Abstract
Background: Esophageal cancer (ESCA) is a major cause of cancer-related mortality worldwide. Altered fatty acid metabolism is a hallmark of cancer. However, studies on the roles of fatty acid metabolism-related genes (FRGs) in ESCA remain limited. Method: We identified differentially expressed FRGs (DE-FRGs). Then, the DE-FRGs prognostic model was constructed and validated using a comprehensive analysis. Moreover, the correlation between the risk model and clinical characteristics was investigated. A nomogram for predicting survival was established and evaluated. Subsequently, the difference in tumor microenvironment (TME) was compared between two risk groups. The sensitivity of key DE-FRGs to chemotherapeutic interventions and their correlation with immune cells were investigated. Finally, DEGs between two risk groups were measured and the prognostic value of key DE-FRGs in ESCA was confirmed in other databases. Results: A prognostic model was constructed based on seven selected DEG-FRGs. TNM staging and CD8+ T cells were significantly correlated with high-risk groups. Low-risk groups exhibited more infiltrated M0 macrophages, an activation of type II interferon (IFN-γ) responses, and were found to be more suitable for immunotherapy. Seven key DE-FRGs with prognostic value were found to be considerably influenced by different chemotherapy drugs. Conclusion: A prognostic model based on seven DE-FRGs may efficiently predict patient prognosis and immunotherapy response, helping to develop individualized treatment strategies in ESCA.
Keywords: esophageal cancer (ESCA), The Cancer Genome Atlas (TCGA), fatty acid metabolism, prognosis, immunotherapy response
1. Introduction
ESCA is the eighth most common cancer worldwide causing the sixth highest cancer-related mortality rates [1]. Despite the range of available treatment options, the overall five-year survival rate remains less than 20%, with most deaths being associated with distant metastases and the emergence of resistance to chemoradiotherapy [2,3]. Therefore, exploration of novel therapeutic options and development of prognostic models for the management of ESCA are crucial.
The reprogramming of energy metabolism is a hallmark of cancer development, promoting cell growth and proliferation [4,5]. Accumulating evidence suggests that fatty acid metabolism plays a crucial role in metabolic reprogramming, affecting cell membrane formation, energy storage, and the production of signaling molecules [6,7]. Previous study has reported that fatty-acid-metabolism-related genes are associated with malignancy, prognosis, and immune phenotype in gliomas [8]. In cervical cancer patients, enhanced lipolysis and fatty acid synthesis promote lymphatic spread via the activation of nuclear factor kB (NF-kB) signaling [4,9]. Activated fatty acid oxidation improves the survival of acute myeloid leukemia cells [10]. In addition to influencing the effectiveness of chemo- and radiation therapy, altered fatty acid metabolism has also been suggested to effect responses to immunotherapy [11,12]. Previous study has indicated that fatty acids can affect the phenotype and functionality of infiltrating immune cells, potentially causing immunosuppression [13]. However, the prognostic relevance and potential therapeutic significance of genes involved in fatty acid metabolism, particularly in the context of immunotherapy, has never been explored among ESCA patients.
In this study, we identified differentially expressed FRGs (DE-FRGs). Then, the DE-FRGs prognostic model was constructed based on univariate Cox analysis, LASSO regression analysis, and multivariate Cox analysis. A receiver operating characteristic (ROC) curve was drawn to validate the accuracy of this model. Moreover, the correlation between the model and clinical characteristics was evaluated. A nomogram for predicting survival was established and evaluated. Subsequently, the difference in immune cell infiltration, immune function and immunotherapy response were compared between high- and low-risk groups. The sensitivity of key DE-FRGs to chemotherapeutic interventions and their correlation with immune cells were investigated. Finally, DEGs between two risk groups were identified and the prognostic value of key DE-FRGs in ESCA was confirmed in other databases.
2. Materials and Methods
2.1. Identification of Fatty-Acid-Metabolism-Related Genes
ESCA-related RNA sequencing data and corresponding clinical data, including information of age, gender, tumor stage, and survival information were downloaded from TCGA Database (https://portal.gdc.cancer.gov/ 15 March 2022). A total of 171 samples were obtained from TCGA database, including 160 tumor samples and 11 normal samples. A list of genes related to fatty acid metabolism (FRGs) was compiled using the Molecular Signature Database (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb 14 March 2022), including the Kyoto Encyclopedia of Genes and Genomes (KEGG), the Reactome fatty acid metabolism, and the Hallmark fatty acid metabolism datasets, followed by the elimination of duplicate transcripts/genes [14]. The expression of fatty acid metabolism-related genes (FRGs) in ESCA were obtained from TCGA database using “Limma” R package. The differentially expressed fatty acid metabolism-related genes (DE-FRGs) in ESCA were identified using “Limma” and “pheatmap” R package. The criteria for identifying DE-FRGs were fold change (FC) >1.5 and false discovery rate (FDR) of <0.05 [15]. The details of clinical information can be found in Supplementary Tables S1 and S2.
2.2. Construction and Evaluation of a Predictive Risk Score Model
Univariate Cox regression analysis was performed to identify DE-FRGs associated with prognosis, using the “survival” and “survminer” packages in R. Additional LASSO regression analysis was conducted to narrow further the field of key DE-FRGs. Subsequently, multivariate Cox regression analysis was performed to develop a prognostic risk score model for predicting overall survival (OS) of ESCA patients. ESCA patients were divided into high-risk (n = 80) and low-risk (n = 80) groups based on the median risk score. Kaplan–Meier (KM) survival curves were plotted to analyze the difference in overall survival (OS) and progression free survival (PFS) of patients assigned into the high-risk and low-risk groups. Finally, the receiver operating characteristic (ROC) curve was drawn and the area under the curve (AUC) for 1-, 3-, and 5-year OS was calculated through the “survival ROC” package in R to assess the predictive accuracy of the prognostic risk score model. A p-value < 0.05 was used as the filter condition.
2.3. The Association between Risk Score and Clinical Characteristics
The Limma R package was utilized to explore the association between risk score and clinical characteristics, including age, gender, and TNM pathology stage [14]. This was followed by univariate and multivariate Cox regression analyses to identify independent prognostic factors using the “survival” package (p < 0.05) [16].
2.4. Construction of a Nomogram for Patients with ESCA
A nomogram consisting of age, gender, pathologic staging, and prognostic risk score model was constructed using the “survival”, “regplot” and “rms” packages, to predict the likelihood of 1-, 3-, and 5-year survival of the studied ESCA patients. Then, independent prognostic analysis was performed to evaluate whether nomogram could independently predict patient prognosis. ROC curves and calibration curves were plotted to confirm the prediction accuracy of the nomogram [17].
2.5. Association between the Risk Model and Immune Parameters
The extent of immune cell infiltration was compared between the high- and low-risk groups using the CIBERSORT algorithms [18] and potential differences in immunologic functioning were evaluated using the “Limma”, “GSVA”, “GSEABase”, “ggpubr”, and “reshape2” R packages [19]. Finally, the tumor immune dysfunction and exclusion (TIDE) algorithm (http://tide.dfci.harvard.edu/login/ 18 March 2022) was applied to predict the effects of the observed changes on potential immunotherapy responses in the high- and low-risk groups [20].
2.6. GSCA Analysis
Gene Set Cancer Analysis (GSCA, http://bioinfo.life.hust.edu.cn/GSCA/#/ 20 June 2022) is an online analysis tool for genomic, pharmacogenomic, and immunogenomic gene expression analyses in cancer [21]. We used GSCA to analyze the correlations between the expression of key biomarkers, immune cells, and drug sensitivity.
2.7. A Protein–Protein Interaction Network of DEGs in Groups of Different Risk Score Groups
DEGs in 160 ESCA patients between two different risk groups were analyzed using the Limma R package (|logFC| > 1, FDR < 0.05) to identify DEGs [4]. Gene Ontology (GO) enrichment analyses of the detected DEGs was then performed using the “clusterProfiler” R package [22]. A protein–protein interaction (PPI) network of DEGs was then constructed and visualized via the STRING database (STRING, https://string-db.org/ 17 March 2022) and the Cytoscape software (version: 3.7.2), identifying hub genes using cytoHubba [4]. According to the median expression value of the hub genes, all samples were divided into low- and high-expression groups. Kaplan–Meier analysis was carried out to compare the survival characteristics between the two groups [23]. Finally, the correlation between immune cell infiltration and prognostically relevant DE-FRGs was analyzed [24].
2.8. Validation of the Expression and Prognostic Value of Seven FRGs
UALCAN (http://ualcan.path.uab.edu/index.html 28 June 2022) is a comprehensive interactive web resource for analyzing cancer OMICS data (TCGA, MET500, CPTAC, and CBTTC), allowing users to identify biomarkers or to perform in silico validation of potential genes of interest. It uses graphical representations of gene expression profiles of protein-coding, miRNA-coding, and lincRNA-coding genes and combines these with survival information [25]. UALCAN database was used to confirm the expression difference of key DE-FRGs and investigate whether their expression was correlated with survival differences between the two groups.
