Abstract
Swine-origin (variant) H1 influenza A viruses associated with numerous human infections in North America in recent years have been extensively studied in vitro and in mammalian models to determine their pandemic potential. However, limited information is available on Eurasian avian-like lineage variant H1 influenza viruses. In 2015, A/Hunan/42443/2015 virus was isolated from a child in China with a severe infection. Molecular analysis revealed that this virus possessed several key virulence and human adaptation markers. Similar to what was previously observed in C57BL/6J mice, we report here that in the BALB/c mouse model, A/Hunan/42443/2015 virus caused more severe morbidity and higher mortality than did North American variant H1 virus isolates. Furthermore, the virus efficiently replicated throughout the respiratory tract of ferrets and exhibited a capacity for transmission in this model, underscoring the need to monitor zoonotic viruses that cross the species barrier as they continue to pose a pandemic threat.
Keywords: Influenza, Ferret, H1N1, Pathogenesis, Variant virus
1. Introduction
Influenza A virus (IAV) is an important respiratory pathogen that continually causes a substantial public health burden worldwide. With respect to H1 subtype viruses, three genetic lineages of IAV currently circulate in swine. The first swine lineage, referred to as the classical swine lineage, likely emerged prior to the 1918 H1N1 Spanish flu pandemic virus (Smith et al., 2009). The second lineage endemic in swine, human seasonal-like, resulted from independent introductions of human seasonal influenza viruses into swine populations in Europe, North America, and South America. The third lineage, Eurasian avian-like (EA), resulted from introduction of avian influenza in swine in Europe which subsequently spread into Asia (Anderson et al., 2016). Swine influenza viruses occasionally cross the species barrier and infect humans. These isolates are uniquely referred to as variant influenza viruses and are identified by adding a letter “v” to the virus subtype (CDC, 2018). Zoonotic infections are of concern from a public health standpoint as variant influenza viruses often are antigenically divergent from seasonal influenza viruses, hence may not be covered by seasonal vaccination (WHO, 2017). Several H1v infections have been reported in North America, Europe, and Asia (Myers et al., 2007; Shu et al., 2012; Zhu et al., 2016; Wang et al., 2013; de Jong et al., 1988; Yang et al., 2012; Qi et al., 2013; Shinde et al., 2009; Winter et al., 2013; Pulit-Penaloza et al., 2018a, 2018b). A cycle of human-to-swine transmission, subsequent evolution by drift and shift in swine, followed by swine-to-human transmission is evident as many of the recently isolated swine viruses, including variant strains, contained genes of 2009 H1N1 pandemic (H1N1pdm09) viruses (Pulit-Penaloza et al., 2018a, 2018b; Sun et al., 2016; Qiao et al., 2014; He et al., 2018).
EA swine H1N1 virus was first detected in 1979 in Europe. Since then, it has been circulating in multiple Eurasian countries while occasionally causing human infections (Myers et al., 2007; Wang et al., 2013; Yang et al., 2012; Qi et al., 2013). The dominant EA swine lineage viruses in China have evolved through genetic reassortment and drift into several genotypes (Yang et al., 2016). In 2015, a novel reassortant influenza H1N1v virus, A/Hunan/42443/2015 (Hunan/42443), was isolated from a 30-month-old boy in Hunan Province, China. The child developed severe complications including pneumonia, respiratory failure, acute respiratory distress syndrome, and heart failure, before recovering from infection. Despite no evidence of human-to-human transmission (Zhu et al., 2016), pooled human sera poorly reacted with the virus and a candidate vaccine virus was generated for this virus strain (WHO, 2017). The virus contained EA H1N1 lineage HA, NA, and M genes, H1N1pdm09 origin PB1, PB2, PA and NP genes, and a classical swine lineage H1N1 NS gene. Hunan/42443 virus displayed higher infectivity and virulence in a C57BL/6J mouse model as compared to recently isolated North American H1N1v viruses (Zhu et al., 2016). In this study, we evaluated pathogenesis and transmissibility of this virus using BALB/c mice and ferrets. Hunan/42443 virus replicated efficiently in BALB/c mice without prior adaptation and resulted in 100% mortality, unlike other H1v viruses, which typically possess mild virulence in this species (Pulit-Penaloza et al., 2018a). Replication was also evident throughout the respiratory tract of ferrets and the virus was readily transmitted among co-housed ferrets but transmitted less efficiently through the air. Our findings highlight the pandemic potential and public health risk associated with swine influenza viruses that are capable of jumping the species barrier to cause human infections.
2. Materials and methods
Viruses.
Virus stocks of A/Hunan/42443/2015 H1N1v virus were propagated in MDCK cells at 37 °C for 48 h (Balish et al., 2013). Pooled cell supernatants were clarified by centrifugation, and frozen in aliquots at −80 °C. Virus titers in each stock were determined by standard plaque assay in MDCK cells (Maines et al., 2005). The stock viruses were sequenced and real-time RT-PCR exclusivity tests were performed to ensure no contamination of other subtypes of influenza A viruses. All work was conducted in a BSL3-enhanced laboratory setting.
Mouse experiments.
