Abstract
Type 1 fimbriae can be expressed by most Escherichia coli strains and mediate mannose-sensitive (MS) adherence to mammalian epithelial cells. However, the role of type 1 fimbriae in enteric pathogenesis has been unclear. Expression of type 1 fimbriae in E. coli is phase variable and is associated with the inversion of a short DNA element (fim switch). Forty-six strains of diarrheagenic E. coli were examined for the expression of type 1 fimbriae. Only four of these strains were originally type 1 fimbriated. Seventeen strains, originally nonfimbriated, expressed type 1 fimbriae in association with off-to-on inversion of the fim switch, after serial passages in static culture. The switching frequencies of these strains, from fimbriate to nonfimbriate, were greater than that of the laboratory strain E. coli K-12. None of the 16 strains of serovar O157:H7 or O157:H− expressed type 1 fimbriae after serial passages in static culture. The nucleotide sequence analysis of the fim switch region revealed that all of the O157:H7 and O157:H− strains had a 16-bp deletion in the invertible element, and the fim switch was locked in the “off” orientation. The results suggest that expression of type 1 fimbriae may be regulated differently in different E. coli pathogens causing enteric infections.
Adherence is generally an initial, prerequisite step for some strains of Escherichia coli in successful colonization of a specific host mucosal tissue (10, 19, 25, 39, 44).
Type 1 fimbriae, which are found on the majority of clinical isolates of E. coli (5), bind to mannose-containing receptors on epithelial cells (33) and on leukocytes (2). Type 1 fimbriae may be important in the pathogenesis of urinary tract infections (24, 41) and play an important role in enterobacterial communicability (3). However, the role of type 1 fimbriae in enteric infection remains unclear. A series of investigations using streptomycin-treated mice colonized with a human fecal E. coli isolate demonstrated that type 1 fimbriae are expressed in the intestinal tract, in vivo, and may be involved in the colonization of the intestinal tract by E. coli (22, 23). It has been shown that type 1 fimbriae are excellent immunogens (13, 37). Thus, the capacity to rapidly switch their expression from “on” to “off” would be advantageous to the organism and consequently important in pathogenesis.
Type 1 fimbriae are encoded by a fim gene cluster, including at least nine genes required for its biosynthesis (20, 35), and are composed primarily of the structural subunit, FimA (18). The mannose-sensitive adhesive function is provided by a small amount of the adhesin subunit, FimH, located at the tip of the fimbrial shaft (15). The expression of these fimbriae is phase variable, depending on the orientation of the 314-bp invertible element located between two 9-bp inverted repeats (1). This element contains a promoter which drives the transcription of the fim subunit genes in one orientation (on) but not the other (off). This inversion is catalyzed by two site-specific recombinases, FimB and FimE, encoded upstream of this invertible element. FimB can catalyze inversion in both directions (on to off, off to on), but FimE can catalyze inversion in only one direction (on to off) (12, 29).
In this study, we examined the expression and switching frequencies of type 1 fimbriae of diarrheagenic E. coli strains. We also showed that all of the O157:H7 and O157:H− strains tested in this work abolish expression of type 1 fimbriae due to a 16-bp deletion in the invertible element and the locking of the fim switch in the “off” orientation.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli strains used in this study are described in Table 1. The strains were collected from Japan (Fukuoka, Kumamoto, Osaka, and Tokyo), Thailand, the United States, Canada, Great Britain, and Denmark. Bacteria were grown in Luria-Bertani (LB) broth (40) containing 5 g of sodium chloride (Wako, Osaka, Japan), 5 g of yeast extract (Difco, Detroit, Mich.), and 10 g of tryptone (Difco) per liter and in LB agar, which was LB broth containing 1.5% agar (Wako). To promote expression of type 1 fimbriae, organisms were serially passaged in a static, nonaerated broth culture of brain heart infusion (BHI) broth (Eiken Chemical, Tokyo, Japan) at 37°C for 10 days in small test tubes as described previously (34). Expression of type 1 fimbriae by bacteria was monitored by the mannose-sensitive hemagglutination (MSHA) test.
TABLE 1.
