain therapies. Due to the genetic tractability, similar physiology, low cost, and rich behavioral repertoire, the zebrafish (Danio rerio) is a powerful aquatic model for modeling pain responses. Here, we summarize the molecular machinery of zebrafish responses to painful stimuli, as well as emphasize how zebrafish-based pain models have been successfully used to understand specific molecular, physiological, and behavioral changes following different algogens and/or noxious stimuli (e.g., acetic acid, formalin, histamine, Complete Freund's Adjuvant, cinnamaldehyde, allyl isothiocyanate, and fin clipping). We also discuss recent advances in zebrafish-based studies and outline the potential advantages and limitations of the existing models to examine the mechanisms underlying pain responses from evolutionary and translational perspectives. Finally, we outline how zebrafish models can represent emergent tools to explore pain behaviors and pain-related mood disorders, as well as to facilitate analgesic therapy screening in translational pain research.
Keywords: Non-traditional pain models, zebrafish, noxious stimuli, nociceptors, pain-related behaviors, anti-pain medication screening
1. INTRODUCTION
Pain is a complex process that involves the transduction of nociceptive stimuli culminating in specific behavioral and physiological responses [1]. Chronic pain is an important biomedical and social problem whose management represents a critical unmet biomedical condition [2], necessitating both novel animal experimental models and non-pharmacological and pharmacological therapies (e.g., analgesic drug discovery) [3]. Thus, developing valid and sensitive animal models is a key factor to the elucidation of neural, molecular, and physiological mechanisms involved in pain response [4-6].
Measuring withdrawal thresholds to noxious stimuli has long been used to examine pain responses in vivo [7-9]. These approaches have markedly improved our knowledge of nociception physiology, analgesic drug properties, as well as neurotransmitters (e.g., glutamate) and genes modulated in pain responses [10-13]. Although rodents are widely used in translational pain research [14, 15], developing novel alternative animal experimental models is essential to unravel evolutionarily conserved mechanisms of pain [16, 17] as well as to perform high-throughput in vivo pharmacological screens [18]. Here, we discuss the growing utility of zebrafish models to evaluate specific molecular, physiological, and behavioral phenotypes related to pain responses. We also outline the suitability of zebrafish models to assess some debilitating pain-related mood disorders, as well as cover recent advances and potential limitations of the zebrafish as a cost-effective and translatable model organism in pain research and pharmacological screens.
2. GENERAL MODEL FEATURES
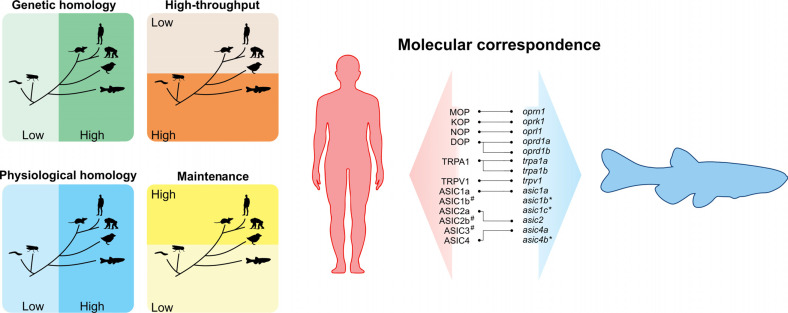
The zebrafish (Danio rerio) is a suitable model organism in genetics, neuroscience, pharmacology, and drug discovery [18-21]. Compared to traditional rodent models, zebrafish exhibit some advantages for basic research, including lower space for maintenance and the large number of offsprings [22, 23]. These features reinforce the growing use of zebrafish as a powerful tool to perform medium-to-high throughput screens cost-effectively [24, 25]. The external fertilization also simplifies the production of transgenic lines, and the presence of translucent embryos facilitates the investigation of potential biomarkers of nociception in vivo (e.g., using gene-expression fluorescent probes) [26-30]. Although the use of zebrafish-based pain models is relatively recent [31], this species shows high sensitivity to various nociceptive stimuli [32-35]. Notably, zebrafish respond to clinically active analgesics, representing a promising model to investigate the molecular basis of human pain related disorders [16, 36-38]. Despite the anatomical differences in the central nervous system (CNS) from humans, teleost fishes display electrical activity and transcriptional changes following noxious stimuli [39-42]. Importantly, zebrafish express genes involved in pain responses [31, 36, 37, 43-50] also presenting specific “protective” pain behaviors when exposed to noxious stimuli, suggesting the existence of central mechanisms underlying pain responses [51]. Fig. (1) highlights important features of zebrafish vs. other model organisms used in pain studies and the molecular correspondence between human and zebrafish orthologs involved in pain responses.
Fig. (1).
General advantages and molecular features of zebrafish in translational pain research. As a vertebrate species, the zebrafish shows a high genetic and physiological homology when compared to mammals, but present a low cost of maintenance and feasibility to high-throughput screens similar to invertebrates (left). Molecular correspondece of the receptors involved in pain responses in humans with their respective genes in zebrafish (right). # are absent in zebrafish. *are splice variants.
3. MOLECULAR MECHANISMS OF PAIN-RELATED RESPONSES
Pain is an individual sensation that mobilizes specific receptors to detect noxious stimuli, playing a pivotal role in survival and well-being of the species [10]. Zebrafish presents a complex physiological system that recognizes and responds to painful stimuli [13, 42, 52], including multiple subtypes of nociceptors already identified with similar organization of nociceptive circuits to those of mammals [36, 51, 53, 54]. Here, we emphasize the basic mechanisms of pain-related responses, focusing on the molecular machinery expressed in zebrafish (e.g., the opioid system, the transient potential receptor family (TRP), the endocannabinoid system, as well as acid-sensitive ion channels (ASIC) and their relevance in the study of pain.
3.1. Opioid System
The opioid system involves multiple receptors, endogenous ligands and natural or synthetic analogs, which play a key role in pain homeostasis, mood, and well-being [55, 56]. In humans, opioid receptors belong to the G protein-coupled receptor (GPCR) superfamily, and three classical opioid receptors have been characterized so far: MOP (μ = mu for morphine; encoded by the OPRM1), KOP (κ = kappa for ketocyclazocine; encoded by the OPRK1), DOP (δ = delta for vas deferens; encoded by the OPRD1), and their respective endogenous ligands; β-endorphin, enkephalin, and dynorphin [57-59]. Although MOP, KOP, and DOP mediate analgesic effects, they rather differ in evoking affective behaviors [60, 61]. While MOP agonists produce euphoria and promote stress coping [62, 63], KOP agonists cause dysphoria and are associated with stress and negative effects [64, 65], and DOP agonists are known to elicit anxiolytic and antidepressant effects [60, 61]. Furthermore, the additional NOP (nociceptin/orphanin FQ) receptor encoded by the OPRL1 gene, a non-opioid member of the opioid receptor family, has also been described in mammals, and its activation with specific ligands shows analgesic effects [66].
The genome sequencing projects allowed a comprehensive identification of the zebrafish orthologs [67, 68], such as zMOP (μ; encoded by the oprm1) [45], zKOP (κ; encoded by the oprk1) [44], and two functional copies of zDOP (δ; encoded by the oprd1a and oprd1b) [69, 70]. Furthermore, zNOP (nociceptin/orphanin FQ; encoded by oprl1) receptor has also been characterized [71]. Here, we will use MOP, DOP, KOP, and NOP for mammals’ opioid receptors, as well as zMOP, zKOP, zDOPa/b, and zNOP to describe their corresponding zebrafish opioid receptors.
In zebrafish, opioid peptide precursors include two proenkephalin genes (penka and penkb), two proopiomelanocortin genes (pomca and pomcb), one prodynorphin gene (pdyn), and two pronociceptin genes (pnoca and pnocb). Penka codes four Met-enkephalins (ME), one Met-enkephalin-Ile (MEI), and one Met-enkephalin-Asp (MED) [31, 67, 72]. Penkb codes four ME, one Leu-enkephalin (LE), and one Met-enkephalin-Gly-Tyr (MEGY) [73]. Pomca codes for adrenocorticotropin (ACTH), γ-lipotropin (γ-LPH), β-melanotropin (β-MSH), and β-endorphin (β-END) while pomcb codes for only α-melanotropin (α-MSH) and β-END [74]. Interestingly, ACTH and α-MSH are released after hypothalamic-pituitary-interrenal axis (HPI; homologous to mammalian hypothalamic-pituitary-adrenal axis) activation following a noxious stimulus in zebrafish [75]. While ACTH stimulates the synthesis and cortisol release [76], α-MSH is responsible for camouflage response in larval zebrafish [77]. Because both molecules are released under aversive conditions, a potential involvement of camouflage in stress- and pain-responses should not be ruled out.
Pdyn codes for Ile-enkephalin, the neoendorphins (α-neoendorphin and β-neoendorphin), dynorphins A and B [78]. Furthermore, both pnoca and pnocb encode two nociceptin peptides, a nociceptin orthologous to the mammalian which presents the classical ‘opioid message’ (-Try-Gly-Gly-Phe-) in its N-terminus, and another ‘nociceptin-like’ peptide, which is lost in mammals and has low homology to mammalian nociceptin [67, 73].
The pharmacological properties of the zebrafish opioid receptors have been extensively characterized [79, 80]. While zMOP, zKOP, and zDOPa/b display high affinity toward both nonselective opioid ligands [3H]-diprenorphine and [3H]-bremazocine in saturation binding assays [44, 55, 70], they show lower affinity for highly selective ligands in mammals ([d-Pen2,d-Pen5]enkephalin [3H]-DPDPE, [3H]-[D-Ala2, NMe-Phe4, Gly5-ol]-enkephalin (DAMGO), and [3H]-U69,593) [81]. Like mammals, [35S]-GTPγS binding assays demonstrate that all classical zebrafish opioid receptors act via Gi (inhibitory) protein-coupled receptors after binding agonist ligands [44, 45, 69], suggesting that the information could be transduced by the same type of mammalian GPCR.
3.1.1. zMOP
Currently, MOP is considered the primary target for painkillers [82], although various adverse side effects (e.g., tolerance, physical dependence, and addiction) limit their effective medical use [60]. zMOP receptors are widely distributed in the zebrafish brain, and similar to their human homologs, they are highly expressed in regions involved in analgesia and reward [83]. Although zMOP shows 74% of genetic similarity related to MOP, the genomic structure of zMOP presents a stop codon on exon 3 (and not on exon 4, as occurs in mammals) [55], suggesting some difference in behavioral effects and regulatory mechanisms mediated by zMOP [55]. For example, while naloxone alone does not affect anxiety in rodents [84] and non-human primates [85], this antagonist induces anxiety-like behaviors in adult zebrafish [86].
Similar to the mammalian ortholog, the zMOP molecular structure is well conserved in both transmembrane domain and intracellular loop [45]. However, zMOP presents low conservation in the extracellular loop as well as in the carboxyl- and amino-terminal, compared to MOP [45]. Although differences in the extracellular loop can determine the selectivity of ligands, zMOP displays similar pharmacological profile and conserved functions compared to the mammalian counterparts [33, 34, 87-89].
Binding experiments using [3H]-diprenorphine demonstrate that naloxone displays highest affinity toward zMOP, followed by β-END = morphine > MEGY > ME > LE [72]. Likewise, dynorphin A and nociceptin can also bind to zMOP [73, 78]. While endomorphins exhibit partial agonistic properties at the zMOP, competition-binding assays in zebrafish brain membranes show that [3H] diprenorphine binds to zMOP with a nanomolar range affinity, which is antagonized by naloxone [55, 81]. Notably, zMOP also shows analgesic properties, since morphine prevents pain behaviors in both adult and larvae zebrafish, in inflammatory and visceral pain models [32-34, 87, 89]. Like in humans, adverse effects can also be observed after zMOP activation with different agonists, and include sedation (mitragynine and morphine) [90, 91], reduced gut mobility (loperamide) [92], and addiction (morphine) [93], suggesting evolutionarily conserved biological functions related to pain and mediated by these receptors.
3.1.2. zKOP
Similar to the stimulation of other opioid receptors, the activation of KOP produces analgesia in mammals [94, 95]. However, the administration of KOP agonists into the spinal cord or intravenously antagonizes morphine-induced analgesia [96]. Clinical trials reveal psychotomimetic and hallucinogenic effects caused by KOP agonists [97]. Interestingly, a biphasic effect was observed after salvinorin A (a KOP selective agonist) injection in zebrafish; while low doses (0.1 or 0.2 μg/kg) evoke rewarding effects and excitation, high doses (5 or 10 μg/kg) cause aversive effects and hallucinogenic-like behavior [98].
zKOP presents 70% genetic similarity compared to its mammalian counterpart [44]. This receptor is well conserved in both the transmembrane domain and intracellular loop, and less conserved in the extracellular loop and the carboxyl- and amino-terminal compared to KOP [44]. Interestingly, the third extracellular loop of zKOP (determinant for the binding selectivity in mammals) displays high similarity to MOP, which may explain why morphine can bind zKOP [44]. Moreover, binding experiments using [3H]-diprenorphine demonstrate that non-specific δ-κ agonist/µ antagonist bremazocine displays highest affinity toward zKOP, followed by dynorphin A = naloxone > D-Arg dynorphin A > morphine [44]. Likewise, both nociceptin and nociceptin- like peptides can also bind zKOP [73]. Because of the presence of a positive-charged residue His294, which is not present in the human receptor extracellular loop, the specific KOP ligand [3H]-U69,593 does not bind zKOP [44]. Zebrafish endogenous opioid MEGY is unable to bind zKOP [99], unlike the mammalian counterpart peptide MERF, which binds the three classical opioids (MOP, DOP, and KOP) [100]. While there is no direct evidence of analgesic effects following zKOP stimulation in zebrafish-based pain models, the structural similarities between zKOP and MOP could imply such a possibility.
3.1.3. zDOP
Although the most prescribed opioids (e.g., morphine, fentanyl, codeine) mainly target MOP receptors, the selective activation of DOP has great potential for chronic pain treatment [101, 102]. Since anxiety and mood disorders are commonly associated with chronic pain, the ability of DOP to cause anxiolytic- and antidepressant-like side effects are desirable [103]. In humans, DOP is widely distributed in the brain, specifically in the periaqueductal gray region, the rostroventral medulla, the cerebral cortex, and the amygdala [104]. In zebrafish, two functional copies of DOP were characterized: zDOPa and zDOPb [69, 70]. Similarly, zDOPa is also widespread in the CNS, from the telencephalon to the spinal cord, including areas involved in analgesia [105]. Moreover, homology analysis revealed 66% of genetic similarity comparing zDOPa to human counterparts [69]. The zDOPa is well conserved in both the transmembrane domain and intracellular loop, being less conserved in the extracellular loop and the carboxyl- and amino-terminal than DOP [69]. In mammals, the third extracellular loop of opioid receptors is a determinant for selective binding [106]. Interestingly, two Arg residues in the third extracellular loop are substituted by a Lys, which may explain why DPDPE, the prototypical ligand for the DOP, has a low affinity for the zDOPa binding site [107]. Pharmacological characterization shows that zDOPa binds the non-selective opioid antagonist [3H]diprenorphine with high affinity, followed by bremazocine > β-END > naloxone > morphine > DEDPE [108].
zDOPb is also widespread in the CNS, being more expressed in the dorsal telencephalic area, hypothalamus, reticular formation, facial lobe, and cerebellum than zDOPa [70]. zDOPb presents 65% of genetic similarity and is well conserved in both transmembrane domain and intracellular loop, as well as less conserved in the extracellular loop and the carboxyl- and amino-terminal compared to DOP [70]. Interestingly, zDOPb displays lower or no affinity toward highly selective DOP ligands due to the presence of positive and negative charges in both second and third extracellular loops [70]. While both zDOPa and zDOPb are homologs with high sequence similarity (71%) [70], they present some differences. For example, an amino acid is substituted in the protein sequence of these receptors (Glu112 replaces Asn115 in zDOPa or by Gly117 in zDOPb), which increases the peptide MEGY affinities for zDOPb [99]. Overall, both zDOPb and zDOPa show similar pharmacological properties and conserved functions, compared to their respective mammalian counterparts, whereas some differences in ligand selectivity also occur.
