Abstract
Preventive strategies involving the use of pneumococcal conjugate vaccines (PCVs) are known to drastically reduce pneumococcal disease. However, PCV vaccination has been plagued with serotype replacement by non-PCV serotypes. In this study, we describe the prevalence and molecular characteristics of non-PCV13 serotypes (non-vaccine serotypes, NVTs) from pneumococcal carriage isolates obtained from children < 5 years old in Cape Coast, Ghana, after PCV introduction. The isolates were subjected to antibiotic susceptibility testing and multilocus sequence typing (MLST), and molecular techniques were used to detect the presence of virulence genes. Serotypes 11A, 13, 15B, 23B, and 34 formed the top five of the 93 NVT isolates. As such, 20 (21.5%), 49 (48.4%), and 70 (74.3%) isolates were non-susceptible to penicillin, tetracycline, and cotrimoxazole, respectively. Sixteen (17.2%) multidrug-resistant isolates were identified. However, non-susceptibility to ceftriaxone and erythromycin was low and all isolates were fully susceptible to levofloxacin, linezolid, and vancomycin. Whereas pcpA, pavB, lytA, and psrP genes were detected in nearly all serotypes, pilus islet genes were limited to serotypes 11A, 13, and 23B. MLST for predominant serotype 23B isolates revealed three known and seven novel sequence types (STs). ST172 and novel ST15111 were the most dominant and both STs were related to PMEN clone Columbia23F-26 (ST338). In conclusion, non-PCV13 serotype 23B was the most prevalent, with characteristics of rapid clonal expansion of ST172 and ST15111, which are related to international clones of the pneumococcus. Continuous monitoring of NVTs in Ghana is, therefore, essential, as they have the potential to cause invasive disease, show high antibiotic resistance, and attenuate the effects of PCV vaccination.
Keywords: Streptococcus pneumoniae 1, non-PCV13 serotypes 2, virulence genes 3, antibiotic susceptibility 4, MLST 5
1. Introduction
Invasive diseases, such as meningitis and bacteremia, caused by Streptococcus pneumoniae, are a leading cause of mortality in children under five years, predominantly in low- and middle-income countries (LMIC). The introduction of pneumococcal conjugate vaccines (PCVs) is estimated to have reduced pneumococcal deaths by 51% by the end of 2015 [1]. Both PCV10 and PCV13 are included in vaccination schedules in childhood immunization programs in several countries. Post-PCV vaccination data have shown a drastic decline in invasive pneumococcal disease globally [2,3]. Although these PCVs have offered protection against vaccine serotypes (VTs), they have contributed to the insurgence of non-PCV serotypes. The increase in non-PCV serotypes is of concern, especially when these serotypes show resistance to antibiotics commonly used in treating invasive pneumococcal disease (IPD) [4,5]. Hence, the quest to develop higher-valency PCVs has been initiated. Interestingly, in 2021, the Food and Drugs Administration (FDA) in the United States of America approved the use of PCV20 (PREVNAR20) and PCV15 (VAXNEUVANCE) in adults 18 years old and above [6,7]. Subsequently, the European Medicines Agency (EMA) also approved the marketing of PCV20 (APEXXNAR) and PCV15 (VAXNEUVANCE) in Europe [8,9].
However, the phenomenon of serotype replacement by NVT is likely to continue, even with the future introduction of these higher-valency PCVs. Therefore, alternatives to PCVs have been proposed, including pneumococcal protein-based vaccine candidates (PBVCs). The proteins in these PBVCs must be conserved in all pneumococci in order to offer protection across all serotypes. Some PBVCs that have been proposed and are currently at different stages of clinical trials include pneumococcal choline-binding protein A (PcpA), pneumolysin (Ply), autolysin A (LytA), pneumococcal serine-rich repeat protein (PsrP), and pilus type 1 [10]. Common non-PCV serotypes that have been reported globally include serotypes 11A, 13, 15A, 15B,19B, 23A, 23B, and 34 [4,11,12]. In addition, some of these NVTs have been implicated in IPD [13,14].
With support from Gavi, The Vaccine Alliance, LMICs, including Ghana, have introduced PCV13 as part of their childhood immunization program. In 2012, Ghana introduced PCV13 vaccination among children following the 3+0 vaccine dosing schedule. However, there are concerns about the 3+0 vaccine dosing schedule, as many countries that implemented this schedule have reported increased NVTs and high residual carriage of VTs [15] compared to high-income countries that implemented the 2+1 vaccine dosing schedule [16,17,18].
Different NVTs have been reported in invasive disease based on geographical locations. In addition, there are indications of expansion of lineages of these NVTs causing invasive disease [13,19]. Assessing the dynamics of NVTs, their contribution to invasive disease as well as their clonal expansion is important to understand the overall impact of PCV vaccination. Such molecular epidemiological data on NVTs are limited in Ghana, although some post-vaccination carriage studies have been performed [20]. However, these studies were performed in the capital city of Ghana. In this study, we describe the antibiotic susceptibility patterns and molecular characteristics, including the detection of virulence genes and genetic relatedness of pneumococcal NVT obtained from children <5 years of age in Cape Coast, Ghana.
2. Materials and Methods
2.1. Pneumococcal Isolates and Serotyping
In February 2018, a total of 513 children under five years was enrolled into the pneumococcal carriage study. We selected children from kindergartens and childhood immunization centers in Cape Coast, Central region, Ghana. The isolation and detection of pneumococci were performed as described [11,21]. Briefly, nasopharyngeal swabs were cultured on sheep blood agar supplemented with 5 µg/mL gentamicin. Presumptive pneumococci were identified by optochin and bile solubility tests.
2.2. Antibiotic Susceptibility Testing
Susceptibility testing was determined using the disc diffusion method in accordance with the Clinical and Laboratory Standards Institute (CLSI) guidelines [22]. The isolates were tested for susceptibility to levofloxacin (5 μg), vancomycin (30 μg), linezolid (30 μg), clindamycin (2 μg), erythromycin (15 μg), tetracycline (30 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), and ceftriaxone (30 μg). Oxacillin (1 μg) discs were used to screen for penicillin non-susceptibility. Antibiotic discs (Thermo Fisher, Germany) were applied on the agar plates and incubated at 37 °C in 5% CO2 for 18–24 h. S. pneumoniae ATCC 49619 was included in each test batch as a control strain. Penicillin MIC test strips (Liofilchem) were used to measure the minimum inhibitory concentration (MIC) of oxacillin-resistant isolates. Isolates with MIC ≤ 0.06 μg/mL, 0.12 μg/mL, and ≥2 μg/mL were defined as penicillin-susceptible, penicillin-non-susceptible, and penicillin-resistant pneumococci, respectively. Isolates resistant to three or more classes of antibiotics were classified as multidrug resistant (MDR).
