Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is the causative agent of coronavirus disease 2019 (COVID‐19), the most consequential pandemic of this century, threatening human health and public safety. SARS‐CoV‐2 has been continuously evolving through mutation of its genome and variants of concern have emerged. The World Health Organization R&D Blueprint plan convened a range of expert groups to develop animal models for COVID‐19, a core requirement for the prevention and control of SARS‐CoV‐2 pandemic. The animal model construction techniques developed during the SARS‐CoV and MERS‐CoV pandemics were rapidly deployed and applied in the establishment of COVID‐19 animal models. To date, a large number of animal models for COVID‐19, including mice, hamsters, minks and nonhuman primates, have been established. Infectious diseases produce unique manifestations according to the characteristics of the pathogen and modes of infection. Here we classified animal model resources around the infection route of SARS‐CoV‐2, and summarized the characteristics of the animal models constructed via transnasal, localized, and simulated transmission routes of infection.
Keywords: animal model, characteristics, establishment, SARS‐CoV‐2
Animal models play crucial roles in the pathogenic mechanisms and potential interventions. In the current review, we summarized the development and characteristics of the animal models around transnasal infection, localized infection and simulated‐transmission route infection.

1. CLINICAL FEATURES OF COVID‐19
SARS‐CoV‐2 is the etiological agent of COVD‐19 and has caused a devastating pandemic. SARS‐CoV‐2 presents with upper and lower respiratory symptoms with the most common clinical signs being respiratory symptoms such as cough, sputum, sore throat, runny nose, earache, wheezing, and chest pain. In addition to the lungs, the virus damages other tissues, including heart, gastrointestinal tract, and central nervous system and can manifest as abdominal pain, vomiting, diarrhea, anosmia, and delusions. Up to 80% of patients with laboratory‐confirmed cases have mild to moderate disease. 1 , 2 , 3 , 4 However, people over 60 years of age and those with preexisting diseases (including hypertension, diabetes, cardiovascular disease, chronic respiratory disease, and cancer) are at the highest risk of severe disease and death, with a mortality rate of 2%–5% for the disease in the early stages of COVID‐19 outbreak. To date, most patients have asymptomatic infections or are mildly ill and the current mortality rate is approximately 1%. 4 , 5 , 6 In addition, patients may experience varying degrees of sequelae after recovery, known as ‘post‐acute or long COVID’, with the most commonly reported symptoms including dyspnea, cough, palpitations, anosmia, loss of taste, anxiety, depression, concentration/memory problems, and chronic fatigue. 7 , 8 , 9 , 10 At the present time, most of the clinical features have been reproduced in animal models, but the comorbidities and the sequelase after recovery have not been fully stimulated in animal models for COVID‐19.
2. ANIMAL MODELS ARE AN IMPORTANT TOOL FOR INFECTIOUS DISEASE RESEARCH
In the 19th century Robert Koch used rabbits and mice to successfully construct the first animal model of anthrax and summarized the golden rule of infectious disease pathogen identification, Koch's law, laying the foundation for the establishment of a systematic approach to pathogen biology. In addition, through animal experiments, Louis Pasteur confirmed that the rabies virus mainly invades the nervous system, and subsequently, developed a rabies vaccine. 11 In 1933, scientists successfully isolated the influenza A virus using a ferret model. 12 Animal model research has made outstanding contributions in clarifying pathogenesis and transmission mechanisms and screening effective vaccine and drugs and has played an important role in combating infectious diseases. At the beginning of the COVID‐19 outbreak, the etiology, pathogenesis, and transmission mechanism of the disease were unknown and no drugs were available. The WHO Blueprint Group established an ad hoc expert working group focused on COVID‐19 disease modeling (WHO‐COM). COVID‐19 animal models were developed with the aim of clarifying the pathogenic agent of COVID‐19, describing the pathogenesis and transmission mechanism of the emerging pathogen, analyzing the host immune response to the pathogen, and identifying molecular targets of pathogen infection. These results could then contribute to screening for effective vaccines and treatments against SARS‐CoV‐2 and promote a swifter application of effective drugs, antibodies, and vaccines.
3. ESTABLISHMENT AND CHARACTERISTICS OF ANIMAL MODELS FOR COVID‐19
The key to animal model preparation is animal selection. The scale of animal resources for establishing animal models of respiratory infectious diseases was initially established by comparative medical analysis based on the anatomical structure and physiological functions of the respiratory system. Bioinformatics analysis was used to identify angiotensin‐converting enzyme II (ACE2) as the functional receptor for SARS‐CoV‐2, 13 and this was used to identify the animal species range that could be established as model target groups. Genetically modified animals with human receptors were established in model target groups, and non‐human primates (rhesus monkeys, crab‐eating monkeys, African green monkeys), hamsters, mink, and cats whose ACE2 receptors have homology to human receptors were identified as possible animal models of SARS‐CoV‐2. 14 , 15 , 16 , 17 Infectious diseases are characterized by unique manifestations based on the biological features of the pathogen and modes of infection 18 ; therefore, this paper will present a classification of animal models based on the mode of infection of COVID‐19.
3.1. Animal models via intranasal infection
SARS‐CoV‐2 propagates by interpersonal transmission and continuous circulation thereafter. Since SARS‐CoV‐2 is a respiratory virus, intranasal challenge is the first choice for researchers aiming to establish animal models for COVID‐19. This method starts the life cycle of the respiratory virus in the nasal cavity, thus simulating the process of natural infection. In addition, the process of intranasal infection is easier than other methods, simple, rapid, stable, and noninvasive to the animals, with low equipment requirements, and is applied in models using small animals such as mice and hamsters.
3.1.1. Adenovirus‐transducted mouse models with intranasal infection
Based on the experience of animal model development for MERS‐CoV, 19 , 20 , 21 investigators used adenovirus or adenovirus‐associated vector (AAV) to deliver exogenous human ACE2 (hACE2) into mice via the intranasal route to establish transiently transfected mouse models. 22 , 23 , 24 BALB/c and C57BL/6 mice administered with Ad5‐hACE2 were intranasally infected with 105 plaque‐forming units (PFU). SARS‐CoV‐2 exhibited ruffled fur, hunching, and difficulty breathing accompanied by weight loss after infection. SARS‐CoV‐2 replication peaked in the lungs 1–2 days post‐infection (dpi) and then declined with time; histopathology showed inflammatory cell infiltration and congestive edema in lung tissue. 22 Most AAV‐constructed mouse models lacked clinical signs but showed viral replication in bronchial and alveolar epithelial cells and lung histopathology indicative of mild pneumonia. These models can make all mouse strains susceptible to SARS‐CoV‐2 and thereby could address the urgent need for a COVID‐19 animal model at the beginning of the COVID‐19 epidemic; however, the infected models only showed mild symptoms, with a short viral maintenance, and no extra‐pulmonary manifestations of COVID‐19.
