Abstract
Griseofulvin is an antifungal polyketide metabolite produced mainly by ascomycetes. Since it was commercially introduced in 1959, griseofulvin has been used in treating dermatophyte infections. This fungistatic has gained increasing interest for multifunctional applications in the last decades due to its potential to disrupt mitosis and cell division in human cancer cells and arrest hepatitis C virus replication. In addition to these inhibitory effects, we and others found griseofulvin may enhance ACE2 function, contribute to vascular vasodilation, and improve capillary blood flow. Furthermore, molecular docking analysis revealed that griseofulvin and its derivatives have good binding potential with SARS-CoV-2 main protease, RNA-dependent RNA polymerase (RdRp), and spike protein receptor-binding domain (RBD), suggesting its inhibitory effects on SARS-CoV-2 entry and viral replication. These findings imply the repurposing potentials of the FDA-approved drug griseofulvin in designing and developing novel therapeutic interventions. In this review, we have summarized the available information from its discovery to recent progress in this growing field. Additionally, explored is the possible mechanism leading to rare hepatitis induced by griseofulvin. We found that griseofulvin and its metabolites, including 6-desmethylgriseofulvin (6-DMG) and 4- desmethylgriseofulvin (4-DMG), have favorable interactions with cytokeratin intermediate filament proteins (K8 and K18), ranging from −3.34 to −5.61 kcal mol−1. Therefore, they could be responsible for liver injury and Mallory body (MB) formation in hepatocytes of human, mouse, and rat treated with griseofulvin. Moreover, the stronger binding of griseofulvin to K18 in rodents than in human may explain the observed difference in the severity of hepatitis between rodents and human.
Keywords: drug repurposing, dermatophytic fungi, griseofulvin, griseofulvin derivatives, gsf gene cluster, polyketide compound, SARS-CoV-2, spindle microtubule
1. Introduction
Griseofulvin (C17H17ClO6) is a natural product that was first discovered and isolated from Penicillium griseofulvum in the 1939. In addition to Penicillium, griseofulvin may be isolated from other genera of ascomycetes including Xylaria flabelliformis, Abieticola koreana, and Stachybotrys levispora [1,2,3,4]. Griseofulvin has a wide range of applications, from agriculture to medicine. In agriculture, griseofulvin is used as a crop protectant to prevent fungal colonization and infection [5]. In medicine, griseofulvin has been widely used as an antifungal drug and in treating ringworm and dermatophyte infections in humans and animals [6,7] due to its low toxicity. In cancer research, griseofulvin has shown inhibitory effects on cancer cell division and may induce cell death through interaction with the mitotic spindle microtubule [8]. Furthermore, griseofulvin may inhibit hepatitis C virus replication by interfering with microtubule polymerization in human cells [9].
Griseofulvin has long been known to cause vasodilation and improve capillary blood flow [10,11]. A new look at this old drug in our study revealed that griseofulvin has a good binding potential with the SARS-CoV-2 main protease and with the human ACE2 receptor [12]. ACE2 facilitates the breakdown of ANG II which reduce blood pressure and inflammation [13]. Our molecular docking results suggest a promising direction in drug repurposing to enhance ACE2 breakdown to relieve COVID symptoms [12].
In this review article, we summarized the history of griseofulvin research since its discovery to the recent findings that suggest a promising future for the clinical application of this fungal secondary metabolite. We also studied the griseofulvin interaction with cytoplasmic intermediate filament proteins using molecular docking to find the binding affinity toward keratin 8 and 18 in human, rat, and mouse. Our findings revealed that griseofulvin has good binding potential with K18 in rat, followed by mouse, which helps explain the significance of mild liver problems in human and the more serious problems in mice.
2. Discovery of Griseofulvin and Its Biosynthetic Gene Cluster in Fungi
In 1939, griseofulvin was first isolated and is still widely used as an antifungal mainly caused by dermatophytes [6,7]. Over the next decade of research, Brian and Hemming (1947) discovered a metabolite of Penicillium janczawski that could cause hyphal waving of plant pathogen Botrytis allii and named it the ‘curling factor’ [14]. This substance was later shown to be biologically and chemically identical to the product of P. griseofulvum which was identified and isolated by Oxford et al. (1939) under the name of griseofulvin [15].
Griseofulvin is produced from many species of the genus Penicillium, which is one of the most important fungal taxa frequently found around the world [2,3,4,16,17,18,19,20,21,22,23,24,25,26,27]. Following P. griseofulvum, P. lanosocoeruleum, a plant pathogen, is one of the most significant fungi that has recently been shown to produce griseofulvin [28]. Further, Abieticola koreana, Stachybotrys levispor, and Xylaria flabelliformis (previously known as X. cubensis) were also shown to produce griseofulvin [2,3,4]. Of note, fungal endophytes such as Xylaria are abundant in plant tissue that produce griseofulvin as an antifungal against plant pathogenic fungi [1,29,30].
Griseofulvin is a polyketide derived from acetyl and malonyl CoA precursor molecules to form dehydrogriseofulvin. The A-B-C ring structures of griseofulvin were first discovered by oxidative degradation [31]. Findings of Brich et al. (1958) showed that 6-methylsalicylic acid (6-MSA), the carbon skeleton of griseofulvin, is assembled from one acetyl CoA and three molecules of malonyl CoA [32,33]. The biosynthetic structure of griseofulvin was elucidated by 13C-NMR study using singly and doubly labeled acetate [34], and the crystal structure of griseofulvin was revealed by X-ray crystallography and UV spectroscopy [35,36,37].
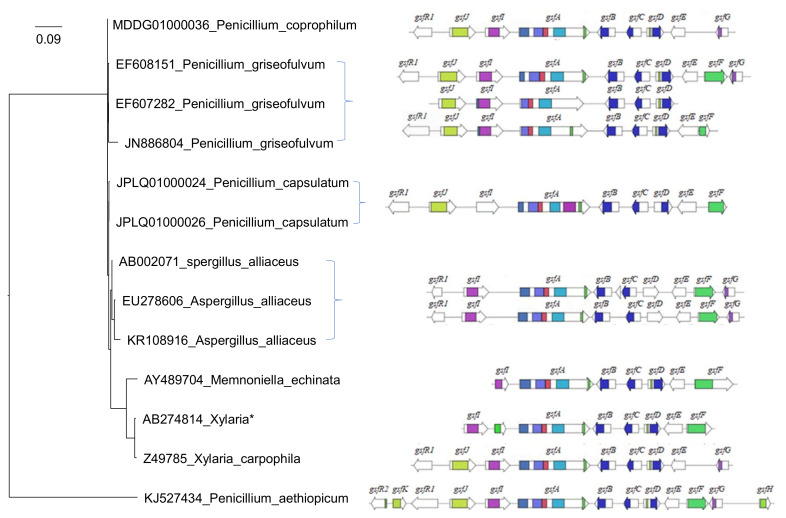
In 2010, the griseofulvin biosynthetic gene cluster (gsf BGC) responsible for griseofulvin production in P. aethiopicum was fully discovered through shotgun sequencing and bioinformatics mining (Figure 1) [16]. Recently, we performed a comparative genome study of more than 266 fungal species and showed that Memnoniella echinate, with only seven out of the thirteen putative gsf cluster genes (Figure 1), can still synthesize Griseofulvin [38]. A phylogenetic analysis reconstructed with 18S rRNA sequences shows the global relationships among whole gsf BGCs of 13 surveyed fungal species including P. aethiopicum, X. flabelliformis, M. echinata, P. griseofulvum, P. vulpinum, P. coprophilum, and A. alliaceus (Figure 1). The 18S rRNA sequences do not have strain information, so there is no one to one correspondence between the leaves and the gene clusters when multiple strains are involved.
Figure 1.
Griseofulvin biosynthetic gene cluster in P. aethiopicum and other fungal species The arrows show the gsf gene candidates and their orientations in griseofulvin biosynthesis gene cluster. The phylogenetic tree is reconstructed with 18S rRNA sequences (in the format of accession_species) with the PhyML software after alignment with MAFFT. The gsf genes for Xlaria* is X. flabelliformis which does not have an 18S rRNA sequence. X. angulosa was used instead. P. aethiopicum has only 33 nt of the 18S rRNA in contrast to other sequences that are ~1700 nt long.