2.3. Serotyping and Molecular Characterization of Pneumococcal Strains
DNA was extracted from the pneumococcal isolates using the manufacturer’s instruction in the QIAamp DNA Mini Kit (Qiagen Diagnostics GmbH, Hilden, Germany). The extracted DNA was used as template DNA for all molecular tests. Primers and protocols described earlier [23] were used in a multiplex PCR (mPCR) to deduce pneumococcal serotypes. The serotypes were further confirmed using the Quellung reaction performed at the German National Reference Center for Streptococci, Aachen, Germany.
2.4. Virulence Gene Determination
Pneumococcal virulence genes including lytA, pavB, pcpA, psrP, the pilus islets (PI) PI-1, and PI-2 were amplified with primers described earlier [11]. In brief, DNA templates from pneumococcal isolates were used in conventional PCR to amplify the virulence genes. Each reaction mixture contained 1 μL DNA extract, 1 μL of 25 mM MgCl2 (Roth), 1 μL of 5 mM dNTPs (Thermofisher), 2.5 μL of 10X Dream buffer (Thermofisher), 1 μL of the respective primers, 0.5 μL of DreamTaq DNA polymerase (Thermofisher), and nuclease-free water to make up 25 μL end volume. The peqSTAR 2X thermocycler (VWR) was used under these conditions: 4 min at 94 °C followed by 30 cycles composed of 30 sec at 94 °C, 30 s at 55 °C, 1–3 min at 72 °C (varies based on expected amplicon size), and 5 min at 72 °C. A 0.8% agarose gel was used to visualize the PCR fragments.
2.5. Multilocus Sequence Typing (MLST)
The internal fragments of the seven housekeeping genes aroE, gdh, gki, recP, spi, xpt, and ddl were amplified by PCR and sequenced. We used primers described in the PubMLST [24] database and alternative primers described by the CDC [23]. Amplifications for all genes were carried out with approximately 1 μL of 100 ng of DNA template, 1 μL of 25 mM MgCl2 (Roth), 1 μL of 5 mM dNTPs (Thermofisher), 5 μL of 10X Dream buffer (Thermofisher), 1 μL of the respective primers, and 0.5 μL of DreamTaq DNA polymerase (Thermofisher), and nuclease-free water in a 50 μL reaction mixture.
Thermocycling conditions were 4 min hold at 94 °C, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 60 s, and a final extension at 72 °C for 5 min. The reaction mixture was separated by electrophoresis on a 0.8% agarose gel and visualized under UV illumination with a gel documentation system. The amplified genes were sequenced in both directions at Macrogen, Inc. (Amsterdam, The Netherlands). Sequence analysis was performed with DNADynamo (Blue Tractor Software Ltd, North Wales, UK). The consensus sequences were submitted to the PubMLST database and each gene sequence was assigned an existing or novel allele type number and sequence type (ST) numbers prescribed by the database. The relatedness of STs was analyzed using the goeBURST program [25]. Cluster analysis of related sequence types was grouped into clonal complexes (CCs) and illustrated using minimum spanning and neighbor-joining trees in the PHYLOViZ program.
2.6. Statistical Analysis
Data were analyzed using GraphPad Prism (GraphPad software®) version 5 (Dotmatics, San Diego, CA, USA) and Statistical Package for Social Sciences (SPSS) software® (version 21) (IBM, New York, NY, USA). Categorical data were expressed as proportions and compared using the Chi-square test or Fisher’s exact test (two-tailed) where necessary.
2.7. Ethical Approval
The institutional review board of the University of Cape Coast granted ethical approval for this study (UCCIRB/EXT/2017/21). In addition to parental consent, all the children voluntarily participated in this study.
3. Results
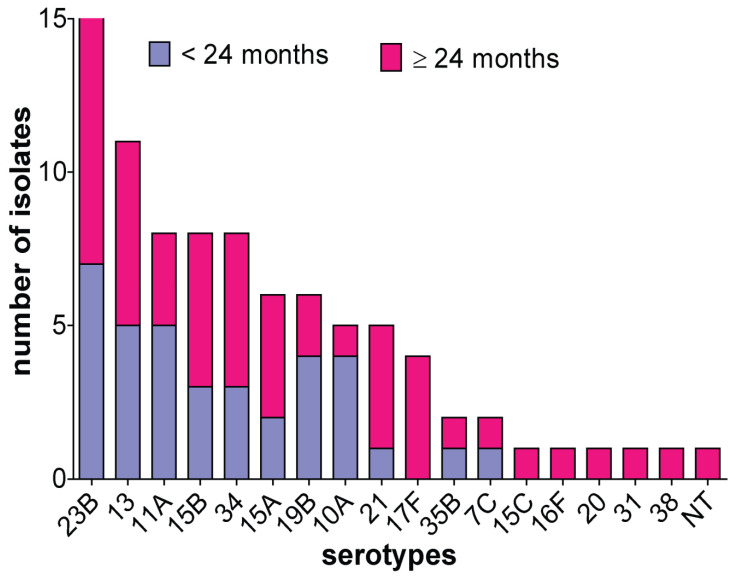
A total of 93 NVT pneumococcal strains was isolated from the nasopharynx of 53 (57%) females and 40 (43%) male children under five years of age. More than half (n = 57, 61.3%) of the isolates were detected in children ≥24 months (Figure 1). A total of 17 different NVTs with one non-typeable strain was identified. The predominant serotypes were serotypes 23B (n = 22, 23.7%), 13 (n = 11, 11.8%), 11A, 15B and 34 (n = 8, 8.6%), and 15A and 19B (n = 6, 6.5%).
Figure 1.
Non-PCV13 serotype distribution by age group. Age group ≤24 months includes children aged 6 months to 24 months and children 25 months to 59 months are included in the ≥24 months age group. NT, non-typeable.
