Abstract
Study Design:
Narrative review.
Objectives:
This review aims to present current applications of machine learning (ML) in spine domain to clinicians.
Methods:
We conducted a comprehensive PubMed search of peer-reviewed articles that were published between 2006 and 2020 using terms (spine, spinal, lumbar, cervical, thoracic, machine learning) to examine ML in spine. Then exclude research of other domain, case report, review or meta-analysis, and which without available abstract or full text.
Results:
Total 1738 articles were retrieved from database, and 292 studies were finally included. Key findings of current applications were compiled and summarized in this review. Main clinical applications of those techniques including image processing, diagnosis, decision supporting, operative assistance, rehabilitation, surgery outcomes, complications, hospitalization and cost.
Conclusions:
ML had achieved excellent performance and hold immense potential in spine. ML could help clinical staff to improve medical level, enhance work efficiency, and reduce adverse events. However more randomized controlled trials and improvement of interpretability are essential to clinicians accepting models’ assistance in real work.
Keywords: machine learning, artificial intelligence, deep learning, spine, current applications
Introduction
Due to popularity of soft computing approaches, artificial intelligence (AI) had made great impact on every aspect of daily life. AI techniques are revolutionizing medical domain by performing complex and huge computational tasks. Nowadays AI had made major progress in healthcare administration, clinical decision support, patient monitoring and healthcare interventions.1,2 As the most promising branch of AI, machine learning (ML) automatically predict outputs based on features of inputs through algorithms.3,4 ML have natural advantage of handling big data comparing to traditional statistical methods. They have more superior accuracy and repeatability than conventional models and even expert operators. ML could provide subtle information, which cannot detected by eye in desired image tasks. 5 In the era of big data, ML will dramatically improve diagnostic accuracy and prognosis.3,6
Publications about applications of ML in spine significant increased recently. However, the development of those techniques for spine are still in infancy. Before clinicians adopt ML in the practical work, preclinical steps are raising their attention, establishing elementary cognition and getting involved in research. We aim to introduce current applications of ML in spine to clinicians. Then identify opportunities and utilization potentiality of future research.
Methods
We conducted a comprehensive PubMed search of peer-reviewed articles that were published between 2006 and 2020 using terms (spine, spinal, lumbar, cervical, thoracic, machine learning) to examine ML in spine. Then exclude research of other domain, case report, review or meta-analysis, and which without available abstract or full text.
Results
Total 1738 articles were retrieved from database, and 292 studies were finally included. Count of articles significantly increased in past 3 years, that indicates growing interest of researchers. Key findings of current applications were compiled and summarized in this review.
Machine Learning
In 1959, Arthur Samuel defined “machine learning” as giving computers the ability to learn without being explicitly programmed. 7 Simply put, ML is that algorithms get the ability of making decisions or predictions through learning data. Models present an analysis and generate desired outputs basing on inputs features. Main ML tasks contain classification, regression, clustering, and dimensionality reduction. ML process comprised by data collection, data preprocessing, feature engineering, model selection, training, model evaluation and optimization. Data collection is an important step, as quality and quantity of data directly affect outcome. Secondly, data preprocessing is to make data usable for computation, improve accuracy, and shorten calculation process. Feature engineering is creating features from raw data using domain knowledge. That largely determines the final algorithms performance, which can be broadly classified as feature extraction, feature construction and feature selection. Next, dataset is divided into training set, cross validation set, and testing set. Models are built and trained on training sets. Then models are scored in cross validation set for selecting better one. And various evaluation methods should be brought to assess models. For optimization, the most basic method is gradient descent algorithm, which minimize loss function through iterating.
ML was classified into supervised learning, unsupervised learning and reinforcement learning according to different forms. Supervised learning is that a learner describes the input-output relationship based on labeled input variables with a grounded truth. 8 A model analyzed the training data to synthesize the pattern between independent variables and dependent variables. Then the testing dataset is to be predicted. In contrast, the unsupervised learner describes relationship of input-output basing on unlabeled inputs. 9 Algorithm analyses input data features to identify clusters of data. 10 Reinforcement learning is that the learner constantly interacts with environment to find the best strategy through trial and error for maximizing rewards. 11
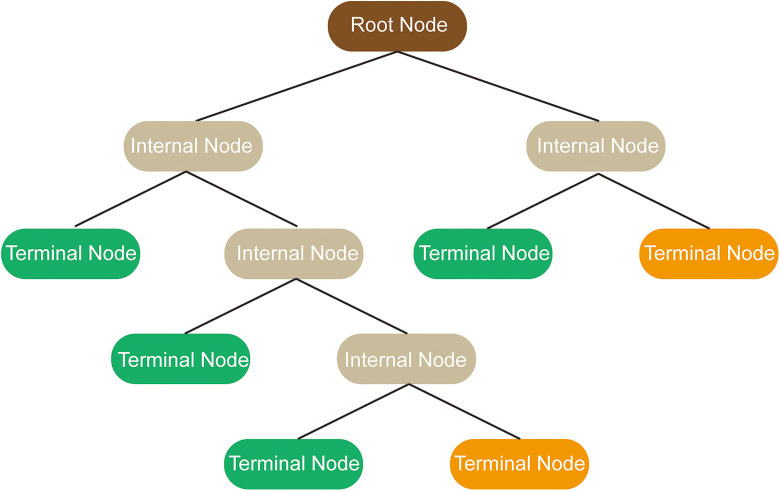
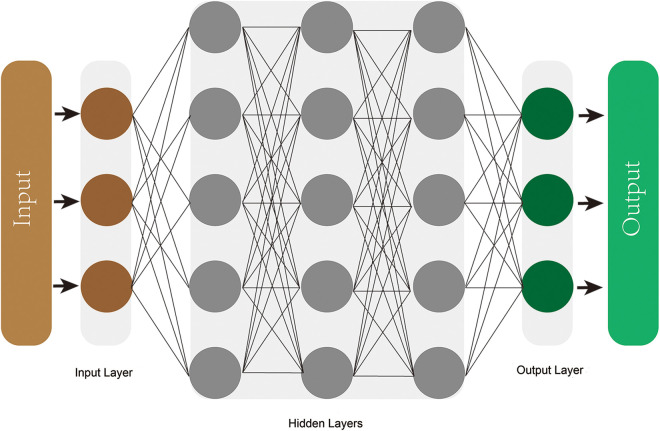
Three common ML models are decision tree learning, support vector machines (SVMs), and artificial neural networks (ANNs). Classification and regression decision tree (CART) implements a classification or a regression task, which is more visible and easier to understand than other modalities. The tree comprises internal nodes (conditions), branches (decisions), and leaves (ends), that is not computationally intensive and therefore suitable for big data5,12 (Figure 1). SVMs accomplishes classification tasks by creating a maximum margin hyperplane between 2 outcomes, or regression tasks by plotting a best-fit plane 13 (Figure 2). ANNs is a deep machine learner inspired by how neurons connected and interacted in brain. 10 Constructing in Hebbian learning, single neurons comprises an entire network, where information flows from the inputs layers to the outputs layers through multiple hidden layers, operating by weights 5 (Figure 3).
Figure 1.
Schematic representation of decision tree learning algorithm.
Figure 2.
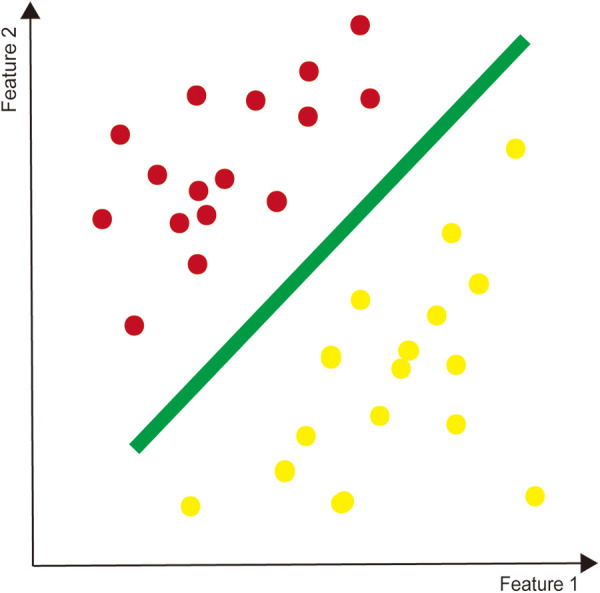
The simple support vector machine as a binary classification.
Figure 3.
The deep artificial neural network comprised by input layer, output layer, and multiple hidden layers.
Underfitting and overfitting are 2 classical problems in ML. If a model is too simple to fully learn features of training data, it will present low accuracy both on the training set and test set, which is called underfitting. Overfitting is that a model performs well on training set, but poor on test data, since adjusting too closely to former. The model memorizes specific training observations, but cannot actually extract generalizable relationships between variables. 14 Common solutions of overfitting are increasing sample, regularization, and dropout technique.