3. Biosynthesis of Griseofulvin
3.1. Gene Cluster Evolution Hypothesis
Genes producing secondary metabolites in fungi are frequently linked and located in biosynthetic gene clusters (BGCs) [39]. It has been hypothesized that horizontal gene transfer (HGT) plays a vital role in this phenomenon. This is so because genes involved in fungal metabolic pathways are frequently physically clustered, increasing the possibility of pathway transfers occurring in a single event. There is evidence that sterigmatocystin (ST), a highly toxic secondary metabolite that serves as a precursor to aflatoxins (AF), was horizontally transferred from Aspergillus [40]. However, it is believed that the metabolic gene clusters may have evolved from bacteria as almost half of the bacterial genes are located in an operon, a group of genes under the control of a promoter, and it may be essential to control the expression of downstream genes [41]. Further investigations have revealed that the gene clusters are usually located at the end of the chromosomes and flanked by mobile genetic elements (MGEs) that facilitate gene transfer within genomes and between species [41,42,43].
3.2. Griseofulvin Biosynthetic Gene Cluster
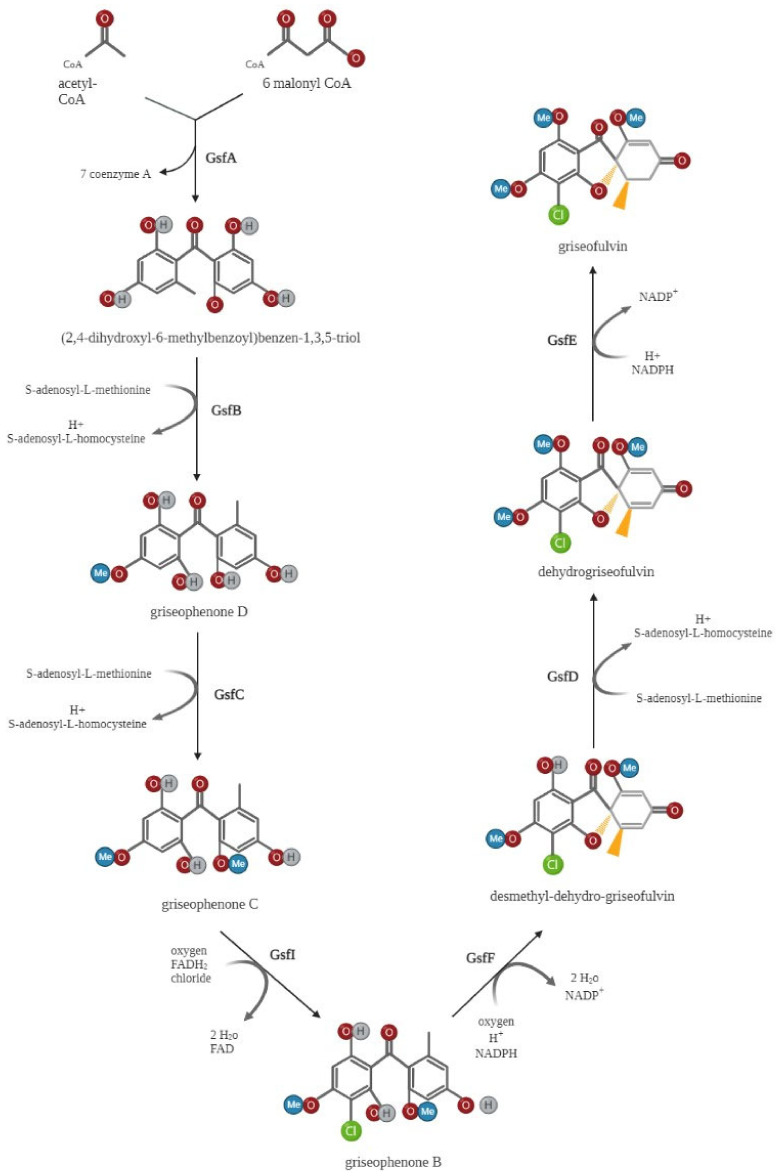
In 2010, the putative griseofulvin gene cluster containing the PKS genes gsfA–gsfK, gsfR1, and gsfR2 was first identified through shotgun sequencing and bioinformatic mining in P. aethiopicum [16]. Later, single-gene deletion and biochemical test procedures were used to investigate the role of these gsf genes in griseofulvin biosynthesis [44], with the key findings summarized in Figure 2. GsfA initiates the biosynthesis of griseofulvin by combining one acetyl CoA and six malonyl CoA molecules to generate heptaketide backbone benzophenone 5a. GsfA is a nonreducing polyketide synthase (NR-PKS) that consists of several functional domains, including starter unit ACP transacylase (SAT), ketosynthase (KS), malonyl-CoA-ACP transacylase (AT), product template (PT), and an acyl carrier protein (ACP). The GsfA PT domain may mediate the cyclization of aromatic rings to form the intermediate benzophenone. The intermediate griseophenone C is produced from the methylation of phenols on benzophenone 5a by the O-methyltransferases (gsfB and gsfC). Following, griseophenone C is chlorinated to convert into griseophenone B by the halogenase gsfI, and the grisan core is produced from griseophenone B by the phenol oxidative activity of gsf F. The grisan core is finally transformed into griseofulvin by two further processes, methylation at 5-OH mediated by gsf D and enoyl redcution catalyzed by gsf E (Figure 2) [44].
Figure 2.
Griseofulvin biosynthetic pathway. The nonreducing polyketide synthase gsfA initiates the synthesis of griseofulvin by combining one acetyl-CoA and six malonyl-CoA units to form the heptaketide backbone of benzophenone 5a. The O-methyltransferases (gsfB and gsfC) then methylate phenols on benzophenone 5a to generate the intermediate griseophenone C. Following this process, griseophenone C is chlorinated by the halogenase gsfI to produce griseophenone B, and griseophenone B is then converted to the grisan core by phenol oxidative activity of gsfF. The grisan core is ultimately converted into griseofulvin by two further processes: enoyl reduction catalyzed by gsfE and methylation at 5-OH mediated by gsfD. Created by Biorender.com.
Not all gsf genes are essential for griseofulvin biosynthesis and function. For instance, the deletion of gsfA or gsfI disrupts the production of griseofulvin since both proteins are essential for the gsf biosynthetic pathway [16,44,45]. Additionally, the deletion of gsfK had no impact on biosynthesis in P. aethiopicum [44]. Further, when compared to P. aethiopicum, P. griseofulvum lacks gsfK, gsfH, and gsfR2 in the gsf BGC [45].
We recently conducted a comparative genome study in which the gsf BGC of 12 fungal genomes was identified, annotated, and compared. In this study, in addition to the Penecillium species, gsf BGC was found for the first time in Aspergillus alliaceus, Aspergillus burnettii, and Memnoniella echinata. Overall, our findings suggest that a total of seven genes (gsfA–gsfF, gsfI) out of the thirteen in P. aethiopicum were conserved by the majority of the genomes investigated and no gene rearrangements were reported at the gsf BGC [38].
3.3. Genetic Regulation of Griseofulvin Biosynthesis
The gsfR1 and gsfR2 encode for putative transcription factors in the griseofulvin gene cluster. However, their functions in the gsf gene cluster remain debatable. The gsfR1 gene may not only impact griseofulvin biosynthesis but also serves as a critical regulator of other secondary metabolisms [46], such as patulin produced by P. griseofulvum [45]. Indeed, the gsfR1 gene deletion increased griseofulvin biosynthesis by increasing gsfA gene expression when tested on apples and PDA medium in vitro, suggesting gsfR1 as a negative regulator of the cluster. However, in different culture conditions, such as high concentrations of nitrogen and complex sugars, and on media supplemented with peptone, the gsfR1 positively regulates griseofulvin biosynthesis [46]. These results indicate that gsfR1 can respond differently depending on external stimulation, particularly nitrogen and carbon availability. Surprisingly, the gene gsfR1 was missing in the recently released genome of griseofulvin producer Xylaria flabelliformis [2], indicating that griseofulvin biosynthesis could be regulated independently of gsfR1 action.