3.1. Antibiotics Susceptibility Patterns of NVT Isolates
Table 1 shows the antibiotic susceptibility patterns among the different serotypes in the study. All isolates were fully susceptible to vancomycin, levofloxacin, and linezolid. Furthermore, susceptibilities towards ceftriaxone, erythromycin, clindamycin, and chloramphenicol were >86% (Table 1). However, the data showed a marked non-susceptibility towards tetracycline 40 (43.0%) and cotrimoxazole 70 (75.4%). In contrast, penicillin non-susceptibility was relatively low, with 19 (20.4%) and 1 (1.1%) of the isolates being intermediate and resistant, respectively. Sixteen (17.2%) isolates were MDR, of which serotype 23B was the most prevalent. Furthermore, MDR to ≥4 different antibiotics was seen in five (5.4%) of the isolates and was limited to serotypes 23B, 35B, and 38, respectively.
Table 1.
Distribution of virulence genes and antibiotic non-susceptibility patterns of non-PCV13 serotypes in Cape Coast, Ghana.
| Pneumococcal Isolates | Antibiotic Non-Susceptibility | Virulence Genes | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Serotypes | Number | CRO | ERY | CLI | TET | CHL | COT | PEN | MDR | pcpA | psrP | PI-1 | PI-2 |
| 23B | 22 | 0 | 4.5 | 4.5 | 50 | 12.5 | 86.4 | 72.7 | 45.5 | 95.5 | 36.4 | 0 | 4.5 |
| 13 | 11 | 0 | 0 | 0 | 72.7 | 27.3 | 72.7 | 0 | 0 | 100 | 90.9 | 9.1 | 0 |
| 11A | 8 | 0 | 0 | 0 | 12.5 | 0 | 87.5 | 12.5 | 0 | 100 | 87.5 | 12.5 | 0 |
| 15B | 8 | 0 | 0 | 0 | 25 | 12.5 | 100 | 0 | 0 | 100 | 75 | 0 | 0 |
| 34 | 8 | 0 | 0 | 0 | 12.5 | 12.5 | 37.5 | 0 | 0 | 75 | 87.5 | 0 | 0 |
| 15A | 6 | 16.7 | 0 | 0 | 83.3 | 16.7 | 100 | 16.7 | 50 | 83.3 | 66.7 | 0 | 0 |
| 19B | 6 | 0 | 0 | 0 | 83.3 | 0 | 66.6 | 0 | 0 | 100 | 83.3 | 0 | 0 |
| 10A | 5 | 20 | 0 | 0 | 0 | 0 | 60 | 0 | 0 | 100 | 20 | 0 | 0 |
| 21 | 5 | 0 | 0 | 0 | 100 | 0 | 100 | 0 | 0 | 80 | 100 | 0 | 0 |
| 17F | 4 | 0 | 0 | 0 | 25 | 0 | 25 | 0 | 0 | 100 | 0 | 0 | 0 |
| 35B | 2 | 0 | 100 | 100 | 100 | 0 | 0 | 50 | 100 | 100 | 100 | 0 | 0 |
| 7C | 2 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 |
| 15C | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 0 | 0 | 0 |
| 16F | 1 | 100 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 100 | 100 | 0 | 0 |
| 20 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 | 0 | 0 |
| 31 | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 100 | 100 | 0 | 0 |
| 38 | 1 | 0 | 100 | 0 | 100 | 0 | 100 | 100 | 100 | 100 | 100 | 0 | 0 |
| NT | 1 | 0 | 0 | 0 | 100 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 93 (100) | 3 (3.2) | 4 (4.3) | 3 (3.2) | 45 (48.4) | 7 (7.5) | 70 (74.3) | 20 (21.5) | 16 (17.2) | 85 (91.4) | 61 (65.6) | 2 (2.2) | 1 (1.1) |
Percentage of isolates (%), presence of virulence genes is expressed in percentages. CRO, ceftriaxone; ERY, erythromycin; CLI, clindamycin; TET, tetracycline; CHL, chloramphenicol; COT, cotrimoxazole; PEN, Penicillin. Antibiotic non-susceptibility (includes intermediate and full resistance isolates). All the isolates were susceptible to vancomycin, levofloxacin, and linezolid. Multidrug resistant (MDR), ≥3 classes of antibiotics. pcpA, pneumococcal choline-binding protein A; psrP, Pneumococcal serine-rich repeat protein; PI-1, Pilus islet 1; PI-2, Pilus islet 2.
3.2. Characterization of Pneumococcal Virulence Genes