3.1.4. zNOP
Although NOP does not bind the opioid antagonist naloxone (traditionally used to discriminate opioid receptors), NOP is classified as a non-opioid member of the opioid receptor family by the International Union of Basic and Clinical Pharmacology (IUPHAR) [66]. In humans, NOP is widely distributed in both central and peripheral nervous systems [109]. Interestingly, the activation of NOP evokes distinct behavioral responses: a classical antinociceptive effect when spinally activated [110, 111], and an anti-opioid effect when NOP is supraspinally activated [112, 113].
In zebrafish, zNOP is highly expressed in the CNS and intestine, being less expressed in the peripheral nervous system [71]. Although zNOP displays 58-59% of genetic similarity related to mammalian NOP [71], their functionality may differ from the respective mammalian counterpart. Indeed, zNOP binds not only human nociceptin but also both zebrafish dynorphin A (with greater affinity) and mammalian dynorphin A, as well as naloxone (order of affinity: zebrafish dynorphin A > nociceptin > bremazocine = norbinaltorphimine = mammalian dynorphin A > naloxone) [71]. Moreover, amino acid residues that are important for ligand binding at the NOP transmembrane domains (TM) (Phe220 and Phe224 in TM5, Asp130 and Tyr131 in TM3, and Trp276 in TM6) are highly conserved in the zNOP [71]. Interestingly, some residues conserved in EL2 (extracellular loop 2; region responsible by interaction with ligands) of NOP are replaced by residues involved in KOP recognition [71]. This change may explain why zNOP shows a preference for the KOP-selective ligands. Ultimately, zebrafish models may help elucidate the role of NOP in the sensory perception of pain.
3.2. TRP Family
The transient receptor potential (TRP) channel family acts as molecular sensors, responding to a wide variety of stimuli (e.g., changes in pH, chemical agents, temperature, and osmolarity) [114, 115]. Structurally, a TRP channel contains six putative transmembrane domains (TM1-6) assembled as tetramers to form cation-permeable pores between TM5 and TM6 [115, 116]. In humans, TRP channels include 28 members divided into six subfamilies, classified as canonical (TRPC), vanilloid (TRPV), ankyrin (TRPA1), melastatin (TRPM), polycystin (TRPP), and mucolipin (TRPML) [117]. Interestingly, zebrafish express all TRP subfamilies (trpc [118], trpv [119], trpa [120], trpm [121], trpml [122], and trpp [123]). Moreover, zebrafish have a single NompC (trpn), which is absent in humans [37]. Because TRP is considered the most important ion channel family that detects and transmits noxious stimuli, in this topic, here we summarize key relevant findings related to the trpa and trpv subfamilies and their importance in pain responses in zebrafish.
3.2.1. Trpa1a, Trpa1b and Trpv1
The peripheral and central nociceptive systems of zebrafish are similar to those of mice and humans [124] and the nociceptive function of TRPA1 is remarkably conserved across animal species [125, 126]. However, unlike in humans and rodents, the zebrafish genome encodes two TRPA1 genes (trpa1a and trpa1b) [119]. Notably, both trpa1a and trpa1b are activated by human TRPA-agonists (allyl isothiocyanate, cinnamaldehyde, diallyl disulfide, acrolein, and 4-hydroxynonenal) [37], and antagonized by HC-030031, a competitive or negative allosteric modulator of human TRPA1 [124]. However, HC-030031 failed to show any inhibitory effects on trpa1b activated by cinnamaldehyde in a heterologous expression system using Xenopus laevis oocytes [127]. Therefore, the hypothesis that HC-030031 antagonizes trpa1a but not trpa1b, cannot be discarded.
Unlike other vertebrates (e.g., chick, frog, and mice) where TRPA1 is responsible for detecting noxious chemicals and harmful temperatures [125], mounting evidence suggests that trpa1a mediates chemical sensing, whereas trpa1b responds to thermal stimuli [37, 128], albeit a mutation on trpa1b reduces or abolishes the hyperlocomotion induced by chemical algogens [37]. Both trpa1a and trpa1b are expressed in sensory neurons and also found in different regions and structures. While trpa1a is predominantly expressed in sensory ganglia innervating visceral organs, trpa1b is expressed in sensory neurons that innervate the skin and cranial sensory ganglia [37]. These expression patterns suggest that trpa1b can be activated by both internal and external stimuli, whereas trpa1a probably detects mainly internal stimuli.
In mammals, TRPV1 is a polymodal receptor, which acts as a sensor for noxious heat (> 42°C) and contributes to painful disorders (e.g., diabetic-induced neuropathic pain, cancer pain, and inflammatory pain) [129]. TRPV1 is also sensitive to acidic solutions (pH < 6.5), food ingredients (capsaicin) [130], lipid derivatives (prostaglandin E2) [131], endocannabinoid, anandamide [132], and toxins (e.g., vanillotoxins and resiniferatoxin) [133]. Interestingly, zebrafish express a single trpv1-like ortholog receptor that could be derived from evolutionary precursors of tetrapod TRPV1 and TRPV2 [119]. Similar to the mammalian ortholog, trpv1 acts as a heat-sensitive channel (325°C), although it presents a lower sensitivity than that of the mammalian counterpart (342°C) [36]. Notably, 5 dpf zebrafish larvae avoid temperatures rearing above 31.5°C and below 24.5°C [134], characterizing thermal hyperalgesia. Importantly, thermal hyperalgesia is a common symptom of individuals suffering from inflammatory pain, which could support the face validity of this animal model. Also, the thermal aversion is prevented by an analgesic (ibuprofen) [134], reinforcing the predictive validity of this model.
Because amino acid residues critical to TRPV1 activation by capsaicin (Ser-512 and Thr-550) are different in trpv1 (Thr-480 and Ile-518), zebrafish larvae do not present behavioral changes after capsaicin administration via water immersion [36]. However, pain behaviors can be observed after a single capsaicin administration into the lips of adult zebrafish and classical TRPV1 antagonists prevent these responses [135-138]. Similar results are observed after capsaicin administration into the lips of Atlantic cod, another teleost fish [139]. In zebrafish larvae, trpv1 is expressed in the early stage (24 hours post fertilization, hpf), along the lateral line ganglion neurons, which do not have direct sensory properties [140]. In the first essay, capsaicin was administered into the water, which may explain why zebrafish larvae do not show aversive behaviors. Furthermore, a feasible interaction between trpv1 and trpa1a/trpa1b in a capsaicin-induced pain behavior cannot be discarded since, like mammals, trpv1 and trpa1a/trpa1b are co-expressed in dorsal root ganglion neurons [37], suggesting a coordinated activation of these receptors. Further studies are necessary to investigate the role of capsaicin in zebrafish-based pain models.
In general, although trpv1, trpa1a, and trpa1b have changed through vertebrate evolution, they modulate a similar behavioral repertoire compared to mammals. Moreover, differences in temperature threshold (3 25°C for zebrafish and 3 42°C in mammals) are clearly associated with the adaptation of these organisms to their respective habitats. Finally, as many aspects of the zebrafish TRPA1 and TRPV1 orthologs remain unclear, more studies are necessary to elucidate the trpv1-, trpa1a-, and trpa1b- mediated nociception.
3.3. The Endocannabinoid System
The endocannabinoid system plays a key role in various biological processes, such as energy metabolism, inflammation, pain transmission, and synaptic neurotransmission [141]. In general, the endocannabinoid system is composed of cannabinoid receptors, endogenous cannabinoid ligands (anandamide and 2-arachidonoylglycerol), and enzymes involved in the synthesis or degradation of ligands [142]. Cannabinoids exert their pharmacological effects through the activation of at least two distinct cannabinoid receptors, CB1 and CB2, although mounting evidence supports the existence of other receptors, such as GPR55 [143]. Both CB1 and CB2 receptors are presynaptic Gi protein-coupled receptors with seven transmembrane domains that are differentially distributed in the CNS and peripheral tissues [144]. While CB1 receptor is highly expressed in the CNS (substantia nigra, globus pallidus, hippocampus, and cerebellum, with little expression in the brainstem) [145], the CB2 receptor is mainly located in immune cells peripherally [146], although it is also expressed in brain neurons (cerebellum, cortex, hippocampus, thalamus and hypothalamus, and ventral tegmental area) [147, 148]. Activation of CB1 receptors causes a robust effect on cognition, reward, and anxiety [149, 150]. On the other hand, the activation of CB2 receptors leads to immunosuppression, which limits inflammation and associated tissue damage [151]. The endocannabinoid system also modulates nociceptive responses. For example, in the peripheral system, CB1 receptors are expressed mainly in the nerve terminals and control neuronal responses [152]. Moreover, CB1 receptors are also expressed in both large and small-diameter fibers (e.g., C-fibers) and inhibit the release of neurotransmitters involved in pain transmission [153-155]. Likewise, peripheral CB2 receptors reduce the release of pronociceptive molecules from immune cells and keratinocytes [152]. Additionally, cannabinoids also activate pain receptors, such as TRPV1, suggesting the use of such molecules as potential analgesics [156].
The endocannabinoid system of zebrafish is evolutionarily conserved. Basically, zebrafish possess the same receptors (cb1 and cb2 receptors share ~ 73% and 39% of genetic similarity compared to the human orthologs, respectively) [157, 158], ligands, and metabolic enzymes [159]. However, the zebrafish genome encodes two cb2 receptor genes: cb2a and cb2b (sharing a sequence identity of 98% between them) [158]. In zebrafish, cb1 mRNA has been detected in whole-body lysates by quantitative real-time reverse transcriptase polymerase chain reactions (qRT-PCRs) as early as the 3-somite stage, with the transcript exhibiting an increase through the first 15 days post-fertilization [160]. The zebrafish cb1 receptors are found in the telencephalon, hypothalamus, tegmentum, and anterior hindbrain by 2 dpf, and their expression remains similar into adulthood, paralleling the mammalian counterpart [157]. On the other hand, cb2a/b are abundant in immune system cells and their activation shows anti-inflammatory effects [158]. Moreover, cb2a/b mRNA has been detected by qRT-PCR in adult zebrafish brain, intestine, retina, gills, heart, pituitary, and spleen [158]. Although both zebrafish cb2a and cb2b do not have an amino acid residue responsible for ligand affinity to WIN55212-2 (Leu175 replaces Phe197 in zebrafish cb2a/b) [161], this radiolabeled synthetic cannabinoid (sCB) can bind to its targets in the hypothalamus, optic tectum, and telencephalon in adult zebrafish brain slices [162]. Similarly, binding assays demonstrate that the endocannabinoid (anandamide) and sCB (HU-210, WIN55212-2, and CP55940) interact with receptors in adult brain homogenates [158]. Cannabinoids also show analgesic properties, since cannabidiol, a phytocannabinoid, prevents hypolocomotion induced by acetic acid in zebrafish larvae [163].
Finally, since cb1 receptors are localized in pain-processing areas of the brain and spinal cord, and their exogenous stimulation promotes antinociception [164], zebrafish could be a powerful tool to access the interaction between opioids and cannabinoid receptors [165]. For example, low doses of cannabinoid prevented antinociceptive tolerance to morphine in animal models of neuropathic pain [166]. Additionally, pretreatment with a non-analgesic dose of tetrahydrocannabinol evokes up to a 22-fold increase in morphine-induced analgesia [167], even though the analgesic effects of cannabinoid products are relatively low [154]. Notably, CB2 receptors can also mediate antinociception and may be promising targets for pain therapy due to their relatively lower CNS expression, which can result in reduced psychotropic effects [164]. Overall, although more studies are necessary, the endocannabinoid system could be a relevant molecular pathway to study pain processes in zebrafish.
3.4. zASIC
Acid-sensing ion channels (ASICs) are excitatory receptors for extracellular H+ [168]. Their functions include peripheral perception of pain, synaptic transmission, and mechanosensation [169]. In mammals, there are four ASICs genes encoding six subunits (ASIC1a [170], ASIC1b [171], ASIC2a [172], ASIC2b [173], ASIC3 [174], and ASIC4 [175]), which act as homo- or hetero-oligomeric assemblies of individual subunits [176, 177]. In humans, ASICs are widely distributed in both central and peripheral nervous systems, but have also been detected in non-neuronal tissues, including testis (ASIC3) [178], pituitary gland (ASIC4) [179], lung epithelial cells (ASIC3) [180], and bone (ASIC1-3) [181]. In the peripheral nervous system, ASIC channels are found predominantly in small-diameter sensory neurons involved in pain [171].
In zebrafish, six ASICs have already been identified (asic1a, asic1b, asic1c, asic2, asic4a, and asic4b) [182]. Phylogenetic analyses show that asic1a, asic2, and asic4a are orthologous to mammals, whereas asic1b, asic1c, and asic4b are splice variants [182]. Although, zebrafish ASICs are broadly expressed in the CNS (starting between 24 and 48 hours post fertilization), only asic1a is expressed in the trigeminal ganglion (region responsible for carrying somatosensory information from the head) [182]. Both asic1a and asic1c are expressed in sensory neurons and may contribute to mechanical stimuli [182]. Although both asic4a and asic4b share ~68% of genetic similarity [171], only asic4a is gated by extracellular H+ (pH ~5.8) [182]. Importantly, understanding how different ASICs expressed in zebrafish contribute to specific behavioral responses still requires further scrutiny, and the use of specific transgenic lines or mutants can provide relevant findings in this field.
