Abstract
Background
Antimicrobial resistance (AMR) is a public health issue with significant impact on health care. Antibiogram development and deployment is a key strategy for managing and preventing AMR. Our objective was to develop an Ontario antibiogram as part of a larger provincial initiative aimed at advancing antimicrobial stewardship in the province.
Methods
As part of a voluntary provincial online survey, antibiogram data from 100 of 201 (49.8%) Ontario hospitals were collected and included. All hospitals in Ontario were eligible to participate except those providing only mental health or ambulatory services. Weighted provincial and regional antibiotic susceptibilities (percentages) were conducted using descriptive statistical analyses, and an interactive antibiogram spreadsheet was developed. Respondent-identified barriers to collecting and interpreting antibiogram data are presented descriptively.
Results
There was wide regional variability in antimicrobial-resistant organisms across Ontario. Provincial methicillin-resistant Staphylococcus aureus prevalence was 24.6%, ranging from 5.9% to 43.7% regionally. Provincial Escherichia coli resistance to ceftriaxone and ciprofloxacin was 13.8% (regional range 6.0%–25.1%) and 22.5% (regional range 9.8–37.8%), respectively. Klebsiella spp resistance to ceftriaxone and ciprofloxacin was similar across all health regions, with overall provincial rates of 7.5% and 5.6%, respectively.
Conclusions
We have demonstrated that integrating hospital AMR tracking and reporting as part of a larger voluntary provincial antimicrobial stewardship program initiative is a feasible approach to capturing AMR data. The provincial antibiogram serves as a benchmark for the current state of AMR provincially and across health regions.
Key words: antibiogram, antimicrobial resistance, antimicrobial stewardship, microbiology, susceptibility
Abstract
Historique
La résistance antimicrobienne (RAM) est un enjeu sanitaire aux conséquences importantes sur les soins. La création et le déploiement d’antibiogrammes sont une stratégie essentielle pour gérer et prévenir la RAM. Les chercheurs s’étaient donné l’objectif de créer un antibiogramme ontarien dans le cadre d’une initiative provinciale plus vaste visant à faire progresser la gestion antimicrobienne dans la province.
Méthodologie
Dans le cadre d’un sondage provincial volontaire en ligne, les chercheurs ont colligé et inclus les données d’antibiogrammes de 100 des 201 hôpitaux ontariens (49,8 %). Tous les hôpitaux de l’Ontario étaient admissibles à participer, sauf ceux qui ne donnaient que des services en santé mentale ou des services ambulatoires. Les chercheurs ont établi les susceptibilités antibiotiques provinciales et régionales pondérées (en pourcentage) d’après les analyses statistiques descriptives et ont créé un chiffrier interactif de l’antibiogramme. Ils ont fait une interprétation descriptive des obstacles indiqués par les participants à la collecte et à l’interprétation des données de l’antibiogramme.
Résultats
La variabilité régionale des organismes résistants aux antimicrobiens est importante en Ontario. La prévalence de Staphylococcus aureus résistant à la méthicilline s’élevait à 24,6 %, et variait entre 5,9 % et 43,7 % selon les régions. La résistance provinciale de l’Escherichia coli à la ceftriaxone et à la ciprofloxacine correspondait à 13,8 % (plage régionale de 6,0 % à 25,1 %) et à 22,5 % (plage régionale de 9,8 % à 37,8 %), respectivement. La résistance des espèces de Klebsiella à la ceftriaxone et à la ciprofloxacine était semblable dans toutes les régions sanitaires, les taux provinciaux globaux s’établissant à 7,5 % et 5,6 %, respectivement.
Conclusion
Les auteurs ont démontré que l’intégration d’une fonction de traçage et de déclaration de la RAM aux hôpitaux dans le cadre d’un plus vaste programme provincial de gestion antimicrobienne volontaire est une démarche faisable pour saisir les données de RAM. L’antibiogramme provincial sert de référence pour obtenir un portrait à jour de la RAM dans la province et les régions sanitaires.
