Abstract
A panel of 30 previously characterized strains representing five genomovars from the Burkholderia cepacia complex (E. Mahenthiralingam, T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme, J. Clin. Microbiol. 38:910–913, 2000) were examined for their iron protoporphyrin IX-binding ability. These included B. cepacia genomovars I and III and B. stabilis (formerly B. cepacia genomovar IV), B. multivorans (formerly B. cepacia genomovar II), and B. vietnamiensis (formerly B. cepacia genomovar V). Cells were exposed to μ-oxo bisheme of iron protoporphyrin IX (μ-oxo dimers) and examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under nonreducing, nondenaturing conditions for the presence of heme-binding proteins using tetramethylbenzidine-H2O2 staining. Seven of the 30 strains, each belonging to B. cepacia genomovar III and designated epidemic (in possessing the B. cepacia epidemic strain marker), expressed a 96- to 100-kDa heme-binding protein which was located in the outer membrane. The heme-binding protein of B. cepacia genomovar III epidemic strain C5424 bound iron(III) protoporphyrin IX in both the monomeric and μ-oxo bisheme forms. Cells of all strains grown on Columbia agar bound iron protoporphyrin IX in the μ-oxo bisheme (dimeric) form. There were no statistical differences between the five genomovars, or those possessing the heme-binding protein, in their μ-oxo bisheme-binding ability. Possession of the outer membrane heme-binding protein may be a pathogenicity trait in enabling the bacterium to withstand oxidative stresses in inflammatory exudates in the lung and may aid identification of invasive epidemic strains of B. cepacia.
Burkholderia cepacia is an opportunistic gram-negative pathogen which can colonize the respiratory airways in patients with cystic fibrosis (CF). Chronic microbial colonization is the major cause of morbidity and mortality in these patients, who have impaired mucociliary clearance. Although not all strains of B. cepacia are epidemic, some of them can be easily transmitted from person to person and are characterized as highly epidemic (13). Patients becoming colonized with epidemic strains develop the so-called cepacia syndrome, a necrotizing pneumonia with fever and bacteremia, which leads to a rapid and fatal clinical deterioration (15, 17).
The B. cepacia complex comprises at least five genomovars, including B. cepacia genomovars I and III, B. multivorans (formerly genomovar II), B. stabilis (formerly genomovar IV), and B. vietnamiensis (formerly genomovar V) (45, 46). A number of pathogenic factors which may contribute to tissue damage and lung pathogenesis during infection have been attributed to such strains (see reference 13 for a review). These include possession of mucin sulfatase activity (18), which may render highly sulfated (and normally protective) respiratory mucins of CF patients more susceptible to bacterial degradation, increasing substrate availability and providing binding sites for bacterial adherence and colonization. Lipopolysacharide (LPS) from B. cepacia can stimulate larger amounts of tumor necrosis factor alpha than LPS from other CF pathogens such as Pseudomonas aeruginosa (37, 49). Macrophage and monocyte superoxide generation in response to infection (1) aids killing of phagocytosed bacteria, and its increased production, as a result of B. cepacia LPS-mediated priming (16), is thought to play a considerable role in disease pathology in CF patients (6). Other virulence factors include hemeolytic, proteolytic, and phospholipase C activities (11, 21, 29, 33, 48) and the production of iron-binding siderophores (10, 30, 42, 43).
The ability to avoid neutrophil surveillance and oxidant killing is, however, a major factor in colonization and infection in the CF lung, and it is noteworthy that a melanin-like pigment which functions as a scavenger of superoxide radicals during the respiratory burst has been characterized from an isolate of B. cepacia genomovar III (50). In a preliminary survey using laser Raman microscopy and pyridine-hemochrome assays (R. Withnall, J. W. Smalley, J. Silver, and C. A. Hart, unpublished data), we have detected iron(III) protoporphyrin IX [Fe(III)PPIX] on the surface of epidemic, melanin-like pigment-producing strains of B. cepacia when the strains are grown on blood agar. Iron protoporphyrin IX accumulation by the periodontal pathogen Porphyromonas gingivalis is responsible for the black pigmentation during growth on blood agar (40). The major heme species in the pigment is the μ-oxo bisheme (dimeric) form of Fe(III)PPIX, [Fe(III)PPIX]2O (40), a structure involving two Fe(III)PPIX molecules joined by an oxygen atom interbridge (31, 38). Formation of μ-oxo bisheme through the reaction of hemoglobin-derived Fe(II)PPIX monomers with oxygen is considered to be an oxidative buffer through which dioxygen and reactive oxygen species are eliminated to generate an impervious cell surface heme layer (40). The accumulation of μ-oxo bisheme and monomeric Fe(III)PPIX is an important pathogenicity factor for P. gingivalis, as both soluble and cell surface aggregated forms protect cells against hydrogen peroxide (41) by virtue of their inherent catalase activity.