It is noteworthy that the gsfR2 gene in P. griseofulvum was found in a separate genomic region of the biosynthetic gene cluster, in contrast to P. aethiopicum [45] and gsfR2 deletion did not affect griseofulvin biosynthesis, suggesting that this gene is probably involved in a different pathway or another secondary metabolite biosynthesis [46].
3.4. Environmental Factors in Producing Griseofulvin
The gene cluster expression is mostly synchronized in response to environmental conditions. Several factors appear to influence producing griseofulvin, of which organic compounds are one of the most critical environmental components [47]. The yield of griseofulvin increases in rich media with glucose, acetate, and succinate. Of note is that a nitrogen concentration of less than 0.04 g% or more than 0.4 g% may inhibit the griseofulvin biosynthesis [48]. Furthermore, the ATP/ADP ratio is a significant parameter of cellular energy status that regulates griseofulvin synthesis; the more the energy level, the more griseofulvin production [49,50]. Another important factor is pH. The highest level of griseofulvin yields in a pH ranging from 5.5 to 6 [51].
Secondary metabolites are mainly synthesized during the stationary-phase growth by microorganisms to improve their ability to thrive in diverse environments [52,53]. It may be assumed that the advantages of griseofulvin production are to defend against other fungal species and reduce competition. Even though only species that encounter such competition would preserve the griseofulvin gene cluster in their genomes, given the cost of time and energy. Thus, when the beneficial effects of producing griseofulvin outweigh the energy cost of biosynthesis, the organism would have increased fitness, according to the natural selection theory [54,55]. One example is the fungal endophyte, Xylaria flabelliformis, which is commonly found within plant tissue and the synthesis of griseofulvin as an antifungal against plant pathogenic fungi [2].
4. Physiochemical Properties of Griseofulvin
Griseofulvin is a fungal secondary polyketide metabolite that is soluble in ethanol and methanol but has poor solubility in water [6]. One notable feature of griseofulvin is its ability to tolerate heat stress and maintain its function at a high temperature of 121 °C without losing functional properties [47]. These properties are important to understand their medical applications because they affect absorption, transportation, excretion and degradation.
5. Medical Applications
5.1. Antifungal Application
The first use of griseofulvin in medicine can be traced back to treating ringworm infections caused by Microsporum canis in guinea pigs [56]. Further investigations revealed its effect on not only the genus Microsporum but Trichophyton and Epidermophyton [57]. Griseofulvin was approved by Food and Drug Administration (FDA) in 1959, and it is still in use today and is considered one of the most widely used treatments for dermatophytes fungal infections in humans and animals.
5.2. Non-fungal Inflammatory Diseases
Further studies reported the effectiveness of griseofulvin in the treatment of non-fungal skin inflammation diseases, including lichen planus [58] and chronic purpuric dermatosis [59], suggesting that it may also have anti-inflammatory and immunomodulatory potential. This is while, earlier studies have shown that griseofulvin is effective in treating of the shoulder-hand syndrome [60], and a few other inflammatory, rheumatic conditions, such as posttraumatic reflex dystrophies and scapulohumeral periarthritis [61].
5.3. Cardiovascular Applications
It has been shown that intravenous administration of griseofulvin improves coronary blood flow and decreases blood pressure, indicating its peripheral vasodilation effects. In addition, griseofulvin increases myocardial heart rate by directly acting on the myocardium and vascular smooth muscle rather than through central nervous systems or humoral mechanisms [10,11].
5.4. Antitumor Applications
Griseofulvin has gained attention for its potential application in cancer chemotherapy because of its low toxicity. In tumor cell lines, antifungal drug griseofulvin inhibits tumor growth and several forms of cancer cell proliferation by suppressing spindle microtubule dynamics, mitotic arrest, and cell death in multipolar spindles, but not in fibroblasts and keratinocytes containing normal centrosome composition. Indeed, griseofulvin binds to the αβ intra-dimer tubulin interface and induces mitotic arrest by a variety of mitotic abnormalities, such as misaligned chromosomes and multipolar spindles, resulting in cells containing fragmented nuclei. These cells exhibited increased p53 accumulation in the nucleus, indicating that the cells undergo apoptosis [8,62].
5.5. Antiviral Applications
Griseofulvin was used for zoster-associated pain relief and prevented the development of further lesions within two days of starting treatment [63]. Further studies have shown that griseofulvin suppresses the hepatitis C virus (HCV) replication by arresting the human cell cycle at the G2/M phase and acting on microtubule polymerization but does not affect HCV internal ribosome entry site (IRES) dependent translation. A synergistic inhibitory effect of griseofulvin and interferon alpha (IFNα) was also noted in Huh7/Rep-Feo (hepatoma cell line containing HCV 1b replicons that expresses a chimeric protein consisting of neomycin phosphotransferase and firefly luciferase) cells [9].
6. Pharmacokinetics of Griseofulvin
Griseofulvin is commonly used orally, although poor intestinal absorption may lead to failure in treatment. A spectrophotofluorometric study showed that griseofulvin absorption and effectiveness are dramatically increased with the high-fat diet [64]. Indeed, the oral bioavailability of griseofulvin-loaded liposomes may be enhanced significantly by high drug encapsulation efficiency and small liposome size [65].
Griseofulvin absorption is greatest from the duodenum and least from the stomach, but colon absorption is hardly noticeable [66]. In addition, a higher level of griseofulvin was found in the lung after intravenous administration, while the liver had a significantly higher level after oral administration [67]. Furthermore, griseofulvin can be secreted through sweat glands and appears in the keratinized layer within four hours after a single-dose administration to inhibit fungal growth [68]. The treatment response time depends on the keratin thickness at the injection site. Accordingly, healing fungal toe-nail infections may take longer than hair or skin infections [69].
The biological half-life of griseofulvin is 9 to 21 hours in the blood [70], and it is excreted as 6-desmethylgriseofulvin (6-DMG) and 4- desmethylgriseofulvin (4-DMG) metabolites in urine and feces after being metabolized by the liver microsomal enzyme system [66].
7. Characterized Interactions between Griseofulvin and Cellular Components
Griseofulvin enters the dermatophyte through energy-dependent transport processes, binds to the fungal microtubules, interfering the microtubule function, thus inhibiting mitosis [71]. The primary mechanism by which griseofulvin inhibits mitosis at metaphase is to suppress spindle microtubule (MT) dynamics by acting directly at the plus end to increase the stability and suppress the shortening rate at the MT plus ends [72]. Griseofulvin has two possible binding sites in tubulin based on docking studies; one of which overlaps with the Taxol, a chemotherapy medication to treat cancer, the binding site at the beta-tubulin H6-H7 hoop, and the other is located at the interface of αβ tubulin [64]. It has been shown that griseofulvin at a concentration of 30 to 60 μM induced both mitotic G2/M arrest and apoptosis in a human malignant cell line (HL-60) using activation of NF-κB pathway with cell division cycle protein (cdc)2 activation and phosphorylation of B-cell lymphoma (Bcl)2 proteins that regulate cell death [73]. Notably, the β-tubulin gene expression has been reported to decrease after griseofulvin treatment in T. rubrum [74].
Further studies have shown that it can be considered a potential therapeutic option for colon and breast cancer by an inhibitory effect on centrosomal clustering activity by altering the interphase microtubule stability. The findings strongly argued that mitotic irregularities and nuclear localization of p53 in human breast cancer MCF-7 cells are induced by kinetic inhibition of microtubule dynamics [8,62]. Likewise, griseofulvin inhibits hepatitis C virus replication by disrupting microtubule polymerization and interrupting the G2/M phase in human cells [9].
Additionally, griseofulvin has been shown effective for inflammatory skin diseases through inducing inhibitory effects on vascular cell adhesion molecule 1 (VCAM-1) in both TNF-alpha and IL-1 stimulated human dermal microvascular endothelial cells (HDMEC) [75].