Image Processing and Diagnosis
Image processing
Intention of developing ML is to reduce manual labor and save time for tasks needed judgment.15,16 Localization, segmentation, grading, calculating parameters of image are time-consuming works, which should be accurately performed by algorithm. Several models accurately detected and quantitative measurement, comparable with human-generated segmentations. Rak et al proposed a novel approach to automatically segment vertebrae in three-dimensional MRI. 17 That CNN combined with a graph cut formulation based on encoding swaps, which segments multiple vertebrae without risking ambiguous segmentations of adjacent vertebrae. 17 FU-Net combined traditional region-based level set with CNN to accurate segment vertebrae, and outperformed other techniques. 18 Overall, deep learning pipelines for segmenting vertebrae and intervertebral discs (IVDs) on MRI provided high accuracy and eliminated laborious manual labeling.19,20
Paugam et al used supervised neural networks to perform single-class or multi-class segmenting the grey and white matter of spinal cord on MRI, which showed good results on small amounts of data. 21 In addition, convolutional neural networks (CNNs) model showed state-of-the-art performance for automated lesion segmentation after spinal cord injury (SCI). 22 Volumes extracted from lesion segmentation were significantly associated with patient motor scores, that will potentially help to advance modernized SC MR image analysis for both research and clinical application. 22 Likewise, ML algorithms were trained to segment lumbar spinal canal areas on axial views of lumbar MRI. 23 And deep multiscale multitask learning network (DMML-Net) achieved highly accurate localization and grading of neural foramina, surrounding vertebra and discs for the pathogenesis-based diagnosis of lumbar neural foraminal stenosis. 24
Besides segmentation, models were also developed for automatically identifying organ and lesion. Jimenez-Pastor et al introduced a 2-stage decision forests and morphological image processing technique to automatically detect and identify vertebral bodies from arbitrary field-of-view body CT scans. 25 Vertebrae automatic detection network achieved satisfying accuracy and precision on CT or unannotated MRI.26,27 An automated framework could detect myelopathic areas, combining diffusion tensor imaging (DTI) metrics with SVM. 28 Another random forest (RF) model 29 had potential benefit of diagnosing and locating the level of cervical injury in CM, using time-frequency components of somatosensory evoked potentials.
Algorithms had automatic measured parameters of spine, comparable to manual measurements. There were fully automatic algorithms to evaluate of lumbar lordosis and Cobb angle for scoliosis.30,31 A CNN model was able to determine the spine shape and calculate posture parameters in biplanar radiographs, which could act on intervention in deformities or degeneration. 32 Another novel cascade amplifier regression network robustly auto-mated quantitative measure spine on T1-weighted MR. 33
Diagnosis
In the outpatient primary care setting, a general practitioner must determine to refer a patient to spinal subspecialist for consultation when discover specific radiographic findings, however large proportion of referrals ultimately fail to meet criteria for surgical intervention.34,35
ML could provide auxiliary information to optimize the referral process and save time of patients and surgeons. 34 Deep learning had been reported to successfully apply for diagnosis of liver masses, parkinsonian disorders, hip fractures and estimating bone age.36-40 Combining multiple data, ML can provide accurate diagnostic predictions and risk warnings. With increasing applications of CNNs to radiological imaging, AI was expected to gradually change clinical practice. 41
ML obtained fine diagnostic accuracy in disease prediction and differentiation, mainly running through radiological imaging. A deep neural network (DNN) had predicted cervical myelopathy (CM) based on MRI. 42 CNNs had well differentiated spinal schwannomas and meningiomas as well as tuberculous and pyogenic spondylitis using MRI.43,44 Likewise, ML methods had been developed for preoperative differentiation of cancer based on 3D CT, enhanced CT, and enhanced MRI.15,45-47
Similarly, clinical and radiographic data-driven ML models showed excellent prediction of radiographic progression in axial spondyloarthritis (axSpA).48,49
ML models could obtain features that cannot be detected by naked eye. Texture features of IVDs and endplate zones are different between people with and without low back pain (LBP), that may not be consistently discovered by clinicians. 50 RF and linear mixed-effect model analysis identified texture features from lumbar MRI to assist surgeons recognizing people with LBP. 50 Intermittent claudication may cause by multiple diseases, like lumber spinal canal stenosis (LSS) and peripheral arterial disease. Texture analysis with ML offered highly reproducible quantitative parameters that increase accuracy for severe LSS detection. A decision tree classifier revealed higher performances for LSS grading compared to qualitative assessments using the reference cut-off cross-sectional area (CSA). 51
ML also added extra information from other disease examination or routine screening. These features may overlook by specialist who just focus on their domain. Additional health assessment will enhance healthcare, save payment and decrease examinational radiation. Several tools tended to be excellent approaches for distinguishing osteoporosis, such as metabolites, low-frequency guided waves, ocal classification of CT textures, segments of DXA images, and spine radiographs.52-61 Currently a few deep learning models predicted high-risk osteoporosis populations using spine X-ray. 62 The best method employed VGGnet for feature extraction and RF training. 62 Yasaka et al 63 operated a CNNs with lumbar vertebrae CT to output BMD, which significantly correlated with DXA’s outcome and perform better than vertebrae CT values. Models could be applied to predict osteoporosis through abdominal CT or chest CT that widely used in daily clinical practice.63,64 Likewise, SVM classifiers could use dual-energy X-ray absorptiometry (DEXA) studies to identify lumbar spine fractures routinely. 65 Jamaludin et al developed an automatic ML model to accurately identify and quantify scoliosis from total body DXA. 66
Besides intuitive radiomics, ML combining various auxiliary examination can provide accurate diagnosis. LBP has physiological relationships with abnormal muscle activation. 67 Clinically interpretable models produced good to excellent predictive capability to LBP using functional kinematic and electromyography (EMG) variables with FDboost. 67 Another SVM analyzed two-dimensional walking motion, and classified the underlying disease of the intermittent claudication with 79.7% accuracy. 68 Also SVM had applied to distinguish chronic lumbar radiculopathy on electroencephalography (EEG). 69 In addition, ML may offer the initial suspected diagnosis based on a simple test which is more available in regions with limited medical resources. 70 Objective functional impairment (OFI) could provide hints for the suspected cause of back and leg pain, for which Staartjes et al carried out five-repetition sit-to-stand test combining with a ML algorithm. 71 Tan et al 72 developed and validated a natural language processing (NLP) system to identify 26 findings related to LBP from x-ray and MR radiology reports. Furthermore, deep learning algorithms were devised for automatic scoliosis screening and classifications, using unclothed back images. 73
Treatment
ML models had been put into treatment decision-making process, such as surgery level, instrumented vertebra and correction range. SpineNet automatically generated MRI grading to predict the surgical level of single-level decompression. 74 It calculated an aggregate score of grading for the following: central canal stenosis, disc narrowing, disc degeneration, spondylolisthesis, upper/lower endplate morphologic changes, and upper/lower marrow changes. For adult spinal deformity correction, an ANN successfully mimicked lead surgeons’ decision making in selection of the upper instrumented vertebra. 75 In another literature, Shen et al 76 presented a new classifier to assist surgeons optimizing surgical plan and therapeutic management. The method classified adolescent idiopathic scoliosis (AIS) using a fuzzy clustering algorithm and 3D parameters describing characteristics of deformity. 76 More recently, a decision tree identified optimal range of spontaneous lumbar Cobb correction in Lenke 1 AIS. 77 Also, a semi-automatic method had been built to classify scoliosis severity and treatment group with 3D markerless surface topography scans. 78 And approaches were proposed to predict curve shape variation, curve types and progression in AIS.79-81
Recently scholars built models to automatically provide measurements of implants, placement trajectories, pedicle screw planning and surgical navigation.82-84 An automated pedicle detector provided robust quasi-automated pedicle localization by calculating vertebral axial rotation values in frontal radiographs of scoliosis with minimal user intervention. 85 That was useful to vertebral rotation estimation and 3D spine reconstructions. Another intraoperative 3D pedicle screws navigation system’s accuracy was 86.1%. 83 von Atzigen et al proposed a purely ML marker-less surgical navigation to bending rod implants. 86 This method required significantly less time than the marker-based benchmark navigation approach needed to contact with anatomy and achieved better or comparable accuracy. 86 Other pullout strength models had predicted combination of density, insertion depth, and insertion angle for the chosen range, for understanding pedicle screw pullout and pre-surgical planning.87,88 Besides, deep learning-based framework could automatically adjust the C-arm pose to a desired standard projection from the first X-ray, and localize needle target for epidural needle placement in ultrasound images.89,90
AI combining with virtual reality simulation provide safer training and objective assessment of surgical skills, leading to improved patient care. ANNs will gain insight into the importance of virtual reality surgical simulators for surgical training. Mirchi et al performed ANNs to distinguish safety metrics in a virtual reality-simulated anterior cervical discectomy scenario. 91 An ML tool assessed surgical expertise in a virtual reality spine procedure, and potentially apply to ensuring surgeons’ technical competency in future. 92 Additionally, identifying patterns of surgical practice will be an important step to understand surgical processes. Researchers built a framework to automatically identify practical patterns for discriminating different experience surgeon from surgery recordings. 93
In rehabilitation of SCI patients, ML framework could support researchers and clinicians for selection of epidural stimulation parameters. 94 During electrically evoked contractions, SVM increased safety by adapting the functional electrical stimulation parameters in motor complete SCI individuals. 95 Another three-steps ML model provided monitor of tenodesis grasp based on egocentric video at home, that implied remote cSCI therapeutic guidance. 96
Prognosis
Clinical outcomes
Preoperative prediction for clinical outcomes could enhance informed patient consent, reduce drug consumption, promote recovery and personalize shared decision-making.97,98 ML algorithms indicated that lower preoperative PROMIS scores, fewer comorbidities, and certain sociodemographic factors increased the likelihood of achieving minimal clinically important difference (MCID), which helps surgeons to determine the appropriateness and timing of surgery. 99 Algorithms had been explored for analyzing of multiple data(patient demographics, clinical presentation, DTI maps) to determine the prognosis in CM.100, 101 A few models were designed for predicting outcome like, VAS, ODI, mJOA, and invasiveness score based on preoperative factors in lumbar disc herniation (LDH) or LBP patients. 14 , 102-105 Similarly, ML meaningful predicted survival outcomes of spinopelvic chondrosarcoma, ependymoma, malignant peripheral nerve sheath tumor and spinal metastases patients.106-110 In spinal metastatic disease, SORG algorithms had been externally validated for survival prediction.111, 112
Complications
Adverse events (AEs) following spine surgery negatively impact patients, surgeons, and the health care system. Therefore, it is critical to build predictive models for investigating factors associating with AEs and develop risk stratification strategies. 113 Researchers presented a set of predictive models for postoperative AEs. They accounted for patient-, diagnosis-, and procedure-related factors which could contribute to timely intervene, patient-counseling, accurate risk adjustment, and quality metrics. Algorithms accurately predicted the risk of proximal junctional kyphosis and spinopelvic compensation for long level fusion surgery. 114 115 ANN and LR models achieved better performance than the ASA classification for predicting complications of ACDF. 116 ML were more accurate than ASA scores for complications risk factor analysis. 117 118 Also, Natural language processing (NLP) algorithms automatically detected incidental durotomies, intraoperative vascular injury, and postoperative wound infection in lumbar surgery.119-121 As a result, ML algorithms could provide prediction of AEs, improve risk stratification and help guide the surgical decision-making process in spine surgery. 122
Hospitalization and Cost
Hospital readmission and prolonged length of stay (LOS) will bear a great burden to the healthcare system. ML improved understanding of predicting readmission after spinal surgery.123, 124 Risk features includes returning to operating room, septic shock and superficial surgical site infection. 123 A RF model suggested demographic features may contribute more readmission risk than perioperative variables. 125 Similarly, models recommended that non-electively admitting and staying in ICU need additional attention to avoid unanticipated prolonged LOS following spine surgery 126
Likewise, ML was employed to predict the medical costs of spinal fusion, and inform hospital strategy to increase the financial management efficiency.127, 128 Agglomerative hierarchical clustering was applied to identity factors associated with higher 2-year post-surgical costs, containing greater utilization of antidepressants, opioids, and behavioral health services. 129
Discussion
Although ML models has got fine outcome currently, there is still more effort need to make for ML development in the practical spine work. Several limits hinder the application of ML. First, single kind data and small size sample cannot well represent complex disease. Then traditional models may have similarly or even better predictive ability than deep learning in the low order of magnitude data. But deep learning has superiority in processing big data and figuring out complex relationships between variables. 130 More advanced algorithms should be leaded in this field. Finally, we discuss the outlook and challenge of ML.