Expression of heme-binding proteins (HBPs) leading to heme binding and accumulation is associated with virulence in other gram-negative pathogens (24). The detection of cell-associated free Fe(III)PPIX (Withnall et al., unpublished data) prompted us to investigate heme binding and the possibility that HPBs are expressed by isolates of B. cepacia known to cause disease in humans. The ability to generate, bind, and accumulate Fe(III)PPIX from heme proteins would be an advantage to B. cepacia in subverting neutrophil-derived peroxide in the CF lung during episodes of inflammation. In this study we examined a well-characterized panel of clinically important representative isolates of the B. cepacia complex (26, 46) for the ability to bind μ-oxo bisheme in vitro. These strains were also examined for presence of HBPs using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and tetramethylbenzidine-H2O2 staining.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The panel of strains used has been extensively characterized (26, 46) and was kindly provided by J. Govan, Department of Medical Microbiology, The University of Edinburgh. These strains are listed in Table 1. All 30 strains were maintained by routine subculture on 5% (vol/vol) horse blood agar (blood agar base no. 2; LAB M, Bury, United Kingdom), from which they were subcultured onto Columbia agar (Oxoid Ltd., Basingstoke, United Kingdom). Lawn growths from the Columbia agar plates, after subculture at least three times to minimize carryover of any heme-containing components, were harvested with plastic sterile loops after 3 days of growth in air at 37°C, and the cells were washed by suspension in 0.14 M NaCl–0.1 M Tris (pH 7.4) (NaCl-Tris buffer) and mild agitation for 2 min using a water bath sonicator. After pelleting by centrifugation at 13,000 × g for 10 min at 20°C, the supernatants were removed and the cells were resuspended in a small volume of the same buffer and frozen at −40°C until required. For some experiments cells were grown on M9 minimal salts medium agar (Sigma Chemicals Ltd., Poole, United Kingdom) supplemented with 0.5% (wt/vol) glucose but containing no FePPIX. Cells were subcultured three times on this solid medium before lawn growths were harvested and treated as described above for the Columbia agar-grown cells.
TABLE 1.
Panel of strains from the B. cepacia complex
| Species | Strain | Genomovar | Sourcea | BCESMb | CblAc | HBPd |
|---|---|---|---|---|---|---|
| B. vietnamiensis | LMG 16232 | V | CF, Sweden | − | − | |
| B. multivorans | C5393 | II | CF, Canada | − | − | |
| B. multivorans | C1576 | II | CF-e, United Kingdom | − | − | |
| B. cepacia | CEP511 | III | CF-e, Australia | + | − | + |
| B. vietnamiensis | FC441 | V | CGD, Canada | − | − | |
| B. vietnamiensis | PC259 | V | CF, United States | − | − | |
| B. multivorans | C1962 | II | Clinic, United Kingdom | − | − | |
| B. multivorans | 249-2 | II | Laboratory, United States | − | − | |
| B. multivorans | ATCC 17616 | II | Soil, United States | − | − | |
| B. multivorans | CF-A1-1 | II | CF-e, United Kingdom | − | − | |
| B. vietnamiensis | LMG 10929 | V | Rice, Vietnam | − | − | |
| B. cepacia | C5424 | III | CF-e, Canada | + | + | + |
| B. cepacia | J2315 | III | CF-e, United Kingdom | + | + | + |
| B. cepacia | CEP509 | I | CF, Australia | − | − | |
| B. multivorans | LMG 13010 | II | CF, Belgium | − | − | |
| B. cepacia | C6433 | III | CF, Canada | + | − | + |
| B. stabilis | LMG 18888 | IV | Clinical, Belgium | − | − | |
| B. cepacia | ATCC 17765 | III | UTI, United Kingdom | + | − | |
| B. stabilis | C7322 | IV | CF, Canada | − | − | |
| B. cepacia | PC184 | III | CF-e, United States | + | − | + |
| B. cepacia | ATCC 17759 | I | Soil, Trinidad | − | − | |
| B. cepacia | J415 | III | CF, United Kingdom | − | − | + |
| B. cepacia | BC-7 | III | CF-e, Canada | + | + | + |
| B. cepacia | LMG 17997 | I | UTI, Sweden | − | − | |
| B. cepacia | ATCC 25416 | I | Onion, United States | − | − | |
| B. stabilis | LMG 14294 | IV | CF, Belgium | − | − | |
| B. cepacia | K56-2 | III | CF-e, Canada | + | + | |
| B. stabilis | LMG 14086 | IV | Respirator, United Kingdom | − | − | |
| B. multivorans | JTC | II | CGD, United States | − | − | |
| B. cepacia | C1394 | III | CF-e, United Kingdom | + | − |
Fe(III)PPIX preparations.
Bovine FePPIX chloride [hemin; Fe(III)PPIX.Cl] (Sigma Chemicals Ltd.) was initially dissolved in 0.14 M NaCl–0.1 M Tris (pH 9.8) to give a 1 mM stock solution. Preparation of the μ-oxo bisheme was achieved by addition of dilute HCl to reduce the pH of the stock solution to 7.5, at which the Fe(III)PPIX exists predominantly in the μ-oxo bisheme (dimeric) form, [Fe(III)PPIX]2O (31, 38). Monomeric hematin [Fe(III)PPIX.OH.H2O] was made by lowering the pH of the hemin stock solution to 6.8.
Assay for μ-oxo bisheme binding.