8. Changes in Keratins 8 and 18 in Griseofulvin-Induced Liver Injury
Because of the high concentration of griseofulvin in the liver as the major detoxification site, it has also been reported to cause occasional hepatitis [76,77]. While griseofulvin-caused hepatitis is extremely rare in human, Mallory bodies (MBs) in hepatocytes associated with griseofulvin treatment were frequently reported in rodents [78,79]. The intermediate filament proteins (IFs), such as cytoplasmic keratins 8 and 18 (K8/18), are one of the most important components of a Mallory body [80]. These intermediate filament proteins are believed to play an important role in hepatocyte protection against toxic or mechanical stress, and regulation of cell response to injuries, cell growth, and death [81,82]. K8 and 18 are present in a 1:1 ratio under normal circumstances [83], but liver diseases such as alcoholic hepatitis, hepatic porphyria, and copper metabolism increase the K8/K18 ratio to more than 1, consequently forming the K8/18-containing aggregates known as Mallory bodies, which impairs centrosome and microtubule network function and results in cell death [84,85,86,87]. Likewise, treatment of mice with griseofulvin-containing diet induces the Mallory body formation likely caused by overexpression of Krt8 (Keratin 8) gene [88], and griseofulvin binding to K8 and K18 in hepatocytes, which result in changing the keratin solubility, keratin phosphorylation on specific sites, and causing protein misfolding [81]. Misfolded proteins are refolded with help of cellular chaperone proteins or tagged by ubiquitin for degradation [89]. These findings suggest the binding potential of griseofulvin with cytoskeletons, especially keratin intermediate filaments proteins K8, and K18.
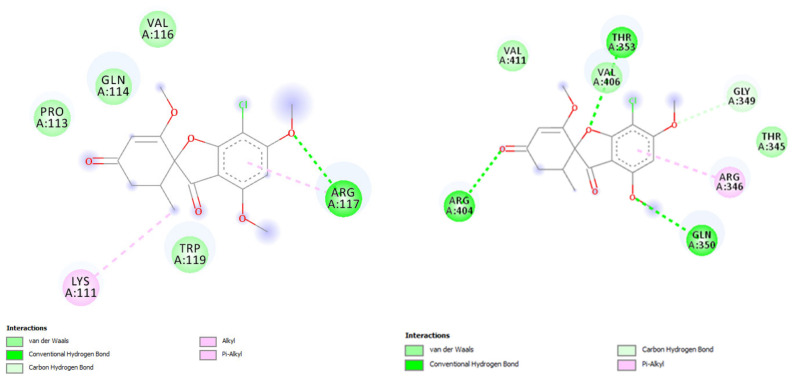
Here, we studied the griseofulvin interaction with K8 and K18 in human, rat, and mouse using molecular docking (Table 1), conducted in AutoDock 4 [90]. The 3D protein structures of the K8 and K18 were downloaded from the RCSB Protein Data Bank (PDB) (https://www.rcsb.org/) or AlphaFold database (https://alphafold.ebi.ac.uk/). The interactions between the receptors and the griseofulvin were visualized using Discovery Studio Visualizer v.20 (https://discover.3ds.com/discovery-studio-visualizer-download). Keratin 8 (Krt8) and keratin 18 (Krt18) genes reside on chromosome 12 (GenBank accession NC_000012) in human, chromosome 15 (NC_000081) in mouse (Mus musculus), and chromosome 7 (NC_051342) in rat (Rattus norvegicus). The Krt8 sequences, after alignment with MAFFT, have 493 aligned sites of which human and mouse K8 differ by 44 aa (89% identity, 86% similarity), human and rat K8 differ by 44 aa (89% identity, 86% similarity) and mouse and rat K8 differ by 27 aa (94% identity, 90% similarity). The alignment of the three Krt18 amino acid sequences have 432 sites, of which human and mouse K18 differ by 48 aa (86% identity, 90% similarity), human and rat K18 differ by 57 aa (84% identity, 89% similarity) and mouse and rat K18 differ by 16 aa (96% identity, 96% similarity). The molecular docking results revealed the K8 residues of Ile 63, Gln 85, and Lys 92 form H bonds with the oxyl groups of griseofulvin, while the K18 residues of Arg 117, Gln 243, Gln 350, Thr 353, and Arg 404 are involved in H-bond interactions (Figure 3). The griseofulvin illustrated low binding energy toward the K8, ranging from −3.17 to −4.6 kcal mol−1. However, a higher binding energy toward K18 was observed in rat and mouse with −5.23 and −5.54 kcal mol−1 respectively (Table 1). In conclusion, the best griseofulvin interaction was found with Keratin 18 in rat, followed by mouse, based on the lowest binding energy and highest number of hydrogen bonds. This differential binding to K8 and K18 may disrupt the 1:1 ratio between the two and contribute to liver problems. The stronger binding of griseofulvin to K18 in rodents than in human may also explain the observed difference in the severity of hepatitis between rodents and humans.
Table 1.
Molecular docking details of griseofulvin interaction with keratin 8 (K8), and keratin 18 (K18) in human, rat, and mouse.
| Keratin | Species | PDB or Alpha Fold IDs | ΔGbinding
(Kcal/mol) |
Hydrogen Bond |
Hydrophobic Interaction |
|---|---|---|---|---|---|
| K8 | Human | 3K3C | −3.17 | ILE 63 | GLY 61, GLY 62, THR 64 |
| 7K3X | −3.2 | ILE 63 | GLY 61 | ||
| 7K3Y | −3.61 | - | SER 58 | ||
| P05787F1 | −4.6 | GLN 85, LYS 92 | ARG 88, LYS 92 | ||
| Mouse | P11679F1 | −4.1 | - | LYS 420, THR 421 | |
| Rat | Q10758F1 | −4.55 | - | SER 15, GLY 16, PRO 17 | |
| K18 | Human | A0A024RAY2F1 | −4.72 | - | LEU 87, ARG 90, SER 93, TYR94 |
| P05783F1 | −4.99 | GLN 243 | SER 242, GLN 243, ASP 244, LYS 247, ILE 248, ASP 251 |
||
| Mouse | P05784F1 | −5.54 | ARG 117 | LYS 111, ARG 117 | |
| Rat | Q5BJY9F1 | −5.23 | GLN 350, THR 353, ARG 404 | ARG 346, GLY 349 |
Figure 3.
2D binding model of griseofulvin docked into the active site of keratin 18 (K18) in mouse (left) and rat (right). The dotted lines show the key interacting residues of the K18 toward griseofulvin as a ligand. The green, light green, and pink dotted lines show conventional hydrogen bonds, van der Waals, alkyl or Pi-alkyl interactions, respectively.
Amiodarone (Am), an antiarrhythmic medication used to treat and prevent a number of types of cardiac dysrhythmias [91], was used as positive control in this study, because it could be associated with liver injury and Mallory body formation in hepatocytes [92,93]. The molecular docking analysis showed that Arg90 is the only residue that can form hydrogen bonds in the binding cavity of K18, while the docking pose of amiodarone with K8 revealed three hydrogen bonds interactions with amino acids Gly 61 and Ile 63 in human (Table 2). Amiodarone showed the lowest free energy values (−4.68 and −4.85 kcal mol−1), corresponding to the high affinity binding site, for human K18. However, the amiodarone exhibited higher binding affinities, with binding energies of −4.14 and −3.64 kcal mol−1, to K8 than K18 of mouse and rat respectively.
Table 2.
Molecular docking details of Amiodarone interaction with Keratin 8 (K8), and Keratin 18 (K18) in Human, Rat, and Mouse.
| Keratin | Species | PDB or Alpha Fold IDs | ΔGbinding
(Kcal/mol) |
Hydrogen Bond |
Hydrophobic Interaction |
|---|---|---|---|---|---|
| K8 | Human | 3K3C | −3.38 | GLY 61, ILE 63 | MET 60, GLY 61, ILE 63 |
| 7K3X | >0 | - | - | ||
| 7K3Y | >0 | - | - | ||
| P05787F1 | −4.1 | - | MET 406, SER 410, HIS 412 | ||
| Mouse | P11679F1 | -4.14 | - | MET 412, MET 415, ILE 417 | |
| Rat | Q10758F1 | −3.64 | ARG 49 | SER 51, LEU 52, PHE 55 | |
| K18 | Human | A0A024RAY2F1 | −4.69 | ARG 90 | LEU 87, ARG 90, LEU 91, TYR 94 |
| P05783F1 | −4.85 | - | LEU 87, ARG 90, LEU 91, TYR 94 | ||
| Mouse | P05784F1 | −2.51 | - | GLU 414 | |
| Rat | Q5BJY9F1 | −0.21 | - | ARG 374, GLU 378 |
We also studied the molecular interactions of griseofulvin metabolites, including 6-desmethylgriseofulvin (6-DMG) and 4- desmethylgriseofulvin (4-DMG) [66], with K8 and K18 in human, mouse, and rat. Our in silico analysis exhibited slightly higher affinities for 6-DMG and 4-DMG metabolites than griseofulvin to mouse and rat K18. In addition, the number of hydrophobic and hydrogen bonds between griseofulvin metabolites and cytokeratin proteins (K8 and K18) were greater than griseofulvin in human, mouse, and rat. These findings suggest that griseofulvin metabolites have favorable interactions with keratins 8 and 18 (K8 and K18), and therefore they could be responsible for liver injury and Mallory body formation in hepatocytes of human and rodents treated with griseofulvin. The molecular docking details of 4-DMG and 6-DMG with K8 and K18 are shown in Table 3 and Table 4 respectively.