The lytA and pavB genes were identified in all pneumococcal isolates; however, the pilus islets (PI-1 and PI-2) were limited to serotypes 13, 11A, and 23B, respectively (Table 1). In contrast, pcpA and psrP genes were detected among the different serotypes in these proportions: 85 (91.4%) and 61 (65.6%), respectively.
3.3. Phylogeny of NVT Serotype 23B
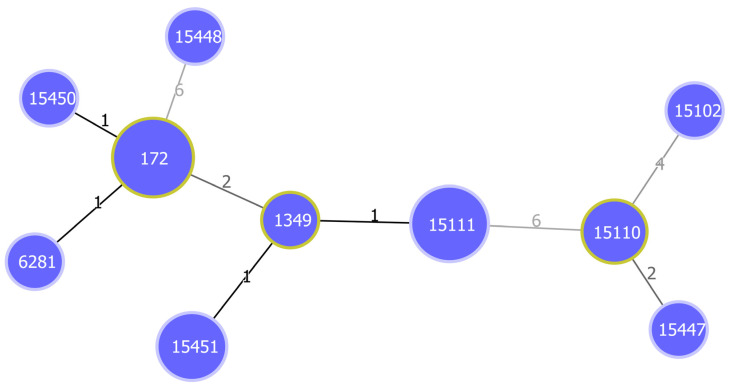
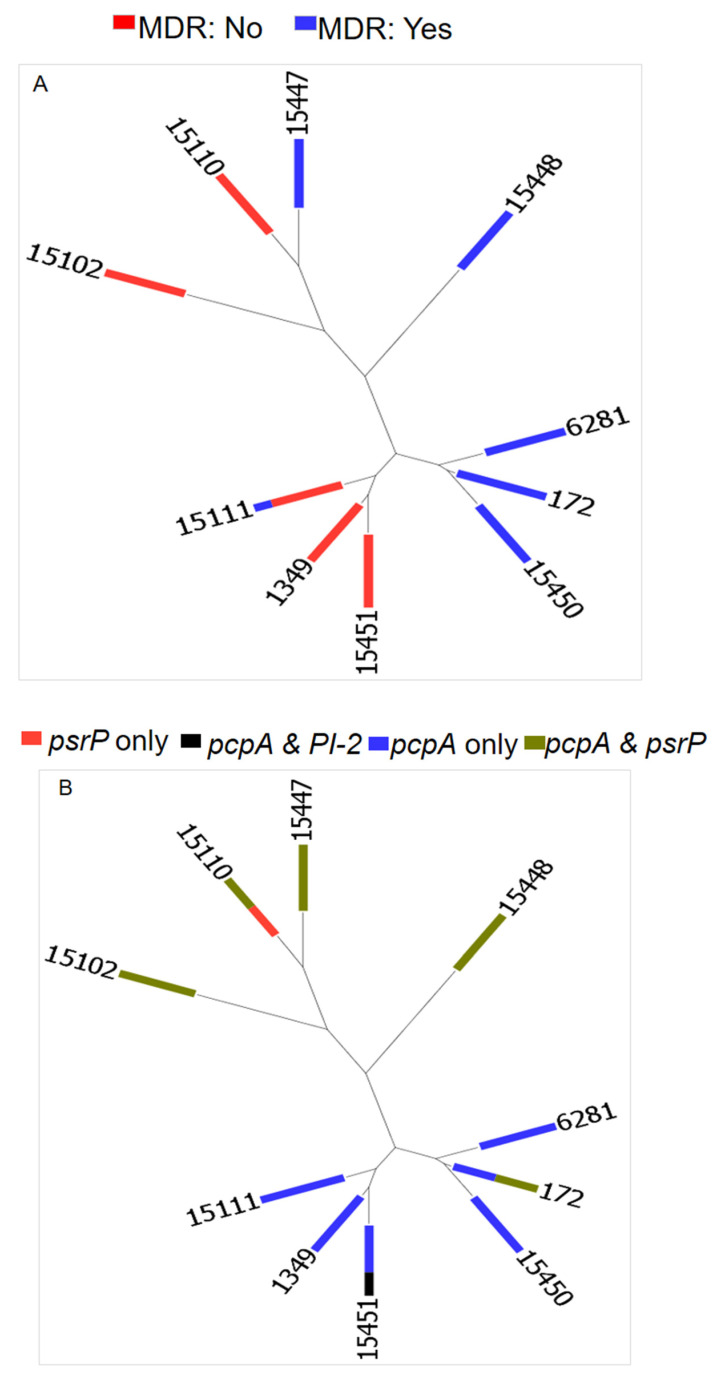
MLST was performed on the most prevalent NVTs (serotype 23B) to determine their genetic profiles. The MLST of the 22 serotype 23B isolates revealed three known STs and seven novel STs, containing 8 and 14 isolates, respectively (Table 2). The most common STs were ST172 (n = 6, 27.3%) and novel ST15111 (n = 5, 22.7%). The goeBurst algorithm (Figure 2) identified two notable clonal complexes, namely CC1 (ST172, ST6281, and ST15450) and CC2 (ST1349, ST15111, and ST15451). MDR was associated with both novel and already known STs (Figure 3A). Unlike the pcpA, psrP, lytA, and pavB virulence genes relating to the various STs, the PI-1 gene was not associated with any ST and PI-2 was present in ST15451 (Figure 3B).
Table 2.
MLST of serotype 23B pneumococcal isolates.
| Isolate ID | aroE | gdh | gki | recP | spi | xpt | ddl | ST |
|---|---|---|---|---|---|---|---|---|
| S119, S110, S158, H174, S258, S357 | 7 | 13 | 8 | 6 | 25 | 6 | 8 | 172 |
| S114 | 7 | 13 | 8 | 6 | 25 | 246 | 8 | 6281 |
| S311 | 7 | 19 | 8 | 6 | 25 | 6 | 8 | 15450 |
| S1 | 12 | 5 | 4 | 10 | 15 | 155 | 9 | 15102 |
| S152, S239 | 12 | 5 | 4 | 18 | 474 | 4 | 31 | 15110 |
| S200, S207, S214, S238, S361 | 12 | 13 | 8 | 6 | 3 | 6 | 8 | 15111 |
| S35 | 18 | 13 | 8 | 6 | 3 | 6 | 8 | 1349 |
| H58, S266, S306 | 18 | 13 | 8 | 6 | 3 | 21 | 8 | 15451 |
| S156 | 2 | 5 | 36 | 18 | 474 | 4 | 31 | 15447 |
| S237 | 1 | 43 | 41 | 18 | 13 | 37 | 8 | 15448 |
Bold numbers: novel STs.
Figure 2.
Minimum spanning tree (MST) showing the genetic relationship between STs obtained from MLST of serotype 23B strains. The tree was generated using the goeBurst algorithm in PHYLOViZ software. The diameters of nodes are proportional to the number of isolates. Founder STs have yellow color around their nodes. Branch labels correspond to the number of allelic variations between STs; branch lengths are not to scale.
Figure 3.
Relationship between STs, multidrug resistance and virulence genes. The neighbor-joining tree (NJT) was generated using PHYLOViZ. (A) Multidrug-resistance (MDR) relationship among the different STs. MDR was detected in both new and existing STs. (B) Distribution of virulence genes by STs. There was a fair distribution of all the virulence genes among the identified STs except the pilus islet 2, which was limited to only ST15451.
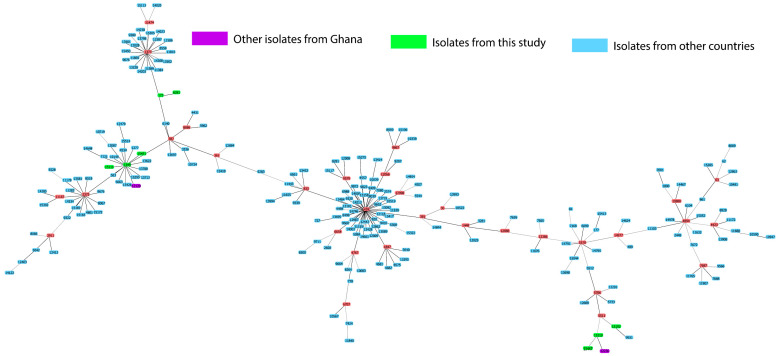
Figure 4 shows a comparison of the genetic relatedness of the serotype 23B isolates from this study to other serotype 23B isolates in the PubMLST database. Two isolates previously reported from Ghana were among the serotype 23B collection in the PuBMLST database. The novel ST15110 from this study varied at one locus from ST12236, found in a serotype 23B nasopharyngeal isolate reported from Ghana in 2011. In addition, the predominant ST172 varied at one locus from emerging clone ST1373l.
Figure 4.