Mots-clés : antibiogramme, gestion antimicrobienne, microbiologie, résistance antimicrobienne, susceptibilité
Introduction
Antimicrobial resistance (AMR) is an important public health issue with a significant impact on health care. In Canada, approximately 26% of infections are caused by antimicrobial-resistant organisms (AROs), accounting for 5,400 deaths annually and $1.4 billion in health care system costs each year. By 2050, the rate of resistance is expected to grow to 40%, resulting in 13,700 increased deaths and $7.6 billion in increased health care costs (1). Since 2013, antimicrobial stewardship programs (ASPs) have been a required organizational practice (ROP) for Canadian inpatient health care institutions, with the goal of promoting appropriate antibiotic use to optimize patient outcomes while limiting undesirable consequences, including AMR (2).
One important antimicrobial stewardship strategy for monitoring AMR and highlighting local susceptibility data is the development and deployment of an antibiogram (3,4). An antibiogram is a summary of the cumulative susceptibility of bacterial isolates to antibiotics during a specified time period; it is a useful tool for trending local resistance patterns and for guiding empiric antibiotic choices. Because of differences in patient populations, international travel (5), antibiotic prescribing practices, infection control practices (6), and patient complexity, susceptibilities reported in antibiograms may vary from one institution or region to another.
In Ontario, although there are several mandatory and voluntary reporting mechanisms for AMR and hospital-associated infection data (7), there is currently no comprehensive provincial repository of hospital antibiotic resistance data.
Our objective was to develop an Ontario antibiogram as part of a larger provincial initiative aimed at advancing antimicrobial stewardship in the province. We describe efforts made to maximize hospital participation in our voluntary survey, analyses to estimate provincewide AMR rates, and development of a provincial antibiogram tool for data dissemination.
Methods
Setting
This project was carried out in Ontario, Canada, by Public Health Ontario (PHO), an agency with a mandate to provide scientific and technical expertise to improve public health in the province.
Survey design
The 2018 Ontario ASP Landscape Survey is the second iteration of a voluntary online survey of ASPs in acute care, inpatient rehabilitation, and complex continuing care (CCC) facilities developed by the ASP team at PHO (8). With this iteration of the survey, total hospital antimicrobial use for 2017 and antimicrobial susceptibility data were requested as additional items. Internal and external reviews of our survey by clinicians involved in hospital ASPs (pharmacists, physicians, and program leads) ensured that survey questions were clear and clinically relevant.
Our survey included a brief description of antibiograms and their main functions, as well as instructions relating to the collection and reporting of antimicrobial susceptibility data. Respondents were asked to provide their most recent hospital-specific antibiograms (based on data from 2014 or later) in Microsoft Excel, Microsoft Word, or PDF format, with the understanding that the data submitted would be included in the provincial antibiogram. Survey questions pertaining to antimicrobial susceptibility data are available in Supplemental Appendix A.
Distribution
The voluntary online survey was open for 4 weeks from September 25, 2018, to October 24, 2018, and administered via an in-house online platform (Surveys@PHO).
Targeted email distribution lists were used, and the survey was addressed to the clinician most responsible for antimicrobial stewardship at each hospital corporation, based on a PHO database of ASP stakeholders and a provincial email network of antimicrobial stewardship pharmacists (Antimicrobial Stewardship Hospital Pharmacists of Ontario Network). We followed up with non-responders via email, telephone reminders, or both 2–3 weeks after survey launch. There were no monetary incentives for participation.
Eligibility
All hospitals in Ontario were eligible to complete the survey except those that provide only mental health or ambulatory services because ASPs are currently not an Accreditation Canada ROP for these types of institutions.
Analysis
Descriptive statistical analyses for the antibiogram component of the survey were conducted using Microsoft Excel (Version 2013; Microsoft Corp., Redmond, WA). Weighted provincial and regional percent susceptibilities for each antibiotic and organism combination are calculated using percent susceptibility and isolate numbers provided by each participating hospital. Inpatient hospital bed numbers from the Ministry of Health are used to determine the proportion of Ontario acute care beds represented by the provincial antibiogram. Respondent-identified barriers to collecting and interpreting antibiogram data are presented descriptively.