All animal experiments were performed under the guidance of the Centers for Disease Control and Prevention’s Institutional Animal Care and Use Committee and were conducted in an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited animal facility. Eleven 6–8 week old female BALB/c mice (Charles River Laboratories, Wilmington, MA) were anesthetized intraperitoneally with 0.2 ml of 2,2,2-tribromoethanol in tert-amyl alcohol (Avertin; Acros Organics) and inoculated intranasally (i.n.) with 50 μl of 5 log10 plaque forming units (PFU) of Hunan/42443 virus diluted in phosphate-buffered saline (PBS). Five mice were monitored for 14 days post-inoculation (p.i.) for clinical signs of infection and loss of body weight. On days 3 and 6 p.i., 3 mice each were euthanized for determination of viral titers in nose and lung tissues by plaque assay (Maines et al., 2005). Mice that lost ≥25% of pre-inoculation body weight were humanely euthanized.
Ferret experiments.
Six-month old male Fitch ferrets (Triple F Farms, Sayre, PA), serologically negative for currently circulating influenza viruses, were housed in Duo-Flo Bioclean mobile units (Lab Products Incorporated, Seaford, DE) during experimentation. Six ferrets were inoculated i.n. with 6 log10 PFU of Hunan/42443 virus diluted in 1 mL of PBS. The following day, contact was established by placing a serologically naive ferret in the same cage as each inoculated ferret (three ferret pairs), and respiratory droplet contact was established by placing a naïve ferret in a cage with a perforated side-wall adjacent to the same type cage housing an inoculated ferret (three ferret pairs) (Maines et al., 2006). Clinical signs of infection were monitored for 14 days p.i. and nasal wash samples were collected every two days from inoculated and contact ferrets for virus titer determination. Three additional inoculated ferrets were euthanized on day 3 p.i. for the assessment of virus replication in tissues as previously described (Maines et al., 2005). Convalescent sera collected from all surviving ferrets 3 weeks p.i. or post-contact (p.c.) were tested by hemagglutination inhibition assay using homologous virus and 0.5% turkey red blood cells to determine seroconversion (Stephenson et al., 2003).
3. Results
The Hunan/42443 H1N1v virus was previously shown to replicate efficiently in C57BL/6J mouse lungs without prior adaptation and to cause fatal infection at high inoculum doses (5–6 log10 TCID50) (Zhu et al., 2016), suggesting enhanced virulence of this H1N1v virus compared to most North American variant influenza viruses and EA lineage swine isolates, which were previously tested in BALB/c mice (Pulit-Penaloza et al., 2018a; He et al., 2018; Yang et al., 2016; Belser et al., 2010). Because there are genetic differences and immune variations among inbred mouse strains that have the potential to affect the responses to influenza virus infection (Sellers et al., 2012), we chose to build upon previous findings by characterizing Hunan/42443 virus using a BALB/c mouse model. The mice were inoculated i.n. with 50 μl of 5 log10 PFU of virus and assessed for morbidity, mortality, and viral replication. In agreement with the data in C57BL/6J mice, all infected BALB/c mice exhibited pronounced weight loss by day 8 p.i. and were euthanized (Fig. 1A). The virus replicated efficiently and to high titers throughout the murine respiratory tract. During the acute phase of infection, the average titers were 7.4 log10 PFU/ml on day 3 p.i. and 6.1 log10 PFU/ml on day 6 p.i. in mouse lungs, and 4.6 log10 PFU/ml on day 3 p.i. and 3.7 log10 PFU/ml on day 6 p.i. in nose tissue (Fig. 1B). The majority of previously tested North American variant influenza viruses and EA lineage swine isolates did not cause mortality in BALB/c mice, with a few exceptions where ≤80% mortality was observed (Pulit-Penaloza et al., 2018a; He et al., 2018; Yang et al., 2016; Belser et al., 2010). Our findings indicate enhanced virulence of Hunan/42443 virus the BALB/c mouse model, compared to previous isolates.
Fig. 1. Pathogenicity of A/Hunan/42443/2015 A(H1N1v) influenza virus in BALB/c mice.