1. Data
The clinical dataset is typical multimodal heterogeneous data including, clinical indicators, images, genetic data and biomarkers. Doctors need to cautiously offer a proposal in view of individual difference and various comorbidity. However, studies mainly based on one kind data like pharmaceutical, clinical or imaging data.74, 102, 131 For instance, predication from pharmaceutical data couldn’t achieve clinical causes of risk factors without complete clinical, symptomatic and imaging data. 131 In another article for postoperative endpoints, predictive tool did not consider features such as quality of life and objective functional impairment, anxiety and depression, severity of stenosis, comorbidities, or neurological deficits. 97 In a word, based on single kind of data models was hard to be fully convinced by decision makers, and provide substantial assistance. It is essential that combining multimodal heterogeneous data to improve value of outcome. In the future, deep learning will establish a connection of radiomics, proteomics, and metabolomics. 132 Another common issue is small size of sample, which is expediently obtained to medical staff. With many variables, department level data may lead to overfitting and poor performance. Multicenter data is crucial to ML performance. Also, single center analysis needs external validation from multiple institutions to guard against institutional biases. 133
2. Transfer learning:
In medical domain, lacking huge training data is a prominent problem. Main cause are expensively professional preprocessing tasks and privacy protection. Fortunately, we can build a model using annotated data that similar with target data, and then employ transfer learning methods. Transfer learning is that applying models learned from old field, in a new field through similarities of data, tasks, or models. 134 The meaningful approach can utilize existing large image datasets to perform image analysis and obtain even faster convergence and higher accuracy than training from scratch.135, 136 This process identifies and integrates one or more types of domain knowledge relating to the designated task for improving deep learning models performance. 137 Recently, transfer learning was employed in deep learning tasks, as pathology image analysis and COVID-19 screening.138, 139
Unsupervised domain adaptation (UDA) is a type of transfer learning with restrictive conditions. It is necessary that source domain and target domain have same label space, feature space, and conditional probability distribution. With labels only from the source domain, UDA could promote neural networks performance on target domain. That especially suits for medical field, as quite labor-intensive annotating and commonly lack of annotated datasets. 137 The effectiveness of UDA had been proved in chest X-ray segmentation. 140
In addition, treating issues as many single tasks (diagnosis or prognosis) will ignore the correlation information of tasks. Multi-task learning (MTL), another domain of transfer learning, is a suitable solution to this problem. The approach is learning multiple related tasks together and sharing information, they learned. Main goal of MTL is to improve generalization ability by using specific domain’s information in multiple related tasks training. 141 Extra information can enhance performance of the current task, such as generalization accuracy, learning speed, and comprehensibility. Pan et al presented a multi-task disease progression model based on hierarchical attention mechanism, that focus on different features and medical records. 142 In the future, it is significant that a comprehensive deep learning model simultaneously work out spine diagnosis and prognosis tasks.
For privacy protection, the federated transfer learning (FTL) is a method that different participants first respectively train models on their own data. Then encrypt models and data for avoiding disclosure of participants’ privacy. On this basis, these models are joint trained to get the final optimal model, which will be returned to each participant. 143 The FTL framework could improve original ML algorithms, that has been verified on public datasets, such as NUS-WIDE and Default-Credit datasets. 144 Also, privacy-preserving FTL had been proposed to extract common discriminative information from multi-subject EEG data for EEG classification. 145
3. Disease progression models:
Spinal degenerative diseases, main component of spinal diseases, are typical chronic diseases. Models were necessary to gather data at different time points from time series database, obtain pattern of disease progression and make accurate prediction, in order to timely intervention and avoiding progression or deterioration of diseases. Disease progression models (DPMs) is employed to characterize the course of disease progression from longitudinal health records, for early detection and precision care at the appropriate time. 146 DPMs had been applied in progression of Alzheimer’s disease (AD), and breast cancer lung metastasis.147-149 For instance, Hidden Markov models (HMMs) could infer discrete latent states and transitions between inferred states from time-varying multivariate data. 146 Kwon et al used HMMs and its variants to discover disease states and make inferences of health states for chronic patients. 146 For doctors understanding patient status and progression patterns, they developed DPVis incorporating HMMs to seamlessly integrates HMMs’ parameters and outcomes into interpretable and interactive visualizations. 146 Oxtoby et al used 2 generative data-driven DPMs to extract patterns of observable biomarker changes in dominantly-inherited AD. 149 Their models reveal probabilistic sequences of biomarker abnormality and estimates of biomarker trajectories through a non-parametric differential equation model from a cross-section of short-term longitudinal data.
4. Incremental learning:
AI has a main criticism of catastrophic forgetting, that is when moving from 1 task to another, models perform well on the new task, but underfit the old one. It is due to original data memory is overwritten by new one. We hope models get ability of gradually sustained accumulating knowledge like human. Incremental learning is constantly learning knowledge from new data and preserving most of previous knowledge. 150 The learning could avoid historical data occupying storage space. On the other hand, it makes full use of previous results, and thus significantly saves time for new training. This learning is mainly applied for big data, especial suitable to medical field. Common methods of incremental learning including feature extraction, fine-tuning, joint training, and knowledge distillation. 151 Recently incremental learning has been widely employed in domains, such as muscle activity and kinematics, nonconvulsive epileptic seizure, atrial fibrillation, and segmentation anatomical structures.152-155
5. Outlook and challenge:
In the past 2 decades, ML field had been driven by huge development of computing power. Currently, the photonic quantum computer, Jiuzhang, is faster than using the state-of-the-art simulation strategy and supercomputers by a factor of ∼1014 on boson sampling task. 156 Although, it cannot be used for other calculation, and still far from practical application at present. Someday in the future, quantum computer will produce computing forces beyond classical computers, process big data with less time and energy dissipation, and extend the scope of ML.
Although achievement of universal health coverage (UHC) was policy priority and ultimate goal for both countries and global institutions, estimated by current projections about 3.1 billion people will still lacking UHC effective coverage in 2023. 157 Several determinants hindered UHC progress, like wealth inequality, race, gender, caste discrimination, air pollution, lack of water and sanitation facilities. 158 There are a large number of disadvantaged groups hard to achieve good-quality healthcare and afford the corresponding cost. In the COVID-19 pandemic era, the global economy is entering a major crisis. COVID-19 pandemic decreases limited medical resources and capacity of government financial support. Meanwhile, home quarantine leads to tremendous sicken people cannot getting treatment for common disease. Patients and accompanying members pay more time and expenditure on virus screening in questing outpatient service. In this special period, illness, losing job and financial hardship are making matters worse especially for impoverished people. That lead to poor members even harder getting good-quality healthcare than at ordinary times. ML may be powerful approaches to enhance poor peoples’ healthcare, save government medical financial support, and promote the UHC progress. A study introduced clinical decision support system (CDSS) support self-referral for patients with LBP and further referral by healthcare professionals, that has the potential to decrease the current long waiting lines in healthcare. 159 Popularization of ML will be a solution for poor people achieving daily healthcare and confronting major health event. If they could use an app to get a preliminary screening of disease, they will get the correct registration of outpatient and even the initial diagnosis at first time. These mobile applications could save time, avoid wrong registration, decrease risk of delaying disease, and relieve pressure of limited medical resources. Indeed, they could be totally free to poor members, which will make significant impact on UHC. With lowest cost, disadvantaged groups could achieve medical service approaching expert level. In someday, patients could achieve meaningful advice from ML, including what disease may have, where to get medical service, and which suitable expert to perform the operation. These models may remove disequilibrium of medical resources. They could provide contribution to rational allocation of government medical expenditures, reducing the risk of dissemination disease, saving insurance expenses of countries and companies.
