Abstract
The global demand for oilseeds is increasing along with the human population. The family of Brassicaceae crops are no exception, typically harvested as a valuable source of oil, rich in beneficial molecules important for human health. The global capacity for improving Brassica yield has steadily risen over the last 50 years, with the major crop Brassica napus (rapeseed, canola) production increasing to ~72 Gt in 2020. In contrast, the production of Brassica mustard crops has fluctuated, rarely improving in farming efficiency. The drastic increase in global yield of B. napus is largely due to the demand for a stable source of cooking oil. Furthermore, with the adoption of highly efficient farming techniques, yield enhancement programs, breeding programs, the integration of high-throughput phenotyping technology and establishing the underlying genetics, B. napus yields have increased by >450 fold since 1978. Yield stability has been improved with new management strategies targeting diseases and pests, as well as by understanding the complex interaction of environment, phenotype and genotype. This review assesses the global yield and yield stability of agriculturally important oilseed Brassica species and discusses how contemporary farming and genetic techniques have driven improvements.
Keywords: Brassica, oilseed, mustard, yield, security, genotype and phenotype
1. Introduction
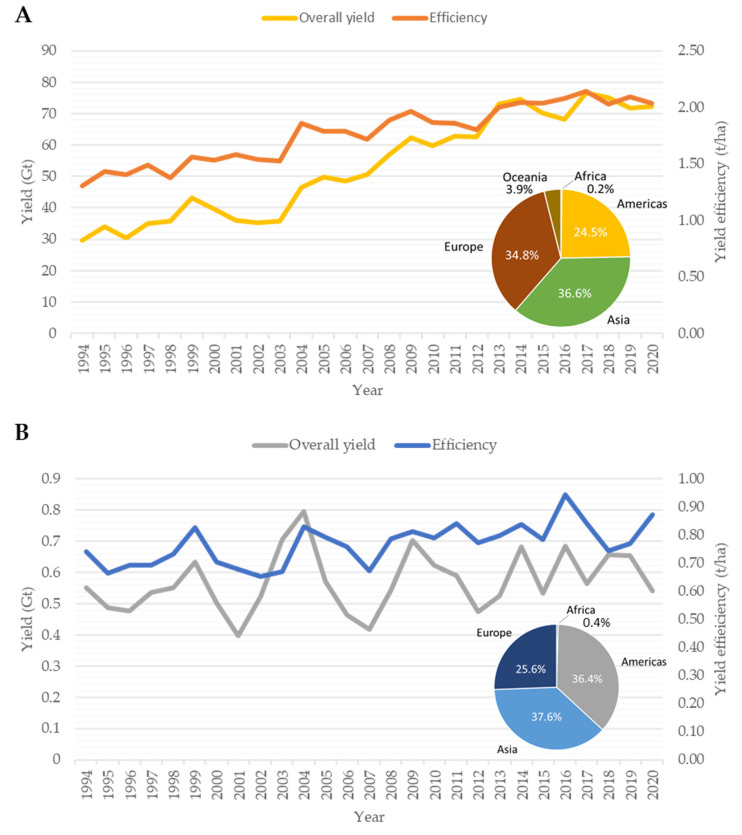
The Brassicaceae family consists of 4636 accepted taxa, divided into 340 genera and 52 tribes [1,2]. Brassica is considered the most important as it contains many of the economically important crops such as oilseeds and condiment varieties, or cruciferous vegetables [3]. Despite the extreme phenotypic variation between Brassica spp. [3,4], only a select few are commonly cultivated. For example, the mustard crops Brassica carinata (Ethiopian mustard), B. nigra (black mustard), B. alba (white mustard) and B.juncea (brown mustard) are typically grown for the production of condiments due to the taste of their oils ranging from sweet to spicy. Whereas B. napus (rapeseed, oilseed rape, canola) is grown as a major source of oilseed for the production of edible vegetable oil [5], as the derived oil has healthy characteristics such as having less than 2% erucic acid and less than 30 µmol g-1 of aliphatic glucosinolates in the meal [6]. Spurred on by the constant need for improvements, breeding programs have developed new varieties with altered oilseed characteristics to fit certain niches. This has in turn provided farmers with a suite of cultivars to choose from, each yielding seeds with different health benefits or industrial uses, for example oilseed with no erucic acid, low levels of gluconisolates, high levels of antioxidants (phenolic compounds), varying vitamin content (C, B9 and K) and the inclusion/exclusion of lutein [7]. Furthermore, with the development of B. napus hybrid lines, such as the dual-purpose winter-hybrid and high stability oil varieties, the annual global production has steadily increased each year [8]. In 1979, the global B. napus crop yield was recorded to be ~154.2 Mt [9], in 1994, the global yield increased to 34.1 Gt, with no commercially available hybrid varieties. In 2020, the global yield was recorded to be 72.37 Gt with the majority being hybrid varieties (Figure 1A) [10]. In contrast to B. napus, the global yield of Brassica mustard crops has been highly variable (Figure 1B). In 1994, the global average yield of mustards was 552.3 Mt, with the following years showing steady increase, until 1999, where global yield plummeted. Thereafter, global yield peaked intermittently at 2004 (796.7 Mt), 2009 (704.1 Mt), 2014 (682.2 Mt) and 2016 (685.9 Mt) (Figure 1B). Over the last 26 years, Brassica mustard crop yield has decreased by ~2.1% indicating the crop may be grown in non-optimal environments or in volatile socio-economic climates [10].
Figure 1.
Global yield, land usage and national contribution summary of major oilseed and mustard Brassica crops. (A) Brassica napus (B) Brassica carinata, Brassica nigra and Brassica alba.
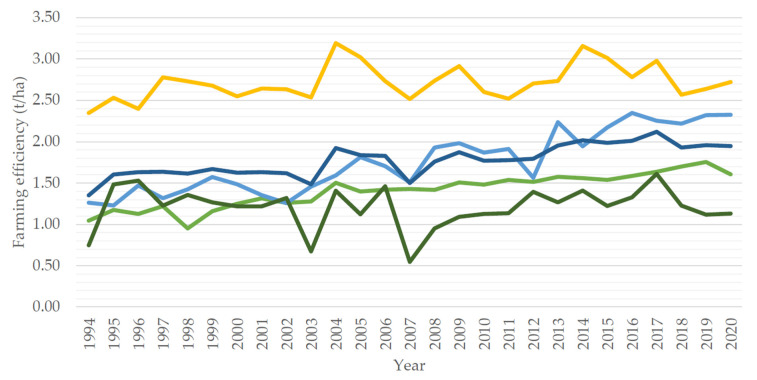
The global production of Brassica mustard crops is expected to increase due to national renewable energy directives being established. For example, B. napus was the only oilseed crop grown in Brazil for decades due to its ability to grow well under the tropical climate, however, with the national drive for production of biodiesel, several B. juncea and B. rapa varieties were found to be better alternatives due to their oilseed properties (such as higher erucic acid content (22:1) [11]. In contrast, B. napus and B. carinata are prioritized as sources of oil for biofuel in the US, UK and the EU [12,13,14] as central Europe is well suited for growing winter varieties of B. napus [8]. For example, in a ten-year average (2010–2020), the EU achieved the highest farming efficiency (yield/area used) of ~2.76 t/ha (Figure 2) globally, whereas Canada and Asia averaged ~2.11 t/ha and 1.59 t/ha, respectively. The main contributor to the difference is climate; the EU has an extended growing period (particularly long day photoperiods) and growing seasons compared to Asia and Canada [15,16].
Figure 2.
Global and national farming efficiency of B. napus oilseed production over 26 years. Yellow—Europe, Light blue—Canada, Dark blue—Global, Light green—Asia, Dark green—Australia.
National socio-economic position and climate are major drivers for yield stability. For example, in 2017, the EU shifted their focus from growing B. napus and instead began relying on imports from Australia and Canada, after both were able to meet the strict EU Renewable Energy Directive greenhouse gas criterion [17]. This led to a significant decrease in production (2.98–2.54 t/ha). However, both drivers can be overcome through by developing new and improved crop management/enhancement programs. China’s production of B. napus maintained a slow and steady increase between 2005 and 2015, thereafter, with the years following (2015–2020) seeing a greater increase in farming efficiency, resulting in the national best farming efficiency of 1.753 t/ha (Figure 2) [10]. This shift indicates a concerted effort towards improving yield from given area farmed, rather than relying on increasing the area farmed for increasing yield due to space limitations.
Several other nations have also begun investing in oilseed enhancement programs. For example, the Canola Council of Canada has projected an overall increase in B. napus yield from ~21 Mt in 2020 to ~25 Mt in 2025 as per their enhancement programs to meet global demand [18,19]. Furthermore, the Australian Field Applied Research (AFAR) initiative showed that Australia is capable of producing B. napus yields in excess of 6 t/ha [20] nearly doubling Europe’s efficiency. This is an amazing feat, as Australia’s production of oilseed crop has consistently been the lowest globally since 2006 due to the extreme climate, such as a short growing season, high temperatures and low rainfall. Climate is a significant constraint and often dictates which crop can be grown. For example, the preferred oilseed crop in India is B. juncea due to consistent rainfall, whereas B. napus is often selected for its drought tolerance [15]. The yield of B. juncea in the region has increased from 0.76 Mt in 1950, to 7.98 Mt in 2020, however, it still does not meet the domestic demand for edible oils.