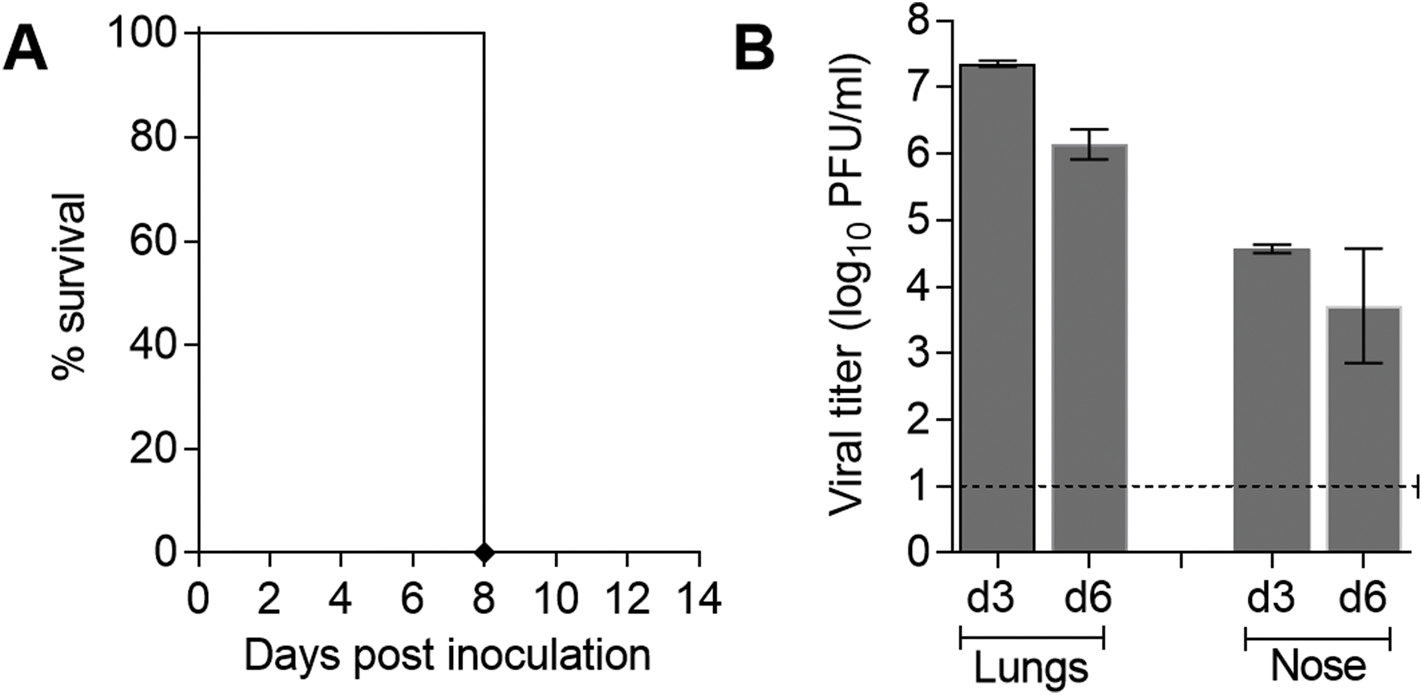
Eleven mice were inoculated with 5.0 log10 PFU of Hunan/42443 virus. Five mice were monitored for signs of morbidity and mortality for 14 days p.i.; percent survival is shown (A). Lung and nose tissues were collected from the remaining mice for titer determination by plaque assay on days 3 (n = 3) and 6 (n = 3) p.i. (B). Mean titers are expressed as log10 PFU/ml ± SD. The limit of detection is 10 PFU (dashed line).
Unlike mice, ferrets offer many advantages as a small mammalian model for the study of influenza viruses, including close physiologic similarity of human and ferret respiratory tracts, distribution of sialic acid receptors, and generally comparable clinical signs and transmissibility of influenza virus as in humans. The use of the ferret model has been indispensable in studies to evaluate the pandemic potential of novel influenza viruses (Belser et al., 2016). Ferrets that were inoculated i.n. with 6 log10 PFU of Hunan/42443 virus exhibited moderate signs of infection, including weight loss (13.5% mean maximum), transient fever (1.5 °C mean rise above baseline), and the majority of inoculated ferrets displayed nasal discharge and sneezing (Table 1). Similar to what was observed in mice, Hunan/42443 virus was found throughout the ferret respiratory tract on day 3 p.i., with mean titers of 4.6 log10 PFU/ml in nasal turbinates, 2.0 log10 PFU/g in tracheal tissue and 3.8 log10 PFU/g in lung tissue (Fig. 2). Virus was not detected outside of the respiratory tract, except for the detection of infectious virus in the olfactory bulb and brain of 1 out of 3 ferrets. The viral load and distribution of Hunan/42443 virus in tissues was comparable with North American H1N1v influenza viruses and Asian swine isolates tested in the ferret model (Pulit-Penaloza et al., 2018a; Yang et al., 2016).
Table 1.
Pathogenicity and transmission of A/Hunan/42443/2015 A (H1N1v) virus in ferrets.
| Ferret groupa | Virus in NWb | Seroconversionc | % Weight lossd | Fever (°C)e | Nasal discharge | Sneezing | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| # ferrets | Virus titer | # ferrets | HI titer | |||||
|
| ||||||||
| Inoculated | 6/6 | 5.8 ± 0.8 | 6/6 | 320–2560 | 13.5 (6/6) | 1.5 (6/6) | 6/6 | 4/6 |
| Contact (DCT) | 3/3 | 5.9 ± 0.6 | 3/3 | 1280–2560 | 7.5 (3/3) | 1.2 (3/3) | 2/3 | 0/3 |
| Contact (RDT) | 2/3 | 6.2 ± 0.9 | 2/3 | 640–1280 | 11.6 (2/3) | 1.5 (2/3) | 1/3 | 0/3 |
Ferrets were intranasally inoculated or exposed to virus in a direct contact transmission (DCT) model or a respiratory droplet transmission (RDT) model.
Number of ferrets with detectable virus in nasal wash (NW) samples and mean peak titer (log10 PFU/ml) ± SD is shown.
Number of ferrets that seroconverted by day 20 (contact ferrets) and 21 (inoculated ferrets) to homologous virus in a hemagglutination inhibition (HI) assay.
Percentage mean maximum weight loss is shown with the number of ferrets displaying weight loss in parentheses.
Mean maximum rise in temperature relative to baseline (38.4–39.3 °C) is shown with the number of ferrets displaying fever in parentheses.
Fig. 2. Detection of A/Hunan/42443/2015 A(H1N1v) influenza virus in ferret tissues.
