Abstract
KIF1A is a neuron-specific member of the kinesin-3 family of microtubule (MT) plus-end-directed motor proteins. It powers the migration of nuclei in differentiating brain stem cells and the transport of synaptic precursors and dense core vesicles in axons. Its dysfunction causes severe neurodevelopmental and neurodegenerative diseases termed KIF1A-associated neurological disorders (KAND). KAND mutations span the entirety of the KIF1A protein sequence, of which the majority are located within the motor domain and are thus predicted to affect the motor’s motility and force-generating properties. Unfortunately, the molecular etiologies of KAND remain poorly understood, in part because KIF1A’s molecular mechanism remains unclear. Here, we describe detailed methods for how to express a tail-truncated dimeric KIF1A in E. coli cells and provide step-by-step protocols for performing single-molecule studies with total internal reflection fluorescence microscopy and optical tweezers assays, which, when combined with structure-function studies, help to decipher KIF1A’s molecular mechanism.
Keywords: Kinesin, KIF1A, Single-molecule, Molecular cloning, E. coli, Recombinant protein expression, His-tag purification, TIRF, Optical tweezers
1. Introduction
KIF1A is a neuron-specific member of the kinesin-3 family of microtubule (MT) plus-end-directed motor proteins [1, 2] and powers the migration of nuclei in differentiating brain stem cells [3, 4] and the transport of synaptic precursors and dense core vesicles in axon terminals [2, 5-9]. Its dysfunction leads to severe neurodevelopmental and neurodegenerative diseases termed KIF1A-associated neurological disorders (KAND) [10, 11], including progressive spastic paraplegias, microcephaly, encephalopathies, intellectual disability, autism, autonomic and peripheral neuropathy, optic nerve atrophy, cerebral and cerebellar atrophy, and others [10-52]. KAND mutations, which are inherited variants or de novo mutations, span the entirety of the KIF1A protein sequence [10]. The majority of the mutations are located within the motor domain (MD or ‘head’) [10] and are thus predicted to affect the motor’s motility properties, whereas mutations located outside the MD are likely involved in mediating dimerization, autoinhibition, and/or cargo binding [12]. Our own work [10, 53] and the work of others [54, 55] has shown that the KAND MD mutations that have been investigated so far indeed affect the motor’s ability to generate force and movement.
Through the patient advocacy group KIF1A.org more than 400 families with children with KIF1A mutations have come together to improve the lives of those affected by KAND and to accelerate drug discovery. Unfortunately, the molecular etiologies of KAND remain poorly understood, in part because KIF1A’s molecular mechanism remains unclear. For example, KIF1A is extremely fast and superprocessive (the motor can take more than a 1000 steps along the MT before dissociating), but, somewhat counterintuitively, gives up easily under load. These behaviors distinguish KIF1A from the founding member of the kinesin family kinesin-1, however, how KIF1A achieves this unique set of properties is unknown. In addition, while most mutations occur in KIF1A’s MD [10], high-resolution structures of the KIF1A-MT complex have not been resolved. In order to understand the molecular etiology of KAND, knowledge of the nucleotide state-dependent structures of KIF1A and how disease mutations affect KIF1A’s conformational states is required. Such knowledge will aid in the development of therapeutics for these neurological disorders, for which, at present, no treatments exist.
Various protein expression systems, including eukaryotic and prokaryotic organisms, have been used to produce recombinant proteins. Among these systems, the E. coli-based expression system provides the means for fast and reliable protein production. We use this system to express a recombinant tail-truncated dimerized KIF1A, for both the wild-type (WT) and KAND mutant proteins [10, 53]. Mutagenesis and structure-function studies on recombinant proteins in combination with in vitro single-molecule assays have been invaluable in providing insights into the molecular mechanisms of MT-associated motors proteins [56-68]. In this chapter, we provide detailed protocols for the expression and purification of recombinant dimeric KIF1A and describe how to perform single-molecule total internal reflection fluorescence (TIRF) microscopy and optical tweezers assays to study the motility and force generation of the purified KIF1A along MTs.
2. Materials
2.1. Construct Generation
Primers (IDTDNA).
KOD hot start DNA polymerase kit (Millipore Sigma, #71086).
NucleoSpin® gel and PCR cleanup kit (Takara, #740609).
A plasmid containing KIF1A(aa1-393)-leucine zipper (AddGene, #61665).
A PCR thermocycler.
pSNAP-tag® (T7)-2 (New England BioLabs, #N9181S).
pSNAPf (New England BioLabs, #N9183S).
NdeI (New England BioLabs, #R0111S, 20 U/μL).
EcoRI-HF (New England BioLabs #R3101S, 20 U/μL).
10× CutSmart buffer (New England BioLabs, usually supplied with restriction enzymes).
rSAP (shrimp alkaline phosphatase) (New England BioLabs, #M0371S, 1 U/μL).
Tris (tris(hydroxymethyl)aminomethane) buffer (1 M, pH 7.6): Add 60.5 g of Tris base and 300 mL ddH2O to a bottle, adjust the pH to 7.6 using 1 N HCl. Fill the solution with ddH2O to final 500 mL, store at room temperature.
EDTA (ethylenediaminetetraacetic acid) solution (0.5 M, pH 8.0): Add 27.9 g EDTA·2H2O and 100 mL ddH2O to a bottle, use NaOH to adjust to pH to 8.0 (EDTA is not soluble in water unless the pH is adjusted), then fill the solution with water to final 150 mL, store at room temperature, or 4 °C for long-term storage.
TE buffer: 10 mM Tris, 1 mM EDTA, pH 7.6.
Agarose.
SYBR safe DNA gel stain (ThermoFisher Scientific, #S33102, 1000×).
A gel electrophoresis system (Thermal Owl™ EasyCast™ B1A mini gel electrophoresis system).
5× Orange G loading buffer (0.125% (w/v) Orange G, 50% (v/v) glycerol, 2.5× TBE buffer): In a 15-mL falcon tube, mix 12.5 mg Orange G, 5 mL glycerol, and 5 mL of 5× TBE buffer, fill with ddH2O to 10 mL, aliquot and store at 4 °C, or −20 °C for long-term storage.
1 Kb Plus DNA Ladder (ThermoFisher Scientific, #10787018).
5× TBE buffer (450 mM Tris, 450 mM boric acid, 10 mM EDTA, pH 8.3): Add 54 g Tris base, 27.5 g boric acid, 20 mL of 5 M EDTA to a bottle, fill to 1 L, nutate or heat to dissolve the solid. If there is solid precipitating out after several months, microwave the solution to redissolve the precipitation.
A spectrophotometer.
10× T4 DNA ligase buffer (New England BioLabs, supplied with T4 DNA ligase).
T4 DNA ligase (New England BioLabs, #M0202S, 400 U/μL).
NEB® Turbo competent E. coli (high efficiency) (New England BioLabs, #C2984H).
SOC media (New England BioLabs, supplied with E. coli competent cells).
Carbenicillin (100 mg/mL): Dissolve 1 g of carbenicillin in 10 mL ddH2O, filter the solution using a 0.22 μm Millex-GS syringe filter unit (Sigma Millipore, #SLGS033SS). Store at −20 °C.
LB/carbenicillin agar plate (50 μg/mL carbenicillin): Autoclave 7 g LB agar (Difco™ LB agar, Lennox, BD Diagnostic System, # DF0402-17-0) in 200 mL ddH2O. When the solution is slightly cooled, add 100 μL of 100 mg/mL carbenicillin, mix well. Pour the solution into 100 mm × 20 mm petri dishes and allow the solution to cool and solidify. Store the plates upside down at 4 °C.
LB media: Suspend 20 g of LB broth (Difco™ LB Broth, Lennox, BD Diagnostic System, #DF0402-07-0) in 1 L ddH2O. Autoclave and store at room temperature.
A temperature-controlled shaker.
PureYield™ plasmid miniprep system (Promega, #A1223).