Development of provincial antibiogram tool
We conducted a comprehensive review and consultation process with ASP experts, including infectious diseases physicians, microbiologists, pharmacists, and project managers, to determine the most clinically relevant format for an interactive provincial antibiogram tool. General feedback on the design and development of the tool was also obtained from the Antimicrobial Stewardship Advisory Committee at PHO, which has representation from infection prevention and control specialists and public health professionals in addition to front-line ASP clinicians.
We reviewed hospital antibiograms in their originally submitted format and manually entered them into Microsoft Excel to create an interactive pivot table. When available, we included hospital-wide inpatient microbiological data from all clinical specimens collected in calendar year 2014 or more recent years. When multiple antibiograms were provided for a specific hospital site, we included only the most recent antibiogram. In situations in which antibiograms for all clinical isolates were unavailable, urine and non-urine or blood and non-blood specimens were used. Because our focus was hospital-wide inpatient data, ward- and outpatient-specific antibiograms were excluded. Hospitals combining inpatient and outpatient specimens were included if they could not be stratified further. We included data for two northern hospitals that provide laboratory services for a large number of outpatient clinics; however, isolate numbers were adjusted to reflect an estimate of the proportion of inpatient specimens. Antibiograms that did not include a total number of isolates were excluded because this was required for weighting of provincial and regional susceptibilities. When possible, we followed up with respondents via email to obtain missing information before making the decision to exclude.
Inferred susceptibility was applied on the basis of feedback from microbiologists. Different phenotypes of bacterial species were combined wherever possible (ie, Staphylococcus aureus includes methicillin-susceptible S. aureus and methicillin-resistant S. aureus [MRSA]); however, because of variations in susceptibility reporting, Escherichia coli (E. coli) was separated into three categories: all isolates, extended-spectrum beta-lactamase (ESBL), and non-ESBL. Coagulase-negative Staphylococcus species were grouped together, with the exception of Staphylococcus lugdunensis.
We performed user testing and collaborated with PHO communication and technical design specialists to optimize the functionality and usability of the tool. We also developed technical notes to assist end users in navigating and interpreting the information presented in the tool.
Results
Study participation
The overall response rate to the 2018 Ontario ASP Landscape Survey was 55.1% (70/127 hospital corporation respondents). The majority of responding corporations (64/70; 91.4%) indicated they have an institutional antibiogram from 2014 or later. The respondents who did not have an antibiogram cited limited resources (3), lack of in-house expertise (ie, no microbiologist on site) (1), not enough samples (1), and use of a local antibiogram produced by a nearby corporation (1). Of the respondents who reported having an antibiogram, three-quarters (48/64; 75.0%) indicated that it was developed in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (9); the majority of remaining respondents indicated that they were not sure (15/64; 23.4%) whether it followed these standards. A small proportion of the corporations currently share their antibiograms publicly on the Internet (10/64; 15.6%).
Hospital representation
Among the 127 eligible corporations, there were 201 eligible hospital sites; 105 (52.2%) submitted antibiograms (Figure 1). Microbiological data from 100/201 (49.8%) of Ontario hospitals were included in the development of the provincial antibiogram tool, which represents 61.0% (16,313/26,740) of inpatient beds in the province. Regional representation varied widely from 100% (5/5 sites) in the Mississauga Halton health region to 0% (0/9 sites) in the Erie St. Clair health region (Table 1). Large community hospitals had the highest participation rate (56/92 sites; 60.9%), contributing more than half of the antibiograms that were included in the Ontario Antibiogram. Small community hospitals had the lowest participation rate (20/59 sites; 33.9%) (Table 2).