Bacterial strains used in this study
| Strain | Serotype | Description | MSHAa | 16-bp deletion in invertible elementb | Source or referencec |
|---|---|---|---|---|---|
| STEC strains | |||||
| 96-7 | O157:H7 | ST1, ST2 | − | + | FIHES |
| 96-24 | O157:H7 | ST1, ST2 | − | + | FIHES |
| 98E1 | O157:H7 | ST1, ST2 | − | + | FIHES |
| 98E2 | O157:H7 | ST2 | − | + | FIHES |
| 98E3 | O157:H7 | ST1, ST2 | − | + | FIHES |
| 98E5 | O157:H7 | ST1, ST2 | − | + | FIHES |
| CL-49 | O157:H7 | ST2 | − | + | 6 |
| 86-24 | O157:H7 | ST2 | − | + | 14 |
| 467 | O157:H7 | ST2 | − | + | FCIHE |
| 468 | O157:H7 | ST2 | − | + | FCIHE |
| Sakai | O157:H7 | ST1, ST2 | − | + | 31 |
| E333 | O157:H− | ST2 | − | + | FCIHE |
| 98E6 | O157:H− | ST2 | − | + | FIHES |
| E32511 | O157:H− | ST2 | − | + | 28 |
| 97E12 | O26:H11 | ST1 | + | − | FIHES |
| 97E16 | O26:H11 | ST1 | + | − | FIHES |
| 97E2 | O111:H− | ST1 | + | − | FIHES |
| 97E22 | O111:H− | ST1 | + | − | FIHES |
| Non-ST-producing O157 strains | |||||
| FSE5 | O157:H7 | − | + | FIHES | |
| FSE13 | O157:H7 | − | + | FIHES | |
| 96-39 | O157:H45 | +* | − | FIHES | |
| 96-11 | O157:H45 | + | − | FIHES | |
| EPEC strains | |||||
| 1646-3 | O55:H− | +* | − | IMSUT | |
| 2 | O55:H6 | + | − | SSI | |
| F76193 | O55:H6 | − | − | SSI | |
| 85/91-5 | O55:H7 | +* | − | SSI | |
| 70 | O55:H7 | + | − | SSI | |
| F57076 | O55:H7 | + | − | SSI | |
| PE163 | O55:H7 | + | − | OPIPH | |
| PE182 | O55:H7 | + | − | OPIPH | |
| 807-257 | O55:H7 | + | − | OPIPH | |
| F43341 | O111:H12 | + | − | SSI | |
| 118 | O111:H12 | + | − | SSI | |
| TEC1101 | O55:H10 | − | ND | TMRLPH | |
| TEC1117 | O55:H10 | − | ND | TMRLPH | |
| E98445 | O55:H10 | − | ND | TMRLPH | |
| ETEC strains | |||||
| BK005 | − | ND | RIMDOU | ||
| CH5667 | + | − | CSTRI | ||
| CH28991 | +* | − | CSTRI | ||
| O22 | + | − | This laboratory | ||
| O29 | − | ND | This laboratory | ||
| EAggEC strains | |||||
| TL100 | − | ND | 51 | ||
| 17-2 | + | ND | 46 | ||
| E2 | − | − | 47 | ||
| 253-1-1 | + | − | RIMCJ | ||
| 452-1-1 | − | ND | RIMCJ | ||
| Laboratory strain (DH5α) | + | − | 40 |
Determined by hemagglutination of guinea pig erythrocytes with or without 2% d-mannose. +, hemagglutinating strain after serial passages; +*, originally hemagglutinating strain (at start of this study).
ND, not determined; no amplification in PCR with primers used in this study.
FIHES, Fukuoka Institute of Health and Environmental Sciences; FCIHE, Fukuoka City Institute for Hygiene and Environment; IMSUT, The Institute of Medical Science, The University of Tokyo; SSI, The State Serum Institute, Copenhagen, Denmark; OPIPH, The Osaka Prefectural Institute of Public Health; TMRLPH, The Tokyo Metropolitan Research Laboratory of Public Health; RIMDOU, The Research Institute for Microbial Diseases, Osaka University; CSTRI, The Chemo-Sero-Therapeutic Research Institute, Kumamoto, Japan; RIMCJ, The Research Institute, International Medical Center of Japan.
Hemagglutination assay.
MSHA was determined in phosphate-buffered saline (PBS) with a 2% (wt/vol) suspension of guinea pig erythrocytes, with or without 1% (wt/vol) d-mannose (Sigma, St. Louis, Mo.). Twenty microliters of the erythrocyte suspension with or without d-mannose was placed on a glass slide, and an equal volume of a bacterial suspension (108 CFU) was added. The slide was gently rotated for 2 min while monitoring for visible HA was conducted.
PCR amplification of the invertible element and its flanking regions.
Chromosome DNA was isolated by using a GenomicPrep Cells and Tissue DNA Isolation Kit according to the manufacturer's instructions (Amersham Pharmacia Biotech, Piscataway, N.J.). PCR amplifications, for the total volume of 100 μl, contained 1× standard PCR buffer (Promega, Madison, Wis.), 2.5 mM of MgCl2, 200 μM each of the four deoxyribonucleotides, 0.25 μM each primer, 2.5 μg of chromosome DNA, and 2.5 U of Taq DNA polymerase (Promega). PCR was carried out for a total of 30 cycles, each consisting of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and extension at 72°C for 1 min, with a final extension of 5 min at 72°C, using a PC-700 thermal cycler (ASTEC, Fukuoka, Japan). Oligonucleotides used in PCR amplification were as follows. P-3 (5′GTGCATCGAAATATTCGCCATACT-3′) (5′-biotin labeled) and P-2 (5′ACGTCCCTGAACCTGGGTAGGTTA-3′) were used for sequencing analysis, and non-biotin-labeled P-3 and P-4 (5′-AGTCGTTCTGTACACTTTGTTTTG-3′) were used for the orientation analysis of the invertible element. All synthetic oligonucleotides were purchased from Nippon Flour Mills (Tokyo, Japan).
Determination of invertible element orientation.
The orientation in the chromosome of the invertible DNA element containing the fimA promoter was determined using a PCR-based assay. This method exploited a restriction fragment length dimorphism, which arises out of the orientation-dependent location of a unique SnaBI restriction site within the amplified DNA (12) (see Fig. 1A).