As improvement of external performance and interpretability, CNNs are anticipated to help radiologists maximize the value of extracted image, achieve diagnostic excellence, enhance interrater reliability, and improve workflow for more timely recommendations. 41 ML could also assist surgeons to decide preoperative patient-specific planning and implant, for enhancing patient healthcare. 160 The computer assisted navigation systems will be an essential tool to reduce adverse event, save operative time, and surmount the technical barrier. ML tools could bring the state-of-the-art technology to the remote region. An ideal single platform should affords central control over functions, like data pre-processing, data governance, regulatory requirements, and operational interaction with existing electronic health record systems. 16 ML has the capacity to primarily generate structured data from raw electronic health records, thus it could have a strong impact on hospital analytics, epidemiological studies, and systematic reviews. 161
Although AI had tremendous development in healthcare, only few applications had been actually implemented evaluated at the frontlines of clinical practice. 162 The rigorous and comprehensive evaluation of clinical AI needs to be improved by more robust randomized controlled trials in the future. 162 There were several challenges need to overcome in the further practical application of ML. Black box is that, most AI technologies operate largely by an opaque logic, which are hardly understandable to users. This ethical challenge limits marketing approval of AI in medicine. For instance, who is responsible for misdiagnosis among doctors, system and AI manufacturers. 163 The local interpretable model-agnostic explanations is a mean to improve the interpretability of model. 164 Another important hurdle is that the lack of standardized regulations related to the application of AI in medicine. 165 Besides, reward hacking means that machines find ways to achieve outcomes that circumvent rules or cheat the system. 166
Conclusions
ML had achieved excellent performance and hold immense potential in spine. ML could help clinical staff to improve medical level, enhance work efficiency, and reduce adverse events. However more randomized controlled trials and improvement of interpretability are essential to clinicians accepting models’ assistance in real work.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: GuanRui Ren  https://orcid.org/0000-0003-2438-8081
https://orcid.org/0000-0003-2438-8081
References
- 1.Reddy S, Fox J, Purohit MP. Artificial intelligence-enabled healthcare delivery. J Royal Soc Med. 2019;112(1):22–28. doi:10.1177/0141076818815510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gambhir S, Malik SK, Kumar Y. Role of soft computing approaches in healthcare domain: a mini review. J Med Syst. 2016;40(12):287. doi:10.1007/s10916-016-0651-x [DOI] [PubMed] [Google Scholar]
- 3.Jordan MI, Mitchell TM. Machine learning: trends, perspectives, and prospects. Science (New York, NY). 2015;349(6245):255–260. doi:10.1126/science.aaa8415 [DOI] [PubMed] [Google Scholar]
- 4.Chang M, Canseco JA, Nicholson KJ, Patel N, Vaccaro AR. The role of machine learning in spine surgery: the future is now. Front Surg. 2020;7:54. doi:10.3389/fsurg.2020.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galbusera F, Casaroli G, Bassani T. Artificial intelligence and machine learning in spine research. JOR Spine. 2019;2(1):e1044. doi:10.1002/jsp2.1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Obermeyer Z, Emanuel EJ. Predicting the future—big data, machine learning, and clinical medicine. N Eng J Med. 2016;375(13):1216–1219. doi:10.1056/NEJMp1606181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuel AL. Some studies in machine learning using the game of checkers. IBM J Res Dev. 1959;3(3):211–229. [Google Scholar]
- 8.Panchmatia JR, Visenio MR, Panch T. The role of artificial intelligence in orthopaedic surgery. Br J Hosp Med (London, England: 2005). 2018;79(12):676–681. doi:10.12968/hmed.2018.79.12.676 [DOI] [PubMed] [Google Scholar]
- 9.Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med. 2019;25(1):44–56. doi:10.1038/s41591-018-0300-7 [DOI] [PubMed] [Google Scholar]
- 10.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi:10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 11.Mnih V, Kavukcuoglu K, Silver D, et al. Human-level control through deep reinforcement learning. Nature. 2015;518(7540):529–533. doi:10.1038/nature14236 [DOI] [PubMed] [Google Scholar]
- 12.Krzywinski M, Altman N. Classification and regression trees. Nat Methods. 2017;14(8):757–758. doi:10.1038/nmeth.4370 [Google Scholar]
- 13.Noble WS. What is a support vector machine? Nat Biotechnol. 2006;24(12):1565–1567. doi:10.1038/nbt1206-1565 [DOI] [PubMed] [Google Scholar]
- 14.Staartjes VE, de Wispelaere MP, Vandertop WP, Schröder ML. Deep learning-based preoperative predictive analytics for patient-reported outcomes following lumbar discectomy: feasibility of center-specific modeling. Spine J. 2019;19(5):853–861. doi:10.1016/j.spinee.2018.11.009 [DOI] [PubMed] [Google Scholar]
- 15.Yin P, Mao N, Zhao C, et al. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol. 2019;29(4):1841–1847. doi:10.1007/s00330-018-5730-6 [DOI] [PubMed] [Google Scholar]
- 16.Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20(5):e262–e273. doi:10.1016/s1470-2045(19)30149-4 [DOI] [PubMed] [Google Scholar]
- 17.Rak M, Steffen J, Meyer A, Hansen C, Tönnies KD. Combining convolutional neural networks and star convex cuts for fast whole spine vertebra segmentation in MRI. Comput Methods Programs Biomed. 2019;177:47–56. doi:10.1016/j.cmpb.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 18.Rehman F, Ali Shah SI, Riaz MN, Gilani SO, Faiza R. A region-based deep level set formulation for vertebral bone segmentation of osteoporotic fractures. J Digit Imag. 2020;33(1):191–203. doi:10.1007/s10278-019-00216-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuang X, Cheung JP, Wu H, Dokos S, Zhang T. MRI-SegFlow: a novel unsupervised deep learning pipeline enabling accurate vertebral segmentation of MRI images. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual International Conference; July 20, 2020. Montréal, Québec, Canada; 2020;2020:1633-1636. doi:10.1109/embc44109.2020.9175987 [DOI] [PubMed] [Google Scholar]
- 20.Li X, Dou Q, Chen H, et al. 3D multi-scale FCN with random modality voxel dropout learning for intervertebral disc localization and segmentation from multi-modality MR images. Med Image Anal. 2018;45:41–54. doi:10.1016/j.media.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 21.Paugam F, Lefeuvre J, Perone CS, et al. Open-source pipeline for multi-class segmentation of the spinal cord with deep learning. Magn Reson Imaging. 2019;64:21–27. doi:10.1016/j.mri.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCoy DB, Dupont SM, Gros C, et al. Convolutional neural network-based automated segmentation of the spinal cord and contusion injury: deep learning biomarker correlates of motor impairment in acute spinal cord injury. Am J Neuroradiol. 2019;40(4):737–744. doi:10.3174/ajnr.A6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaonkar B, Villaroman D, Beckett J, et al. Quantitative analysis of spinal canal areas in the lumbar spine: an imaging informatics and machine learning study. AJNR Am J Neuroradiol. 2019;40(9):1586–1591. doi:10.3174/ajnr.A6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han Z, Wei B, Leung S, Nachum IB, Laidley D, Li S. Automated pathogenesis-based diagnosis of lumbar neural foraminal stenosis via deep multiscale multitask learning. Neuroinformatics. 2018;16(3-4):325–337. doi:10.1007/s12021-018-9365-1 [DOI] [PubMed] [Google Scholar]
- 25.Jimenez-Pastor A, Alberich-Bayarri A, Fos-Guarinos B, et al. Automated vertebrae localization and identification by decision forests and image-based refinement on real-world CT data. La Radiologia Medica. 2020;125(1):48–56. doi:10.1007/s11547-019-01079-9 [DOI] [PubMed] [Google Scholar]
- 26.Zhou Y, Liu Y, Chen Q, Gu G, Sui X. Automatic lumbar MRI detection and identification based on deep learning. J Digit Imag. 2019;32(3):513–520. doi:10.1007/s10278-018-0130-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao H, Mesfin A, Luo J. Joint vertebrae identification and localization in spinal CT images by combining short- and long-range contextual information. IEEE Trans Med Imag. 2018;37(5):1266–1275. doi:10.1109/tmi.2018.2798293 [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Hu Y, Shen Y, Li H. Classification of diffusion tensor metrics for the diagnosis of a myelopathic cord using machine learning. Int J Neural Syst. 2018;28(2):1750036. doi:10.1142/s0129065717500368 [DOI] [PubMed] [Google Scholar]