3.1.2. Genetically modified mice models with intranasal infection
Genetically engineered mice have stable inheritance of engineered changes. A genetically modified hACE2 mouse model was achieved using different specific promoters to transfer hACE2 into mice. hACE2‐ICR mice were inoculated intranasally with SARS‐CoV‐2 virus at a dose of 105 TCID50, and weight loss, viral RNA levels, and virus replication were subsequently monitored in lung tissue. Lung histopathology indicated moderate to mild interstitial pneumonia, and SARS‐CoV‐2‐specific IgG could be monitored at 21 dpi. 25 hACE2‐KI/NIFDC transgenic mice, where endogenous mouse ACE2 was replaced with hACE2 using CRISPR/Cas9 technology, were intranasally inoculated with 4 × 105 PFU SARS‐CoV‐2 and exhibited weight loss with high viral levels in lung, trachea, and brain tissue. Lung tissue showed interstitial pneumonia including inflammatory cell infiltration, alveolar septal thickening, and vascular damage. 26 These two strategies are murine promoter‐driven animal models, both of which can be infected with SARS‐CoV‐2 through the natural infection route and enable live viruses to be isolated in the lung tissue. These animals developed moderate to mild pneumonia in the lung tissue with mild clinical signs and no mortality, and thus can mimic the symptoms seen in patients with clinical COVID‐19 with mild disease and be used to study pathogenesis and test therapeutics. K18 promoter‐hACE2 transgenic mice (K18‐hACE2) were highly susceptible to SARS‐CoV‐2 and exhibited significant weight loss, even becoming moribund after intranasal inoculation with SARS‐CoV‐2. Viral RNA was expressed in several tissues including lung, heart, brain, kidney, spleen, and duodenum, but the viral load in brain tissue was more prominent. The lung tissue of K18‐hACE2 mice showed mildly focal inflammation. 23 , 27 , 28 , 29 Lung and brain are the main target organs of SARS‐CoV‐2 in CAG promoter‐hACE2 transgenic mice (CAG‐hACE2) inoculated intranasally, and acute lung injury occurs in early infection due to dramatically elevated cytokine and chemokine levels, although with untypical histopathological features. 30 , 31 A proportion of HFH4‐hACE2 mice, in which the disease is regulated by the lung ciliary epithelial cell‐specific HFH4/FOXJ1 promoter, experienced significant weight loss and death after SARS‐CoV‐2 infection. Although the lung tissue of these mice showed typical interstitial pneumonia, the complementary metrics of pulmonary obstruction and bronchoconstriction, including PenH and Rpef, were maintained at normal levels. 32 hACE2 is widely distributed in the heterologous promoter‐associated hACE2 mouse model, thereby affecting the cellular tropism of SARS‐CoV‐2. Enrichment of SARS‐CoV‐2 in brain tissue from dead animals who exhibited significant neurological symptoms or neuronal damage indicates that respiratory infection was likely not a major driver of mortality. In addition, ACE2 is highly expressed in patients with comorbidities such as hypertension, cardiovascular disease, and diabetes, and this model ignores the complex role of hACE2 expression, which may enhance viral susceptibility and induce the development of severe COVID‐19 disease.
3.1.3. Alternative virus strains in intranasally infected animal models
The animal models established above were developed using wild virus strains. Mouse‐adapted or ‐modified viruses also can be used to establish animal models. Successive adaptive passages of SARS‐CoV‐2 in mice generate mouse‐adapted strains, whose pathogen‐host interactions closely resemble those found naturally in mice. An intranasally inoculated SARS‐CoV‐2 mouse‐adapted strain, MASCp6, showed no obvious clinical signs, although the presence of viral RNA was monitored in the lung, trachea, heart, liver, spleen, brain tissue, and feces, and mild to moderate interstitial pneumonia was observed in lung tissue; moreover, aged mice experienced more severe lung tissue damage. 33 MASCp36‐infected mice experienced binary clinical phenotypes, including ruffled fur, hunching, and difficulty breathing; high viral loads were evident in the lung and trachea, with the lung tissue developing necrotizing pneumonia and extensive diffuse alveolar damage, hyaline membrane formation, and pulmonary fibrosis, and multi‐organ damage occurred in extra‐pulmonary organs such as the spleen and kidney. 34 Mutations in virus‐adapted strain MASCp6‐36 (N501Y/Q493H/K417N) may be key in enabling SARS‐CoV‐2 to break through host restriciton. Furthermore, the mouse‐adapted strain WBP‐1 with Q493K and Q498H mutations in RBD (receptor binding domain) can successfully infected BALB/c mice, resulting in severe interstitial pneumonia. 35 In keeping with this, the investigators introduced the mutant site Q498Y/P499T into the RBD through reverse genetic techniques to establish a virus replacement strain (SARS‐CoV‐2 MA). SARS‐CoV‐2 MA (at 105 PFU) was then administered to BALB/c mice via the intranasal route. No obvious clinical signs were observed, and the virus was cleared from the lung tissue within 4 days while the indicators of lung function (Penh and Rpef) were abnormal, manifesting as mild pneumonia. 36 MA‐10‐infected mice showed significant weight loss on intranasal infection, developed diffuse alveolar injury with alveolar septal neutrophil infiltration accompanied by hyaline membrane formation in early infection. 37 Substitution strain‐infected animal models enable SARS‐CoV‐2 infection to break the species restrictiveness, and the viral pathogenicity may gradually increase with the increasing adaptation over subsequent generations; this can simulate clinical mild to severe disease in patients and highlights the need to monitor the mutation sites that may occur during the natural course of an epidemic of SARS‐CoV‐2. Meanwhile, animal models established by virus substitution strains can be used to assess the role of host genetics and antiviral defense genes in virus pathogenesis and to test countermeasures. However, these models do not necessarily fully reflect the infection characteristics of wild‐type viruses. 38
3.1.4. Spontaneous animal models in intranasal infection
Hamsters are widely used in the study of respiratory infections. 39 , 40 All Syrian hamster models of COVID‐19 are inoculated transnasally. Infected hamsters display clearly apparent symptoms, such as lethargy, ruffled fur, hunching, and shortness of breath. 41 , 42 Viral load can be detected in the respiratory tract (nose, turbinates, trachea, and lung tissue) as well as in extra‐pulmonary tissues (brain, heart, liver, spleen, lymph nodes, intestines, kidneys, adrenal glands, and reproductive organs), with diffuse alveolar damage, 43 severe interstitial pneumonia, and various degrees of multi‐organ lesions in hamsters 3–7 days after infection. The hamster model can also display other important clinical features, such as anosmia, neurotropism, and vascular inflammation, found in patients with COVID‐19. 44 , 45 , 46 The virus persists in the tissue for a longer time when compared with infections in mouse tissue, and pathological changes in tissues and organs can still be examined at approximately 18 days in the hamster model, allowing this model to be used for studies on the sequelae and assessment of therapeutic measures during the recovery period after the course of COVID‐19 disease. 43 The hamster model of COVID‐19 pneumonia can result in different phenotypes depending on age, sex, and strain. 47 , 48 , 49 Currently, the Syrian hamster model is rapidly becoming a widely used animal model for SARS‐CoV‐2 studies, including preclinical studies on the effectiveness of antiviral drugs 50 , 51 , 52 , 53 and vaccines 54 and evaluating the efficacy of surgical masks in blocking virus transmission. 55 However, hamster models also have limitations: the models lack the common extra‐pulmonary manifestations of COVID‐19 in human patients and may be unsuitable for simulating severe disease in patients with COVID‐19 comorbidities. Moreover, this model lacks advanced tools for immunology studies of, for example, the relationship between the severity of lung pathology and mild to moderate clinical signs.