Genetic relatedness of serotype 23B isolates from this study compared with serotype 23B isolates from the PubMLST database. PubMLST database (accessed on 29 September 2020). Green color represents STs reported from this study, blue color, STs from other countries, purple color, STs of other serotype 23B strains from Ghana.
4. Discussion
Non-vaccine serotypes (NVTs) have emerged in countries where PCV vaccination has been introduced [15,19,20,26,27]. It is likely that NVTs were present prior to PCV introduction and have expanded to fill the niches vacated by VTs due to vaccine-induced selective pressure [27]. It is, therefore, important to continuously monitor and characterize these NVTs to fully understand their epidemiology and evolution.
Serotypes 11A, 13, 15A, 15B, 19B, and 23B are among the most common NVTs identified in this study. The emergence of replacement serotypes similar to those from this study has been reported in Ghana and several African countries that have introduced PCV in their routine immunization program [26,28,29]. Although some of these NVTs are considered to be of low potential to cause IPD, serotypes, such as 15B/C, 23B, 35B, and 38, are already associated with IPD in many countries [13,27,30]. Interestingly, the aforementioned serotypes have also been reported to be causal pathogens of pneumococcal disease among children under five years old in Ghana [27,31]. In this study, none of the isolates expressed serotype 12F, a non-PCV13 serotype that was implicated in a recent pneumococcal meningitis outbreak in Ghana and also found in IPD among children under five years [27,31].
Antibiotic-resistant pneumococcal isolates causing IPD declined after PCV introduction, partly due to the reduction in VTs, which were mostly associated with antibiotic resistance. However, the emergence of antibiotic-resistant NVTs identified in this study and similar to reports from previous studies emphasizes the need to maintain surveillance on NVTs [29,30]. There is a likelihood of these antibiotic-resistant NVTs progressing from carriage to cause IPD, which could eventually eliminate the full benefit of PCVs. Penicillin is the drug of choice for treating pneumococcal infections, yet increased penicillin resistance has been observed in populations where penicillin has been used extensively. Implementation of PCVs led to a drastic decline in penicillin resistance among pneumococcal isolates [11,20,32]. This decline in penicillin resistance among pneumococcal isolates causing IPD has been indicated as a positive effect of PCV vaccination [32]. In this study, we identified a relatively low resistance to penicillin, which contrasts with findings from other studies [27,33]. It is not surprising to observe penicillin resistance in NVTs, as this development could be due to the acquisition of resistance genes from other alpha-hemolytic streptococci present in the nasopharynx.
Although antibiotic resistance was low towards erythromycin, levofloxacin, and ceftriaxone, marked resistance to cotrimoxazole and tetracycline was noted. In contrast, a decline in cotrimoxazole resistance was reported among invasive pneumococcal isolates from children under five in Ghana [26,27]. However, our results are in accordance with previous post-PCV carriage studies conducted in Ghana [20]. It is noteworthy that similar antibiotic-resistance patterns were observed in the pre-vaccination era [34,35]. Therefore, the marked antibiotic resistance seen among the NVTs could be attributed to the acquisition of resistance genes and the uncontrolled use and abuse of antibiotics by the general public. Furthermore, cotrimoxazole continues to be administered in the treatment of common respiratory infections among children in Ghana and also as prophylaxis in HIV treatment [27,36]. Our results show that serotypes 15A, 23B, and 35B were multidrug resistant and this finding is in agreement with previous studies [33]. Other studies [27,33] that identified similar trends attributed the MDR seen in these serotypes to their genetic complexities. In this regard, these serotypes could become dominant strains with limited treatment options in the future.
In this study, we showed that all the NVT isolates possessed lytA and pavB genes, with >65% testing positive for pcpA and psrP genes. The proteins encoded by these genes are, hence, highly conserved in our NVT strain collection, similar to what was detected among VTs [11]. This finding suggests that these proteins cover multiple serotypes and could offer protection when included in protein-based vaccines. In contrast, the detection of pilus islets was very low and limited to serotypes 11A, 13, and 23B. Although previous studies identified pilus islets predominantly in VTs [37], recent studies [38] have also demonstrated the emergence of piliation among NVTs. The presence of pilus islets has been linked to antibiotic resistance and it is, therefore, not surprising that piliated serotypes from this study show antibiotic resistance. It is, therefore, important to monitor piliation in NVTs as they could expand and emerge in IPD and become associated with treatment difficulties as well.
Previous post-PCV data from Ghana [20] identified serotype 23B as a dominant strain, a finding which concurs with our findings. However, data on the molecular characteristics of this emerging serotype are lacking in Ghana. In this study, we, therefore, explored the genetic background of 22 serotype 23B isolates. The most frequently occurring ST was ST172, which is associated with serotypes 23F and 23B, with the majority of the isolates expressing penicillin non-susceptibility [26,27]. ST172 seems to be a global clone as it has been reported by several African countries, the USA, Australia, Europe, Asia, and the Middle East [24]. It is, however, interesting to note that ST172 is a single locus variant (SLV) of ST338 (Colombia23F-26), a PMEN clone that has been linked to the global spread of penicillin resistance [39]. Hence, it is not surprising to see that the study isolates of ST172 were all penicillin resistant. Because vaccines induce a serotype-specific immune response, some serotypes switch their capsules in order to evade the immune system. It is, therefore, possible that serotype 23B strains from this study associated with ST172 could have evolved from serotype 23F. A study in the USA reported that serotype 23A belonging to ST172 shared the same genetic background as serotype 23F strains, suggesting an expansion of the clone as well as a capsular switch [40].
The other known STs have been reported solely by South Africa (ST6281), whereas ST1349 has been reported primarily from Europe and the USA [24], with >90% of the isolates presenting as serotype 23B and being penicillin non-susceptible. This observation suggests the dissemination of these clones beyond their previously identified geographical areas. On the one hand, ST6281 is associated with serotype 23F [24]. However, in our study, ST6281 is of serotype 23B, demonstrating yet another natural capsule switch event. This event could occur as a result of the high homology between serotypes 23F and 23B [41]. Other studies have reported such events between serotypes 6A and 6C [42]. On the other hand, ST1349 is a double locus variant (DLV) of ST338 (Colombia23F-26), with a serotype 23F genetic background. The similarities in the genetic backgrounds observed between serotypes 23F and 23B suggest a possibility of clonal expansion of the previously existing clones, which could have been present in the pre-vaccination era. In addition, findings from a recent study suggest that closely related serotypes, such as (6B/6C, 15B/15C, 35F/35D), could switch their capsules and yet maintain the same ST over a period [43].