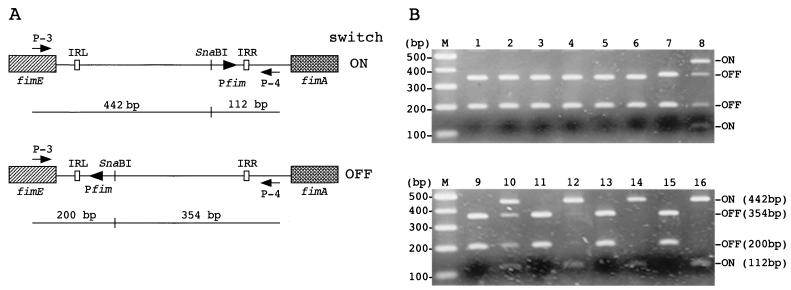
FIG. 1.
Orientation of the switch-regulating phase variation in E. coli. (A) Schematic representation of the fim switch showing the positions of primers and restriction sites used in this study. The orientation of the fim switch can be determined by combined PCR and restriction analysis, since SnaBI cuts asymmetrically. IRL and IRR, left and right inverted repeats, respectively. The fimA promoter (Pfim) is indicated. (B) Agarose gel (2%) electrophoresis of SnaBI digests of PCR products. Lanes M, molecular weight markers; lanes 1 and 2, 96-7 (O157:H7); lanes 3 and 4, CL-49 (O157:H7); lanes 5 and 6, E333 (O157:H−); lanes 7 and 8, 96-11 (O157:H45); lanes 9 and 10, 97E12 (O26:H11); lanes 11 and 12, 97E2 (O111:H−); lanes 13 and 14, 70 (O55:H7); lanes 15 and 16, DH5α. Lanes 1, 3, 5, 7, 9, 11, 13, and 15 are from strains amplified at the start of this study (MSHA−); lanes 2, 4, and 6 (MSHA−) and lanes 8, 10, 12, 14, and 16 (MSHA+) are from strains amplified after 10 static serial passages.
Bacteria (108 CFU) were harvested from a cell suspension before and after being serially passaged 10 times. This suspension was boiled for 10 min to release genomic DNA. An upstream sequence of the fimA gene, including the fim switch region, was amplified with oligonucleotides P-3 and P-4 to generate a 554-bp DNA product for the K-12 strain JE2571 (18). Amplified products (2 μl; approximately 0.1 μg) were digested with 5 U of SnaBI (TAKARA, Shiga, Japan). Digested PCR products were resolved on 2% agarose gels. Phase-oh populations of bacteria yielded two DNA fragments of 442 and 112 bp, whereas phase-off populations yielded two fragments of 200 and 354 bp (Fig. 1). Mixed populations of both phase “on” and phase “off” contained a mixture of all four fragments.
DNA sequencing analysis.
After PCR amplifications, 40 μl of Dynabeads streptavidin (Dynal, Oslo, Norway) was added to an equal volume of PCR products, and amplified DNA was collected in a magnet stand (Dynal). After separation of DNA strands by addition of 40 μl of 0.1 N NaOH, sense strands were collected in a magnet stand. DNA sequencing analysis was performed with a Sequencing PRO Kit (Toyobo, Tokyo, Japan) using single-stranded DNA, primer P-4, and the radioactive deoxyribonucleotide [α-33P]dCTP (Amersham Pharmacia Biotech). Sequencing reactions were carried out according to the manufacturer's instructions.
Measurement of frequency of production of MSHA− colonies by the MSHA+ colony on plate culture.
After promotion of type 1 fimbriation by 10 passages in a static culture, the cell suspension was serially diluted in PBS and spread on an LB plate. After overnight growth, three individual blocks of agar, each bearing a single MSHA+ colony, were cut out, transferred to 5 ml of PBS in separate test tubes, and agitated vigorously. Bacterial-cell suspensions were serially diluted in PBS and spread on the LB plates. The MSHA statuses of 50 randomly selected colonies were determined after overnight growth. The switching frequencies were determined as the means of frequencies from the three separate cultures, as described previously (11).
Phylogenetic characterization.
Phylogenetic analysis was conducted with the nucleotide sequence of the 314-bp invertible element. A phylogenetic tree based on DNA sequences of this element was constructed by using the CLUSTAL program (version 1.5) (45).
RESULTS
Expression of type 1 fimbriae and orientation of the fim switch.
Expression of type 1 fimbriae was monitored by MSHA. At the start of this study, only four strains (96-39, 1646-3, 85/91-5, and CH28991) exhibited the MSHA-positive phenotype. After 2 to 8 consecutive passages in static broth, 17 strains expressed type 1 fimbriae. None of the O157:H7 or O157:H− strains produced type 1 fimbriae; neither did F76193 (O55:H6), TEC1101, TEC1117, and E98445 (O55:H10), BK005 and O29 (enterotoxigenic E. coli [ETEC]), or TL100, E2, and 452-1-1 (enteroaggregative E. coli [EAggEC]) (Table 1).