Binding of μ-oxo bisheme was carried out as previously described (40), with a slight modification. Suspensions of Columbia agar-grown cells, standardized to give ≈1 mg of protein ml−1, were incubated with μ-oxo bisheme (50 μM) in a volume of 1 ml for 30 min at 37°C on an orbital shaker at 90 rpm. After centrifugation, the cell pellets were resuspended in 1 ml of NaCl-Tris buffer, and the concentration of cell surface-bound [Fe(III)PPIX]2O was measured by pyridine-hemochrome assay. This assay is based on the generation of the Fe(II)PPIX-bispyridine complex (with absorbance maxima at 419, 525, and 555 nm) resulting from the reaction of Fe(III)PPIX with pyridine in the presence of sodium dithionite. Cell suspensions (≈1 mg of protein ml−1) were agitated in a sonicating water bath in 1 ml of 0.14 M NaCl–0.1 M Tris-HCl (pH 7.4) containing freshly prepared Na2S2O4 (10 mM) and 1 M pyridine. Preliminary experiments indicated that maximum pyridine-hemochrome production occurred after 10 min. Bovine hemin [Fe(III)PPIX.Cl; Sigma Chemicals Ltd.] initially dissolved in 0.14 M NaCl–0.1 M Tris (pH 10), which was then adjusted to pH 7.4, was used as a Fe(III)PPIX standard. A419 values were used to calculate the concentration of Fe(III)PPIX in each sample, and assays were carried out in triplicate. Cell suspensions incubated in NaCl-Tris buffer plus 10 mM sodium dithionite for 10 min were used to correct for background absorbance. The amounts of heme bound by the strains from each genomovar [expressed as Fe(III)PPIX monomer] were plotted using the GraphPad Prism and analyzed using the unpaired t test.
Detection of HPBs.
HBPs were identified on SDS-polyacrylamide gels as previously described (39) using tetramethylbenzidine-H2O2 staining, which detects the presence of heme-associated peroxidase activity (9, 45). Briefly, whole-cell samples (≈1 mg of protein) from growth on Columbia agar were exposed to μ-oxo bisheme (4 μM) in NaCl-Tris buffer for 30 min at 37°C. Excess unbound heme was removed from the cell pellets by washing three times in the NaCl-Tris buffer, and the cells were solubilized at 37°C for 1 h in nonreducing sample application buffer and electrophoresed on 10% polyacrylamide gels. Each gel track was loaded with a nominal 200 μg of protein. The gels were stained and developed with tetramethylbenzidine-H2O2. Duplicate samples, electrophoresed under the same conditions, were stained for protein with Coomassie blue. In some experiments cells grown on M9 minimal salts agar were exposed to Fe(III)PPIX in the μ-oxo bisheme and monomeric forms before electrophoresis and tetramethylbenzidine-H2O2 staining.
Outer membrane extraction.
Outer membranes were extracted by EDTA shearing according to the method of Mansheim and Kasper (28). Briefly, 3-day lawn growths from Columbia agar were suspended in 0.01 M phosphate-buffered 0.14 M NaCl (pH 7.3), mixed with an equal volume of 0.02 mM Na3EDTA in the same buffer, and incubated for 30 min at 37°C. The suspension was syringed twice through a 25-gauge needle. The cells were pelleted by centrifugation at 20,000 × g for 30 min at 4°C, and the outer membrane-containing supernatant was dialyzed first against the same buffer for 4 h and then against repeated changes of distilled-deionized water for a further 20 h. The outer membrane was recovered by freeze-drying.
Spectrophotometry.
UV-visible spectrophotometry was performed using an Ultrospec 2000 scanning spectrophotometer (Amersham-Pharmacia-Biotech Ltd., St. Albans, United Kingdom) using plastic 1-ml microcuvettes (BDH Ltd., Poole, United Kingdom) with a 1-cm path length.
Protein assay.
Whole-cell protein was measured using the Lowry method with bovine serum albumin (Sigma Chemicals Ltd.) as a standard.
RESULTS
Detection of HPBs.