Despite the possible change in the capsule, the genetic background remains highly identical to that of serotype 23F of ST338 (Colombia23F-26). Out of the seven novel STs, three were SLVs of known STs, indicating a form of clonal expansion of their founding ancestor. In addition, two new STs were DLVs of the PMEN clone ST338 (Colombia23F-26), which supports the evidence of penicillin resistance seen among these STs. It is interesting to note that some of the novel STs already expressed MDR. Therefore, the expansion of these clones in the future could be problematic as treatment options may be limited.
The only pilus possessing serotype 23B strain belonged to the novel ST15451, showing resistance to penicillin but not being multidrug resistant. Similarly, the presence of PI-2 among pneumococcal strains in Canada did not influence their propensity to drug resistance [44]. The other virulence genes (pcpA and psrP) were evenly distributed among all the STs, except PI-1, which was not detected among any of the STs. This suggests that including PcpA and PsrP in protein-based vaccines could be beneficial in the long term to fight emerging NVTs.
We compared the STs of our serotype 23B with STs of all serotype 23B present in the PubMLST database. Interestingly, the STs identified in this study did not cluster around the predominant clone ST439, which has been reported to circulate in Europe. ST439 is the major clone associated with serotype 23B strains circulating in Germany [13]. However, the predominant clone ST172 found in this study appeared as an SLV of ST1373, which is a subgroup founder that has been reported mainly by the USA. The two other subgroup founders associated with STs from this study were ST1349 and ST5511. These STs (ST1373, ST1349, and ST5511) are quite distant from the dominant clone ST439.
5. Conclusions
To conclude, this study describes the serotype distribution of NVTs observed in pneumococcal carriage isolates from vaccinated children less than five years of age in Cape Coast, Ghana. Despite the recorded low antibiotic resistance to penicillin, marked resistance towards cotrimoxazole and tetracycline was observed. The studied virulence genes were present among all the NVTs, except for the pilus islets, which were limited to specific serotypes. ST172, an SLV of PMEN clone ST338 (Colombia23F-26), was the predominant clone among the serotype 23B isolates. This underscores the need for continuous monitoring of NVTs, their molecular characteristics, and antibiotic susceptibility patterns in carriage and IPD.
Acknowledgments
We would like to thank Birgit Rietow and Gerhard Burchhardt (Department of Molecular Genetics and Infection Biology, University of Greifswald, Germany) for providing technical support.
Author Contributions
Conceptualization, R.O.M., M.R.A. and S.H.; methodology, R.O.M., S.A.A. and D.C.S.; formal analysis, R.O.M., M.R.A., M.P.G.v.d.L., J.A.B. and G.G.; investigation, R.O.M., M.R.A., S.A.A., D.C.S. and M.P.G.v.d.L.; resources, S.H. and M.P.G.v.d.L.; writing—original draft preparation, R.O.M.; writing—review and editing, S.H., M.P.G.v.d.L., G.G. and M.R.A.; visualization, R.O.M. and M.R.A.; supervision, S.H.; project administration, S.H.; funding acquisition, R.O.M. and S.H. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
Data set for this study is available upon request from the authors.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Funding Statement
This study was supported by the Ghana Scholarship Secretariat and the German Academic Exchange Service (DAAD) as a joint grant scholarship and part of the Ph.D. thesis of Richael O. Mills, funding program/-ID: Research Grants—Doctoral Programs in Germany, 2016/17 (57251550), ST32. The work was further supported by the Bundesministerium für Bildung und Forschung (BMBF- Zwanzig20 -InfectControl 2020—project VacoME—FKZ 03ZZ0816A to SH). The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wahl B., O’Brien K.L., Greenbaum A., Majumder A., Liu L., Chu Y., Lukšić I., Nair H., McAllister D.A., Campbell H., et al. Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: Global, regional, and national estimates for 2000–15. Lancet Glob. Health. 2018;6:e744–e757. doi: 10.1016/S2214-109X(18)30247-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richter S.S., Diekema D.J., Heilmann K.P., Dohrn C.L., Riahi F., Doern G.V. Changes in pneumococcal serotypes and antimicrobial resistance after introduction of the 13-valent conjugate vaccine in the United States. Antimicrob. Agents Chemother. 2014;58:6484–6489. doi: 10.1128/AAC.03344-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan J., Nguyen C.D., Dunne E.M., Kim Mulholland E., Mungun T., Pomat W.S., Rafai E., Satzke C., Weinberger D.M., Russell F.M. Using pneumococcal carriage studies to monitor vaccine impact in low- and middle-income countries. Vaccine. 2019;37:6299–6309. doi: 10.1016/j.vaccine.2019.08.073. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.K., Yun K.W., Choi E.H., Kim S.J., Lee S.Y., Lee H.J. Changes in the Serotype Distribution among Antibiotic Resistant Carriage Streptococcus pneumonia isolates in Children after the Introduction of the Extended-Valency Pneumococcal Conjugate Vaccine. J. Korean Med. Sci. 2017;32:1431. doi: 10.3346/jkms.2017.32.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe Y.J., Lee H.J., Lee H., Oh C.E., Cho E.Y., Choi J.H., Kang H.M., Yoon I.A., Jung H.J., Choi E.H. Emergence of antibiotic-resistant non-vaccine serotype pneumococci in nasopharyngeal carriage in children after the use of extended-valency pneumococcal conjugate vaccines in Korea. Vaccine. 2016;34:4771–4776. doi: 10.1016/j.vaccine.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 6.Administration U.S.F.D. VAXNEUVANCE. [(accessed on 12 August 2021)]; Available online: https://www.fda.gov/vaccines-blood-biologics/vaccines/vaxneuvance.
- 7.Administration U.S.F.D. PREVNAR20. [(accessed on 12 August 2021)]; Available online: https://www.fda.gov/vaccines-blood-biologics/vaccines/prevnar-20.
- 8.European Commission Approves Merck’s VAXNEUVANCE™ (Pneumococcal 15-Valent Conjugate Vaccine) for Individuals 18 Years of Age and Older. [(accessed on 30 May 2022)]. Available online: https://www.merck.com/news/european-commission-approves-mercks-vaxneuvance-pneumococcal-15-valent-conjugate-vaccine-for-individuals-18-years-of-age-and-older/
- 9.European Medicines Agency Approves Pfizer’s 20-Valent Pneumococcal Conjugate Vaccine Against Invasive Pneumococcal Disease and Pneumonia in Adults. [(accessed on 30 May 2022)]. Available online: https://www.pfizer.com/news/press-release/press-release-detail/european-medicines-agency-approves-pfizers-20-valent#.