4. COMPARISON OF THE EXISTING ADULT ZEBRAFISH-BASED PAIN MODELS
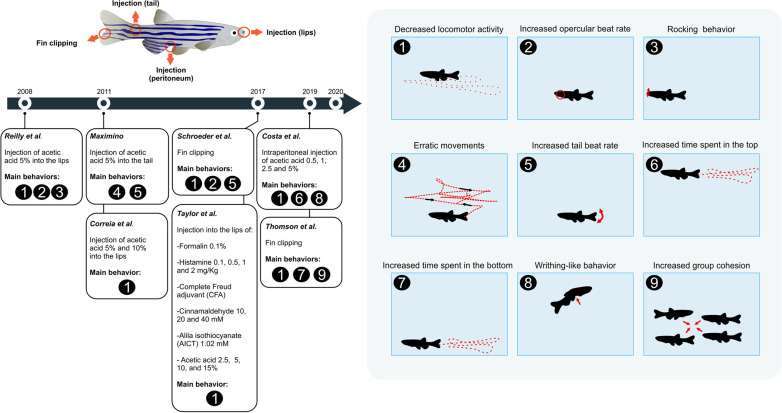
Animal experimental models are essential to characterize the functionality of distinct classes of nociceptors across taxa [51]. Mounting evidence supports that zebrafish express the necessary cellular components to recognize nociceptive agents and display aberrant behaviors in responses to algogens [13, 42, 52], also see Fig. (2) describing major methodological approaches and behavioral phenotypes of adult zebrafish related to pain responses. Because pain perception in animal models is subjective, the recognition of pain-related phenotypes is a cornerstone for understanding how noxious stimuli influence animal physiology and behavior [183, 184]. Here, we summarize the main experimental approaches to access pain behavior in zebrafish.
Fig. (2).
Common experimental protocols used to assess pain in adult zebrafish with the respective behavioral endpoints measured in different studies depicted in timeline perspective. References: Reilly et al. [42], Maximino [52], Correia et al. [196], Schroeder et al. [185], Taylor et al. [33], Costa et al. [34], Thomsom et al. [35].
4.1. Fin Clipping
The fin clipping is a surgical procedure that removes the caudal posterior tissue, injuring nociceptive fibers and triggering pain-like responses [185]. The fin clip procedure increases the opercular- and tail-beating, the bottom-dwelling, as well as decreases locomotion [35, 87]. These pain responses are entirely prevented after lidocaine or morphine administration [35, 87, 185], supporting a robust nociceptive response in zebrafish.
4.2. Cinnamaldehyde
Widely used in murine models, cinnamaldehyde has been recently used in zebrafish as a nociceptive agent [33]. This TRPA1 agonist acts differently on zebrafish trpa1a and trpa1b receptors [127]. While trpa1a is desensitized in two applications of cinnamaldehyde (even at low concentrations), trpa1b shows uniform currents after cinnamaldehyde applications [127]. Notably, cinnamaldehyde administered into the lips (40 mM) or the tail (0.003 mM) causes hypolocomotion in zebrafish [33], supporting a well-conserved biological response even when administered in different routes and doses.
4.3. Allyl Isothiocyanate
Another TRPA1 agonist used to induce pain in zebrafish is allyl isothiocyanate (AITC; commonly known as mustard oil). While AITC consistently increases Ca2+ concentration in both trpa1a and trpa1b, this response is prevented by a mammalian TRPA1 antagonist (ruthenium red), suggesting that both zebrafish TRPA1 paralogs can be activated by AITC [37]. Furthermore, a single injection of AITC (1.02 M) causes a remarkable hypolocomotion in adult zebrafish [33]. Although a reduction in distance traveled has been commonly described as pain behavior in zebrafish, there are no data showing a preventive effect of analgesics in this model, therefore, further scrutiny is required.
4.4. Complete Freund's Adjuvant
Commonly used to study chronic pain in preclinical research [186], the Complete Freund's Adjuvant (CFA) is composed of inactivated mycobacteria in which are recognized by immune cells, through Toll-like receptor 4 (TRL4) [187], leading to persistent inflammation and pain-like behaviors. To date, there is a single report using CFA to access pain-like behavior in zebrafish [33]. Like in mammals, a single CFA intraperitoneal administration causes a persistent inflammation in zebrafish, starting 24 hours post-injection and persisting for at least 48 hours. CFA injection results in decreased swimming activity, which can be prevented by morphine [33]. Because depression and chronic pain often correlate in mammals [188], assessing whether the CFA administration serves as a tool to access both pain- and comorbid depression-like responses in zebrafish could be interesting in the future.
4.5. Histamine
Histamine is involved in several regulatory mechanisms, including allergic reactions and pain [189]. In general, the zebrafish histaminergic system resembles that of other vertebrates [190]. In adult zebrafish, histamine (1 and 2 mg/kg) injected into the lips causes hypolocomotion [33], similar to what occurs in zebrafish larvae [191]. Although the histaminergic system in zebrafish is well described, relatively little is known about its role in zebrafish pain responses.
4.6. Formalin Test
Unlike other approaches, the formalin test is characterized by two different stages of pain-like responses [192]. In mammals, the first stage (neurogenic) begins immediately after the injection lasting for 3-5 min, triggered by chemical stimulation of nociceptors (C fibers). The second stage (inflammatory) begins at 15-20 min and lasts for ~20-40 min, as multiple pro-inflammatory mediators are released (e.g., histamine, prostaglandins, and serotonin) [193]. Formalin (0.1%) injected into the tail impairs the locomotion of adult zebrafish in both neurogenic (0-5min) and inflammatory (15-30 min) stages, which are prevented by analgesics (morphine and indomethacin) [32]. Although formalin is a TRPA1 agonist in mammals, there is no evidence that formalin activates the same receptor in zebrafish.
4.7. Acetic Acid
Widely used in rodents to access pain-like phenotypes [194, 195], the acetic acid administration in zebrafish evokes robust behavioral responses. For example, acid acetic injected into the lips (2.5, 5, 10, and 15%) reduces locomotion, increases the opercular beat rate, and promotes 'rocking' behavior (Fig. 2, moving side to side) in zebrafish, as well as causes them to rub their lips against the walls or gravel [33, 42, 196]. When administered near the adipose fin, acetic acid (1%) increases both erratic movements and tail beating [52]. Moreover, zebrafish display a writhing-like behavior following a single acetic acid (2.5 and 5%) intraperitoneal injection, analogous to writhing response in rodents [34, 197]. Importantly, all behaviors described above can be prevented by clinical analgesics, supporting their pharmacological validity to measure pain-like responses.
Indeed, zebrafish-based pain models are becoming promising strategies as first-choice tools for drug screening. However, future experiments aiming to improve the use of zebrafish in translational pain research are needed. For example, there are no methodological approaches to access allodynia, as well as neuropathic pain in zebrafish so far. These models help validate the use of zebrafish as a potential organism to test novel analgesic drugs, fostering the creation of more sensitive, complex, and objective tools in preclinical research, complementing the existent rodent approaches.
5. PAIN-RELATED MOOD DISORDERS
Chronic pain is a global problem that affects millions of people and is linked to significant morbidity, limited mobility, and social isolation [198, 199]. Moreover, chronic pain is also associated to different emotional disorders, including anxiety and depression [200-202]. Considering the complexity of these emotions, anxiety-related disorders and major depression overlap in symptoms and comorbidity, including cognitive, affective, physical, and behavioral deficits [203, 204]. Additionally, anxiety-related disorders and depression are associated with increased pain perception [205]. In humans, pharmacological agents can be used for treating both chronic pain and emotional disorders [206, 207], whereas animal models serve as important tools to understand the evolutionarily conserved mechanisms underlying these conditions [208, 209]. Evidence shows that neurotransmitters systems (e.g., dopaminergic and serotonergic systems) are involved in CNS disorders and pain-regulatory processes [210, 211]. In addition to traditional rodent models, zebrafish serve as a promising tool to investigate the molecular basis of pain and emotional disorders due to high genetic homology and neurochemical conservation when compared to humans [212, 213]. Furthermore, several behavioral paradigms have been validated in zebrafish to assess anxiety- and depression-like phenotypes (e.g., the novel tank and the light-dark tests), as well as pain behavior [52]. Importantly, although changes in the mean distance traveled or altered swim velocity may indicate pain-like conditions, these endpoints may also reflect other behaviors in zebrafish (e.g., anxiety-, non-specific sedation- and/or depressive-like states) [214]. Indeed, locomotor-related parameters represent an overall state of “well-being” and should not be merely described as unique measures of nociception [34]. In line with this, locomotor activity should be tested in the presence of analgesics, aiming to improve predictive validity. Using complementary behavioral endpoints to assess pain responses in adult zebrafish more accurately should also be considered.
Zebrafish is sensitive to various anxiolytics and anti-pain medications [34, 215] and present well-characterized behaviors (e.g., relatively complex swimming activity, as well as high social interaction). Pharmacological experiments revealed that acute morphine (2 mg/L) exposure for 20 min promotes anxiolytic-like behaviors, while 5 mg/L naloxone increases anxiety-like responses in the novel tank diving test [86]. Altogether, these data implicate opioids in the modulation of anxiety-like behaviors, reinforcing the existence of useful behavioral repertoires in zebrafish for modeling emotional disorders and pain-related phenotypes [212, 214].
6. ADVANTAGES AND LIMITATIONS OF ZEBRAFISH MODELS IN ANTI-PAIN MEDICATION SCREENING
6.1. Comparison between Larvae and Adult Zebrafish
To understand the molecular and physiological mechanisms underlying nociception in vertebrates, both larvae and adult zebrafish have become tractable systems in translational pain research. For example, adult fish show reduced activity and swimming behavior in response to a range of potentially painful laboratory procedures that are ameliorated by several drugs with analgesic properties, including local anaesthetics, non-steroidal anti-inflammatory drugs, and opioids [35, 87]. Studies utilizing < 5 dpf larvae have demonstrated the same results with a reduction in activity with selected drugs, preventing painful responses [89]. These findings confirm the validity of replacing adult zebrafish with younger animals. Moreover, a large number of larvae can be assessed simultaneously in a high-throughput manner (25-96 in well plates) compared to multiple adults being assessed at once. Furthermore, transparent larvae can be imaged in real-time to investigate CNS changes in relation to innocuous and potentially painful stimuli and whether anaesthetic or analgesic drugs can reduce or prevent pain-related brain activity. Although zebrafish larvae have less sophisticated behaviors than adults and a CNS yet to be fully matured [216], their simple and transparent brain allows for the investigation of the brain circuitry involved in pain responses during early developmental stages. The precise CNS processing of painful stimuli in zebrafish is yet to be elucidated, but their utility can help target specific brain areas involved in mammalian pain processing and assist in the search for novel analgesic compounds. However, caution should be applied to the use of < 5 dpf larvae in terms of the ethics of animal use. The 3Rs (Reduction, Replacement, Refinement) and ethical review should apply to all animal studies, especially regarding the number of animals used in experiments [217]. Researchers should use the minimum number of larvae to achieve the research objectives, and should also apply refinement by using the least invasive techniques and safeguarding welfare.
6.2. Limitations
Zebrafish models possess some limitations in the translatability of their data due to certain differences from mammals, such as in metabolic physiology (cold-blooded fish vs. warm-blooded mammals), brain development, and anatomy (e.g., zebrafish lack cortex) [218], In addition, zebrafish also present some limitations in neuroscience research. For example, the genome duplication event in teleost fishes [219] complicates genetic analyses of pain responses since some pain-related genes may exist in two copies or be missing in zebrafish. Because of its small size, the long-term monitoring of endocrine levels from blood (e.g., glucose levels) is also problematic as it is difficult to obtain a sufficient amount of blood without euthanizing the animal [220].
6.3. Advantages
Zebrafish models have been used to study CNS pharmacological modulation by conventional and non-conventional therapies (e.g., Traditional Chinese Medicine) [221], reinforcing its potential for the development of pharmacological and non-pharmacological therapies for CNS disorders [222] and pain. Another advantage in zebrafish compared to other animal models (e.g., rodents) in pharmacological screening is its high performance. For instance, using zebrafish larvae, one may test over 10000 drugs behaviorally in a single study [223], which may help understand behavioral effects of analgesic therapies, molecular mechanisms, and neural circuits involved in pain response. Zebrafish present a high degree (~70%) of genetic homology when compared to humans [224], as well as offer several CNS genetic models [225], which reinforce this aquatic model as a powerful species to explore genetic modulation of pain and related disorders. Importantly, zebrafish have sophisticated behaviors, which can be easily assessed using automated video-tracking systems to perform 3D reconstructions of the swimming traces in adult specimens [214]. Although the development of novel automated systems to measure behavioral activities simultaneously in a group of subjects is necessary, the use of existing video-tracking technologies may increase the efficiency and speed of time-intensive manual coding to measure pain-related responses in zebrafish. This strategy not only minimizes human bias, but also allows an in-depth investigation of specific behavioral responses in fish treated with different algogens in a high-throughput and reliable manner.
CONCLUSION
In conclusion, zebrafish have been considered an emergent tool to investigate the neurobehavioral basis of pain due to their numerous practical advantages over traditional (rodent) models. This species is space-efficient and enables easy experimental manipulations with a relatively lower cost of maintenance [23]. Some features such as the high reproduction rate, external fertilization, transparency of embryos, and rapid development [226], may foster studying epigenetic mechanisms of pain and probing how painful stimuli during early development affect fish as adults. Multiple transgenic lines created by gene-editing (e.g., CRISPR-Cas9 or transcription activator-like effector nucleases/TALENS) methods have been generated in zebrafish [227, 228]. For example, the expression of mutant SCN9A, a voltage-gated sodium channel, represents a model to study small-fiber neuropathy and potential analgesic properties of novel compounds in zebrafish [229]. Moreover, zebrafish show high plasticity of CNS [230]. Prominent cell proliferation can occur in different CNS regions of zebrafish, such as the telencephalon and the spinal cord [231, 232], helping to elucidate how adult neurogenesis modulates pain responses, learning, and emotional behaviors, complementing the existing rodent approaches. Finally, the existence of relatively complex behavioral responses [214], as well as multiple strain-, sex- and individual differences in zebrafish, offers further important practical and conceptual advantages to elucidate the link between pain and affective disorders in this aquatic species.
Zebrafish also represent an important model organism in the discovery of novel treatments and therapeutics for pain [124, 134]. However, our present understanding of zebrafish pain-related phenotypes and their molecular mechanisms remains limited (Table 1). One key question to be solved is the precise CNS processing of ascending nociceptive inputs and descending control of pain. This knowledge helps target specific brain areas in the pursuit of discovering new means of reducing pain in biomedical and veterinary studies. Replacing adult zebrafish with young larval forms can increase the throughput of experiments and enhance the rapid testing of pharmacological agents. This could lead to greater translatability of drug efficacy from laboratory models to clinical testing. The use of artificial intelligence monitoring systems is growing in the behavioral analysis of animals [233, 234]. Not only does this provide a means of automatically measuring behaviors that cannot be done by the human observer but it also eliminates any human error or bias and can provide a more subtle means of measuring changes in pain behavior in response to treatment and application of pain-relieving drugs. Furthermore, to combat the reproducibility crisis that we are currently experiencing in the field, standard pain assessment protocols should be developed so that all laboratories are following the same methods. For experimental studies employing invasive techniques where the pain is not the objective of that study, the development of pain management protocols for zebrafish would be a major step forward in the refinement of zebrafish use. Importantly, animal models are suitable tools to investigate the neurobehavioral mechanisms of pain, aiming to develop safer and more effective pharmacological treatments. Mounting evidence which is only briefly discussed here implicates the use of non-traditional zebrafish models as a fruitful strategy to generate important translational insights into molecular, physiological, and neurobehavioral mechanisms of pain.
Table 1.