A mink model of intranasal infection has also been constructed, and infected mink displayed significant weight loss, respiratory distress (manifested as infiltrative pneumonia), and even death. 56 The nasal cavity of infected mink was filled with mucus‐purulent secretions, and viral RNA was detected in the nasal turbinates, soft palate, tonsils, all lung lobes and submandibular lymph nodes, and trachea. In addition, SARS‐CoV‐2 was widely distributed in extra‐respiratory tissues (cardiovascular, hepatobiliary, urinary, endocrine, digestive and immune systems), and multi‐organ and systemic lesions were present with a significant increase in inflammatory response, consistent with reports of clinically critical patients. 57 The mink model of infection presents moderate to severe pneumonia with diffuse alveolar damage, thrombosis, and pathological damage that appears very similar to that seen in humans and thus can mimic clinical patients with moderate to severe pneumonia. Our study found that SARS‐CoV‐2‐infected mink appear to have similar lipidomic and metabolomic gene expression differences to patients with severe COVID‐19 and provide evidence for melatonin as a potential therapeutic approach against COVID‐19 57 ; additionally, mink models can be used to assess neutralizing antibody responses against SARS‐CoV‐2 variants. 58 , 59
Cats intranasally infected with SARS‐CoV‐2 showed no obvious clinical signs and did not develop lower respiratory pathology associated with human disease. 60 , 61 , 62 Viral loads were monitored in the upper and lower respiratory tract and intestine in 6‐ to 9‐month‐old cats inoculated intranasally with the SARS‐CoV‐2. 63 Intestinal lesions showed multifocal inflammatory cell infiltrates in the submucosa and muscularis of the intestine, and respiratory lesions mainly concentrated in the upper respiratory tract were closely associated with viral enrichment in the trachea. 43 The cat model can therefore be used in the study of SARS‐CoV‐2 transmission.
3.1.5. Animal models constructed based on comorbidities
To address the complex disease course of patients with underlying disorders such as hypertension, diabetes, and cardiovascular disease during SARS‐CoV‐2 infection and to select appropriate life‐saving treatment, animal models with compound diseases have been developed. hACE2 transgenic mice were used to create a hypertension model via high salt (1% NaCl) feeding and osmotic micropump subcutaneous infusion of angiotensin II (Ang II, 500 ng/mg/d) for 3 weeks. These mice were then infected with SARS‐CoV‐2. Compared with control mice, hypertensive mice showed a higher viral load in the lungs and developed pulmonary edema with hemorrhage. Lung pathology progressed into severe interstitial pneumonia with thickened alveolar septa. 64 The cardiovascular disease co‐infection model increased the risk of myocardial injury on intranasal infection; meanwhile, ob/ob mice infected with SARS‐CoV‐2 intranasally showed a higher blood glucose and lower insulin response, and displayed more severe pneumonia phentypes. 65 Similarly, diabetic mice (BKS‐leprdb, db/db) with SARS‐CoV‐2 infection exhibited higher viral loads and more severe respiratory tissue damage. 66 In addition, Western diet‐fed hamsters had more pronounced weight loss and higher viral loads in lung tissue after infection. 67 The presence of a prior comorbidity is a major risk factor for poor prognosis of COVID‐19. Therefore, establishing an animal model of risk factor‐based co‐infection that mimics the pathological features of co‐infection to explore the pathogenesis of disease after SARS‐CoV‐2 infection provides a vehicle to study clinically complex disease and to evaluate specific and effective therapeutic measures for severe COVID‐19. However, comorbidities are difficult to model within animal experiments, except those in mice, and additional comorbidities associated with severe COVID‐19 (including platelet reaction and coagulation dysfunction) have not been studied in animal models.
3.1.6. Animal models of immunodeficiency
To date, the short viral maintenance time present in the existing animal models cannot reproduce the ongoing immunopathology of patients with severe disease. Animal models in which the virus replicates to sufficiently high titers for extended periods of time without causing severe pathology are therefore required. Immunosuppressed hamster models were prepared by intraperitoneal injection of cyclophosphamide at a dose of 70 mg/kg from the day of attack and every 3 days until the end of the test. Immunocompromised hamsters experienced prolonged sustained weight loss, slow weight recovery, and suppression of lung tissue innate immune and inflammatory pathway genes, including IFNB, IFNL, NK cell activation, and hypercytokinemia. 68 In addition, viral clearance of lung tissue in Ad5‐hACE2‐transduced STAT1 −/− C57BL/6 or IFNAR −/− C57BL/6 mice with SARS‐CoV‐2 infection was delayed. 22 Moreover, in infected AAV‐hACE2 transduced and humanized MISTRG6 mice high viral titers and RNA levels were prolonged for at least 35 dpi. 69 The establishment and study of humanized or immunodeficient mouse models should facilitate the identification of viruses or host factors to improve the understanding of the molecular mechanisms underlying SARS‐CoV‐2 lung pathogenesis and help to identify potential therapeutic targets for the treatment of SARS‐CoV‐2‐related lung diseases.