The expression of type 1 fimbriae is phase variable and depends on the orientation of an invertible 314-bp DNA switch (1). In E. coli strain K-12, an asymmetric SnaBI restriction site within the 314-bp invertible element, upstream of the fimA gene, allows for the determination of the orientation of the switch responsible for phase variation (12). When DNAs were amplified by PCR with one set of primer sequences upstream of the E. coli K-12 fimA gene including an invertible sequence and then digested with SnaBI, DNA from MSHA− strains showed fragments of 354 and 200 bp, representing the “off” orientation, while MSHA+ strains showed fragments of 442 and 112 bp, representing the “on” orientation (Fig. 1A and Fig. 1B, lanes 15 and 16). DNA from MSHA− strains (before and after passages) amplified with the same primers and digested with SnaBI indicated that fim switches were in the “off” orientation (Fig. 1B, lanes 1, 2, 3, 4, 5, 6, 7, 9, 11, and 13). Restriction analysis of PCR products from the strains which became MSHA+ after serial passages showed the presence of all four SnaBI bands, indicating that MSHA+ strains were composed of a mixture of MSHA+ and MSHA− cells. (Fig. 1B, lanes 8, 10, 12, and 14).
These results confirm that the expression of type 1 fimbriae and the orientation of the fim switch are related as described previously (1).
DNA sequencing analysis.
PCR products flanking the switch region in the phase-off orientation were directly sequenced. Sequences are shown in Fig. 2. The PCR products of eight strains (TEC1101, TEC1117, E98445, BK005, O29, TL100, 17-2, and 452-1-1) were not amplified, and DNA sequences of these strains could not be determined.
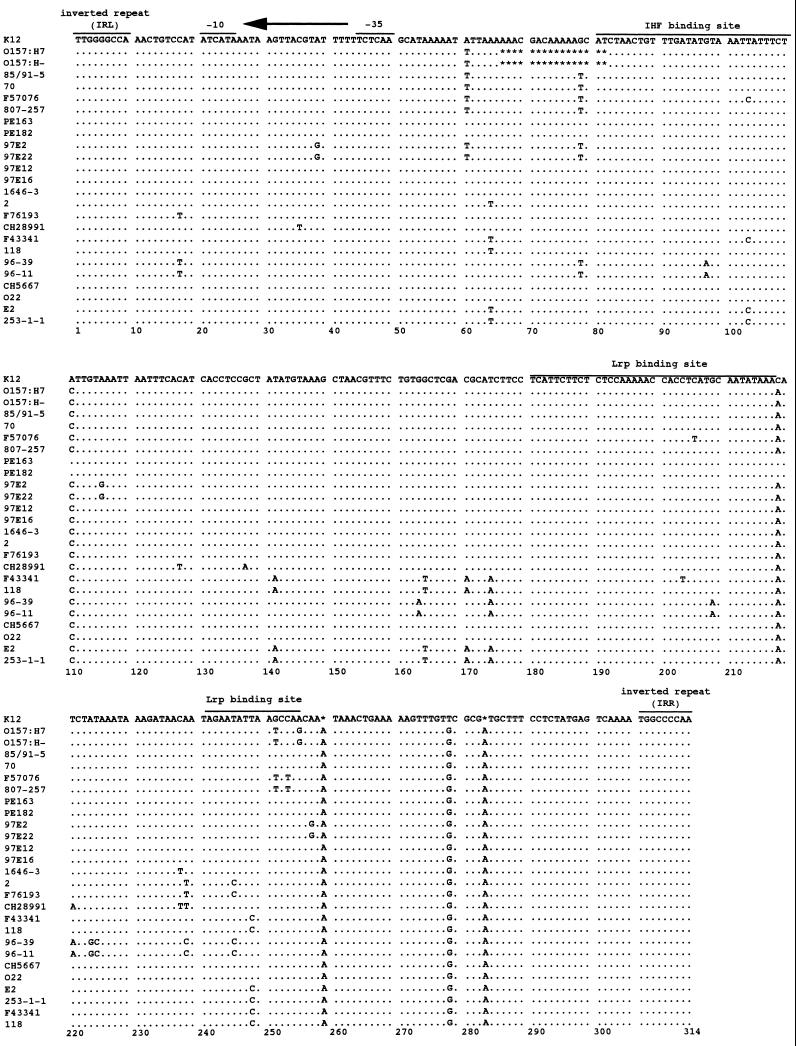
FIG. 2.
DNA sequence comparison of the invertible element of E. coli K-12 (GenBank accession number X00981) and strains used in this study. O157:H7 and O157:H− strains have completely identical sequences. Dots indicate identity; stars indicate gaps. These sequences are shown in phase-off orientation. The inverted repeats that border the fim switch and the fimA promoter (1) are overlined and allow for the indication of the direction of transcription. Binding sites for IHF (4) and Lrp (38) are also shown.
All 16 strains of serovar O157:H7 or O157:H− had perfectly identical sequences and had a unique 16-bp deletion (bp 66 to 81) in the switch region (Fig. 2). This deletion included 2 bp (bp 80 to 81) of a putative binding site of integration host factor (IHF), which is required for inversion of the fim switch (4). This deletion was not observed in the other strains tested in this study.
DNA sequence comparison of invertible elements of E. coli K-12 and clinical isolates used in this study revealed that nucleotide changes (A to C at bp 110; C to A at bp 218, and T to G at bp 278) and insertions (A at bp 259; A at bp 283) were observed in most of the pathogenic strains (Fig. 2).