Figure 1:
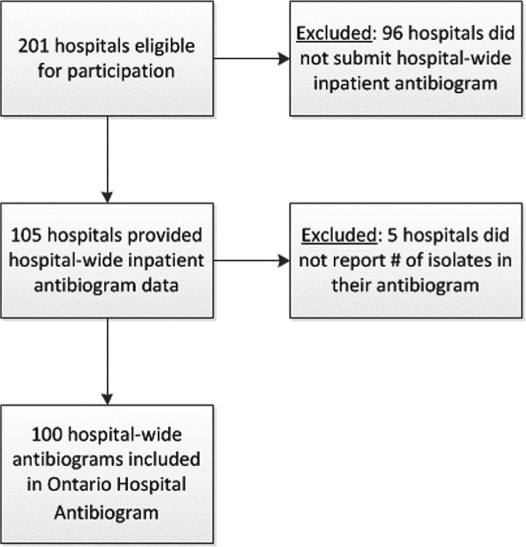
Flowchart for inclusion of hospital microbiological data into the Ontario Hospital Antibiogram
Table 1:
Representation of antibiogram data by health region
| Health region | No./n (% provincial) | |
|---|---|---|
| Hospital participation | Hospital beds represented | |
| Central | 5/8 (2.5) | 1,487/1,857 (5.6) |
| Central East | 8/13 (4.0) | 1,591/2,128 (5.9) |
| Central West | 2/3 (1.0) | 730/820 (2.7) |
| Champlain | 5/23 (2.5) | 398/3,268 (1.5) |
| Erie St. Clair | 0/9 (0.0) | 0/1,271 (0.0) |
| Hamilton Niagara Haldimand Brant | 12/17 (6.0) | 2,206/2,672 (8.2) |
| Mississauga Halton | 5/5 (2.5) | 1,554/1,554 (5.8) |
| North East | 7/29 (3.5) | 884/1,685 (3.3) |
| North Simcoe Muskoka | 2/6 (1.0) | 414/688 (1.5) |
| North West | 2/15 (1.0) | 115/794 (0.4) |
| South East | 8/13 (4.0) | 720/1,076 (2.7) |
| South West | 19/29 (9.5) | 566/2,246 (2.1) |
| Toronto Central | 19/22 (9.5) | 5,118/5,669 (19.1) |
| Waterloo Wellington | 6/9 (3.0) | 530/1,012 (2.0) |
| Ontario total | 100/201 (49.8) | 16,313/26,740 (61.0) |
Table 2:
Representation of antibiogram data by hospital type
| Hospital type | No./n (% provincial) | |
|---|---|---|
| Hospital participation | Hospital beds represented | |
| Acute teaching | 14/26 (7.0) | 4,428/7,903 (16.6) |
| Large community | 56/92 (27.9) | 9,318/13,307 (34.8) |
| Small community | 20/59 (10.0) | 555/1,556 (2.1) |
| Complex continuing care and rehabilitation | 10/24 (5.0) | 2,012/3,974 (7.5) |
| All hospitals | 100/201 (49.8) | 16,313/26,740 (61.0) |
The majority (73/100 sites; 73.0%) of hospital antibiograms were based on microbiological data from 2017 or later. Nine sites provided data from individual years 2014 to 2016 (one from 2014, two from 2015, and six from 2016). Eighteen hospitals, including one corporation consisting of five sites in the South West region, provided data from multiple years.
Provincial antibiogram tool
This Ontario Hospital Antibiogram is part of the Ontario ASP Comparison Tool, which is an online, publicly available interactive tool that summarizes structural and strategic components of participating hospital ASPs across the province. The provincial antibiogram itself is an interactive spreadsheet that allows users to view aggregate cumulative susceptibility data using various filters such as Gram stain and geographical region.
Between May 27, 2019, and December 23, 2019, the online ASP Comparison Tool has been accessed more than 2,100 times by more than 1,600 unique users from across 37 countries. The tool can be accessed via the following link: https://www.publichealthontario.ca/en/health-topics/antimicrobial-stewardship/asp-comparison-tool.
Antimicrobial resistance in Ontario
Focusing on AROs, the overall MRSA prevalence in Ontario was 24.6% (5,827/23,695) for all clinical isolates of S. aureus. We found wide variability in MRSA rates across the province, ranging from 5.9% (62/1,052) (95% CI 5.2% to 6.6%) in the Central West Region to as high as 43.7% (485/1,109) (95% CI 32.0% to 55.5%) in the North West Region (Figure 2).