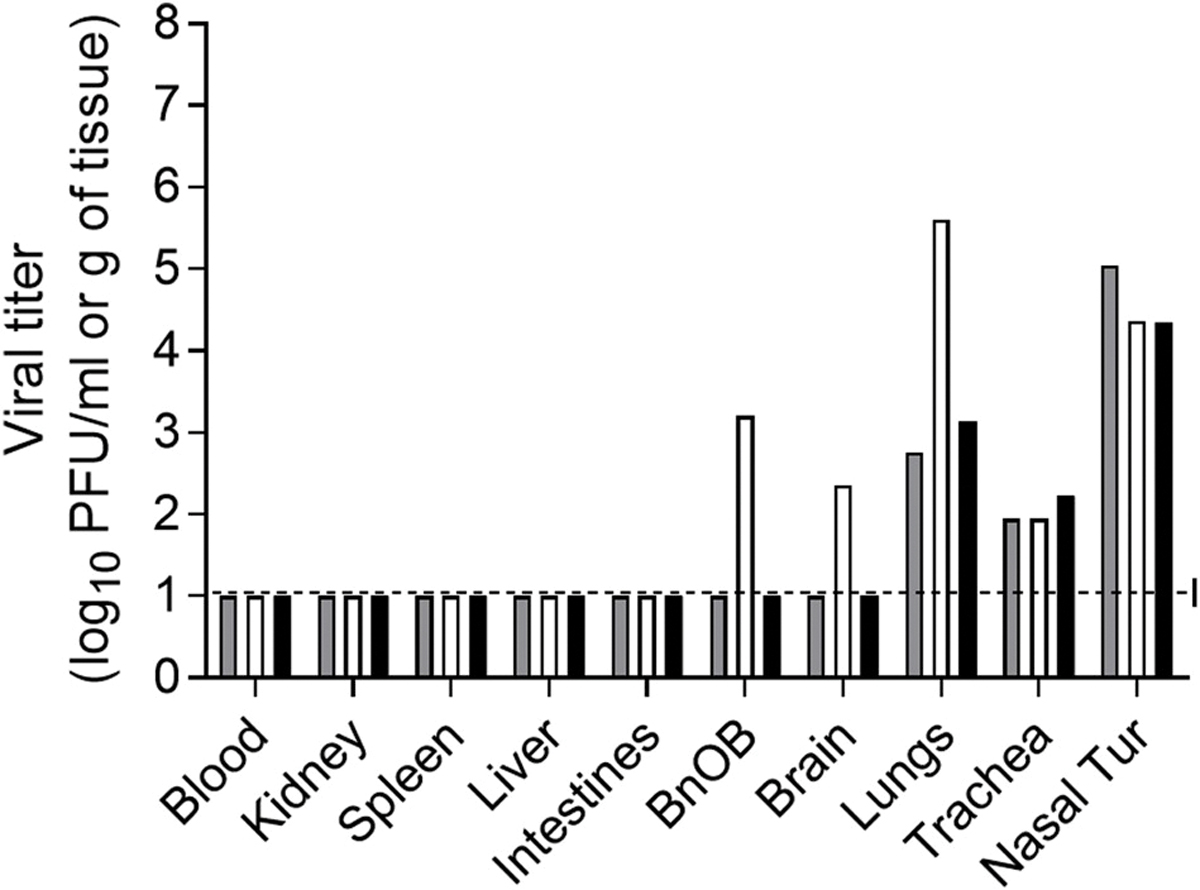
Viral titers in tissues collected on day 3 post inoculation with 6.0 log10 PFU of virus. Blood and nasal turbinate (Nasal Tur) viral titers are presented as log10 PFU/ml and kidney, spleen, liver, intestines (pooled duodenum, jejuno-ileal loop, and descending colon), olfactory bulb (BnOB), brain (pooled anterior and posterior brain), lungs (each lobe sampled and pooled), and trachea are presented as log10 PFU/g of tissue. Bars represent individual ferrets and are expressed as log10 PFU/ml. The limit of detection is 10 PFU (dashed line).
North American H1N1 swine-origin influenza viruses have been shown to efficiently transmit among cohoused ferrets, but with varying efficiency through the air via respiratory droplets or droplet nuclei (Pulit-Penaloza et al., 2018a, 2018b; Barman et al., 2012). Similarly, Asian swine influenza isolates have been shown to possess the ability to transmit among ferrets via air with varying efficiencies (Yang et al., 2016; Pascua et al., 2012), but transmissibility of Hunan/42443 virus in ferrets has not been previously tested. Transmission experiments were performed by housing a naïve ferret in the same cage as an inoculated ferret to test transmission between animals that are in direct contact (3 ferret pairs). This model provides the opportunity for transmission to occur via contact or inhalation (Fig. 3A). Transmission experiments were also performed by placing a naïve ferret in an adjacent cage allowing for air exchange through perforated cage side walls (3 ferret pairs) which prevents transmission from occurring due to direct or indirect contact (Fig. 3B). Nasal wash specimens were collected every other day from both inoculated and contact ferrets to measure the level of infectious virus. Hunan/42443 virus replicated well in all 6 inoculated animals; peak nasal wash titers observed on day 1 or 3 p.i. ranged from 5.0 to 6.7 log10 PFU/mL (Fig. 3, left side of panels). Efficient transmission was observed between ferrets that were in direct contact as evidenced by the presence of virus in nasal wash samples from all contact ferrets as early as day 1 p.c. and seroconversion on day 20 p.c. (Fig. 3A, Table 1). In the respiratory droplet transmission model, 2 out of 3 contact ferrets became productively infected and shed comparable levels of virus to those found in the inoculated ferrets (6.2 log10 compared to 5.8 log10 PFU/ml; Fig. 3B, Table 1). Seroconversion was observed 20 days p.c. only in the two animals that shed virus (Table 1). These findings indicate that, although Hunan/42443 virus displays enhanced virulence in the mouse model, it requires further adaptation to present a pandemic threat.