2.2. Mutagenesis
Q5® site-directed mutagenesis Kit (New England Biolabs, #E0554S).
Primers for mutagenesis (IDTDNA).
DMSO.
NEB® Turbo competent E. coli (high efficiency) (see Subheading 2.1).
LB/carbenicillin agar plate (see Subheading 2.1).
SOC media (see Subheading 2.1).
2.3. E. coli Growth for Protein Expression
BL21(DE3)CodonPlus RILP chemical competent cells (Agilent, #230280).
SOC media (see Subheading 2.1).
Chloramphenicol (34 mg/mL): Dissolve 34 mg chloramphenicol in ethanol, then filter the solution using a 0.2 μm filter. Store at −20 °C.
Carbenicillin (100 mg/mL) (see Subheading 2.1).
LB/chloramphenicol/carbenicillin agar plate (17 μg/mL chloramphenicol, 25 μg/mL carbenicillin): Suspend 7 g LB agar in 200 mL ddH2O and autoclave. When the solution is slightly cooled, add 50 μL of 34 mg/mL chloramphenicol and 50 μL of 100 mg/mL carbenecillin. Pour the solution into 100 mm × 20 mm petri dishes and allow the solution to cool and solidify. Store the plates upside down at 4 °C.
TB media (1.2% (w/v) peptone, 2.4% (w/v) yeast extract): Dissolve 4.8 g peptone (Gibco™ Bacto™ peptone, Fisher Scientific, # DF0118-17-0) and 9.6 g yeast extract (Gibco™ Bacto™ yeast extract, #DF0127-17-9) in 380 mL ddH2O, autoclave the media.
20× phosphate glycerol buffer (8% (v/v) glycerol, 0.34 M KH2PO4, 1.44 M K2HPO4): Add 40 mL glycerol, 23.1 g KH2PO4, 164 g K2HPO4·3H2O to a bottle, fill ddH2O to final 500 mL, autoclave and store at room temperature.
A temperature-controlled shaker.
IPTG (isopropyl β-d-1-thiogalactopyranoside) solution (100 mM): Dissolve 1 g in 42 mL ddH2O, filter the solution using a 0.22 μm Millex-GS syringe filter unit. Aliquot and store at −20 °C.
450-mL centrifuge tubes.
A refrigerated centrifuge.
B-PER™ complete bacterial protein extraction reagent (ThermoFisher Scientific, #89821).
MgCl2 solution (1 M): Dissolve 4.76 g of MgCl2 in 50 mL ddH2O, store at room temperature.
EGTA solution (0.5 M, pH 8.0): Add 28.5 g EGTA and 100 mL to a bottle, use NaOH to adjust to pH to 8.0 (EGTA is not soluble in water unless the pH is adjusted), then fill with water to final 150 mL, store at room temperature.
DTT (dithiothreitol) solution (1 M): Dissolve 1.54 g DTT in 10 mL ddH2O, aliquot and store at −20 °C.
ATP (adenosine 5′-triphosphate) solution (100 mM): Dissolve 50.7 mg of ATP in 1 mL ddH2O, aliquot and store at −20 ° C.
PMSF (phenylmethylsulfonyl fluoride) solution (100 mM in isopropanol): Dissolve 0.174 g of PMSF in 10 mL isopropanol, aliquot and store at −20 °C.
2.4. Protein Purification
Beckman Coulter Optima TLX-120 ultracentrifuge.
TLA 110 fixed-angle rotor (Beckman Coulter, #366735).
Open-top thickwall polycarbonate tube (13 mm × 56 mm, 3.2 mL, Beckman Coulter, #362305).
cOmplete™ His-tag purification Ni-NTA resin (Roche, #5893682001).
HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) solution (1 M, pH 7.2): Add 119.2 g of HEPES and 400 mL ddH2O to a bottle, adjust the pH using NaOH pellet to pH 7.2, fill the solution to 500 mL, store at 4 °C.
KCl solution (1 M): Dissolve 37.7 g of KCl in 400 mL ddH2O, fill with ddH2O to 500 mL, store at room temperature.
MgCl2 solution (1 M) (see Subheading 2.3).
EGTA solution (0.5 M, pH 8.0) (see Subheading 2.3).
DTT solution (1 M) (see Subheading 2.3).
ATP solution (100 M) (see Subheading 2.3).
PMSF solution (100 M) (see Subheading 2.3).
Pluronic™ F-127 solution (10% (w/v)): Add 5 g of Pluronic F-127 and 10 mL ddH2O to a tube, nutate at room temperature until the solid is completely dissolved, store at 4 °C.
Wash buffer (WB): 50 mM HEPES, 300 mM KCl, 2 mM MgCl2, 1 mM EGTA, 10% (v/v) glycerol, 1 mM DTT, 0.1 mM ATP, 1 mM PMSF, 0.1% (w/v) Pluronic F-127.
SNAP-tag fluorophore ligand solution (5 mM in DMSO): Dissolve SNAP-tag fluorophore ligand (New England Biolabs) in DMSO to final 5 mM, store at −20 °C.
Imidazole (2 M, pH 8.0): Add 20.4 g imidazole, 50 mL 1 N HCl, 50 mL ddH2O in a bottle, use HCl to adjust the pH to 8.0. Fill the solution to 150 mL, store at 4 °C.
Elution buffer (EB): 50 mM HEPES, 300 mM KCl, 2 mM MgCl2, 1 mM EGTA, 10% (v/v) glycerol, 1 mM DTT, 0.1 mM ATP, 1 mM PMSF, 0.1% (w/v) Pluronic F-127, 250 mM imidazole.
2.5. MT-Binding and - Release Assay
PIPES (piperazine-N,N'-bis(2-ethanesulfonic acid)) solution (0.5 M, pH 6.8): Add 75.6 g of PIPES and 400 mL ddH2O to a bottle, use NaOH pellets to adjust the pH to 6.8. Fill the solution to 500 mL, store at 4 °C.
MgCl2 solution (1 M) (see Subheading 2.3).
EGTA solution (0.5 M, pH 8.0) (see Subheading 2.3).
BRB80G10: PIPES 80 mM, MgCl2 2 mM, EGTA 1 mM, 10% (v/v) glycerol, pH 6.8.
Tubulin (porcine brain, unlabeled, 10 mg/mL) solution: Dissolve one vial (1 mg) of lyophilized tubulin (Cytoskeleton, #T238P-B) in 100 μL of BRB80G10 with 1 mM DTT, aliquot to 5 μL/aliquot, flash freeze and store at −80 °C.
GTP (guanosine 5′-triphosphate) solution (100 mM): Dissolve 52 mg of GTP in 1 mL ddH2O, aliquot and store at −20 °C.
Open-top thickwall polycarbonate tube (7 × 20 mm, 230 μL volume, suitable for rotor TLA-100) (Beckman Coulter, #343775).
BRB80G60: 80 mM PIPES, 2 mM MgCl2, 1 mM EGTA, 60% (v/v) glycerol, pH 6.8.
DTT (1 M) (see Subheading 2.3).
Taxol solution (10 mM in DMSO): Dissolve 25 mg in 2.927 mL DMSO, aliquot and store at −20 °C.
Beckman Coulter Optima TLX-120 ultracentrifuge.
Zeba™ spin desalting columns (40K MWCO, 0.5 mL, ThermoFisher Scientific, #87766).
AMP·PNP (adenylyl-imidodiphosphate) solution (100 mM): Dissolve 25 mg AMP·PNP in 0.5 mL ddH2O, aliquot and store at −20 °C.
TLA-100 fixed-angle rotor (Beckman Coulter, #343837).
High-salt release buffer: 80 mM PIPES, 2 mM MgCl2, 1 mM EGTA, 10% (v/v) glycerol, 300 mM KCl, pH 6.8.
2.6. TIRF Assay
Coverslips (No. 1.5H, 170 ± 5 μm thickness, 18 mm × 18 mm) (Carl Zeiss, #474030-9000-000).