Yield and yield stability of Brassica oilseed and mustard crops currently relies on the optimization of agronomic practices by enhancement programs, creating new varieties through breeding programs and improving management programs to prevent disease and pest incursions. All these programs have gained significant momentum with the integration of molecular resources such as characterization of germplasm, breeding pedigrees and identification of trait-associated loci in breeding programs. Karim et al. 2014., showed that high yielding, short duration B. napus lines with up to 4.6 times higher yields can be rapidly created when crossed with hybrid Brassica rapa and/or Brassica oleracea [16]. However, targeting a particular phenotypic trait such as yield, or oilseed content is not always straightforward. Crop genotypes do not always correlate with crop phenotype. For example, quantitative disease resistance in B. napus has been shown to improve yield stability by providing enhanced immunity against several pathogens such as Leptosphearia maculans (blackleg), Sclerotinia sclerotium (Sclerotinia stem rot) and Plasmodiophora brassicae (clubroot), however the underlying genes have yet to be identified due to the multi-genic nature of quantitative resistance, despite substantial genomic resources being available such as reference genomes or pangenomes [5,21,22,23,24]. Furthermore, phenotypic traits often dictate regional viability as the main influence of yield security are stressors such as biotic (disease and pests) and abiotic (temperature, salinity, pH, heavy metals and hydration) stress. More emphasis has been applied to targeting biotic stress resistance phenotypes as unmanaged biotic stress leads to yield loss. Annually, B. napus yield is affected by 10–20% loss in UK and Canada and up to 90% in Australia, due to L. maculans, the causal agent of blackleg [25,26,27]. As such, to aid in improving Brassica crop yield and yield security, this review will assess current farming and disease management programs; introduce the gap between phenotype and genotype; discuss the underlying genetics of yield; and discuss studies that have adopted molecular technology to develop new and improved Brassica crops.
Enhancing Crop Yield and Yield Stability with Farming Techniques and Management Plans
Crop yield and yield stability has been influenced by human intervention for centuries. However, in the future, crop yield and stability will become more affected by the consequences of human intervention. Ray et al. 2019., performed an extensive study of ten global crops, including B. napus, relating observed yield to observed weather from 1974–2013 and found that climate change may have already affected canola production. Since the 1970s, the growing season temperature has increased by 1.2 °C and this change has likely affected canola production globally [28]. As a result, the study found that the mean production of canola in western and eastern Europe decreased by 11.4%, North and Central America decreased by 0.4%, whereas in northern Europe, Asia and Oceania it increased by 3.1%, 5.9% and 0.6%, respectively. However, the declining trend of B. napus production does not continue past 2013, with the following years (2013–2020) of global production increasing both in yield (averaging ~72 Gt) (Figure 1A) and efficiency (averaging ~2.06 t/ha, Figure 2) [10]. This is most likely due to the rapidly developing molecular resources available (discussed later in Underlying genetics dictate crop yield), integrated farming strategies and adaptive management programs. Several studies have shown through integrating molecular resources and farming techniques, crop yield, yield stability and yield quality can be secured [29,30,31,32,33].
Environmental conditions are the greatest influencers of biomass accumulation, yield during the growth period [6] and may influence plant immunity [34,35,36,37,38,39]. The critical period is the phase of growth in which abiotic stresses have the greatest influence on yield [40]. Kirkegaard et al. 2018., showed that the critical period for canola is between 100 °Cd to 400 °Cd after the start of flowering. The critical period for the other Brassica oilseed crops is yet to be established. The second major period is the seed-filling period, also known as grain-fill. The environmental conditions during this time have been found to influence seed size and oil content [6]. Furthermore, the harvested yield is dictated by monitoring the seed branches and meristem for colour change; Graham et al. 2017., found that once 60–80% of all pod branches have changed colour the canola should be harvested to achieve maximum yield [41]. Along with time of harvesting, plant density also dictates maximum yield and harvestability [42,43,44,45,46,47,48]. The angle of the lowest branches decreases as row spacing increases for B. napus. As such, not only does high density improve total crop yield, it also improves the actual yield captured through mechanical harvesting [43]. In contrast, in B. carinata, row spacing affected seed and oil yield, branch production and the number of pods per plant [46], implying plant density must be carefully evaluated for climate and oilseed crop used.
Plant density differs by climate. In China, the optimum density for B. napus is 58.5 × 104 plants/ha [45,49]; in Europe, the optimum density is ~80–150 × 104 plants/ha [50,51]; in Canada, the optimum density is ~50–80 × 104 plants/ha [51]; and in Australia (Western Australia), the optimum density is 25–35 × 104 plants/ha [52]. Furthermore, Ming et al. 2017, showed that typically with high density plant trials, traits associated with rapid leaf senescence (green leaf index, chlorophyll content and malondialdehyde accumulation) are increased, seed pod traits (pod area index, pod photosynthesis and radiation use efficiency) were increased and root associated traits (root length, root tips, root surface area and root volume) were decreased.
Lastly, yield security of Brassica oilseed crops is a major concern. Andert et al. 2021., performed a case study of East-German canola farmers and found that on-field data showed yield instability for winter hybrid varieties, which in turn, may cause farmers to begin growing less canola overall [53]. The concern is borne mainly out of future risk from insects and diseases causing irreparable damage to their seasonal yields. As such, disease management strategies are critical for yield stability. A survey of over 100 growers and agronomists established that the best approach towards disease management involved the integration of farming techniques, managing genetic diversity, and careful use of fungicides [25]. Integrated strategies for improving yield with respect to farming techniques has been reviewed extensively [6,25,51]. In summary, maximum grain yield from Brassica oilseed crops is dictated by (1) environmental conditions during the critical period, (2) plant density and (3) disease management strategies; grain quality is dictated by: (1) the environmental conditions during the pod-fill stage and (2) genetic diversity. As such, the complex interaction between genotype, phenotype and the environment together, drive grain yield capacity, quality and stability.
2. Bridging the Genetic and Phenotypic Gap
Determining the underlying genetic mechanisms driving crop yield is a complex task, considering the interactions between genetic (G), environmental (E) and GxE forces that may affect crop performance [54,55,56]. Multi-environment studies can be more powerful in detecting smaller effect quantitative trait loci (QTL) controlling complex traits such as yield, allowing for the identification of QTL of pleiotropic effect or QTL that suffer significant effects due to GxE interaction [54,56,57]. For instance, a study using segregating B. napus populations showed that 81.5% of the QTL linked to yield were pleiotropic [58]. In B. napus, a multi-environment analysis of seed composition traits observed that most QTL suffered from GxE effects, with a major QTL qWIE_N9 associated with seed pigment in over five environments [56]. Another study identified a QTL region on chromosome A09 linked to variation in days to flowering, seed yield and plant height under water limited conditions [58]. However, measuring phenotypic traits of hundreds of plants growing in multiple location breeding trials is costly and time consuming, restricting the number of traits measured and frequency of measurements through crop development.
High-throughput phenotyping (HTP) has emerged as an alternative to manual phenotypic trait measurement, accelerating the process of collecting phenotype information through remote and proximal sensors to measure a variety of plant traits [59,60,61]. HTP sensors can be deployed on stationary platforms at a greenhouse or attached to ground and aerial vehicles in the field. These platforms enable the non-destructive measurement of phenotypic traits, which can be used to identify genetic variants associated with improved crop performance and tolerance to environmental stress [62,63,64]. The use of HTP platforms is less labor-intensive than manually measuring traits and prevents the introduction of biases due to human error or sampling methods [65]. Several international groups have invested in the development of HTP centres that either provide the structure for the collection of HTP datasets or publishes their datasets, such as the Australian Plants Phenomics Facility, International Maize and Wheat Improvement Center (CIMMYT), the Genomes to Fields Initiative and TERRA-REF [66,67]. For instance, the TERRA-REF project collects multiple image types of the plants throughout their development along with environmental data, agronomic information and genomic sequences of hundreds of plant varieties [67]. The in-depth measurements of plant development through HTP offered by TERRA-REF offers a great opportunity to uncover genes linked to crop yield and a better understanding of their combined impact on plant response to environmental conditions. Nonetheless, even less comprehensive HTP datasets offer an advantage over manual phenotype measurement methods as the images collected can be re-analysed and shared with other researchers for further investigation [68].
A wide range of sensors is available for monitoring specific plant traits under field or controlled conditions such as RGB, multispectral and hyperspectral cameras, infrared thermal or LiDAR sensors. Infrared thermography images of the plant canopy contribute to determining crop water stress [69] and assist in identifying QTL associated with stomatal density and canopy temperature in Setaria [70] https://sciwheel.com/work/citation?ids=13155099&pre=&suf=&sa=0, accessed on 4 September 2022. Multispectral and hyperspectral cameras have been widely employed to obtain quantitative measurements of the canopy reflectance throughout plant development, supporting the identification of potassium deficiency and green peach aphid susceptibility [71], classification of fungal infection severity in B. napus seeds [72] and seed pod maturity [73]. Besides tracking specific crop traits that would be difficult to track manually, such as measuring canola flower numbers in the field to predict yield [74,75], HTP platforms can substantially expand the temporal resolution and number of traits monitored to identify genetic variation linked to increased crop performance. For instance, an HTP study using weekly images of maize identified candidate genes associated with regulating plant architecture at early development [76]. Similarly, daily HTP measurements of 477 B. napus genotypes revealed multiple medium and small effect QTL were associated with early plant growth, most of which were active during short phases of the development [77]. Dynamic QTLs were also observed in a B. napus trial that monitored 43 traits across twelve time points, reporting that only 35% of the QTL identified were present on all time points [78]. These studies indicate the need for stage-specific investigations that uncover transient QTL that may play a role in the plant’s early vigour [79].