3.1.7. Animal models for comparison of VOC pathogenicity
With the accumulation of SARS‐CoV‐2 mutations, the virus has evolved into many variants. Mouse models are widely used to assess the virulence and pathogenicity of VOC. In K18‐hACE2 mice infected intranasally with wild‐type (WT), Alpha, Beta, Delta, and Omicron variants, the Omicron‐infected mice showed slight clinical signs and the highest survival rate compared with WT‐ and Delta‐infected mice and had decreased viral load in the nasal turbinates and lung tissue. Lung histopathological changes were also less severe in mice with Omicron infection. 70 , 71 In addition, hamsters infected with Omicron had delayed and mild weight loss and significantly lower clinical scores and viral load titers in lung tissue compared with Delta‐infected hamsters. The Omicron virus strain was cleared from hamster lung tissue and trachea and inflammatory cytokine and chemokine levels returned to normal at 7 dpi. Moreover, compared with the severe diffuse bronchial and alveolar inflammatory infiltrates in mice with Delta infection, lung injury in hamsters with Omicron infection was less severe and recovery was faster. 72 However, current studies on the comparative pathogenicity of VOCs are limited by variation in experimental settings and viral strains, which has a significant impact on the experimental results. Comparative analysis studies are required using different animal model resources and different mutant strains under the same experimental conditions.
Mice models are widely used in studies of immunology and lung injury for a broader understanding of several different aspects of COVID‐19. The mice models for COVID‐19 are summarized in Figure 1.
FIGURE 1.
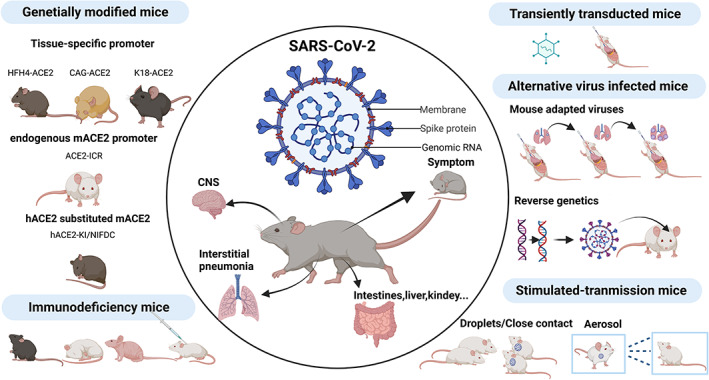
Mouse models for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2)
3.2. Locating infection
Most animal models with transnasal infection show mild to moderate interstitial pneumonia, whereas acute respiratory distress syndrome (ARDS), the pathological feature of severe COVID‐19, is rare.
Unlike the intranasally infected cat model, SARS‐CoV‐2 transtracheally inoculated domestic cats experience clinical signs of lethargy, shortness of breath, and cough; lung tissue showed edema, multifocal alveolar damage and necrosis, and perivascular inflammatory cell infiltration accompanied with the appearance of hyaline membranes. 73 The cat model obtained by intratracheal inoculation showed clinical disease features consistent with the early exudative phase of acute COVID‐19. It is noteworthy that despite the transtracheal localization of lung tissue infection, high viral loads were not evident in lung tissue, and the pathogenesis of the cat model of transtracheal infection still needs to be further explored.
To directly mimic the characteristics of pneumonia, large animals such as non‐human primates, which have similar anatomical structure and physiological characteristics to humans, are used with transtracheal infection to prepare pneumonia models. Most non‐human primate models have been infected with combined mucosal exposure inoculations (intratracheal, intranasal, and conjunctival), and only two studies have used the transtracheal route alone. However, rhesus monkeys can be successfully infected and showed patient‐like histopathological changes, suggesting that transtracheal infection is an important route of SARS‐CoV‐2 infection. Rhesus macaques with intratracheal SARS‐CoV‐2 infection developed pulmonary infiltration visible on X‐rays, which is a hallmark of infection in humans 74 , 75 , 76 ; viral load was detected in nasopharyngeal swabs, viral shedding was monitored in the lower respiratory tract, and lung tissue showed mild to moderate interstitial pneumonia. 77 The most common findings reported using the non‐human primate model were lung consolidation, edema, and hemorrhage. A proportion of models showed vascular changes, 78 , 79 , 80 epithelial pathology, 81 , 82 , 83 or raised biomarker levels associated with coagulation, thrombosis, and vascular disease, and upregulated pro‐inflammatory cytokines. 78 , 82 , 83 , 84 , 85 , 86 Meanwhile, the age dependence of SARS‐CoV‐2 infection was reproduced in a rhesus macaque model. Adult monkeys aged 3–5 years and elderly rhesus monkeys aged 15 years were transtracheally infected with 106 TCID50 SARS‐CoV‐2 HB‐01. 77 SARS‐CoV‐2 replicated in the upper and lower respiratory tracts and peripheral blood lymphocytes decreased, and while young monkeys developed typical interstitial pneumonia, old monkeys exhibited diffuse severe interstitial pneumonia, similar to the clinical presentation of COVID‐19 patients. Non‐human primates are close to humans physiologically, genetically, and immunologically, 87 and the moderate disease observed in rhesus macaques is thus in line with the majority of human COVID‐19 infections; the rhesus macaque model is therefore an ideal experimental animal for the development of the COVID‐19 vaccine. The rhesus macaque model has been used to evaluate the immunogenicity and protective efficacy of inactivated, DNA, and vector vaccines, to comparatively analyze the protective efficiency of vaccines against different VOCs after immunization, and to explore the effectiveness of sequential immunization and polyvalent vaccines, thus laying the foundation for the translational application of vaccines and epidemic control. 88 , 89 , 90 , 91 , 92 However, use of non‐human primates has limitations such as high animal cost, low breeding efficiency, large body size, complicated operational management, and large individual differences, which make it difficult to conduct studies with large sample sizes. In addition, studies to explore comorbidities such as diabetes and cardiovascular diseases using non‐human primate models are lacking, and some clinical symptoms of patients (e.g. thromboembolism) are rare in non‐human primate models, which thus need further exploration and optimization.
