Abstract
Background
Chronic Helicobacter pylori infection may induce gastric intestinal metaplasia (IM). We compared anti-H. pylori antibody profiles between IM cases and non-atrophic gastritis (NAG) controls.
Methods
We evaluated humoral responses to 1528 H. pylori proteins among a discovery set of 50 IM and 50 NAG using H. pylori protein arrays. Antibodies with ≥ 20% sensitivity at 90% specificity for either group were selected and further validated in an independent set of 100 IM and 100 NAG using odds ratios (OR). A validated multi-signature was evaluated using the area under the receiver operating characteristics curve (AUC) and net reclassification improvement (NRI).
Results
Sixty-two immunoglobulin (Ig) G and 11 IgA antibodies were detected in > 10%. Among them, 22 IgG and 6 IgA antibodies were different between IM and NAG in the discovery set. Validated antibodies included 11 IgG (anti-HP1177/Omp27/HopQ [OR = 8.1, p < 0.001], anti-HP0547/CagA [4.6, p < 0.001], anti-HP0596/Tipα [4.0, p = 0.002], anti-HP0103/TlpB [3.8, p = 0.001], anti-HP1125/PalA/Omp18 [3.1, p = 0.001], anti-HP0153/RecA [0.48, p = 0.03], anti-HP0385 [0.41, p = 0.006], anti-HP0243/TlpB [0.39, p = 0.016], anti-HP0371/FabE [0.37, p = 0.017], anti-HP0900/HypB/AccB [0.35, p = 0.048], and anti-HP0709 [0.30, p = 0.003]), and 2 IgA (anti-HP1125/PalA/Omp18 [2.7, p = 0.03] and anti-HP0596/Tipα [2.5, p = 0.027]). A model including all 11 IgG antibodies (AUC = 0.81) had better discriminated IM and NAG compared with an anti-CagA only (AUC = 0.77) model (NRI = 0.44; p = 0.001).
Conclusions
Our study represents the most comprehensive assessment of anti-H. pylori antibody profiles in IM. The target antigens for these novel antibodies may act together with CagA in the progression to IM. Along with other biomarkers, specific H. pylori antibodies may identify IM patients, who would benefit from surveillance.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00535-022-01933-0.
Keywords: Intestinal metaplasia, H. pylori, NAPPA, Serology, Premalignant lesions
Introduction
Helicobacter pylori-driven gastric carcinogenesis is a multistep process with well-defined histological stages, variably progressing from chronic non-atrophic gastritis (NAG) to atrophic gastritis with or without gastric intestinal metaplasia (IM), dysplasia, and cancer [1]. At the stage of atrophy, the H. pylori infection burden is reduced due to chronic inflammation [2]. A decrease in bacterial stimuli may explain a reduced humoral response against H. pylori in IM and gastric cancer.
Gastric IM is a heterogeneous precancerous lesion with variable prevalence across populations. The estimated overall annual risk of gastric cancer in patients with IM in endoscopy-based studies is 0.34% [3]. International guidelines recommend endoscopic surveillance of patients with advanced IM as defined by histological features and anatomical location [4]. There is a need for non-invasive tests to identify these high-risk individuals, especially in resource-limited settings.
We recently developed and validated an H. pylori array to assess anti-H. pylori humoral response profiles that scan ~ 90% of the complete bacterial proteome [5]. In this study, we used H. pylori arrays on samples from IM patients and NAG controls to evaluate the potential utility of specific anti-H. pylori antibodies as a non-endoscopic diagnosis. Our study will also advance our understanding of the etiological role of H. pylori in the progression to IM. To further explain the dynamics of H. pylori antibodies, we also compared immunoproteomic profiles of current and past H. pylori infections.