- 29.Cui H, Wang Y, Li G, Huang Y, Hu Y. Exploration of cervical myelopathy location from somatosensory evoked potentials using random forests classification. IEEE Trans Neural Syst Rehabil Eng. 2019;27(11):2254–2262. doi:10.1109/tnsre.2019.2945634 [DOI] [PubMed] [Google Scholar]
- 30.Cho BH, Kaji D, Cheung ZB, et al. Automated measurement of lumbar lordosis on radiographs using machine learning and computer vision. Global Spine J. 2020;10(5):611–618. doi:10.1177/2192568219868190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Xu Q, Leung S, Chung J, Chen B, Li S. Accurate automated Cobb angles estimation using multi-view extrapolation net. Med Image Anal. 2019;58:101542. doi:10.1016/j.media.2019.101542 [DOI] [PubMed] [Google Scholar]
- 32.Galbusera F, Niemeyer F, Wilke HJ, et al. Fully automated radiological analysis of spinal disorders and deformities: a deep learning approach. Eur Spine J. 2019;28(5):951–960. doi:10.1007/s00586-019-05944-z [DOI] [PubMed] [Google Scholar]
- 33.Pang S, Su Z, Leung S, et al. Direct automated quantitative measurement of spine by cascade amplifier regression network with manifold regularization. Med Image Anal. 2019;55:103–115. doi:10.1016/j.media.2019.04.012 [DOI] [PubMed] [Google Scholar]
- 34.Wilson B, Gaonkar B, Yoo B, et al. Predicting spinal surgery candidacy from imaging data using machine learning. Neurosurgery. 2021;89(1):116–121. doi:10.1093/neuros/nyab085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deis N, Findlay JM. Appropriateness of lumbar spine referrals to a neurosurgical service. Can J Neurol Sci. 2010;37(6):843–848. doi:10.1017/s0317167100051544 [DOI] [PubMed] [Google Scholar]
- 36.Urakawa T, Tanaka Y, Goto S, Matsuzawa H, Watanabe K, Endo N. Detecting intertrochanteric hip fractures with orthopedist-level accuracy using a deep convolutional neural network. Skeletal Radiol. 2019;48(2):239–244. doi:10.1007/s00256-018-3016-3 [DOI] [PubMed] [Google Scholar]
- 37.Kiryu S, Yasaka K, Akai H, et al. Deep learning to differentiate parkinsonian disorders separately using single midsagittal MR imaging: a proof of concept study. Eur Radiol. 2019;29(12):6891–6899. doi:10.1007/s00330-019-06327-0 [DOI] [PubMed] [Google Scholar]
- 38.Derkatch S, Kirby C, Kimelman D, Jozani MJ, Davidson JM, Leslie WD. Identification of vertebral fractures by convolutional neural networks to predict nonvertebral and hip fractures: a registry-based cohort study of dual X-ray absorptiometry. Radiology. 2019;293(2):405–411. doi:10.1148/radiol.2019190201 [DOI] [PubMed] [Google Scholar]
- 39.Yasaka K, Akai H, Abe O, Kiryu S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology. 2018;286(3):887–896. doi:10.1148/radiol.2017170706 [DOI] [PubMed] [Google Scholar]
- 40.Larson DB, Chen MC, Lungren MP, Halabi SS, Stence NV, Langlotz CP. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2018;287(1):313–322. doi:10.1148/radiol.2017170236 [DOI] [PubMed] [Google Scholar]
- 41.Yasaka K, Abe O. Deep learning and artificial intelligence in radiology: current applications and future directions. PLoS Med. 2018;15(11):e1002707. doi:10.1371/journal.pmed.1002707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hopkins BS, Weber KA, II, Kesavabhotla K, Paliwal M, Cantrell DR, Smith ZA. Machine learning for the prediction of cervical spondylotic myelopathy: a post hoc pilot study of 28 participants. World Neurosurg. 2019;127:e436–e442. doi:10.1016/j.wneu.2019.03.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maki S, Furuya T, Horikoshi T, et al. A deep convolutional neural network with performance comparable to radiologists for differentiating between spinal schwannoma and meningioma. Spine. 2020;45(10):694–700. doi:10.1097/brs.0000000000003353 [DOI] [PubMed] [Google Scholar]
- 44.Kim K, Kim S, Lee YH, Lee SH, Lee HS, Kim S. Performance of the deep convolutional neural network based magnetic resonance image scoring algorithm for differentiating between tuberculous and pyogenic spondylitis. Sci Rep. 2018;8(1):13124. doi:10.1038/s41598-018-31486-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin P, Mao N, Zhao C, Wu J, Chen L, Hong N. A triple-classification radiomics model for the differentiation of primary chordoma, giant cell tumor, and metastatic tumor of sacrum based on T2-weighted and contrast-enhanced T1-weighted MRI. J Magn Reson Imag. 2019;49(3):752–759. doi:10.1002/jmri.26238 [DOI] [PubMed] [Google Scholar]
- 46.Lang N, Zhang Y, Zhang E, et al. Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn Reson Imag. 2019;64:4–12. doi:10.1016/j.mri.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mehta SD, Sebro R. Random forest classifiers aid in the detection of incidental osteoblastic osseous metastases in DEXA studies. Int J Comput Assist Radiol Surg. 2019;14(5):903–909. doi:10.1007/s11548-019-01933-1 [DOI] [PubMed] [Google Scholar]
- 48.Joo YB, Baek IW, Park YJ, Park KS, Kim KJ. Machine learning-based prediction of radiographic progression in patients with axial spondyloarthritis. Clin Rheumatol. 2020;39(4):983–991. doi:10.1007/s10067-019-04803-y [DOI] [PubMed] [Google Scholar]
- 49.Joo YB, Baek IW, Park KS, Tagkopoulos I, Kim KJ. Novel classification of axial spondyloarthritis to predict radiographic progression using machine learning. Clin Exp Rheumatol. 2020;39(3):508–518. [PubMed] [Google Scholar]
- 50.Abdollah V, Parent EC, Dolatabadi S, et al. Texture analysis in the classification of T(2) weighted magnetic resonance images in persons with and without low back pain. J Orthop Res. 2020. doi:10.1002/jor.24930 [DOI] [PubMed] [Google Scholar]
- 51.Huber FA, Stutz S, Vittoria de Martini I, et al. Qualitative versus quantitative lumbar spinal stenosis grading by machine learning supported texture analysis-experience from the LSOS study cohort. Eur J Radiol. 2019;114:45–50. doi:10.1016/j.ejrad.2019.02.023 [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Yan D, Zhao A, et al. Discovery of potential biomarkers for osteoporosis using LC-MS/MS metabolomic methods. Osteoporos Int. 2019;30(7):1491–1499. doi:10.1007/s00198-019-04892-0 [DOI] [PubMed] [Google Scholar]
- 53.Vogl F, Friesenbichler B, Hüsken L, Kramers-de Quervain IA, Taylor WR. Can low-frequency guided waves at the tibia paired with machine learning differentiate between healthy and osteopenic/osteoporotic subjects? A pilot study. Ultrasonics. 2019;94:109–116. doi:10.1016/j.ultras.2018.11.012 [DOI] [PubMed] [Google Scholar]
- 54.Valentinitsch A, Trebeschi S, Kaesmacher J, et al. Opportunistic osteoporosis screening in multi-detector CT images via local classification of textures. Osteoporos Int. 2019;30(6):1275–1285. doi:10.1007/s00198-019-04910-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nam KH, Seo I, Kim DH, Lee JI, Choi BK, Han IH. Machine learning model to predict osteoporotic spine with Hounsfield units on lumbar computed tomography. J Korean Neurosurg Soc. 2019;62(4):442–449. doi:10.3340/jkns.2018.0178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krishnaraj A, Barrett S, Bregman-Amitai O, et al. Simulating dual-energy X-ray absorptiometry in CT using deep-learning segmentation cascade. J Am Coll Radiol. 2019;16(10):1473–1479. doi:10.1016/j.jacr.2019.02.033 [DOI] [PubMed] [Google Scholar]
- 57.Hussain D, Han SM. Computer-aided osteoporosis detection from DXA imaging. Comput Methods Programs Biomed. 2019;173:87–107. doi:10.1016/j.cmpb.2019.03.011 [DOI] [PubMed] [Google Scholar]
- 58.Dimai HP, Ljuhar R, Ljuhar D, et al. Assessing the effects of long-term osteoporosis treatment by using conventional spine radiographs: results from a pilot study in a sub-cohort of a large randomized controlled trial. Skeletal Radiol. 2019;48(7):1023–1032. doi:10.1007/s00256-018-3118-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kavitha MS, An SY, An CH, et al. Texture analysis of mandibular cortical bone on digital dental panoramic radiographs for the diagnosis of osteoporosis in Korean women. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015;119(3):346–356. doi:10.1016/j.oooo.2014.11.009 [DOI] [PubMed] [Google Scholar]
- 60.Yoo TK, Kim SK, Kim DW, et al. Osteoporosis risk prediction for bone mineral density assessment of postmenopausal women using machine learning. Yonsei Med J. 2013;54(6):1321–1330. doi:10.3349/ymj.2013.54.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kavitha MS, Asano A, Taguchi A, Kurita T, Sanada M. Diagnosis of osteoporosis from dental panoramic radiographs using the support vector machine method in a computer-aided system. BMC Med Imaging. 2012;12:1. doi:10.1186/1471-2342-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee S, Choe EK, Kang HY, Yoon JW, Kim HS. The exploration of feature extraction and machine learning for predicting bone density from simple spine X-ray images in a Korean population. Skeletal Radiol. 2020;49(4):613–618. doi:10.1007/s00256-019-03342-6 [DOI] [PubMed] [Google Scholar]