Fig. 3. Transmissibility of A/Hunan/42443/2015 A(H1N1v) influenza virus in ferrets.
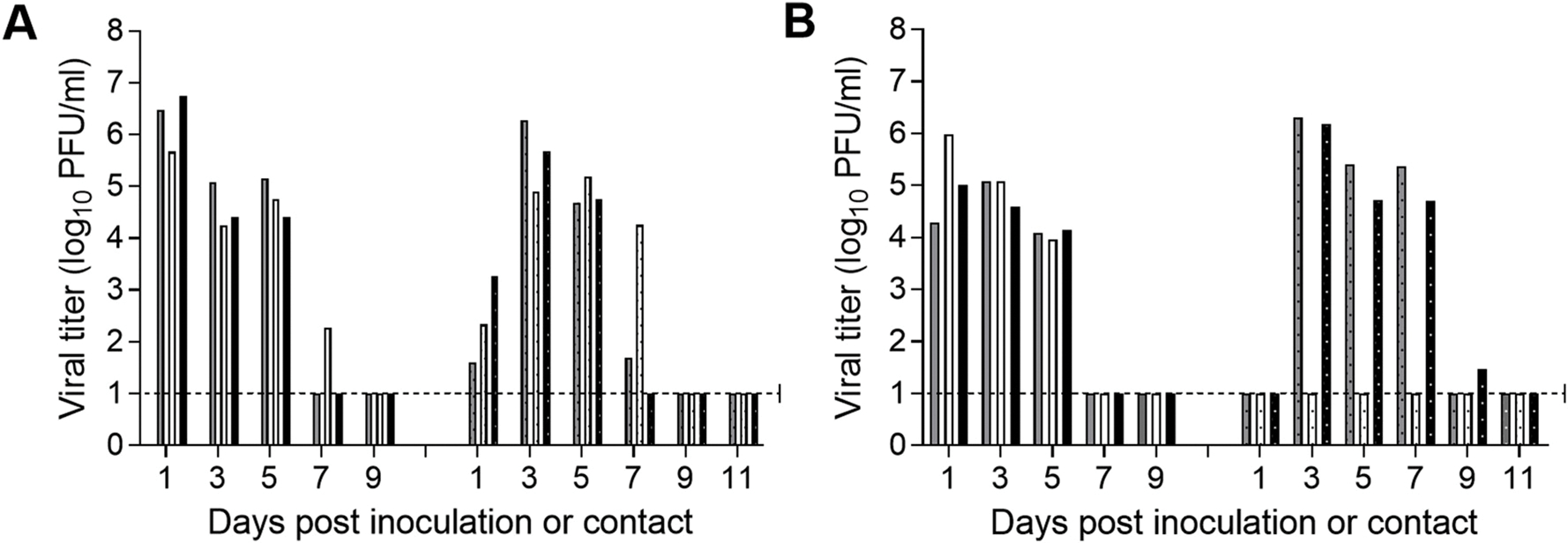
Six ferrets were inoculated i.n. with 6.0 log10 PFU of Hunan/42443 virus. The following day, a naïve ferret was placed in each of the three cages housing an inoculated ferret for the direct contact transmission model (A) and in an adjacent cage to each of the 3 remaining animals for the respiratory droplet transmission model (B). Nasal wash specimens were collected from both the inoculated (left side of each panel) and contact ferrets (right side of each panel) every other day post inoculation or post contact for titration by plaque assay. Titers of individual ferrets are expressed as log10 PFU/ml. The limit of detection is 10 PFU (dashed line).
4. Discussion
Pigs represent important hosts for influenza viruses with pandemic potential. Susceptibility of swine to both human and avian influenza viruses, and continuous virus evolution via both genetic reassortment and drift, results in the expansion of swine influenza virus diversity (Lewis et al., 2016). Swine influenza viruses that occasionally cross the species barrier and infect humans are of particular public health concern as they often are antigenically distinct from human seasonal viruses and can have a great impact on public health if they gain the ability to efficiently spread from person-to-person (CDC, 2018). In 2015, A/Hunan/42443/2015, an H1N1v virus that was isolated from a child with severe pneumonia, raised concern partly due to its ability to cause severe morbidity and mortality in the C57BL/6J mouse model (Zhu et al., 2016), similar to what was observed in BALB/c mice infected with reconstructed 1918 virus (Tumpey et al., 2005). In this study, we further characterized the Hunan/42443 virus by performing a risk assessment of the pathogenicity in a different mouse model. Due to genetic differences between mouse strains, C57BL/6J mice predominantly induce Th-1 (cellular), while BALB/c mice induce Th-2 (humoral) immune responses; the severity of influenza virus infection differs for some virus strains depending on the mouse model used (Otte and Gabriel, 2011). Despite the differences in mouse strains, our findings in the BALB/c model were in agreement with previous observations in C57BL/6J mice; as mice inoculated i.n. with a comparable dose (5 log10 PFU) of Hunan/42443 virus displayed enhanced morbidity including ruffed fur, hunched posture, and severe weight loss and were humanely euthanized on day 8 post inoculation. Overall, the magnitude of viral replication and mortality rates in BALB/c mice inoculated with Hunan/42443 virus were higher as compared to other North American and EA lineage swine H1N1 isolates described previously (Pulit-Penaloza et al., 2018a, 2019; He et al., 2018).