HNO3 (25% (v/v)): Slowly add 107 mL of 70% (v/v) HNO3 to 193 mL ddH2O, do not add water into the acid.
NaOH solution (2 M): Dissolve 24 g of NaOH in 300 mL ddH2O, keep in a plastic bottle at room temperature, do not store the strong base in a glass bottle.
Fisherbrand™ Superfrost™ disposable microscope slides (Pre-cleaned, 75 mm × 25 mm × 1 mm) (Fisher Scientific, #12-550-123).
Tubulin (porcine brain, unlabeled, 10 mg/mL) (see Subheading 2.5).
Biotinylated tubulin solution (porcine brain, 1 mg/mL): Dissolve one vial (20 μg) of lyophilized biotinylated tubulin (Cytoskeleton, #T333P-A) in 20 μL of BRB80G10 with 1 mM DTT, aliquot to 2 μL/aliquot, flash freeze and store at −80 °C.
Fluorescent-labeled tubulin solution (porcine brain, 1 mg/mL): Preparation is the same as in “biotinylated tubulin solution,” use fluorescently labeled lyophilized tubulin (Cytoskeleton) instead of lyophilized biotinylated tubulin.
Biotinylated BSA (bovine serum albumin) solution (5 mg/mL in BRB80G10): Dissolve 25 mg biotinylated BSA (Thermo-Fisher Scientific, #29130) in 5 mL of BRB80G10, aliquot and flash freeze, store at −20 °C. The working concentration is 0.5 mg/mL.
Streptavidin (1 mg/mL in BRB80G10): Dissolve 1 mg streptavidin (Promega, #Z7041) in 1 mL of BRB80G10, aliquot and flash freeze, store at −20 °C. The working concentration is 0.25 mg/mL.
HME30G10: HEPES 30 mM, MgCl2 2 mM, EGTA 1 mM, 10% (v/v) glycerol, pH 7.2.
α-casein solution (25 mg/mL in HME30G10): Add 1 g of α-casein (Sigma, #C6780) and 40 mL HME30G10 in a 50-mL conical tube, nutate at 4 °C for several hours until all the α-casein has dissolved. Aliquot, flash freeze, and store at −20 °C.
BSA (50 mg/mL in BRB80G10): Dissolve 0.5 g of BSA in 10 mL BRB80G10, aliquot, flash freeze, and store at −20 °C.
Pluronic F-127 (10% (v/v)) (see Subheading 2.4).
Blocking buffer (BB): PIPES 80 mM, MgCl2 2 mM, EGTA 1 mM, 10 μM taxol, 1 mM DTT, 0.5% Pluronic F-127, 2 mg/mL BSA, 1 mg/mL α-casein, pH 6.8.
Motility buffer (MB): HEPES 50 mM, KCl 60 mM, EGTA 1 mM, 10 μM taxol, 1 mM DTT, 0.5% Pluronic F-127, 50 mM glucose, 2 mg/mL BSA, 1 mg/mL α-casein, pH 7.2.
ATP (100 mM) (see Subheading 2.3).
d-Biotin solution (ThermoScientific, #B20656, 50 mM).
Glucose oxidase solution (10 μM): Dissolve 4.8 mg glucose oxidase (Sigma, #G2133) in 3 mL of HEPES 50 mM with 50% glycerol, store at −20 °C. Working concentration is 100 nM.
Catalase solution (150 μM): Dissolve 100 mg catalase (Sigma, #C40) in 2.67 mL of HEPES 50 mM with 50% glycerol, store at −20 °C. Working concentration is 1.5 μM.
Gloxy: mix equivalent volume of 10 μM glucose oxidase and 150 μM catalase, store at −20 °C.
Vacuum grease.
2.7. Optical Tweezers Assay
Polybead® carboxylate microspheres (0.50 μm, 2.5% (w/v), Polysciences, #09836-15).
Activation buffer (AB): 100 mM NaCl, 10 mM MES, pH 6.0.
EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide).
Sulfo-NHS (N-Hydroxysulfosuccinimide).
Coupling buffer (CB): 100 mM Na3PO4, 0.1% Pluronic F-127, pH 7.4.
Anti-GFP antibody (total 100 μg in PBS buffer).
BSA solution (50 mg/mL in BRB80G10) (see Subheading 2.6).
Wash buffer (WB): 30 mM HEPES, 2 mM MgCl2, 1 mM EGTA, 0.1% Pluronic F-127, pH 7.2.
Storage buffer (SB): 30 mM HEPES, 150 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.1% Pluronic F-127, 10% glycerol, pH 7.2.
α-casein solution (25 mg/mL in HME30G10) (see Subheading 2.3).
HME30G10: HEPES 30 mM, MgCl2 2 mM, EGTA 1 mM, 10% (v/v) glycerol, pH 7.2.
DTT solution (1 M) (see Subheading 2.3).
DMSO.
EZ-link™ sulfo-NHS-biotin solution (100 mM in DMSO) (ThermoFisher Scientific, #21217): Dissolve 50 mg of sulfo-NHS-biotin in 1.129 mL DMSO, aliquot, flash freeze, and store at −80 °C.
Amicon ultra-0.5 mL centrifugal filters, 10 kDa (Millipore, #UFC501024).
Blocking buffer (BB): PIPES 80 mM, MgCl2 2 mM, EGTA 1 mM, 10 μM taxol, 1 mM DTT, 0.5% Pluronic F-127, 1 mg/mL α-casein, pH 6.8.
Streptavidin (1 mg/mL in BRB80G10) (see Subheading 2.6).
MTs (0.2 mg/mL in BRB80G10 with 1 mM DTT and 10 μM taxol).
Motility buffer (MB): HEPES 50 mM, KCl 60 mM, MgCl2 mM, EGTA 1 mM, 10 μM taxol, 1 mM DTT, 0.5% Pluronic F-127, 1 mg/mL α-casein, pH 6.8.
ATP (100 mM) (see Subheading 2.3).
d-biotin solution (50 mM) (see Subheading 2.6).
Gloxy (see Subheading 2.6).
Vacuum grease.
3. Methods
3.1. KIF1A Construct Generation
This section describes the method of inserting a KIF1A construct (aa1-393 with a leucine zipper (LZ)) into a vector backbone bearing a C-terminal SNAPf-EGFP-6His tag, utilizing restriction enzymes. This method can be applied to the insertion of any gene of interest into a vector backbone.
If unspecified, primers are ordered from IDTDNA; the PCR reactions are performed using a KOD hot start DNA polymerase kit following the recommended protocol provided by the vendor; cleanup of the PCR products is done using a NucleoSpin® Gel and a PCR Clean-Up kit following the recommended protocol provided by the vendor.
We will first describe the generation of a KIF1A(1-393)-LZ DNA fragment with NdeI and EcoRI restriction sites plus an 8 bp overhang using PCR and a KIF1A-containing plasmid as template, followed by the insertion of the obtained PCR product into a custom-made vector backbone containing C-terminal MCS-SNAPf-EGFP-6His (see Note 1) using standard restriction enzyme (RE)-based molecular cloning methods.
Digest the PCR insert and the vector backbone separately in a 50 μL reaction: mix 20 μL PCR product or backbone (preferably >100 ng/μL), 1 μL NdeI, 1 μL EcoRI-HF, 5 μL 10× CutSmart buffer, and 23 μL ddH2O (see Note 2). For the vector digestion, add additionally 2.5 μL of rSAP to remove the phosphate groups from the backbone to prevent the backbone from recircularizing (see Note 3). Incubate the mixtures at 37 °C for 1 h to digest the DNA, and then heat the solution to deactivate the restriction enzymes at 65 °C for 20 min (see Note 4).
Purify the digested DNA using the PCR cleanup kit and eluted the DNA with 30 μL ddH2O or TE buffer (see Note 5).