3. Results
3.1. Identifying Fatty-Acid-Metabolism-Related DEGs in ESCA Samples
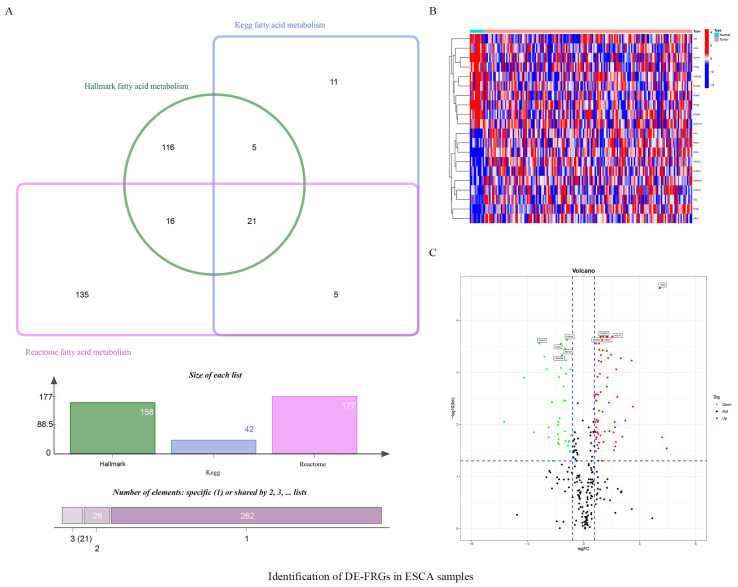
The database search-derived lists of fatty acid metabolism-related genes were intersected, and duplicates were eliminated. A total of 309 genes were identified as having a known or proposed role in fatty acid metabolism (Figure 1A). Of these, 108 were differentially expressed between ESCA samples and healthy samples in the TCGA database (Figure 1B,C; Table 1).
Figure 1.
Identification of DE-FRGs in ESCA samples. (A) 309 fatty acid metabolism-related genes from Kegg, Hallmark and Reactome. (B) Heatmap display the upregulated and downregulated fatty acid metabolism−related DEGs (top 10, respectively, p < 0.05). (C) Volcano plot of fatty acid metabolism-related DEGs (only displaying 10 DE-FRGs).
Table 1.
Identification of differentially expressed fatty acid metabolism-related genes.
| Gene | logFC | p-Value | FDR |
|---|---|---|---|
| FADS2 | 1.6647 | 0.0022 | 0.0090 |
| HACD1 | −0.8386 | 0.0109 | 0.0258 |
| PTGES2 | 0.8793 | 0.0001 | 0.0011 |
| HPGD | −1.9718 | 0.0001 | 0.0009 |
| PTGES | 2.5728 | 0.0001 | 0.0006 |
| MAOA | −0.9333 | 0.0001 | 0.0011 |
| CD36 | −0.6685 | 0.0053 | 0.0161 |
| ELOVL2 | 1.7260 | 0.0084 | 0.0217 |
| GPX1 | 0.6824 | 0.0031 | 0.0114 |
| ENO3 | 0.7959 | 0.0125 | 0.0288 |
| CEL | 4.4503 | 0.0128 | 0.0291 |
| ACBD6 | 0.8421 | 0.0000 | 0.0004 |
| MORC2 | 0.9267 | 0.0000 | 0.0002 |
| UBE2L6 | 1.4763 | 0.0000 | 0.0005 |
| CYP2C9 | −1.3856 | 0.0050 | 0.0153 |
| SLC25A1 | 0.6648 | 0.0006 | 0.0034 |
| ALDH1A1 | −1.6541 | 0.0256 | 0.0499 |
| PRKAA2 | −1.3722 | 0.0091 | 0.0227 |
| ACOT7 | 1.5324 | 0.0000 | 0.0002 |
| ABCD1 | 1.0133 | 0.0012 | 0.0057 |
| GLUL | −1.6938 | 0.0007 | 0.0037 |
| ACOT9 | 0.6684 | 0.0071 | 0.0193 |
| NCAPH2 | 0.8264 | 0.0000 | 0.0003 |
| HACD2 | 0.7813 | 0.0004 | 0.0027 |
| OSTC | 0.5871 | 0.000 | 0.0028 |
| FADS1 | 1.6133 | 0.0046 | 0.0142 |
| HMGCS2 | −0.9822 | 0.0038 | 0.0136 |
| ALOX15 | 4.2060 | 0.0063 | 0.0177 |
| CPOX | 0.9419 | 0.0001 | 0.0006 |
| ACSM6 | −2.6558 | 0.0043 | 0.0142 |
| NTHL1 | 0.9232 | 0.0000 | 0.0005 |
| MIX23 | 1.0243 | 0.0000 | 0.0004 |
| HSPH1 | 1.3146 | 0.0000 | 0.0005 |
| PTGDS | −0.7081 | 0.0160 | 0.0339 |
| CYP2C8 | −4.2436 | 0.0021 | 0.0089 |
| PTGES3 | 1.0439 | 0.0000 | 0.0002 |
| CPT1B | 0.9034 | 0.0055 | 0.0163 |
| ADH1C | −1.3389 | 0.0071 | 0.0193 |
| BLVRA | 0.6532 | 0.0152 | 0.0327 |
| SUCLG2 | −1.3197 | 0.0002 | 0.0012 |
| THRSP | −1.3148 | 0.0099 | 0.0242 |
| ACOX3 | −0.5875 | 0.0115 | 0.0266 |
| ELOVL3 | 2.6467 | 0.0009 | 0.0046 |
| TDO2 | 4.0796 | 0.0000 | 0.0000 |
| ACADS | −0.9901 | 0.0007 | 0.0035 |
| YWHAH | 0.7439 | 0.0002 | 0.0012 |
| AMACR | −0.7589 | 0.0080 | 0.0209 |
| PCCA | −0.9226 | 0.0001 | 0.0006 |
| ODC1 | 1.8059 | 0.0005 | 0.0030 |
| ALAD | −1.2068 | 0.0000 | 0.0003 |
| CA2 | −1.8684 | 0.0031 | 0.0114 |
| RDH11 | 0.6085 | 0.0004 | 0.0027 |
| LDHA | 1.2471 | 0.0003 | 0.0019 |
| ACADSB | −1.1380 | 0.0000 | 0.0005 |
| ACAT1 | −1.2122 | 0.0001 | 0.0008 |
| GABARAPL1 | −0.9593 | 0.0080 | 0.0209 |
| ALOX5AP | 1.0724 | 0.0086 | 0.0219 |
| NSDHL | 0.7857 | 0.0020 | 0.0085 |
| FMO1 | 2.2368 | 0.0019 | 0.0084 |
| ACAT2 | 0.7943 | 0.0049 | 0.0152 |
| GPX4 | 0.6893 | 0.0018 | 0.0082 |
| ACSS1 | −0.7269 | 0.0150 | 0.0324 |
| ETFDH | −0.8984 | 0.0000 | 0.0002 |
| ACSM3 | −1.3758 | 0.0026 | 0.0102 |
| ACBD7 | 1.6988 | 0.0113 | 0.0266 |
| AUH | −0.8033 | 0.0083 | 0.0215 |
| H2AZ1 | 1.2866 | 0.0000 | 0.0002 |
| SMS | 0.9141 | 0.0004 | 0.0024 |
| ELOVL5 | 1.3837 | 0.0037 | 0.0135 |
| ALDOA | 0.7531 | 0.0023 | 0.0090 |
| CYP4B1 | −1.2830 | 0.0017 | 0.0076 |
| ACO2 | −0.7125 | 0.0001 | 0.0009 |
| MDH2 | 0.6924 | 0.0004 | 0.0026 |
| PPT1 | 1.4317 | 0.0000 | 0.0002 |
| PON2 | 1.7812 | 0.0000 | 0.0002 |
| PSME1 | 0.9799 | 0.0000 | 0.0002 |
| PTGS2 | 1.6912 | 0.0031 | 0.0114 |
| HSD17B10 | 0.6922 | 0.0007 | 0.0035 |
| ACLY | 1.2460 | 0.0000 | 0.0004 |
| HMGCS1 | 0.8293 | 0.0089 | 0.0225 |