Switching frequency of MSHA+ to MSHA− on plate culture.
The frequencies of MSHA+-to-MSHA− switching were measured for strains expressing the MSHA+ phenotype. Data are shown in Table 2. The pathogenic strains expressing the MSHA+ phenotype showed MSHA+-to-MSHA− changes on plate culture; however, the K-12 strain DH5α did not produce any MSHA− colonies.
TABLE 2.
On-to-off switching in strains used in this study
| Strain | Serotype | Switching frequencya |
|---|---|---|
| STEC strains | ||
| 97E12 | O26:H11 | 0.101 |
| 97E16 | O26:H11 | 0.147 |
| 97E2 | O111:H− | 0.168 |
| 97E22 | O111:H− | 0.154 |
| Non-ST-producing O157 strains | ||
| 96-39 | O157:H45 | 0.110 |
| 96-11 | O157:H45 | 0.101 |
| EPEC strains | ||
| 1646-3 | O55:H− | 0.186 |
| 2 | O55:H6 | 0.035 |
| 85/91-5 | O55:H7 | 0.134 |
| 70 | O55:H7 | 0.021 |
| F57076 | O55:H7 | 0.101 |
| PE163 | O55:H7 | 0.001 |
| PE182 | O55:H7 | 0.029 |
| 807-257 | O55:H7 | 0.077 |
| F43341 | O111:H12 | 0.077 |
| 118 | O111:H12 | 0.134 |
| ETEC strains | ||
| CH5667 | 0.027 | |
| CH28991 | 0.001 | |
| O22 | 0.004 | |
| EAggEC strains | ||
| 17-2 | 0.022 | |
| 253-1-1 | 0.026 | |
| K-12 strain (DH5α) | 0b |
Switching frequency per cell per generation. Rates were determined as the means of the frequencies calculated from three separate colonies based on a method described previously (11).
The transition rates from fimbriate to nonfimbriate of the K-12 strain CSH50 were reported as 0.001 per cell per generation (7).
The frequency of MSHA+-to-MSHA− phenotype switching was high (>0.1 per cell per generation) in non-O157 Shiga-like toxin-producing E. coli (STEC) strains. Enteropathogenic E. coli (EPEC) strains showed both higher and lower frequencies than STEC strains. The switching frequencies for ETEC and EAggEC strains were lower than those for other pathogenic E. coli strains.
Phylogenetic analysis based on sequences of the switching element.
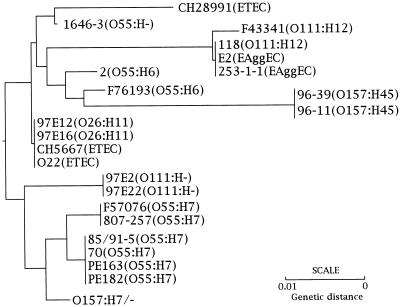
A phylogenetic tree based on the DNA sequences of the fim switch region was constructed (Fig. 3). Strains of the same serotypes were clustered into closely related or identical branches.
FIG. 3.
Phylogenetic tree showing the genetic relationship among the strains used in this study on the basis of DNA sequences of invertible elements. The phylogenetic tree was constructed with CLUSTAL, version 1.5 (45). The program initially calculates pairwise similarity scores based on the method of Wilbur and Lipman (50). Based on these scores, the program constructs a phylogenetic tree using the average linkage cluster analysis method (43).
This phylogenetic tree indicates that the O157:H7, O157:H−, and O55:H7 strains were clustered into a single branch, while O111:H− strains formed another distinct branch. The O26:H11 strains and strains CH5667 and O22 (ETEC) formed a third branch. The other strains represented a fourth branch. The O157:H45 and O55:H6 strains were clustered together and were related to the O111:H12 and EAggEC strains, but they were quite divergent from the O157:H7 and O55:H7 strains.
DISCUSSION
We investigated the expression of type 1 fimbriae in E. coli isolates from gastrointestinal tract infections. Of the 46 strains used in this study, only 4 strains were originally type 1 fimbriated. It has been shown that certain growth conditions favor the isolation of fimbriate bacteria (growth in static broth, anaerobic growth), whereas other conditions favor afimbriate bacteria (exponential growth in well-aerated broth, growth on agar) (34). After serial passages in static culture, 17 strains expressed type 1 fimbriae in association with fim switch off-to-on inversion (Table 1 and Fig. 1). None of the 16 strains belonging to serotype O157:H7 or O157:H− expressed type 1 fimbriae after more than 20 passages in static culture (Table 1). Frequencies of switching from an MSHA+ to an MSHA− phenotype were greater in EPEC and STEC strains than in the K-12 strain (Table 2).
Oral challenge of humans with EPEC strain E2348/69 resulted in an immune response to EPEC type 1 fimbriae (16). Type 1 fimbriae might be expressed during the course of EPEC infections. The type 1 fimbriated strain was more susceptible to phagocytosis than the nonfimbriated strain (32), and type 1 fimbriae were found to be a good immunogen (21, 37). There is no consensus, on the role of type 1 fimbriae, either in susceptibility to phagocytosis or in pathogenicity. Type 1 fimbriae may be disadvantageous for bacteria in the presence of phagocytes, but other reports indicate that they could be advantageous for bacteria which are entering the circulatory system or are located within phagocytes (17, 27).