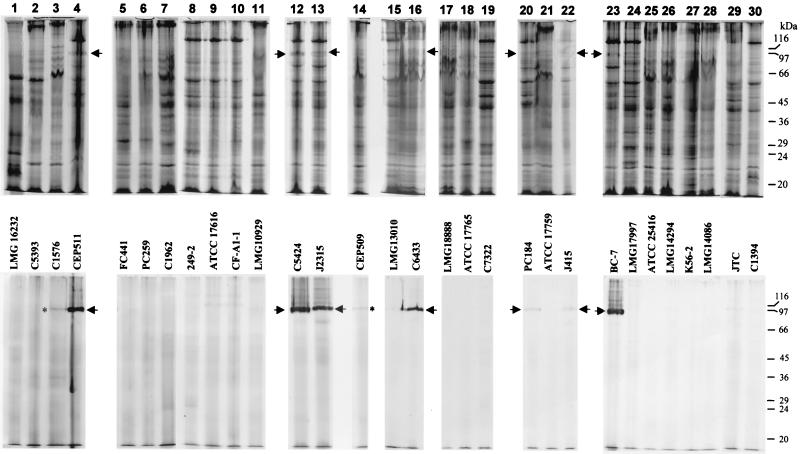
To identify HPBs, whole-cell samples were exposed to μ-oxo bisheme, solubilized at 37°C, and electrophoresed under nonreducing conditions, and the gels were stained with tetramethylbenzidine-H2O2. While the Coomassie blue-stained proteins were less distinct using these solubilization and electrophoretic conditions (Fig. 1, top row), HPBs were clearly detected after staining with tetramethylbenzidine-H2O2. Intensely stained bands were revealed as the main staining component in strains CEP511, C5424, J2315, C6433, and BC-7 (Fig. 1, bottom row, lanes 4, 12, 13, 16, and 23, respectively), while less intensely tetramethylbenzidine-stained bands were obtained for strains PC184 and J415 (Fig. 1, bottom row, lanes 20 and 22, respectively). The Rfs of these bands varied between 0.20 and 0.18, corresponding to molecular masses of 96 to 100 kDa. Diffuse Coomassie blue-stained proteins with the same Rfs as those of the tetramethylbenzidine-stained bands were observed in these strains (Fig. 1, top row, arrows). The faint tetramethylbenzidine-positive staining observed in strains C1576 and CEP509 (Fig. 1, bottom row, lanes 3 and 14, asterisks) was due to sample carryover from adjacent HPB-positive tracks. These were confirmed as HPB-negative strains after reelectrophoresis and staining (data not shown). All of the isolates staining positive for the HPB belonged to B. cepacia genomovar III. We observed that there was no direct correlation between the amount of Coomassie blue staining of the HBPs and tetramethylbenzidine-H2O2 staining. The reason for this is not known, but it is likely that the presence of bound heme masks the uptake of Coomassie blue by the HBP or that the HBPs from different strains have different heme- to protein-binding stoichiometries.
FIG. 1.
SDS-PAGE on 10% polyacrylamide gels of cells grown on Columbia agar, exposed to μ-oxo bisheme (4 μM) for 30 min, and solubilized under nonreducing, nondenaturing conditions. (Top row) Separated proteins were stained with Coomassie blue. (Bottom row) Gels stained with tetramethylbenzidine-H2O2 to reveal HPBs. Arrows indicate HBPs; asterisks indicate staining from sample carryover from adjacent lanes. Each gel lane was loaded with a nominal 200 μg of protein. The mobilities of standard molecular mass markers are indicated.
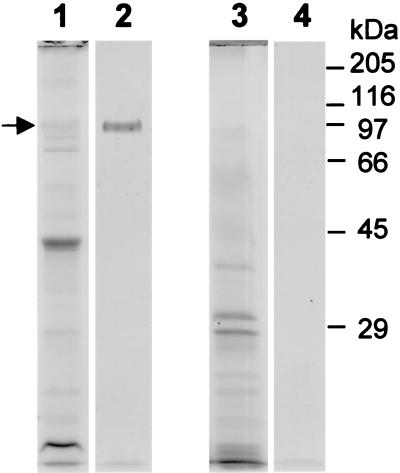
Further investigations were carried out on the HPB-positive strain C5424. The electrophoretic mobility of the HBP was unaltered following sample solubilization under denaturing conditions (100°C for 5 min), with or without 50 mM dithiothreitol (data not presented). However, as previously observed for the HPBs of P. gingivalis (39), tetramethylbenzidine staining of the B. cepacia HPB was abrogated by heating at 100°C alone or by incubation at 37°C with 50 mM dithiothreitol, treatments which would disrupt the heme-protein interaction and destroy the H2O2 (45). Outer membranes from B. cepacia (genomovar III) strain C5424 and B. vietnamiensis strain FC441 (HBP negative) were prepared by EDTA shearing, electrophoresed under nonreducing conditions, and stained for protein and for HPBs. This revealed the presence of a diffuse Coomassie blue-stained band with a molecular mass of 97 kDa and a tetramethylbenzidine-stained component with the same mobility in strain C5424. These components were not present in strain FC441 (Fig. 2).
FIG. 2.
SDS-PAGE of outer membranes from B. cepacia genomovar III strain C5424 (lanes 1 and 2) and B. vietnamiensis strain FC441 (lanes 3 and 4). Outer membranes were solubilized at 37°C and electrophoresed under nonreducing conditions. Proteins in lanes 1 and 3 were stained with Coomassie blue, while those in lanes 2 and 4 were stained with tetramethylbenzidine-H2O2. The HPB is marked with an arrow. Mobilities of standard molecular mass markers are indicated.
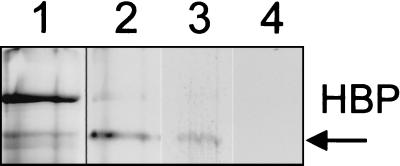
Tetramethylbenzidine-positive staining of HPBs was also observed in cells which had not been exposed to added μ-oxo bisheme (data not shown), suggesting that heme acquisition from heme-containing growth medium components had occurred during culture on Columbia agar. This possibility was supported by the detection of free FePPIX (by pyridine-hemochrome assay) in suspensions of these cells. The heme-binding ability of the protein was confirmed by experiments carried out on cells of strain C5424 grown on M9 minimal salts medium containing no FePPIX. Coomassie blue staining revealed the presence of a 97-kDa component (Fig. 3, lane 1), but this protein was not stained with tetramethylbenzidine-H2O2 unless the cells were exposed to either monomeric hematin or μ-oxo bisheme (Fig. 3, lanes 2 and 3, respectively).