- 10.Odutola A., Ota M.O.C., Antonio M., Ogundare E.O., Saidu Y., Owiafe P.K., Worwui A., Idoko O.T., Owolabi O., Kampmann B., et al. Immunogenicity of pneumococcal conjugate vaccine formulations containing pneumococcal proteins, and immunogenicity and reactogenicity of co-administered routine vaccines—A phase II, randomised, observer-blind study in Gambian infants. Vaccine. 2019;37:2586–2599. doi: 10.1016/j.vaccine.2019.03.033. [DOI] [PubMed] [Google Scholar]
- 11.Mills R.O., Abdullah M.R., Akwetey S.A., Sappor D.C., Cole I., Baffuor-Asare M., Bolivar J.A., Gamez G., van der Linden M.P.G., Hammerschmidt S. Post-Vaccination Streptococcus pneumoniae Carriage and Virulence Gene Distribution among Children Less Than Five Years of Age, Cape Coast, Ghana. Microorganisms. 2020;8:1987. doi: 10.3390/microorganisms8121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindstrand A., Galanis I., Darenberg J., Morfeldt E., Naucler P., Blennow M., Alfven T., Henriques-Normark B., Ortqvist A. Unaltered pneumococcal carriage prevalence due to expansion of non-vaccine types of low invasive potential 8years after vaccine introduction in Stockholm, Sweden. Vaccine. 2016;34:4565–4571. doi: 10.1016/j.vaccine.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Van der Linden M., Perniciaro S., Imohl M. Increase of serotypes 15A and 23B in IPD in Germany in the PCV13 vaccination era. BMC Infect. Dis. 2015;15:207. doi: 10.1186/s12879-015-0941-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan S.L., Barson W.J., Lin P.L., Romero J.R., Bradley J.S., Tan T.Q., Pannaraj P.S., Givner L.B., Hulten K.G. Invasive Pneumococcal Disease in Children’s Hospitals: 2014–2017. Pediatrics. 2019;144:e20190567. doi: 10.1542/peds.2019-0567. [DOI] [PubMed] [Google Scholar]
- 15.Heinsbroek E., Tafatatha T., Phiri A., Swarthout T.D., Alaerts M., Crampin A.C., Chisambo C., Mwiba O., Read J.M., French N. Pneumococcal carriage in households in Karonga District, Malawi, before and after introduction of 13-valent pneumococcal conjugate vaccination. Vaccine. 2018;36:7369–7376. doi: 10.1016/j.vaccine.2018.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lovlie A., Vestrheim D.F., Aaberge I.S., Steens A. Changes in pneumococcal carriage prevalence and factors associated with carriage in Norwegian children, four years after introduction of PCV13. BMC Infect. Dis. 2020;20:29. doi: 10.1186/s12879-019-4754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spijkerman J., van Gils E.J.M., Veenhoven R.H., Hak E., Yzerman F., van der Ende A., Wijmenga-Monsuur A.J., van den Dobbelsteen G.P.J.M., Sanders E.A.M. Carriage ofStreptococcus pneumoniae3 Years after Start of Vaccination Program, the Netherlands. Emerg. Infect. Dis. 2011;17:584–591. doi: 10.3201/eid1704.101115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson H.L., Deloria-Knoll M., Levine O.S., Stoszek S.K., Freimanis Hance L., Reithinger R., Muenz L.R., O’Brien K.L. Systematic Evaluation of Serotypes Causing Invasive Pneumococcal Disease among Children Under Five: The Pneumococcal Global Serotype Project. PLoS Med. 2010;7:e1000348. doi: 10.1371/journal.pmed.1000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kavalari I.D., Fuursted K., Krogfelt K.A., Slotved H.C. Molecular characterization and epidemiology of Streptococcus pneumoniae serotype 24F in Denmark. Sci. Rep. 2019;9:5481. doi: 10.1038/s41598-019-41983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dayie N., Tettey E.Y., Newman M.J., Bannerman E., Donkor E.S., Labi A.K., Slotved H.C. Pneumococcal carriage among children under five in Accra, Ghana, five years after the introduction of pneumococcal conjugate vaccine. BMC Pediatr. 2019;19:316. doi: 10.1186/s12887-019-1690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satzke C., Turner P., Virolainen-Julkunen A., Adrian P.V., Antonio M., Hare K.M., Henao-Restrepo A.M., Leach A.J., Klugman K.P., Porter B.D., et al. Standard method for detecting upper respiratory carriage of Streptococcus pneumoniae: Updated recommendations from the World Health Organization Pneumococcal Carriage Working Group. Vaccine. 2013;32:165–179. doi: 10.1016/j.vaccine.2013.08.062. [DOI] [PubMed] [Google Scholar]
- 22.CLSI . M100 Performance Standards for Antimicrobial Susceptibility Testing. 27th ed. Volume 37 Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2017. [Google Scholar]
- 23.Streptococcus Laboratory Resources and Protocols. [(accessed on 6 February 2018)]; Available online: https://www.cdc.gov/streplab/pneumococcus/resources.html.