We found that the clinical signs and symptoms of infection displayed in ferrets inoculated with Hunan/42443 virus, including transient fever, weight loss, sneezing and rhinorrhea, along with the distribution of virus replication throughout the ferret respiratory tract, were comparable to previous reports for many swine-origin H1 viruses (Pulit-Penaloza et al., 2018a, 2018b; Yang et al., 2016; Barman et al., 2012). Unlike in mice, Hunan/42443 virus did not display enhanced virulence and did not cause mortality in ferrets. The virus transmitted between all three ferret pairs when placed in direct contact, with naïve animals becoming infected within 24h of contact, demonstrating a high efficiency of transmission in this setting. However, in a more stringent respiratory droplet setting, the virus transmitted less efficiently, with 2 out of 3 contact ferrets shedding virus by day 3 p.c. These results indicate that Hunan/42443 virus possesses the ability to transmit between ferrets through the air, similar to North American swine influenza viruses isolated from humans (Pulit-Penaloza et al., 2019).
Surveillance for known genetic markers of mammalian adaptation can be helpful in predicting the pathogenicity, transmissibility, and public health risk associated with novel viruses. The PB2 subunit of the RNA polymerase complex is a major virulence and host range determinant of influenza viruses. Amino acid substitutions in PB2 including E627K, D701N, and G590S/Q591R have been demonstrated to be critical for increased polymerase activity, and enhanced viral replication and transmissibility in mammalian models (Steel et al., 2009; Liu et al., 2012; Mehle and Doudna, 2009; Van Hoeven et al., 2009). In general, the H1N1pdm09-origin PB2 gene lacks characteristic human/mammalian amino acid mutations, such as E627K and D701 N, and rather possesses the compensatory amino acids 590S and 591R. The PB2 gene of Hunan/42443 virus is closely related to that of H1N1pdm09 virus (Zhu et al., 2016), the sequence (GISAID database ID, 206573) lacks 627K and 701 N markers, but possesses 591R, which could explain the efficient replication of Hunan/42443 virus in the mammalian models examined here. Two additional adaptation markers, T271A and T588I, are observed in the Hunan/42443 virus PB2 sequence. The T271A substitution was previously shown to increase polymerase activity in mammalian cells, leading to enhanced replication in mouse lungs and severe lung pathology (Bussey et al., 2010). In contrast, the T588I mutation, in addition to increased polymerase activity in mammalian cells, has been shown to mediate suppression of interferon-beta expression (Zhao et al., 2014). Although swine influenza viruses do not typically cause high mortality in mice, both the Hunan/42443 virus and a previously described swine H1N1v virus, A/Ohio/09/2015 (Pulit-Penaloza et al., 2018a), caused ≥80% mortality and both viruses contained 271A and 588I residues in the PB2 protein. This finding suggests that substitutions at these positions may play a role in the enhanced morbidity phenotype observed with Hunan/42443 virus in the murine model.
Each human infection caused by a swine-origin influenza virus provides another opportunity for these viruses to further adapt and become capable of sustained transmission in the human population; therefore, it is important to closely monitor novel swine influenza viruses to gain a greater understanding of the molecular adaptations necessary for sustained transmission in susceptible populations. EA lineage influenza viruses from zoonotic sources can spread readily to other continents, as recently demonstrated by the spread of H5Nx and canine influenza viruses (Pulit-Penaloza et al., 2015, 2017; Hon et al., 2015; Lee et al., 2017), underscoring the necessity to perform risk assessments of influenza viruses of multiple lineages from diverse geographic areas. Furthermore, the information in this and other similar studies aid in the characterization of emerging influenza viruses and in the generation of candidate vaccine viruses, which collectively contribute to the worldwide efforts for pandemic preparedness.
Acknowledgements
We are grateful to Dr. Dayan Wang at the Institute for Viral Disease Control and Prevention, Chinese Center for Disease Control and Prevention, for access to virus. We thank the Comparative Medicine Branch for excellent care of the animals used in this study. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
References
- Anderson TK, Macken CA, Lewis NS, Scheuermann RH, Van Reeth K, Brown IH, Swenson SL, Simon G, Saito T, Berhane Y, Ciacci-Zanella J, Pereda A, Davis CT, Donis RO, Webby RJ, Vincent AL, 2016. A phylogeny-based global nomenclature system and automated annotation tool for H1 hemagglutinin genes from swine influenza a viruses. mSphere 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balish AL, Katz JM, Klimov AI, 2013. Influenza: propagation, quantification, and storage. Curr Protoc Microbiol Chapter 15 Unit 15G 1. [DOI] [PubMed] [Google Scholar]
- Barman S, Krylov PS, Fabrizio TP, Franks J, Turner JC, Seiler P, Wang D, Rehg JE, Erickson GA, Gramer M, Webster RG, Webby RJ, 2012. Pathogenicity and transmissibility of North American triple reassortant swine influenza A viruses in ferrets. PLoS Pathog. 8, e1002791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Wadford DA, Pappas C, Gustin KM, Maines TR, Pearce MB, Zeng H, Swayne DE, Pantin-Jackwood M, Katz JM, Tumpey TM, 2010. Pathogenesis of pandemic influenza A (H1N1) and triple-reassortant swine influenza A (H1) viruses in mice. J. Virol. 84, 4194–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Eckert AM, Tumpey TM, Maines TR, 2016. Complexities in ferret influenza virus pathogenesis and transmission models. Microbiol. Mol. Biol. Rev. 80, 733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T, 2010. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J. Virol. 84, 4395–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC Information on swine influenza/variant influenza virus. https://www.cdc.gov/flu/swineflu/index.htm, Accessed date: December 2018.