Prepare 50 mL of 0.8% (w/v) agarose gel with 5 μL of SYBR Safe dyes in a gel cast. Add 10 μL of 5× Orange G loading buffer to the cleaned-up DNA, and carefully load the samples as well as the DNA ladder into the wells. Run the gel at 100 V in 0.5× TBE buffer for 45 min up to an hour, then carefully cut out the correct bands.
Purify the DNA using the PCR cleanup kit and measure the concentration either using a spectrophotometer or on a DNA gel using a known standard.
Calculate the DNA molarity based on the DNA’s molecular weight, then prepare a ligation reaction to ligate the DNA accordingly. A typical ligation reaction is 10 μL volume, containing 0.02 pmol vector, 0.06 pmol insert DNA, 1 μL 10× T4 DNA ligase buffer, 0.5 μL T4 DNA ligase, and ddH2O (see Note 6). Incubate the ligation mixture at room temperature for 10 min (see Note 7).
To transform E. coli competent cells, thaw 10 μL of NEB Turbo chemical competent cells on ice for 10 min (see Note 8), then add 2 μL of the ligation mixture and flick the tube several times. Incubate the cells on ice for 30 min, then heat shock them for 30 s at 42 °C. Cool the cells on ice for 5 min, then add 190 μL of SOC media to the cells. Shake the cells at 37 °C for 1 h vigorously to allow recovery. Spread the whole culture onto a LB/carbenicillin agar plate and incubate at 37 °C overnight with the agar side facing up.
If colonies appear, pick 2–3 colonies and inoculate each colony in 3 mL LB media with 1.5 μL of 100 mg/mL carbenicilin, shake at 37 °C for 4–5 h or overnight.
Purify the plasmids using PureYield™ plasmid miniprep system according to the vendor’s protocol.
To verify the plasmids, prepare the following 25 μL digestion reaction for each plasmid as well as the backbone vector as control: 5 μL plasmid, 0.5 μL NdeI, 0.5 μL EcoRI-HF, 2.5 μL 10× CutSmart buffer, and 16.5 μL ddH2O. Incubate at 37 °C for 30 min to up to an hour, and then load a 5 μL sample with 2 μL 5× Orange G loading buffer and 3 μL ddH2O (total volume of 10 μL) on a 0.8% agarose gel. The correct plasmids will result in a band for the insert, while an incorrect plasmid and the control vector will not. The plasmid that has the insert can be further confirmed by DNA sequencing.
3.2. Mutagenesis
The results of mutagenesis of kinesin motors have been broadly studied in vitro, in vivo, and in silico [69, 70]. Mutagenesis and structure-function studies provide insights into the molecular mechanisms that underlie the motion and force generation of kinesin motors. Here, we use the Q5® Site-Directed Mutagenesis Kit. In principle, the Q5® high-fidelity DNA polymerase is used to polymerase the template vector with primers carrying the mutations, and subsequently DpnI is used to degrade the template vector, while kinase and ligase are used to phosphorylate and ligate the PCR product into a circular vector, which can be transformed into competent E. coli cells.
Primers are designed to have melting temperature of about 60 °C.
The PCR reaction is assembled following the vender’s protocol. 5% (v/v) DMSO is added to the PCR reaction to reduce complex DNA structures, which helps the DNA polymerase to progress.
After the PCR reaction, set up a 5 μL KLD reaction following the vendor’s recommendation. 2 μL of the KLD reaction mixture is then used for the transformation of E. coli cells. The steps for the transformation of 10 μL NEB Turbo chemical competent cells, the growth of the E. coli cells, and the purification of the plasmids are the same as described above (Subheading 3.1).
After the plasmids are obtained, verify the mutations via DNA sequencing.
3.3. E. coli Growth for Protein Expression
This section describes the methods of transforming and growing E. coli cells, the induction of protein expression, and the harvesting of the cells.
Day 1
Thaw 10 μL of BL21-CodonPlus (DE3)-RILP chemical competent cells on ice for 10 min, add 1 μL of the plasmid, flick the tube several times, and incubate the tube on ice for 30 min.
Heat shock the cells for 30 s at 42 °C and recover on ice for 5 min. Afterwards, add 90 μL SOC media to the tube and spread the cells onto a LB/chloramphenicol/carbenicillin plate, and incubate at 37 °C overnight.
Day 2
Pick a single colony and inoculate it in 0.95 mL of TB with 0.05 mL of 20× phosphate glycerol buffer, 50 μg/mL chloramphenicol, and 50 μg/mL carbenicillin. Shake at 37 °C overnight at 200 rpm.
Day 3
Inoculate 380 mL of TB with 20 mL of 20× phosphate glycerol buffer with the overnight 1 mL culture without adding extra antibiotics. Shake at 37 °C for 5 h. Cool down the culture on ice for 1 h, then add IPTG to final 0.1 mM. Shake at 18 °C overnight at 200 rpm.
Day 4
Harvest the cells by centrifugation for 10 min at 3000 rcf at 4 °C. Discard the supernatant and resuspend the pellet in 5 mL of B-PER complete bacterial protein extraction reagent with 4 mM MgCl2, 2 mM EGTA, 2 mM DTT, 0.2 mM ATP, and 2 mM PMSF. Flash freeze and store at −80 °C (see Note 9).
3.4. Protein Purification
The purification of KIF1A outlined below follows the standard Histag-based purification method.
Thaw the frozen pellet completely at 37 °C and nutate at room temperature for 20 min.
Cool a Dounce homogenizer on ice and homogenize the lysate on ice for ten strokes.
Fill the 3.2-mL open-top thickwall polycarbonate tubes with the lysate, balance the tubes and move them into the TLA-110 fixed-angle rotor, and clear the lysate by centrifugation at 80,000 rpm (average 260,000 × g, k-factor 28) for 10 min at 4 °C in a Beckman tabletop ultracentrifuge.
In the meantime, wash 0.5 mL Ni-NTA resin with 2 × 1 mL wash buffer (WB) in a 15-mL empty column. Once the WB is drained, cap the bottom of the column and load the lysate carefully on top of the resin. Let the lysate flow through the resin and wash the resin with 10 mL WB.
(Optional) If a labeling of the tagged protein with a SNAP-tag ligand (dye) is desired, after the lysate has flown through the resin, wash the resin with 2 × 1 mL WB. Leave ~100 μL solution on top of the resin, then cap the column. Add the SNAP-tag ligand to a final 10 μM concentration and gently shake the column to mix the resin. Incubate at room temperature for 10 min, shake the column 2–3 times during the incubation time. Then wash the resin with 4 × 2 mL WB (see Note 10).
Elute the protein by adding 0.3 mL elution buffer (EB) for each collected fraction, then aliquot the collected solution into 50 μL per aliquot and flash freeze. Store the protein at −80 °C.
3.5. MT-Binding and - Release Assay
The MT-binding and -release assay is used to remove inactive motors: The motors that are capable of binding to MTs are pelleted with the MTs in the presence of AMP·PNP, while the motors that have lost the ability to bind MTs will remain in the supernatant. The pellet is then resuspended in a buffer containing ATP and high salt, which causes the release the functional motors from the MTs into the supernatant, while the motors that are incapable of hydrolyzing ATP will remain bound to MTs. If a KAND mutation impairs the ATPase activity of the motor, this step should be skipped.
3.5.1. Preparation of MTs
Thaw one aliquot of unlabeled tubulin, add 0.55 μL of 10 mM GTP, and incubate at 37 °C for 15 min to polymerize the tubulin.
Add 0.6 μL of 100 μM taxol (in DMSO), mix well, and incubate for another 15 min at 37 °C to stabilize the MTs.
In a 230 μL open-top thickwall polycarbonate tube, add 60 μL BRB80 with 60% glycerol, 1 mM DTT, and 10 μM taxol (cushion), then carefully lay the MT solution on top of the cushion.
Centrifuge at 80,000 rpm (average 250,000 × g, k-factor 10) for 5 min in a TLA-100 fixed-angle rotor in a Beckman tabletop ultracentrifuge at room temperature.