HTP platforms have the potential to provide ample information regarding the plant phenotype; however, efficient data collection and processing are considered a key constraint for breeding [60,80]. Image data collection and processing protocol must be carefully designed to avoid biases due to lightning and other environmental conditions, mainly if the measured phenotypic traits are based on the tissue spectral reflectance [81,82]. In addition, there is a need to adapt conventional GWAS and GS methods or implement machine/deep learning models to incorporate the rich information from HTP into the genotype selection pipeline [60]. Machine/deep learning currently presents competitive results for predicting phenotypic traits based on genomic data for GS [83], multi-trait and multi-environment prediction [84,85]. Machine/deep learning has the advantage of automatically extracting features from complex data, building an abstract representation of their relationship regarding the prediction target, which is particularly suited to image dataset analysis [86]. Recent studies have applied machine/deep learning to integrate HTP, environmental and genetic data for selecting varieties under field trial. For example, a study on wheat used generalized Poisson regression, a statistical machine learning method, to merge hyperspectral images with environmental and genetic data to predict count phenotypes for GS [87]. Another study used canopy temperature and vegetation indices for GS of wheat, highlighting that adding the HTP features increased the yield prediction by 70% in genomic models [88]. In maize, it was shown that a multimodal deep learning model using multispectral images and genomic data could accurately identify 75% high yield plots at an early developmental stage [89]. The use of HTP also allows monitoring of the phenotypic traits during the plant development under multi-environment trials, helping the identification of varieties better adapted to the changing environmental conditions. Leveraging phenotypic and genomic datasets using the appropriate analysis tools have the potential to accelerate the breeding of higher-yielding B. napus varieties. Observing the impact of genomic variation on crop yield can be facilitated by the use of HTP, but a comprehensive database on B. napus genetic resources is required to design future resilient crops.
3. Underlying Genetics Dictate Plant Yield
In order to improve yield in plants, one must study the underlying genetic mechanisms and related mechanisms, such as plant architecture. Plant architecture (PA) refers to the three-dimensional organisation of the plant, including its morphological characteristics [90,91]. PA modifications are fundamental for high-yield breeding, often linked with a crop’s adaptive ability and yield potential, such as seed oil content [92], silique number, number of seeds per silique and seed weight [93]. Additionally, traits such as plant height, biomass yield and flowering time indirectly influence seed yield [94]. There have been many studies in B. napus linking PA with yield. For example, a gene knockout experiment showed that a stop codon mutation in the meristem identity gene APETALA1 affected flower morphology, PA and yield [95]. Additionally, a study in B. rapa showed plant yield was significantly correlated with PA-related traits such as main inflorescence length, branch height and branch segment [96].
Identifying yield-related traits has been the focus of many genetic studies in B. napus. These studies primarily focus on identifying QTL and single nucleotide polymorphisms (SNPs) related to yield and PA. For example, genetic mapping of different genotypes of B. napus and genetic mapping identified 190 PA-related genes for 91 unique PA QTL and 2350 yield loci-pairs [96]. In a separate study, a map comprising 7716 DArTseq markers was created from a population of 145 B. napus lines and identified 20 QTL associated with flowering time and grain yield. Twenty-two putative candidate genes for flowering time and grain yield were identified in the QTL region [97]. Raboanatahiry et al. 2018., aligned 972 QTL for seed-yield and yield-related traits in B. napus onto one genetic map and identified 92 regions where 198 QTL overlapped. The study showed that the regions identified could be used to select for desired traits. Additionally, 147 candidate genes potentially influencing PA and yield were identified [94]. A SNP array-based genetic map was used to identify 695 QTL for 14 traits, including PA, flowering, silique and other seed-related traits, in B. juncea. It also showed that epistasis among loci plays an important role in controlling heterosis in yield of B. juncea [98].
Many studies in B. napus have used GWAS to identify yield-related genetic variants. Using data from 520 B. napus accessions, GWAS for seven yield-determining traits; main inflorescence pod number, branch pod number, pod number per plant, seed number per pod, thousand seed weight, main inflorescence yield, and branch yield, identified 128 SNPs and 14 candidate genes for yield improvement [99]. GWAS was also used to identify candidate genes associated with stress tolerance, oil content, seed quality and ecotype improvement for 588 accessions of B. napus [100]. Another study used genotyping by sequencing to screen 125 accessions of B. napus and identified 85,126 SNPs for GWAS, directly associating 18 SNPs with seed yield and another 61 SNPs with yield-related traits [101]. By understanding the genetic control of PA in crops, more efficient breeding strategies to improve crop yield can be developed [102].
A pangenome is the collection of all genes within a species, first coined by Tettlin et al. in 2005 to describe the gene diversity in Streptococcus agalactiae [103]. Pangenomes consist of a core genome, containing the sequences shared between all individuals of a species, and the accessory genome (also known as the dispensable or variable genome), which contains the genes that are not found in all individuals. In the last couple of decades, pangenomes have been constructed for various bacteria, fungi, animals and plants, including B. oleracea [104], B. rapa [105] and B. napus [106]. Pangenomes can be assembled in one of three ways: de novo sequencing and comparison; iterative mapping and assembly; and graph-based assembly. These methods have been extensively covered in other reviews, detailing the construction, benefits and disadvantages of each method [107,108].
Unlike single reference genomes, pangenomes allow the capture of sequences affected by structural variation such as presence/absence variation (PAVs) or copy number variations (CNVs), that may affect agronomically important traits such as disease resistance and yield [109,110]. In B. oleracea, use of the pangenome has identified multiple genes coding for resistance against other abiotic factors such as drought [111]. Similarly, pangenomes have identified important presence/absence variations (PAVs) in the Brassica genus. In B. oleracea and Brassica macrocarpa, a pangenome was used to identify PAVs associated with disease resistance, secondary metabolites and flowering time [104]. In B. napus, the pangenome identified PAVs associated with flowering time, silique length, seed weight and flowering time [106] as well as PAVs and SNPs associated with disease resistance [112,113]. In B. rapa, a pangenome was used to identify PAVs associated with flowering time, stress resistance and lignin formation [114]. Pangenomes can be used for further study of QTL and SNPs by acting as detailed references for trait-mapping tools such as GWAS, allowing for improved studies of genetic variation.
Another way to study candidate genes in Brassicas is to construct pangenomes based on specific functional traits. Trait pangenomes aim to describe the landscape of genetic variation related to a trait and investigate the impact of genetic variation. Trait-specific pangenomes have been used to study resistance gene analogs in B. oleracea and B. napus [111,113,115] and have been employed as a reference for resistance gene cloning [116]. Trait pangenomes could help in further dissecting the genetic variability associated with yield under certain conditions, such as low phosphorous deficiency in B. napus [117]. Pangenomes have only been introduced to Brassica research recently and use of pangenomics in Brassica breeding is still in its infancy. However, further understanding of the genetics underlying variation can lead to the development of molecular markers that can be used to predict the location of desirable crop traits and marker-assisted breeding for improvement of crop yield in Brassicas.
4. Developing New Phenotypes via Genome Editing Technology
Genome editing presents an opportunity to rapidly introduce or manipulate specific traits of interest that may not be present in the existing gene pool of elite crop varieties, or that may be difficult and time consuming to introduce through traditional introgression breeding approaches. The advent of clustered regularly interspaced short palindromic repeats systems associated with Cas enzymes (CRISPR/Cas) has opened the doors for widespread editing in oilseed Brassicas, which greatly benefit from sharing homologs of well characterised genes associated with yield in Arabidopsis. However, the high gene homology and copy number of the allotetraploid Brassicas, such as B. napus and B. juncea, present a challenge in terms of editing accuracy, in particular single base editing [118], and in gene redundancy via multiple homologs, requiring all genes homologous to the target to be modified to produce a reliable phenotype [119]. Polygenic traits will require a more intricate multiplex approach, targeting more than one locus in a single round of editing. Yield is one such trait that is complex and will likely benefit from a multiplex approach in order to achieve rapid improvement [120]. Although hybrid-CRISPR Cas enzymes have been designed for targeting multiple loci for inactivation or activation [121,122], the technology has yet to be applied in Brassica. In Brassica oilseeds, yield remains poorly characterised on the molecular level, and knowledge of the genetic mechanisms controlling seed size and oil content is scarce [120]. Hence, many of the traits currently targeted for yield improvement are those that are closely related to yield, or those that are indirectly related, but highly correlated. For example, current research has mainly focused on increasing seed number through changes to silique structure and improving harvestability through a reduction in plant heigh and an increase in plant density [118]. In addition, other traits such as pod shattering resistance [123,124] and flowering time [125], which significantly influence yield, have been investigated.
Not surprisingly, B. napus has received the most attention of all the oilseed Brassicas when it comes to the use of editing for yield improvement. Yang et al. were the first to examine multilocular siliques in B. napus using CRISPR/Cas9 and introduced mutations to CLV pathway genes, resulting in an increased number of multilocular siliques. Multilocular siliques were found to contain more seeds than regular bilocular siliques, and seed weight per silique was increased by 74% in the mutants [126]. A more recent study induced mutations in BnD14, a strigolactone receptor, to change architectural traits in B. napus relating to yield [127]. Knockout lines for BnD14 displayed dwarfed phenotypes, which are easier to harvest, with prolific branching and nearly 40% more flowers than their WT counterparts. To combat pod shattering, Zaman et al., knocked out homeologs of SHATTERPROOF1/2, a class of genes involved in the pod dehiscence regulatory pathway, creating a transgene-free line that was approximately 10 times more resistant to pod shattering when measured on the pod-shattering resistance index [127]. These examples represent a growing list of studies (list shown in Table 1) that successfully exploit genome editing via CRISPR systems to functionally characterise and manipulate key genes associated with yield or yield-related traits in B. napus. Although examples of editing for yield improvement in B. juncea are yet to be seen, several potential targets have been identified and characterised, such as genes involved with multilocular silique development [128,129], and provide a guide for future editing efforts.