Table 3.
Molecular docking details of 4-desmethylgriseofulvin (4-DMG) interaction with Keratin 8 (K8), and Keratin 18 (K18) in Human, Rat, and Mouse.
| Keratin | Species | PDB or Alpha Fold IDs | ΔGbinding
(Kcal/mol) |
Hydrogen Bond |
Hydrophobic Interaction |
|---|---|---|---|---|---|
| K8 | Human | 3K3C | −3.34 | GLY 62, THR 64 | GLY 61, GLY 62 |
| 7K3X | −3.34 | GLY 61, ILE 63 | GLY 61, ILE 63 | ||
| 7K3Y | −3.8 | SER 58 | SER 58, ALA 57 | ||
| P05787F1 | −5.43 | THR 64, ALA 65 | THR 64, ALA 65,VAL 66, THR 67 | ||
| Mouse | P11679F1 | −4.14 | THR 419, LYS 420, THR 421 | THR 421, THR 422 | |
| Rat | Q10758F1 | −3.93 | SER 21, ARG 23 | ALA 19, PHE 20, SER 21 | |
| K18 | Human | A0A024RAY2F1 | −4.48 | ARG 90 | ARG 90 |
| P05783F1 | −4.96 | SER 242, ASP 244 | GLN 243, LYS 247, ASP 251 | ||
| Mouse | P05784F1 | −5.28 | GLN 114, ARG 117 | LYS 111, PRO 113, GLN 114, ARG 117 | |
| Rat | Q5BJY9F1 | −5.22 | THR 353, ARG 404 | ARG 346, GLN 350, VAL 411 |
Table 4.
Molecular docking details of 6-desmethylgriseofulvin (6-DMG) interaction with Keratin 8 (K8), and Keratin 18 (K18) in Human, Rat, and Mouse.
| Keratin | Species | PDB or Alpha Fold IDs | ΔGbinding
(Kcal/mol) |
Hydrogen Bond |
Hydrophobic Interaction |
|---|---|---|---|---|---|
| K8 | Human | 3K3C | −4.04 | GLY 62, THR 64 | GLY 61, GLY 62 |
| 7K3X | −3.71 | ILE 63 | ILE 63 | ||
| 7K3Y | −3.95 | GLY 56, SER 58 | GLY 56, SER 58 | ||
| P05787F1 | −4.85 | LEU 72 | LEU 72, PRO 75, VAL 77 | ||
| Mouse | P11679F1 | −4.29 | GLN 134, SER 138 | LEU 131, GLN 134, GLN 135, SER 138 | |
| Rat | Q10758F1 | −4.50 | SER 21 | ARG 18, ALA 19 | |
| K18 | Human | A0A024RAY2F1 | −4.88 | TYR94 | LEU 91, TYR 94 |
| P05783F1 | −4.81 | GLU 236 | SER230, SER 231, LEU 233, VAL 235, GLU 236 | ||
| Mouse | P05784F1 | −5.61 | LEU 108, TPR 119 | LYS 111, PRO 113, VAL 116 | |
| Rat | Q5BJY9F1 | −5.33 | ARG 346, GLN 350, ARG 404 | ARG 346, GLN 350, VAL 411 |
9. Repurposing of Griseofulvin
Computational drug repurposing accelerates drug development and reduces the time, effort, and expenses associated with the drug discovery process [94,95]. In our recent study, griseofulvin and its derivatives were repurposed as potent inhibitors of SARS-CoV-2 entry and viral replication. The inhibitory effects of griseofulvin and its derivatives on essential SARS-CoV-2 main protease, RNA-dependent RNA polymerase (RdRp), spike protein receptor-binding domain (RBD), and angiotensin-converting enzyme 2 (ACE2) from humans were investigated using molecular docking and molecular dynamics simulation. Our findings showed the activity of these compounds against SARS-CoV-2 target proteins. The highest docking score with the most hydrogen bond interactions to COVID-19 main protease was observed for griseofulvin. However, griseofulvin was also shown to interact favorably with ACE2, RBD, and RdRb proteins [12].
In addition to these inhibitory effects, griseofulvin may enhance ACE2 function to regulate the renin-angiotensin-aldosterone system (RAAS). ACE converts angiotensin I (ANG I) to angiotensin II (ANG II), which increases blood pressure and causes inflammation. ACE2 facilitates the breakdown of ANG II which lowers blood pressure and inflammation while maintaining homeostasis and preventing tissue damage [13]. Inhibition of ACE2 function caused by SARS-CoV-2 binding to the ACE2 receptor leads to an increase in ANG II protein and its deleterious effects. It is well-known that griseofulvin has vasodilation and increased capillary blood flow effects [10,11], although the molecular mechanism by which griseofulvin affects blood pressure remains unclear. Results from our molecular docking study put forward the hypothesis that griseofulvin binds to ACE2 and competes against SARS-CoV-2 binding to the extracellular domain of ACE2, therefore improving ACE2 vasodilation and tissue protection functions [12]. These findings suggest that griseofulvin and its derivatives might be considered when developing future SARS-CoV-2 treatment options.
10. Conclusions
Since the discovery of the FDA-approved drug griseofulvin, practical clinical information and applications have been well established in the literature. Nevertheless, computational and bioinformatics analysis have promoted a revolution in scientific discovery, in which repurposing and developing new indications for already approved drugs gained tremendous interest during past years. Griseofulvin has proven to be a safe therapeutic agent in managing and suppressing dermatophyte infections, however its potential to manage cancer, hepatitis C, and SARS-CoV-2 have been predicted in theory and are under experimental investigations. Although it is well documented that griseofulvin interacts with spindle microtubules, our molecular docking study showed its binding potential toward keratin intermediate filament proteins in human, rat, and mouse, which explains more serious liver problems in rodents than humans. Here, we also reviewed the earliest to the most current knowledge about griseofulvin from a medical perspective. Further studies on griseofulvin may reveal new applications, design and development in future therapeutic interventions.
Acknowledgments
The authors would like to thank the academic editors and referees for their helpful comments that significantly improved the presentation of the study.
Author Contributions
P.A. designed the study, collected the data, and wrote the original draft of the manuscript. M.M. conducted molecular docking. Y.W. and X.X. reviewed and edited the manuscript. X.X. designed, supervised, and coordinated the study. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that there is no conflict of interest.