Remove the solution completely, then wash the pellet three times with 20 μL BRB80G10 with 1 mM DTT and 10 μM taxol. Resuspend the pellet carefully in 10 μL of the same buffer, which yields 10 μL of 5 mg/mL MTs. Store the MT solution at room temperature in the dark.
3.5.2. MT-Binding and - Release Assay
Thaw an aliquot of the frozen KIF1A stock quickly in a warm water bath.
Use a desalting column to exchange the buffer to BRB80G10 with 1 mM DTT, 1 mM AMP·PNP, and 10 μM taxol at 4 °C, following the vendor’s protocol (see Note 11).
Warm the protein solution to room temperature, add 3–5 μL MT solution (see Note 12), and incubate at room temperature for 2 min.
In a 230 μL open-top thickwall polycarbonate tube, carefully lay the solution on top of 100 μL BRB80G60 with 1 mM DTT and 10 μM taxol.
Centrifuge the sample at 45,000 rpm (average 78,000 × g, k-factor 32) for 10 min in a TLA-100 fixed-angle rotor in a Beckman tabletop ultracentrifuge at room temperature.
Carefully remove the supernatant, then wash the pellet three times with 20 μL BRB80G10 with 1 mM DTT and 10 μM taxol.
Resuspend the solution in 50 μL of high-salt release buffer with 1 mM DTT, 3 mM ATP, and 10 μM taxol.
Centrifuge the sample at 40,000 rpm (average 62,000 × g, k-factor 41) for 5 min.
Move the supernatant into a 0.5-mL tube and keep it on ice, then aliquot the supernatant to 2 μL per aliquot and flash freeze in liquid nitrogen. Store the samples at −80 °C. The protein should be stable for at least 2–3 months.
3.6. TIRF Assay
The total internal reflection fluorescence (TIRF) microscopy assay is one of the assays that are routinely used to study kinesins both in vitro and in vivo. For in vitro assay, MTs are firmly attached to a coverslip in a flow chamber, then motors are introduced into the chamber with ATP. Motor properties, such as its velocity, processivity (run length), and MT on-rates, can be extracted from the acquired movies.
3.6.1. Coverslip Cleaning
Arrange coverslips on a coverslip holder and submerge the holder in 25% (v/v) HNO3 for 15 min.
Wash the holder twice with ddH2O, then submerge it in 2 M NaOH for 2–5 min.
Wash six times with ddH2O, then dry the coverslips on a heat block (at least 90 °C) for 30 min. Store at 4 °C.
3.6.2. Microscope Slide Assembly
Arrange two ~5 mm-wide strips of Parafilm in parallel with a ~5 mm gap in between on a disposable microscope slide.
Use a small cylinder (e.g., a 1 mL plastic pipette tip) to burnish the film onto the glass. Rub with even pressure across both strips several times to make good adhesion.
Use the pincers on a pair of sharp-nosed forceps to “pick” at the paper backing on one of the stripes of Parafilm until it begins to separate. Once the backing is separated from the Parafilm, use the tweezers to pull it off completely. Remove the backing from both pieces of Parafilm.
Carefully place a coverslip on top of the parafilm, then push down lightly in a few spots with a finger to help them stick.
Pick up the whole slide chamber with the tweezers, cover-slip-side-up. With a heat gun on the table pointing upward, turn on “high.” Hold the center of the slide over the heat gun, about ½ inch away (or place the slide chamber on a heat block). As the Parafilm heats, it becomes transparent. Wait ~1 s after this happens and remove the side quickly.
Hold the slide firmly with forceps (or set on a clean surface). Then quickly use the back of the tweezers to lightly press or tap on the cover slip regions above the Parafilm. Set the slide chamber aside to cool (Parafilm will turn opaque). The created flow chamber holds a ~10 μL volume.
3.6.3. MT Preparation
Mix 2 μL of 1 mg/mL biotinylated tubulin and 2 μL of 1 mg/mL fluorescently labeled tubulin with 2 μL of 10 mg/mL tubulin on ice, then polymerize the MTs as describe in Subheading 3.5.1. After centrifugation, resuspend the pellet in 120 μL of BRB80G10 with 1 mM DTT and 10 μM taxol, which will result in a final MT concentration of 0.2 mg/mL. Store at room temperature in the dark.
3.6.4. Slide Preparation
All incubation steps are performed at room temperature in a humidity chamber (see Note 13). Flow 10 μL of 0.5 mg/mL BSA-biotin into the chamber and incubate for 5 min.
Wash with 3 × 20 μL BB and incubate for 5 min.
Flow in 10 μL of 0.25 mg/mL streptavidin and incubate for 5 min.
Wash the chamber with 3 × 20 μL BB.
Dilute 0.5 μL of 0.2 mg/mL biotinylated MTs in 20 μL BB. Completely remove the solution from the chamber and flow the diluted MTs into the chamber. To achieve that a large fraction of the MTs is aligned with the flow direction, wash the chamber immediately with 3 × 20 μL BB; if a higher density of MTs on the surface is preferred, incubate the MTs in the chamber for 1–2 min before washing with 3 × 20 μL BB.
Exchange the buffer in the chamber with 2 × 20 μL MB.
Mix 1 μL of 100 mM ATP, 1 μL of 50 mM biotin, 1 μL of gloxy, and 46 μL of MB (as KIF1A is “sticky” and has a tendency to bind to the cover glass, increase the BSA concentration to 5 mg/mL if a significant amount of motors is bound to the cover glass surface instead of the MTs), then add 1 μL of an appropriately diluted motor stock and mix well. Flow 2 × 20 μL of the mixture into the chamber.
Seal the chamber using vacuum grease.
3.6.5. TIRF Imaging
Mount the slide on a TIRF microscope and adjust the laser power to obtain a good signal-to-noise ratio without causing a significant photo bleaching of the fluorophores. Acquisition times of 100–200 ms per frame (or faster) are preferred as KIF1A is relatively fast.
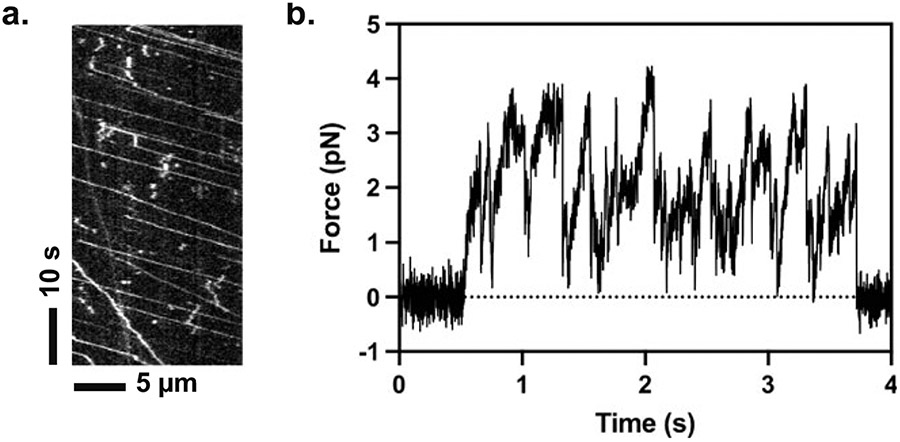
Analyze the velocity and processivity either using ImageJ or a custom-written software [61]. Figure 1a depicts an example kymograph of KIF1A molecules moving alone an MT.
Fig. 1.
(a) A kymograph example showing processive motion of KIF1A (Homo sapiens 1-393)-LZ. (b) An example optical trapping trace showing the force generation of a single KIF1A (Homo sapiens 1-393)-LZ molecule
3.7. Optical Tweezers Assay
Optical tweezers has been applied to study kinesin motors for more than 30 years [71-73]. It is a powerful tool to measure the biophysical properties of cytoskeleton motor proteins [57-59, 74-76]. Here, we describe how to set up and perform an optical tweezers assay to investigate KIF1A force generation. GFP-tagged KIF1A motors are bound to 500-nm diameter polystyrene beads that are coated with anti-GFP antibody and BSA. An individual bead is then trapped by the focused laser trapping beam and placed on top of an MT immobilized on the coverslip. Movement and force generation will be observed if a KIF1A motor is bound to the bead surface. Using this assay, the motor’s force-dependent properties can be determined, such as maximal force generation, load-dependent velocity and dwell time, and step sizes.
