Abstract

Antimicrobial resistance is a global health threat that is exacerbated by the overuse and misuse of antibiotics in medicine and agriculture. As an alternative to conventional antimicrobial drugs, phage therapy involves the treatment of infected patients with a bacteriophage that naturally destroys bacterial pathogens. With the re-emergence of phage therapy, novel tools are needed to study phages. In this work we set out to screen and isolate peptide candidates that bind to phages and act as affinity tags. Such peptides functionalized with an imaging agent could serves as versatile tools for tracking and imaging of phages. Specifically, we screened a phage display library for peptides that bind to the Good Vibes phage (GV), which lyses the bacterial pathogen Pseudomonas aeruginosa. Isolated monoclonal library phages featured a highly conserved consensus motif, LPPIXRX. The corresponding peptide WDLPPIGRLSGN was synthesized with a GGGSK linker and conjugated to cyanine 5 or biotin. The specific binding of the LPPIXRX motif to GV in vitro was confirmed using an enzyme-linked immunosorbent assay. We demonstrated imaging and tracking of GV in bacterial populations using the fluorescent targeting peptide and flow cytometry. In conclusion, we developed fluorescent labeled peptides that can bind to bacteriophage GV specifically, which may enable real-time analysis of phage in vivo and monitor the efficacy of phage therapy.
Introduction
Antimicrobial resistance (AMR) complicates treatment of serious infections with conventional antibiotics.1 The World Health Organization (WHO) has declared AMR as one of the top 10 global public health threats,2 with the potential to cause more deaths than major diseases such as HIV and malaria, especially in low-resource settings.3 In the US alone, more than 2.8 million infections with AMR pathogens are reported every year, leading to more than 35,000 deaths.4 AMR is exacerbated by the misuse and overuse of antibiotics in medicine and agriculture, and the lack of novel antibiotics in the drug discovery and development pipeline suggests that the AMR crisis will worsen over time.5
Pseudomonas aeruginosa is one of the leading nosocomial AMR pathogens.3,6 This Gram-negative bacterium is an opportunistic pathogen that rarely infects healthy individuals but is a grave threat to immunocompromised patients.7 Antimicrobials are the gold standard to treat P. aeruginosa infections, but this creates selective pressure that favors the emergence of AMR strains.8 AMR P. aeruginosa can exclude, metabolize or export first-line antibiotics making the infections much harder to treat.9,10
Phage therapy is an alternative to conventional antibiotics involving the use of bacteriophages that naturally infect pathogenic bacteria and kill them.11 Interest in phage therapeutics has been rekindled by the AMR crisis.12 Like other viruses, phages are abundant and ubiquitous nucleoprotein structures comprising a DNA or RNA genome encased in a proteinaceous capsid.13 Phage infection is specific to a particular species or even strains of bacteria, killing the cells by lysis or initiating a latent infection known as lysogeny.14 Phage therapy effectively controls bacterial infections, including P. aeruginosa,15,16 and has been used under emergency authorization to save patients infected with multidrug-resistant bacteria.17−19 The recognition of the potential impact of phage therapy on MDR infections led to the foundation of the first phage therapy center in the US, the Center for Innovative Phage Applications and Therapeutics (IPATH) at UC San Diego.
With the re-emergence of phage therapy, there is an urgent need for novel tools, such as affinity tags, that allow to study phages in the preclinical or clinical setting. For example, surveillance of administered phages in vivo is challenging. Unlike small molecule antibiotics whose concentration can be assessed in blood, intravenous administrated phages are rapidly cleared so that their replicates and concentrations are difficult to evaluate at the infection site.20−22 Another unique challenge is that phages replicate in the patient. Therefore, direct phage labeling with quantum dots, radioisotopes and fluorochromes can be used to monitor the distribution of injected phage, but progeny phage generated following the infection of bacteria cannot be detected.23,24 For these reasons there is a need for novel reagents that allow to study phages in cells and in vivo for imaging and quantification.
Here, we describe an alternative approach to label phage by introducing a fluorophore-conjugated peptide that binds noncovalently to the phage surface. We identified peptides that bind specifically to the Good Vibes phage (GV), which infects P. aeruginosa. The peptides were isolated from a phage display library by three rounds of biopanning. Monoclonal library phages featured a highly conserved consensus motif (LPPIXRX) in the peptide sequences. The corresponding peptide was synthesized, labeled with biotin or cyanine 5 (Cy5); the binding specificity of the peptide for GV was evaluated by enzyme-linked immunosorbent assay (ELISA). Finally, we used flow cytometry to confirm that the Cy5-labeled peptide preferentially bound to GV attached to the surface of P. aeruginosa cells.