- de Jong JC, Paccaud MF, de Ronde-Verloop FM, Huffels NH, Verwei C, Weijers TF, Bangma PJ, van Kregten E, Kerckhaert JA, Wicki F, et al. , 1988. Isolation of swine-like influenza A(H1N1) viruses from man in Switzerland and The Netherlands. Ann. Inst. Pasteur. Virol. 139, 429–437. [DOI] [PubMed] [Google Scholar]
- He P, Wang G, Mo Y, Yu Q, Xiao X, Yang W, Zhao W, Guo X, Chen Q, He J, Liang M, Zhu J, Ding Y, Wei Z, Ouyang K, Liu F, Jian H, Huang W, Garcia-Sastre A, Chen Y, 2018. Novel triple-reassortant influenza viruses in pigs, Guangxi, China. Emerg. Microb. Infect. 7, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon S, Ip MKT, Crespo Rocio, Paul Kohrs, Paul DeBruyn, Mansfield Kristin G., Baszler Timothy, Badcoe Lyndon, Bodenstein Barbara, Shearn-Bochsler Valerie, Killian Mary Lea, Pedersen Janice C., Hines Nichole, Gidlewski Thomas, DeLiberto Thomas, Jonathan M, Sleeman, 2015. Novel eurasian highly pathogenic avian influenza a H5 viruses in wild birds, Washington, USA, 2014. EID Journal 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Bertran K, Kwon JH, Swayne DE, 2017. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 18, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, consortium E, Webby RJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH, Vincent AL, 2016. The global antigenic diversity of swine influenza A viruses. Elife 5, e12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Qiao C, Marjuki H, Bawa B, Ma J, Guillossou S, Webby RJ, Richt JA, Ma W, 2012. Combination of PB2 271A and SR polymorphism at positions 590/591 is critical for viral replication and virulence of swine influenza virus in cultured cells and in vivo. J. Virol. 86, 1233–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, Chittaganpitch M, Waicharoen S, Nguyen DT, Nguyen T, Nguyen HH, Kim JH, Hoang LT, Kang C, Phuong LS, Lim W, Zaki S, Donis RO, Cox NJ, Katz JM, Tumpey TM, 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79, 11788–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, Mai le Q, Sedyaningsih ER, Harun S, Tumpey TM, Donis RO, Cox NJ, Subbarao K, Katz JM, 2006. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc. Natl. Acad. Sci. U. S. A. 103, 12121–12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A, Doudna JA, 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106, 21312–21316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers KP, Olsen CW, Gray GC, 2007. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis. 44, 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte A, Gabriel G, 2011. 2009 pandemic H1N1 influenza A virus strains display differential pathogenicity in C57BL/6J but not BALB/c mice. Virulence 2, 563–566. [DOI] [PubMed] [Google Scholar]
- Pascua PN, Song MS, Lee JH, Baek YH, Kwon HI, Park SJ, Choi EH, Lim GJ, Lee OJ, Kim SW, Kim CJ, Sung MH, Kim MH, Yoon SW, Govorkova EA, Webby RJ, Webster RG, Choi YK, 2012. Virulence and transmissibility of H1N2 influenza virus in ferrets imply the continuing threat of triple-reassortant swine viruses. Proc. Natl. Acad. Sci. U. S. A. 109, 15900–15905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Sun X, Creager HM, Zeng H, Belser JA, Maines TR, Tumpey TM, 2015. Pathogenesis and transmission of novel highly pathogenic avian influenza H5N2 and H5N8 viruses in ferrets and mice. J. Virol. 89, 10286–10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Simpson N, Yang H, Creager HM, Jones J, Carney P, Belser JA, Yang G, Chang J, Zeng H, Thor S, Jang Y, Killian ML, Jenkins-Moore M, Janas-Martindale A, Dubovi E, Wentworth DE, Stevens J, Tumpey TM, Davis CT, Maines TR, 2017. Assessment of molecular, antigenic, and pathological features of canine influenza A(H3N2) viruses that emerged in the United States. J. Infect. Dis. 216, S499–S507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Pappas C, Belser JA, Sun X, Brock N, Zeng H, Tumpey TM, Maines TR, 2018a. Comparative in vitro and in vivo analysis of H1N1 and H1N2 variant influenza viruses isolated from humans between 2011 and 2016. J. Virol. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Jones J, Sun X, Jang Y, Thor S, Belser JA, Zanders N, Creager HM, Ridenour C, Wang L, Stark TJ, Garten R, Chen LM, Barnes J, Tumpey TM, Wentworth DE, Maines TR, Davis CT, 2018b. Antigenically diverse swine origin H1N1 variant influenza viruses exhibit differential ferret pathogenesis and transmission phenotypes. J. Virol. 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Belser JA, Tumpey TM, Maines TR, 2019. Sowing the seeds of a pandemic? Mammalian pathogenicity and transmissibility of H1 variant influenza viruses from the swine reservoir. Trav. Med. Infect. Dis. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Cui L, Jiao Y, Pan Y, Li X, Zu R, Huo X, Wu B, Tang F, Song Y, Zhou M, Wang H, Cardona CJ, Xing Z, 2013. Antigenic and genetic characterization of a European avian-like H1N1 swine influenza virus from a boy in China in 2011. Arch. Virol. 158, 39–53. [DOI] [PubMed] [Google Scholar]