Table 1.
Recent examples of yield-related trait improvement using CRISPR/Cas9 in Brassica napus.
| Trait | Target Gene | Arabidopsis Homolog | Mutant Phenotype | Reference |
|---|---|---|---|---|
| Flowering time | BnaSVP | Short Vegetative Phase (SVP) | 40–50% decrease in time until flowering | [133] |
| Plant height, internode length and number of branches | BnD14 | DWARF14 (D14) | 34% reduction in plant height, 200% increase in branch number and 37% increase in total flowers per plant | [127] |
| Plant height and branch angle | BnaA03.BP | BREVIPEDICELLUS (BP) | ~16% reduction in plant height and branch angle reduced from 84° to 14° | [134] |
| Pod shattering resistance | BnSHP1/BnSHP2 homeologs | SHATTERPROOF1/2 | ~10 times more resistant to pod shattering | [135] |
| Number of seeds per silique | BnaEOD3 | ENHANCER3 OF DA1 (EOD3) | Shorter silique length and smaller seeds, but 42% increase of number of seeds per silique | [136] |
| Pod shattering resistance | BnJAG.A02, BnJAG.C02, BnJAG.C06, BnJAG.A07, BnJAG.A08 | JAGGED (JAG) | 2 times more resistant to pod shattering | [123] |
| Multilocular silique development |
BnA04.CLV3, BnC04.CLV3, BnC02.CLV3 |
CLAVATA3 (CLV3) | 74% increase in seed weight per silique | [126] |
| Plant height, primary branch number and silique number | BnaMAX1 | More Axillary Growth (MAX) | ~35% reduction in plant height, 3 times more primary branches and ~65% increase in total silique number | [137] |
As we start to understand the complex mechanisms underpinning yield and identify yield-related genes in oilseed Brassicas, exponentially improving their yield through genome editing is only just out of reach. Advances in CRISPR systems in terms of simplicity and accuracy is a driving force propelling CRISPR into the mainstream. Novel CRISPR techniques, such as the recently proposed and highly sophisticated CRISPR-Combo system, which simultaneously enables gene editing and gene activation at multiple loci, will likely spearhead the race to high-yielding genome edited crops [122], as well as DNA-free gene editing technology which has been dubbed ‘the way of true plant editing’ [130,131,132]. These systems will become necessary in future for the development of new and improved crops that have improved yield and yield stability.
5. Conclusions
Crop yield and yield stability have been the focus of agriculture for thousands of years. However, with realization of climate change and the predicted increase in world population by 2050, concerted efforts are necessary to meet growing demands. As an example of this demand, B. napus production has increased from 154.2 Mt in 1979 to 72.37 Gt in 2020 and is now only second to soybean as a source of oilseed. Although primarily grown as a source of vegetable oil or mustard oil, B. napus (and to a lesser extent B. carinata), B. juncea and B. rapa have also been identified as excellent sources of biofuel, applying more pressure for studies to maximise yields. The majority of (if not all) studies that are associated with maximising yield involve genetics and/or gene-editing technology as it is a proven and rapid method for yield improvement. The global FE (t/ha) of B. napus pre-genetics age (<2003, before the sequencing of the human genome) was 1.48 t/ha, compared to now (2004–2020) which is 1.94 t/ha. Despite the adoption of genetic resources, the global yield of Brassica crops is insufficient for current and future demands, in particular mustard crops, however, this is more likely due to yield instability. An investigation of global and national FE clearly shows fluctuations for both B. napus and Brassica mustard crops in recent years, which can cause repercussions towards the supply chain of these crops, for example, farmers losing confidence in their crops. As such, research studies investigating the influence of environment, phenotype and genotype on crop yield and yield stability are required to produce reliable crops for farmers to meet demands.
Studies have shown that environmental conditions are the greatest influencers of biomass accumulation, furthermore, climate dictates the particular genotypes and phenotypes that can thrive in any one region. High-throughput phenotyping technology allows for the rapid analysis and characterization of new and potential Brassica cultivars of their ability to grow well in a region. As an alternative to manual phenotyping, HTP technology accelerates the process of collecting phenotype information through remote and proximal sensors to measure a variety of plant traits all of which are catalogued and integrated. Coupled to deep/machine learning technology, HTP can quickly establish the optimal phenotype (highest yield) for the region. However, HTP is limited by the availability of genomic resources, such as QTL, GWAS and SLAF-seq data associated with plant architecture. As such, genetic studies are critical. However, the underlying genes that are associated with phenotypic variations exhibited by plants are great and nearly impossible to identify by genomics alone. Hence, pangenomic analysis has been developed as a strategy to capture the genetic diversity to bridge the gap between genotype to phenotype. Many Brassica crops have pangenomes available such as B. napus, B. rapa and B. oleracea, providing unique insights into DNA polymorphisms, structural variation and patterns for genome duplication. The combination of genomics/pangenomics and phenotypic studies has enhanced the prediction of molecular markers associated with specific crop traits for crop yield and yield stability. Furthermore, like HTP technology leading to improved phenotyping capabilities; gene-editing technology has enhanced genotyping by rapidly characterizing yield-associated candidate genes and creating new genotypes. It is predicted that DNA-free gene-editing technology will lead to the development of non-GMO plant genotypes that can rapidly be produced and adopted by farmers.
The combination of environment, phenotype and genotype dictate yield and yield stability. As such, developing methodologies to understand these factors and their complex interactions are critical for ensuring future demand is met. This review shows how farming practice and management programs, HTP, genomics, pangenomics and gene editing technology have influenced the production and security of Brassica oilseed and mustard crops. Only through continued improvement and innovation of these resources can yield increase and be secured.
Acknowledgments
W.J.W.T would like to acknowledge the support of the Grains Research and Development Corporation.
Author Contributions
Draft manuscript was written by J.D.Z., C.T.F., M.F.D. and W.J.W.T. Editing and finalisation of the manuscript was done by all aforementioned authors, D.E. and J.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This study did not produce any data, however, any data described in ‘Introduction’ can be found in FAOSTAT (https://www.fao.org/faostat/en/#home, accessed on 4 September 2022).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Australian Research Council projects DP200100762, and DP210100296 and the Grains Research and Development Corporation (UWA1905-006RTX).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Francis A., Lujan-Toro B.E., Warwick S.I., Macklin J.A., Martin S.L. Update on the Brassicaceae species checklist. Biodivers. Data J. 2021;9:e58773. doi: 10.3897/BDJ.9.e58773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen H., Wang T., He X., Cai X., Lin R., Liang J., Wu J., King G., Wang X. BRAD V3. 0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022;50:D1432–D1441. doi: 10.1093/nar/gkab1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng F., Wu J., Wang X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014;1:14024. doi: 10.1038/hortres.2014.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng F., Sun R., Hou X., Zheng H., Zhang F., Zhang Y., Liu B., Liang J., Zhuang M., Liu Y. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016;48:1218–1224. doi: 10.1038/ng.3634. [DOI] [PubMed] [Google Scholar]
- 5.Neik T.X., Amas J., Barbetti M., Edwards D., Batley J. Understanding host–pathogen interactions in Brassica napus in the omics era. Plants. 2020;9:1336. doi: 10.3390/plants9101336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirkegaard J.A., Lilley J.M., Berry P.M., Rondanini D.P. Crop Physiology Case Histories for Major Crops. Elsevier; Amsterdam, The Netherlands: 2021. Canola; pp. 518–549. [Google Scholar]
- 7.Farnham M., Wilson P., Stephenson K., Fahey J. Genetic and environmental effects on glucosinolate content and chemoprotective potency of broccoli. Plant Breed. 2004;123:60–65. doi: 10.1046/j.0179-9541.2003.00912.x. [DOI] [Google Scholar]
- 8.Wittkop B., Snowdon R., Friedt W. Status and perspectives of breeding for enhanced yield and quality of oilseed crops for Europe. Euphytica. 2009;170:131–140. doi: 10.1007/s10681-009-9940-5. [DOI] [Google Scholar]
- 9.Shahidi F. Canola and Rapeseed. Springer; Berlin/Heidelberg, Germany: 1990. Rapeseed and canola: Global production and distribution; pp. 3–13. [Google Scholar]
- 10.FAOSTAT Food and Agriculture Organization of the United Nations. 2022. [(accessed on 7 July 2022)]. Available online: https://www.fao.org/faostat/en/#home.