Materials and Methods
Preparation of Good Vibes (GV) Phage
Wastewater from the Tijuana River, San Diego, CA (32°33′24.7″N, 117°07′09.7″W) was centrifuged (4000g, 10 min, 4 °C) to remove soil particles and other debris. The supernatant was passed through a 0.45-μm filter and added to an overnight P. aeruginosa culture (incubated overnight at 37 °C in lysogeny broth (LB), shaking at 200 rpm). The culture was diluted OD = 0.2, which represents the stationary phase. The centrifugation, filtration, and inoculation steps were repeated daily for the next 5 days. On the final day, the sample was centrifuged and filtered as above, and 4 μL of the filtrate was spotted onto a P. aeruginosa lawn and incubated at 37 °C overnight to form plaques.
The plaques were picked and suspended in phosphate-buffered saline (PBS) as 10-fold serial dilutions. We plated 100 μL of the 10–5 and 10–6 dilutions with 100 μL P. aeruginosa (OD = 0.2) and 3–4 mL of warm top agar. At these dilutions, well-separated plaques were able to form in the whole-plate assays, allowing us to distinguish between phage morphologies. Plates were left in the incubator overnight at 37 °C. The following day individual plaques were identified, picked, and suspended in PBS. The assays were carried out three times to ensure that all morphologies were identical, thus representing homogeneous phages.
The phages were harvested and purified by picking plaques from the whole-plate assays and suspending them in 200 μL PBS. We then mixed 200 μL of an overnight P. aeruginosa culture (OD = 0.2) with 25 mL of LB and incubated for 20 min at 37 °C, shaking at 200 rpm. We added 25 μL of 0.001 M MgCl2, 25 μL of 0.001 M CaCl2, and 200 μL of the phage suspension and incubated overnight as above. The next day the culture was centrifuged (4000g, 30 min, 4 °C) and passed through two 0.45-μm filters to remove bacterial cells before storage at 4 °C. Titers of the stock were determined by serial dilution using the whole-plate plaque assay method described above.
Isolation of GV-Binding Peptides
GV-binding peptides (GVBPs) were isolated using a PhD-12 Phage Display Peptide Library Kit (New England Biolabs) as previously described, with slight modifications.25 Each well of a Nunc Maxisorp flat-bottom 96-well plate was coated with 1010 phage per unit (pfu) GV overnight at 4 °C. Three rounds of affinity selection were carried out to enrich for GVBPs by increasing the stringency of selection in each round. This was achieved by washing with Tris-buffered saline (TBS) containing increasing concentrations of Tween-20 (TBST) from 0.1% to 0.3% to 0.5%. The enriched phages were eluted and amplified according to the manufacturer’s protocol.
Characterization of GV
GV phages were characterized by transmission electron microscopy (TEM) as previously reported.26 Briefly, 5 μL of 0.2 mg/mL GV phages was diluted in Milli-Q water and was adsorbed to Formvar/carbon-coated 400 mesh copper grids (Electron Microscopy Science) for 2 min. The grid was washed with 5 μL of water for 1 min followed by adsorption of 5 μL of 2% (w/v) uranyl acetate (Fisher Scientific) for 2 min. Solution was removed from the grid by blotting with filter paper. TEM grids were imaged with FEI Tecnai G2 Spirit transmission microscope at 80 kV.
Spot Test Assay
Overnight cultures of P. aeruginosa were incubated in fresh 2× yeast extract tryptone (YT) medium at 37 °C until the OD reached ∼0.2. We then mixed 200 μL of the culture with 3–5 mL warm soft agar (0.5% w/v) and layered it onto the solid agar plate. The soft agar was allowed to solidify for 15 min under flowing air. We then spotted 10 μL of the phage dilution onto the agar plate and air-dried for another 15 min. The plate was then inverted and incubated at 37 °C overnight. The presence of clear zones at the spotting sites was recorded the next day.
Whole Genome Sequencing of the GV Phage
Phage DNA was extracted from 100 μL of high-titer phage lysates using the Qiagen DNeasy Blood and Tissue Kit. DNA quantification was performed and standardized using the Qubit dsDNA HS assay. DNA preparation and sequencing was performed using the Illumina Nextera DNA FlexKit and Adapter Indexes followed by whole genome sequencing using the lab’s MiSeq sequencing platform. Sequencing reads were downloaded from Illumina Basespace, then trimmed for length and quality, and assembled de novo using CLC Genomics Workbench 9.
Polyclonal ELISA
A Nunc Maxisorp flat-bottom 96-well plate was coated with 6 × 109 pfu GV per well (in TBS, pH 8) and incubated overnight at 4 °C. Plates coated with 2% (w/v) bovine serum albumin (BSA) were used as negative controls. The next day, the plates were blocked with 2% (w/v) BSA at room temperature for 1 h, shaking at 800 rpm. The plates were then washed with 0.1% TBST (3 × 1 min) before adding 20 μL of amplified phage from each biopanning cycle to each well in 5% (w/v) BSA. After further incubation at room temperature for 1 h, shaking at 800 rpm, the plates were washed with 0.5% TBST (3 × 5 min) before adding 100 μL of horseradish peroxidase (HRP)-conjugated anti-M13 monoclonal antibody (Abcam ab50370, diluted 1:500) and incubating at room temperature for 1 h, shaking at 800 rpm. After further washes in 0.5% TBST (3 × 5 min), we added 100 μL of the tetramethylbenzidine (TMB) substrate (Thermo Fisher Scientific) to each well. The plates were incubated in the dark for 10 min, and the absorbance was measured at 370 nm using an Infinite 200 Rx plate reader (Tecan Life Sciences) with 25 flashes in 96-well flat-bottom plate mode.