Funding Statement
This work is supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant to X.X. [RGPIN/2018-03878]. Y.W. was supported by an NSERC postdoctoral fellowship.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Whalley A. The xylariaceous way of life. Mycol. Res. 1996;100:897–922. doi: 10.1016/S0953-7562(96)80042-6. [DOI] [Google Scholar]
- 2.Mead M.E., Raja H.A., Steenwyk J.L., Knowles S.L., Oberlies N.H., Rokas A. Draft genome sequence of the griseofulvin-producing fungus xylaria flabelliformis strain G536. Microbiol. Resour. Announc. 2019;8:e00890-19. doi: 10.1128/MRA.00890-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee H.B., Mun H.Y., Nguyen T.T.T., Kim J.-C., Stone J.K. Abieticola koreana Gen. Et Sp. Nov.; A griseofulvin-producing endophytic xylariaceous ascomycete from korea. Mycotaxon. 2016;131:749–764. doi: 10.5248/131.749. [DOI] [Google Scholar]
- 4.Ribeiro A.I., Costa E.S., Thomasi S.S., Brandão D.F.R., Vieira P.C., Fernandes J.O.B., Forim M.R., Ferreira A.G., Pascholati S.R.F., Gusmão L.F.P. Biological and chemical control of sclerotinia sclerotiorum using stachybotrys levispora and its secondary metabolite griseofulvin. J. Agric. Food Chem. 2018;66:7627–7632. doi: 10.1021/acs.jafc.7b04197. [DOI] [PubMed] [Google Scholar]
- 5.Dayan F.E., Cantrell C.L., Duke S.O. Natural products in crop protection. Bioorganic Med. Chem. 2009;17:4022–4034. doi: 10.1016/j.bmc.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 6.Oxford A.E., Raistrick H., Simonart P. Studies in the biochemistry of micro-organisms: Griseofulvin, c17h17o6cl, a metabolic product of penicillium griseo-fulvum dierckx. Biochem. J. 1939;33:240. doi: 10.1042/bj0330240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert D.R., Siegle R.J., Camisa C. Griseofulvin and ketoconazole in the treatment of dermatophyte infections. Int. J. Dermatol. 1989;28:300–304. doi: 10.1111/j.1365-4362.1989.tb01348.x. [DOI] [PubMed] [Google Scholar]
- 8.Rebacz B., Larsen T.O., Clausen M.H., Rønnest M.H., Löffler H., Ho A.D., Krämer A. Identification of griseofulvin as an inhibitor of centrosomal clustering in a phenotype-based screen. Cancer Res. 2007;67:6342–6350. doi: 10.1158/0008-5472.CAN-07-0663. [DOI] [PubMed] [Google Scholar]
- 9.Jin H., Yamashita A., Maekawa S., Yang P., He L., Takayanagi S., Wakita T., Sakamoto N., Enomoto N., Ito M. Griseofulvin, an oral antifungal agent, suppresses hepatitis c virus replication In Vitro. Hepatol. Res. 2008;38:909–918. doi: 10.1111/j.1872-034X.2008.00352.x. [DOI] [PubMed] [Google Scholar]
- 10.Rubin A.A. Coronary vascular effects of griseofulvin. JAMA. 1963;185:971–972. doi: 10.1001/jama.1963.03060120081033. [DOI] [PubMed] [Google Scholar]
- 11.Aldinger E.E. Cardiovascular effects of griseofulvin. Circ. Res. 1968;22:589–593. doi: 10.1161/01.RES.22.5.589. [DOI] [PubMed] [Google Scholar]
- 12.Aris P., Mohamadzadeh M., Wei Y., Xia X. In Silico molecular dynamics of griseofulvin and its derivatives revealed potential therapeutic applications for COVID-19. Int. J. Mol. Sci. 2022;23:6889. doi: 10.3390/ijms23136889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Atlas S.A. The Renin-angiotensin aldosterone system: Pathophysiological role and pharmacologic inhibition. J. Manag. Care Pharm. 2007;13:9–20. doi: 10.18553/jmcp.2007.13.s8-b.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brian P., Hemming H. Production of antifungal and antibacterial substances by fungi; preliminary examination of 166 strains of fungi imperfecti. Microbiology. 1947;1:158–167. doi: 10.1099/00221287-1-2-158. [DOI] [PubMed] [Google Scholar]
- 15.Grove J.F., Macmillan J., Mulholland T., Rogers M.T. Griseofulvin. Part I. J. Chem. Soc. 1952;759:3949–3958. doi: 10.1039/jr9520003949. [DOI] [Google Scholar]
- 16.Chooi Y.-H., Cacho R., Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem. Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang H.-Y., Zhang Q., Li H., Gao J.-M. Antimicrobial and allelopathic metabolites produced by Penicillium brasilianum. Nat. Prod. Res. 2015;29:345–348. doi: 10.1080/14786419.2014.940347. [DOI] [PubMed] [Google Scholar]
- 18.Roullier C., Bertrand S., Blanchet E., Peigné M., Robiou Du Pont T., Guitton Y., Pouchus Y.F., Grovel O. Time dependency of chemodiversity and biosynthetic pathways: An lc-ms metabolomic study of marine-sourced Penicillium. Mar. Drugs. 2016;14:103. doi: 10.3390/md14050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jadulco R., Edrada R.A., Ebel R., Berg A., Schaumann K., Wray V., Steube K., Proksch P. New communesin derivatives from the fungus Penicillium sp. derived from the mediterranean sponge Axinella verrucosa. J. Nat. Prod. 2004;67:78–81. doi: 10.1021/np030271y. [DOI] [PubMed] [Google Scholar]
- 20.Petit K., Mondeguer F., Roquebert M., Biard J., Pouchus Y. Detection of griseofulvin in a marine strain of Penicillium waksmanii by ion trap mass spectrometry. J. Microbiol. Methods. 2004;58:59–65. doi: 10.1016/j.mimet.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Xue C., Li T., Deng Z., Fu H., Lin W. Janthinolide Ab, Two New 2, 5-piperazinedione derivatives from the endophytic Penicillium janthinellum isolated from the soft coral dendronephthya sp. Die Pharm. 2006;61:1041–1044. [PubMed] [Google Scholar]
- 22.Clarke S., Mckenzie M. Penicillium sclerotigenum: A new source of griseofulvin. Nature. 1967;213:504–505. doi: 10.1038/213504b0. [DOI] [PubMed] [Google Scholar]
- 23.Wang L., Zhou H.-B., Frisvad J.C., Samson R.A. Penicillium persicinum, a new griseofulvin, chrysogine and roquefortine c producing species from qinghai province, china. Antonie Van Leeuwenhoek. 2004;86:173–179. doi: 10.1023/B:ANTO.0000036140.86059.51. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen J.C., Grijseels S., Prigent S., Ji B., Dainat J., Nielsen K.F., Frisvad J.C., Workman M., Nielsen J. Global analysis of biosynthetic gene clusters reveals vast potential of secondary metabolite production in Penicillium species. Nat. Microbiol. 2017;2:17044. doi: 10.1038/nmicrobiol.2017.44. [DOI] [PubMed] [Google Scholar]
- 25.Frisvad J.C., Samson R.A. Polyphasic taxonomy of Penicillium subgenus Penicillium. a guide to identification of food and air-borne terverticillate penicillia and their mycotoxins. Stud. Mycol. 2004;49:1–174. [Google Scholar]
- 26.Larsen T.O., Smedsgaard J., Nielsen K.F., Hansen M.E., Frisvad J.C. Phenotypic taxonomy and metabolite profiling in microbial drug discovery. Nat. Prod. Rep. 2005;22:672–695. doi: 10.1039/b404943h. [DOI] [PubMed] [Google Scholar]
- 27.Samson R.A., Pitt J.I. Modern Concepts in Penicillium and Aspergillus Classification. Springer Science & Business Media; Berlin/Heidelberg, Germany: 2013. [Google Scholar]
- 28.Allawi M.Y.A., Al-Taee W.S.Q. Investigation of a new local isolate of Penicillium lanosocoeruleum that produces the antifungal griseofulvin. Pak. J. Med. Health Sci. 2022;16:380. [Google Scholar]
- 29.Park J.-H., Choi G.-J., Lee S.-W., Lee H.-B., Kim K.-M., Jung H.-S., Jang K.-S., Cho K.-Y., Kim J.-C. Griseofulvin from xylaria sp. strain f0010, an endophytic fungus of abies holophylla and its antifungal activity against plant pathogenic fungi. J. Microbiol. Biotechnol. 2005;15:112–117. [Google Scholar]