Type 1 fimbriae might be disadvantageous for E. coli strains which colonize the mucosa by an attaching and effacing (A/E) lesion, because they induce an immune response (16). In the present study, STEC strains other than O157:H7 and O157:H− serotypes produced type 1 fimbriae after serial passages in static culture. Serogroup O157:H7 strains are most prevalent among STEC strains. The type 1 afimbriated situation might be advantageous for O157:H7 strains.
Sherman et al. (42) and Durno et al. (6) reported that the E. coli O157:H7 strain CL-49 expressed type 1 fimbriae after serial passages in static culture and could adhere to human and rabbit epithelial cells, while binding could not be demonstrated for type 1 afimbriated O157:H7 strains. However, we could not isolate any type 1 fimbriated O157:H7 or O157:H− strains, after serial passages of 16 O157:H7 and O157:H− strains, including strain CL-49, in static culture. DNA sequence analysis revealed that all 16 strains had a 16-bp deletion in the invertible element and that the fim switch was locked in the “off” orientation. It would be of interest to elucidate the mechanisms of the type 1 fimbrial expression of CL-49 in the studies cited above.
Li et al. (26) demonstrated that a 16-bp sequence 5′ to fimA was absent in O157 strains and suggested that a PCR assay using the primer flanking the 16-bp deletion offers a simple, rapid, and reliable means to detect E. coli strains of the O157:H7 serotype. However, they did not investigate if fimbrial expression was affected by this 16-bp deletion.
Enami et al. (9) reported that expression of type 1 fimbriae was not observed in verotoxin-producing E. coli O157, though the genetic mechanism for the lack of expression of type 1 fimbriae was not studied.
We demonstrated that the 16-bp deletion in the fim switching region is a possible molecular mechanism responsible for the lack of type 1 fimbrial expression of serovars O157:H7 and O157:H−. Pathogenic strains have obtained virulence genes such as stx or eae in the case of STEC. Such bacteria may abandon or destroy some gene systems while acquiring others. Type 1 fimbrial expression might be subject to such alterations in the genetic determinant in O157:H7 strains.
A phylogenetic tree was constructed based on the DNA sequence of the fim switching region (Fig. 3). Strikingly, the DNA sequence used as a basis for construction of the phylogenetic tree with the strains used in this study almost perfectly reflected the clonal relationships among E. coli strains associated with enteric disease, as defined by Whittam et al. (49), who compared the mutilocus enzyme profiles of E. coli strains. We are convinced that the ubiquity of type 1 fimbriae and the moderate evolutionary divergence of their DNA sequences make them very useful molecular chronometers for phylogenetic analyses.
It has not been known whether E. coli type 1 fimbriae play a direct role in enteric pathogenesis (8, 22, 23, 30). Type 1 fimbrial adhesin specifically binds mannose, which is ubiquitous in mammalian cell membranes. Thus, these structures have the potential to attach to a wide variety of host cells. It is possible that the environment within the host induces a transient expression of fimbrial structures. It has also been reported that type 1 fimbriae or MSHA fimbriae are required for biofilm formation in E. coli (36) and Vibrio cholerae (48). Type 1 fimbriae may promote survival in aquatic environments, such as wells and ponds, by allowing fimbriated cells to form biofilms at the water-air interface, thereby contributing to bacterial survival and the outbreak of disease.
ACKNOWLEDGMENTS
We thank Bernt Eric Uhlin for critical reading of the manuscript and L. Saza for improving the language of this paper.
This work was supported by Grants-in-Aid for Scientific Research (B) from the Ministry of Education, Science, Culture and Sports of Japan and by grants from Kurozumi Medical Foundation.