FIG. 3.
Cells of HBP-positive B. cepacia strain C5424 (genomovar III) grown on M9 minimal salts medium without FePPIX. Lanes: 1, cells stained with Coomassie blue; 2, cells exposed to monomeric hematin; 3, cells exposed to μ-oxo bisheme; 4, cells not exposed to heme prior to electrophoresis. Lanes 2, 3, and 4 show the results of staining with tetramethylbenzidine-H2O2. Each lane was loaded with a nominal 200 μg of protein.
Cellular μ-oxo bisheme binding.
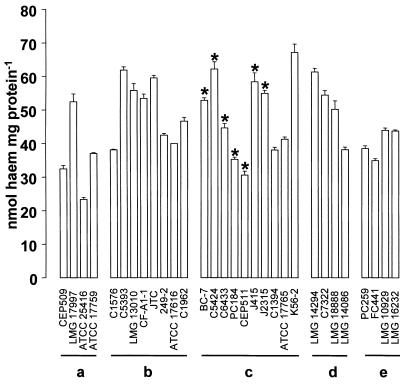
All strains tested bound FePPIX in the μ-oxo bisheme form (Fig. 4). The amounts of heme bound [expressed as Fe(III)PPIX monomer] varied between 23.3 and 67.3 nmol ml−1 mg of cell protein−1. There was no statistical difference in the mean amounts of heme bound between the strains expressing the HPB and isolates from the other strains grouped as genomovars.
FIG. 4.
Binding of μ-oxo bisheme in vitro by the panel of strains. Suspensions of cells (≈ 1 mg of cell protein) were incubated with 50 μM μ-oxo bisheme in 0.14 M NaCl–0.1 M Tris-HCl (pH 7.5) for 30 min at 37°C. The excess unbound heme was removed by washing three times in the same buffer, and the cell-bound heme was quantified by pyridine-hemochrome assay. The amounts of bound heme are expressed as nanomoles of heme monomer per milligram of cell protein. (a) B. cepacia genomovar I; (b) B. multivorans; (c) B. cepacia genomovar III; (d) B. stabilis; (e) B. vietnamiensis. Asterisks indicate possession of HBPs. Error bars indicate standard deviations.
DISCUSSION
In this study we have demonstrated binding of μ-oxo bisheme and the presence of an outer membrane HBP in some bacteria in the B. cepacia complex. HBPs with molecular masses of 96 to 100 kDa were detected by SDS-PAGE and tetramethylbenzidine-H2O2 staining in 7 out of 10 strains belonging to B. cepacia genomovar III. These proteins were not heat modifiable, nor could they be dissociated into subunits by treatment with dithiothreitol. Investigations with B. cepacia genomovar III strain C5424 revealed that the HPB was present in the outer membrane. HBPs have not been reported previously for B. cepacia, although an 80-kDa outer membrane heme receptor protein (34) and a multimeric bacterioferritin composed of 17- to19-kDa subunits with both iron- and heme-binding activities (32) have been observed in P. aeruginosa. Outer membrane HPBs similar in size to those detected in this study have been characterized from Neisseria gonorrhoeae (22), Neisseria meningitidis (23), Shigella flexneri, and enteroinvasive Escherichia coli (44). It is not yet known whether the B. cepacia HBP is solely cell associated or is also secreted, although a lower-molecular-mass extracellular HPB (≈22 kDa) has been observed in P. aeruginosa (25).
The function of the B. cepacia HPB is, as yet, unclear. Expression of HPBs is associated with virulence in other gram-negative pathogens (24). In the heme-pigmenting species Yersinia pestis and Aeromonas salmonicida, cell surface expression of HPBs correlates with virulence and both the binding and accumulation of FePPIX (20, 35). In P. gingivalis, a 32-kDa outer membrane HPB expressed under heme limitation (5, 39) acts as a receptor for subsequent translocation of FePPIX across the outer membrane (4) to be used as an iron source (3). Although B. cepacia produces siderophores (10, 30, 42, 43), it is not known whether this species has an absolute requirement for intact iron porphyrins or whether they are utilized as a source of ferric ions. The in vivo expression of an HBP may function both for reception of monomeric and/or μ-oxo dimeric Fe(III)PPIX under conditions of heme scarcity and as a seed to initiate cell-surface heme deposition.
It is noteworthy that six out of seven of the HBP-positive strains were isolated from epidemic spreads, possessed the B. cepacia epidemic strain marker (BCESM) (27), and were associated with severe pulmonary disease in CF patients (14). The HBP-positive strain J415, however, was not associated with patient-to-patient spread (12) and did not possess the BCESM, whereas strain ATCC 17765, which was isolated from a urinary tract infection, did carry the BCESM.