Figure 2:
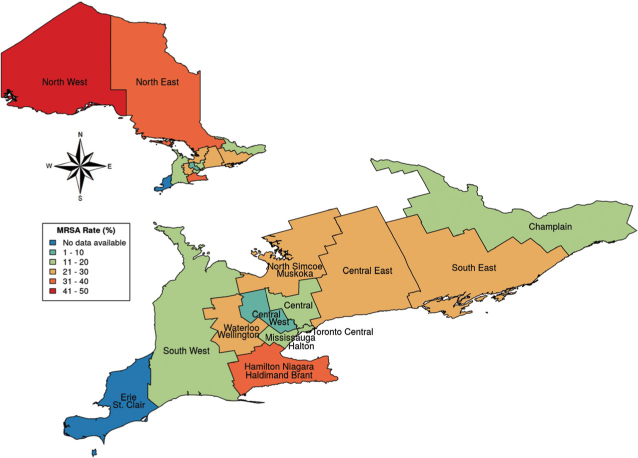
2014–2017 provincial antibiogram: MRSA rate variation across Ontario
MRSA = Methicillin-resistant Staphylococcus aureus
The provincial resistance rates for E. coli were 13.8% (7,797/56,479) for ceftriaxone and 22.5% (12,247/54,451) for ciprofloxacin (Figure 3). The regional variability in E. coli resistance to ceftriaxone in the inpatient hospital setting ranged from 6.0% (47/787) in the North West Region to 25.1% (2,400/9,543) (95% CI 23.7% to 26.6%) in the Central West Region (Figure 3). We also found wide variability in the resistance pattern of E. coli isolates to ciprofloxacin, ranging from only 9.8% (119/1,214) (95% CI 5.1% to 14.4%) in the North West Region to as high as 37.8% (1,805/4,779) (95% CI 33.9% to 41.6%) in the Central West Region (Figure 3).
Figure 3:
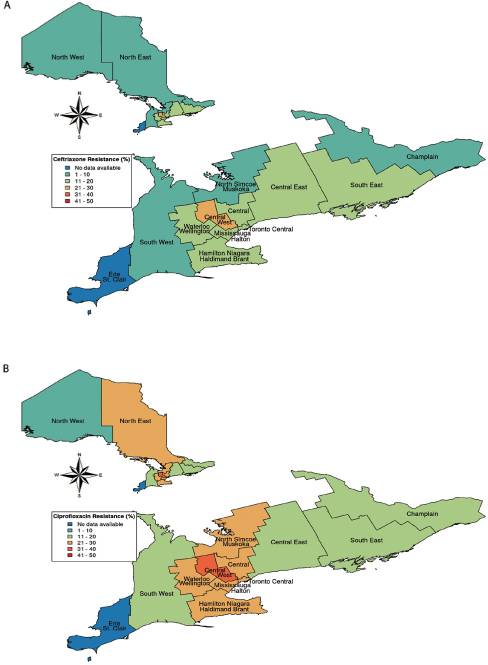
Escherichia coli resistance across Ontario. This figure shows 2014–2017 provincial resistance patterns for E. coli resistance against (a) ceftriaxone and (b) ciprofloxacin
The Klebsiella spp inpatient resistance rates for both ceftriaxone and ciprofloxacin were similar across all health regions, with overall provincial rates of 7.5% (951/12,661) and 5.6% (712/12,597), respectively. Provincial and regional isolate numbers for MRSA, E. coli, and Klebsiella spp are available in Supplemental Appendix B.
Interpretation
Aggregate cumulative susceptibility data at a regional level are beneficial for improved surveillance of AMR and to support the development of regional empiric antibiotic prescribing guidelines, particularly for smaller facilities with a low number of isolates. The Ontario Hospital Antibiogram summarizes provincial and regional cumulative antimicrobial susceptibility data for a range of organisms, accounting for almost two-thirds of hospital inpatient beds in the province. This complements what is known from national surveillance efforts such as the Canadian Antimicrobial Resistance Alliance (CARA) and the Canadian Nosocomial Infection Surveillance Program (CNISP), who conduct national surveillance of AMR using data from a smaller number of sentinel tertiary acute-care hospitals across Canada (10,11), which is not representative of most Ontario hospitals. For example, CARA conducts national AMR surveillance studies such as the Canadian Ward Surveillance Study using data from 10 to 15 Canadian tertiary centres (12), whereas our Ontario Hospital Antibiogram is based on data collected from 100 hospital sites representing 61% of inpatient hospital beds in the province.