- 24.Source of Isolates Submitted to the Streptococcus pneumoniae Database. [(accessed on 3 May 2020)]. Available online: https://pubmlst.org/spneumoniae/
- 25.PHYLOViZ. [(accessed on 18 May 2020)]. Available online: https://www.phyloviz.net/
- 26.Renner L.A., Usuf E., Mohammed N.I., Ansong D., Dankwah T., Kusah J.T., Owusu S.K., Awunyo M., Arhin B., Addo Y., et al. Hospital-based Surveillance for Pediatric Bacterial Meningitis in the Era of the 13-Valent Pneumococcal Conjugate Vaccine in Ghana. Clin. Infect. Dis. 2019;69:S89–S96. doi: 10.1093/cid/ciz464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boettiger D.C., Law M.G., Sohn A.H., Davies M.-A., Wools-Kaloustian K., Leroy V., Yotebieng M., Vinikoor M., Vreeman R., Amorissani-Folquet M., et al. Temporal Trends in Co-trimoxazole Use Among Children on Antiretroviral Therapy and the Impact of Co-trimoxazole on Mortality Rates in Children Without Severe Immunodeficiency. J. Pediatr. Infect. Dis. Soc. 2019;8:450–460. doi: 10.1093/jpids/piy087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birindwa A.M., Emgard M., Norden R., Samuelsson E., Geravandi S., Gonzales-Siles L., Muhigirwa B., Kashosi T., Munguakonkwa E., Manegabe J.T., et al. High rate of antibiotic resistance among pneumococci carried by healthy children in the eastern part of the Democratic Republic of the Congo. BMC Pediatr. 2018;18:361. doi: 10.1186/s12887-018-1332-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skosana Z., Von Gottberg A., Olorunju S., Mohale T., Du Plessis M., Adams T., Mbelle N. Non-vaccine serotype pneumococcal carriage in healthy infants in South Africa following introduction of the 13-valent pneumococcal conjugate vaccine. S. Afr. Med. J. Suid-Afrik. Tydskr. Vir Geneeskd. 2021;111:143–148. doi: 10.7196/SAMJ.2021.v111i2.14626. [DOI] [PubMed] [Google Scholar]
- 30.Metcalf B.J., Gertz R.E., Gladstone R.A., Walker H., Sherwood L.K., Jackson D., Li Z., Law C., Hawkins P.A., Chochua S., et al. Strain features and distributions in pneumococci from children with invasive disease before and after 13-valent conjugate vaccine implementation in the USA. Clin. Microbiol. Infect. 2016;22:60.e69. doi: 10.1016/j.cmi.2015.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kwambana-Adams B.A., Asiedu-Bekoe F., Sarkodie B., Afreh O.K., Kuma G.K., Owusu-Okyere G., Foster-Nyarko E., Ohene S.-A., Okot C., Worwui A.K., et al. An outbreak of pneumococcal meningitis among older children (≥5 years) and adults after the implementation of an infant vaccination programme with the 13-valent pneumococcal conjugate vaccine in Ghana. BMC Infect. Dis. 2016;16:575. doi: 10.1186/s12879-016-1914-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dewe T.C.M., D’Aeth J.C., Croucher N.J. Genomic epidemiology of penicillin-non-susceptible Streptococcus pneumoniae. Microb. Genom. 2019;5:mgen000305. doi: 10.1099/mgen.0.000305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yahiaoui R.Y., Bootsma H.J., den Heijer C.D.J., Pluister G.N., John Paget W., Spreeuwenberg P., Trzcinski K., Stobberingh E.E. Distribution of serotypes and patterns of antimicrobial resistance among commensal Streptococcus pneumoniae in nine European countries. BMC Infect. Dis. 2018;18:440. doi: 10.1186/s12879-018-3341-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mills R.O., Twum-Danso K., Owusu-Agyei S., Donkor E.S. Epidemiology of pneumococcal carriage in children under five years of age in Accra, Ghana. Infect. Dis. 2015;47:326–331. doi: 10.3109/00365548.2014.994185. [DOI] [PubMed] [Google Scholar]
- 35.Dayie N.T., Arhin R.E., Newman M.J., Dalsgaard A., Bisgaard M., Frimodt-Moller N., Slotved H.C. Multidrug-Resistant Streptococcus pneumoniae Isolates from Healthy Ghanaian Preschool Children. Microb. Drug Resist. 2015;21:636–642. doi: 10.1089/mdr.2014.0314. [DOI] [PubMed] [Google Scholar]
- 36.Mwenya D.M., Charalambous B.M., Phillips P.P., Mwansa J.C., Batt S.L., Nunn A.J., Walker S., Gibb D.M., Gillespie S.H. Impact of cotrimoxazole on carriage and antibiotic resistance of Streptococcus pneumoniae and Haemophilus influenzae in HIV-infected children in Zambia. Antimicrob. Agents Chemother. 2010;54:3756–3762. doi: 10.1128/AAC.01409-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hjalmarsdottir M.A., Petursdottir B., Erlendsdottir H., Haraldsson G., Kristinsson K.G. Prevalence of pilus genes in pneumococci isolated from healthy preschool children in Iceland: Association with vaccine serotypes and antibiotic resistance. J. Antimicrob. Chemother. 2015;70:2203–2208. doi: 10.1093/jac/dkv096. [DOI] [PubMed] [Google Scholar]
- 38.Kawaguchiya M., Urushibara N., Aung M.S., Shinagawa M., Takahashi S., Kobayashi N. Serotype distribution, antimicrobial resistance and prevalence of pilus islets in pneumococci following the use of conjugate vaccines. J. Med. Microbiol. 2017;66:643–650. doi: 10.1099/jmm.0.000479. [DOI] [PubMed] [Google Scholar]
- 39.Pneumococcal Molecular Epidemiology Network. [(accessed on 15 June 2020)]. Available online: https://www.pneumogen.net/pmen/
- 40.Pai R., Gertz R.E., Whitney C.G., Beall B. Clonal Association between Streptococcus pneumoniae Serotype 23A, Circulating within the United States, and an Internationally Dispersed Clone of Serotype 23F. J. Clin. Microbiol. 2005;43:5440–5444. doi: 10.1128/JCM.43.11.5440-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravenscroft N., Omar A., Hlozek J., Edmonds-Smith C., Follador R., Serventi F., Lipowsky G., Kuttel M.M., Cescutti P., Faridmoayer A. Genetic and structural elucidation of capsular polysaccharides from Streptococcus pneumoniae serotype 23A and 23B, and comparison to serotype 23F. Carbohydr. Res. 2017;450:19–29. doi: 10.1016/j.carres.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 42.Sabharwal V., Stevenson A., Figueira M., Orthopoulos G., Trzciński K., Pelton S.I. Capsular switching as a strategy to increase pneumococcal virulence in experimental otitis media model. Microbes Infect. 2014;16:292–299. doi: 10.1016/j.micinf.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Rose M.A., Laurenz M., Sprenger R., Imöhl M., van der Linden M. Nasopharyngeal Carriage in Children After the Introduction of Generalized Infant Pneumococcal Conjugate Vaccine Immunization in Germany. Front. Med. 2021;8:1509. doi: 10.3389/fmed.2021.719481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Golden A.R., Adam H.J., Karlowsky J.A., Baxter M., Nichol K.A., Martin I., Demczuk W., Van Caeseele P., Gubbay J.B., Lefebvre B., et al. Molecular characterization of predominant Streptococcus pneumoniae serotypes causing invasive infections in Canada: The SAVE study, 2011–2015. J. Antimicrob. Chemother. 2018;73:vii20–vii31. doi: 10.1093/jac/dky157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data set for this study is available upon request from the authors.