- 11.Bassegio D., Zanotto M.D. Growth, yield, and oil content of Brassica species under Brazilian tropical conditions. Bragantia. 2020;79:203–212. doi: 10.1590/1678-4499.20190411. [DOI] [Google Scholar]
- 12.Song X., Wei Y., Xiao D., Gong K., Sun P., Ren Y., Yuan J., Wu T., Yang Q., Li X. Brassica carinata genome characterization clarifies U’s triangle model of evolution and polyploidy in Brassica. Plant Physiol. 2021;186:388–406. doi: 10.1093/plphys/kiab048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ban Y., Khan N.A., Yu P. Nutritional and metabolic characteristics of Brassica carinata co-products from biofuel processing in dairy cows. J. Agric. Food Chem. 2017;65:5994–6001. doi: 10.1021/acs.jafc.7b02330. [DOI] [PubMed] [Google Scholar]
- 14.Taylor D.C., Falk K.C., Palmer C.D., Hammerlindl J., Babic V., Mietkiewska E., Jadhav A., Marillia E.F., Francis T., Hoffman T. Brassica carinata—A new molecular farming platform for delivering bio-industrial oil feedstocks: Case studies of genetic modifications to improve very long-chain fatty acid and oil content in seeds. Biofuels Bioprod. Biorefin. 2010;4:538–561. doi: 10.1002/bbb.231. [DOI] [Google Scholar]
- 15.Jat R., Singh V., Sharma P., Rai P. Oilseed brassica in India: Demand, supply, policy perspective and future potential. OCL. 2019;26:8. doi: 10.1051/ocl/2019005. [DOI] [Google Scholar]
- 16.Karim M.M., Siddika A., Tonu N.N., Hossain D.M., Meah M.B., Kawanabe T., Fujimoto R., Okazaki K. Production of high yield short duration Brassica napus by interspecific hybridization between B. oleracea and B. rapa. Breed. Sci. 2014;63:495–502. doi: 10.1270/jsbbs.63.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tunbridge K., Goddard N., Eady S. ‘Green’ canola secures $1 billion EU trade, in Ground Cover. 2018, Grains Research and Development Corporation. GroundCover™ Issue: 133 March–April 2018. [(accessed on 4 September 2022)]. Available online: https://grdc.com.au/resources-and-publications.
- 18.Rashid M.H., Liban S.H., Zhang X., Parks P.S., Borhan H., Fernando W.G. Comparing the effectiveness of R genes in a 2-year canola—Wheat rotation against Leptosphaeria maculans, the causal agent of blackleg disease in Brassica species. Eur. J. Plant Pathol. 2022;163:573–586. doi: 10.1007/s10658-022-02498-7. [DOI] [Google Scholar]
- 19.Morrison M., Harker K., Blackshaw R., Holzapfel C., O’donovan J. Canola yield improvement on the Canadian Prairies from 2000 to 2013. Crop Pasture Sci. 2016;67:245–252. doi: 10.1071/CP15348. [DOI] [Google Scholar]
- 20.GRDC Hyper Yielding Canola Breaks 5 Tonne Yield Target. Grains Research & Development Corporation. 2022. South Australia. [(accessed on 28 June 2022)]. Available online: https://groundcover.grdc.com.au/crops/oilseeds/hyper-yielding-canola-breaks-5-tonne-yield-target.
- 21.Amas J., Anderson R., Edwards D., Cowling W., Batley J. Status and advances in mining for blackleg (Leptosphaeria maculans) quantitative resistance (QR) in oilseed rape (Brassica napus) Theor. Appl. Genet. 2021;134:3123–3145. doi: 10.1007/s00122-021-03877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubbard M., Peng G. Quantitative resistance against an isolate of Leptosphaeria maculans (blackleg) in selected Canadian canola cultivars remains effective under increased temperatures. Plant Pathol. 2018;67:1329–1338. doi: 10.1111/ppa.12832. [DOI] [Google Scholar]
- 23.Raman R., Taylor B., Marcroft S., Stiller J., Eckermann P., Coombes N., Rehman A., Lindbeck K., Luckett D., Wratten N. Molecular mapping of qualitative and quantitative loci for resistance to Leptosphaeria maculans causing blackleg disease in canola (Brassica napus L.) Theor. Appl. Genet. 2012;125:405–418. doi: 10.1007/s00122-012-1842-6. [DOI] [PubMed] [Google Scholar]
- 24.Wagner G., Laperche A., Lariagon C., Marnet N., Renault D., Guitton Y., Bouchereau A., Delourme R., Manzanares-Dauleux M.J., Gravot A. Resolution of quantitative resistance to clubroot into QTL-specific metabolic modules. J. Exp. Bot. 2019;70:5375–5390. doi: 10.1093/jxb/erz265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van de Wouw A.P., Marcroft S.J., Sprague S.J., Scanlan J.L., Vesk P.A., Idnurm A. Epidemiology and management of blackleg of canola in response to changing farming practices in Australia. Australas. Plant Pathol. 2021;50:137–149. doi: 10.1007/s13313-020-00767-9. [DOI] [Google Scholar]
- 26.Fitt B.D., Brun H., Barbetti M., Rimmer S. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Sustainable strategies for managing Brassica napus (oilseed rape) resistance to Leptosphaeria maculans (phoma stem canker) 2006. [(accessed on 4 September 2022)]. pp. 3–15. Available online: https://repository.rothamsted.ac.uk/
- 27.Zhang X., Fernando W. Insights into fighting against blackleg disease of Brassica napus in Canada. Crop Pasture Sci. 2018;69:40–47. doi: 10.1071/CP16401. [DOI] [Google Scholar]
- 28.Ray D.K., West P.C., Clark M., Gerber J.S., Prishchepov A.V., Chatterjee S. Climate change has likely already affected global food production. PLoS ONE. 2019;14:e0217148. doi: 10.1371/journal.pone.0217148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ichihashi Y., Date Y., Shino A., Shimizu T., Shibata A., Kumaishi K., Funahashi F., Wakayama K., Yamazaki K., Umezawa A., et al. Multi-omics analysis on an agroecosystem reveals the significant role of organic nitrogen to increase agricultural crop yield. Proc. Natl. Acad. Sci. USA. 2020;117:14552–14560. doi: 10.1073/pnas.1917259117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wassermann B., Rybakova D., Müller C., Berg G. Harnessing the microbiomes of Brassica vegetables for health issues. Sci. Rep. 2017;7:17649. doi: 10.1038/s41598-017-17949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scossa F., Alseekh S., Fernie A.R. Integrating multi-omics data for crop improvement. J. Plant Physiol. 2021;257:153352. doi: 10.1016/j.jplph.2020.153352. [DOI] [PubMed] [Google Scholar]
- 32.Lavarenne J., Guyomarc’h S., Sallaud C., Gantet P., Lucas M. The spring of systems biology-driven breeding. Trends Plant Sci. 2018;23:706–720. doi: 10.1016/j.tplants.2018.04.005. [DOI] [PubMed] [Google Scholar]
- 33.Snowdon R.J., Iniguez Luy F.L. Potential to improve oilseed rape and canola breeding in the genomics era. Plant Breed. 2012;131:351–360. doi: 10.1111/j.1439-0523.2012.01976.x. [DOI] [Google Scholar]
- 34.Kissoudis C., Chowdhury R., van Heusden S., van de Wiel C., Finkers R., Visser R.G., Bai Y., van der Linden G. Combined biotic and abiotic stress resistance in tomato. Euphytica. 2015;202:317–332. doi: 10.1007/s10681-015-1363-x. [DOI] [Google Scholar]
- 35.Sewelam N., Oshima Y., Mitsuda N., Ohme-Takagi M. A step towards understanding plant responses to multiple environmental stresses: A genome-wide study. Plant Cell Environ. 2014;37:2024–2035. doi: 10.1111/pce.12274. [DOI] [PubMed] [Google Scholar]
- 36.Saijo Y., Loo E.P.i. Plant immunity in signal integration between biotic and abiotic stress responses. New Phytol. 2020;225:87–104. doi: 10.1111/nph.15989. [DOI] [PubMed] [Google Scholar]
- 37.Ku Y.-S., Sintaha M., Cheung M.-Y., Lam H.-M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2018;19:3206. doi: 10.3390/ijms19103206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dresselhaus T., Hückelhoven R. Biotic and Abiotic Stress Responses in Crop Plants. Multidisciplinary Digital Publishing Institute; Basel, Switzerland: 2018. [Google Scholar]
- 39.Zhou J.-M., Zhang Y. Plant immunity: Danger perception and signaling. Cell. 2020;181:978–989. doi: 10.1016/j.cell.2020.04.028. [DOI] [PubMed] [Google Scholar]
- 40.Robertson D.W. Studies on the critical period for applying irrigation water to wheat. 1934. [(accessed on 4 September 2022)]. Available online: https://agris.fao.org/agris-search/search.do?recordID=US201300674080.
- 41.Graham R., Jenkins L., Hertel K., Brill R., McCaffery D., Graham N. Re-evaluating seed colour change in canola to improve harvest management decisions. In Proceedings of the “Doing More with Less” 18th Australian Agronomy Conference 2017, Ballarat, VIC, Australia, 24–28 September 2017. Australian Society of Agronomy Inc. [(accessed on 4 September 2022)]; Available online: https://www.dpi.nsw.gov.au/__data/assets/pdf_file/0003/1258851/SRR20-web.pdf.