3.3. Animal models for simulated transmission
SARS‐CoV‐2 is zoonotic virus, and it is important to explore its transmission potential for epidemic prevention and control and therefore to establish animal transmission models. hACE2 mice were inoculated intranasally with SARS‐CoV‐2 and placed in the same experimental cage with wild mice at 1 dpi, and after cohabitation, weight loss, positive pharyngeal‐anal swab virus RNA and specific antibody levels were monitored in the wild mice. 93 In addition, SARS‐CoV‐2 has been shown to infect cohabiting hamsters, and pathological features developed similar to those with direct infections. 94 The above model simulated the close contact route, confirming that SARS‐CoV‐2 could be transmitted by direct contact. The placement of wild hACE2 mice in a unidirectional airflow transmission cage resulted in several wild mice becoming positive for antibodies to SARS‐CoV‐2. 93 When uninfected cats were placed in cages adjacent to infected cats, viral RNA was detected in the nasal turbinates, soft palate, tonsils, and trachea in the uninfected cats, and all cats produced neutralizing antibodies, confirming that SARS‐CoV‐2 could be transmitted by droplet transmission. 95 In addition, the SARS‐CoV‐2 tandem transmission model was established by serial passaging via cohousing a naive cat with infected or exposed cats in a ventilated isolator for 2 days, to explore the transmission ability of the virus over different generations in the cat population. This study confirmed that serial passaging of the virus between cats dramatically attenuated the viral transmissibility and pathogenicity. 43
Non‐human primates are rarely used for transmission studies because of the limitations of the confinement facilities. The possibility of oculoconjunctival and gastrointestinal transmission have been explored using rhesus macaque models. After oculoconjunctival infection, the virus was mainly distributed to the nasal lacrimal gland and ocular tissues, the nose, pharynx, lower lobe of the left lung, tonsils, stomach, duodenum, and ileum. The virus was successfully isolated from the lower lobe of the left lung, with mild localized interstitial pneumonia in the lung tissue. However, compared with intratracheal inoculation, viruses were abundant in the nasolacrimal system, and lung pathology displayed mild localized lesions after transconjunctival inoculation, 96 which indicated that SARS‐CoV‐2 may be a transmitted via the oculoconjunctival route. However, no viral load or SARS‐CoV‐2‐specific antibodies were detected in monkeys infected through the gastrointestinal route, and the possibility of SARS‐CoV‐2 transmission through the digestive tract route still needs further exploration.
SARS‐CoV‐2 infection via the intratracheal or intraocular route in non‐human primate models requires direct contact mucosal infection. To mimic the natural route of infection, African green monkeys, rhesus macaques, and cynomolgus macaques were exposed to SARS‐CoV‐2 by the aerosol route, and, consistent with the results of infection via the transtracheal route, all animals showed respiratory abnormalities, virus shedding, and mild to moderate respiratory disease after aerosol infection. 97 The animal models established by simulating the transmission route further have clarified the transmission route of SARS‐CoV‐2, although most animal models established are based on transmission routes using the original SARS‐CoV‐2 strain. Hamster models have been used to verify the transmission of the D614G variant, 98 , 99 and studies on the transmission of VOCs in animals in the presence of vaccine‐induced immunity are needed for comparative analysis. In addition, current transmission animal models ignore risk factors such as age and gender complications, and the transmission models need improvement for further exploration.
4. CONCLUSION AND FUTURE COURSE
The currently established animal models – non‐human primate, hamster, mouse, mink, and cat – have made significant contributions to the exploration of the pathogenesis and transmission mechanism of SARS‐CoV‐2. The hACE2 mouse model helped clarify the pathogen and receptor and determined the pathological characteristics of COVID‐19 pneumonia, whereas hamster models with long disease duration have simulated the recovery period of the disease. The rhesus macaque, hamster, mouse, cat, and mink models have clarified that SARS‐CoV‐2 can be transmitted by contact, droplet, restricted aerosol, and conjunctival transmission. The monkey and hamster models have also been used to explore the risk of ‘reinfection’, which has important implications for patient management, serotherapy, and vaccine design during recovery. 81 , 100 The mouse, hamster, and monkey models have been used to evaluate the efficacy of various drugs, vaccines, and antibodies, 101–103 and vaccination duration, laying the foundation for the selection and application of immunization strategies.
However, animal models for COVID‐19 cannot fully reproduce all key feature of severe COVID‐19. The mouse model shows mild to moderate pathological changes after infection and cannot simulate the disease characteristics of patients with severe clinical disease; several monkey and hamster models show severe disease and can simulate the disease manifestations of patients with severe disease; hamster and mink models can show systemic multi‐organ damage and can simulate the disease characteristics of patients with severe COVID‐19. Moreover, features such as thrombosis and neurological damage sequelae that appear in COVID‐19 patients are rarely seen in commonly used animal models, and the interaction between the virus and the host is unclear due to the lack of reagents, which makes it difficult to conduct in‐depth studies on the pathogenesis and immune mechanisms. In addition, the accumulation of viral mutations may lead to the emergence of new receptors for SARS‐CoV‐2. Therefore, the construction of animal models still needs further improvement and exploration.
Development of immunodeficient animals or experimentally induced immunodeficient animals through pharmacological intervention, establishment of humanized/gene knockout integrated animal models, or establishment of multigene target mouse models that accurately summarize the biological systems of human infectious diseases needs to be further explored to provide support for multi‐dimensional analysis of infectious disease pathogenesis, new drug development and safety evaluation.
AUTHOR CONTRIBUTIONS
Feifei Qi and Chuan Qin wrote the original draft of the manuscript and revised the manuscript. All the authors read and approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors declared no conflict of interest. Chuan Qin is an Editorial Board member of AMEM and a co‐author of this article. To minimize bias, she was excluded from all editorial decision‐making related to the acceptance of this article for publication.
Supporting information
Data S1
ACKNOWLEDGMENT
None.