3.7.1. Coating of Polystyrene Microspheres
This section describes how to covalently coat carboxyl polystyrene beads with an antibody of choice (see Note 14) and BSA via EDC/sulfo-NHS coupling. If unspecified, the centrifugation steps are performed at 4 °C. The beads are resuspended in solution by briefly vortexing and sonicating the solution after each centrifugation step.
Bead preparation: In a 1.5-mL tube, add 200 μL AB to 75 μL of 2.5% w/v 0.5 μm carboxyl beads and centrifuge at 10,000 rcf for 4 min. Carefully remove the supernatant and then resuspend the beads in 200 μL AB. Repeat the wash step twice, each time with 200 μL AB. After the last wash, resuspend the beads in 200 μL AB.
Surface activation: Weight 2 mg EDAC and 4 mg sulfo-NHS and transfer to a 1.5-mL tube. Add the bead solution to the tube, vortex to mix well. Rotate the tube at room temperature for 30 min. Centrifuge the tube at 10,000 rcf for 4 min, remove the supernatant, and then resuspend the beads in 200 μL CB. Wash the beads three more times with 200 μL CB. After the last wash, resuspend the beads in 100 μL CB.
Bead coating: Mix 50 μL of 2 mg/mL anti-GFP antibody (see Note 15) with 4 μL of 50 mg/mL BSA. Add the protein solution to the beads and rotate at room temperature for 1 h. Add an additional 3 μL of BSA to the solution to further block the bead surfaces and rotate at room temperature for another 3 h. Leave the beads in the fridge overnight. On the second day, centrifuge the tube to remove the supernatant. Resuspend the coated beads in wash buffer (WB) and wash the beads with 200 μL WB four times. After the last wash, resuspend the beads in 400 μL SB with 1 mg/mL BSA, which results in a final bead concentration is 0.5% (w/v). Store the beads at 4 °C (do not freeze).
3.7.2. Labeling of α-Casein with Sulfo-NHS-Biotin for MT Immobilization
This step is to generate non-specifically labeled biotinylated α-casein for immobilizing MTs on the cover glass surface via streptavidin (see Note 16).
Mix 20 μL of 100 mM sulfo-NHS-biotin in DMSO with 200 μL of 25 mg/mL α-casein and nutate at room temperature for 1 h to label α-casein with biotin.
Move the solution to an Amicon ultra-0.5 mL 10 kDa centrifugal filter unit and fill the tube up to 500 μL with HME30G10 with 1 mM DTT.
Spin the tube at 14,000 rcf for 5 min at 4 °C. Repeat the wash step five times. The solution has a volume of ~200 μL after the last wash step.
Aliquot, flash freeze and store at −20 °C.
3.7.3. Coverslip Cleaning and Slide Assembly
See Subheadings 3.6.1 and 3.6.2.
3.7.4. MT Preparation
See Subheading 3.6.3.
3.7.5. Slide Preparation
To immobilize MTs on the coverslips, we use a biotin-casein-strep-tavidin-biotin-MT linkage. The surface is passivated by α-casein and Pluronic F-127.
Dilute 0.25 μL of biotin-α-casein in 9.75 μL of HME30G10, flow the diluted solution into an assembled flow chamber and incubate at room temperature for 10 min in a humidity chamber (see Note 17).
Wash the chamber with 3 × 20 μL BB, then incubate the slide in the humidity chamber for 10 min to passivate the surface.
Completely remove the solution from the chamber using vacuum. Dilute 2.5 μL of 1 mg/mL streptavidin in 7.5 μL of BRB80G10, flow the streptavidin solution into the chamber, and incubate for 5 min.
Completely remove the solution from the chamber using vacuum. Add 0.5 μL of 0.2 mg/mL MT to 20 μL of BB and flow the diluted MT solution into the chamber. Immediately wash the chamber with 2 × 20 μL BB to cause the MTs to align with the flow direction.
Incubate 0.4 μL of antibody-coated beads with 1 μL of appropriately diluted motor stock in MB on ice for 10 min.
Add 0.8 μL of 100 mM ATP, 0.8 μL of 50 mM biotin, and 0.8 μL gloxy to 36.2 μL MB, then mix well. Add the mixture to the bead-protein mix and mix well.
Flow 2 × 20 μL of the mixture into the flow chamber and seal the chamber using vacuum grease.
3.7.6. Optical Tweezers Assay
Mount the slide chamber onto the optical tweezers microscope.
Find a straight MT using the fluorescence signal of the MTs or interference reflection microscopy (IRM) [77-79].
Place a bead on top of the center of the MT and wait for an appropriate time to determine if movement and force generation can be observed (see Note 18); dilute the motor concentration until less than 30% of the trapped beads display movement.
Record the data and further analyze the data using the software of your choice (see Note 19). Figure 1b depicts an example trace of KIF1A’s force generation.
4. Notes
SNAPf is a kinetically faster version of SNAP-tag [80]. To achieve fast labeling, the SNAP-tag in pSNAP-tag® (T7)-2 was swapped with SNAPf using the pSNAPf vector as template. The C-terminus EGFP-6His was then added behind the SNAPf-tag. EGFP is used for the coupling to the trapping beads, while 6His serves as the purification tag.
Any pair of commercially available restriction enzymes of choice could be used, however, be sure to check if the two enzymes are compatible. For example, if both enzymes are provided by NEB, use the NEB double digest finder (http://nebcloner.neb.com/#!/redigest) to check if the two enzymes can be used simultaneously, and if so, which is the optimal buffer for double digestion.
If one obtains a high percentage of false positive colonies, it is an indication that the rSAP is losing its activity.
Similar to the double digestion, be sure to check the correct heat deactivation temperature for both enzymes. Some enzymes may require higher temperatures than 65 °C.
The reaction mixture can be directly loaded to the agarose gel without the cleaning step. However, keep in mind that the DNA band tends to run slower due to the high salt concentration, which may cause the molecular weight to appear not to align with the corresponding band of the DNA ladder.
Usually, a 1:3 ratio of vector:DNA works well for most applications. However, if difficulties are encountered, one can vary the ratio to determine more optimal conditions.
If no colony appears, the ligation is likely not efficient. If this is the case, either increase the concentration of T4 DNA ligase, supplement fresh ATP, or increase the incubation time to 1 h or overnight at room temperature.
Commercially available E. coli competent cells usually have a high competent efficiency. The tube with the E. coli cells can therefore be thawed, divided into 10 μL aliquots, and frozen again for future use. Here, we use NEB® Turbo competent E. coli (High Efficiency) for its high transformation efficiency and fast growth rate.
If one decides to perform the purification on the same day, it is still recommended to go through the freeze-thaw cycle, which aids the lysis of the cells.
As the SNAP-tag ligand tends to be sticky, it is important to thoroughly wash the resin to remove the residual ligands.
It is important to remove the salt when performing an MT-binding and -release assay with kinesin motors. The high salt concentration in the elution buffer from the last kinesin purification step will prevent kinesin-MT binding.
The amount of MTs added depends on the protein concentration. If the protein concentration is low, add less MTs.
A humidity chamber can be created by using an empty pipette tip box. Fill the bottom with ddH2O, and keep the lid closed.
Here, we use an anti-GFP antibody that was generated for us by YenZym Antibodies. For this, we express the recombinant enhanced GFP mutant 3 in E. coli bacteria and send the purified protein to YenZym Antibodies, who produces the rabbit anti-GFP serum. We have purified the anti-GFP antibodies from the rabbit antiserum and verified their functionality in previous optical trapping studies [53, 56-59, 81, 82]. However, any antibody could be used if it has a sufficiently high specificity and affinity, such as anti-StrepII-tag antibodies. Alternatively, the trapping beads could be covalently coated with streptavidin for the binding to biotinylated proteins.