- 42.Siles L., Hassall K.L., Sanchis-Gritsch C., Eastmond P.J., Kurup S. Uncovering the ideal plant ideotype for maximising seed yield in Brassica napus. bioRxiv. 2020 doi: 10.1101/2020.12.04.411371. [DOI] [Google Scholar]
- 43.Kuai J., Sun Y., Zuo Q., Huang H., Liao Q., Wu C., Lu J., Wu J., Zhou G. The yield of mechanically harvested rapeseed (Brassica napus L.) can be increased by optimum plant density and row spacing. Sci. Rep. 2015;5:18835. doi: 10.1038/srep18835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian C., Zhou X., Liu Q., Peng J., Zhang Z., Song H., Ding Z., Zhran M.A., Eissa M.A., Kheir A.M. Increasing yield, quality and profitability of winter oilseed rape (Brassica napus) under combinations of nutrient levels in fertiliser and planting density. Crop Pasture Sci. 2020;71:1010–1019. doi: 10.1071/CP20328. [DOI] [Google Scholar]
- 45.Li M., Naeem M.S., Ali S., Zhang L., Liu L., Ma N., Zhang C. Leaf senescence, root morphology, and seed yield of winter oilseed rape (Brassica napus L.) at varying plant densities. BioMed Res. Int. 2017;2017:8581072. doi: 10.1155/2017/8581072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulvaney M.J., Leon R.G., Seepaul R., Wright D.L., Hoffman T.L. Brassica carinata seeding rate and row spacing effects on morphology, yield, and oil. Agron. J. 2019;111:528–535. doi: 10.2134/agronj2018.05.0316. [DOI] [Google Scholar]
- 47.Li X., Li Z., Xie Y., Wang B., Kuai J., Zhou G. An improvement in oilseed rape (Brassica napus L.) productivity through optimization of rice-straw quantity and plant density. Field Crops Res. 2021;273:108290. doi: 10.1016/j.fcr.2021.108290. [DOI] [Google Scholar]
- 48.Siles L., Hassall K.L., Sanchis Gritsch C., Eastmond P.J., Kurup S. Uncovering Trait Associations Resulting in Maximal Seed Yield in Winter and Spring Oilseed Rape. Front. Plant Sci. 2021;12:1901. doi: 10.3389/fpls.2021.697576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leach J.E., Stevenson H.J., Rainbow A.J., Mullen L.A. Effects of high plant populations on the growth and yield of winter oilseed rape (Brassica napus) J. Agric. Sci. 1999;132:173–180. doi: 10.1017/S0021859698006091. [DOI] [Google Scholar]
- 50.Rathke G.W., Behrens T., Diepenbrock W. Integrated nitrogen management strategies to improve seed yield, oil content and nitrogen efficiency of winter oilseed rape (Brassica napus L.): A review. Agric. Ecosyst. Environ. 2006;117:80–108. doi: 10.1016/j.agee.2006.04.006. [DOI] [Google Scholar]
- 51.Zheng M., Terzaghi W., Wang H., Hua W. Integrated strategies for increasing rapeseed yield. Trends Plant Sci. 2022;27:742–745. doi: 10.1016/j.tplants.2022.03.008. [DOI] [PubMed] [Google Scholar]
- 52.French R.J., Seymour M., Malik R.S. Plant density response and optimum crop densities for canola (Brassica napus L.) in Western Australia. Crop Pasture Sci. 2016;67:397–408. doi: 10.1071/CP15373. [DOI] [Google Scholar]
- 53.Andert S., Ziesemer A., Zhang H. Farmers’ perspectives of future management of winter oilseed rape (Brassica napus L.): A case study from north-eastern Germany. Eur. J. Agron. 2021;130:126350. doi: 10.1016/j.eja.2021.126350. [DOI] [Google Scholar]
- 54.Zhang Y., Li Y.-X., Wang Y., Liu Z.-Z., Liu C., Peng B., Tan W.-W., Wang D., Shi Y.-S., Sun B.-C., et al. Stability of QTL Across Environments and QTL-by-Environment Interactions for Plant and Ear Height in Maize. Agric. Sci. China. 2010;9:1400–1412. doi: 10.1016/S1671-2927(09)60231-5. [DOI] [Google Scholar]
- 55.Xie Y., Xu J., Tian G., Xie L., Xu B., Liu K., Zhang X. Unraveling yield-related traits with QTL analysis and dissection of QTL × environment interaction using a high-density bin map in rapeseed (Brassica napus. L) Euphytica. 2020;216:171. doi: 10.1007/s10681-020-02708-5. [DOI] [Google Scholar]
- 56.Yu B., Boyle K., Zhang W., Robinson S.J., Higgins E., Ehman L., Relf-Eckstein J.-A., Rakow G., Parkin I.A.P., Sharpe A.G., et al. Multi-trait and multi-environment QTL analysis reveals the impact of seed colour on seed composition traits in Brassica napus. Mol. Breed. 2016;36:111. doi: 10.1007/s11032-016-0521-8. [DOI] [Google Scholar]
- 57.Alimi N.A., Bink M.C.A.M., Dieleman J.A., Magán J.J., Wubs A.M., Palloix A., van Eeuwijk F.A. Multi-trait and multi-environment QTL analyses of yield and a set of physiological traits in pepper. TAG Theor. Appl. Genet. 2013;126:2597–2625. doi: 10.1007/s00122-013-2160-3. [DOI] [PubMed] [Google Scholar]
- 58.Raman H., Raman R., Pirathiban R., McVittie B., Sharma N., Liu S., Qiu Y., Zhu A., Kilian A., Cullis B., et al. Multienvironment QTL analysis delineates a major locus associated with homoeologous exchanges for water-use efficiency and seed yield in canola. Plant Cell Environ. 2022;45:2019–2036. doi: 10.1111/pce.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hall R.D., D’Auria J.C., Ferreira A.C.S., Gibon Y., Kruszka D., Mishra P., Van de Zedde R. High-throughput plant phenotyping: A role for metabolomics? Trends Plant Sci. 2022;27:549–563. doi: 10.1016/j.tplants.2022.02.001. [DOI] [PubMed] [Google Scholar]
- 60.Araus J.L., Kefauver S.C., Vergara-Díaz O., Gracia-Romero A., Rezzouk F.Z., Segarra J., Buchaillot M.L., Chang-Espino M., Vatter T., Sanchez-Bragado R., et al. Crop phenotyping in a context of global change: What to measure and how to do it. J. Integr. Plant Biol. 2022;64:592–618. doi: 10.1111/jipb.13191. [DOI] [PubMed] [Google Scholar]
- 61.Shakoor N., Lee S., Mockler T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017;38:184–192. doi: 10.1016/j.pbi.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Atieno J., Colmer T.D., Taylor J., Li Y., Quealy J., Kotula L., Nicol D., Nguyen D.T., Brien C., Langridge P., et al. Novel salinity tolerance loci in chickpea identified in glasshouse and field environments. Front. Plant Sci. 2021;12:667910. doi: 10.3389/fpls.2021.667910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomelin P., Bonneau J., Brien C., Suchecki R., Baumann U., Kalambettu P., Langridge P., Tricker P., Fleury D. The wheat Seven in absentia gene is associated with increases in biomass and yield in hot climates. J. Exp. Bot. 2021;72:3774–3791. doi: 10.1093/jxb/erab044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tran B.T.T., Cavagnaro T.R., Jewell N., Brien C., Berger B., Watts-Williams S.J. High-throughput phenotyping reveals growth of Medicago truncatula is positively affected by arbuscular mycorrhizal fungi even at high soil phosphorus availability. Plants People Planet. 2021;3:600–613. doi: 10.1002/ppp3.10101. [DOI] [Google Scholar]
- 65.Duddu H.S.N., Johnson E.N., Willenborg C.J., Shirtliffe S.J. High-Throughput UAV Image-Based Method Is More Precise Than Manual Rating of Herbicide Tolerance. Plant Phenomics. 2019;2019:6036453. doi: 10.34133/2019/6036453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lawrence-Dill C.J., Schnable P.S., Springer N.M. Idea factory: The maize genomes to fields initiative. Crop Sci. 2019;59:1406–1410. doi: 10.2135/cropsci2019.02.0071. [DOI] [Google Scholar]
- 67.LeBauer D., Burnette M., Fahlgren N., Kooper R., McHenry K., Stylianou A. What Does TERRA-REF’s High Resolution, Multi Sensor Plant Sensing Public Domain Data Offer the Computer Vision Community?; Proceedings of the IEEE/CVF International Conference on Computer Vision; Montreal, BC, Canada. 11 October 2021; pp. 1409–1415. [DOI] [Google Scholar]
- 68.Danilevicz M.F., Bayer P.E., Nestor B.J., Bennamoun M., Edwards D. Resources for image-based high-throughput phenotyping in crops and data sharing challenges. Plant Physiol. 2021;187:699–715. doi: 10.1093/plphys/kiab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou Z., Majeed Y., Diverres Naranjo G., Gambacorta E.M.T. Assessment for crop water stress with infrared thermal imagery in precision agriculture: A review and future prospects for deep learning applications. Comput. Electron. Agric. 2021;182:106019. doi: 10.1016/j.compag.2021.106019. [DOI] [Google Scholar]
- 70.Prakash P.T., Banan D., Paul R.E., Feldman M.J., Xie D., Freyfogle L., Baxter I., Leakey A.D.B. Correlation and co-localization of QTL for stomatal density, canopy temperature, and productivity with and without drought stress in Setaria. J. Exp. Bot. 2021;72:5024–5037. doi: 10.1093/jxb/erab166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Severtson D., Callow N., Flower K., Neuhaus A., Olejnik M., Nansen C. Unmanned aerial vehicle canopy reflectance data detects potassium deficiency and green peach aphid susceptibility in canola. Precis. Agric. 2016;17:659–677. doi: 10.1007/s11119-016-9442-0. [DOI] [Google Scholar]
- 72.Senthilkumar T., Jayas D.S., White N.D.G. Detection of different stages of fungal infection in stored canola using near-infrared hyperspectral imaging. J. Stored Prod. Res. 2015;63:80–88. doi: 10.1016/j.jspr.2015.07.005. [DOI] [Google Scholar]