- 63.Yasaka K, Akai H, Kunimatsu A, Kiryu S, Abe O. Prediction of bone mineral density from computed tomography: application of deep learning with a convolutional neural network. Eur Radiol. 2020;30(6):3549–3557. doi:10.1007/s00330-020-06677-0 [DOI] [PubMed] [Google Scholar]
- 64.Pan Y, Shi D, Wang H, et al. Automatic opportunistic osteoporosis screening using low-dose chest computed tomography scans obtained for lung cancer screening. Eur Radiol. 2020;30(7):4107–4116. doi:10.1007/s00330-020-06679-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehta SD, Sebro R. Computer-aided detection of incidental lumbar spine fractures from routine Dual-Energy X-Ray Absorptiometry (DEXA) studies using a support vector machine (SVM) classifier. J Digit Imaging. 2020;33(1):204–210. doi:10.1007/s10278-019-00224-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jamaludin A, Fairbank J, Harding I, et al. Identifying scoliosis in population-based cohorts: automation of a validated method based on total body dual energy X-ray absorptiometry scans. Calcif Tissue Int. 2020;106(4):378–385. doi:10.1007/s00223-019-00651-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liew BXW, Rugamer D, De Nunzio AM, Falla D. Interpretable machine learning models for classifying low back pain status using functional physiological variables. Eur Spine J. 2020;29(8):1845–1859. doi:10.1007/s00586-020-06356-0 [DOI] [PubMed] [Google Scholar]
- 68.Watanabe T, Yoneyama T, Toribatake Y, Hayashi H. Main disease classification of intermittent claudication via L1-regularized SVM. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual International Conference; July 3, 2013. Japan: 2013;2013:6409-6412. doi:10.1109/embc.2013.6611021 [DOI] [PubMed] [Google Scholar]
- 69.Levitt J, Edhi MM, Thorpe RV, et al. Pain phenotypes classified by machine learning using electroencephalography features. NeuroImage. 2020;223:117256. doi:10.1016/j.neuroimage.2020.117256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Munakomi S. Letter to the editor. Reappraising role of clinical evaluations in degenerative lumbar spine pathologies. J Neurosurg Spine. 2019:1–2. doi:10.3171/2018.10.Spine181282 [DOI] [PubMed] [Google Scholar]
- 71.Staartjes VE, Quddusi A, Klukowska AM, Schröder ML. Initial classification of low back and leg pain based on objective functional testing: a pilot study of machine learning applied to diagnostics. Eur Spine J. 2020;29(7):1702–1708. doi:10.1007/s00586-020-06343-5 [DOI] [PubMed] [Google Scholar]
- 72.Tan WK, Hassanpour S, Heagerty PJ, et al. Comparison of natural language processing rules-based and machine-learning systems to identify lumbar spine imaging findings related to low back pain. Acad Radiol. 2018;25(11):1422–1432. doi:10.1016/j.acra.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang J, Zhang K, Fan H, et al. Development and validation of deep learning algorithms for scoliosis screening using back images. Commun Biol. 2019;2:390. doi:10.1038/s42003-019-0635-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roller BL, Boutin RD, O’Gara TJ, et al. Accurate prediction of lumbar microdecompression level with an automated MRI grading system. Skeletal Radiol. 2021;50(1):69–78. doi:10.1007/s00256-020-03505-w [DOI] [PubMed] [Google Scholar]
- 75.Lafage R, Ang B, Alshabab BS, et al. Predictive model for selection of upper treated vertebra using a machine learning approach. World Neurosurg. 2020;146:e225–e232. doi:10.1016/j.wneu.2020.10.073 [DOI] [PubMed] [Google Scholar]
- 76.Shen J, Parent S, Wu J, et al. Towards a new 3D classification for adolescent idiopathic scoliosis. Spine Deform. 2020;8(3):387–396. doi:10.1007/s43390-020-00051-2 [DOI] [PubMed] [Google Scholar]
- 77.Pasha S, Mac-Thiong JM. Defining criteria for optimal lumbar curve correction following the selective thoracic fusion surgery in Lenke 1 adolescent idiopathic scoliosis: developing a decision tree. Eur J Orthop Surg Traumatol. 2020;30(3):513–522. doi:10.1007/s00590-019-02596-z [DOI] [PubMed] [Google Scholar]
- 78.Rothstock S, Weiss HR, Krueger D, Paul L. Clinical classification of scoliosis patients using machine learning and markerless 3D surface trunk data. Med Biol Eng Comput. 2020;58(12):2953–2962. doi:10.1007/s11517-020-02258-x [DOI] [PubMed] [Google Scholar]
- 79.García-Cano E, Arámbula Cosío F, Duong L, et al. Prediction of spinal curve progression in adolescent idiopathic scoliosis using random forest regression. Comput Biol Med. 2018;103:34–43. doi:10.1016/j.compbiomed.2018.09.029 [DOI] [PubMed] [Google Scholar]
- 80.García-Cano E, Arámbula Cosío F, Duong L, et al. Dynamic ensemble selection of learner-descriptor classifiers to assess curve types in adolescent idiopathic scoliosis. Med Biol Eng Comput. 2018;56(12):2221–2231. doi:10.1007/s11517-018-1853-9 [DOI] [PubMed] [Google Scholar]
- 81.Kadoury S, Mandel W, Roy-Beaudry M, Nault ML, Parent S. 3-D morphology prediction of progressive spinal deformities from probabilistic modeling of discriminant manifolds. IEEE Trans Med Imag. 2017;36(5):1194–1204. doi:10.1109/tmi.2017.2657225 [DOI] [PubMed] [Google Scholar]
- 82.Siemionow K, Luciano C, Forsthoefel C, Aydogmus S. Autonomous image segmentation and identification of anatomical landmarks from lumbar spine intraoperative computed tomography scans using machine learning: a validation study. J Craniovertebr Junction Spine. 2020;11(2):99–103. doi:10.4103/jcvjs.JCVJS_37_20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Burström G, Buerger C, Hoppenbrouwers J, et al. Machine learning for automated 3-dimensional segmentation of the spine and suggested placement of pedicle screws based on intraoperative cone-beam computer tomography. J Neurosurg Spine. 2019;31(1):147–154. doi:10.3171/2018.12.Spine181397 [DOI] [PubMed] [Google Scholar]
- 84.Esfandiari H, Newell R, Anglin C, Street J, Hodgson AJ. A deep learning framework for segmentation and pose estimation of pedicle screw implants based on C-arm fluoroscopy. Int J Comput Assist Radiol Surg. 2018;13(8):1269–1282. doi:10.1007/s11548-018-1776-9 [DOI] [PubMed] [Google Scholar]
- 85.Ebrahimi S, Gajny L, Vergari C, Angelini ED, Skalli W. Vertebral rotation estimation from frontal X-rays using a quasi-automated pedicle detection method. Eur Spine J. 2019;28(12):3026–3034. doi:10.1007/s00586-019-06158-z [DOI] [PubMed] [Google Scholar]
- 86.von Atzigen M, Liebmann F, Hoch A, et al. HoloYolo: a proof-of-concept study for marker-less surgical navigation of spinal rod implants with augmented reality and on-device machine learning. Int J Med Robot. 2020;17(1):e2184. doi:10.1002/rcs.2184 [DOI] [PubMed] [Google Scholar]
- 87.Khatri R, Varghese V, Sharma S, Kumar GS, Chhabra HS. Pullout strength predictor: a machine learning approach. Asian Spine J. 2019;13(5):842–848. doi:10.31616/asj.2018.0243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Varghese V, Krishnan V, Kumar GS. Evaluating pedicle-screw instrumentation using decision-tree analysis based on pullout strength. Asian Spine J. 2018;12(4):611–621. doi:10.31616/asj.2018.12.4.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kausch L, Thomas S, Kunze H, et al. Toward automatic C-arm positioning for standard projections in orthopedic surgery. Int J Comput Assist Radiol Surg. 2020;15(7):1095–1105. doi:10.1007/s11548-020-02204-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pesteie M, Lessoway V, Abolmaesumi P, Rohling RN. Automatic localization of the needle target for ultrasound-guided epidural injections. IEEE Trans Med Imag. 2018;37(1):81–92. doi:10.1109/tmi.2017.2739110 [DOI] [PubMed] [Google Scholar]
- 91.Mirchi N, Bissonnette V, Ledwos N, et al. Artificial neural networks to assess virtual reality anterior cervical discectomy performance. Oper Neurosurg (Hagerstown, Md). 2020;19(1):65–75. doi:10.1093/ons/opz359 [DOI] [PubMed] [Google Scholar]
- 92.Bissonnette V, Mirchi N, Ledwos N, Alsidieri G, Winkler-Schwartz A, Del Maestro RF. Artificial intelligence distinguishes surgical training levels in a virtual reality spinal task. J Bone Joint Surg Am. 2019;101(23):e127. doi:10.2106/jbjs.18.01197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Forestier G, Petitjean F, Senin P, Riffaud L, Henaux PL, Jannin P. Finding discriminative and interpretable patterns in sequences of surgical activities. Artif Intell Med. 2017;82:11–19. doi:10.1016/j.artmed.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 94.Mesbah S, Gonnelli F, Angeli CA, El-Baz A, Harkema SJ, Rejc E. Neurophysiological markers predicting recovery of standing in humans with chronic motor complete spinal cord injury. Sci Rep. 2019;9(1):14474. doi:10.1038/s41598-019-50938-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naeem J, Hamzaid NA, Islam MA, Azman AW, Bijak M. Mechanomyography-based muscle fatigue detection during electrically elicited cycling in patients with spinal cord injury. Med Biol Eng Comput. 2019;57(6):1199–1211. doi:10.1007/s11517-019-01949-4 [DOI] [PubMed] [Google Scholar]