Selected open questions in zebrafish-based pain research.
| Questions |
|---|
| · Can zebrafish present a relatively complex central processing associated with emotional responses of pain? |
| · Which areas of the zebrafish CNS are involved in processing painful stimuli? |
| · How important is the descending control of pain, given the empirical results for stress induced analgesia in zebrafish? |
| · Given the evidence for pain in zebrafish should we implement pain management protocols in the laboratory employing invasive techniques where pain is not the objective? |
| · Can we develop standard protocols for investigating pain in zebrafish to increase reproducibility within and between laboratories? |
| · When using non-protected or non-regulated larval zebrafish (typically <5dpf) ethical review and implementation of 3Rs frameworks should be considered as important tools to ensure humane treatment of the experimental fish. What is the best strategy to implement this aspect practically? |
| · How much the environment (e.g., pH, temperature, salinity) can influence in zebrafish pain response? |
| · How much can stress (e.g., experimental manipulation) influence in zebrafish pain response? |
| · What is the most appropriate behavioral test to assess zebrafish pain phenotypes? |
| · How to distinguish between a stress and a painful behavioral response in zebrafish? |
| · How translational to human is the zebrafish pharmacological and non-pharmacological analgesic response? |
| · Are there differences across strain and age in zebrafish pain response? |
ACKNOWLEDGEMENTS
The authors thank the financial support from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)-Finance Code 001, and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS). AVK collaboration is supported by Sirius University, Sochi, Russia.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
F.V.C., V.A.Q., and L.C.R. received CAPES fellowship. D.B.R. and A.R.S. are recipients of CNPq research productivity grant. D.B.R. research is also supported by PROEX/CAPES (process number 23038.005450/2020-19) and Programa PQ-Gaúcho FAPERGS (process number 19/2551-0001764-2) fellowship grants. A.V.K. is the Chair of the International Zebrafish Neuroscience Research Consortium (ZNRC). His research is supported by the Russian Science Foundation (RSF) grant 20-65-46006. L.U.S. is convenor of the FELASA working group producing a report on Pain Management in Zebrafish and is a member of the NC3Rs (UK) expert panel on zebrafish welfare. The funders did not influence writing and submission of this manuscript.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Garcia-Larrea L., Bastuji H. Pain and consciousness. Prog. Neuropsychopharm. Biol. Psychiat. 2018;87(Pt B):193–199. doi: 10.1016/j.pnpbp.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Mogil J.S., Davis K.D., Derbyshire S.W. The necessity of animal models in pain research. Pain. 2010;151(1):12–17. doi: 10.1016/j.pain.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 3.Yekkirala A.S., Roberson D.P., Bean B.P., Woolf C.J. Breaking barriers to novel analgesic drug development. Nat. Rev. Drug Discov. 2017;16(8):545–564. doi: 10.1038/nrd.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piel M.J., Kroin J.S., van Wijnen A.J., Kc R., Im H.J. Pain assessment in animal models of osteoarthritis. Gene. 2014;537(2):184–188. doi: 10.1016/j.gene.2013.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackburn-Munro G. Pain-like behaviours in animals-how human are they? Trends Pharmacol. Sci. 2004;25(6):299–305. doi: 10.1016/j.tips.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 6.Tracey I., Mantyh P.W. The cerebral signature for pain perception and its modulation. Neuron. 2007;55(3):377–391. doi: 10.1016/j.neuron.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Gregory N.S., Harris A.L., Robinson C.R., Dougherty P.M., Fuchs P.N., Sluka K.A. An overview of animal models of pain: disease models and outcome measures. J. Pain. 2013;14(11):1255–1269. doi: 10.1016/j.jpain.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobinski F., Teixeira J.M., Sluka K.A., Santos A.R.S. Interleukin-4 mediates the analgesia produced by low-intensity exercise in mice with neuropathic pain. Pain. 2018;159(3):437–450. doi: 10.1097/j.pain.0000000000001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martins D.F., Martins T.C., Batisti A.P., Dos Santos Leonel L., Bobinski F., Belmonte L.A.O., Mazzardo-Martins L., Cargnin-Ferreira E., Santos A.R.S. Long-Term Regular Eccentric Exercise Decreases Neuropathic Pain-like Behavior and Improves Motor Functional Recovery in an Axonotmesis Mouse Model: the Role of Insulin-like Growth Factor-1. Mol. Neurobiol. 2018;55(7):6155–6168. doi: 10.1007/s12035-017-0829-3. [DOI] [PubMed] [Google Scholar]
- 10.Basbaum A.I., Bautista D.M., Scherrer G., Julius D. Cellular and molecular mechanisms of pain. Cell. 2009;139(2):267–284. doi: 10.1016/j.cell.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coderre T.J., Melzack R. The contribution of excitatory amino acids to central sensitization and persistent nociception after formalin-induced tissue injury. J. Neurosci. 1992;12(9):3665–3670. doi: 10.1523/JNEUROSCI.12-09-03665.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woolf C.J. What is this thing called pain? J. Clin. Invest. 2010;120(11):3742–3744. doi: 10.1172/JCI45178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malafoglia V., Bryant B., Raffaeli W., Giordano A., Bellipanni G. The zebrafish as a model for nociception studies. J. Cell. Physiol. 2013;228(10):1956–1966. doi: 10.1002/jcp.24379. [DOI] [PubMed] [Google Scholar]
- 14.Meotti F.C., Coelho I dos S., Santos A.R. The nociception induced by glutamate in mice is potentiated by protons released into the solution. J. Pain. 2010;11(6):570–578. doi: 10.1016/j.jpain.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Stevenson G.W., Bilsky E.J., Negus S.S. Targeting pain-suppressed behaviors in preclinical assays of pain and analgesia: effects of morphine on acetic acid-suppressed feeding in C57BL/6J mice. J. Pain. 2006;7(6):408–416. doi: 10.1016/j.jpain.2006.01.447. [DOI] [PubMed] [Google Scholar]
- 16.Sneddon L.U., Braithwaite V.A., Gentle M.J. Novel object test: examining nociception and fear in the rainbow trout. J. Pain. 2003;4(8):431–440. doi: 10.1067/S1526-5900(03)00717-X. [DOI] [PubMed] [Google Scholar]
- 17.Du X., Yuan B., Wang J., Zhang X., Tian L., Zhang T., Li X., Zhang F. Effect of heat-reinforcing needling on serum metabolite profiles in rheumatoid arthritis rabbits with cold syndrome. Zhongguo Zhenjiu. 2017;37(9):977–983. doi: 10.13703/j.0255-2930.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 18.MacRae C.A., Peterson R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015;14(10):721–731. doi: 10.1038/nrd4627. [DOI] [PubMed] [Google Scholar]
- 19.Stewart A.M., Braubach O., Spitsbergen J., Gerlai R., Kalueff A.V. Zebrafish models for translational neuroscience research: from tank to bedside. Trends Neurosci. 2014;37(5):264–278. doi: 10.1016/j.tins.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fontana B. D., Mezzomo N. J., Kalueff A. V., Rosemberg D. B. The developing utility of zebrafish models of neurological and neuropsychiatric disorders: A critical review. Exp Neurol. 2018;299(Pt A):157–171. doi: 10.1016/j.expneurol.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Kalueff A.V., Stewart A.M., Gerlai R. Zebrafish as an emerging model for studying complex brain disorders. Trends Pharmacol. Sci. 2014;35(2):63–75. doi: 10.1016/j.tips.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoo J.Y., Kumari Y., Shaikh M.F., Hue S.M., Goh B.H. Zebrafish: A Versatile Animal Model for Fertility Research. BioMed Res. Int. 2016;2016:9732780. doi: 10.1155/2016/9732780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Avdesh A., Chen M., Martin-Iverson M.T., Mondal A., Ong D., Rainey-Smith S., Taddei K., Lardelli M., Groth D.M., Verdile G., Martins R.N. Regular care and maintenance of a zebrafish (Danio rerio) laboratory: an introduction. J. Vis. Exp. 2012;(69):e4196. doi: 10.3791/4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart A.M., Gerlai R., Kalueff A.V. Developing highER-throughput zebrafish screens for in-vivo CNS drug discovery. Front. Behav. Neurosci. 2015;9:14. doi: 10.3389/fnbeh.2015.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kithcart A., MacRae C.A. Using zebrafish for high-throughput screening of novel cardiovascular Drugs. JACC Basic Transl. Sci. 2017;2(1):1–12. doi: 10.1016/j.jacbts.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X., Gays D., Santoro M.M. Transgenic zebrafish. Methods Mol. Biol. 2016;1464:107–114. doi: 10.1007/978-1-4939-3999-2_10. [DOI] [PubMed] [Google Scholar]
- 27.Kawakami K., Asakawa K., Hibi M., Itoh M., Muto A., Wada H. Gal4 driver transgenic zebrafish: powerful tools to study developmental biology, organogenesis, and neuroscience. Adv. Genet. 2016;95:65–87. doi: 10.1016/bs.adgen.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Rougeot J., Torraca V., Zakrzewska A., Kanwal Z., Jansen H.J., Sommer F., Spaink H.P., Meijer A.H. RNAseq profiling of leukocyte populations in zebrafish larvae reveals a cxcl11 chemokine gene as a marker of macrophage polarization during mycobacterial infection. Front. Immunol. 2019;10:832. doi: 10.3389/fimmu.2019.00832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu G.E., Park C.S., Cho H.J., Ha T.H., Bae J., Kwon O.S., Lee J.S., Lee C.S. Fluorescent polydopamine nanoparticles as a probe for zebrafish sensory hair cells targeted in vivo imaging. Sci. Rep. 2018;8(1):4393. doi: 10.1038/s41598-018-22828-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu C.X., Li C.Y., Hu C.C., Wang Y., Lin J., Jiang Y.H., Li Q., Xu X. CRISPR/Cas9-induced shank3b mutant zebrafish display autism-like behaviors. Mol. Autism. 2018;9:23. doi: 10.1186/s13229-018-0204-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Nunez V., Rodríguez R.E. The zebrafish: a model to study the endogenous mechanisms of pain. ILAR J. 2009;50(4):373–386. doi: 10.1093/ilar.50.4.373. [DOI] [PubMed] [Google Scholar]
- 32.Magalhães F.E.A., de Sousa C.Á.P.B., Santos S.A.A.R., Menezes R.B., Batista F.L.A., Abreu A.O., de Oliveira M.V., Moura L.F.W.G., Raposo R.D.S., Campos A.R. Adult zebrafish (Danio rerio): An alternative behavioral model of formalin-induced nociception. Zebrafish. 2017;14(5):422–429. doi: 10.1089/zeb.2017.1436. [DOI] [PubMed] [Google Scholar]
- 33.Taylor J.C., Dewberry L.S., Totsch S.K., Yessick L.R., DeBerry J.J., Watts S.A., Sorge R.E. A novel zebrafish-based model of nociception. Physiol. Behav. 2017;174:83–88. doi: 10.1016/j.physbeh.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Costa F.V., Rosa L.V., Quadros V.A., Santos A.R.S., Kalueff A.V., Rosemberg D.B. Understanding nociception-related phenotypes in adult zebrafish: Behavioral and pharmacological characterization using a new acetic acid model. Behav. Brain Res. 2019;359:570–578. doi: 10.1016/j.bbr.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 35.Thomson J.S., Al-Temeemy A.A., Isted H., Spencer J.W., Sneddon L.U. Assessment of behaviour in groups of zebrafish (Danio rerio) using an intelligent software monitoring tool, the chromatic fish analyser. J. Neurosci. Methods. 2019;328:108433. doi: 10.1016/j.jneumeth.2019.108433. [DOI] [PubMed] [Google Scholar]
- 36.Gau P., Poon J., Ufret-Vincenty C., Snelson C.D., Gordon S.E., Raible D.W., Dhaka A. The zebrafish ortholog of TRPV1 is required for heat-induced locomotion. J. Neurosci. 2013;33(12):5249–5260. doi: 10.1523/JNEUROSCI.5403-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prober D.A., Zimmerman S., Myers B.R., McDermott B.M., Jr, Kim S.H., Caron S., Rihel J., Solnica-Krezel L., Julius D., Hudspeth A.J., Schier A.F. Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function. J. Neurosci. 2008;28(40):10102–10110. doi: 10.1523/JNEUROSCI.2740-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dooley K., Zon L.I. Zebrafish: a model system for the study of human disease. Curr. Opin. Genet. Dev. 2000;10(3):252–256. doi: 10.1016/S0959-437X(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 39.Rose J.D. The Neurobehavioral Nature of Fishes and the Question of Awareness and Pain. Fish. Sci. 2002;10(1):1–38. [Google Scholar]
- 40.Braithwaite V.A., Boulcott P. Pain perception, aversion and fear in fish. Dis. Aquat. Organ. 2007;75(2):131–138. doi: 10.3354/dao075131. [DOI] [PubMed] [Google Scholar]
- 41.Nordgreen J., Horsberg T.E., Ranheim B., Chen A.C. Somatosensory evoked potentials in the telencephalon of Atlantic salmon (Salmo salar) following galvanic stimulation of the tail. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2007;193(12):1235–1242. doi: 10.1007/s00359-007-0283-1. [DOI] [PubMed] [Google Scholar]
- 42.Reilly S.C., Quinn J.P., Cossins A.R., Sneddon L.U. Behavioural analysis of a nociceptive event in fish: Comparisons between three species demonstrate specific responses. Appl. Anim. Behav. Sci. 2008;114:248–259. doi: 10.1016/j.applanim.2008.01.016. [DOI] [Google Scholar]
- 43.Sanchez-Simon F.M., Rodriguez R.E. Developmental expression and distribution of opioid receptors in zebrafish. Neuroscience. 2008;151(1):129–137. doi: 10.1016/j.neuroscience.2007.09.086. [DOI] [PubMed] [Google Scholar]