REFERENCES
- 1.Abraham J M, Freitag C S, Clements J R, Eisenstein B I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bar-Shavit Z, Ofek I, Goldman R, Mirelman D, Sharon N. Mannose residues on phagocytes as receptors for the attachment of Escherichia coli and Salmonella Typhi. Biochem Biophys Res Commun. 1977;78:455–460. doi: 10.1016/0006-291x(77)91276-1. [DOI] [PubMed] [Google Scholar]
- 3.Bloch C A, Stocker B A, Orndorff P E. A key role for type 1 pili in enterobacterial communicability. Mol Microbiol. 1992;6:697–701. doi: 10.1111/j.1365-2958.1992.tb01518.x. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield I C, Kulasekara D H, Eisenstein B I. Integration host factor stimulates both FimB- and FimE-mediated site-specific DNA inversion that controls phase variation of type 1 fimbriae expression in Escherichia coli. Mol Microbiol. 1997;23:705–717. doi: 10.1046/j.1365-2958.1997.2241615.x. [DOI] [PubMed] [Google Scholar]
- 5.Duguid J P, Old D C. Adhesive properties of Enterobacteriaceae. In: Beachey E H, editor. Bacterial adherence. London, United Kingdom: Chapman and Hall, Ltd.; 1980. pp. 185–217. [Google Scholar]
- 6.Durno C, Soni R, Sherman P. Adherence of vero cytotoxin-producing Escherichia coli serotype O157:H7 to isolated epithelial cells and brush border membranes in vitro: role of type 1 fimbriae (pili) as a bacterial adhesin expressed by strain CL-49. Clin Investig Med. 1989;12:194–200. [PubMed] [Google Scholar]
- 7.Eisenstein B I. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science. 1981;214:337–339. doi: 10.1126/science.6116279. [DOI] [PubMed] [Google Scholar]
- 8.Elliott S J, Kaper J B. Role of type 1 fimbriae in EPEC infections. Microb Pathog. 1997;23:113–118. doi: 10.1006/mpat.1997.0135. [DOI] [PubMed] [Google Scholar]
- 9.Enami M, Nakasone N, Honma Y, Kakinohana S, Kudaka J, Iwanaga M. Expression of type I pili is abolished in verotoxin-producing Escherichia coli O157. FEMS Microbiol Lett. 1999;179:467–472. doi: 10.1111/j.1574-6968.1999.tb08764.x. [DOI] [PubMed] [Google Scholar]
- 10.Gaastra W, de Graaf F K. Host-specific fimbrial adhesins of noninvasive enterotoxigenic Escherichia coli strains. Microbiol Rev. 1982;46:129–161. doi: 10.1128/mr.46.2.129-161.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gally D L, Bogan J A, Eisenstein B I, Blomfield I C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–6193. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gally D L, Leathart J, Blomfield I C. Interaction of FimB and FimE with the fim switch that controls the phase variation of type 1 fimbriae in Escherichia coli K-12. Mol Microbiol. 1996;21:725–738. doi: 10.1046/j.1365-2958.1996.311388.x. [DOI] [PubMed] [Google Scholar]
- 13.Girón J A, Ho A S, Schoolnik G K. An inducible bundle-forming pilus of enteropathogenic Escherichia coli. Science. 1991;254:710–713. doi: 10.1126/science.1683004. [DOI] [PubMed] [Google Scholar]
- 14.Griffin P M, Ostroff S M, Tauxe R V, Greene K D, Wells J G, Lewis J H, Blake P A. Illnesses associated with Escherichia coli O157:H7 infections. A broad clinical spectrum. Ann Intern Med. 1988;109:705–712. doi: 10.7326/0003-4819-109-9-705. [DOI] [PubMed] [Google Scholar]
- 15.Hanson M S, Brinton C C., Jr Identification and characterization of E. coli type-1 pilus tip adhesion protein. Nature. 1988;332:265–268. doi: 10.1038/332265a0. [DOI] [PubMed] [Google Scholar]
- 16.Karch H, Heesemann J, Laufs R, Kroll H P, Kaper J B, Levine M M. Serological response to type 1-like somatic fimbriae in diarrheal infection due to classical enteropathogenic Escherichia coli. Microb Pathog. 1987;2:425–434. doi: 10.1016/0882-4010(87)90049-0. [DOI] [PubMed] [Google Scholar]
- 17.Keith B R, Harris S L, Russell P W, Orndorff P E. Effect of type 1 piliation on in vitro killing of Escherichia coli by mouse peritoneal macrophages. Infect Immun. 1990;58:3448–3454. doi: 10.1128/iai.58.10.3448-3454.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klemm P. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Nucleotide sequence and primary structure of the protein. Eur J Biochem. 1984;143:395–399. doi: 10.1111/j.1432-1033.1984.tb08386.x. [DOI] [PubMed] [Google Scholar]
- 19.Klemm P. Fimbrial adhesions of Escherichia coli. Rev Infect Dis. 1985;7:321–340. doi: 10.1093/clinids/7.3.321. [DOI] [PubMed] [Google Scholar]
- 20.Klemm P, Christiansen G. Three fim genes required for the regulation of length and mediation of adhesion of Escherichia coli type 1 fimbriae. Mol Gen Genet. 1987;208:439–445. doi: 10.1007/BF00328136. [DOI] [PubMed] [Google Scholar]
- 21.Korhonen T K, Rhen M. Bacterial fimbriae as vaccines. Ann Clin Res. 1982;14:272–277. [PubMed] [Google Scholar]
- 22.Krogfelt K A. Bacterial adhesion: genetics, biogenesis, and role in pathogenesis of fimbrial adhesins of Escherichia coli. Rev Infect Dis. 1991;13:721–735. doi: 10.1093/clinids/13.4.721. [DOI] [PubMed] [Google Scholar]
- 23.Krogfelt K A, McCormick B A, Burghoff R L, Laux D C, Cohen P S. Expression of Escherichia coli F-18 type 1 fimbriae in the streptomycin-treated mouse large intestine. Infect Immun. 1991;59:1567–1568. doi: 10.1128/iai.59.4.1567-1568.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 25.Levine M M. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 26.Li B, Koch W H, Cebula T A. Detection and characterization of the fimA gene of Escherichia coli O157:H7. Mol Cell Probes. 1997;11:397–406. doi: 10.1006/mcpr.1997.0132. [DOI] [PubMed] [Google Scholar]