Monoclonal ELISA
The protocol was similar to the polyclonal ELISA. We added 100 μL of amplified phage from the previous biopanning cycle to each well, followed by incubation at room temperature for 1 h, shaking at 800 rpm. Monoclonal phages differing in absorbance between GV and BSA by at least 0.3 units were isolated for DNA sanger sequencing (Eurofins Genomics).
Cross-Reactivity Assay
The protocol was similar to the polyclonal ELISA. We coated wells with 10 μg cowpea mosaic virus (CPMV) or 1010 GV particles to assess their binding activity.
Synthesis and Testing of GV-Binding Peptides
The linear peptides GVBP-biotin (H2N-WDLPPIGRLSGNGGGSK/biotin/-CO2H) and GVBP-FITC (H2N-WDLPPIGRLSGNGGGSK/Cy5/-CO2H) were prepared by solid phase peptide synthesis (GenScript) with a purity of 75%. Peptide sequence H2N-WDLPPIGRLSGN-CO2H was obtained from the most prevalent monoclonal phages in terms of hits. We added the C-terminal linker GGGS to improve flexibility as well as a lysine residue to provide a side-chain amide bond that could be used to attach cyanine 5 (Cy5) by addition or biotin by substitution.
GVBP-biotin ELISA
GV (1, 10, or 65 μg) was coated onto Nunc Maxisorp flat-bottom 96-well plates and incubated for 1 h at room temperature, shaking at 400 rpm. All incubation and washing steps were carried out at room temperature, shaking at 400 rpm, unless otherwise stated. The wells were blocked for 1 h with 300 μL of 5% (w/v) BSA followed by washing once with 0.1% PBST for 5 min. We then added 0.01 μg of GVBP-biotin peptide to the wells and incubated for 1 h. The wells were then washed three times with 0.1% PBST for 5 min each before adding streptavidin HRP conjugate (ab7403) diluted 1:10,000 and incubating for 30 min. The wells were washed another three times as above before adding 100 μL of TMB substrate and incubating for 5–10 min in the dark. The reaction was stopped by adding 50 μL of 2 M H2SO4, and the absorbance was measured at 450 nm using an Infinite 200 Rx plate reader as described above.
GVBP-Cy5 ELISA
GV (0.1, 1, 10, or 63 μg) was coated onto Corning Costar 96-well white solid plates and incubated for 1 h at room temperature, shaking at 400 rpm. All incubation and washing steps were carried out at room temperature, shaking at 400 rpm, unless otherwise stated. The wells were blocked for 1 h with 300 μL of 5% (w/v) BSA followed by washing once with 0.1% PBST for 5 min. We then added 0.05 μg of GVBP-Cy5 peptide and incubated for 1 h. The wells were washed three times with 0.1% PBST for 5 min each before adding 100 μL of distilled water. The fluorescence was measured using an Infinite 200 Rx plate reader with excitation and emission wavelengths of 647 and 665 nm, respectively, and a gain of 50.
SYBR Tagging of GVER2738 Monoclonal Phage
We mixed 200 μg of the GVER3738 monoclonal phage with 0.5 μL of SYBR safe DNA gel stain (APExBIO) and topped up to 500 μL with distilled water. After mixing on a rotator for 15 min at room temperature and passing through a 0.45-μm PES filter to remove unbound SYBR dye, the protein concentration was quantified using the Pierce BCA protein assay kit (Thermo Fisher Scientific). We fractionated 10 μg of GVER2738 with and without SYBR by 1.2% (w/v) TAE agarose gel electrophoresis for 35 min at 110 V to assess the intercalation of the SYBR stain. The same gel was stained with Coomassie Brilliant Blue to show the colocalization of SYBR-stained nucleic acid and phage coat proteins.
Competitive ELISA
The protocol was similar to the polyclonal ELISA with slight modifications to obtain the KD value. We coated the wells with 5 μg of GV, then GVER2738 (1, 0.5, 1, 2, 3, 4, or 5 μg) was added to the wells together with GVER2738-SYBR (1, 0.5, 1, 2, 3, 4, or 5 μg). Bound GVER2738-SYBR was detected using an Infinite 200 Rx plate reader as described above.