In our unpublished survey (Withnall et al., unpublished data), using Raman spectroscopy and pyridine-hemochrome assays, we have detected cell-associated Fe(III)PPIX in 26 out of the present 30 strains when the strains were grown on blood agar. In the present study, strains of all species examined bound μ-oxo bisheme {i.e., [Fe(III)PPIX]2O} in vitro. However, we found no quantitative differences between the five genomovars, or those possessing HBPs, in their ability to bind this heme species from a fixed single concentration of the ligand, suggesting that the μ-oxo bisheme binding may have been nonspecific. It is not known, however, if epidemic HBP-expressing strains accumulate more heme than HBP-negative strains during growth on blood-containing media (where hemoglobin is the major heme source) or whether they display differences in their avidity for heme compared to HBP-negative strains. More-detailed analyses are needed to determine the cellular affinities for FePPIX and the contribution made by the HBP to heme binding.
It is noteworthy that the HPB of B. cepacia genomovar III strain C5424 was able to bind both monomeric hematin [Fe(III)PPIX.OH.H2O] and the μ-oxo bisheme, as determined by SDS-PAGE. Binding of Fe(III)PPIX in either the μ-oxo bisheme or monomeric form would advantage B. cepacia by aiding subversion of neutrophil oxidant-killing mechanisms, as both of these heme species possess intrinsic catalase activity (19). The ability to bind monomeric Fe(III)PPIX may be an advantage during colonization and infection of the lung by epidemic strains. Endobronchial pHs of between 6.58 and 6.62 have been recorded in both normal subjects and those with gram-negative pneumonia and chronic lung disease (2). These acidic pHs would favor the existence of the monomeric form of Fe(III)PPIX, which exists in a pH-dependent equilibrium with the μ-oxo bisheme (31, 38). In addition, any FePPIX bound to the cell surface or the HBP in the μ-oxo bisheme (dimeric) form would dissociate at these lower pHs to give the monomeric species, which is more catalase active (19). One important property of both monomeric and dimeric hemes is their ability to aggregate (7). Bacterial cell surface heme aggregates would provide both a physical and chemical barrier resistant to H2O2 and other reactive oxidants. Both soluble and cell surface-aggregated μ-oxo bisheme can protect P. gingivalis cells against hydrogen peroxide (41) through catalase activity while concomitantly acting, via porphyrin ring destruction (8), as a sacrificial oxidizable substrate in the presence of H2O2. Thus, expression of an HPB may allow B. cepacia to acquire protective cell surface hemes to subvert the effects of neutrophil-derived H2O2 during inflammatory episodes, a factor which may also aid colonization and infection. The detection of the HBP may serve both as a useful taxonomic characteristic and as an adjunct in establishing the epidemic status of clinical isolates of B. cepacia.
REFERENCES
- 1.Allen R C. Phagocytic leukocyte oxygenation activities and chemiluminescence: a kinetic approach to analysis. Methods Enzymol. 1986;133:449–493. doi: 10.1016/0076-6879(86)33085-4. [DOI] [PubMed] [Google Scholar]
- 2.Bodem C R, Lampton L M, Miller D P, Tarka E F, Everett E D. Endobronchial pH relevance to aminoglycoside activity in gram-negative bacillary pneumonia. Am Rev Respir Dis. 1983;127:39–41. doi: 10.1164/arrd.1983.127.1.39. [DOI] [PubMed] [Google Scholar]
- 3.Bramanti T E, Holt S C. Role of porphyrins and host iron transport proteins in regulation of growth in Porphyromonas gingivalis W50. J Bacteriol. 1991;173:7330–7339. doi: 10.1128/jb.173.22.7330-7339.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bramanti T E, Holt S C. Localization of a Porphyromonas gingivalis 26-kilodalton heat-modifiable, heme-regulated surface protein which translocates across the outer membrane. J Bacteriol. 1992;174:5827–5839. doi: 10.1128/jb.174.18.5827-5839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramanti T E, Holt S C. Hemin uptake in Porphyromonas gingivalis: Omp26 is a hemin-binding surface protein. J Bacteriol. 1993;175:7413–7420. doi: 10.1128/jb.175.22.7413-7420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown R K, Kelly F J. Role of free radicals in the pathogenesis of cystic fibrosis. Thorax. 1994;49:738–742. doi: 10.1136/thx.49.8.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown S B, Dean T C, Jones P. Aggregation of ferrihaems. Dimerization and protolytic equilibria of protoferrihaem and deuteroferrihaem in aqueous solution. Biochem J. 1970;117:733–739. doi: 10.1042/bj1170733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown S B, Jones P. Reactions between haemin and hydrogen peroxide. 2. Destructive oxidation of haemin. Biochem J Trans Faraday Soc. 1968;64:994–998. [Google Scholar]