| ECI2 | −1.3581 | 0.0021 | 0.0089 |
| METAP1 | 0.6304 | 0.0040 | 0.0138 |
| APEX1 | 0.8167 | 0.0001 | 0.0006 |
| MIF | 1.2857 | 0.0002 | 0.0013 |
| ADSL | 0.9045 | 0.0000 | 0.0002 |
| SCD | 1.3604 | 0.0007 | 0.0038 |
| RDH16 | 1.6334 | 0.0139 | 0.0315 |
| PRXL2B | 0.7951 | 0.0256 | 0.0499 |
| IL4I1 | 1.9874 | 0.0000 | 0.0005 |
| ACADL | −3.1819 | 0.0002 | 0.0013 |
| PTGIS | −1.8784 | 0.0062 | 0.0177 |
| ACAD11 | 0.7438 | 0.0103 | 0.0249 |
| ACACB | −2.0952 | 0.0000 | 0.0005 |
| ADH1B | −2.3779 | 0.0000 | 0.0003 |
| ENO2 | 1.1322 | 0.0040 | 0.0138 |
| NBN | 0.6371 | 0.0022 | 0.0090 |
| LGALS1 | 1.7481 | 0.0001 | 0.0009 |
| GAPDHS | 2.0802 | 0.0059 | 0.0173 |
| SLC27A2 | 0.9832 | 0.0256 | 0.0499 |
| HSP90AA1 | 1.2248 | 0.0000 | 0.0002 |
| MLYCD | −0.9958 | 0.0000 | 0.0003 |
| SLC25A17 | 0.6682 | 0.0000 | 0.0003 |
| BCKDHB | −1.0404 | 0.0006 | 0.0034 |
| DBI | 0.6881 | 0.0062 | 0.0177 |
| GPD2 | 0.6431 | 0.0045 | 0.0142 |
| S100A10 | 0.7390 | 0.0046 | 0.0142 |
| HSD17B7 | 0.8224 | 0.0011 | 0.0054 |
| CYP4A11 | −1.0684 | 0.0003 | 0.0019 |
3.2. Establishing and Validating a Prognostic FRG Signature
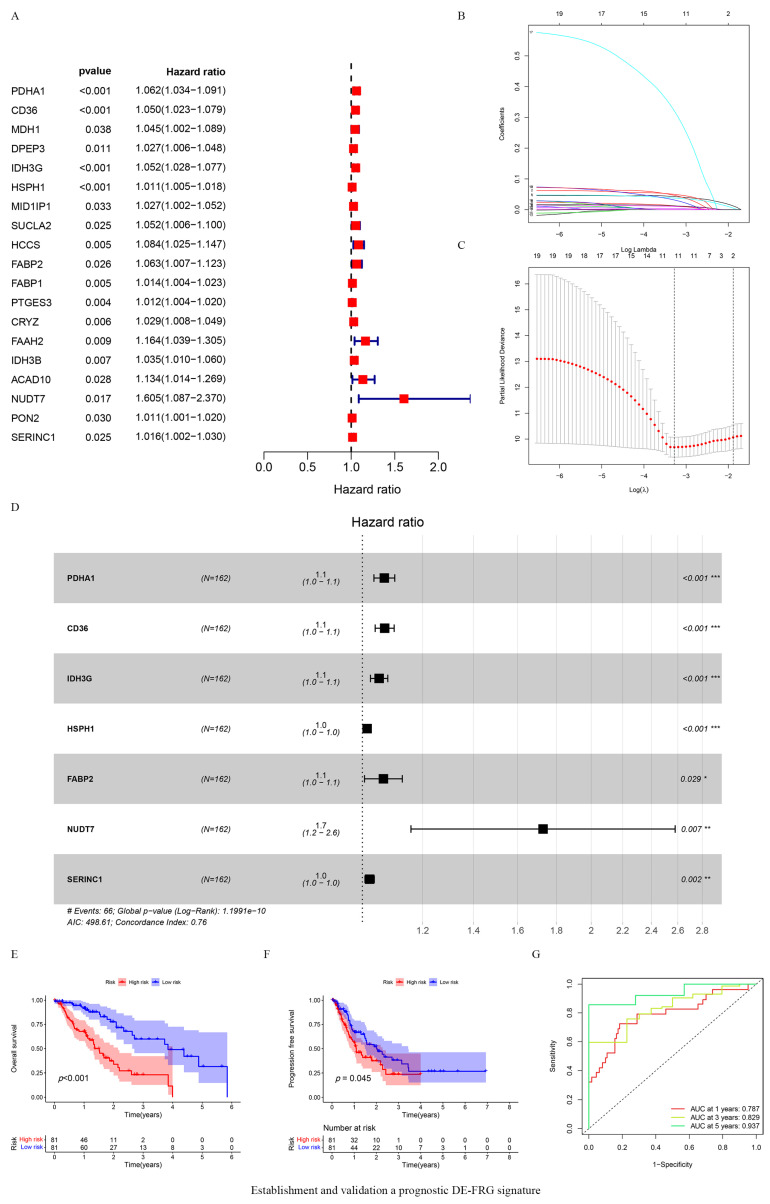
Univariate Cox regression analysis indicated that 19 differentially expressed DE-FRGs (DE-FRGs) were associated with different clinical outcomes (p < 0.05) (Figure 2A). LASSO analyses were used to further analyze 19 DE-FRGs to preventing overfitting. This additional analysis identified a signature, consisting of 11 genes, that showed altered expression levels depending on clinical prognosis (Figure 2B,C). Subsequently, seven DE-FRGs were identified as potential prognostic-related biomarkers of ESCA patients using multivariate Cox regression analysis (Figure 2D). Kaplan–Meier analysis indicated that compared with the low-risk group, the high-risk group was significantly associated with poor OS and PFS (p < 0.05) (Figure 2E,F). Furthermore, time-dependent ROC curves were constructed and the corresponding area under the curve (AUCs) figures were calculated to assess the predictive power of the prognostic risk model. The AUC values for 1-, 3-, and 5-year OS were 0.787, 0.829, and 0.937, respectively (Figure 2G).
Figure 2.
Establishment and validation a prognostic DE-FRG signature. (A) 19 genes were associated with the clinical outcomes of ESCA patients based on univariate Cox regression analysis; Red squares represnt Hazard Ratio (HR) value. (B) LASSO coefficient profiles of the 19 selected fatty acid metabolic genes; (C) The best parameter (lambda) in the LASSO-Cox model; the red dots indicates the partial probability of deviation values, the gray lines indicates standard error (SE). (D) 7 genes were eventually identified as prognosis related biomarkers based on multivariate Cox regression analysis; ESCA patients were divided into high-risk (n = 80) and low-risk (n = 80) groups based on the median risk score. * p < 0.05, ** p < 0.01, *** p < 0.001. Black squares represent Hazard Ratio (HR) value. (E,F) Kaplan–Meier survival analysis was performed to assess the difference in OS and PFS between the high-risk and low-risk group; (G) ROC curves of risk model for predicting overall survival at 1, 3, and 5 years.
3.3. Association between Risk Score and Clinical Characteristics
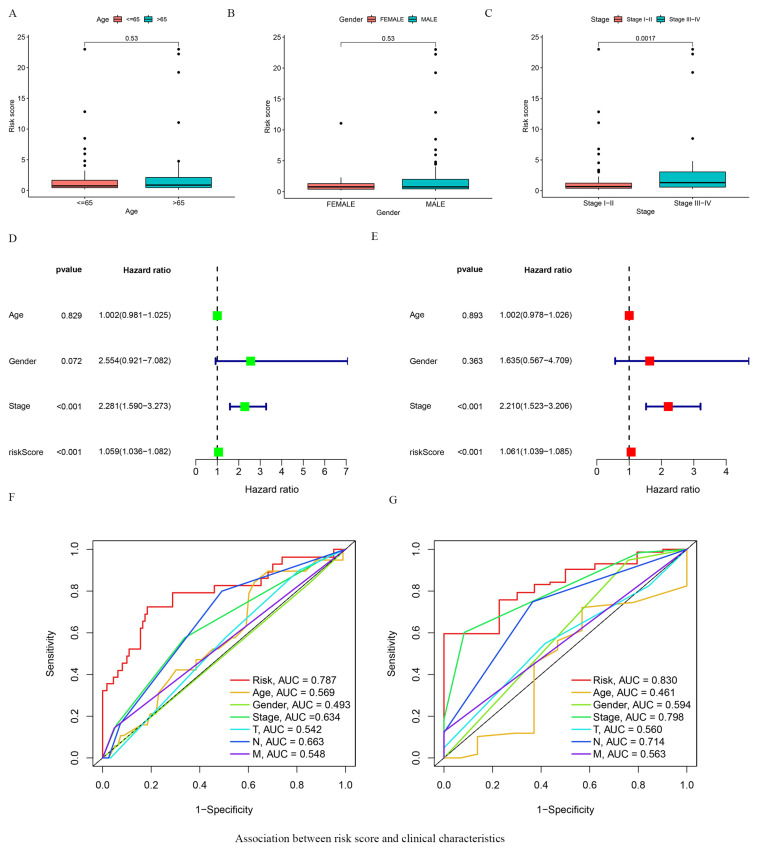
We analyzed the association between patient clinical characteristics and the gene expression-based risk score. This analysis showed no association between the age or gender of the patients and their corresponding risk scores (Figure 3A,B). However, a higher risk score correlated with significantly more advanced pathologic stage and T stage of the tumor (p = 0.0017, Figure 3C, Figure S1). Univariate and multivariate Cox regression analysis revealed that our risk score and the clinical T stage could serve as independent prognostic factors (p < 0.001) (Figure 3D,E). ROC curve analysis further supported the high sensitivity and specificity of the predictive score (Figure 3F,G).