- 96.Dousty M, Zariffa J. Tenodesis grasp detection in egocentric video. IEEE J Biomed Health Inform. 2020;25(5):1463–1470. doi:10.1109/jbhi.2020.3003643 [DOI] [PubMed] [Google Scholar]
- 97.Siccoli A, de Wispelaere MP, Schröder ML, Staartjes VE. Machine learning-based preoperative predictive analytics for lumbar spinal stenosis. Neurosurg Focus. 2019;46(5):E5. doi:10.3171/2019.2.Focus18723 [DOI] [PubMed] [Google Scholar]
- 98.André A, Peyrou B, Carpentier A, Vignaux JJ. Feasibility and assessment of a machine learning-based predictive model of outcome after lumbar decompression surgery. Global Spine J. 2020:2192568220969373. doi:10.1177/2192568220969373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karhade AV, Fogel HA, Cha TD, et al. Development of prediction models for clinically meaningful improvement in PROMIS scores after lumbar decompression. Spine J. 2020;21(3);397–404. doi:10.1016/j.spinee.2020.10.026 [DOI] [PubMed] [Google Scholar]
- 100.Merali ZG, Witiw CD, Badhiwala JH, Wilson JR, Fehlings MG. Using a machine learning approach to predict outcome after surgery for degenerative cervical myelopathy. PLoS One. 2019;14(4):e0215133. doi:10.1371/journal.pone.0215133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jin R, Luk KD, Cheung JPY, Hu Y. Prognosis of cervical myelopathy based on diffusion tensor imaging with artificial intelligence methods. NMR Biomed. 2019;32(8):e4114. doi:10.1002/nbm.4114 [DOI] [PubMed] [Google Scholar]
- 102.Wirries A, Geiger F, Hammad A, Oberkircher L, Blümcke I, Jabari S. Artificial intelligence facilitates decision-making in the treatment of lumbar disc herniations. Eur Spine J. 2020. doi:10.1007/s00586-020-06613-2 [DOI] [PubMed] [Google Scholar]
- 103.De Silva T, Vedula SS, Perdomo-Pantoja A, et al. SpineCloud: image analytics for predictive modeling of spine surgery outcomes. J Med Imaging (Bellingham, Wash). 2020;7(3):031502. doi:10.1117/1.Jmi.7.3.031502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pedersen CF, Andersen M, Carreon LY, Eiskjær S. Applied machine learning for spine surgeons: predicting outcome for patients undergoing treatment for lumbar disc herniation using PRO data. Global Spine J. 2020. doi:10.1177/2192568220967643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Campagner A, Berjano P, Lamartina C, Langella F, Lombardi G, Cabitza F. Assessment and prediction of spine surgery invasiveness with machine learning techniques. Comput Biol Med. 2020;121:103796. doi:10.1016/j.compbiomed.2020.103796 [DOI] [PubMed] [Google Scholar]
- 106.Wei D, Nistal DA, Sobotka S, Martini M, Hawks C, Jenkins AL. III. New predictive index for survival in symptomatic spinal metastases. World Neurosurg. 2019;123:e133–e140. doi:10.1016/j.wneu.2018.11.088 [DOI] [PubMed] [Google Scholar]
- 107.Ryu SM, Seo SW, Lee SH. Novel prognostication of patients with spinal and pelvic chondrosarcoma using deep survival neural networks. BMC Med Inform Decis Mak. 2020;20(1):3. doi:10.1186/s12911-019-1008-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yan P, Huang R, Hu P, et al. Nomograms for predicting the overall and cause-specific survival in patients with malignant peripheral nerve sheath tumor: a population-based study. J Neuro-oncol. 2019;143(3):495–503. doi:10.1007/s11060-019-03181-4 [DOI] [PubMed] [Google Scholar]
- 109.Ryu SM, Lee SH, Kim ES, Eoh W. Predicting survival of patients with spinal ependymoma using machine learning algorithms with the SEER database. World Neurosurg. 2018. doi:10.1016/j.wneu.2018.12.091 [DOI] [PubMed] [Google Scholar]
- 110.DiSilvestro KJ, Veeramani A, McDonald CL, et al. Predicting post-operative mortality following metastatic intraspinal neoplasm excision: development of a novel machine learning approach. World Neurosurg. 2020;146:e917–e924. doi:10.1016/j.wneu.2020.11.037 [DOI] [PubMed] [Google Scholar]
- 111.Karhade AV, Ahmed AK, Pennington Z, et al. External validation of the SORG 90-day and 1-year machine learning algorithms for survival in spinal metastatic disease. Spine J. 2020;20(1):14–21. doi:10.1016/j.spinee.2019.09.003 [DOI] [PubMed] [Google Scholar]
- 112.Bongers MER, Karhade AV, Villavieja J, et al. Does the SORG algorithm generalize to a contemporary cohort of patients with spinal metastases on external validation? Spine J. 2020;20(10):1646–1652. doi:10.1016/j.spinee.2020.05.003 [DOI] [PubMed] [Google Scholar]
- 113.Han SS, Azad TD, Suarez PA, Ratliff JK. A machine learning approach for predictive models of adverse events following spine surgery. Spine J. 2019;19(11):1772–1781. doi:10.1016/j.spinee.2019.06.018 [DOI] [PubMed] [Google Scholar]
- 114.Peng L, Lan L, Xiu P, et al. Prediction of proximal junctional kyphosis after posterior scoliosis surgery with machine learning in the Lenke 5 adolescent idiopathic scoliosis patient. Front Bioeng Biotechnol. 2020;8:559387. doi:10.3389/fbioe.2020.559387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee NJ, Sardar ZM, Boddapati V, et al. Can machine learning accurately predict postoperative compensation for the uninstrumented thoracic spine and pelvis after fusion from the lower thoracic spine to the sacrum? Global Spine J. 2020:2192568220956978. doi:10.1177/2192568220956978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Arvind V, Kim JS, Oermann EK, Kaji D, Cho SK. Predicting surgical complications in adult patients undergoing anterior cervical discectomy and fusion using machine learning. Neurospine. 2018;15(4):329–337. doi:10.14245/ns.1836248.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kim JS, Merrill RK, Arvind V, et al. Examining the ability of artificial neural networks machine learning models to accurately predict complications following posterior lumbar spine fusion. Spine. 2018;43(12):853–860. doi:10.1097/brs.0000000000002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim JS, Arvind V, Oermann EK, et al. Predicting surgical complications in patients undergoing elective adult spinal deformity procedures using machine learning. Spine Deform. 2018;6(6):762–770. doi:10.1016/j.jspd.2018.03.003 [DOI] [PubMed] [Google Scholar]
- 119.Karhade AV, Bongers MER, Groot OQ, et al. Natural language processing for automated detection of incidental durotomy. Spine J. 2020;20(5):695–700. doi:10.1016/j.spinee.2019.12.006 [DOI] [PubMed] [Google Scholar]
- 120.Karhade AV, Bongers MER, Groot OQ, et al. Development of machine learning and natural language processing algorithms for preoperative prediction and automated identification of intraoperative vascular injury in anterior lumbar spine surgery. Spine J. 2020. doi:10.1016/j.spinee.2020.04.001 [DOI] [PubMed] [Google Scholar]
- 121.Karhade AV, Bongers MER, Groot OQ, et al. Can natural language processing provide accurate, automated reporting of wound infection requiring reoperation after lumbar discectomy? Spine J. 2020;20(10):1602–1609. doi:10.1016/j.spinee.2020.02.021 [DOI] [PubMed] [Google Scholar]
- 122.Fatima N, Zheng H, Massaad E, Hadzipasic M, Shankar GM, Shin JH. Development and validation of machine learning algorithms for predicting adverse events after surgery for lumbar degenerative spondylolisthesis. World Neurosurg. 2020;140:627–641. doi:10.1016/j.wneu.2020.04.135 [DOI] [PubMed] [Google Scholar]
- 123.Hopkins BS, Yamaguchi JT, Garcia R, et al. Using machine learning to predict 30-day readmissions after posterior lumbar fusion: an NSQIP study involving 23,264 patients. J Neurosurg Spine. 2019:1–8. doi:10.3171/2019.9.Spine19860 [DOI] [PubMed] [Google Scholar]
- 124.Jain D, Durand W, Burch S, Daniels A, Berven S. Machine learning for predictive modeling of 90-day readmission, major medical complication, and discharge to a facility in patients undergoing long segment posterior lumbar spine fusion. Spine. 2020;45(16):1151–1160. doi:10.1097/brs.0000000000003475 [DOI] [PubMed] [Google Scholar]
- 125.Martini ML, Neifert SN, Oermann EK, et al. Machine learning with feature domains elucidates candidate drivers of hospital readmission following spine surgery in a large single-center patient cohort. Neurosurgery. 2020;87(4):E500–E510. doi:10.1093/neuros/nyaa136 [DOI] [PubMed] [Google Scholar]
- 126.Martini ML, Neifert SN, Gal JS, Oermann EK, Gilligan JT, Caridi JM. Drivers of prolonged hospitalization following spine surgery: a game-theory-based approach to explaining machine learning models. J Bone Joint Surg Am. 2020;103(1):64–73. doi:10.2106/jbjs.20.00875 [DOI] [PubMed] [Google Scholar]
- 127.Kuo CY, Yu LC, Chen HC, Chan CL. Comparison of models for the prediction of medical costs of spinal fusion in Taiwan diagnosis-related groups by machine learning algorithms. Healthcare Inform Res. 2018;24(1):29–37. doi:10.4258/hir.2018.24.1.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Karnuta JM, Golubovsky JL, Haeberle HS, et al. Can a machine learning model accurately predict patient resource utilization following lumbar spinal fusion? Spine J. 2020;20(3):329–336. doi:10.1016/j.spinee.2019.10.007 [DOI] [PubMed] [Google Scholar]