- 30.Petrini O. Microbial Ecology of Leaves. Springer; Berlin/Heidelberg, Germany: 1991. Fungal endophytes of tree leaves. [Google Scholar]
- 31.Grove J.F., Macmillan J., Mulholland T., Rogers M.T. Griseofulvin. Part IV. Structure. J. Chem. Soc. 1952;762:3977–3987. doi: 10.1039/jr9520003977. [DOI] [Google Scholar]
- 32.Birch A., Massy-Westropp R., Rickards R., Smith H. Studies in relation to biosynthesis. Part XIII. Griseofulvin. J. Chem. Soc. 1958;66:360–365. doi: 10.1039/jr9580000360. [DOI] [Google Scholar]
- 33.Birch A., Donovan F. Studies In relation to biosynthesis. I. Some possible routes to derivatives of orcinol and phloroglucinol. Aust. J. Chem. 1953;6:360–368. doi: 10.1071/CH9530360. [DOI] [Google Scholar]
- 34.Simpson T.J., Holker J.S. 13c-Nmr studies on griseofulvin biosynthesis and acetate metabolism in penicillium patulum. Phytochemistry. 1977;16:229–233. doi: 10.1016/S0031-9422(00)86791-6. [DOI] [Google Scholar]
- 35.Nirmala K., Sakegowda D., Duax W.L. Crystal structure of griseofulvin. J. Crystallogr. Spectrosc. Res. 1982;12:415–423. [Google Scholar]
- 36.Brown W., Sim G. Fungal metabolites. Part I. The stereochemistry of griseofulvin: X-ray analysis of 5-bromogriseofulvin. J. Chem. Soc. 1963;193:1050–1059. doi: 10.1039/jr9630001050. [DOI] [Google Scholar]
- 37.Levine S.G., Hicks R.E. The conformation of griseofulvin. Application of an nmr shift reagent. Tetrahedron Lett. 1971;12:311–314. doi: 10.1016/S0040-4039(01)96427-5. [DOI] [Google Scholar]
- 38.Aris P., Yan L., Wei Y., Chang Y., Shi B., Xia X. Conservation of griseofulvin genes in the gsf gene cluster among fungal genomes. G3. 2021;2:jkab399. doi: 10.1093/g3journal/jkab399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cimermancic P., Medema M.H., Claesen J., Kurita K., Brown L.C.W., Mavrommatis K., Pati A., Godfrey P.A., Koehrsen M., Clardy J., et al. Insights into Secondary Metabolism from a Global Analysis of Prokaryotic Biosynthetic Gene Clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slot J.C., Rokas A. Horizontal transfer of a large and highly toxic secondary metabolic gene cluster between fungi. Curr. Biol. 2011;21:134–139. doi: 10.1016/j.cub.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 41.Dobrindt U., Hochhut B., Hentschel U., Hacker J. Genomic Islands in Pathogenic and Environmental Microorganisms. Nat. Rev. Microbiol. 2004;2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 42.Keller N.P., Turner G., Bennett J.W. Fungal Secondary Metabolism—From Biochemistry to Genomics. Nat. Rev. Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 43.Wisecaver J.H., Slot J.C., Rokas A. The Evolution of Fungal Metabolic Pathways. PLoS Genet. 2014;10:e1004816. doi: 10.1371/journal.pgen.1004816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cacho R.A., Chooi Y.-H., Zhou H., Tang Y. Complexity generation in fungal polyketide biosynthesis: A spirocycle-forming p450 in the concise pathway to the antifungal drug griseofulvin. Acs Chem. Biol. 2013;8:2322–2330. doi: 10.1021/cb400541z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Banani H., Marcet-Houben M., Ballester A.-R., Abbruscato P., González-Candelas L., Gabaldón T., Spadaro D. Genome sequencing and secondary metabolism of the postharvest pathogen penicillium griseofulvum. BMC Genom. 2016;17:19. doi: 10.1186/s12864-015-2347-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valente S., Cometto A., Piombo E., Meloni G.R., Ballester A.-R., González-Candelas L., Spadaro D. Elaborated regulation of griseofulvin biosynthesis in penicillium griseofulvum and its role on conidiation and virulence. Int. J. Food Microbiol. 2020;328:108687. doi: 10.1016/j.ijfoodmicro.2020.108687. [DOI] [PubMed] [Google Scholar]
- 47.Wright J.M. The production of antibiotics in soil: II. Production of griseofulvin by penicillium nigricans. Ann. Appl. Biol. 1955;43:288–296. doi: 10.1111/j.1744-7348.1955.tb02477.x. [DOI] [Google Scholar]
- 48.Soloveva N., Malkov M. Kil® N, Gi; Golubeva, La: Role of nitrogen nutrition in the cultivation of penicillium nigricans producing griseofulvin on a synthetic medium. Antibiot. 1972;9:104–106. [Google Scholar]
- 49.Rogal I., Malkov M. Effect Of The cultivation temperature on the dynamics of the atp and adp levels and on the growth of p. nigricans thom. strains in the presence of various carbon sources. Antibiotiki. 1978;23:971. [PubMed] [Google Scholar]
- 50.Dasu V.V., Panda T. Studies on production of griseofulvin. Bioprocess Eng. 1999;21:489–495. doi: 10.1007/s004490050707. [DOI] [Google Scholar]
- 51.Alan R., Rodger C., Primrose F.T. Production of Griseofulvin in Low Nitrogen Level Medium. 2843527A. U.S. Patent. 1958 July 15;
- 52.Niu G., Tan H. Biosynthesis and Regulation of Secondary Metabolites in Microorganisms. Sci China Life Sci. 2013;56:581–583. doi: 10.1007/s11427-013-4501-5. [DOI] [PubMed] [Google Scholar]
- 53.Vining L. Functions of Secondary Metabolites. Annu. Rev. Microbiol. 1990;44:395–427. doi: 10.1146/annurev.mi.44.100190.002143. [DOI] [PubMed] [Google Scholar]
- 54.Hurst L. Genetics and the understanding of selection. Nat. Rev. Genet. 2009;10:83–93. doi: 10.1038/nrg2506. [DOI] [PubMed] [Google Scholar]
- 55.Kimura M. The Neutral Theory of Molecular Evolution and the World View of the Neutralists. Genome. 1989;31:24–31. doi: 10.1139/g89-009. [DOI] [PubMed] [Google Scholar]
- 56.Gentles J. Experimental ringworm in guinea pigs: Oral treatment with griseofulvin. Nature. 1958;182:476–477. doi: 10.1038/182476a0. [DOI] [PubMed] [Google Scholar]
- 57.Gupta A.K., Adam P., Dlova N., Lynde C.W., Hofstader S., Morar N., Aboobaker J., Summerbell R.C. Therapeutic options for the treatment of tinea capitis caused by trichophyton species: Griseofulvin versus the new oral antifungal agents, terbinafine, itraconazole, and fluconazole. Pediatr. Dermatol. 2001;18:433–438. doi: 10.1046/j.1525-1470.2001.01978.x. [DOI] [PubMed] [Google Scholar]
- 58.Sehgal V., Bikhchandani R., Koranne R., Nayar M., Saxena H. Histopathological Evaluation of griseofulvin therapy in lichen planus. Dermatology. 1980;161:22–27. doi: 10.1159/000250326. [DOI] [PubMed] [Google Scholar]
- 59.Tamaki K., Yasaka N., Osada A., Shibagaki N., Furue M. Successful Treatment of pigmented purpuric dermatosis with griseoiulvin. Br. J. Dermatol. 1995;132:159–160. doi: 10.1111/j.1365-2133.1995.tb08650.x. [DOI] [PubMed] [Google Scholar]
- 60.Cohen A., Goldman J., Daniels R., Kanenson W. Treatment of shoulder-hand syndrome with griseofulvin. J. Am. Med. Assoc. 1960;173:542–543. doi: 10.1001/jama.1960.73020230041009. [DOI] [PubMed] [Google Scholar]
- 61.Serre H., Simon L. Action Therapeutique inattendue en rhumatologie dun antibiotique antifongique-la griseofulvine. Presse Med. 1962;70:2263. [Google Scholar]
- 62.Rathinasamy K., Jindal B., Asthana J., Singh P., Balaji P.V., Panda D. Griseofulvin stabilizes microtubule dynamics, activates p53 and inhibits the proliferation of mcf-7 cells synergistically with vinblastine. BMC Cancer. 2010;10:213. doi: 10.1186/1471-2407-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Livingston W.A. Griseofulvin in treatment of zoster. Arch. Dermatol. 1965;92:761. doi: 10.1001/archderm.1965.01600180153035. [DOI] [Google Scholar]