- 27.Lock R, Dahlgren C, Linden M, Stendahl O, Svensbergh A, Ohman L. Neutrophil killing of two type 1 fimbria-bearing Escherichia coli strains: dependence on respiratory burst activation. Infect Immun. 1990;58:37–42. doi: 10.1128/iai.58.1.37-42.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.March S B, Ratnam S. Latex agglutination test for detection of Escherichia coli serotype O157. J Clin Microbiol. 1989;27:1675–1677. doi: 10.1128/jcm.27.7.1675-1677.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClain M S, Blomfield I C, Eisenstein B I. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–5314. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCormick B A, Franklin D P, Laux D C, Cohen P S. Type 1 pili are not necessary for colonization of the streptomycin-treated mouse large intestine by type 1-piliated Escherichia coli F-18 and E. coli K-12. Infect Immun. 1989;57:3022–3029. doi: 10.1128/iai.57.10.3022-3029.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michino H, Araki K, Minami S, Takaya S, Sakai N, Miyazaki M, Ono A, Yanagawa H. Massive outbreak of Escherichia coli O157:H7 infection in schoolchildren in Sakai City, Japan, associated with consumption of white radish sprouts. Am J Epidemiol. 1999;150:787–796. doi: 10.1093/oxfordjournals.aje.a010082. [DOI] [PubMed] [Google Scholar]
- 32.Mizunoe Y, Matsumoto T, Haraoka M, Sakumoto M, Kubo S, Kumazawa J. Effect of pili of Serratia marcescens on superoxide production and phagocytosis of human polymorphonuclear leukocytes. J Urol. 1995;154:1227–1230. [PubMed] [Google Scholar]
- 33.Ofek I, Beachey E H. Mannose binding and epithelial cell adherence of Escherichia coli. Infect Immun. 1978;22:247–254. doi: 10.1128/iai.22.1.247-254.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Old D C, Duguid J P. Selective outgrowth of fimbriate bacteria in static liquid medium. J Bacteriol. 1970;103:447–456. doi: 10.1128/jb.103.2.447-456.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orndorff P E, Falkow S. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J Bacteriol. 1984;159:736–744. doi: 10.1128/jb.159.2.736-744.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pratt L A, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 37.Rene P, Dinolfo M, Silverblatt F J. Serum and urogenital antibody responses to Escherichia coli pili in cystitis. Infect Immun. 1982;38:542–547. doi: 10.1128/iai.38.2.542-547.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roesch P L, Blomfield I C. Leucine alters the interaction of the leucine-responsive regulatory protein (Lrp) with the fim switch to stimulate site-specific recombination in Escherichia coli. Mol Microbiol. 1998;27:751–761. doi: 10.1046/j.1365-2958.1998.00720.x. [DOI] [PubMed] [Google Scholar]
- 39.Sack R B. Enterotoxigenic Escherichia coli: identification and characterization. J Infect Dis. 1980;142:279–286. doi: 10.1093/infdis/142.2.279. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Schaeffer A J, Schwan W R, Hultgren S J, Duncan J L. Relationship of type 1 pilus expression in Escherichia coli to ascending urinary tract infections in mice. Infect Immun. 1987;55:373–380. doi: 10.1128/iai.55.2.373-380.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherman P, Soni R, Petric M, Karmali M. Surface properties of the Vero cytotoxin-producing Escherichia coli O157:H7. Infect Immun. 1987;55:1824–1829. doi: 10.1128/iai.55.8.1824-1829.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. New York, N.Y: Freeman; 1973. [Google Scholar]
- 44.Stamm W E, Hooton T M, Johnson J R, Johnson C, Stapleton A, Roberts P L, Moseley S L, Fihn S D. Urinary tract infections: from pathogenesis to treatment. J Infect Dis. 1989;159:400–406. doi: 10.1093/infdis/159.3.400. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vial P A, Robins-Browne R, Lior H, Prado V, Kaper J B, Nataro J P, Maneval D, Elsayed A, Levine M M. Characterization of enteroadherent-aggregative Escherichia coli, a putative agent of diarrheal disease. J Infect Dis. 1988;158:70–79. doi: 10.1093/infdis/158.1.70. [DOI] [PubMed] [Google Scholar]
- 47.Wai S N, Takade A, Amako K. The hydrophobic surface protein layer of enteroaggregative Escherichia coli strains. FEMS Microbiol Lett. 1996;135:17–22. doi: 10.1111/j.1574-6968.1996.tb07960.x. [DOI] [PubMed] [Google Scholar]
- 48.Watnick P I, Fullner K J, Kolter R. A role for the mannose-sensitive hemagglutinin in biofilm formation by Vibrio cholerae El Tor. J Bacteriol. 1999;181:3606–3609. doi: 10.1128/jb.181.11.3606-3609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whittam T S, Wolfe M L, Wachsmuth I K, O/rskov F, O/rskov I, Wilson R A. Clonal relationships among Escherichia coli strains that cause hemorrhagic colitis and infantile diarrhea. Infect Immun. 1993;61:1619–1629. doi: 10.1128/iai.61.5.1619-1629.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilbur W J, Lipman D J. Rapid similarity searches of nucleic acid and protein data banks. Proc Natl Acad Sci USA. 1983;80:726–730. doi: 10.1073/pnas.80.3.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto T, Endo S, Yokota T, Echeverria P. Characteristics of adherence of enteroaggregative Escherichia coli to human and animal mucosa. Infect Immun. 1991;59:3722–3739. doi: 10.1128/iai.59.10.3722-3739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]