The volume can vary as long as the total amount of antibody is 100 μg. The antibody can be in PBS or HEPES buffer as long as the buffer pH is above 7 and the buffer doesn’t have primary amine-containing components such as Tris.
One can use commercially available biotin-BSA as a substitute, however, we found that commercial biotin-BSA sometimes causes the beads to stick to the cover glass surface.
One should empirically determine the minimum concentration of biotin-α-casein that allows firm attachment of the MTs.
The waiting time correlates with the diameter of the trapped bead. For small beads, such as 500 nm-diameter beads, force generation is typically observed within 10 s if the bead is bound to a motor; for larger beads (e.g., 1 μm), it can take significantly more time to observe force generation events.
We use a custom-written MATLAB program to visualize and analyze optical trapping data. However, other software packages can be used as well, such as IDL or Python. Python is an open-source language, which is widely used in scientific research.
Acknowledgments
L. Rao and A. Gennerich are supported by National Institutes of Health grants R01GM098469 and R01NS114636.
References
- 1.Miki H, Setou M, Kaneshiro K, Hirokawa N (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci U S A 98:7004–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N (1995) The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81:769–780 [DOI] [PubMed] [Google Scholar]
- 3.Carabalona A, Hu DJ, Vallee RB (2016) KIF1A inhibition immortalizes brain stem cells but blocks BDNF-mediated neuronal migration. Nat Neurosci 19:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsai J-W, Lian W-N, Kemal S, Kriegstein AR, Vallee RB (2010) Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat Neurosci 13:1463–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkus RV, Klyachko O, Horiuchi D, Dickson BJ, Saxton WM (2008) Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Mol Biol Cell 19:274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall DH, Hedgecock EM (1991) Kinesin-related gene unc-104 is required for axonal transport of synaptic vesicles in C. elegans. Cell 65:837–847 [DOI] [PubMed] [Google Scholar]
- 7.Lo KY, Kuzmin A, Unger SM, Petersen JD, Silverman MA (2011) KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett 491:168–173 [DOI] [PubMed] [Google Scholar]
- 8.Yonekawa Y et al. (1998) Defect in synaptic vesicle precursor transport and neuronal cell death in KIF1A motor protein-deficient mice. J Cell Biol 141:431–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zahn TR et al. (2004) Dense core vesicle dynamics in Caenorhabditis elegans neurons and the role of kinesin UNC-104. Traffic 5:544–559 [DOI] [PubMed] [Google Scholar]
- 10.Boyle L et al. (2021) Genotype and defects in microtubule-based motility correlate with clinical severity in KIF1A associated neurological disorder. Hum Genet Genomics Adv 2:100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicita F et al. (2021) Heterozygous KIF1A variants underlie a wide spectrum of neurodevelopmental and neurodegenerative disorders. J Med Genet 58:475. [DOI] [PubMed] [Google Scholar]
- 12.Gabrych DR, Lau VZ, Niwa S, Silverman MA (2019) Going too far is the same as falling short(dagger): kinesin-3 family members in hereditary spastic paraplegia. Front Cell Neurosci 13:419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y et al. (2020) A rare KIF1A missense mutation enhances synaptic function and increases seizure activity. Front Genet 11:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John A, Ng-Cordell E, Hanna N, Brkic D, Baker K (2021) The neurodevelopmental spectrum of synaptic vesicle cycling disorders. J Neurochem 157:208. [DOI] [PubMed] [Google Scholar]
- 15.Van Beusichem AE et al. (2020) Mobility characteristics of children with spastic paraplegia due to a mutation in the KIF1A gene. Neuropediatrics 51:146–153 [DOI] [PubMed] [Google Scholar]
- 16.Riviere JB et al. (2011) KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet 89:219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erlich Y et al. (2011) Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res 21:658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamdan FF et al. (2011) Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 88:306–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klebe S et al. (2012) KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet 20:645–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okamoto N et al. (2014) KIF1A mutation in a patient with progressive neurodegeneration. J Hum Genet 59:639–641 [DOI] [PubMed] [Google Scholar]
- 21.Jamuar SS, Walsh CA (2014) Somatic mutations in cerebral cortical malformations. N Engl J Med 371:2038. [DOI] [PubMed] [Google Scholar]
- 22.Lee JR et al. (2015) De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum Mutat 36:69–78 [DOI] [PubMed] [Google Scholar]
- 23.Esmaeeli Nieh S et al. (2015) De novo mutations in KIF1A cause progressive encephalopathy and brain atrophy. Ann Clin Transl Neurol 2:623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ylikallio E et al. (2015) Dominant transmission of de novo KIF1A motor domain variant underlying pure spastic paraplegia. Eur J Hum Genet 23:1427–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Citterio A et al. (2015) Variants in KIF1A gene in dominant and sporadic forms of hereditary spastic paraparesis. J Neurol 262:2684–2690 [DOI] [PubMed] [Google Scholar]
- 26.Ohba C et al. (2015) De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J Hum Genet 60:739–742 [DOI] [PubMed] [Google Scholar]
- 27.Megahed H et al. (2016) Utility of whole exome sequencing for the early diagnosis of pediatric-onset cerebellar atrophy associated with developmental delay in an inbred population. Orphanet J Rare Dis 11:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hotchkiss L et al. (2016) Novel de novo mutations in KIF1A as a cause of hereditary spastic paraplegia with progressive central nervous system involvement. J Child Neurol 31:1114–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iqbal Z et al. (2017) Targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. PLoS One 12:e0174667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasegawa A et al. (2017) Co-existence of spastic paraplegia-30 with novel KIF1A mutation and spinocerebellar ataxia 31 with intronic expansion of BEAN and TK2 in a family. J Neurol Sci 372:128–130 [DOI] [PubMed] [Google Scholar]
- 31.Krenn M et al. (2017) Hereditary spastic paraplegia caused by compound heterozygous mutations outside the motor domain of the KIF1A gene. Eur J Neurol 24:741–747 [DOI] [PubMed] [Google Scholar]
- 32.Cheon CK et al. (2017) Autosomal dominant transmission of complicated hereditary spastic paraplegia due to a dominant negative mutation of KIF1A, SPG30 gene. Sci Rep 7:12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roda RH, Schindler AB, Blackstone C (2017) Multigeneration family with dominant SPG30 hereditary spastic paraplegia. Ann Clin Transl Neurol 4:821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Travaglini L et al. (2018) The impact of next-generation sequencing on the diagnosis of pediatric-onset hereditary spastic paraplegias: new genotype-phenotype correlations for rare HSP-related genes. Neurogenetics 19:111–121 [DOI] [PubMed] [Google Scholar]
- 35.Demily C et al. (2018) Additive effect of variably penetrant 22q11.2 duplication and pathogenic mutations in autism Spectrum disorder: to which extent does the tree hide the forest? J Autism Dev Disord 48:2886–2889 [DOI] [PubMed] [Google Scholar]
- 36.Dong F, Costigan DC, Howitt BE (2019) Targeted next-generation sequencing in the detection of mismatch repair deficiency in endometrial cancers. Mod Pathol 32:252–257 [DOI] [PubMed] [Google Scholar]
- 37.Samanta D, Gokden M (2019) PEHO syndrome: KIF1A mutation and decreased activity of mitochondrial respiratory chain complex. J Clin Neurosci 61:298–301 [DOI] [PubMed] [Google Scholar]