Flow Cytometry
An overnight culture of P. aeruginosa was diluted 100-fold in fresh medium. The culture was incubated at 37 °C to reach OD600 ∼ 0.2 before adding ∼1011 pfu of GV to the cells and incubating at 37 °C for 15 min. The cells were pelleted by centrifugation (6000g, 15 min, at room temperature), washed once with 800 μL TM buffer (10 mM Tris base, 5 μM CaCl2, 10 mM MgSO4, pH 7.4), and resuspended in 200 μL of the same buffer. We added GVBP-Cy5 (2 × 1013 units) and incubated the mixture for 15 min at room temperature before centrifugal washing three times with 0.1% PBST (6000 g, 5 min, at room temperature). The pellet was resuspended in 200 μL TM buffer and the binding of GVBP-Cy5 to GV was analyzed using an Accuri C6 Plus flow cytometer (BD Biosciences). A total of 30,000 events were collected for each analysis.
Results and Discussion
Isolation, Preparation, And Characterization of Good Vibes Phage (GV)
We isolated a lytic phage that infected P. aeruginosa and named it Good Vibes (GV) phage. Morphological analysis by transmission electron microscopy (TEM) placed the phage in the family Myoviridae, featuring an icosahedral head, contractile tail, and tail fibers connecting the base plate (Figure 1A). The spot test assay confirmed the lytic activity of GV by producing clear plaques on a bacterial lawn of P. aeruginosa (Figure 1B). Whole-genome sequencing of GV revealed that the total length of its linear genomic DNA was 65.8 kbp consisting of 24% A, 28.6% C, 23.7% G, and 23.7% T, with 92 predicted coding sequences (Figure 1C).
Figure 1.

Characterization of Good Vibes phage (GV). (A) Transmission electron micrograph of negatively stained GV. (B) Spot test assay of GV using the double-layer agar technique. (C) Complete genome map of GV visualized using the SnapGene Viewer.
Isolation of GV-Binding Peptides
We carried out three rounds of biopanning with a PhD-12 Phage Display Peptide Library Kit to isolate GV-binding peptides (GVBPs). The stringency was increased in every round to remove nonspecific binders and to enrich for monoclonal library phages against GV, which was confirmed by the increasing absorbance signal detected by polyclonal ELISA (Figure 2A). Forty monoclonal phages from the third round were randomly picked for monoclonal ELISA against the target (GV) and BSA as a negative control (Figure 2B). Monoclonal library phages with an absorbance difference of 1.0 were selected for Sanger sequencing.
Figure 2.
Detection of GV-binding monoclonal phages by ELISA. (A) Polyclonal ELISA of enriched binders from each round against GV. (B) Monoclonal ELISA of 40 monoclonal phages against GV (red dots) and BSA (negative control, green dots).
The main advantage of phage display technology is that the genotype (DNA sequence) and the phenotype (displayed peptide) are directly linked, allowing us to determine the nature of the peptide binders by DNA sequencing.27 We identified nine unique peptide sequences enriched against GV (Figure 3A). The most prevalent peptide sequence (WDLPPIGRLSGN) accounted for 50% of the sequenced phages. We also observed the consensus motif LPPI in most of the peptide sequences (Figure 3A, bold red). Peptide sequence alignment revealed strong conservation, with a consistency score of at least 77 out of 100 (Figure 3B). The major hit WDLPPIGRLSGN was therefore chosen for synthesis and further characterization.
Figure 3.
Analysis of the monoclonal phages that bind GV. (A) Sequences of GV-binding peptides with a highly conserved motif shown in bold red. (B) Heat map showing the cross-reactivity of GV monoclonal phages against GV (target), MAT (same family as GV), cowpea mosaic virus (CPMV, unrelated plant virus), and bovine serum albumin (BSA). (C) Sequence alignment of all peptides from GV monoclonal phages using the T-coffee multiple sequence alignment server (https://tcoffee.crg.eu/). (D) SAROTUP analysis (http://i.uestc.edu.cn/sarotup/cgi-bin/TUPScan.pl) of the potential GV-binding peptide WDLPPIGRLSGN. Number in brackets represents the probability in percentage.
Cross-reactivity assays, in which the nine unique monoclonal phages were screened against the target (GV), Matera (MAT, also representing the family Myoviridae), cowpea mosaic virus (CPMV, a plant virus), and BSA as a negative control, showed that monoclonal phage displaying peptides WDLPPIGRLSGN or THLPPIMRNLQF produced a strong signal against both GV and MAT, but not against CPMV or BSA (Figure 3C). Cross-reaction to MAT was anticipated because MAT and GV are closely related and share a high degree of structural similarity. SAROTUP analysis predicted that the WDLPPIGRLSGN peptide is not polystyrene binder and contains no target-unrelated motifs (Figure 3D). However, monoclonal phages displaying this peptide were predicted to be fast growing, suggesting the phages have a higher infection rate or secretion rate but may not display a target-specific binder.28,29
Validation of Monoclonal Phage GVER2738 Binding to GV
The binding of monoclonal phage GVER2738 to GV was confirmed by ELISA. We showed that monoclonal phages displaying the peptide WDLPPIGRLSGN bound more strongly to GV and MAT than the unrelated virus TRI-180 and the negative control BSA (Figure 4A). GVER2738 also bound more strongly to GV (our target) than the closely related virus MAT. We also compared the binding activity of GVER2738 and empty M13 phage (M13KE). Monoclonal phages displaying peptide WDLPPIGRLSGN bound more strongly than M13KE to GV and MAT (Figure 4B). Again, GVER2738 also bound more strongly to GV than MAT. These results confirmed that the binding of GVER2738 to GV and MAT reflected the recognition of the displayed peptide rather than the M13 capsid.