Internationally, the Center for Disease Dynamics, Economics & Policy has developed ResistanceMap, which is a web-based collection of data visualization tools allowing for interactive exploration of AMR trends across 46 countries (13); however, regional resistance rates at the level of the province or state are not presented. ResistanceOpen is a global map of AMR that presents resistance rates within a 25-mile radius of a selected location, based on aggregated publicly available and user-submitted antibiograms (14). Alberta Health Services has partnered with microbiology labs to make antibiograms across their province publicly available online; however, the current tool does not allow for easy comparisons across zones (15).
In Ontario, the private laboratory LifeLabs publishes annual antibiograms stratified by Local Health Integration Network; however, these data are limited to outpatients and long-term care (LTC) residents only (16). In addition, PHO and the Institute for Quality Management in Healthcare (IQMH) have established a partnership to conduct an annual surveillance of AROs in laboratories and hospitals. As part of this collaboration, they have released their annual Laboratory and Hospital Survey Report highlighting the resistance patterns of common hospital pathogens (17). All 77 bacteriology laboratories in Ontario participated in this survey, demonstrating differences in their lab practices, including procedures for MRSA and ESBL screening. Compared with our data that consist of hospital-wide clinical isolates only, the data from the IQMH Laboratory and Hospital Survey Report combines both clinical and surveillance isolates, which are reported as a rate per admission. They also note that data on ESBL Enterobacteriaceae were requested at the specimen level; thus, duplicate specimens submitted for a single patient were included. However, similar to our results, their survey reported a noticeable regional variation in pathogen susceptibility across the province.
Overall, in terms of provincial MRSA trends (see Supplemental Appendix B), the IQMH Laboratory and Hospital Survey Report found the highest rates of MRSA in the North West, Hamilton Niagara Haldimand Brant (HNHB), and North East regions (17). These higher rates were also reported by LifeLabs for the North West and HNHB regions for outpatient isolates, in addition to the Erie St. Clair region, for which we were not able to interpret data (as a result of lack of a reported number of isolates) (16). The LifeLabs LTC data, however, showed higher MRSA rates across most health regions, with an overall provincial rate of 30.7% (16). This may be due to higher patient complexity, exposure to antibiotics, and receipt of health care services.
Our provincial inpatient resistance rates for E. coli for ceftriaxone and ciprofloxacin were slightly higher than the rates of 9.6% and 17.7%, respectively, reported by IQMH in 2017 (17) and also higher than the national rates of 8.5% and 18.4%, respectively, reported by CNISP sentinel sites in 2016 (11). Although LifeLabs also reported similar rates of ceftriaxone (7.5%) and ciprofloxacin (18.6%) resistance in outpatient E. coli isolates, they found higher rates of 18.7% and 48.8%, respectively, from LTC specimens (16). Similar to that of MRSA, this pattern of resistance across health sectors may be due to increased patient complexity and previous exposure to health care services and antibiotics, but additional work is required to determine these differences.
The E. coli resistance rates for ceftriaxone reported by LifeLabs for patients in LTC homes were slightly higher than the inpatient hospital rates across most health regions, with the exception of the Central West and Waterloo Wellington regions, which were similar. In the LTC setting, E. coli resistance rates for ciprofloxacin were higher across all health regions, ranging from 41% in the Central East region to 57% in the Central region (16) (see Supplemental Appendix B).
Provincial inpatient Klebsiella spp resistance rates were similar to those reported by IQMH and by LifeLabs in both the LTC and outpatient settings (16,17).
The Ontario Hospital Antibiogram has significant potential to enhance the monitoring of antibiotic resistance and advance antimicrobial stewardship efforts in the province by creating a benchmark for current AMR and highlighting provincial and regional variations in resistance patterns. More important, it also provides a foundation for advancing ASP strategies such as the development of guidance for empiric antibiotic choices based on regional resistance patterns, which is particularly helpful for smaller, less-resourced hospitals that currently do not have their own institutional antibiograms. In addition, we have shown that integrating hospital AMR tracking and reporting as part of a larger voluntary provincial ASP initiative is a feasible approach to capturing AMR data that have historically been underrepresented from national surveillance initiatives.