- 9.Daldal F, Cheng S, Applebaum J, Davidson E, Prince R C. Cytochrome c2 is not essential for photosynthetic growth of Rhodopseudomonas capsulata. Proc Natl Acad Sci USA. 1986;83:2012–2016. doi: 10.1073/pnas.83.7.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling P, Chan M, Cox A D, Sokol P A. Siderophore production by cystic fibrosis isolates of Burkholderia cepacia. Infect Immun. 1998;66:874–877. doi: 10.1128/iai.66.2.874-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gessner A R, Mortensen J E. Pathogenic factors of Pseudomonas cepacia isolates from patients with cystic fibrosis. J Med Microbiol. 1990;33:115–120. doi: 10.1099/00222615-33-2-115. [DOI] [PubMed] [Google Scholar]
- 12.Glass S, Govan J R W. Pseudomonas cepacia—fatal pulmonary infection in a patient with cystic fibrosis. J Infect. 1986;13:157–158. doi: 10.1016/s0163-4453(86)92953-1. [DOI] [PubMed] [Google Scholar]
- 13.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Govan J R W, Hughes J E, Vandamme P. Burkholderia cepacia: medical, taxonomic and ecological issues. J Med Microbiol. 1996;45:395–407. doi: 10.1099/00222615-45-6-395. [DOI] [PubMed] [Google Scholar]
- 15.Govan J R W, Nelson J W. Microbiology of cystic fibrosis. Br Med Bull. 1992;48:912–930. doi: 10.1093/oxfordjournals.bmb.a072585. [DOI] [PubMed] [Google Scholar]
- 16.Hughes J E, Stewart J, Barclay G R, Govan J R W. Priming of neutrophil respiratory burst activity by lipopolysaccharide from Burkholderia cepacia. Infect Immun. 1997;65:4282–4287. doi: 10.1128/iai.65.10.4281-4287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isles A, Maclusky I, Corey M, Gold R, Prober C, Flemming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 18.Jansen H-J, Hart C A, Rhodes J M, Saunders J R, Smalley J W. A novel mucin-sulphatase activity found in Burkholderia cepacia and Pseudomonas aeruginosa. J Med Microbiol. 1999;48:551–557. doi: 10.1099/00222615-48-6-551. [DOI] [PubMed] [Google Scholar]
- 19.Jones P, Robson T, Brown S B. The catalase activity of ferrihaems. Biochem J. 1973;135:353–359. doi: 10.1042/bj1350353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kay W W, Phipps B M, Ishiguro E E, Trust T J. Porphyrin binding by the surface array virulence protein of Aeromonas salmonicida. J Bacteriol. 1985;164:1332–1336. doi: 10.1128/jb.164.3.1332-1336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kooi C, Cox A, Darling P, Sokol P A. Neutralizing antibodies to an extracellular Pseudomonas cepacia protease. Infect Immun. 1994;62:2811–2817. doi: 10.1128/iai.62.7.2811-2817.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee B C. Isolation of haemin-binding proteins of Neisseria gonorrhoeae. J Med Microbiol. 1992;36:121–127. doi: 10.1099/00222615-36-2-121. [DOI] [PubMed] [Google Scholar]
- 23.Lee B C. Isolation and characterization of haemin-binding proteins from Neisseria meningitidis. Microbiology. 1994;140:1473–1480. doi: 10.1099/00221287-140-6-1473. [DOI] [PubMed] [Google Scholar]
- 24.Lee B C. Quelling the red menace: haem capture by bacteria. Mol Microbiol. 1995;18:383–390. doi: 10.1111/j.1365-2958.1995.mmi_18030383.x. [DOI] [PubMed] [Google Scholar]
- 25.Létoffé S, Redeker V, Wandersman C. Isolation and characterization of an extracellular haem-binding protein from Pseudomonas aeruginosa that shares function and sequence homology with the Serratia marcescens HasA haemophore. Mol Microbiol. 1998;28:1223–1234. doi: 10.1046/j.1365-2958.1998.00885.x. [DOI] [PubMed] [Google Scholar]
- 26.Mahenthiralingam E, Coenye T, Chung J W, Speert D P, Govan J R W, Taylor P, Vandamme P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J Clin Microbiol. 2000;38:910–913. doi: 10.1128/jcm.38.2.910-913.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahenthiralingam E, Simpson D A, Speert D P. Identification of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J Clin Microbiol. 1997;35:808–816. doi: 10.1128/jcm.35.4.808-816.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansheim B J, Kasper D L. Purification and immunochemical characterization of the outer membrane complex of Bacteroides melaninogenicus subspecies asaccharolyticus. J Infect Dis. 1977;135:787–799. doi: 10.1093/infdis/135.5.787. [DOI] [PubMed] [Google Scholar]
- 29.McKevitt A I, Woods D E. Characterization of Pseudomonas cepacia isolates from patients with cystic fibrosis. J Clin Microbiol. 1984;19:291–293. doi: 10.1128/jcm.19.2.291-293.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer J-M, Hohnadel D, Halle F. Cepabactin from Pseudomonas cepacia, a new type of siderophore. J Gen Microbiol. 1989;135:1479–1487. doi: 10.1099/00221287-135-6-1479. [DOI] [PubMed] [Google Scholar]
- 31.Miller J R, Taies J A, Silver J. Mossbauer and spectroscopic studies on substituted tetraphenylporphyrinato iron (III) complexes in aqueous solutions and the formation of the μ-oxo-bridged species. Inorg Chim Acta. 1987;138:205–214. [Google Scholar]