Figure 4.
Analysis of GVER2738 binding activity by ELISA. (A) The binding of monoclonal phage GVER2738 to the target GV, the closely related virus MAT, the unrelated virus GVTRI-180 (Siphoviridae), and BSA (negative control), as determined by ELISA. (B) The binding of monoclonal phage GVER2738 and the empty phage M13KE to GV and MAT. Data are means ± standard deviations (n = 3, one-way ANOVA; **p < 0.01).
Next, we carried out a competitive binding assay between monoclonal phage GVER2738 and GVER2738 intercalated with SYBR dye (GVER2738-SYBR) with GV as the target (Figure 5A). The same amount of GV was coated onto the plates and the same amount of GVER2738-SYBR was used in each assay. By increasing the amount of GVER2738, fewer binding sites on the GV surface were available for its competitor GVER2738-SYBR, resulting in a weaker SYBR signal with the increasing amount of GVER2738. The signal from the wells coated with GV was much stronger than that from the negative control wells (coated with BSA), indicating a specific noncovalent interaction between the peptides displayed on the monoclonal phage and GV. The KD value of the binding with GV was ∼12 times higher than BSA. To confirm these results, we synthesized the WDLPPIGRLSGN peptide, which we describe as the GV-binding peptide (GVBP), with two separate C-terminal modifications: biotin (GVBP-biotin) and Cy5 (GVBP-Cy5). We confirmed by ELISA that the intensity of signals representing bound GVBP-biotin and GVBP-Cy5 became stronger with increasing amounts of GV coating the plates (Figure 5B and C).
Figure 5.
Analysis of GVER2738 binding activity by competitive ELISA. (A) Competitive ELISA between GVER2738 and GVER2738-SYBR against GV (target) and GVBSA (negative control). Agarose gel shows the intercalation of SYBR with GV under UV light. The same gel was stained with Coomassie Brilliant Blue to show the colocalization of phage nucleic acid and coat proteins. (B) ELISA showing the binding of GVBP-biotin to GV and (C) GVBP-Cy5 to GV. Data are means ± standard deviations (n = 3, one-way ANOVA; *p < 0.05).
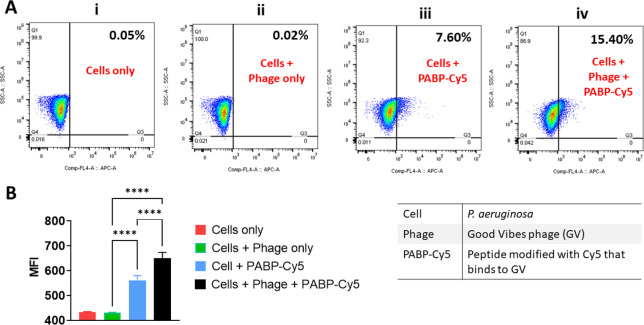
Flow Cytometry Analysis of GVBP-Cy5 Binding to GV
The binding of GVBP-Cy5 to GV in a population of P. aeruginosa cells was investigated by flow cytometry (Figure 6A). The adherence of GVBP-Cy5 on GV bound to P. aeruginosa cells can be detected by flow cytometry using the APC channel. Higher GVBP-Cy5 bound to GV leads to higher APC shift compared to population without GVBP-Cy5. The population containing both cells and phages (Figure 6A, panel (iv)) showed the highest APC+ shift of 15.4% in the presence of GVBP-Cy5, although the population containing cells without phages also showed a moderate shift of 7.6% when the peptide was added, indicating that the peptide binds nonspecifically to the bacterial cells (Figure 6A, panel (iii)). These results correlated with the median fluorescence intensity (MFI) values (Figure 6B). The population containing cells and phages plus GVBP-Cy5 generated the strongest fluorescent signal, indicating the binding of GVBP-Cy5 to phage particles. A weaker signal was detected in the absence of phage. The binding of GVBP-Cy5 to the phage therefore increases the APC+ signal.
Figure 6.
Analysis of GVBP-Cy5 binding by flow cytometry. (A) Scatter plots showing percent of APC+ cells by gating on P. aeruginosa cells. Mean APC+ population is shown as an inset. (B) Histogram showing median fluorescence intensity (MFI) values of APC+ cells. Data are means ± standard deviations (n = 3, one-way ANOVA; ****p < 0.0001).