Limitations
Although the Ontario Hospital Antibiogram is an important first step to tracking and reporting provincial AMR data, several challenges were associated with the development process. Because variability in regional representativeness may limit generalizability and applicability of our results to clinicians, we attempted to decrease the burden of participating hospitals by accepting data in several formats. We accepted information from multiple years (ie, 2014 and later) and summarized isolates collected from various anatomical sites (ie, all clinical isolates, urine only, blood only, etc), at various levels of stratification (ie, hospital-wide, wards only, ICU only, etc), and from various patient populations (ie, inpatient only, and all patients when we were not able to distinguish inpatient from outpatient data). That said, for sites that provided ward-specific antibiograms and hospital-wide antibiograms, we only included data from the hospital-wide antibiograms.
In addition to the variability of the data received from each hospital, individual microbiology laboratories that perform susceptibility testing and reporting also have differing practices. In Ontario, most laboratories currently follow IQMH external quality assurance programs for the province of Ontario and CLSI guidelines (9,18). However, lab procedures, including antimicrobial susceptibility testing, are far from standardized in the province. Reasons for this lack of standardization include differences in available resources and variability in the tests that are performed. For example, community labs will complete primary susceptibilities and identification, but if additional information is needed, the specimen is sent to a reference laboratory for further work-up.
Although 75.0% of participating hospitals that submitted data indicated that their antibiograms were developed in accordance with CLSI guidelines, which includes eliminating repeat isolates, we did not formally assess this because such an analysis was beyond the scope of this study. However, other jurisdictions have previously demonstrated that full adherence to CLSI guidelines for hospital antibiograms is uncommon, and future work should focus on identifying opportunities for improvement (19).
Finally, we also acknowledge that the tool contains only information about hospitals that voluntarily responded to the 2018 Ontario ASP Landscape survey, provided antibiogram data, and consented to sharing their responses publicly on the PHO website. In this case, we found that large community and acute teaching hospitals had the highest participation rate; however, small community and CCC and rehabilitation hospitals were less likely to contribute microbiological data. Although we did not formally explore reasons for not sharing an antibiogram, some of the issues provided by respondents included time constraints, competing clinical priorities, and data-sharing requirements.
Future directions
The provincial antibiogram serves as a benchmark for the current state of AMR provincially and across health regions. We believe that apart from the content within the tool itself, the interactive functionality was essential in making it relevant to a variety of stakeholders with an interest in ASPs, ranging from front-line clinicians to ASP leaders and decision makers in government, and to other health care system partners. The presence of variability in susceptibility across regions supports the ongoing use of such a tool.
Since launching the Ontario Antibiogram in May 2019, additional microbiological data from hospitals have been uploaded to the tool as a result of targeted outreach in an effort into increase participation. Our next steps include collecting additional antibiogram information with the next iteration of the ASP Landscape Survey, expected to be launched in Fall–Winter 2020–21, and more comprehensive evaluation of the tool to determine potential future enhancements. Collaboration with the Public Health Agency of Canada would enable a wider reach and potentially an extension of this initiative to the rest of the nation. In the future, we plan to streamline the data collection process for any updates to the tool and to encourage consistency and standardization of laboratory testing and reporting.
Ethics Approval:
N/A
Informed Consent:
N/A
Funding:
No funding was received for this article.
Disclosures:
The authors have nothing to disclose.
Peer Review:
This manuscript has been peer reviewed.