- 44.Alvarez F.A., Rodriguez-Martin I., Gonzalez-Nuñez V., Marrón Fernández de Velasco E., Gonzalez Sarmiento R., Rodríguez R.E. New kappa opioid receptor from zebrafish Danio rerio. Neurosci. Lett. 2006;405(1-2):94–99. doi: 10.1016/j.neulet.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 45.Barrallo A., González-Sarmiento R., Alvar F., Rodríguez R.E. ZFOR2, a new opioid receptor-like gene from the teleost zebrafish (Danio rerio). Brain Res. Mol. Brain Res. 2000;84(1-2):1–6. doi: 10.1016/S0169-328X(00)00152-2. [DOI] [PubMed] [Google Scholar]
- 46.Dymowska A.K., Boyle D., Schultz A.G., Goss G.G. The role of acid-sensing ion channels in epithelial Na+ uptake in adult zebrafish (Danio rerio). J. Exp. Biol. 2015;218(Pt 8):1244–1251. doi: 10.1242/jeb.113118. [DOI] [PubMed] [Google Scholar]
- 47.Viña E., Parisi V., Abbate F., Cabo R., Guerrera M.C., Laurà R., Quirós L.M., Pérez-Varela J.C., Cobo T., Germanà A., Vega J.A., García-Suárez O. Acid-sensing ion channel 2 (ASIC2) is selectively localized in the cilia of the non-sensory olfactory epithelium of adult zebrafish. Histochem. Cell Biol. 2015;143(1):59–68. doi: 10.1007/s00418-014-1264-4. [DOI] [PubMed] [Google Scholar]
- 48.van der Vaart M., Spaink H.P., Meijer A.H. Pathogen recognition and activation of the innate immune response in zebrafish. Adv. Hematol. 2012;2012:159807. doi: 10.1155/2012/159807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watzke J., Schirmer K., Scholz S. Bacterial lipopolysaccharides induce genes involved in the innate immune response in embryos of the zebrafish (Danio rerio). Fish Shellfish Immunol. 2007;23(4):901–905. doi: 10.1016/j.fsi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 50.Levanti M., Randazzo B., Viña E., Montalbano G., Garcia-Suarez O., Germanà A., Vega J.A., Abbate F. Acid-sensing ion channels and transient-receptor potential ion channels in zebrafish taste buds. Ann. Anat. 2016;207:32–37. doi: 10.1016/j.aanat.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 51.Sneddon L.U., Braithwaite V.A., Gentle M.J. Do fishes have nociceptors? Evidence for the evolution of a vertebrate sensory system. Proc. Biol. Sci. 2003;270(1520):1115–1121. doi: 10.1098/rspb.2003.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maximino C. Modulation of nociceptive-like behavior in zebrafish (Danio rerio) by environmental stressors. Psychol. Neurosci. 2011;4(1):149–155. doi: 10.3922/j.psns.2011.1.017. [DOI] [Google Scholar]
- 53.Caron S.J., Prober D., Choy M., Schier A.F. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development. 2008;135(19):3259–3269. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan Y.A., Choy M., Prober D.A., Schier A.F. Robo2 determines subtype-specific axonal projections of trigeminal sensory neurons. Development. 2012;139(3):591–600. doi: 10.1242/dev.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marron Fdez de Velasco E., Law P.Y., Rodríguez R.E. Mu opioid receptor from the zebrafish exhibits functional characteristics as those of mammalian mu opioid receptor. Zebrafish. 2009;6(3):259–268. doi: 10.1089/zeb.2009.0594. [DOI] [PubMed] [Google Scholar]
- 56.Chu Sin Chung P., Kieffer B.L. Delta opioid receptors in brain function and diseases. Pharmacol. Ther. 2013;140(1):112–120. doi: 10.1016/j.pharmthera.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rachinger-Adam B., Conzen P., Azad S.C. Pharmacology of peripheral opioid receptors. Curr. Opin. Anaesthesiol. 2011;24(4):408–413. doi: 10.1097/ACO.0b013e32834873e5. [DOI] [PubMed] [Google Scholar]
- 58.Corbett A.D., Henderson G., McKnight A.T., Paterson S.J. 75 years of opioid research: the exciting but vain quest for the Holy Grail. Br. J. Pharmacol. 2006;147(Suppl. 1):S153–S162. doi: 10.1038/sj.bjp.0706435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stevens C.W. The evolution of vertebrate opioid receptors. Front. Biosci. 2009;14:1247–1269. doi: 10.2741/3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valentino R.J., Volkow N.D. Untangling the complexity of opioid receptor function. Neuropsychopharmacology. 2018;43(13):2514–2520. doi: 10.1038/s41386-018-0225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lutz P.E., Kieffer B.L. Opioid receptors: distinct roles in mood disorders. Trends Neurosci. 2013;36(3):195–206. doi: 10.1016/j.tins.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loh H.H., Liu H.C., Cavalli A., Yang W., Chen Y.F., Wei L.N. mu Opioid receptor knockout in mice: effects on ligand-induced analgesia and morphine lethality. Brain Res. Mol. Brain Res. 1998;54(2):321–326. doi: 10.1016/S0169-328X(97)00353-7. [DOI] [PubMed] [Google Scholar]
- 63.Matthes H.W., Maldonado R., Simonin F., Valverde O., Slowe S., Kitchen I., Befort K., Dierich A., Le Meur M., Dollé P., Tzavara E., Hanoune J., Roques B.P., Kieffer B.L. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383(6603):819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 64.Ma J., Zhang Y., Kalyuzhny A.E., Pan Z.Z. Emergence of functional delta-opioid receptors induced by long-term treatment with morphine. Mol. Pharmacol. 2006;69(4):1137–1145. doi: 10.1124/mol.105.019109. [DOI] [PubMed] [Google Scholar]
- 65.Zhu Y., King M.A., Schuller A.G., Nitsche J.F., Reidl M., Elde R.P., Unterwald E., Pasternak G.W., Pintar J.E. Retention of supraspinal delta-like analgesia and loss of morphine tolerance in delta opioid receptor knockout mice. Neuron. 1999;24(1):243–252. doi: 10.1016/S0896-6273(00)80836-3. [DOI] [PubMed] [Google Scholar]
- 66.Lambert D.G. The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat. Rev. Drug Discov. 2008;7(8):694–710. doi: 10.1038/nrd2572. [DOI] [PubMed] [Google Scholar]
- 67.Sundström G., Dreborg S., Larhammar D. Concomitant duplications of opioid peptide and receptor genes before the origin of jawed vertebrates. PLoS One. 2010;5(5):e10512. doi: 10.1371/journal.pone.0010512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dreborg S., Sundström G., Larsson T.A., Larhammar D. Evolution of vertebrate opioid receptors. Proc. Natl. Acad. Sci. USA. 2008;105(40):15487–15492. doi: 10.1073/pnas.0805590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barrallo A., González-Sarmiento R., Porteros A., García-Isidoro M., Rodríguez R.E. Cloning, molecular characterization, and distribution of a gene homologous to delta opioid receptor from zebrafish (Danio rerio). Biochem. Biophys. Res. Commun. 1998;245(2):544–548. doi: 10.1006/bbrc.1998.8496. [DOI] [PubMed] [Google Scholar]
- 70.Pinal-Seoane N., Martin I.R., Gonzalez-Nuñez V., Marron Fernandez de Velasco E., Alvarez F.A., Sarmiento R.G., Rodriguez R.E. Characterization of a new duplicate delta-opioid receptor from zebrafish. J. Mol. Endocrinol. 2006;37(3):391–403. doi: 10.1677/jme.1.02136. [DOI] [PubMed] [Google Scholar]
- 71.Rivas-Boyero A.A., Herrero-Turrión M.J., Gonzalez-Nunez V., Sánchez-Simón F.M., Barreto-Valer K., Rodríguez R.E. Pharmacological characterization of a nociceptin receptor from zebrafish (Danio rerio). J. Mol. Endocrinol. 2011;46(2):111–123. doi: 10.1530/JME-10-0130. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez-Nunez V., Jimenez González A., Barreto-Valer K., Rodríguez R.E. In vivo regulation of the μ opioid receptor: role of the endogenous opioid agents. Mol. Med. 2013;19:7–17. doi: 10.2119/molmed.2012.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez-Nunez V., Gonzalez-Sarmiento R., Rodríguez R.E. Cloning and characterization of a full-length pronociceptin in zebrafish: evidence of the existence of two different nociceptin sequences in the same precursor. Biochim. Biophys. Acta. 2003;1629(1-3):114–118. doi: 10.1016/j.bbaexp.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Gonzalez-Nunez V., Gonzalez-Sarmiento R., Rodríguez R.E. Identification of two proopiomelanocortin genes in zebrafish (Danio rerio). Brain Res. Mol. Brain Res. 2003;120(1):1–8. doi: 10.1016/j.molbrainres.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 75.Wagle M., Mathur P., Guo S. Corticotropin-releasing factor critical for zebrafish camouflage behavior is regulated by light and sensitive to ethanol. J. Neurosci. 2011;31(1):214–224. doi: 10.1523/JNEUROSCI.3339-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Demin K.A., Taranov A.S., Ilyin N.P., Lakstygal A.M., Volgin A.D., de Abreu M.S., Strekalova T., Kalueff A.V. Understanding neurobehavioral effects of acute and chronic stress in zebrafish. Stress. 2021;24(1):1–18. doi: 10.1080/10253890.2020.1724948. [DOI] [PubMed] [Google Scholar]
- 77.Sheets L., Ransom D.G., Mellgren E.M., Johnson S.L., Schnapp B.J. Zebrafish melanophilin facilitates melanosome dispersion by regulating dynein. Curr. Biol. 2007;17(20):1721–1734. doi: 10.1016/j.cub.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gonzalez-Nuñez V., Marrón Fernández de Velasco E., Arsequell G., Valencia G., Rodríguez R.E. Identification of dynorphin a from zebrafish: a comparative study with mammalian dynorphin A. Neuroscience. 2007;144(2):675–684. doi: 10.1016/j.neuroscience.2006.09.028. [DOI] [PubMed] [Google Scholar]
- 79.Bao W., Volgin A.D., Alpyshov E.T., Friend A.J., Strekalova T.V., de Abreu M.S., Collins C., Amstislavskaya T.G., Demin K.A., Kalueff A.V. Opioid neurobiology, neurogenetics and neuropharmacology in zebrafish. Neuroscience. 2019;404:218–232. doi: 10.1016/j.neuroscience.2019.01.045. [DOI] [PubMed] [Google Scholar]
- 80.Demin K.A., Meshalkina D.A., Kysil E.V., Antonova K.A., Volgin A.D., Yakovlev O.A., Alekseeva P.A., Firuleva M.M., Lakstygal A.M., de Abreu M.S., Barcellos L.J.G., Bao W., Friend A.J., Amstislavskaya T.G., Rosemberg D.B., Musienko P.E., Song C., Kalueff A.V. Zebrafish models relevant to studying central opioid and endocannabinoid systems. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2018;86:301–312. doi: 10.1016/j.pnpbp.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 81.González-Núñez V., Barrallo A., Traynor J.R., Rodríguez R.E. Characterization of opioid-binding sites in zebrafish brain. J. Pharmacol. Exp. Ther. 2006;316(2):900–904. doi: 10.1124/jpet.105.093492. [DOI] [PubMed] [Google Scholar]
- 82.Chan H.C.S., McCarthy D., Li J., Palczewski K., Yuan S. Designing safer analgesics via μ-opioid receptor pathways. Trends Pharmacol. Sci. 2017;38(11):1016–1037. doi: 10.1016/j.tips.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sivalingam M., Ogawa S., Parhar I.S. Mapping of morphine-induced OPRM1 gene expression pattern in the adult zebrafish brain. Front. Neuroanat. 2020;14:5. doi: 10.3389/fnana.2020.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Belzung C., Agmo A. Naloxone potentiates the effects of subeffective doses of anxiolytic agents in mice. Eur. J. Pharmacol. 1997;323(2-3):133–136. doi: 10.1016/S0014-2999(97)00142-8. [DOI] [PubMed] [Google Scholar]
- 85.Kalin N.H., Shelton S.E., Barksdale C.M. Opiate modulation of separation-induced distress in non-human primates. Brain Res. 1988;440(2):285–292. doi: 10.1016/0006-8993(88)90997-3. [DOI] [PubMed] [Google Scholar]
- 86.Stewart A., Wu N., Cachat J., Hart P., Gaikwad S., Wong K., Utterback E., Gilder T., Kyzar E., Newman A., Carlos D., Chang K., Hook M., Rhymes C., Caffery M., Greenberg M., Zadina J., Kalueff A.V. Pharmacological modulation of anxiety-like phenotypes in adult zebrafish behavioral models. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35(6):1421–1431. doi: 10.1016/j.pnpbp.2010.11.035. [DOI] [PubMed] [Google Scholar]
- 87.Deakin A.G., Buckley J., AlZu’bi H.S., Cossins A.R., Spencer J.W., Al’Nuaimy W., Young I.S., Thomson J.S., Sneddon L.U. Automated monitoring of behaviour in zebrafish after invasive procedures. Sci. Rep. 2019;9(1):9042. doi: 10.1038/s41598-019-45464-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Magalhães F.E.A., Batista F.L.A., Lima L.M.G., Abrante I.A., Batista F.L.A., Abrante I.A., de Araújo J.I.F., Santos S.A.A.R., de Oliveira B.A., Raposo R.D.S., Campos A.R. Adult zebrafish (Danio rerio) as a model for the study of corneal antinociceptive compounds. Zebrafish. 2018;15(6):566–574. doi: 10.1089/zeb.2018.1633. [DOI] [PubMed] [Google Scholar]
- 89.Lopez-Luna J., Al-Jubouri Q., Al-Nuaimy W., Sneddon L.U. Reduction in activity by noxious chemical stimulation is ameliorated by immersion in analgesic drugs in zebrafish. J. Exp. Biol. 2017;220(Pt 8):1451–1458. doi: 10.1242/jeb.146969. [DOI] [PubMed] [Google Scholar]
- 90.Khor B.S., Jamil M.F., Adenan M.I., Shu-Chien A.C. Mitragynine attenuates withdrawal syndrome in morphine-withdrawn zebrafish. PLoS One. 2011;6(12):e28340. doi: 10.1371/journal.pone.0028340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cachat J., Canavello P., Elegante M., Bartels B., Hart P., Bergner C., Egan R., Duncan A., Tien D., Chung A., Wong K., Goodspeed J., Tan J., Grimes C., Elkhayat S., Suciu C., Rosenberg M., Chung K.M., Kadri F., Roy S., Gaikwad S., Stewart A., Zapolsky I., Gilder T., Mohnot S., Beeson E., Amri H., Zukowska Z., Soignier R.D., Kalueff A.V. Modeling withdrawal syndrome in zebrafish. Behav. Brain Res. 2010;208(2):371–376. doi: 10.1016/j.bbr.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 92.Shi Y., Zhang Y., Zhao F., Ruan H., Huang H., Luo L., Li L. Acetylcholine serves as a derepressor in Loperamide-induced Opioid-Induced Bowel Dysfunction (OIBD) in zebrafish. Sci. Rep. 2014;4:5602. doi: 10.1038/srep05602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bretaud S., Li Q., Lockwood B.L., Kobayashi K., Lin E., Guo S. A choice behavior for morphine reveals experience-dependent drug preference and underlying neural substrates in developing larval zebrafish. Neuroscience. 2007;146(3):1109–1116. doi: 10.1016/j.neuroscience.2006.12.073. [DOI] [PubMed] [Google Scholar]