Figure 3.
Association between risk score and clinical characteristics. (A–C) The association of risk score and clinicopathological features, including age (A), gender (B), and tumor stage (C); The circle represent each individual. (D) The univariate Cox regression analysis of clinical parameters in patients with ESCA; (E) The multivariate Cox regression analysis of clinical parameters in patients with ESCA; (F) ROC curves show 1-year survival in ESCA patients based on risk score and multiple clinical features; (G) ROC curves display 3-year survival in ESCA patients based on risk score and multiple clinical features. We excluded patients with deficient clinical information and 124 patients were retained for analysis.Red and green square represent HR value.
3.4. Establishing and Evaluating a Nomogram for Predicting Survival
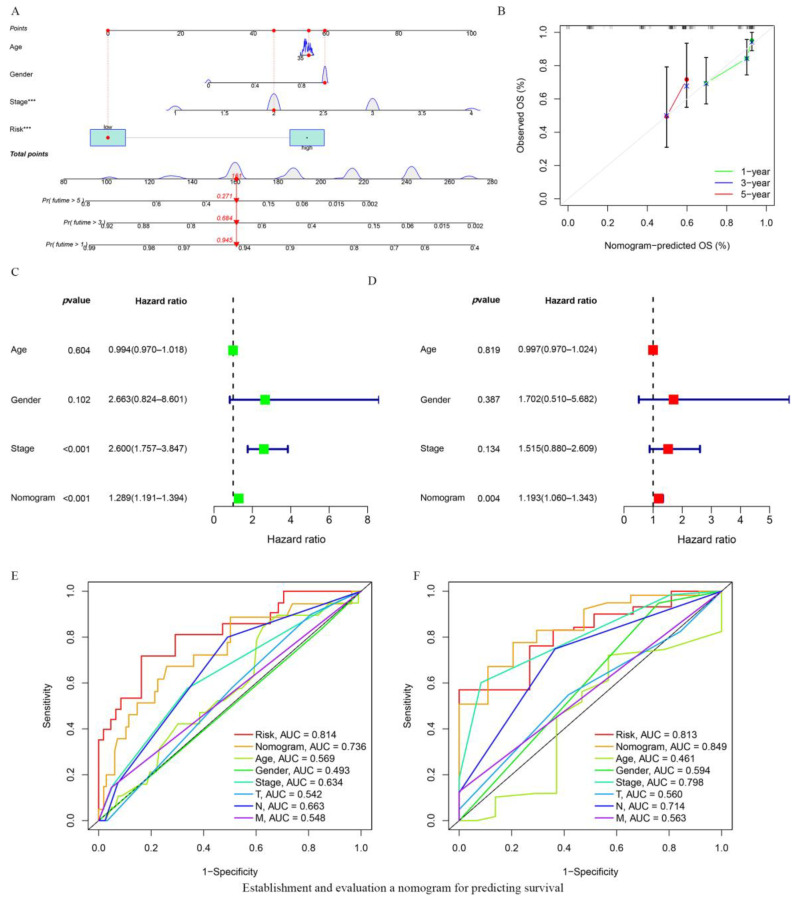
A nomogram, integrating age, gender, TNM stage of the tumor, and the risk score model was established to predict OS in ESCA patients (Figure 4A). The calibration curves at 1 year, 3 years, and 5 years indicated that the nomogram could accurately predict the OS of ESCA patients (Figure 4B). Based on univariate Cox regression analysis the nomogram model and the TNM tumor stage could predict the prognosis of the patients independently of each other or other clinical parameters (Figure 4C). Multivariate Cox regression analysis showed that the nomogram model was an independent prognostic factor with an HR of 1.193 (95% CI = 1.060–1.343) (Figure 4D). ROC analysis also revealed that the nomogram could predict the OS for ESCA patients with remarkable accuracy (AUC: 1 year = 0.736, 3 years = 0.849; Figure 4E,F).
Figure 4.
Establishment and evaluation a nomogram for predicting survival. (A) Nomogram predicting OS of ESCA patients. (B) The calibration plots of the nomogram. The x axis is nomogram-predicted survival, and the y axis is actual survival. (C) Univariate Cox regression analysis of the nomogram. (D) Multivariate Cox regression analysis of the nomogram. Red and green square represent HR value. (E,F) ROC curves at 1 and 3 years showing the predictive ability of the nomogram.
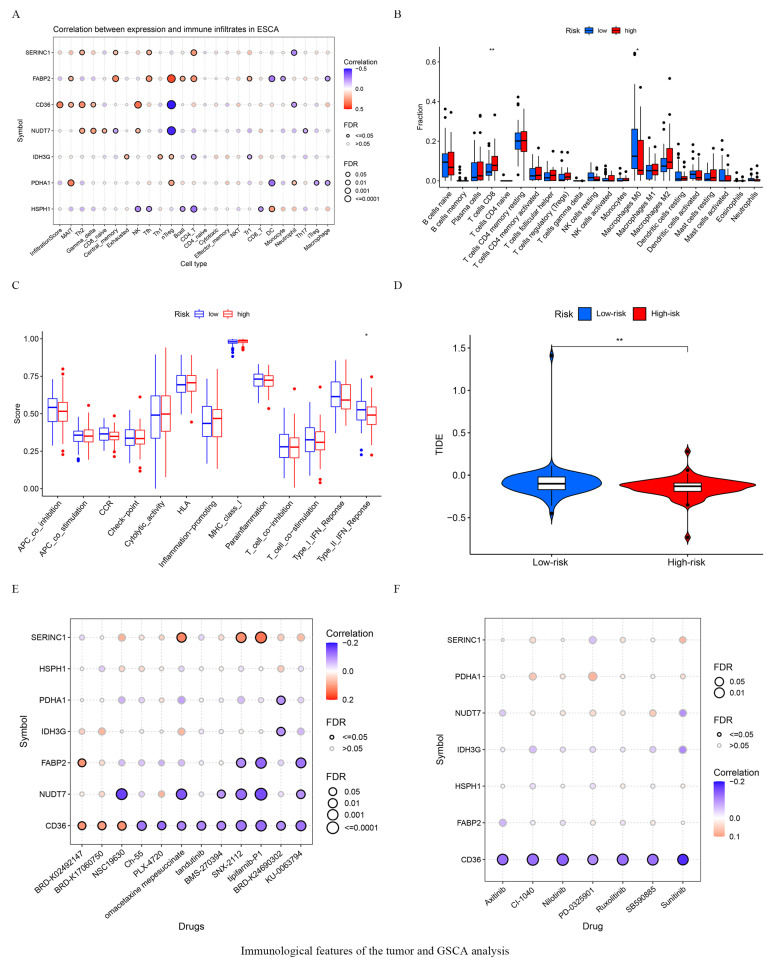
3.5. Immunological Features of the Tumor and GSCA Analysis
The expression of seven key FRGs showed strong correlation with the infiltration of the tumors with immune-activating cells (Figure 5A). The ratios of 22 distinct immune cell types in the tumors of high- and low-risk ESCA patients is shown in Figure 5B. The number of infiltrating CD8+ T cells was higher in the high-risk group, while M0 macrophages were more predominant in the low-risk patients (Figure 5B). In terms of immune function, our results also showed that type II IFN production was significantly higher in the low-risk group (Figure 5C). Furthermore, significantly higher TIDE scores were seen in the low-risk than group, indicating that potentially the high-risk group could benefit more from receiving immunotherapy (Figure 5D). We also explored the correlation between the expression of the seven key FRGs and drug sensitivity, defined as IC50 values, based on Spearman’s correlation analysis. Our results revealed that most drugs effected the seven key FRGs, suggesting that these molecules could be exploited as potential therapeutic drug targets in the management of ESCA (Figure 5E,F).
Figure 5.
Immunological features of the tumor and GSCA analysis. (A) The correlation between 7 key genes and infiltrated immune cells. Different color circle denote different p value, the smaller the p value, the larger the circle. (B) The ratios of 22 immune cell types in high- and low-risk patients. The black circle represents a single individual. (C) Immune function difference in high- and low-risk group in ESCA patients. (D) Comparison of Immunotherpy response between high- and low-risk patients. (E) Correlation between CTRP drug sensitivity and 7 key gene expressions. (F) Correlation between GDSC drug sensitivity and 7 key gene expressions. * p < 0.05, ** p < 0.01.