- Qiao C, Liu L, Yang H, Chen Y, Xu H, Chen H, 2014. Novel triple reassortant H1N2 influenza viruses bearing six internal genes of the pandemic 2009/H1N1 influenza virus were detected in pigs in China. J. Clin. Virol. 61, 529–534. [DOI] [PubMed] [Google Scholar]
- Sellers RS, Cli?ord CB, Treuting PM, Brayton C, 2012. Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice. Vet Pathol 49, 32–43. [DOI] [PubMed] [Google Scholar]
- Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L, 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N. Engl. J. Med. 360, 2616–2625. [DOI] [PubMed] [Google Scholar]
- Shu B, Garten R, Emery S, Balish A, Cooper L, Sessions W, Deyde V, Smith C, Berman L, Klimov A, Lindstrom S, Xu X, 2012. Genetic analysis and antigenic characterization of swine origin influenza viruses isolated from humans in the United States, 1990–2010. Virology 422, 151–160. [DOI] [PubMed] [Google Scholar]
- Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peiris JS, Guan Y, 2009. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 106, 11709–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Mubareka S, Palese P, 2009. Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLoS Pathog. 5, e1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson I, Wood JM, Nicholson KG, Zambon MC, 2003. Sialic acid receptor specificity on erythrocytes affects detection of antibody to avian influenza haemagglutinin. J. Med. Virol. 70, 391–398. [DOI] [PubMed] [Google Scholar]
- Sun YF, Wang XH, Li XL, Zhang L, Li HH, Lu C, Yang CL, Feng J, Han W, Ren WK, Tian XX, Tong GZ, Wen F, Li ZJ, Gong XQ, Liu XM, Ruan BY, Yan MH, Yu H, 2016. Novel triple-reassortant H1N1 swine influenza viruses in pigs in Tianjin, Northern China. Vet. Microbiol. 183, 85–91. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Basler CF, Aguilar PV, Zeng H, Solorzano A, Swayne DE, Cox NJ, Katz JM, Taubenberger JK, Palese P, Garcia-Sastre A, 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310, 77–80. [DOI] [PubMed] [Google Scholar]
- Van Hoeven N, Pappas C, Belser JA, Maines TR, Zeng H, Garcia-Sastre A, Sasisekharan R, Katz JM, Tumpey TM, 2009. Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc. Natl. Acad. Sci. U. S. A. 106, 3366–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DY, Qi SX, Li XY, Guo JF, Tan MJ, Han GY, Liu YF, Lan Y, Yang L, Huang WJ, Cheng YH, Zhao X, Bai T, Wang Z, Wei HJ, Xiao N, Shu YL, 2013. Human infection with Eurasian avian-like influenza A(H1N1) virus, China. Emerg. Infect. Dis. 19, 1709–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2017. Antigenic and Genetic Characteristics of Zoonotic Influenza Viruses and Development of Candidate Vaccine Viruses for Pandemic Preparedness. https://www.who.int/influenza/vaccines/virus/characteristics_virus_vaccines/en/, Accessed date: November 2018.
- Winter AL, Eshaghi A, Farrell DJ, King A, Li A, Li Y, Gubbay JB, 2013. Variant influenza A (H1N1) virus infection in Canada. J. Clin. Virol. 57, 279–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Qiao C, Tang X, Chen Y, Xin X, Chen H, 2012. Human infection from avian-like influenza A (H1N1) viruses in pigs, China. Emerg. Infect. Dis. 18, 1144–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen Y, Qiao C, He X, Zhou H, Sun Y, Yin H, Meng S, Liu L, Zhang Q, Kong H, Gu C, Li C, Bu Z, Kawaoka Y, Chen H, 2016. Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 113, 392–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yi C, Zhao L, Wang S, Zhou L, Hu Y, Zou W, Chen H, Jin M, 2014. PB2–588I enhances 2009 H1N1 pandemic influenza virus virulence by increasing viral replication and exacerbating PB2 inhibition of beta interferon expression. J. Virol. 88, 2260–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhang H, Xiang X, Zhong L, Yang L, Guo J, Xie Y, Li F, Deng Z, Feng H, Huang Y, Hu S, Xu X, Zou X, Li X, Bai T, Chen Y, Li Z, Li J, Shu Y, 2016. Reassortant eurasian avian-like influenza A(H1N1) virus from a severely ill child, hunan Province, China, 2015. Emerg. Infect. Dis. 22, 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]