- 94.Bhargava H.N., Thorat S.N. Effect of dizocilpine (MK-801) on analgesia and tolerance induced by U-50,488H, a kappa-opioid receptor agonist, in the mouse. Brain Res. 1994;649(1-2):111–116. doi: 10.1016/0006-8993(94)91053-7. [DOI] [PubMed] [Google Scholar]
- 95.Yamada H., Shimoyama N., Sora I., Uhl G.R., Fukuda Y., Moriya H., Shimoyama M. Morphine can produce analgesia via spinal kappa opioid receptors in the absence of mu opioid receptors. Brain Res. 2006;1083(1):61–69. doi: 10.1016/j.brainres.2006.01.095. [DOI] [PubMed] [Google Scholar]
- 96.McDonald J., Lambert D. Opioid receptors. Contin. Educ. Anaesth. Crit. Care Pain. 2005;5(1):22–25. doi: 10.1093/bjaceaccp/mki004. [DOI] [Google Scholar]
- 97.Liu-Chen L.Y. Agonist-induced regulation and trafficking of kappa opioid receptors. Life Sci. 2004;75(5):511–536. doi: 10.1016/j.lfs.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 98.Braida D., Limonta V., Pegorini S., Zani A., Guerini-Rocco C., Gori E., Sala M. Hallucinatory and rewarding effect of salvinorin A in zebrafish: kappa-opioid and CB1-cannabinoid receptor involvement. Psychopharmacology (Berl.) 2007;190(4):441–448. doi: 10.1007/s00213-006-0639-1. [DOI] [PubMed] [Google Scholar]
- 99.Gonzalez-Nuñez V., Toth G., Rodríguez R.E. Endogenous heptapeptide Met-enkephalin-Gly-Tyr binds differentially to duplicate delta opioid receptors from zebrafish. Peptides. 2007;28(12):2340–2347. doi: 10.1016/j.peptides.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 100.Mansour A., Hoversten M.T., Taylor L.P., Watson S.J., Akil H. The cloned mu, delta and kappa receptors and their endogenous ligands: evidence for two opioid peptide recognition cores. Brain Res. 1995;700(1-2):89–98. doi: 10.1016/0006-8993(95)00928-J. [DOI] [PubMed] [Google Scholar]
- 101.Kieffer B.L., Gavériaux-Ruff C. Exploring the opioid system by gene knockout. Prog. Neurobiol. 2002;66(5):285–306. doi: 10.1016/S0301-0082(02)00008-4. [DOI] [PubMed] [Google Scholar]
- 102.Gavériaux-Ruff C., Kieffer B.L. Delta opioid receptor analgesia: recent contributions from pharmacology and molecular approaches. Behav. Pharmacol. 2011;22(5-6):405–414. doi: 10.1097/FBP.0b013e32834a1f2c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goldenberg D.L. The interface of pain and mood disturbances in the rheumatic diseases. Semin. Arthritis Rheum. 2010;40(1):15–31. doi: 10.1016/j.semarthrit.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 104.Sharif N.A., Hughes J. Discrete mapping of brain Mu and delta opioid receptors using selective peptides: quantitative autoradiography, species differences and comparison with kappa receptors. Peptides. 1989;10(3):499–522. doi: 10.1016/0196-9781(89)90135-6. [DOI] [PubMed] [Google Scholar]
- 105.Porteros A., García-Isidoro M., Barrallo A., González-Sarmiento R., Rodríguez R.E. Expression of ZFOR1, a delta-opioid receptor, in the central nervous system of the zebrafish (Danio rerio). J. Comp. Neurol. 1999;412(3):429–438. doi: 10.1002/(SICI)1096-9861(19990927)412:3<429:AID-CNE4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 106.Valiquette M., Vu H.K., Yue S.Y., Wahlestedt C., Walker P. Involvement of Trp-284, Val-296, and Val-297 of the human delta-opioid receptor in binding of delta-selective ligands. J. Biol. Chem. 1996;271(31):18789–18796. doi: 10.1074/jbc.271.31.18789. [DOI] [PubMed] [Google Scholar]
- 107.Meng F., Ueda Y., Hoversten M.T., Thompson R.C., Taylor L., Watson S.J., Akil H. Mapping the receptor domains critical for the binding selectivity of delta-opioid receptor ligands. Eur. J. Pharmacol. 1996;311(2-3):285–292. doi: 10.1016/0014-2999(96)00431-1. [DOI] [PubMed] [Google Scholar]
- 108.Rodriguez R.E., Barrallo A., Garcia-Malvar F., McFadyen I.J., Gonzalez-Sarmiento R., Traynor J.R. Characterization of ZFOR1, a putative delta-opioid receptor from the teleost zebrafish (Danio rerio). Neurosci. Lett. 2000;288(3):207–210. doi: 10.1016/S0304-3940(00)01239-8. [DOI] [PubMed] [Google Scholar]
- 109.Mollereau C., Mouledous L. Tissue distribution of the opioid receptor-like (ORL1) receptor. Peptides. 2000;21(7):907–917. doi: 10.1016/S0196-9781(00)00227-8. [DOI] [PubMed] [Google Scholar]
- 110.King M.A., Rossi G.C., Chang A.H., Williams L., Pasternak G.W. Spinal analgesic activity of orphanin FQ/nociceptin and its fragments. Neurosci. Lett. 1997;223(2):113–116. doi: 10.1016/S0304-3940(97)13414-0. [DOI] [PubMed] [Google Scholar]
- 111.Ko M.C., Wei H., Woods J.H., Kennedy R.T. Effects of intrathecally administered nociceptin/orphanin FQ in monkeys: behavioral and mass spectrometric studies. J. Pharmacol. Exp. Ther. 2006;318(3):1257–1264. doi: 10.1124/jpet.106.106120. [DOI] [PubMed] [Google Scholar]
- 112.Zeilhofer H.U., Calò G. Nociceptin/orphanin FQ and its receptor-potential targets for pain therapy? J. Pharmacol. Exp. Ther. 2003;306(2):423–429. doi: 10.1124/jpet.102.046979. [DOI] [PubMed] [Google Scholar]
- 113.Pan Z., Hirakawa N., Fields H.L. A cellular mechanism for the bidirectional pain-modulating actions of orphanin FQ/nociceptin. Neuron. 2000;26(2):515–522. doi: 10.1016/S0896-6273(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 114.Julius D. TRP channels and pain. Annu. Rev. Cell Dev. Biol. 2013;29:355–384. doi: 10.1146/annurev-cellbio-101011-155833. [DOI] [PubMed] [Google Scholar]
- 115.Clapham D.E. TRP channels as cellular sensors. Nature. 2003;426(6966):517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 116.Montell C., Birnbaumer L., Flockerzi V. The TRP channels, a remarkably functional family. Cell. 2002;108(5):595–598. doi: 10.1016/S0092-8674(02)00670-0. [DOI] [PubMed] [Google Scholar]
- 117.Tsagareli M.G., Nozadze I. An overview on transient receptor potential channels superfamily. Behav. Pharmacol. 2020;31(5):413–434. doi: 10.1097/FBP.0000000000000524. [DOI] [PubMed] [Google Scholar]
- 118.Von Niederhäusern V., Kastenhuber E., Stäuble A., Gesemann M., Neuhauss S.C. Phylogeny and expression of canonical transient receptor potential (TRPC) genes in developing zebrafish. Dev. Dyn. 2013;242(12):1427–1441. doi: 10.1002/dvdy.24041. [DOI] [PubMed] [Google Scholar]
- 119.Saito S., Shingai R. Evolution of thermoTRP ion channel homologs in vertebrates. Physiol. Genomics. 2006;27(3):219–230. doi: 10.1152/physiolgenomics.00322.2005. [DOI] [PubMed] [Google Scholar]
- 120.Corey D.P., García-Añoveros J., Holt J.R., Kwan K.Y., Lin S.Y., Vollrath M.A., Amalfitano A., Cheung E.L., Derfler B.H., Duggan A., Géléoc G.S., Gray P.A., Hoffman M.P., Rehm H.L., Tamasauskas D., Zhang D.S. TRPA1 is a candidate for the mechanosensitive transduction channel of vertebrate hair cells. Nature. 2004;432(7018):723–730. doi: 10.1038/nature03066. [DOI] [PubMed] [Google Scholar]
- 121.Kastenhuber E., Gesemann M., Mickoleit M., Neuhauss S.C. Phylogenetic analysis and expression of zebrafish transient receptor potential melastatin family genes. Dev. Dyn. 2013;242(11):1236–1249. doi: 10.1002/dvdy.24020. [DOI] [PubMed] [Google Scholar]
- 122.Benini A., Bozzato A., Mantovanelli S., Calvarini L., Giacopuzzi E., Bresciani R., Moleri S., Zizioli D., Beltrame M., Borsani G. Characterization and expression analysis of mcoln1.1 and mcoln1.2, the putative zebrafish co-orthologs of the gene responsible for human mucolipidosis type IV. Int. J. Dev. Biol. 2013;57(1):85–93. doi: 10.1387/ijdb.120033gb. [DOI] [PubMed] [Google Scholar]
- 123.England S.J., Campbell P.C., Banerjee S., Swanson A.J., Lewis K.E. Identification and expression analysis of the complete family of zebrafish pkd genes. Front. Cell Dev. Biol. 2017;5:5. doi: 10.3389/fcell.2017.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ko M.J., Ganzen L.C., Coskun E., Mukadam A.A., Leung Y.F., van Rijn R.M. A critical evaluation of TRPA1-mediated locomotor behavior in zebrafish as a screening tool for novel anti-nociceptive drug discovery. Sci. Rep. 2019;9(1):2430. doi: 10.1038/s41598-019-38852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Laursen W.J., Anderson E.O., Hoffstaetter L.J., Bagriantsev S.N., Gracheva E.O. Species-specific temperature sensitivity of TRPA1. Temperature (Austin) 2015;2(2):214–226. doi: 10.1080/23328940.2014.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kang K., Pulver S.R., Panzano V.C., Chang E.C., Griffith L.C., Theobald D.L., Garrity P.A. Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature. 2010;464(7288):597–600. doi: 10.1038/nature08848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gupta R., Saito S., Mori Y., Itoh S.G., Okumura H., Tominaga M. Structural basis of TRPA1 inhibition by HC-030031 utilizing species-specific differences. Sci. Rep. 2016;6:37460. doi: 10.1038/srep37460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Oda M., Kurogi M., Kubo Y., Saitoh O. Sensitivities of two zebrafish TRPA1 paralogs to chemical and thermal stimuli analyzed in heterologous expression systems. Chem. Senses. 2016;41(3):261–272. doi: 10.1093/chemse/bjv091. [DOI] [PubMed] [Google Scholar]
- 129.Veldhuis N.A., Poole D.P., Grace M., McIntyre P., Bunnett N.W. The G protein-coupled receptor-transient receptor potential channel axis: molecular insights for targeting disorders of sensation and inflammation. Pharmacol. Rev. 2015;67(1):36–73. doi: 10.1124/pr.114.009555. [DOI] [PubMed] [Google Scholar]
- 130.Caterina M.J., Schumacher M.A., Tominaga M., Rosen T.A., Levine J.D., Julius D. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389(6653):816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- 131.Moriyama T., Higashi T., Togashi K., Iida T., Segi E., Sugimoto Y., Tominaga T., Narumiya S., Tominaga M. Sensitization of TRPV1 by EP1 and IP reveals peripheral nociceptive mechanism of prostaglandins. Mol. Pain. 2005;1:3. doi: 10.1186/1744-8069-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zygmunt P.M., Petersson J., Andersson D.A., Chuang H., Sørgård M., Di Marzo V., Julius D., Högestätt E.D. Vanilloid receptors on sensory nerves mediate the vasodilator action of anandamide. Nature. 1999;400(6743):452–457. doi: 10.1038/22761. [DOI] [PubMed] [Google Scholar]
- 133.Szallasi A., Blumberg P.M. Resiniferatoxin, a phorbol-related diterpene, acts as an ultrapotent analog of capsaicin, the irritant constituent in red pepper. Neuroscience. 1989;30(2):515–520. doi: 10.1016/0306-4522(89)90269-8. [DOI] [PubMed] [Google Scholar]
- 134.Curtright A., Rosser M., Goh S., Keown B., Wagner E., Sharifi J., Raible D.W., Dhaka A. Modeling nociception in zebrafish: a way forward for unbiased analgesic discovery. PLoS One. 2015;10(1):e0116766. doi: 10.1371/journal.pone.0116766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Santos A.L.E., Leite G.O., Carneiro R.F., Roma R.R., Santos V.F., Santos M.H.C., Pereira R.O., Silva R.C., Nagano C.S., Sampaio A.H., Rocha B.A.M., Delatorre P., Campos A.R., Teixeira C.S. Purification and biophysical characterization of a mannose/N-acetyl-d-glucosamine-specific lectin from Machaerium acutifolium and its effect on inhibition of orofacial pain via TRPV1 receptor. Arch. Biochem. Biophys. 2019;664:149–156. doi: 10.1016/j.abb.2019.02.009. [DOI] [PubMed] [Google Scholar]
- 136.do Nascimento J.E.T., de Morais S.M., de Lisboa D.S., de Oliveira Sousa M., Santos S.A.A.R., Magalhães F.E.A., Campos A.R. The orofacial antinociceptive effect of Kaempferol-3-O-rutinoside, isolated from the plant Ouratea fieldingiana, on adult zebrafish (Danio rerio). Biomed. Pharmacother. 2018;107:1030–1036. doi: 10.1016/j.biopha.2018.08.089. [DOI] [PubMed] [Google Scholar]
- 137.Soares I.C.R., Santos S.A.A.R., Coelho R.F., Alves Y.A., Vieira-Neto A.E., Tavares K.C.S., Magalhaes F.E.A., Campos A.R. Oleanolic acid promotes orofacial antinociception in adult zebrafish (Danio rerio) through TRPV1 receptors. Chem. Biol. Interact. 2019;299:37–43. doi: 10.1016/j.cbi.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 138.Batista F.L.A., Lima L.M.G., Abrante I.A., de Araújo J.I.F., Batista F.L.A., Abrante I.A., Magalhães E.A., de Lima D.R., Lima M.D.C.L., do Prado B.S., Moura L.F.W.G., Guedes M.I.F., Ferreira M.K.A., de Menezes J.E.S.A., Santos S.A.A.R., Mendes F.R.S., Moreira R.A., Monteiro-Moreira A.C.O., Campos A.R., Magalhães F.E.A. Antinociceptive activity of ethanolic extract of Azadirachta indica A. Juss (Neem, Meliaceae) fruit through opioid, glutamatergic and acid-sensitive ion pathways in adult zebrafish (Danio rerio). Biomed. Pharmacother. 2018;108:408–416. doi: 10.1016/j.biopha.2018.08.160. [DOI] [PubMed] [Google Scholar]
- 139.Ghysen A., Dambly-Chaudière C. The lateral line microcosmos. Genes Dev. 2007;21(17):2118–2130. doi: 10.1101/gad.1568407. [DOI] [PubMed] [Google Scholar]