- 64.Crounse R. Effective Use of Griseofulvin. Arch. Dermatol. Res. 1963;87:176–178. doi: 10.1001/archderm.1963.01590140038006. [DOI] [PubMed] [Google Scholar]
- 65.Ong S.G.M., Ming L.C., Lee K.S., Yuen K.H. Influence of the encapsulation efficiency and size of liposome on the oral bioavailability of griseofulvin-loaded liposomes. Pharmaceutics. 2016;8:25. doi: 10.3390/pharmaceutics8030025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lin C., Symchowicz S. Absorption, Distribution, Metabolism, and Excretion of Griseofulvin in Man and Animals. Drug Metab. Rev. 1975;4:75–95. doi: 10.3109/03602537508993749. [DOI] [PubMed] [Google Scholar]
- 67.Bedford C., Busfield D., Child K.J., Mac G., Sutherland P., Tomich E.G. Studies on the biological disposition of griseofulvin, an oral antifungal agent. AMA Arch. Dermatol. 1960;81:735–745. doi: 10.1001/archderm.1960.03730050091016. [DOI] [PubMed] [Google Scholar]
- 68.Araujo O.E., Flowers F.P., King M.M. Griseofulvin: A new look at an old drug. DICP. 1990;24:851–854. doi: 10.1177/106002809002400912. [DOI] [PubMed] [Google Scholar]
- 69.Roth J., Frank J. Griseofulvin. Ann. N. Y. Acad Sci. 1960;89:247–253. doi: 10.1111/j.1749-6632.1960.tb20146.x. [DOI] [PubMed] [Google Scholar]
- 70.Develoux M.G. Paper presented at the Annales de dermatologie et de venereologie. Ann. Dermatol. Venereol. 2001;128:910–989. [PubMed] [Google Scholar]
- 71.Gull K., Trinci A.P.J. Griseofulvin Inhibits Fungal Mitosis. Nature. 1973;244:292–294. doi: 10.1038/244292a0. [DOI] [PubMed] [Google Scholar]
- 72.Panda D., Rathinasamy K., Santra M.K., Wilson L. Kinetic suppression of microtubule dynamic instability by griseofulvin: Implications for its possible use in the treatment of cancer. Proc. Natl. Acad. Sci. USA. 2005;102:9878–9883. doi: 10.1073/pnas.0501821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uen Y.H., Liu D.Z., Weng M.S., Ho Y.S., Lin S.Y. Nf-Κb pathway is involved in griseofulvin-induced g2/m arrest and apoptosis in hl-60 cells. J. Cell. Biochem. 2007;101:1165–1175. doi: 10.1002/jcb.21240. [DOI] [PubMed] [Google Scholar]
- 74.Zomorodian K., Uthman U., Tarazooie B., Rezaie S. The effect of griseofulvin on the gene regulation of β-tubulin in the dermatophyte pathogen trichophyton rubrum. J. Infect. Chemother. 2007;13:373–379. doi: 10.1007/s10156-007-0552-5. [DOI] [PubMed] [Google Scholar]
- 75.Asahina A., Tada Y., Nakamura K., Tamaki K. Colchicine and griseofulvin inhibit VCAM-1 expression on human vascular endothelial cells—Evidence for the association of VCAM-1 expression with microtubules. J. Dermatol. Sci. 2001;25:1–9. doi: 10.1016/S0923-1811(00)00097-9. [DOI] [PubMed] [Google Scholar]
- 76.Chiprut R.O., Viteri A., Jamroz C., Dyck W.P. Intrahepatic cholestasis after griseofulvin administration. Gastroenterology. 1976;70:1141–1143. doi: 10.1016/S0016-5085(76)80327-7. [DOI] [PubMed] [Google Scholar]
- 77.Watanabe M., Akagi S., Kohge N., Uchida Y., Nguyen T., Hirakawa K., Fukumoto S. Laparoscopy of griseofulvin-induced liver injury presenting a wide depression. Endoscopy. 1994;26:514–515. doi: 10.1055/s-2007-1009024. [DOI] [PubMed] [Google Scholar]
- 78.Zimmerman H.J. Hepatotoxicity: The Adverse Effects of Drugs and Other Chemicals on the Liver. Lippincott; Philadelphia, PA, USA: 1999. [Google Scholar]
- 79.Moseley R.H. Drug-Induced Liver Disease. 3rd ed. Elsevier; Amsterdam, The Netherlands: 2013. Hepatotoxicity of antimicrobials and antifungal agents. [Google Scholar]
- 80.Zatloukal K., French S.W., Stumptner C., Strnad P., Harada M., Toivola D.M., Cadrin M., Omary M.B. From mallory to mallory–denk bodies: What, how and why? Exp. Cell Res. 2007;313:2033–2049. doi: 10.1016/j.yexcr.2007.04.024. [DOI] [PubMed] [Google Scholar]
- 81.Fortier A.-M., Riopel K., Désaulniers M., Cadrin M. Novel insights into changes in biochemical properties of keratins 8 and 18 in griseofulvin-induced toxic liver injury. Exp. Mol. Pathol. 2010;89:117–125. doi: 10.1016/j.yexmp.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 82.Fortier A.-M., Asselin E., Cadrin M. Keratin 8 and 18 loss in epithelial cancer cells increases collective cell migration and cisplatin sensitivity through claudin1 up-regulation. J. Biol. Chem. 2013;288:11555–11571. doi: 10.1074/jbc.M112.428920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Golob-Schwarzl N., Bettermann K., Mehta A.K., Kessler S.M., Unterluggauer J., Krassnig S., Kojima K., Chen X., Hoshida Y., Bardeesy N.M. High keratin 8/18 ratio predicts aggressive hepatocellular cancer phenotype. Transl. Oncol. 2019;12:256–268. doi: 10.1016/j.tranon.2018.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.French S.W., Bardag-Gorce F., Li J., French B.A., Oliva J. Mallory-denk body pathogenesis revisited. World J. Hepatol. 2010;2:295. doi: 10.4254/wjh.v2.i8.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Redeker A.G., Sterling R.E., Bronow R.S. Effect of griseofulvin in acute intermittent porphyria. JAMA. 1964;188:466–468. doi: 10.1001/jama.1964.03060310066017. [DOI] [PubMed] [Google Scholar]
- 86.Berman A., Franklin R.L. Precipitation of acute intermittent porphyria by griseofulvin therapy. JAMA. 1965;192:1005–1007. doi: 10.1001/jama.1965.03080240075027. [DOI] [PubMed] [Google Scholar]
- 87.Maitra D., Cunha J.B., Elenbaas J.S., Bonkovsky H.L., Shavit J.A., Omary M.B. Porphyrin-induced protein oxidation and aggregation as a mechanism of porphyria-associated cell injury. Cell. Mol. Gastroenterol. Hepatol. 2019;8:535–548. doi: 10.1016/j.jcmgh.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Knasmüller S., Parzefall W., Helma C., Kassie F., Ecker S., Schulte-Hermann R. Toxic Effects of griseofulvin: Disease models, mechanisms, and risk assessment. Crit. Rev. Toxicol. 1997;27:495–537. doi: 10.3109/10408449709078444. [DOI] [PubMed] [Google Scholar]
- 89.Zhang X., Qian S.-B. Chaperone-mediated hierarchical control in targeting misfolded proteins to aggresomes. Mol. Biol. Cell. 2011;22:3277–3288. doi: 10.1091/mbc.e11-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. Autodock4 and autodocktools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rosenbaum M.B., Chiale P.A., Halpern M.S., Nau G.J., Przybylski J., Levi R.J., Lázzari J.O., Elizari M.V. Clinical efficacy of amiodarone as an antiarrhythmic agent. Am. J. Cardiol. 1976;38:934–944. doi: 10.1016/0002-9149(76)90807-9. [DOI] [PubMed] [Google Scholar]
- 92.Poucell S., Ireton J., Valencia-Mayoral P., Downar E., Larratt L., Patterson J., Blendis L., Phillips M.J. Amiodarone-associated phospholipidosis and fibrosis of the liver. Light, immunohistochemical, and electron microscopic studies. Gastroenterology. 1984;86:926–936. [PubMed] [Google Scholar]
- 93.Mason J.W. Amiodarone. N. Engl. J. Med. 1987;316:455–466. doi: 10.1056/NEJM198702193160807. [DOI] [PubMed] [Google Scholar]
- 94.March-Vila E., Pinzi L., Sturm N., Tinivella A., Engkvist O., Chen H., Rastelli G. On the integration of in silico drug design methods for drug repurposing. Front. Pharmacol. 2017;8:298. doi: 10.3389/fphar.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ashburn T.T., Thor K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.