- 32.Moore G R, Kadir F H A, Al-Massad F K, LeBrun N E, Thomsom A J, Greenwood C, Keen J N, Findlay J B C. Structural heterogeneity of Pseudomonas aeruginosa bacterioferritin. Biochem J. 1994;304:493–497. doi: 10.1042/bj3040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson J W, Butler S L, Krieg D P, Govan J W R. Virulence factors of Burkholderia cepacia. FEMS Immunol Med Microbiol. 1994;8:89–98. doi: 10.1111/j.1574-695X.1994.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 34.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 35.Pendrak M L, Perry R D. Proteins essential for expression of the Hms+ phenotype of Yersinia pestis. Mol Microbiol. 1993;8:857–864. doi: 10.1111/j.1365-2958.1993.tb01632.x. [DOI] [PubMed] [Google Scholar]
- 36.Sajjan U S, Sun L, Goldstein R, Forstner J F. Cable (Cbl) type II pili of cystic fibrosis-associated Burkholderia (Pseudomonas) cepacia: nucleotide sequence of the cblA major subunit pilin gene and novel morphology of the assembled appendage fibers. J Bacteriol. 1995;177:1030–1038. doi: 10.1128/jb.177.4.1030-1038.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw D, Poxton I R, Govan J R W. Biological activity of Burkholderia (Pseudomonas) cepacia lipopolysaccharide. FEMS Immunol Med Microbiol. 1995;11:99–106. doi: 10.1111/j.1574-695X.1995.tb00095.x. [DOI] [PubMed] [Google Scholar]
- 38.Silver J, Lukas B. Mössbauer studies on protoporphyrin IX iron(III) solutions. Inorg Chim Acta. 1983;78:219–224. [Google Scholar]
- 39.Smalley J W, Birss A J, McKee A S, Marsh P D. Haemin-binding proteins in Porphyromonas gingivalis W50 grown in the chemostat under haemin-limitation. J Gen Microbiol. 1993;139:2145–2150. doi: 10.1099/00221287-139-9-2145. [DOI] [PubMed] [Google Scholar]
- 40.Smalley J W, Silver J, Marsh P J, Birss A J. The periodontopathogen Porphyromonas gingivalis binds iron protoporphyrin IX in the μ-oxo dimeric form: an oxidative buffer and possible pathogenic mechanism. Biochem J. 1998;331:681–685. doi: 10.1042/bj3310681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smalley J W, Birss A J, Silver J. The periodontal pathogen Porphyromonas gingivalis harnesses the chemistry of the μ-oxo bishaem of iron protoporphyrin IX to protect against hydrogen peroxide. FEMS Microbiol Lett. 2000;183:159–164. doi: 10.1111/j.1574-6968.2000.tb08951.x. [DOI] [PubMed] [Google Scholar]
- 42.Sokol P A. Production and utilization of pyochelin by clinical isolates of Pseudomonas cepacia. J Clin Microbiol. 1986;23:560–562. doi: 10.1128/jcm.23.3.560-562.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokol P A, Lewis C J, Dennis J J. Isolation of a novel siderophore from Pseudomonas cepacia. J Med Microbiol. 1992;36:184–189. doi: 10.1099/00222615-36-3-184. [DOI] [PubMed] [Google Scholar]
- 44.Stugard C E, Daskaleros P A, Payne S M. A 101-kilodalton heme-binding protein associated with Congo red binding and virulence of Shigella flexneri and enteroinvasive Escherichia coli strains. Infect Immun. 1989;57:3534–3539. doi: 10.1128/iai.57.11.3534-3539.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas P E, Ryan D, Levin W. An improved staining procedure for the detection of peroxidase activity of cytochrome P-450 on sodium dodecyl sulphate polyacrylamide gels. Anal Biochem. 1976;75:168–176. doi: 10.1016/0003-2697(76)90067-1. [DOI] [PubMed] [Google Scholar]
- 46.Vandamme P, Holmes B, Vancanneyt H, M, Coeyne T, Hoste B, Coopman R, Revets H, Lauwers S, Gillis M, Kersters K, Govan J R W. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients: proposal of Burkholderia multivorans sp. nov. Int J Syst Bacteriol. 1997;47:1188–1200. doi: 10.1099/00207713-47-4-1188. [DOI] [PubMed] [Google Scholar]
- 47.Vandamme P, Mahenthiralingam E, Holmes B, Coeyne T, Hoste B, De Vos P, Henry D, Speert D P. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV) J Clin Microbiol. 2000;38:1042–1047. doi: 10.1128/jcm.38.3.1042-1047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vasil M L, Krieg D P, Kuhns J S, Ogle J W, Shortridge V D, Weisbeek P. Molecular analysis of hemolytic and phospholipase activities of Pseudomonas cepacia. Infect Immun. 1990;58:4020–4029. doi: 10.1128/iai.58.12.4020-4029.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zughaier S M, Ryley H C, Jackson S K. Lipopolysaccharide (LPS) from Burkholderia cepacia is more active than LPS from Pseudomonas aeruginosa and Stenotrophomonas maltophilia in stimulating tumor necrosis factor alpha from human monocytes. Infect Immun. 1999;67:1505–1507. doi: 10.1128/iai.67.3.1505-1507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zughaier S M, Ryley H C, Jackson S K. A melanin-like pigment purified from an epidemic strain of Burkholderia cepacia attenuates monocyte respiratory burst activity by scavenging superoxide anion. Infect Immun. 1999;67:908–913. doi: 10.1128/iai.67.2.908-913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]