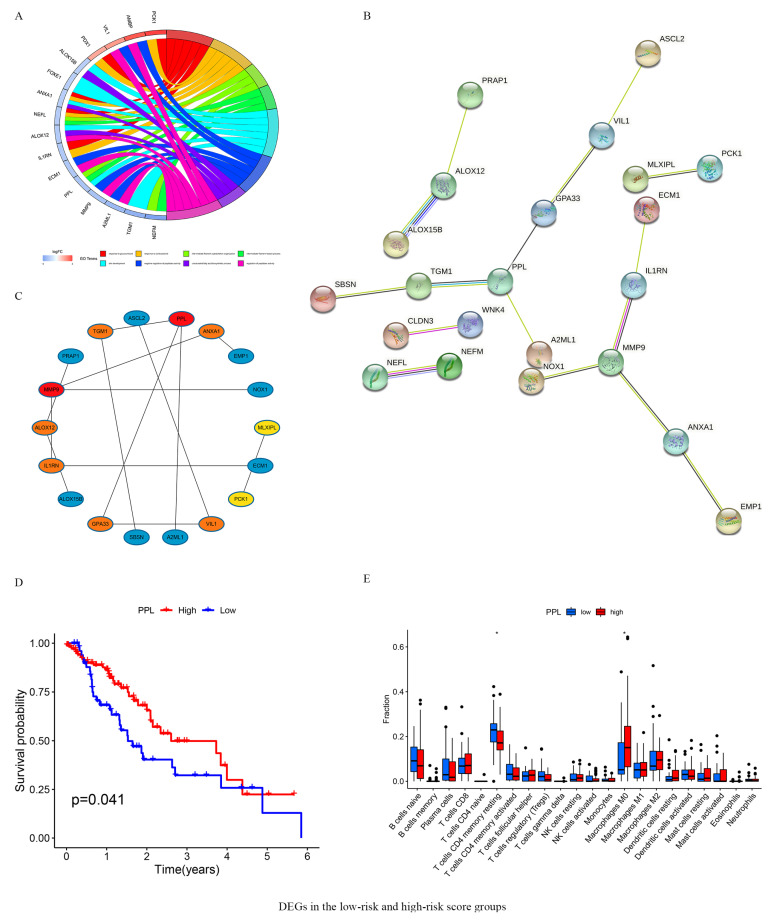
3.6. DEGs in the Low- and High-Risk Score Groups
Using the “Limma” package, we identified 48 genes that were differentially expressed between the high- and low-risk groups (Table 2). GO analyses indicated that these DEGs mainly belonged to the response to glucocorticoid, response to corticosteroid, intermediate filament cytoskeleton organization, intermediate filament-based process, skin development, negative regulation of peptidase activity, unsaturated fatty acid biosynthetic process, and regulation of peptidase activity KEGG pathways (Figure 6A). A PPI network of 48 DEGs was constructed using the STRING database (Figure 6B). The Top 10 genes of the network, including PPL, MMP9, TGM1, ALOX12, ANXA1, VIL1, IL1RN, GPA33, MLXIPL, and PCK1 of the network were selected using the cytoHubba plugin in Cytoscape (Figure 6C). Analyzing these against the survival parameters showed that high expression of PPL was significantly associated with favorable OS of ESCA patients (Figure 6D). The proportions of infiltrating immune cells in samples expressing PPL at high or low levels was analyzed using the “Limma” package. Our result showed that the presence of M0 macrophages was significantly more pronounced in samples with a high PPL high expression. In addition, the proportion of T cell CD4+ resting memory T cells was lower in the immune infiltrates in the high-PPL-expression group (Figure 6E).
Table 2.
Differentially expressed genes between high and low-risk groups.
| Gene | logFC | p-Value | FDR |
|---|---|---|---|
| KRT16P2 | −1.1383 | 0.0004 | 0.0175 |
| RNF225 | −1.5708 | 0.0038 | 0.0488 |
| FOXE1 | −1.1111 | 0.0035 | 0.0477 |
| USH1G | −1.1662 | 0.0024 | 0.0386 |
| BCAT1 | −1.1360 | 0.0015 | 0.0301 |
| MIEN1 | 1.1579 | 0.0012 | 0.0275 |
| MLXIPL | 1.2968 | 0.0012 | 0.0270 |
| PPL | −1.3393 | 0.0006 | 0.0204 |
| TMEM74B | 1.0136 | 0.0003 | 0.0142 |
| NEFM | −2.1322 | 0.0017 | 0.0325 |
| GBP6 | −1.4412 | 0.0018 | 0.0334 |
| KRT16P6 | −1.1958 | 0.0025 | 0.0398 |
| ALOX15B | −1.1045 | 0.0008 | 0.0223 |
| AHNAK2 | −1.0332 | 0.0002 | 0.0111 |
| AMBP | 2.2574 | 0.0031 | 0.0450 |
| MIR559 | 1.1306 | 0.0013 | 0.0275 |
| ALOX12 | −1.2123 | 0.0007 | 0.0212 |
| PCK1 | 2.4876 | 0.0000 | 0.0039 |
| PDX1 | 1.2546 | 0.0005 | 0.0187 |
| PINLYP | −1.1081 | 0.0003 | 0.0151 |
| FAM83C | −1.0035 | 0.0034 | 0.0471 |
| ANXA1 | −1.1213 | 0.0006 | 0.0199 |
| YBX2 | 1.1975 | 0.0000 | 0.0047 |
| MIR3189 | 1.1694 | 0.0028 | 0.0425 |
| NOX1 | 1.1536 | 0.0002 | 0.0125 |
| MMP9 | −1.3834 | 0.0012 | 0.0272 |
| EMP1 | −1.1129 | 0.0009 | 0.0241 |
| SBSN | −1.0531 | 0.0030 | 0.0439 |
| CCL15 | 1.6283 | 0.0036 | 0.0480 |
| ACY3 | 1.2213 | 0.0003 | 0.0153 |
| ASCL2 | 1.4926 | 0.0034 | 0.0467 |
| IL1RN | −1.2627 | 0.0002 | 0.0138 |
| A2ML1 | −1.6120 | 0.0008 | 0.0233 |
| TGM1 | −1.8265 | 0.0014 | 0.0295 |
| NEFL | −1.1713 | 0.0006 | 0.0200 |
| CLRN3 | 1.2620 | 0.0028 | 0.0427 |
| QPRT | 1.2541 | 0.0013 | 0.0275 |
| VIL1 | 1.4509 | 0.0032 | 0.0454 |
| WNK4 | 1.2133 | 0.0034 | 0.0469 |
| GOLT1A | 1.2505 | 0.0006 | 0.0206 |
| ANKRD33B | −1.0516 | 0.0016 | 0.0314 |
| CLDN3 | 1.4046 | 0.0004 | 0.0170 |
| PRAP1 | 1.5993 | 0.0020 | 0.0354 |
| ZBED2 | −1.3404 | 0.0001 | 0.0100 |
| ECM1 | −1.2862 | 0.0022 | 0.0368 |
| GPA33 | 1.5683 | 0.0023 | 0.0380 |
| BCAN-AS1 | 1.1560 | 0.0004 | 0.0175 |
| RNF157 | 1.2480 | 0.0011 | 0.0262 |
Figure 6.
DEGs in the low- and high- risk score groups. (A) The results of GO enrichment analysis on DEGs. (B) PPI network processed by STRING. (C) Identification of 10 hub DEGs using cytoHubba software. (D) Survival analysis for subgroup patients stratified by PPL mRNA expression. (E) The abundance of each infiltrated cell in patients with high- and low PPL mRNA expression. DEGs in 160 ESCA patients between 2 different risk groups were analyzed using the Limma R package (|logFC| > 1, FDR < 0.05). Black circle represent individual values. * p < 0.05.