Qi F, Qin C. Characteristics of animal models for COVID‐19. Anim Models Exp Med.2022;5:401‐409. doi: 10.1002/ame2.12278
REFERENCES
- 1. Sancho Ferrando E, Hanslin K, Hultström M, et al. Soluble TNF receptors predict acute kidney injury and mortality in critically ill COVID‐19 patients: a prospective observational study. Cytokine. 2022;149:155727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeng J, Liu Y, Yuan J, et al. First case of COVID‐19 complicated with fulminant myocarditis: a case report and insights. Infection. 2020;48(5):773‐777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pe F, Mc A, Sd C, et al. Association Between Comorbidities and Death From COVID‐19 in Different Age Groups. Cold Spring Harbor Laboratory Press; 2021. [Google Scholar]
- 4. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan. China JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang N, Liu Y‐N, Bao J, et al. Clinical features and risk factors associated with severe COVID‐19 patients in China. Chin Med J (Engl). 2021;134(8):944‐953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Al‐Aly Z, Xie Y, Bowe B. High‐dimensional characterization of post‐acute sequelae of COVID‐19. Nature. 2021;594(7862):259‐264. [DOI] [PubMed] [Google Scholar]
- 8. Huang C, Huang L, Wang Y, et al. 6‐month consequences of COVID‐19 in patients discharged from hospital: a cohort study. Lancet. 2021;397(10270):220‐232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davis HE, Assaf GS, McCorkell L, et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. 2021;38:101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luciani LG, Gallo F, Malossini G, et al. Urinary frequency as a possible overlooked symptom in COVID‐19 patients: does SARS‐CoV‐2 cause viral cystitis? Eur Urol. 2020;78(3):e129‐e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pasteur L. Methode pour prevenir la rage apres morsure.
- 12. Brockhurst JK, Villano JS. The role of animal research in pandemic responses. Comp Med. 2021;71(5):359‐368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miao J, Chard LS, Wang Z, et al. Syrian hamster as an animal model for the study on infectious diseases. Front Immunol. 2019;10:2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chan JF‐W, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID‐19) in a Golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428‐2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Damas J, Hughes GM, Keough KC, et al. Broad host range of SARS‐CoV‐2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci USA. 2020;117(36):22311‐22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melin AD, Janiak MC, Marrone F, et al. Comparative ACE2 variation and primate COVID‐19 risk. Commun Biol. 2020;3(1):641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bodewes R, Kreijtz JH, van Amerongen G, et al. Pathogenesis of influenza a/H5N1 virus infection in ferrets differs between intranasal and intratracheal routes of inoculation. Am J Pathol. 2011;179(1):30‐36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cockrell AS, Peck KM, Yount BL, et al. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J Virol. 2014;88(9):5195‐5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao J, Li K, Wohlford‐Lenane C, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014;111(13):4970‐4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agrawal AS, Garron T, Tao X, et al. Generation of a transgenic mouse model of Middle East respiratory syndrome coronavirus infection and disease. J Virol. 2015;89(7):3659‐3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun J, Zhuang Z, Zheng J, et al. Generation of a broadly useful model for COVID‐19 pathogenesis, vaccination, and treatment. Cell. 2020;182(3):734‐743.e735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rathnasinghe R, Strohmeier S, Amanat F, et al. Comparison of transgenic and adenovirus hACE2 mouse models for SARS‐CoV‐2 infection. Emerging Microbes Infect. 2020;9(1):2433‐2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Israelow B, Song E, Mao T, et al. Mouse model of SARS‐CoV‐2 reveals inflammatory role of type I interferon signaling. J Exp Med. 2020;217(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bao L, Deng W, Huang B, et al. The pathogenicity of SARS‐CoV‐2 in hACE2 transgenic mice. Nature. 2020;583(7818):830‐833. [DOI] [PubMed] [Google Scholar]
- 26. Sun S‐H, Chen Q, Gu H‐J, et al. A mouse model of SARS‐CoV‐2 infection and pathogenesis. Cell Host Microbe. 2020;28(1):124‐133.e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCray Paul B, Pewe L, Wohlford‐Lenane C, et al. Lethal infection of K18‐hACE2 mice infected with severe acute respiratory syndrome coronavirus. J Virol. 2007;81(2):813‐821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Golden JW, Cline CR, Zeng X, et al. Human angiotensin‐converting enzyme 2 transgenic mice infected with SARS‐CoV‐2 develop severe and fatal respiratory disease. JCI Insight. 2020;5(19). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zheng J, Wong LR, Li K, et al. COVID‐19 treatments and pathogenesis including anosmia in K18‐hACE2 mice. Nature. 2021;589(7843):603‐607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsai C‐Y, Chen C‐Y, Jan J‐T, et al. Sex‐biased response to and brain cell infection by SARS‐CoV‐2 in a highly susceptible human ACE2 transgenic model. bioRxiv. 2004; 2021.2005.2004.441029. [Google Scholar]
- 31. Yang S, Cao L, Xu W, et al. Comparison of model‐specific histopathology in mouse models of COVID‐19. J Med Virol. 2022;94(8):3605‐3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang R‐D, Liu M‐Q, Chen Y, et al. Pathogenesis of SARS‐CoV‐2 in transgenic mice expressing human angiotensin‐converting enzyme 2. Cell. 2020;182(1):50‐58.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gu H, Chen Q, Yang G, et al. Adaptation of SARS‐CoV‐2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369(6511):1603‐1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen Q, Huang X‐Y, Liu Y, et al. Comparative characterization of SARS‐CoV‐2 variants of concern and mouse‐adapted strains in mice. J Med Virol. 2022;94(7):3223‐3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Søli NE, Frøslie A, Aaseth J. The mobilization of copper in sheep by chelating agents. Acta Vet Scand. 1978;19(3):422‐429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dinnon KH 3rd, Leist SR, Schäfer A, et al. A mouse‐adapted model of SARS‐CoV‐2 to test COVID‐19 countermeasures. Nature. 2020;586(7830):560‐566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leist SR, Dinnon KH 3rd, Schäfer A, et al. A mouse‐adapted SARS‐CoV‐2 induces acute lung injury and mortality in standard laboratory mice. Cell. 2020;183(4):1070‐1085.