Data support that P. aeruginosa cells can be tracked and imaged using the identified GVBP. To proceed with in vivo studies, the sensitivity of the approach should be improved; this may be achieved through multivalency therefore introducing avidity effects; e.g., multivalent peptides could be synthesized, and nanoparticle formulations could be utilized to generate high multivalency.
Conclusion
We have successfully isolated 12-mer peptides binding to GV, a phage that infects and lyses the pathogenic bacterium P. aeruginosa. We isolated nine unique peptide sequences using peptide phage display technology, and the consensus motif LPPI was found in most of the peptides following multiple sequence alignment. ELISAs using monoclonal phage GVER2738 and modified GVBPs confirmed that the peptides bind to GV and closely related phage but not to unrelated viruses. Flow cytometry also showed the significant binding of Cy5-labeled GVBP to GV. This preliminary data may facilitate the development of in vivo tracers for the real-time analysis of phage therapy.
Acknowledgments
This work was funded in part by the NSF through the UC San Diego Materials Research Science and Engineering Center (UCSD MRSEC; DMR-2011924) as well as a UC San Diego Galvanizing Engineering in Medicine (GEM) Award. We thank Dr. Steffanie Strathdee, Co-Director for the Center for Innovative Phage Applications & Therapeutics (IPATH) and Professor of the Department of Medicine at UCSD, for helpful discussions.
Author Contributions
S.K.C. designed and performed the experimental work. Z.Z assisted in flow analysis. S.P and E. K. prepared phages and P. aeruginosa. Whole genome sequencing was performed by Pride lab. N.F.S. and R.T.S. conceived the study. N.F.S. oversaw the design and testing. S.K.C. and N.F.S. wrote the manuscript. All authors read and edited the manuscript.
The authors declare no competing financial interest.
References
- How Antibiotic Resistance Happens | CDC. https://www.cdc.gov/drugresistance/about/how-resistance-happens.html (accessed 2022-04-03).
- Antimicrobial resistance. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed 2022-04-03).
- Murray C. J.; Ikuta K. S.; Sharara F.; Swetschinski L.; Robles Aguilar G.; Gray A.; Han C.; Bisignano C.; Rao P.; Wool E.; Johnson S. C.; Browne A. J.; Chipeta M. G.; Fell F.; Hackett S.; Haines-Woodhouse G.; Kashef Hamadani B. H.; Kumaran E. A. P.; McManigal B.; Agarwal R.; Akech S.; Albertson S.; Amuasi J.; Andrews J.; Aravkin A.; Ashley E.; Bailey F.; Baker S.; Basnyat B.; Bekker A.; Bender R.; Bethou A.; Bielicki J.; Boonkasidecha S.; Bukosia J.; Carvalheiro C.; Castañeda-Orjuela C.; Chansamouth V.; Chaurasia S.; Chiurchiù S.; Chowdhury F.; Cook A. J.; Cooper B.; Cressey T. R.; Criollo-Mora E.; Cunningham M.; Darboe S.; Day N. P. J.; De Luca M.; Dokova K.; Dramowski A.; Dunachie S. J.; Eckmanns T.; Eibach D.; Emami A.; Feasey N.; Fisher-Pearson N.; Forrest K.; Garrett D.; Gastmeier P.; Giref A. Z.; Greer R. C.; Gupta V.; Haller S.; Haselbeck A.; Hay S. I.; Holm M.; Hopkins S.; Iregbu K. C.; Jacobs J.; Jarovsky D.; Javanmardi F.; Khorana M.; Kissoon N.; Kobeissi E.; Kostyanev T.; Krapp F.; Krumkamp R.; Kumar A.; Kyu H. H.; Lim C.; Limmathurotsakul D.; Loftus M. J.; Lunn M.; Ma J.; Mturi N.; Munera-Huertas T.; Musicha P.; Mussi-Pinhata M. M.; Nakamura T.; Nanavati R.; Nangia S.; Newton P.; Ngoun C.; Novotney A.; Nwakanma D.; Obiero C. W.; Olivas-Martinez A.; Olliaro P.; Ooko E.; Ortiz-Brizuela E.; Peleg A. Y.; Perrone C.; Plakkal N.; Ponce-de-Leon A.; Raad M.; Ramdin T.; Riddell A.; Roberts T.; Robotham J. V.; Roca A.; Rudd K. E.; Russell N.; Schnall J.; Scott J. A. G.; Shivamallappa M.; Sifuentes-Osornio J.; Steenkeste N.; Stewardson A. J.; Stoeva T.; Tasak N.; Thaiprakong A.; Thwaites G.; Turner C.; Turner P.; van Doorn H. R.; Velaphi S.; Vongpradith A.; Vu H.; Walsh T.; Waner S.; Wangrangsimakul T.; Wozniak T.; Zheng P.; Sartorius B.; Lopez A. D.; Stergachis A.; Moore C.; Dolecek C.; Naghavi M. Global Burden of Bacterial Antimicrobial Resistance in 2019: A Systematic Analysis. Lancet 2022, 399, 629–655. 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- About Antibiotic Resistance | CDC. https://www.cdc.gov/drugresistance/about.html (accessed 2022-04-03).