- 140.Eckroth J.R., Aas-Hansen Ø., Sneddon L.U., Bichão H., Døving K.B. Physiological and behavioural responses to noxious stimuli in the Atlantic cod (Gadus morhua). PLoS One. 2014;9(6):e100150. doi: 10.1371/journal.pone.0100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Basavarajappa B.S., Shivakumar M., Joshi V., Subbanna S. Endocannabinoid system in neurodegenerative disorders. J. Neurochem. 2017;142(5):624–648. doi: 10.1111/jnc.14098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Luchtenburg F.J., Schaaf M.J.M., Richardson M.K. Functional characterization of the cannabinoid receptors 1 and 2 in zebrafish larvae using behavioral analysis. Psychopharmacology (Berl.) 2019;236(7):2049–2058. doi: 10.1007/s00213-019-05193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lauckner J.E., Jensen J.B., Chen H.Y., Lu H.C., Hille B., Mackie K. GPR55 is a cannabinoid receptor that increases intracellular calcium and inhibits M current. Proc. Natl. Acad. Sci. USA. 2008;105(7):2699–2704. doi: 10.1073/pnas.0711278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pertwee R.G., Howlett A.C., Abood M.E., Alexander S.P., Di Marzo V., Elphick M.R., Greasley P.J., Hansen H.S., Kunos G., Mackie K., Mechoulam R., Ross R.A. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB1 and CB2. Pharmacol. Rev. 2010;62(4):588–631. doi: 10.1124/pr.110.003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Herkenham M., Lynn A.B., Little M.D., Johnson M.R., Melvin L.S., de Costa B.R., Rice K.C. Cannabinoid receptor localization in brain. Proc. Natl. Acad. Sci. USA. 1990;87(5):1932–1936. doi: 10.1073/pnas.87.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Munro S., Thomas K.L., Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365(6441):61–65. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- 147.Van Sickle M.D., Duncan M., Kingsley P.J., Mouihate A., Urbani P., Mackie K., Stella N., Makriyannis A., Piomelli D., Davison J.S., Marnett L.J., Di Marzo V., Pittman Q.J., Patel K.D., Sharkey K.A. Identification and functional characterization of brainstem cannabinoid CB2 receptors. Science. 2005;310(5746):329–332. doi: 10.1126/science.1115740. [DOI] [PubMed] [Google Scholar]
- 148.Zhang H.Y., Gao M., Shen H., Bi G.H., Yang H.J., Liu Q.R., Wu J., Gardner E.L., Bonci A., Xi Z.X. Expression of functional cannabinoid CB2 receptor in VTA dopamine neurons in rats. Addict. Biol. 2017;22(3):752–765. doi: 10.1111/adb.12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oleson E.B., Beckert M.V., Morra J.T., Lansink C.S., Cachope R., Abdullah R.A., Loriaux A.L., Schetters D., Pattij T., Roitman M.F., Lichtman A.H., Cheer J.F. Endocannabinoids shape accumbal encoding of cue-motivated behavior via CB1 receptor activation in the ventral tegmentum. Neuron. 2012;73(2):360–373. doi: 10.1016/j.neuron.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rey A.A., Purrio M., Viveros M.P., Lutz B. Biphasic effects of cannabinoids in anxiety responses: CB1 and GABA(B) receptors in the balance of GABAergic and glutamatergic neurotransmission. Neuropsychopharmacology. 2012;37(12):2624–2634. doi: 10.1038/npp.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Muccioli G.G., Lambert D.M. Current knowledge on the antagonists and inverse agonists of cannabinoid receptors. Curr. Med. Chem. 2005;12(12):1361–1394. doi: 10.2174/0929867054020891. [DOI] [PubMed] [Google Scholar]
- 152.Ibrahim M.M., Porreca F., Lai J., Albrecht P.J., Rice F.L., Khodorova A., Davar G., Makriyannis A., Vanderah T.W., Mata H.P., Malan T.P., Jr CB2 cannabinoid receptor activation produces antinociception by stimulating peripheral release of endogenous opioids. Proc. Natl. Acad. Sci. USA. 2005;102(8):3093–3098. doi: 10.1073/pnas.0409888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wilson R.I., Nicoll R.A. Endocannabinoid signaling in the brain. Science. 2002;296(5568):678–682. doi: 10.1126/science.1063545. [DOI] [PubMed] [Google Scholar]
- 154.Hohmann A.G., Herkenham M. Localization of central cannabinoid CB1 receptor messenger RNA in neuronal subpopulations of rat dorsal root ganglia: a double-label in situ hybridization study. Neuroscience. 1999;90(3):923–931. doi: 10.1016/S0306-4522(98)00524-7. [DOI] [PubMed] [Google Scholar]
- 155.Drew G.M., Lau B.K., Vaughan C.W. Substance P drives endocannabinoid-mediated disinhibition in a midbrain descending analgesic pathway. J. Neurosci. 2009;29(22):7220–7229. doi: 10.1523/JNEUROSCI.4362-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Starkus J., Jansen C., Shimoda L.M.N., Stokes A.J., Small-Howard A.L., Turner H. Diverse TRPV1 responses to cannabinoids. Channels (Austin) 2019;13(1):172–191. doi: 10.1080/19336950.2019.1619436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lam C.S., Rastegar S., Strähle U. Distribution of cannabinoid receptor 1 in the CNS of zebrafish. Neuroscience. 2006;138(1):83–95. doi: 10.1016/j.neuroscience.2005.10.069. [DOI] [PubMed] [Google Scholar]
- 158.Rodriguez-Martin I., Herrero-Turrion M.J., Marron Fdez de Velasco E., Gonzalez-Sarmiento R., Rodriguez R.E. Characterization of two duplicate zebrafish Cb2-like cannabinoid receptors. Gene. 2007;389(1):36–44. doi: 10.1016/j.gene.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 159.Krug R.G., II, Clark K.J. Elucidating cannabinoid biology in zebrafish (Danio rerio). Gene. 2015;570(2):168–179. doi: 10.1016/j.gene.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Migliarini B., Carnevali O. A novel role for the endocannabinoid system during zebrafish development. Mol. Cell. Endocrinol. 2009;299(2):172–177. doi: 10.1016/j.mce.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 161.Song Z.H., Slowey C.A., Hurst D.P., Reggio P.H. The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2). Mol. Pharmacol. 1999;56(4):834–840. [PubMed] [Google Scholar]
- 162.Connors K.A., Valenti T.W., Lawless K., Sackerman J., Onaivi E.S., Brooks B.W., Gould G.G. Similar anxiolytic effects of agonists targeting serotonin 5-HT1A or cannabinoid CB receptors on zebrafish behavior in novel environments. Aquat. Toxicol. 2014;151:105–113. doi: 10.1016/j.aquatox.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ellis L.D., Berrue F., Morash M., Achenbach J.C., Hill J., McDougall J.J. Comparison of cannabinoids with known analgesics using a novel high throughput zebrafish larval model of nociception. Behav. Brain Res. 2018;337:151–159. doi: 10.1016/j.bbr.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 164.Walker J.M., Hohmann A.G. Cannabinoid mechanisms of pain suppression. Handb. Exp. Pharmacol. 2005;(168):509–554. doi: 10.1007/3-540-26573-2_17. [DOI] [PubMed] [Google Scholar]
- 165.Crombie K.M., Brellenthin A.G., Hillard C.J., Koltyn K.F. Endocannabinoid and opioid system interactions in exercise-induced hypoalgesia. Pain Med. 2018;19(1):118–123. doi: 10.1093/pm/pnx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Slivicki R.A., Iyer V., Mali S.S., Garai S., Thakur G.A., Crystal J.D., Hohmann A.G. Positive Allosteric modulation of CB1 cannabinoid receptor signaling enhances morphine antinociception and attenuates morphine tolerance without enhancing morphine- induced dependence or reward. Front. Mol. Neurosci. 2020;13:54. doi: 10.3389/fnmol.2020.00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smith F.L., Cichewicz D., Martin Z.L., Welch S.P. The enhancement of morphine antinociception in mice by delta9-tetrahydrocannabinol. Pharmacol. Biochem. Behav. 1998;60(2):559–566. doi: 10.1016/S0091-3057(98)00012-4. [DOI] [PubMed] [Google Scholar]
- 168.Lee C.H., Chen C.C. Roles of ASICs in nociception and proprioception. Adv. Exp. Med. Biol. 2018;1099:37–47. doi: 10.1007/978-981-13-1756-9_4. [DOI] [PubMed] [Google Scholar]
- 169.Schaefer L., Sakai H., Mattei M., Lazdunski M., Lingueglia E. Molecular cloning, functional expression and chromosomal localization of an amiloride-sensitive Na(+) channel from human small intestine. FEBS Lett. 2000;471(2-3):205–210. doi: 10.1016/S0014-5793(00)01403-4. [DOI] [PubMed] [Google Scholar]
- 170.Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. A proton-gated cation channel involved in acid-sensing. Nature. 1997;386(6621):173–177. doi: 10.1038/386173a0. [DOI] [PubMed] [Google Scholar]
- 171.Chen C.C., England S., Akopian A.N., Wood J.N. A sensory neuron-specific, proton-gated ion channel. Proc. Natl. Acad. Sci. USA. 1998;95(17):10240–10245. doi: 10.1073/pnas.95.17.10240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Price M.P., Snyder P.M., Welsh M.J. Cloning and expression of a novel human brain Na+ channel. J. Biol. Chem. 1996;271(14):7879–7882. doi: 10.1074/jbc.271.14.7879. [DOI] [PubMed] [Google Scholar]
- 173.Lingueglia E., de Weille J.R., Bassilana F., Heurteaux C., Sakai H., Waldmann R., Lazdunski M. A modulatory subunit of acid sensing ion channels in brain and dorsal root ganglion cells. J. Biol. Chem. 1997;272(47):29778–29783. doi: 10.1074/jbc.272.47.29778. [DOI] [PubMed] [Google Scholar]
- 174.Waldmann R., Bassilana F., de Weille J., Champigny G., Heurteaux C., Lazdunski M. Molecular cloning of a non-inactivating proton-gated Na+ channel specific for sensory neurons. J. Biol. Chem. 1997;272(34):20975–20978. doi: 10.1074/jbc.272.34.20975. [DOI] [PubMed] [Google Scholar]
- 175.Akopian A.N., Chen C.C., Ding Y., Cesare P., Wood J.N. A new member of the acid-sensing ion channel family. Neuroreport. 2000;11(10):2217–2222. doi: 10.1097/00001756-200007140-00031. [DOI] [PubMed] [Google Scholar]
- 176.Vukicevic M., Kellenberger S. Modulatory effects of acid-sensing ion channels on action potential generation in hippocampal neurons. Am. J. Physiol. Cell Physiol. 2004;287(3):C682–C690. doi: 10.1152/ajpcell.00127.2004. [DOI] [PubMed] [Google Scholar]
- 177.Hesselager M., Timmermann D.B., Ahring P.K. pH Dependency and desensitization kinetics of heterologously expressed combinations of acid-sensing ion channel subunits. J. Biol. Chem. 2004;279(12):11006–11015. doi: 10.1074/jbc.M313507200. [DOI] [PubMed] [Google Scholar]
- 178.Babinski K., Lê K.T., Séguéla P. Molecular cloning and regional distribution of a human proton receptor subunit with biphasic functional properties. J. Neurochem. 1999;72(1):51–57. doi: 10.1046/j.1471-4159.1999.0720051.x. [DOI] [PubMed] [Google Scholar]
- 179.Gründer S., Geissler H.S., Bässler E.L., Ruppersberg J.P. A new member of acid-sensing ion channels from pituitary gland. Neuroreport. 2000;11(8):1607–1611. doi: 10.1097/00001756-200006050-00003. [DOI] [PubMed] [Google Scholar]
- 180.Su X., Li Q., Shrestha K., Cormet-Boyaka E., Chen L., Smith P.R., Sorscher E.J., Benos D.J., Matalon S., Ji H.L. Interregulation of proton-gated Na(+) channel 3 and cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 2006;281(48):36960–36968. doi: 10.1074/jbc.M608002200. [DOI] [PubMed] [Google Scholar]
- 181.Jahr H., van Driel M., van Osch G.J., Weinans H., van Leeuwen J.P. Identification of acid-sensing ion channels in bone. Biochem. Biophys. Res. Commun. 2005;337(1):349–354. doi: 10.1016/j.bbrc.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 182.Paukert M., Sidi S., Russell C., Siba M., Wilson S.W., Nicolson T., Gründer S. A family of acid-sensing ion channels from the zebrafish: widespread expression in the central nervous system suggests a conserved role in neuronal communication. J. Biol. Chem. 2004;279(18):18783–18791. doi: 10.1074/jbc.M401477200. [DOI] [PubMed] [Google Scholar]
- 183.Vierck C.J., Mauderli A.P., Wiley R.G. Assessment of pain in laboratory animals: a comment on Mogil and Crager (2004). Pain. 2005;114(3):520–523. doi: 10.1016/j.pain.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 184.Treede R.D. Assessment of pain as an emotion in animals and in humans. Exp. Neurol. 2006;197(1):1–3. doi: 10.1016/j.expneurol.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 185.Schroeder P.G.S.L.U. Exploring the efficacy of immersion analgesics in zebrafish using an integrative approach. Appl. Anim. Behav. Sci. 2017;187:93–102. doi: 10.1016/j.applanim.2016.12.003. [DOI] [Google Scholar]
- 186.Larson A.A., Brown D.R., el-Atrash S., Walser M.M. Pain threshold changes in adjuvant-induced inflammation: a possible model of chronic pain in the mouse. Pharmacol. Biochem. Behav. 1986;24(1):49–53. doi: 10.1016/0091-3057(86)90043-2. [DOI] [PubMed] [Google Scholar]
- 187.Beggs S., Salter M.W. Microglia-neuronal signalling in neuropathic pain hypersensitivity 2.0. Curr. Opin. Neurobiol. 2010;20(4):474–480. doi: 10.1016/j.conb.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Sheng J., Liu S., Wang Y., Cui R., Zhang X. The link between depression and chronic pain: neural mechanisms in the brain. Neural Plast. 2017;2017:9724371. doi: 10.1155/2017/9724371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Brown R.E., Stevens D.R., Haas H.L. The physiology of brain histamine. Prog. Neurobiol. 2001;63(6):637–672. doi: 10.1016/S0301-0082(00)00039-3. [DOI] [PubMed] [Google Scholar]
- 190.Kaslin J., Panula P. Comparative anatomy of the histaminergic and other aminergic systems in zebrafish (Danio rerio). J. Comp. Neurol. 2001;440(4):342–377. doi: 10.1002/cne.1390. [DOI] [PubMed] [Google Scholar]
- 191.Peitsaro N., Sundvik M., Anichtchik O.V., Kaslin J., Panula P. Identification of zebrafish histamine H1, H2 and H3 receptors and effects of histaminergic ligands on behavior. Biochem. Pharmacol. 2007;73(8):1205–1214. doi: 10.1016/j.bcp.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 192.McNamara C.R., Mandel-Brehm J., Bautista D.M., Siemens J., Deranian K.L., Zhao M., Hayward N.J., Chong J.A., Julius D., Moran M.M., Fanger C.M. TRPA1 mediates formalin-induced pain. Proc. Natl. Acad. Sci. USA. 2007;104(33):13525–13530. doi: 10.1073/pnas.0705924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dubuisson D., Dennis S.G. The formalin test: a quantitative study of the analgesic effects of morphine, meperidine, and brain stem stimulation in rats and cats. Pain. 1977;4(2):161–174. doi: 10.1016/0304-3959(77)90130-0. [DOI] [PubMed] [Google Scholar]