- 73.Singh K.D., Duddu H.S.N., Vail S., Parkin I., Shirtliffe S.J. UAV-Based Hyperspectral Imaging Technique to Estimate Canola (Brassica napus L.) Seedpods Maturity. Can. J. Remote Sens. 2021;47:33–47. doi: 10.1080/07038992.2021.1881464. [DOI] [Google Scholar]
- 74.Sankaran S., Zhang C., Marzougui A., Khot L., Davis J.B., Brown J., Craine W., Hulbery S. Autonomous Air and Ground Sensing Systems for Agricultural Optimization and Phenotyping III. SPIE; Bellingham, WA, USA: 2018. Detection of canola flowering using proximal and aerial remote sensing techniques. [Google Scholar]
- 75.Zhang T., Vail S., Duddu H.S.N., Parkin I.A.P., Guo X., Johnson E.N., Shirtliffe S.J. Phenotyping Flowering in Canola (Brassica napus L.) and Estimating Seed Yield Using an Unmanned Aerial Vehicle-Based Imagery. Front. Plant Sci. 2021;12:686332. doi: 10.3389/fpls.2021.686332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Adak A., Murray S.C., Božinović S., Lindsey R., Nakasagga S., Chatterjee S., Anderson S.L., Wilde S. Temporal Vegetation Indices and Plant Height from Remotely Sensed Imagery Can Predict Grain Yield and Flowering Time Breeding Value in Maize via Machine Learning Regression. Remote Sens. 2021;13:2141. doi: 10.3390/rs13112141. [DOI] [Google Scholar]
- 77.Knoch D., Abbadi A., Grandke F., Meyer R.C., Samans B., Werner C.R., Snowdon R.J., Altmann T. Strong temporal dynamics of QTL action on plant growth progression revealed through high-throughput phenotyping in canola. Plant Biotechnol. J. 2020;18:68–82. doi: 10.1111/pbi.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li H., Feng H., Guo C., Yang S., Huang W., Xiong X., Liu J., Chen G., Liu Q., Xiong L., et al. High-throughput phenotyping accelerates the dissection of the dynamic genetic architecture of plant growth and yield improvement in rapeseed. Plant Biotechnol. J. 2020;18:2345–2353. doi: 10.1111/pbi.13396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Muraya M.M., Chu J., Zhao Y., Junker A., Klukas C., Reif J.C., Altmann T. Genetic variation of growth dynamics in maize (Zea mays L.) revealed through automated non-invasive phenotyping. Plant J. Cell Mol. Biol. 2017;89:366–380. doi: 10.1111/tpj.13390. [DOI] [PubMed] [Google Scholar]
- 80.Araus J.L., Kefauver S.C., Zaman-Allah M., Olsen M.S., Cairns J.E. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci. 2018;23:451–466. doi: 10.1016/j.tplants.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Danilevicz M.F., Gill M., Anderson R., Batley J., Bennamoun M., Bayer P.E., Edwards D. Plant genotype to phenotype prediction using machine learning. Front. Genet. 2022;13:822173. doi: 10.3389/fgene.2022.822173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sulik J.J., Long D.S. Spectral considerations for modeling yield of canola. Remote Sens. Environ. 2016;184:161–174. doi: 10.1016/j.rse.2016.06.016. [DOI] [Google Scholar]
- 83.Montesinos-López O.A., Martín-Vallejo J., Crossa J., Gianola D., Hernández-Suárez C.M., Montesinos-López A., Juliana P., Singh R. New Deep Learning Genomic-Based Prediction Model for Multiple Traits with Binary, Ordinal, and Continuous Phenotypes. G3. 2019;9:1545–1556. doi: 10.1534/g3.119.300585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crossa J., Martini J.W.R., Gianola D., Pérez-Rodríguez P., Jarquin D., Juliana P., Montesinos-López O., Cuevas J. Deep Kernel and Deep Learning for Genome-Based Prediction of Single Traits in Multienvironment Breeding Trials. Front. Genet. 2019;10:1168. doi: 10.3389/fgene.2019.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Montesinos-López O.A., Montesinos-López A., Crossa J., Gianola D., Hernández-Suárez C.M., Martín-Vallejo J. Multi-trait, Multi-environment Deep Learning Modeling for Genomic-Enabled Prediction of Plant Traits. G3. 2018;8:3829–3840. doi: 10.1534/g3.118.200728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.LeCun Y., Bengio Y., Hinton G. Deep learning. Nature. 2015;521:436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 87.Kismiantini, Montesinos-López O.A., Crossa J., Setiawan E.P., Wutsqa D.U. Prediction of count phenotypes using high-resolution images and genomic data. G3. 2021;11:jkab035. doi: 10.1093/g3journal/jkab035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rutkoski J., Poland J., Mondal S., Autrique E., Pérez L.G., Crossa J., Reynolds M., Singh R. Canopy Temperature and Vegetation Indices from High-Throughput Phenotyping Improve Accuracy of Pedigree and Genomic Selection for Grain Yield in Wheat. G3. 2016;6:2799–2808. doi: 10.1534/g3.116.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Danilevicz M.F., Bayer P.E., Boussaid F., Bennamoun M., Edwards D. Maize yield prediction at an early developmental stage using multispectral images and genotype data for preliminary hybrid selection. Remote Sens. 2021;13:3976. doi: 10.3390/rs13193976. [DOI] [Google Scholar]
- 90.Busov V.B., Brunner A.M., Strauss S.H. Genes for control of plant stature and form. New Phytol. 2008;177:589–607. doi: 10.1111/j.1469-8137.2007.02324.x. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y., Li J. Molecular basis of plant architecture. Annu. Rev. Plant Biol. 2008;59:253–279. doi: 10.1146/annurev.arplant.59.032607.092902. [DOI] [PubMed] [Google Scholar]
- 92.Liu S., Fan C., Li J., Cai G., Yang Q., Wu J., Yi X., Zhang C., Zhou Y. A genome-wide association study reveals novel elite allelic variations in seed oil content of Brassica napus. Theor. Appl. Genet. 2016;129:1203–1215. doi: 10.1007/s00122-016-2697-z. [DOI] [PubMed] [Google Scholar]
- 93.Yang P., Shu C., Chen L., Xu J., Wu J., Liu K. Identification of a major QTL for silique length and seed weight in oilseed rape (Brassica napus L.) Theor. Appl. Genet. 2012;125:285–296. doi: 10.1007/s00122-012-1833-7. [DOI] [PubMed] [Google Scholar]
- 94.Raboanatahiry N., Chao H., Dalin H., Pu S., Yan W., Yu L., Wang B., Li M. QTL Alignment for Seed Yield and Yield Related Traits in Brassica napus. Front. Plant Sci. 2018;9:1127. doi: 10.3389/fpls.2018.01127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shah S., Karunarathna N.L., Jung C., Emrani N. An APETALA1 ortholog affects plant architecture and seed yield component in oilseed rape (Brassica napus L.) BMC Plant Biol. 2018;18:380. doi: 10.1186/s12870-018-1606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cai G., Yang Q., Chen H., Yang Q., Zhang C., Fan C., Zhou Y. Genetic dissection of plant architecture and yield-related traits in Brassica napus. Sci. Rep. 2016;6:21625. doi: 10.1038/srep21625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Raman R., Diffey S., Carling J., Cowley R.B., Kilian A., Luckett D.J., Raman H. Quantitative genetic analysis of grain yield in an Australian Brassica napus doubled-haploid population. Crop Pasture Sci. 2016;67:298–307. doi: 10.1071/CP15283. [DOI] [Google Scholar]
- 98.Aakanksha, Yadava S.K., Yadav B.G., Gupta V., Mukhopadhyay A., Pental D., Pradhan A.K. Genetic Analysis of Heterosis for Yield Influencing Traits in Brassica juncea Using a Doubled Haploid Population and Its Backcross Progenies. Front. Plant Sci. 2021;12:721631. doi: 10.3389/fpls.2021.721631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lu K., Peng L., Zhang C., Lu J., Yang B., Xiao Z., Liang Y., Xu X., Qu C., Zhang K., et al. Genome-Wide Association and Transcriptome Analyses Reveal Candidate Genes Underlying Yield-determining Traits in Brassica napus. Front. Plant Sci. 2017;8:206. doi: 10.3389/fpls.2017.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lu K., Wei L., Li X., Wang Y., Wu J., Liu M., Zhang C., Chen Z., Xiao Z., Jian H., et al. Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat. Commun. 2019;10:1154. doi: 10.1038/s41467-019-09134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pal L., Sandhu S.K., Bhatia D., Sethi S. Genome-wide association study for candidate genes controlling seed yield and its components in rapeseed (Brassica napus subsp. napus) Physiol. Mol. Biol. Plants. 2021;27:1933–1951. doi: 10.1007/s12298-021-01060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pickersgill B. Domestication of plants in the Americas: Insights from Mendelian and molecular genetics. Ann. Bot. 2007;100:925–940. doi: 10.1093/aob/mcm193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA. 2005;102:13950. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Golicz A.A., Bayer P.E., Barker G.C., Edger P.P., Kim H., Martinez P.A., Chan C.K.K., Severn-Ellis A., McCombie W.R., Parkin I.A.P., et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat. Commun. 2016;7:13390. doi: 10.1038/ncomms13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cai X., Chang L., Zhang T., Chen H., Zhang L., Lin R., Liang J., Wu J., Freeling M., Wang X. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021;22:166. doi: 10.1186/s13059-021-02383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song J.-M., Guan Z., Hu J., Guo C., Yang Z., Wang S., Liu D., Wang B., Lu S., Zhou R., et al. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants. 2020;6:34–45. doi: 10.1038/s41477-019-0577-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tay Fernandez C.G., Nestor B.J., Danilevicz M.F., Marsh J.I., Petereit J., Bayer P.E., Batley J., Edwards D. Expanding Gene-Editing Potential in Crop Improvement with Pangenomes. Int. J. Mol. Sci. 2022;23:2276. doi: 10.3390/ijms23042276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zanini S.F., Bayer P.E., Wells R., Snowdon R.J., Batley J., Varshney R.K., Nguyen H.T., Edwards D., Golicz A.A. Pangenomics in crop improvement—From coding structural variations to finding regulatory variants with pangenome graphs. Plant Genome. 2021;15:e20177. doi: 10.1002/tpg2.20177. [DOI] [PubMed] [Google Scholar]