- 129.Lerner J, Ruppenkamp J, Etter K, et al. Preoperative behavioral health, opioid, and antidepressant utilization and 2-year costs after spinal fusion-revelations from cluster analysis. Spine. 2020;45(2):E90–E98. doi:10.1097/brs.0000000000003233 [DOI] [PubMed] [Google Scholar]
- 130.Wainberg M, Merico D, Delong A, Frey BJ. Deep learning in biomedicine. Nat Biotechnol. 2018;36(9):829–838. doi:10.1038/nbt.4233 [DOI] [PubMed] [Google Scholar]
- 131.Zhang Y, Fatemi P, Medress Z, et al. A predictive-modeling based screening tool for prolonged opioid use after surgical management of low back and lower extremity pain. Spine J. 2020;20(8):1184–1195. doi:10.1016/j.spinee.2020.05.098 [DOI] [PubMed] [Google Scholar]
- 132.Tătaru OS, Vartolomei MD, Rassweiler JJ, et al. artificial intelligence and machine learning in prostate cancer patient management-current trends and future perspectives. Diagnostics (Basel, Switzerland). 2021;11(2). doi:10.3390/diagnostics11020354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shah AA, Karhade AV, Bono CM, Harris MB, Nelson SB, Schwab JH. Development of a machine learning algorithm for prediction of failure of nonoperative management in spinal epidural abscess. Spine J. 2019;19(10):1657–1665. doi:10.1016/j.spinee.2019.04.022 [DOI] [PubMed] [Google Scholar]
- 134.Pan SJ, Qiang Y. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2010;22(10):1345–1359. [Google Scholar]
- 135.Shin HC, Roth HR, Gao M, et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging. 2016;35(5):1285–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huynh B, Drukker K, Giger MJMP. MO-DE-207B-06: computer-aided diagnosis of breast ultrasound images using transfer learning from deep convolutional neural networks. Am Assoc Phys Med. 2016;43(6):3705–3705. [Google Scholar]
- 137.Xie X, Niu J, Liu X, Chen Z, Tang S, Yu S. A survey on incorporating domain knowledge into deep learning for medical image analysis. Med Image Anal. 2021;69:101985. doi:10.1016/j.media.2021.101985 [DOI] [PubMed] [Google Scholar]
- 138.Wang S, Yang DM, Rong R, et al. Artificial intelligence in lung cancer pathology image analysis. Cancers. 2019;11(11). doi:10.3390/cancers11111673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Elakkiya R, Vijayakumar P, Karuppiah M. COVID_SCREENET: COVID-19 screening in chest radiography images using deep transfer stacking. Inf Syst Front. 2021:1–15. doi:10.1007/s10796-021-10123-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Cheng C, Qi D, Hao C, Heng PA. Semantic-aware generative adversarial nets for unsupervised domain adaptation in chest X-ray segmentation. In: Shi Y, Suk HI, Liu M, eds. International Workshop on Machine Learning in Medical Imaging MLMI Lecture Notes in Computer Science, Vol 11046. Springer, Cham; 2018: 143–151. [Google Scholar]
- 141.Evgeniou T, Pontil M. Regularized multi—task learning. In: Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 22, 2004; Seattle, Washington; 2004:109-117. [Google Scholar]
- 142.Pan Z, Liu N, Zhang W, Wang J. MTHAM: multitask disease progression modeling based on hierarchical attention mechanism. Comput Sci. 2020;47(9):185–189. doi:10.11896/jsjkx.190900001 [Google Scholar]
- 143.Jing Q, Wang W, Zhang J, Tian H, Chen K. Quantifying the Performance of Federated Transfer Learning. ArXiv; 2019. [Google Scholar]
- 144.Liu Y, Kang Y, Xing C, Chen T, Yang QJIS. A Secure Federated Transfer Learning Framework. In: Intelligent Systems. Volume, 35, Issue, 4; July-August 1, 2020; IEEE; 2020. [Google Scholar]
- 145.Ju C, Gao D, Mane R, Tan B, Liu Y, Guan C. Federated transfer learning for EEG signal classification. In: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual International Conference; July 20, 2020; Montreal, QC, Canada; 2020;2020:3040-3045. doi:10.1109/embc44109.2020.9175344 [DOI] [PubMed] [Google Scholar]
- 146.Kwon BC, Anand V, Severson KA, et al. DPVis: visual analytics with hidden Markov models for disease progression pathways. IEEE Trans Vis Comput Graph. 2020. doi:10.1109/tvcg.2020.2985689 [DOI] [PubMed] [Google Scholar]
- 147.Yang L, Yong L, Zhu X, et al. Disease progression model of 4T1 metastatic breast cancer. J Pharmacokinet Pharmacodyn. 2020;47(1):105–116. doi:10.1007/s10928-020-09673-5 [DOI] [PubMed] [Google Scholar]
- 148.Archetti D, Ingala S, Venkatraghavan V, et al. Multi-study validation of data-driven disease progression models to characterize evolution of biomarkers in Alzheimer’s disease. NeuroImage Clin. 2019;24:101954. doi:10.1016/j.nicl.2019.101954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oxtoby NP, Young AL, Cash DM, et al. Data-driven models of dominantly-inherited Alzheimer’s disease progression. Brain. 2018;141(5):1529–1544. doi:10.1093/brain/awy050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Belou Ad Ah E, Popescu A. DeeSIL: deep-shallow incremental learning. In: Proceedings of the European Conference on Computer Vision (ECCV) Workshops; September 8-14, 2018. Munich, Germany; 2018. [Google Scholar]
- 151.Li Z, Hoiem D. Learning without forgetting. IEEE Trans Pattern Anal Mach Intell. 2017;40(12):1–1. [DOI] [PubMed] [Google Scholar]
- 152.Kumarasinghe K, Kasabov N, Taylor D. Brain-inspired spiking neural networks for decoding and understanding muscle activity and kinematics from electroencephalography signals during hand movements. Sci Rep. 2021;11(1):2486. doi:10.1038/s41598-021-81805-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rodríguez Aldana Y, Marañón Reyes EJ, Macias FS, et al. Nonconvulsive epileptic seizure monitoring with incremental learning. Comput Biol Med. 2019;114:103434. doi:10.1016/j.compbiomed.2019.103434 [DOI] [PubMed] [Google Scholar]
- 154.Shi H, Wang H, Qin C, Zhao L, Liu C. An incremental learning system for atrial fibrillation detection based on transfer learning and active learning. Comput Methods Programs Biomed. 2020;187:105219. doi:10.1016/j.cmpb.2019.105219 [DOI] [PubMed] [Google Scholar]
- 155.Ozdemir F, Goksel O. Extending pretrained segmentation networks with additional anatomical structures. Int J Comput Assisted Radiol Surg. 2019;14(7):1187–1195. doi:10.1007/s11548-019-01984-4 [DOI] [PubMed] [Google Scholar]
- 156.Zhong HS, Wang H, Deng YH, et al. Quantum computational advantage using photons. Science (New York, NY). 2020;370(6523):1460–1463. doi:10.1126/science.abe8770 [DOI] [PubMed] [Google Scholar]
- 157.GBD 2019 Universal Health Coverage Collaborators. Measuring universal health coverage based on an index of effective coverage of health services in 204 countries and territories, 1990-2019: a systematic analysis for the global burden of disease study 2019. Lancet (London, England). 2020;396(10258):1250–1284. doi:10.1016/s0140-6736(20)30750-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Bloom DE, Khoury A, Subbaraman R. The promise and peril of universal health care. Science (New York, NY). 2018;361(6404). doi:10.1126/science.aat9644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Nijeweme-d Hollosy WO, van Velsen L, Poel M, Groothuis-Oudshoorn CGM, Soer R, Hermens H. Evaluation of three machine learning models for self-referral decision support on low back pain in primary care. Int J Med Inform. 2018;110:31–41. doi:10.1016/j.ijmedinf.2017.11.010 [DOI] [PubMed] [Google Scholar]
- 160.Lafage R, Pesenti S, Lafage V, Schwab FJ. Self-learning computers for surgical planning and prediction of postoperative alignment. Eur Spine J. 2018;27(Suppl 1):123–128. doi:10.1007/s00586-018-5497-0 [DOI] [PubMed] [Google Scholar]
- 161.Staartjes VE, Stienen MN. Data mining in spine surgery: leveraging electronic health records for machine learning and clinical research. Neurospine. 2019;16(4):654–656. doi:10.14245/ns.1938434.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yin J, Ngiam KY, Teo HH. Role of artificial intelligence applications in real-life clinical practice: systematic review. J Med Internet Res. 2021;23(4):e25759. doi:10.2196/25759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Char DS, Shah NH, Magnus D. Implementing machine learning in health care—addressing ethical challenges. N Eng J Med. 2018;378(11):981–983. doi:10.1056/NEJMp1714229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ribeiro MT, Singh S, Guestrin CJA. Why should I trust you?: explaining the predictions of any classifier. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 13 August, 2016. San Francisco; 2016. [Google Scholar]
- 165.You E, Lin V, Mijovic T, Eskander A, Crowson MG. Artificial intelligence applications in otology: a state of the art review. Otolaryngology Head Neck Surg. 2020;163(6):1123–1133. doi:10.1177/0194599820931804 [DOI] [PubMed] [Google Scholar]
- 166.Challen R, Denny J, Pitt M, Gompels L, Edwards T, Tsaneva-Atanasova K. Artificial intelligence, bias and clinical safety. BMJ Qual Saf. 2019;28(3):231–237. doi:10.1136/bmjqs-2018-008370 [DOI] [PMC free article] [PubMed] [Google Scholar]