- 194.Noriega V., Sierralta F., Poblete P., Aranda N., Sotomayor-Zarate R., Prieto J.C., Miranda H.F. Receptors involved in dexketoprofen analgesia in murine visceral pain. J. Biosci. 2020;45:94. doi: 10.1007/s12038-020-00064-z. [DOI] [PubMed] [Google Scholar]
- 195.Siegmund E., Cadmus R., Lu G. A method for evaluating both non-narcotic and narcotic analgesics. Proc. Soc. Exp. Biol. Med. 1957;95(4):729–731. doi: 10.3181/00379727-95-23345. [DOI] [PubMed] [Google Scholar]
- 196.Correia A.D., Cunha S.R., Scholze M., Stevens E.D. A novel behavioral fish model of nociception for testing analgesics. Pharmaceuticals. 2011;4(4):665–680. doi: 10.3390/ph4040665. [DOI] [Google Scholar]
- 197.Costa F.V., Canzian J., Stefanello F.V., Kalueff A.V., Rosemberg D.B. Naloxone prolongs abdominal constriction writhing-like behavior in a zebrafish-based pain model. Neurosci. Lett. 2019;708:134336. doi: 10.1016/j.neulet.2019.134336. [DOI] [PubMed] [Google Scholar]
- 198.Murray C.J., Lopez A.D. Measuring the global burden of disease. N. Engl. J. Med. 2013;369(5):448–457. doi: 10.1056/NEJMra1201534. [DOI] [PubMed] [Google Scholar]
- 199.Treede R.D., Rief W., Barke A., Aziz Q., Bennett M.I., Benoliel R., Cohen M., Evers S., Finnerup N.B., First M.B., Giamberardino M.A., Kaasa S., Korwisi B., Kosek E., Lavand’homme P., Nicholas M., Perrot S., Scholz J., Schug S., Smith B.H., Svensson P., Vlaeyen J.W.S., Wang S.J. Chronic pain as a symptom or a disease: the IASP Classification of Chronic Pain for the International Classification of Diseases (ICD-11). Pain. 2019;160(1):19–27. doi: 10.1097/j.pain.0000000000001384. [DOI] [PubMed] [Google Scholar]
- 200.Gatchel R.J., Peng Y.B., Peters M.L., Fuchs P.N., Turk D.C. The biopsychosocial approach to chronic pain: scientific advances and future directions. Psychol. Bull. 2007;133(4):581–624. doi: 10.1037/0033-2909.133.4.581. [DOI] [PubMed] [Google Scholar]
- 201.Li Q., Liu S., Zhu X., Mi W., Maoying Q., Wang J., Yu J., Wang Y. Hippocampal PKR/NLRP1 inflammasome pathway is required for the depression-like behaviors in rats with neuropathic pain. Neuroscience. 2019;412:16–28. doi: 10.1016/j.neuroscience.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 202.Parent A.J., Beaudet N., Beaudry H., Bergeron J., Bérubé P., Drolet G., Sarret P., Gendron L. Increased anxiety-like behaviors in rats experiencing chronic inflammatory pain. Behav. Brain Res. 2012;229(1):160–167. doi: 10.1016/j.bbr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Craske M.G., Rauch S.L., Ursano R., Prenoveau J., Pine D.S., Zinbarg R.E. What is an anxiety disorder? Depress. Anxiety. 2009;26(12):1066–1085. doi: 10.1002/da.20633. [DOI] [PubMed] [Google Scholar]
- 204.Malhi G.S., Mann J.J. Depression. Lancet. 2018;392(10161):2299–2312. doi: 10.1016/S0140-6736(18)31948-2. [DOI] [PubMed] [Google Scholar]
- 205.Guimarães F.S.C., Graeff A.P. Modulation of anxiety behaviors by 5-HT-interacting drugs. In: Handbook of Behavioral Neuroscience; Robert, J.; Blanchard, D. C. B.; Griebel, G.; Nutt, D., Eds.; Elsevier. 2008;17:241–268. [Google Scholar]
- 206.Ferjan I., Lipnik-Štangelj M. Chronic pain treatment: the influence of tricyclic antidepressants on serotonin release and uptake in mast cells. Mediators Inflamm. 2013;2013:340473. doi: 10.1155/2013/340473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Dale R., Stacey B. Multimodal treatment of chronic pain. Med. Clin. North Am. 2016;100(1):55–64. doi: 10.1016/j.mcna.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 208.Humo M., Lu H., Yalcin I. The molecular neurobiology of chronic pain-induced depression. Cell Tissue Res. 2019;377(1):21–43. doi: 10.1007/s00441-019-03003-z. [DOI] [PubMed] [Google Scholar]
- 209.Descalzi G., Mitsi V., Purushothaman I., Gaspari S., Avrampou K., Loh Y.E., Shen L., Zachariou V. Neuropathic pain promotes adaptive changes in gene expression in brain networks involved in stress and depression. Sci. Signal. 2017;10(471): ,eaaj1549. doi: 10.1126/scisignal.aaj1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Chesselet M.F., Carmichael S.T. Animal models of neurological disorders. Neurotherapeutics. 2012;9(2):241–244. doi: 10.1007/s13311-012-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Richards E.M., Mathews D.C., Luckenbaugh D.A., Ionescu D.F., Machado-Vieira R., Niciu M.J., Duncan W.C., Nolan N.M., Franco-Chaves J.A., Hudzik T., Maciag C., Li S., Cross A., Smith M.A., Zarate C.A. Jr A randomized, placebo-controlled pilot trial of the delta opioid receptor agonist AZD2327 in anxious depression. Psychopharmacology (Berl.) 2016;233(6):1119–1130. doi: 10.1007/s00213-015-4195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Stewart A.M., Grossman L., Nguyen M., Maximino C., Rosemberg D.B., Echevarria D.J., Kalueff A.V. Aquatic toxicology of fluoxetine: understanding the knowns and the unknowns. Aquat. Toxicol. 2014;156:269–273. doi: 10.1016/j.aquatox.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 213.Wyatt C., Bartoszek E.M., Yaksi E. Methods for studying the zebrafish brain: past, present and future. Eur. J. Neurosci. 2015;42(2):1746–1763. doi: 10.1111/ejn.12932. [DOI] [PubMed] [Google Scholar]
- 214.Kalueff A.V., Gebhardt M., Stewart A.M., Cachat J.M., Brimmer M., Chawla J.S., Craddock C., Kyzar E.J., Roth A., Landsman S., Gaikwad S., Robinson K., Baatrup E., Tierney K., Shamchuk A., Norton W., Miller N., Nicolson T., Braubach O., Gilman C.P., Pittman J., Rosemberg D.B., Gerlai R., Echevarria D., Lamb E., Neuhauss S.C., Weng W., Bally-Cuif L., Schneider H. Towards a comprehensive catalog of zebrafish behavior 1.0 and beyond. Zebrafish. 2013;10(1):70–86. doi: 10.1089/zeb.2012.0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Maximino C., Puty B., Benzecry R., Araújo J., Lima M.G., de Jesus Oliveira Batista E., Renata de Matos Oliveira K., Crespo-Lopez M.E., Herculano A.M. Role of serotonin in zebrafish (Danio rerio) anxiety: relationship with serotonin levels and effect of buspirone, WAY 100635, SB 224289, fluoxetine and para-chlorophenylalanine (pCPA) in two behavioral models. Neuropharmacology. 2013;71:83–97. doi: 10.1016/j.neuropharm.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 216.Singleman C., Holtzman N.G. Growth and maturation in the zebrafish, Danio rerio: a staging tool for teaching and research. Zebrafish. 2014;11(4):396–406. doi: 10.1089/zeb.2014.0976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sneddon L.U., Halsey L.G., Bury N.R. Considering aspects of the 3Rs principles within experimental animal biology. J. Exp. Biol. 2017;220(Pt 17):3007–3016. doi: 10.1242/jeb.147058. [DOI] [PubMed] [Google Scholar]
- 218.Parker M.O., Brock A.J., Walton R.T., Brennan C.H. The role of zebrafish (Danio rerio) in dissecting the genetics and neural circuits of executive function. Front. Neural Circuits. 2013;7:63. doi: 10.3389/fncir.2013.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lu J., Peatman E., Tang H., Lewis J., Liu Z. Profiling of gene duplication patterns of sequenced teleost genomes: evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genomics. 2012;13:246. doi: 10.1186/1471-2164-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lakstygal A.M., de Abreu M.S., Lifanov D., Wappler-Guzetta E.A., Serikuly N., Alpsychov E.T., Wang D., Wang M., Tang Z., Yan N. Zebrafish models of diabetes-related CNS pathogenesis. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;98:48–58. doi: 10.1016/j.pnpbp.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 221.Wang D., Hu G., Wang J., Yan D., Wang M., Yang L., Serikuly N., Alpyshov E., Demin K.A., Galstyan D.S., Amstislavskaya T.G., de Abreu M.S., Kalueff A.V. Studying CNS effects of Traditional Chinese Medicine using zebrafish models. J. Ethnopharmacol. 2021;267:113383. doi: 10.1016/j.jep.2020.113383. [DOI] [PubMed] [Google Scholar]
- 222.de Abreu M.S., Giacomini A.C.V.V., Genario R., Rech N., Carboni J., Lakstygal A.M., Amstislavskaya T.G., Demin K.A., Leonard B.E., Vlok M., Harvey B.H., Piato A., Barcellos L.J.G., Kalueff A.V. Non-pharmacological and pharmacological approaches for psychiatric disorders: Re-appraisal and insights from zebrafish models. Pharmacol. Biochem. Behav. 2020;193:172928. doi: 10.1016/j.pbb.2020.172928. [DOI] [PubMed] [Google Scholar]
- 223.Jordi J., Guggiana-Nilo D., Bolton A.D., Prabha S., Ballotti K., Herrera K., Rennekamp A.J., Peterson R.T., Lutz T.A., Engert F. High-throughput screening for selective appetite modulators: A multibehavioral and translational drug discovery strategy. Sci. Adv. 2018;4(10): ,eaav1966. doi: 10.1126/sciadv.aav1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., Collins J.E., Humphray S., McLaren K., Matthews L., McLaren S., Sealy I., Caccamo M., Churcher C., Scott C., Barrett J.C., Koch R., Rauch G.J., White S., Chow W., Kilian B., Quintais L.T., Guerra-Assunção J.A., Zhou Y., Gu Y., Yen J., Vogel J.H., Eyre T., Redmond S., Banerjee R., Chi J., Fu B., Langley E., Maguire S.F., Laird G.K., Lloyd D., Kenyon E., Donaldson S., Sehra H., Almeida-King J., Loveland J., Trevanion S., Jones M., Quail M., Willey D., Hunt A., Burton J., Sims S., McLay K., Plumb B., Davis J., Clee C., Oliver K., Clark R., Riddle C., Elliot D., Threadgold G., Harden G., Ware D., Begum S., Mortimore B., Kerry G., Heath P., Phillimore B., Tracey A., Corby N., Dunn M., Johnson C., Wood J., Clark S., Pelan S., Griffiths G., Smith M., Glithero R., Howden P., Barker N., Lloyd C., Stevens C., Harley J., Holt K., Panagiotidis G., Lovell J., Beasley H., Henderson C., Gordon D., Auger K., Wright D., Collins J., Raisen C., Dyer L., Leung K., Robertson L., Ambridge K., Leongamornlert D., McGuire S., Gilderthorp R., Griffiths C., Manthravadi D., Nichol S., Barker G., Whitehead S., Kay M., Brown J., Murnane C., Gray E., Humphries M., Sycamore N., Barker D., Saunders D., Wallis J., Babbage A., Hammond S., Mashreghi-Mohammadi M., Barr L., Martin S., Wray P., Ellington A., Matthews N., Ellwood M., Woodmansey R., Clark G., Cooper J., Tromans A., Grafham D., Skuce C., Pandian R., Andrews R., Harrison E., Kimberley A., Garnett J., Fosker N., Hall R., Garner P., Kelly D., Bird C., Palmer S., Gehring I., Berger A., Dooley C.M., Ersan-Ürün Z., Eser C., Geiger H., Geisler M., Karotki L., Kirn A., Konantz J., Konantz M., Oberländer M., Rudolph-Geiger S., Teucke M., Lanz C., Raddatz G., Osoegawa K., Zhu B., Rapp A., Widaa S., Langford C., Yang F., Schuster S.C., Carter N.P., Harrow J., Ning Z., Herrero J., Searle S.M., Enright A., Geisler R., Plasterk R.H., Lee C., Westerfield M., de Jong P.J., Zon L.I., Postlethwait J.H., Nüsslein-Volhard C., Hubbard T.J., Roest Crollius H., Rogers J., Stemple D.L., Rogers J., Stemple D.L. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Cheresiz S.V., Volgin A.D., Kokorina Evsyukova A., Bashirzade A.A.O., Demin K.A., de Abreu M.S., Amstislavskaya T.G., Kalueff A.V. Understanding neurobehavioral genetics of zebrafish. J. Neurogenet. 2020;34(2):203–215. doi: 10.1080/01677063.2019.1698565. [DOI] [PubMed] [Google Scholar]
- 226.Kimmel C.B. Genetics and early development of zebrafish. Trends Genet. 1989;5(8):283–288. doi: 10.1016/0168-9525(89)90103-0. [DOI] [PubMed] [Google Scholar]
- 227.Ata H., Clark K.J., Ekker S.C. The zebrafish genome editing toolkit. Methods Cell Biol. 2016;135:149–170. doi: 10.1016/bs.mcb.2016.04.023. [DOI] [PubMed] [Google Scholar]
- 228.Varshney G.K., Sood R., Burgess S.M. Understanding and Editing the Zebrafish Genome. Adv. Genet. 2015;92:1–52. doi: 10.1016/bs.adgen.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 229.Eijkenboom I., Sopacua M., Otten A.B.C., Gerrits M.M., Hoeijmakers J.G.J., Waxman S.G., Lombardi R., Lauria G., Merkies I.S.J., Smeets H.J.M., Faber C.G., Vanoevelen J.M., Group P.S. Expression of pathogenic SCN9A mutations in the zebrafish: A model to study small-fiber neuropathy. Exp. Neurol. 2019;311:257–264. doi: 10.1016/j.expneurol.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 230.Labusch M., Mancini L., Morizet D., Bally-Cuif L. Conserved and divergent features of adult neurogenesis in zebrafish. Front. Cell Dev. Biol. 2020;8:525. doi: 10.3389/fcell.2020.00525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.McIntosh R., Norris J., Clarke J.D., Alexandre P. Spatial distribution and characterization of non-apical progenitors in the zebrafish embryo central nervous system. Open Biol. 2017;7(2):160312. doi: 10.1098/rsob.160312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Than-Trong E., Bally-Cuif L. Radial glia and neural progenitors in the adult zebrafish central nervous system. Glia. 2015;63(8):1406–1428. doi: 10.1002/glia.22856. [DOI] [PubMed] [Google Scholar]
- 233.Pylatiuk C., Zhao H., Gursky E., Reischl M., Peravali R., Foulkes N., Loosli F. DIY Automated feeding and motion recording system for the analysis of fish behavior. SLAS Technol. 2019;24(4):394–398. doi: 10.1177/2472630319841412. [DOI] [PubMed] [Google Scholar]
- 234.Franco-Restrepo J.E., Forero D.A., Vargas R.A. A review of freely available, open-source software for the automated analysis of the behavior of adult zebrafish. Zebrafish. 2019;16(3):223–232. doi: 10.1089/zeb.2018.1662. [DOI] [PubMed] [Google Scholar]