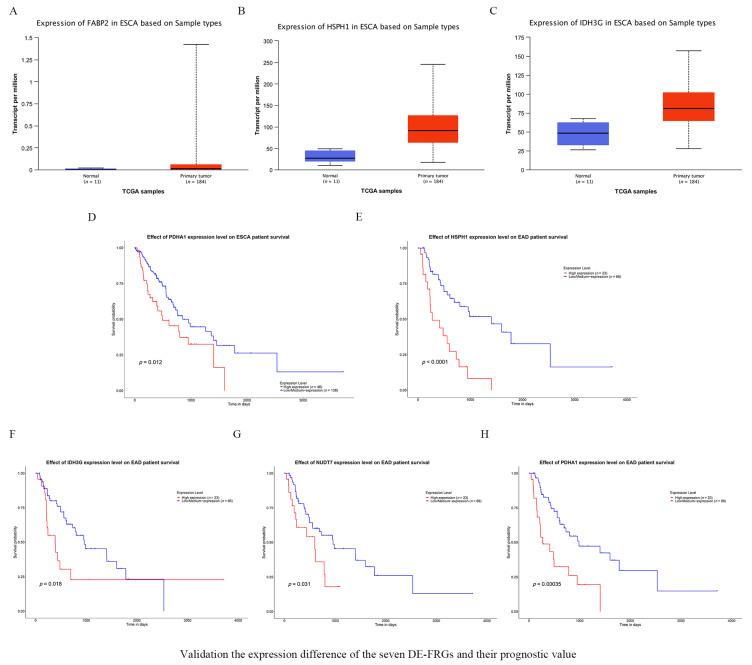
3.7. Validating the Expression of the Seven FRGs and Their Prognostic Value
The UALCAN online database was used to analyze how the expression of key FRGs affected survival times. The results indicated that FABP2, HSPH1, and IDH3G were more abundantly expressed in tumors (Figure 7A–C). Furthermore, a higher abundance of PDHA1 correlated with poor prognosis in ESCA patients (Figure 7D). Similarly, the increased expression of HSPH1, IDH3G, NUDT7 and PDHA1 was associated with shorter OS in the subgroup of patients suffering from esophageal adenocarcinoma (EAD) (Figure 7E–H).
Figure 7.
Validation the expression difference of the seven DE-FRGs and their prognostic value. (A–C) Confirmation of the expression of FRGs in ESCA. (A) FABP2, (B) HSPH1 and (C) IDH3G. (D) High PDHA1expression was correlated with poor OS in ESCA. (E–H) Survival rates were calculated between high and low gene expression groups in EAD patients. (E) HSPH1, (F) IDH3G, (G) NUDT7 and (H) PDHA1. EAD = Esophageal adenocarcinoma.
4. Discussion
There is increasing evidence that metabolic dysregulation plays a critical role in cancer cell growth, proliferation, angiogenesis, and invasiveness [14,26]. Previous observations suggest that abnormal glycolytic metabolism is associated with the physiological behavior of human malignant tumors [27]. In colorectal cancer, abnormal anaerobic metabolic pathways play an important role in the formation of cancer stem-like cells (CSCs), promoting the rapid formation, development, and therapy resistance of these tumors [28,29]. Fatty acid metabolism is involved in cellular energy production, membrane synthesis, and signal transduction pathways relevant to tumorigenesis and development [14,30]. Deregulated anabolism/catabolism of fatty acids may support cancer cell growth [6]. A previous study reported that fatty acid synthesis may promotes esophageal adenocarcinoma [31]. A recent study has shown that loss of FBP1 promotes migration, proliferation and invasion through regulating fatty acid metabolism in ESCA [32].Although several studies focused on the role of fatty acid metabolism in a variety of tumors, this topic remains unclear in ESCA. The identification of key molecular markers related to fatty acid metabolism, together with the exploration of their role in the development of ESCA, could provide novel insights into the biological behavior of these tumors, potentially highlighting new, more effective, therapeutic strategies.
In the present study, we explore the role of DE-FRGs in ESCA. Interestingly, 19 FRGs that were differentially expressed in patients with ESCA showed a correlation with the clinical/biological behavior of the tumors. Via LASSO and multivariate Cox regression analysis, we were able to identify seven key DE-FRGs, including PDHA1, CD36, IDH3G, HSPH1, FABP2, NUDT7 and SERINC1 that may become useful prognostic biomarkers in the clinical management of ESCA patients. Subsequently, a prognostic risk score model was established that was able to divide patients into distinct high- and low-risk groups showing significant differences in OS and DFS. This prognostic risk score was an independent prognostic factor according to univariate and multivariate Cox regression analyses. Furthermore, the predictive potential of this model was confirmed by combination with clinical characteristics (age, gender, and TNM stage of the tumor) in a risk-assessment nomogram. The risk model presented here might help to identify ESCA patients with a poor prognoses, and could be utilized in the management of the disease.
PDHA1(Pyruvate Dehydrogenase E1 Subunit Alpha 1) is a protein-coding gene involved in the pyruvate and thiamine metabolism pathways. In HNSCC cells, LDHA/PDHA1 changes may associated with a broad metabolic reprogramming while intracellular molecules including polyunsaturated fatty acids and nitrogen-metabolism-related metabolites underlie the malignant changes [33]. Previous study has shown that promotion of the tricarboxylic acid cycle (upregulation of PDHA1, PDHB, ACO2, and DLST expression) and inhibition of ME1 expression may inhibit fatty acid synthesis [34]. The inhibition of PDHA1 expression in the LnCap human prostate cancer cells led to the “Warburg effect”, resistance to chemotherapy, improved migration, and increased expression of stem cell markers [35]. In ESCC, the low expression of PDHA1 correlates with poor clinical prognosis and can result in metabolic reprogramming, again leading to the Warburg effect increasing malignant potential [36,37]. Decreased PDHA1 protein expression was also found to predict poor prognosis in gastric cancer [38]. HSPH1 encodes a member of the heat shock protein 70 family. Previously, elevated levels of HSPH1 expression were reported in tissues of HNSC patients. Moreover, the overexpression of HSPH1 was associated with poor overall survival (OS) [39]. HSPH1 is one of the most prominently upregulated proteins in several malignancies, with a well-documented involvement in Wnt- and chronic nuclear factor-kappa B signaling [40,41,42]. It has been suggested that the analysis of HSPH1 expression levels may help in predicting the effectiveness of chemotherapeutic approaches acting on the EGFR-TKI pathway in advanced lung adenocarcinoma [40]. The involvement of HSPH1 in anticancer immunity has also been described [41]. NUDT7, Nudix Hydrolase 7, is an enzyme involved in peroxisomal lipid metabolism. In the liver, upregulated expression of NUDT7 can inhibit peroxisomal fatty acid oxidation [43]. In clear cell renal cell carcinomas the alternative splicing of NUDT7 was found to be correlated with overall survival time [44]. Fatty acid binding proteins (FABPs) are key proteins in lipid transport, which can maintain a steady pool of fatty acids in the epithelium by traffic lipids from the intestinal lumen to enterocytes and bind superfluous fatty acids. As a lipid chaperone, FABP2 can also carry lipophilic drugs to improve targeted transport [45]. During the work presented here we identified and validated the prognostic value of seven key DE-FRGs. Of these FABP2, HSPH1, and IDH3G were upregulated and the high expression of four FRGs, including PDHA1, HSPH1, IDH3G, and NUDT7 was associated with poorer OS. These findings suggest that these key DE-FRGs might have a prognostic value in the assessment of ESCA and could represent potential therapeutic targets.
Previous studies have suggested that prominent CD8+ T cell infiltrates were associated with clinical prognosis and immune responses in ESCA [46,47]. In renal cell cancer an increased CD8+ T-cells to Treg ratio is associated with poor prognosis [48]. We observed an increase in the number of infiltrating CD8+ T cells in the samples of high-risk group ESCA patients, indicating an unfavorable prognosis. In contrast, patients with a low-risk score showed an activated type II IFN response. Based on the TIDE algorithm this may indicate suitability for the immunotherapy. Previous studies demonstrated that type II IFN (IFN-γ) could indirectly regulate PD-L1 levels in small cell lung cancer [49]. It was also suggested that IFN-γ production induced elevated PD-L1-mRNA expression resulting in favorable OS [50]. Indeed IFN-γ production is a key driver of PD-L1 expression in both cancer and host cells, and may improve the likelihood of anti–PD-1 therapies being effective [51]. These previous findings support the notion that the favorable prognosis of patients in the low-risk group may be due to more effective immune responses.
It was reported that the expression of PPL was significantly decreased in esophageal cancer tissues and that the proteins were barely detectable in advanced cancer samples [52]. The experimental knockdown of PPL decreased cellular motility, reduced attachment, and generally inhibited malignant progression [53]. However, this finding contrasts with observations that the high expression of PPL is associated with favorable survival in patients with adenoid cystic carcinomas and sarcomas [54]. Our results showed that the expression of PPL gene was upregulated in low-risk group, and high expression of this gene was associated with better prognoses. This result may further support the hypothesis that the low-risk group was significantly associated with a favorable prognosis.
5. Conclusions
In summary, this first investigation of the role of fatty acid metabolism-related genes in ESCA identified seven genes showing strong association with the prognosis and therapeutic responses to chemotherapy and immunotherapy in this disease. This allowed us to develop a risk score model that could effectively divide patients into high- and low-risk groups. Using this score clinically might aid the development of individualized treatment strategies in the future. However, the prognosis value and immunotherapy effect of seven fatty-acid-metabolism-related genes in patients with esophageal cancer need to be further confirmed in larger clinical studies.