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Daniloski Z, Jordan TX, Ilmain JK, et al. The spike D614G mutation increases SARS‐CoV‐2 infection of multiple human cell types. Elife. 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schaecher SR, Stabenow J, Oberle C, et al. An immunosuppressed Syrian golden hamster model for SARS‐CoV infection. Virology. 2008;380(2):312‐321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwatsuki‐Horimoto K, Nakajima N, Ichiko Y, et al. Syrian hamster as an animal model for the study of human influenza virus infection. J Virol. 2018;92(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imai M, Iwatsuki‐Horimoto K, Hatta M, et al. Syrian hamsters as a small animal model for SARS‐CoV‐2 infection and countermeasure development. Proc Natl Acad Sci USA. 2020;117(28):16587‐16595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rosenke K, Meade‐White K, Letko M, et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS‐CoV‐2 infection. Emerging Microbes Infect. 2020;9(1):2673‐2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bao L, Song Z, Xue J, et al. Susceptibility and attenuated transmissibility of SARS‐CoV‐2 in domestic cats. J Infect Dis. 2021;223(8):1313‐1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Melo GD, Lazarini F, Levallois S, et al. COVID‐19‐related anosmia is associated with viral persistence and inflammation in human olfactory epithelium and brain infection in hamsters. Sci Transl Med. 2021;13(596). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Allnoch L, Beythien G, Leitzen E, et al. Vascular inflammation is associated with loss of aquaporin 1 expression on endothelial cells and increased fluid leakage in SARS‐CoV‐2 infected Golden Syrian hamsters. Viruses. 2021;13(4):639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Becker K, Beythien G, de Buhr N, et al. Vasculitis and neutrophil extracellular traps in lungs of Golden Syrian hamsters with SARS‐CoV‐2. Front Immunol. 2021;12:640842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bertzbach LD, Vladimirova D, Dietert K, et al. SARS‐CoV‐2 infection of Chinese hamsters (Cricetulus griseus) reproduces COVID‐19 pneumonia in a well‐established small animal model. Transbound Emerg Dis. 2021;68(3):1075‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trimpert J, Vladimirova D, Dietert K, et al. The Roborovski dwarf hamster is a highly susceptible model for a rapid and fatal course of SARS‐CoV‐2 infection. Cell Rep. 2020;33(10):108488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yuan L, Zhu H, Zhou M, et al. Gender associates with both susceptibility to infection and pathogenesis of SARS‐CoV‐2 in Syrian hamster. Signal Transduct Target Ther. 2021;6(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fagre AC, Manhard J, Adams R, et al. A potent SARS‐CoV‐2 neutralizing human monoclonal antibody that reduces viral burden and disease severity in Syrian hamsters. Front Immunol. 2020;11:614256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kreye J, Reincke SM, Kornau HC, et al. A therapeutic non‐self‐reactive SARS‐CoV‐2 antibody protects from lung pathology in a COVID‐19 hamster model. Cell. 2020;183(4):1058‐1069.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ye ZW, Yuan S, Chan JF, et al. Beneficial effect of combinational methylprednisolone and remdesivir in hamster model of SARS‐CoV‐2 infection. Emerging Microbes Infect. 2021;10(1):291‐304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yuan S, Yin X, Meng X, et al. Clofazimine broadly inhibits coronaviruses including SARS‐CoV‐2. Nature. 2021;593(7859):418‐423. [DOI] [PubMed] [Google Scholar]
- 54. Yahalom‐Ronen Y, Tamir H, Melamed S, et al. A single dose of recombinant VSV‐∆G‐spike vaccine provides protection against SARS‐CoV‐2 challenge. Nat Commun. 2020;11(1):6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chan JF, Yuan S, Zhang AJ, et al. Surgical mask partition reduces the risk of noncontact transmission in a Golden Syrian hamster model for coronavirus disease 2019 (COVID‐19). Clin Infect Dis. 2020;71(16):2139‐2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shuai L, Zhong G, Yuan Q, et al. Replication, pathogenicity, and transmission of SARS‐CoV‐2 in minks. Natl Sci Rev. 2021;8(3):nwaa291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Song Z, Bao L, Deng W, et al. Integrated histopathological, lipidomic, and metabolomic profiles reveal mink is a useful animal model to mimic the pathogenicity of severe COVID‐19 patients. Signal Transduct Target Ther. 2022;7(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hoffmann M, Zhang L, Krüger N, et al. SARS‐CoV‐2 mutations acquired in mink reduce antibody‐mediated neutralization. Cell Rep. 2021;35(3):109017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bayarri‐Olmos R, Rosbjerg A, Johnsen LB, et al. The SARS‐CoV‐2 Y453F mink variant displays a pronounced increase in ACE‐2 affinity but does not challenge antibody neutralization. J Biol Chem. 2021;296:100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gaudreault NN, Trujillo JD, Carossino M, et al. SARS‐CoV‐2 infection, disease and transmission in domestic cats. Emerging Microbes Infect. 2020;9(1):2322‐2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bosco‐Lauth AM, Hartwig AE, Porter SM, et al. Experimental infection of domestic dogs and cats with SARS‐CoV‐2: pathogenesis, transmission, and response to reexposure in cats. Proc Natl Acad Sci USA. 2020;117(42):26382‐26388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Gaudreault NN, Carossino M, Morozov I, et al. Experimental re‐infected cats do not transmit SARS‐CoV‐2. Emerging Microbes Infect. 2021;10(1):638‐650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shi J, Wen Z, Zhong G, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science. 2020;368(6494):1016‐1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiang X, Li H, Liu Y, et al. The effects of ATIR blocker on the severity of COVID‐19 in hypertensive inpatients and virulence of SARS‐CoV‐2 in hypertensive hACE2 transgenic mice. J Cardiovasc Transl Res. 2022;15(1):38‐48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ma Y, Lu D, Bao L, et al. SARS‐CoV‐2 infection aggravates chronic comorbidities of cardiovascular diseases and diabetes in mice. Animal Model Exp Med. 2021;4(1):2‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tong L, Xiao X, Li M, et al. A glucose‐like metabolite deficient in diabetes inhibits cellular entry of SARS‐CoV‐2. Nat Metab. 2022;4(5):547‐558. [DOI] [PubMed] [Google Scholar]
- 67. Port JR, Adney DR, Schwarz B, et al. Western diet increases COVID‐19 disease severity in the Syrian hamster. bioRxiv. 2021. [Google Scholar]