- 38.Tomaselli PJ et al. (2017) A de novo dominant mutation in KIF1A associated with axonal neuropathy, spasticity and autism spectrum disorder. J Peripher Nerv Syst 22:460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshikawa K et al. (2019) The novel de novo mutation of KIF1A gene as the cause for spastic paraplegia 30 in a Japanese case. eNeurologicalSci 14:34–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volk A, Conboy E, Wical B, Patterson M, Kirmani S (2015) Whole-exome sequencing in the clinic: lessons from six consecutive cases from the clinician’s perspective. Mol Syndromol 6:23–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun M et al. (2019) Targeted exome analysis identifies the genetic basis of disease in over 50% of patients with a wide range of ataxia-related phenotypes. Genet Med 21:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muir AM et al. (2019) Genetic heterogeneity in infantile spasms. Epilepsy Res 156:106181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kashimada A et al. (2019) Genetic analysis of undiagnosed ataxia-telangiectasia-like disorders. Brain and Development 41:150–157 [DOI] [PubMed] [Google Scholar]
- 44.Pennings M et al. (2020) KIF1A variants are a frequent cause of autosomal dominant hereditary spastic paraplegia. Eur J Hum Genet 28:40–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiaojing W et al. (2020) Generation of a human induced pluripotent stem cell line (SDUBMSi001-A) from a hereditary spastic paraplegia patient carrying kif1a c.773C>T missense mutation. Stem Cell Res 43:101727. [DOI] [PubMed] [Google Scholar]
- 46.van de Warrenburg BP et al. (2016) Clinical exome sequencing for cerebellar ataxia and spastic paraplegia uncovers novel gene-disease associations and unanticipated rare disorders. Eur J Hum Genet 24:1460–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.L Raffa et al. (2017) Optic nerve hypoplasia in a patient with a de novo KIF1A heterozygous mutation. Can J Ophthalmol 52:e169–e171 [DOI] [PubMed] [Google Scholar]
- 48.Hosokawa S, Kubo Y, Arakawa R, Takashima H, Saito K (2020) Analysis of spinal muscular atrophy-like patients by targeted resequencing. Brain and Development 42:148–156 [DOI] [PubMed] [Google Scholar]
- 49.Spagnoli C, Rizzi S, Salerno GG, Frattini D, Fusco C (2019) Long-term follow-up until early adulthood in autosomal dominant, complex SPG30 with a novel KIF1A variant: a case report. Ital J Pediatr 45:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurihara M et al. (2020) A novel de novo KIF1A mutation in a patient with autism, hyperactivity, epilepsy, sensory disturbance, and spastic paraplegia. Intern Med 59:839–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nemani T et al. (2020) KIF1A-related disorders in children: a wide spectrum of central and peripheral nervous system involvement. J Peripher Nerv Syst 25:117–124 [DOI] [PubMed] [Google Scholar]
- 52.Langlois S et al. (2016) De novo dominant variants affecting the motor domain of KIF1A are a cause of PEHO syndrome. Eur J Hum Genet 24:949–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Budaitis BG et al. (2021) Pathogenic mutations in the kinesin-3 motor KIF1A diminish force generation and movement through allosteric mechanisms. J Cell Biol 220:e202004227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guedes-Dias P et al. (2019) Kinesin-3 responds to local microtubule dynamics to target synaptic cargo delivery to the presynapse. Curr Biol 29:268–282 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiba K et al. (2019) Disease-associated mutations hyperactivate KIF1A motility and anterograde axonal transport of synaptic vesicle precursors. Proc Natl Acad Sci U S A 116:18429–18434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lam AJ et al. (2021) A highly conserved 310 helix within the kinesin motor domain is critical for kinesin function and human health. Sci Adv 7:eabf1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X, Rao L, Gennerich A (2020) The regulatory function of the AAA4 ATPase domain of cytoplasmic dynein. Nat Commun 11:5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brenner S, Berger F, Rao L, Nicholas MP, Gennerich A (2020) Force production of human cytoplasmic dynein is limited by its processivity. Sci Adv 6:eaaz4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao L, Berger F, Nicholas MP, Gennerich A (2019) Molecular mechanism of cytoplasmic dynein tension sensing. Nat Commun 10:3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rao L, Hulsemann M, Gennerich A (2018) Combining structure-function and single-molecule studies on cytoplasmic dynein. Methods Mol Biol 1665:53–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rao L et al. (2013) The yeast dynein Dyn2-Pac11 complex is a dynein dimerization/processivity factor: structural and single-molecule characterization. Mol Biol Cell 24:2362–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reck-Peterson SL, Vale RD, Gennerich A (2012) Motile properties of cytoplasmic dynein. In: KHAL Amos (ed) Handbook of dynein. Pan Stanford Publishing, Singapore, pp 145–172 [Google Scholar]
- 63.Gennerich A, Reck-Peterson SL (2011) Probing the force generation and stepping behavior of cytoplasmic dynein. Methods Mol Biol 783:63–80 [DOI] [PubMed] [Google Scholar]
- 64.Belyy V, Hendel NL, Chien A, Yildiz A (2014) Cytoplasmic dynein transports cargos via load-sharing between the heads. Nat Commun 5:5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reck-Peterson SL et al. (2006) Single-molecule analysis of dynein processivity and stepping behavior. Cell 126:335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yildiz A, Tomishige M, Vale RD, Selvin PR (2004) Kinesin walks hand-over-hand. Science 303:676–678 [DOI] [PubMed] [Google Scholar]
- 67.Yildiz A et al. (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300:2061–2065 [DOI] [PubMed] [Google Scholar]
- 68.Htet ZM et al. (2020) LIS1 promotes the formation of activated cytoplasmic dynein-1 complexes. Nat Cell Biol 22:518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Woehlke G et al. (1997) Microtubule interaction site of the kinesin motor. Cell 90:207–216 [DOI] [PubMed] [Google Scholar]
- 70.Li M, Zheng W (2011) Probing the structural and energetic basis of kinesin-microtubule binding using computational alanine-scanning mutagenesis. Biochemistry 50:8645–8655 [DOI] [PubMed] [Google Scholar]
- 71.Block SM, Goldstein LS, Schnapp BJ (1990) Bead movement by single kinesin molecules studied with optical tweezers. Nature 348:348–352 [DOI] [PubMed] [Google Scholar]
- 72.Kuo SC, Sheetz MP (1993) Force of single kinesin molecules measured with optical tweezers. Science 260:232–234 [DOI] [PubMed] [Google Scholar]
- 73.Svoboda K, Schmidt CF, Schnapp BJ, Block SM (1993) Direct observation of kinesin stepping by optical trapping interferometry. Nature 365:721–727 [DOI] [PubMed] [Google Scholar]
- 74.Dunn AR, Chuan P, Bryant Z, Spudich JA (2010) Contribution of the myosin VI tail domain to processive stepping and intramolecular tension sensing. Proc Natl Acad Sci U S A 107:7746–7750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rief M et al. (2000) Myosin-V stepping kinetics: a molecular model for processivity. Proc Natl Acad Sci U S A 97:9482–9486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Carter NJ, Cross RA (2005) Mechanics of the kinesin step. Nature 435:308–312 [DOI] [PubMed] [Google Scholar]
- 77.Hirst WG, Kiefer C, Abdosamadi MK, Schaffer E, Reber S (2020) In vitro reconstitution and imaging of microtubule dynamics by fluorescence and label-free microscopy. STAR Protoc 1:100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahamdeh M, Simmert S, Luchniak A, Schaffer E, Howard J (2018) Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy. J Microsc 272:60–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Simmert S, Abdosamadi MK, Hermsdorf G, Schaffer E (2018) LED-based interference-reflection microscopy combined with optical tweezers for quantitative three-dimensional microtubule imaging. Opt Express 26:14499–14513 [DOI] [PubMed] [Google Scholar]
- 80.Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K (2006) Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng Des Sel 19:309–316 [DOI] [PubMed] [Google Scholar]
- 81.Nicholas MP et al. (2015) Control of cytoplasmic dynein force production and processivity by its C-terminal domain. Nat Commun 6:6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nicholas MP et al. (2015) Cytoplasmic dynein regulates its attachment to microtubules via nucleotide state-switched mechanosensing at multiple AAA domains. Proc Natl Acad Sci U S A 112:6371–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]