Acknowledgments
The authors thank Zhong Guo Liang for bioinformatics-related technical support.
Abbreviations
AUCs—areas under the curve; CSCs—cancer-stem-like cells; DEGs—differentially expressed genes; DE-FRGs—differentially expressed fatty-acid-metabolism-related genes; EAD—esophageal adenocarcinoma; ESCA—esophageal cancer; FRGs—fatty-acid-metabolism-related genes; GSCA—gene set cancer analysis; LASSO—least absolute shrinkage and selection operator; OS—overall survival; ROC—receiver operating characteristic; TCGA—The Cancer Genome Atlas; TME—tumor microenvironment; TIDE—tumor immune dysfunction and exclusion.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/vaccines10101721/s1, Figure S1: The association between risk score and clinical characteristics; Supplementary Table S1: Details of clinical characteristic; Supplementary Table S2: Summarized clinical information.
Author Contributions
Y.G. analyzed the data and wrote the main text of the article. S.P. contributed to manuscript revision. Y.K., J.P. and Y.L. participated in preparing Figure 1, Figure 2, Figure 3, Figure 4, Figure 5, Figure 6 and Figure 7 and prepared the tables. H.M. conceived and designed the study. All authors contributed to the article and approved the submitted version. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data of this study were obtained from the TCGA database (https://portal.gdc.cancer.gov/), which is a publicly available database. FRGs are derived from the Molecular Signature Database (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb).
Conflicts of Interest
The authors declare that they have no competing interests.
Funding Statement
This work was supported by the Key research and development program of Shaanxi Province (grant number 2022SF-213).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Li Y., Sebastian Zschaeck Q., Chen S., Chen L., Wu H. Metabolic parameters of sequential 18F-FDG PET/CT predict overall survival of esophageal cancer patients treated with (chemo-) radiation. Radiat. Oncol. 2019;14:35. doi: 10.1186/s13014-019-1236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Peng Z., Liu X.-Y., Cheng Z., Kai W., Song Z. Comprehensive analysis of a new immune-related prognostic signature for esophageal cancer and its correlation with infiltrating immune cells and target genes. Ann. Transl. Med. 2021;9:1576. doi: 10.21037/atm-21-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mao Y., Gao S., Wang Q., Shi X., Li Y., Gao W., Guan F., Li X., Han Y., Liu Y., et al. Analysis of a registry database for esophageal cancer from high-volume centers in China. Dis. Esophagus. 2020;33:doz091. doi: 10.1093/dote/doz091. [DOI] [PubMed] [Google Scholar]
- 4.Ding C., Shan C., Li M., Chen H., Li X., Jin Z. Characterization of the fatty acid metabolism in colorectal cancer to guide clinical therapy. Mol. Ther. Oncolytics. 2021;20:532–544. doi: 10.1016/j.omto.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schulze A., Harris A. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature. 2012;491:364–373. doi: 10.1038/nature11706. [DOI] [PubMed] [Google Scholar]
- 6.Li Z., Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell. Mol. Life Sci. CMLS. 2016;73:377–392. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arkaitz Carracedo L.C.C., Pandolfi P.P. Cancer metabolism: Fatty acid oxidation in the limelight. Nat. Rev. Cancer. 2013;13:227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qi Y., Chen D., Lu Q., Yao Y., Ji C. Bioinformatic Profiling Identifies a Fatty Acid Metabolism-Related Gene Risk Signature for Malignancy, Prognosis, and Immune Phenotype of Glioma. Dis. Markers. 2019;2019:3917040. doi: 10.1155/2019/3917040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C., Liao Y., Liu P., Du Q., Liang Y., Ooi S., Qin S., He S., Yao S., Wang W. FABP5 promotes lymph node metastasis in cervical cancer by reprogramming fatty acid metabolism. Theranostics. 2020;10:6561–6580. doi: 10.7150/thno.44868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tabe Y., Konopleva M., Andreeff M. Fatty Acid Metabolism, Bone Marrow Adipocytes, and AML. Front. Oncol. 2020;10:155. doi: 10.3389/fonc.2020.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han S., Wei R., Zhang X., Jiang N., Fan M., Huang J., Xie B., Zhang L., Miao W., Butler A., et al. CPT1A/2-Mediated FAO Enhancement-A Metabolic Target in Radioresistant Breast Cancer. Front. Oncol. 2019;9:1201. doi: 10.3389/fonc.2019.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu Y., Fabritius M., Ip C. Chemotherapeutic sensitization by endoplasmic reticulum stress: Increasing the efficacy of taxane against prostate cancer. Cancer Biol. Ther. 2009;8:146–152. doi: 10.4161/cbt.8.2.7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corn K., Windham M., Rafat M. Lipids in the tumor microenvironment: From cancer progression to treatment. Prog. Lipid Res. 2020;80:101055. doi: 10.1016/j.plipres.2020.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He D., Cai L., Huang W., Weng Q., Lin X., You M., Liao S. Prognostic value of fatty acid metabolism-related genes in patients with hepatocellular carcinoma. Aging. 2021;13:17847–17863. doi: 10.18632/aging.203288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu D., Zhang P., Li F., Shen Y., Chen H., Feng Y., He A., Wang F. CD138 multiple myeloma cells express high level of CHK1 which correlated to overall survival in MM patient. Aging. 2020;12:23067–23081. doi: 10.18632/aging.104066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z., Du M., Lu L. A Novel 16-Genes Signature Scoring System as Prognostic Model to Evaluate Survival Risk in Patients with Glioblastoma. Biomedicines. 2022;10:317. doi: 10.3390/biomedicines10020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z.-K., Wu K.-F., Zhang R.-Y., Kong L.-M., Shang R.-Z., Lv J.-J., Li C., Lu M., Yong Y.-L., Zhang C., et al. Pyroptosis-Related LncRNA Signature Predicts Prognosis and Is Associated With Immune Infiltration in Hepatocellular Carcinoma. Front. Oncol. 2022;12:794034. doi: 10.3389/fonc.2022.794034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J., Chen W., Chen C., Jie Z., Xiao T. Comprehensive Analysis and Prognosis Prediction of N6-Methyladenosine-Related lncRNAs in Immune Microenvironment Infiltration of Gastric Cancer. Int. J. Gen. Med. 2022;15:2629–2643. doi: 10.2147/IJGM.S349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiang L., Lan C. Significance of a Tumor Mutation Burden Gene Signature with Prognosis and Immune Feature of Gastric Cancer Patients. Int. J. Genom. 2022;2022:1–15. doi: 10.1155/2022/7684606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Cheng Q., Chang K., Bao L., Yi X. Integrated Analysis of Ferroptosis-Related Biomarker Signatures to Improve the Diagnosis and Prognosis Prediction of Ovarian Cancer. Front. Cell Dev. Biol. 2021;9:807862. doi: 10.3389/fcell.2021.807862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu C., Hu F., Xia M., Han L., Zhang Q., Guo A. GSCALite: A web server for gene set cancer analysis. Bioinformatics. 2018;34:3771–3772. doi: 10.1093/bioinformatics/bty411. [DOI] [PubMed] [Google Scholar]
- 22.Xia Q.-D., Sun J.-X., Liu C.-Q., Xu J.-Z., An Y., Xu M.-Y., Liu Z., Hu J., Wang S.-G. Ferroptosis Patterns and Tumor Microenvironment Infiltration Characterization in Bladder Cancer. Front. Cell Dev. Biol. 2022;10:832892. doi: 10.3389/fcell.2022.832892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu Z., Wang X., Xu Y., Li Y., Gong X., Zeng Q., Zhang B., Xi J., Pei X., Yue W., et al. Development and Validation of a Novel Survival Model for Cutaneous Melanoma Based on Necroptosis-Related Genes. Front. Oncol. 2022;12:852803. doi: 10.3389/fonc.2022.852803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei X., Zhang M., Guan B., Chen Q., Dong Z., Wang C. Identification of hub genes associated with prognosis, diagnosis, immune infiltration and therapeutic drug in liver cancer by integrated analysis. Hum. Genom. 2021;15:39. doi: 10.1186/s40246-021-00341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chandrashekar D., Bashel B., Balasubramanya S., Creighton C., Ponce-Rodriguez I., Chakravarthi B., Varambally S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao D., Wu J., Zhao H., Nie X.C. RPP25 as a Prognostic-Related Biomarker That Correlates With Tumor Metabolism in Glioblastoma. Front. Oncol. 2021;11:714904. doi: 10.3389/fonc.2021.714904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei X., Mao T., Li S., He J., Hou X., Li H., Zhan M., Yang X., Li R., Xiao J., et al. DT-13 inhibited the proliferation of colorectal cancer via glycolytic metabolism and AMPK/mTOR signaling pathway. Phytomedicine Int. J. Phytother. Phytopharm. 2019;54:120–131. doi: 10.1016/j.phymed.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Vincent Z., Urakami K., Maruyama K., Yamaguchi K., Kusuhara M. CD133-positive cancer stem cells from Colo205 human colon adenocarcinoma cell line show resistance to chemotherapy and display a specific metabolomic profile. Genes Cancer. 2014;5:250–260. doi: 10.18632/genesandcancer.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeki S.S., Graham T.A., Wright N.A. Stem cells and their implications for colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2011;8:90–100. doi: 10.1038/nrgastro.2010.211. [DOI] [PubMed] [Google Scholar]