- 68. Ramasamy S, Kolloli A, Kumar R, et al. Comprehensive analysis of disease pathology in immunocompetent and immunocompromised hosts following pulmonary SARS‐CoV‐2 infection. Biomedicine. 2022;10(6):1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sefik E, Israelow B, Mirza H, et al. A humanized mouse model of chronic COVID‐19. Nat Biotechnol. 2022;40(6):906‐920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Shuai H, Chan JF, Hu B, et al. Attenuated replication and pathogenicity of SARS‐CoV‐2 B.1.1.529 omicron. Nature. 2022;603(7902):693‐699. [DOI] [PubMed] [Google Scholar]
- 71. Halfmann PJ, Iida S, Iwatsuki‐Horimoto K, et al. SARS‐CoV‐2 omicron virus causes attenuated disease in mice and hamsters. Nature. 2022;603(7902):687‐692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yuan S, Ye ZW, Liang R, et al. Pathogenicity, transmissibility, and fitness of SARS‐CoV‐2 omicron in Syrian hamsters. Science. 2022;377(6604):428‐433. [DOI] [PubMed] [Google Scholar]
- 73. Rudd JM, Tamil Selvan M, Cowan S, et al. Clinical and histopathologic features of a feline SARS‐CoV‐2 infection model are analogous to acute COVID‐19 in humans. Viruses. 2021;13(8):1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Silverstein WK, Stroud L, Cleghorn GE, et al. First imported case of 2019 novel coronavirus in Canada, presenting as mild pneumonia. Lancet. 2020;395(10225):734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Arentz M, Yim E, Klaff L, et al. Characteristics and outcomes of 21 critically ill patients with COVID‐19 in Washington state. Jama. 2020;323(16):1612‐1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Yu P, Qi F, Xu Y, et al. Age‐related rhesus macaque models of COVID‐19. Anim Models Exp Med. 2020;3(1):93‐97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Aid M, Busman‐Sahay K, Vidal SJ, et al. Vascular disease and thrombosis in SARS‐CoV‐2‐infected rhesus macaques. Cell. 2020;183(5):1354‐1366.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Koo BS, Oh H, Kim G, et al. Transient lymphopenia and interstitial pneumonia with Endotheliitis in SARS‐CoV‐2‐infected macaques. J Infect Dis. 2020;222(10):1596‐1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Shan C, Yao YF, Yang XL, et al. Infection with novel coronavirus (SARS‐CoV‐2) causes pneumonia in rhesus macaques. Cell Res. 2020;30(8):670‐677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Deng W, Bao L, Liu J, et al. Primary exposure to SARS‐CoV‐2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818‐823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Munster VJ, Feldmann F, Williamson BN, et al. Respiratory disease in rhesus macaques inoculated with SARS‐CoV‐2. Nature. 2020;585(7824):268‐272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zheng H, Li H, Guo L, et al. Virulence and pathogenesis of SARS‐CoV‐2 infection in rhesus macaques: a nonhuman primate model of COVID‐19 progression. PLoS Pathog. 2020;16(11):e1008949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fahlberg MD, Blair RV, Doyle‐Meyers LA, et al. Cellular events of acute, resolving or progressive COVID‐19 in SARS‐CoV‐2 infected non‐human primates. Nat Commun. 2020;11(1):6078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lu S, Zhao Y, Yu W, et al. Comparison of nonhuman primates identified the suitable model for COVID‐19. Signal Transduct Target Ther. 2020;5(1):157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Singh DK, Singh B, Ganatra SR, et al. Responses to acute infection with SARS‐CoV‐2 in the lungs of rhesus macaques, baboons and marmosets. Nat Microbiol. 2021;6(1):73‐86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Lu YR, Wang LN, Jin X, et al. A preliminary study on the feasibility of gene expression profile of rhesus monkey detected with human microarray. Transplant Proc. 2008;40(2):598‐602. [DOI] [PubMed] [Google Scholar]
- 88. Mercado NB, Zahn R, Wegmann F, et al. Single‐shot Ad26 vaccine protects against SARS‐CoV‐2 in rhesus macaques. Nature. 2020;586(7830):583‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Corbett KS, Flynn B, Foulds KE, et al. Evaluation of the mRNA‐1273 vaccine against SARS‐CoV‐2 in nonhuman primates. N Engl J Med. 2020;383(16):1544‐1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Vogel AB, Kanevsky I, Che Y, et al. BNT162b vaccines protect rhesus macaques from SARS‐CoV‐2. Nature. 2021;592(7853):283‐289. [DOI] [PubMed] [Google Scholar]
- 91. Gao Q, Bao L, Mao H, et al. Development of an inactivated vaccine candidate for SARS‐CoV‐2. Science. 2020;369(6499):77‐81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Deng W, Lv Q, Li F, et al. Sequential immunizations confer cross‐protection against variants of SARS‐CoV‐2, including omicron in rhesus macaques. Signal Transduct Target Ther. 2022;7(1):124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bao L, Gao H, Deng W, et al. Transmission of severe acute respiratory syndrome coronavirus 2 via close contact and respiratory droplets among human angiotensin‐converting enzyme 2 mice. J Infect Dis. 2020;222(4):551‐555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sia SF, Yan LM, Chin AWH, et al. Pathogenesis and transmission of SARS‐CoV‐2 in golden hamsters. Nature. 2020;583(7818):834‐838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Halfmann PJ, Hatta M, Chiba S, et al. Transmission of SARS‐CoV‐2 in domestic cats. N Engl J Med. 2020;383(6):592‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Deng W, Bao L, Gao H, et al. Ocular conjunctival inoculation of SARS‐CoV‐2 can cause mild COVID‐19 in rhesus macaques. Nat Commun. 2020;11(1):4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Johnston SC, Ricks KM, Jay A, et al. Development of a coronavirus disease 2019 nonhuman primate model using airborne exposure. PLOS One. 2021;16(2):e0246366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182(4):812‐827.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhou B, Thao TTN, Hoffmann D, et al. SARS‐CoV‐2 spike D614G change enhances replication and transmission. Nature. 2021;592(7852):122‐127. [DOI] [PubMed] [Google Scholar]
- 100. Chandrashekar A, Liu J, Martinot AJ, et al. SARS‐CoV‐2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812‐817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cao Y, Su B, Guo X, et al. Potent neutralizing antibodies against SARS‐CoV‐2 identified by high‐throughput single‐cell sequencing of convalescent Patients' B cells. Cell. 2020;182(1):73‐84.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Du S, Cao Y, Zhu Q, et al. Structurally resolved SARS‐CoV‐2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy. Cell. 2020;183(4):1013‐1023.e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Deng W, Xu Y, Kong Q, et al. Therapeutic efficacy of Pudilan Xiaoyan Oral liquid (PDL) for COVID‐19 in vitro and in vivo. Signal Transduct Target Ther. 2020;5(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1