- Tacconelli E.; Carrara E.; Savoldi A.; Harbarth S.; Mendelson M.; Monnet D. L.; Pulcini C.; Kahlmeter G.; Kluytmans J.; Carmeli Y.; Ouellette M.; Outterson K.; Patel J.; Cavaleri M.; Cox E. M.; Houchens C. R.; Grayson M. L.; Hansen P.; Singh N.; Theuretzbacher U.; Magrini N.; Aboderin A. O.; Al-Abri S. S.; Awang Jalil N.; Benzonana N.; Bhattacharya S.; Brink A. J.; Burkert F. R.; Cars O.; Cornaglia G.; Dyar O. J.; Friedrich A. W.; Gales A. C.; Gandra S.; Giske C. G.; Goff D. A.; Goossens H.; Gottlieb T.; Guzman Blanco M.; Hryniewicz W.; Kattula D.; Jinks T.; Kanj S. S.; Kerr L.; Kieny M. P.; Kim Y. S.; Kozlov R. S.; Labarca J.; Laxminarayan R.; Leder K.; Leibovici L.; Levy-Hara G.; Littman J.; Malhotra-Kumar S.; Manchanda V.; Moja L.; Ndoye B.; Pan A.; Paterson D. L.; Paul M.; Qiu H.; Ramon-Pardo P.; Rodríguez-Baño J.; Sanguinetti M.; Sengupta S.; Sharland M.; Si-Mehand M.; Silver L. L.; Song W.; Steinbakk M.; Thomsen J.; Thwaites G. E.; van der Meer J. W.; Van Kinh N.; Vega S.; Villegas M. V.; Wechsler-Fördös A.; Wertheim H. F. L.; Wesangula E.; Woodford N.; Yilmaz F. O.; Zorzet A. Discovery, Research, and Development of New Antibiotics: The WHO Priority List of Antibiotic-Resistant Bacteria and Tuberculosis. Lancet. Infect. Dis. 2018, 18, 318–327. 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- Pseudomonas aeruginosa pneumonia - UpToDate. https://www.uptodate.com/contents/pseudomonas-aeruginosa-pneumonia (accessed 2022-04-21).
- Laborda P.; Sanz-García F.; Hernando-Amado S.; Martínez J. L. Pseudomonas Aeruginosa: An Antibiotic Resilient Pathogen with Environmental Origin. Curr. Opin. Microbiol. 2021, 64, 125–132. 10.1016/j.mib.2021.09.010. [DOI] [PubMed] [Google Scholar]
- Paterson D. L.; Rice L. B. Empirical Antibiotic Choice for the Seriously Ill Patient: Are Minimization of Selection of Resistant Organisms and Maximization of Individual Outcome Mutually Exclusive?. Clin. Infect. Dis. 2003, 36, 1006–1012. 10.1086/374243. [DOI] [PubMed] [Google Scholar]
- Pseudomonas aeruginosa | HAI | CDC. https://www.cdc.gov/hai/outbreaks/pseudomonas-aeruginosa.html (accessed 2022-04-21).
- Spagnolo A. M.; Sartini M.; Cristina M. L. Pseudomonas Aeruginosa in the Healthcare Facility Setting. Rev. Med. Microbiol. 2021, 32, 169–175. 10.1097/MRM.0000000000000271. [DOI] [Google Scholar]
- Schooley R. T.; Strathdee S. Treat Phage like Living Antibiotics. Nat. Microbiol. 2020, 5 (3), 391–392. 10.1038/s41564-019-0666-4. [DOI] [PubMed] [Google Scholar]
- d’Herelle F. Annual Graduate Fortnight. Medical and Surgical Aspects of Acute Bacterial Infections, October 20 to 31, 1930: Bacteriophage as a Treatment in Acute Medical and Surgical Infections. Bull. N. Y. Acad. Med. 1931, 7, 329. [PMC free article] [PubMed] [Google Scholar]
- Brives C.; Pourraz J. Phage Therapy as a Potential Solution in the Fight against AMR: Obstacles and Possible Futures. Palgrave Commun. 2020, 6, 1–11. 10.1057/s41599-020-0478-4. [DOI] [Google Scholar]
- Kasman L. M.; Porter L. D. Bacteriophages. Brenner’s Encycl. Genet. Second Ed. 2021, 280–283. [Google Scholar]
- Cafora M.; Deflorian G.; Forti F.; Ferrari L.; Binelli G.; Briani F.; Ghisotti D.; Pistocchi A. Phage Therapy against Pseudomonas Aeruginosa Infections in a Cystic Fibrosis Zebrafish Model. Sci. Reports 2019, 9, 1–10. 10.1057/s41599-020-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Haque A.; Matsuzaki S.; Matsumoto T.; Nakamura S. The Efficacy of Phage Therapy in a Murine Model of Pseudomonas Aeruginosa Pneumonia and Sepsis. Front. Microbiol. 2021, 12, 1698. 10.3389/fmicb.2021.682255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel Phage Therapy Saves Patient with Multidrug-Resistant Bacterial Infection. https://health.ucsd.edu/news/releases/pages/2017-04-25-novel-phage-therapy-saves-patient-with-multidrug-resistant-bacterial-infection.aspx (accessed 2022-05-09).