Animal Studies:
N/A
References
- 1.Council of Canadian Academies. When antibiotics fail: The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada [Internet]. Ottawa: Council of Canadian Academies, 2019. http://www.deslibris.ca/ID/10102747 (April 25, 2020). [Google Scholar]
- 2.Accreditation Canada. Required organizational practices (ROP) handbook 2016. Ottawa: Accreditation Canada, 2015. [Google Scholar]
- 3.Barlam TF, Cosgrove SE, Abbo LM, et al. Implementing an antibiotic stewardship program: Guidelines by the Infectious Diseases Society of America and the Society for Healthcare Epidemiology of America. Clin Infect Dis;62(10):e51–77. 10.1093/cid/ciw118. Medline: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morency-Potvin P, Schwartz DN, Weinstein RA. Antimicrobial stewardship: how the microbiology laboratory can right the ship. Clin Microbiol Rev 2017;30(1):381–407. 10.1128/CMR.00066-16. Medline: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frost I, Van Boeckel TP, Pires J, Craig J, Laxminarayan R. Global geographic trends in antimicrobial resistance: the role of international travel. J Travel Med. 2019;26(8):1–13. 10.1093/jtm/taz036. Medline: [DOI] [PubMed] [Google Scholar]
- 6.Public Health Agency of Canada. Pan-Canadian framework for action on antimicrobial resistance and antimicrobial use. Can Commun Dis Rep. 2017; 43(11):217–9. 10.14745/ccdr.v43i11a01. Medline: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grant J, Saxinger L, Patrick D. Surveillance of antimicrobial resistance and antimicrobial utilization in Canada [Internet]. Winnipeg, MB: National Collaborating Centre for Infectious Diseases; 2014. https://www.deslibris.ca/ID/244350 (April 25, 2020). [Google Scholar]
- 8.Leung V, Wu JHC, Langford BJ, Garber G. Landscape of antimicrobial stewardship programs in Ontario: a survey of hospitals. CMAJ Open 2018;6(1):e71–6. 10.9778/cmajo.20170111. Medline: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. Microbiology standards [Internet]. Annapolis Junction (MD): The Institute; 2021. https://clsi.org/standards/products/microbiology/ (April 25, 2020). [Google Scholar]
- 10.Canadian Antimicrobial Resistance Alliance. CANWARD pathogens [Internet]. n.d. http://can-r.ca/study.php?study=canw2017&year=2017 (April 25, 2020).
- 11.Canadian Nosocomial Infection Surveillance Program. Summary report of healthcare associated infection (HAI), antimicrobial resistance (AMR) and antimicrobial use (AMU) surveillance data from January 1, 2013 to December 31, 2017 [Internet]. Ottawa: Public Health Agency of Canada; 2019. https://www.canada.ca/en/public-health/services/publications/science-research-data/summary-report-healthcare-associated-infection-antimicrobial-resistance-antimicrobial-use-surveillance-data-2013-2017.html (April 25, 2020). [Google Scholar]
- 12.Denisuik AJ, Garbutt LA, Golden AR, et al. Antimicrobial-resistant pathogens in Canadian ICUs: results of the CANWARD 2007 to 2016 study. J Antimicrob Chemother 2019;74(3):645–53. 10.1093/jac/dky477. Medline: [DOI] [PubMed] [Google Scholar]
- 13.Center for Disease Dynamics, Economics & Policy. ResistanceMap [Internet]. Washington (DC): The Center; 2021. https://resistancemap.cddep.org/ (April 25, 2020). [Google Scholar]
- 14.HealthMap. ResistanceOpen [Internet]. https://resistanceopen.org/ (April 25, 2020).
- 15.Alberta Health Services. Laboratory services: antibiograms [Internet]. Edmonton (AB): Alberta Health Services; 2021. https://www.albertahealthservices.ca/lab/Page3294.aspx (April 25, 2020). [Google Scholar]
- 16.LifeLabs. Antibiograms: Ontario. Toronto: LifeLabs; 2021. https://www.lifelabs.com/healthcare-providers/reports/antibiograms/ (April 25, 2020). [Google Scholar]
- 17.Ontario Agency for Health Protection and Promotion (Public Health Ontario), Institute for Quality, Management in Healthcare. Antimicrobial resistance in common hospital pathogens in Ontario: annual laboratory and hospital survey report 2017 [Internet]. Toronto: Queen’s Printer for Ontario; 2019. https://www.publichealthontario.ca/-/media/documents/aro-survey-2017.pdf (Accessed month day, year). [Google Scholar]
- 18.Institute for Quality Management in Healthcare. Institute for Quality Management in Healthcare. Toronto: The Institute; 2021. https://iqmh.org/ (April 25, 2020). [Google Scholar]
- 19.Moehring RW, Hazen KC, Hawkins MR, Drew RH, Sexton DJ, Anderson DJ. Challenges in preparation of cumulative antibiogram reports for community hospitals. J Clin Microbiol 2015;53(9):2977–82. 10.1128/JCM.01077-15. Medline: [DOI] [PMC free article] [PubMed] [Google Scholar]