- 30.Currie E., Schulze A., Zechner R., Walther T.C., Farese R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma S., Zhou B., Yang Q., Pan Y., Yang W., Freedland S., Ding L., Freeman M., Breunig J., Bhowmick N., et al. A Transcriptional Regulatory Loop of Master Regulator Transcription Factors, PPARG, and Fatty Acid Synthesis Promotes Esophageal Adenocarcinoma. Cancer Res. 2021;81:1216–1229. doi: 10.1158/0008-5472.CAN-20-0652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y., Hua R., Li B., Gu H., Sun Y., Li Z. Loss of FBP1 promotes proliferation, migration, and invasion by regulating fatty acid metabolism in esophageal squamous cell carcinoma. Aging. 2020;13:4986–4998. doi: 10.18632/aging.103916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T., Hsieh Y., Huang J., Liu C., Chuang L., Huang P., Kuo T., Chia H., Chou C., Chang C., et al. Determination of Pyruvate Metabolic Fates Modulates Head and Neck Tumorigenesis. Neoplasia. 2019;21:641–652. doi: 10.1016/j.neo.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng M., Han J., Li L., Ma H. Suppression of fat deposition in broiler chickens by (-)-hydroxycitric acid supplementation: A proteomics perspective. Sci. Rep. 2016;6:32580. doi: 10.1038/srep32580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong Y., Li X., Ji Y., Li X., Li Y., Yu D., Yuan Y., Liu J., Li H., Zhang M., et al. Pyruvate dehydrogenase expression is negatively associated with cell stemness and worse clinical outcome in prostate cancers. Oncotarget. 2017;8:13344–13356. doi: 10.18632/oncotarget.14527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong Y., Huang R., Li X., Xu R., Zhou F., Wang J., Fan H., Goscinski M., Zhang M., Wen J.-G., et al. Decreased Expression of PDHE1α Predicts Worse Clinical Outcome in Esophageal Squamous Cell Carcinoma. Anticancer. Res. 2015;35:5533–5538. [PubMed] [Google Scholar]
- 37.Liu L., Cao J., Zhao J., Li X., Suo Z., Li H. PDHA1 Gene Knockout In Human Esophageal Squamous Cancer Cells Resulted In Greater Warburg Effect And Aggressive Features In Vitro And In Vivo. OncoTargets Ther. 2019;12:9899–9913. doi: 10.2147/OTT.S226851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S., Liu D., Zhang X., Zhu X., Lu X., Huang J., Yang L., Wu Y. Low expression of PDHA1 predicts poor prognosis in gastric cancer. Pathol. Res. Pract. 2019;215:478–482. doi: 10.1016/j.prp.2018.12.038. [DOI] [PubMed] [Google Scholar]
- 39.Fan G., Tu Y., Wu N., Xiao H. The expression profiles and prognostic values of HSPs family members in Head and neck cancer. Cancer Cell Int. 2020;20:220. doi: 10.1186/s12935-020-01296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Y., Zhang N., Zhang L., Song Y., Liu J., Yu J., Yang M. Oncogene HSPH1 modulated by the rs2280059 genetic variant diminishes EGFR-TKIs efficiency in advanced lung adenocarcinoma. Carcinogenesis. 2020;41:1195–1202. doi: 10.1093/carcin/bgaa069. [DOI] [PubMed] [Google Scholar]
- 41.Berthenet K., Christophe B., Collura A., Svrcek M., Richaud S., Hammann A., Yousfi S.C.N., Wanherdrick K., Duplomb L. Extracellular HSP110 skews macrophage polarization in colorectal cancer. Oncoimmunology. 2016;5:e1170264. doi: 10.1080/2162402X.2016.1170264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zappasodi R., Ruggiero G., Tortoreto M., Tringali C., Cavanè A., Cabras A.D., Castagnoli L., Venerando B., Zaffaroni N., Gianni A.M., et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood. 2015;125:1768–1771. doi: 10.1182/blood-2014-07-590034. [DOI] [PubMed] [Google Scholar]
- 43.Shumar S., Kerr E., Fagone P., Infante A., Leonardi R. Overexpression of Nudt7 decreases bile acid levels and peroxisomal fatty acid oxidation in the liver. J. Lipid Res. 2019;60:1005–1019. doi: 10.1194/jlr.M092676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T., Zheng W., Chen J., Lin S., Zou Z., Li X., Tan Z. Systematic analysis of survival-associated alternative splicing signatures in clear cell renal cell carcinoma. J. Cell. Biochem. 2020;121:4074–4084. doi: 10.1002/jcb.29590. [DOI] [PubMed] [Google Scholar]
- 45.Huang X., Zhou Y., Sun Y., Wang Q. Intestinal fatty acid binding protein: A rising therapeutic target in lipid metabolism. Prog. Lipid Res. 2022;87:101178. doi: 10.1016/j.plipres.2022.101178. [DOI] [PubMed] [Google Scholar]
- 46.Duan J., Xie Y., Qu L., Wang L., Zhou S., Wang Y., Fan Z., Yang S., Jiao S. A nomogram-based immunoprofile predicts overall survival for previously untreated patients with esophageal squamous cell carcinoma after esophagectomy. J. Immunother. Cancer. 2018;6:100. doi: 10.1186/s40425-018-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao Y., Guo W., Geng X., Zhang Y., Zhang G., Qiu B., Tan F., Xue Q., Gao S., He J. Prognostic value of tumor-infiltrating lymphocytes in esophageal cancer: An updated meta-analysis of 30 studies with 5122 patients. Ann. Transl. Med. 2020;8:822. doi: 10.21037/atm-20-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murakami T., Tanaka N., Takamatsu K., Hakozaki K., Fukumoto K., Masuda T., Mikami S., Kakimi T.S.K., Tsunoda T., Sawada K., et al. Multiplexed single-cell pathology reveals the association of CD8 T-cell heterogeneity with prognostic outcomes in renal cell carcinoma. Cancer Immunol. Immunother. CII. 2021;70:3001–3013. doi: 10.1007/s00262-021-03006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi H., Caeser R., Chavan S., Zhan Y., Chow A., Manoj P., Uddin F., Kitai H., Qu R., Hayatt O., et al. WEE1 inhibition enhances the antitumor immune response to PD-L1 blockade by the concomitant activation of STING and STAT1 pathways in SCLC. Cell Rep. 2022;39:110814. doi: 10.1016/j.celrep.2022.110814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hänze J., Wegner M., Noessner E., Hofmann R., Hegele A. Co-Regulation of Immune Checkpoint PD-L1 with Interferon-Gamma Signaling is Associated with a Survival Benefit in Renal Cell Cancer. Target. Oncol. 2020;15:377–390. doi: 10.1007/s11523-020-00728-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D., Albright A., Cheng J., Kang S., Shankaran V., et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J. Clin. Investig. 2017;127:2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Otsubo T., Hagiwara T., Tamura-Nakano M., Sezaki T., Miyake O., Hinohara C., Shimizu T., Yamada K., Dohi T., Kawamura Y.I. Aberrant DNA hypermethylation reduces the expression of the desmosome-related molecule periplakin in esophageal squamous cell carcinoma. Cancer Med. 2015;4:415–425. doi: 10.1002/cam4.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tonoike Y., Matsushita K., Tomonaga T., Katada K., Tanaka N., Shimada H., Nakatani Y., Okamoto Y., Nomura F. Adhesion molecule periplakin is involved in cellular movement and attachment in pharyngeal squamous cancer cells. BMC Cell Biol. 2011;12:41. doi: 10.1186/1471-2121-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding L.L., Zhang L., Zhang H., Zhang Y., Liu D. Mechanism of periplakin on ovarian cancer cell phenotype and its influence on prognosis. Transl. Cancer Res. 2022;11:1372–1385. doi: 10.21037/tcr-22-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data of this study were obtained from the TCGA database (https://portal.gdc.cancer.gov/), which is a publicly available database. FRGs are derived from the Molecular Signature Database (MSigDB; https://www.gsea-msigdb.org/gsea/msigdb).