- Aslam S.; Lampley E.; Wooten D.; Karris M.; Benson C.; Strathdee S.; Schooley R. T. Lessons Learned From the First 10 Consecutive Cases of Intravenous Bacteriophage Therapy to Treat Multidrug-Resistant Bacterial Infections at a Single Center in the United States. Open Forum Infect. Dis. 2020, 10.1093/ofid/ofaa389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedrick R. M.; Smith B. E.; Cristinziano M.; Freeman K. G.; Jacobs-Sera D.; Belessis Y.; Whitney Brown A.; Cohen K. A.; Davidson R. M.; van Duin D.; Gainey A.; Garcia C. B.; Robert George C. R.; Haidar G.; Ip W.; Iredell J.; Khatami A.; Little J. S.; Malmivaara K.; McMullan B. J.; Michalik D. E.; Moscatelli A.; Nick J. A.; Tupayachi Ortiz M. G.; Polenakovik H. M.; Robinson P. D.; Skurnik M.; Solomon D. A.; Soothill J.; Spencer H.; Wark P.; Worth A.; Schooley R. T.; Benson C. A.; Hatfull G. F. Phage Therapy of Mycobacterium Infections: Compassionate-Use of Phages in Twenty Patients with Drug-Resistant Mycobacterial Disease. Clin. Infect. Dis. 2022, 10.1093/cid/ciac453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhungana G.; Nepal R.; Regmi M.; Malla R. Pharmacokinetics and Pharmacodynamics of a Novel Virulent Klebsiella Phage Kp_Pokalde_002 in a Mouse Model. Front. Cell. Infect. Microbiol. 2021, 11, 731. 10.3389/fcimb.2021.684704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y.; Peng Y.; Zhang Y.; Chen Y.; Zhang C.; Luo X.; Chen Y.; Yuan Z.; Chen J.; Gong Y. Safety and Efficacy of a Phage, Kpssk3, in an in Vivo Model of Carbapenem-Resistant Hypermucoviscous Klebsiella Pneumoniae Bacteremia. Front. Microbiol. 2021, 10.3389/fmicb.2021.613356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ács N.; Gambino M.; Brøndsted L. Bacteriophage Enumeration and Detection Methods. Front. Microbiol. 2020, 11, 2662. 10.3389/fmicb.2020.594868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly K. A.; Waterman P.; Weissleder R. In Vivo Imaging of Molecularly Targeted Phage. Neoplasia 2006, 8, 1011–1018. 10.1593/neo.06610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusckowski M.; Gupta S.; Liu G.; Dou S.; Hnatowich D. J. Investigation of Four 99mTc-Labeled Bacteriophages for Infection Specific Imaging. Nucl. Med. Biol. 2008, 35, 433. 10.1016/j.nucmedbio.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. K.; Steinmetz N. F. Isolation of Cowpea Mosaic Virus-Binding Peptides. Biomacromolecules 2021, 22, 3613–3623. 10.1021/acs.biomac.1c00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan S. K.; Du P.; Ignacio C.; Mehta S.; Newton I. G.; Steinmetz N. F. Biomimetic Virus-Like Particles as Severe Acute Respiratory Syndrome Coronavirus 2 Diagnostic Tools. ACS Nano 2021, 15, 1259–1272. 10.1021/acsnano.0c08430. [DOI] [PubMed] [Google Scholar]
- Wu C. H.; Liu I. J.; Lu R. M.; Wu H. C. Advancement and Applications of Peptide Phage Display Technology in Biomedical Science. J. Biomed. Sci. 2016, 23, 1–14. 10.1186/s12929-016-0223-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas W. D.; Golomb M.; Smith G. P. Corruption of Phage Display Libraries by Target-Unrelated Clones: Diagnosis and Countermeasures. Anal. Biochem. 2010, 407, 237–240. 10.1016/j.ab.2010.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brammer L. A.; Bolduc B.; Kass J. L.; Felice K. M.; Noren C. J.; Hall M. F. A Target-Unrelated Peptide in an M13 Phage Display Library Traced to an Advantageous Mutation in the Gene II Ribosome-Binding Site. Anal. Biochem. 2008, 373, 88–98. 10.1016/j.ab.2007.10.015. [DOI] [PubMed] [Google Scholar]