- 109.Danilevicz M.F., Tay Fernandez C.G., Marsh J.I., Bayer P.E., Edwards D. Plant pangenomics: Approaches, applications and advancements. Curr. Opin. Plant Biol. 2020;54:18–25. doi: 10.1016/j.pbi.2019.12.005. [DOI] [PubMed] [Google Scholar]
- 110.Golicz A.A., Batley J., Edwards D. Towards plant pangenomics. Plant Biotechnol. J. 2016;14:1099–1105. doi: 10.1111/pbi.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bayer P.E., Golicz A.A., Tirnaz S., Chan C.-K.K., Edwards D., Batley J. Variation in abundance of predicted resistance genes in the Brassica oleracea pangenome. Plant Biotechnol. J. 2019;17:789–800. doi: 10.1111/pbi.13015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hurgobin B., Golicz A.A., Bayer P.E., Chan C.K., Tirnaz S., Dolatabadian A., Schiessl S.V., Samans B., Montenegro J.D., Parkin I.A.P., et al. Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol. J. 2018;16:1265–1274. doi: 10.1111/pbi.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dolatabadian A., Bayer P.E., Tirnaz S., Hurgobin B., Edwards D., Batley J. Characterization of disease resistance genes in the Brassica napus pangenome reveals significant structural variation. Plant Biotechnol. J. 2020;18:969–982. doi: 10.1111/pbi.13262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lin K., Zhang N., Severing E.I., Nijveen H., Cheng F., Visser R.G., Wang X., de Ridder D., Bonnema G. Beyond genomic variation--comparison and functional annotation of three Brassica rapa genomes: A turnip, a rapid cycling and a Chinese cabbage. BMC Genom. 2014;15:250. doi: 10.1186/1471-2164-15-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cantila A.Y., Saad N.S.M., Amas J.C., Edwards D., Batley J. Recent Findings Unravel Genes and Genetic Factors Underlying Leptosphaeria maculans Resistance in Brassica napus and Its Relatives. Int. J. Mol. Sci. 2021;22:313. doi: 10.3390/ijms22010313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang Y., Thomas W., Bayer P.E., Edwards D., Batley J. Frontiers in Dissecting and Managing Brassica Diseases: From Reference-Based RGA Candidate Identification to Building Pan-RGAomes. Int. J. Mol. Sci. 2020;21:8964. doi: 10.3390/ijms21238964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi T., Li R., Zhao Z., Ding G., Long Y., Meng J., Xu F., Shi L. QTL for yield traits and their association with functional genes in response to phosphorus deficiency in Brassica napus. PLoS ONE. 2013;8:e54559. doi: 10.1371/journal.pone.0054559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chang T., Guan M., Zhou B., Peng Z., Xing M., Wang X., Guan C. Progress of CRISPR/Cas9 technology in breeding of Brassica napus. Oil Crop Sci. 2021;6:53–57. doi: 10.1016/j.ocsci.2021.03.005. [DOI] [Google Scholar]
- 119.Wells R., Trick M., Soumpourou E., Clissold L., Morgan C., Werner P., Gibbard C., Clarke M., Jennaway R., Bancroft I. The control of seed oil polyunsaturate content in the polyploid crop species Brassica napus. Mol. Breed. 2014;33:349–362. doi: 10.1007/s11032-013-9954-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tian Q., Li B., Feng Y., Zhao W., Huang J., Chao H. Application of CRISPR/Cas9 in Rapeseed for Gene Function Research and Genetic Improvement. Agronomy. 2022;12:824. doi: 10.3390/agronomy12040824. [DOI] [Google Scholar]
- 121.Pan C., Wu X., Markel K., Malzahn A.A., Kundagrami N., Sretenovic S., Zhang Y., Cheng Y., Shih P.M., Qi Y. CRISPR–Act3. 0 for highly efficient multiplexed gene activation in plants. Nat. Plants. 2021;7:942–953. doi: 10.1038/s41477-021-00953-7. [DOI] [PubMed] [Google Scholar]
- 122.Pan C., Li G., Malzahn A.A., Cheng Y., Leyson B., Sretenovic S., Gurel F., Coleman G.D., Qi Y. Boosting plant genome editing with a versatile CRISPR-Combo system. Nat. Plants. 2022;8:513–525. doi: 10.1038/s41477-022-01151-9. [DOI] [PubMed] [Google Scholar]
- 123.Zaman Q.U., Chu W., Hao M., Shi Y., Sun M., Sang S.-F., Mei D., Cheng H., Liu J., Li C. CRISPR/Cas9-Mediated Multiplex Genome Editing of JAGGED Gene in Brassica napus L. Biomolecules. 2019;9:725. doi: 10.3390/biom9110725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhai Y., Cai S., Hu L., Yang Y., Amoo O., Fan C., Zhou Y. CRISPR/Cas9-mediated genome editing reveals differences in the contribution of INDEHISCENT homologues to pod shatter resistance in Brassica napus L. Theor. Appl. Genet. 2019;132:2111–2123. doi: 10.1007/s00122-019-03341-0. [DOI] [PubMed] [Google Scholar]
- 125.Park S.-C., Park S., Jeong Y.J., Lee S.B., Pyun J.W., Kim S., Kim T.H., Kim S.W., Jeong J.C., Kim C.Y. DNA-free mutagenesis of GIGANTEA in Brassica oleracea var. capitata using CRISPR/Cas9 ribonucleoprotein complexes. Plant Biotechnol. Rep. 2019;13:483–489. doi: 10.1007/s11816-019-00585-6. [DOI] [Google Scholar]
- 126.Yang Y., Zhu K., Li H., Han S., Meng Q., Khan S.U., Fan C., Xie K., Zhou Y. Precise editing of CLAVATA genes in Brassica napus L. regulates multilocular silique development. Plant Biotechnol. J. 2018;16:1322–1335. doi: 10.1111/pbi.12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Stanic M., Hickerson N.M., Arunraj R., Samuel M.A. Gene-editing of the strigolactone receptor BnD14 confers promising shoot architectural changes in Brassica napus (canola) Plant Biotechnol. J. 2021;19:639. doi: 10.1111/pbi.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu P., Cao S., Hu K., Wang X., Huang W., Wang G., Lv Z., Liu Z., Wen J., Yi B. Trilocular phenotype in Brassica juncea L. resulted from interruption of CLAVATA1 gene homologue (BjMc1) transcription. Sci. Rep. 2017;7:3498. doi: 10.1038/s41598-017-03755-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gol S., Pena R.N., Rothschild M.F., Tor M., Estany J. A polymorphism in the fatty acid desaturase-2 gene is associated with the arachidonic acid metabolism in pigs. Sci. Rep. 2018;8:14336. doi: 10.1038/s41598-018-32710-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kim S.-G. The way to true plant genome editing. Nat. Plants. 2020;6:736–737. doi: 10.1038/s41477-020-0723-2. [DOI] [PubMed] [Google Scholar]
- 131.Zhang D., Hussain A., Manghwar H., Xie K., Xie S., Zhao S., Larkin R.M., Qing P., Jin S., Ding F. Genome editing with the CRISPR-Cas system: An art, ethics and global regulatory perspective. Plant Biotechnol. J. 2020;18:1651–1669. doi: 10.1111/pbi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gong Z., Cheng M., Botella J.R. Non-GM Genome Editing Approaches in Crops. Front. Genome Ed. 2021;3:817279. doi: 10.3389/fgeed.2021.817279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ahmar S., Zhai Y., Huang H., Yu K., Khan M.H.U., Shahid M., Samad R.A., Khan S.U., Amoo O., Fan C. Development of mutants with varying flowering times by targeted editing of multiple SVP gene copies in Brassica napus L. Crop J. 2022;10:67–74. doi: 10.1016/j.cj.2021.03.023. [DOI] [Google Scholar]
- 134.Fan S., Zhang L., Tang M., Cai Y., Liu J., Liu H., Liu J., Terzaghi W., Wang H., Hua W. CRISPR/Cas9-targeted mutagenesis of the BnaA03. BP gene confers semi-dwarf and compact architecture to rapeseed (Brassica napus L.) Plant Biotechnol. J. 2021;19:2383–2385. doi: 10.1111/pbi.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zaman Q.U., Wen C., Yuqin S., Mengyu H., Desheng M., Jacqueline B., Baohong Z., Chao L., Qiong H. Characterization of SHATTERPROOF Homoeologs and CRISPR-Cas9-Mediated Genome Editing Enhances Pod-Shattering Resistance in Brassica napus L. CRISPR J. 2021;4:360–370. doi: 10.1089/crispr.2020.0129. [DOI] [PubMed] [Google Scholar]
- 136.Khan M.H., Hu L., Zhu M., Zhai Y., Khan S.U., Ahmar S., Amoo O., Zhang K., Fan C., Zhou Y. Targeted mutagenesis of EOD3 gene in Brassica napus L. regulates seed production. J. Cell. Physiol. 2021;236:1996–2007. doi: 10.1002/jcp.29986. [DOI] [PubMed] [Google Scholar]
- 137.Zheng M., Zhang L., Tang M., Liu J., Liu H., Yang H., Fan S., Terzaghi W., Wang H., Hua W. Knockout of two Bna MAX 1 homologs by CRISPR/Cas9-targeted mutagenesis improves plant architecture and increases yield in rapeseed (Brassica napus L.) Plant Biotechnol. J. 2020;18:644–654. doi: 10.1111/pbi.13228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not produce any data, however, any data described in ‘Introduction’ can be found in FAOSTAT (https://www.fao.org/faostat/en/#home, accessed on 4 September 2022).