Materials and methods
Study population
Adults included in this study are part of an endoscopic campaign conducted between April and May 2017 in a high gastric cancer risk area of Chile. Individuals with gastrointestinal symptoms were referred for an upper endoscopy at the Intercultural Hospital of Nueva Imperial. From available patients with histologically confirmed IM and NAG, we randomly selected discovery (50 IM and 50 NAG) and validation (100 IM and 100 NAG) sets (Table 1). A global IM diagnosis was based on five biopsies collected using the Sydney protocol. Approximately 50% of the IM cases in both sets (52% discovery and 47% validation) represent advanced stages (i.e., extension to corpus). IM cases and NAG controls were comparable in H. pylori seropositivity as determined by whole-cell enzyme-linked immunosorbent assay (wcELISA; Biohit), H. pylori positivity by histology (modified Diff-Quick stain; Sydney biopsies; the H. pylori organisms that were dark blue are readily detected because of their spiral or curved rods), and H. pylori positivity by urease (Pronto Dry®; antral biopsy with a final reading at 60 min). CagA seropositivity (CagA-ELISA; Genesis Diagnostics) was higher in IM cases as compared to NAG controls. A past H. pylori infection was defined as a positive result for either wcELISA or CagA-ELISA with a negative result for both histology and urease. A current H. pylori infection was defined as a positive result for either histology or urease, regardless of results on wcELISA and CagA-ELISA. Overall positivity was defined as positivity for either past or current infection.
Table 1.
Selected characteristics of gastric intestinal metaplasia (IM) patients and non-atrophic gastritis (NAG) controls
| Discovery set | Validation set | |||
|---|---|---|---|---|
| IM cases (n = 50) | NAG controls (n = 50) | IM cases (n = 100) | NAG controls (n = 100) | |
| Age in years, median (range) | 60 (35–73) | 55 (40–76) | 59 (39–79) | 54 (39–85) |
| Male sex, n (%) | 26 (52) | 14 (28) | 36 (36) | 23 (23) |
| H. pylori seropositivitya, n (%) | 27 (54) | 28 (56) | 61 (61) | 62 (62) |
| CagA serology, n (%) | 37 (74) | 19 (38) | 74 (74) | 41 (41) |
| H. pylori histologyb, n (%) | 19 (38) | 21 (42) | 47 (47) | 49 (49) |
| H. pylori urease test, n (%) | 21 (42) | 20 (40) | 49 (49) | 48 (48) |
| Overall H. pylori infectionc, n (%) | 43 (86) | 37 (74) | 90 (90) | 86 (86) |
| Past H. pylori infectiond, n (%) | 19 (38) | 15 (30) | 34 (34) | 32 (32) |
| Current H. pylori infectione, n (%) | 24 (48) | 22 (44) | 56 (56) | 54 (54) |
aELISA tested with H. pylori whole-cell lysate (wcELISA)
bModified Diff-Quick stain for H. pylori
cPositivity for either past or current H. pylori infection
dwcELISA( +) or CagA-ELISA( +) with histology(−) and urease(−) among individuals with overall H. pylori infection
eHistology( +) or urease( +), regardless of results on wcELISA and CagA-ELISA among individuals with overall H. pylori infection
The original endoscopy campaign was approved by the Institutional Review Boards (IRB) of the Chilean Pontificia Universidad Catolica (No. 14–280) and the U.S. National Cancer Institute (No. 17CN094). Informed consent was obtained from all participants. This study was exempted by the U.S. National Institutes of Health Office of Human Subjects Research Protection from IRB evaluation.
Selection of H. pylori genes
A total of 1528 full-length H. pylori genes were obtained in Gateway Entry clones from the U.S. Biodefense and Emerging Infections Research Resources Repository, including 1454 genes from the strain 26695 (covering 91% of the full proteome) and 74 genes from the strain J99 (12 genes with homology > 90% between these two strains). CagA was PCR cloned from full-length (P12 strain) and a fragment comprising residues 1 to 884 [26,695 strain] [6]. All genes were transferred into the pANT7-cGST expression vector using recombinational cloning. VacA gene was unavailable in our clone library and hence anti-VacA was not assessed in our study.
Fabrication, expression and probing of H. pylori-NAPPA arrays
We developed a 1528-protein H. pylori-NAPPA array to scan all bacterial antigens. Based on the results of the 1528-protein array in the discovery set, we created a smaller 245-protein H. pylori-NAPPA that includes 62 immunogenic antigens for which antibodies were present in > 10% of IM cases or NAG controls and 183 proteins for which antibodies were absent in either group (0% seroprevalence) to be used for the array data normalization. Both arrays were fabricated as previously reported [5, 7]. Briefly, plasmid DNA for H. pylori expression clones into silicon nano-well substrates using a piezoelectric dispensing system. At the time of usage, arrays were incubated with cell-free protein expression lysates at 30 °C for 2 h and 15 °C for 30 min for in vivo protein expression and in situ capture. Isotype-specific (Immunoglobulin, IgG, and IgA) antibody profiling was performed by incubating the arrays with 1:100 dilution of serum followed by detection with Alexa 647 labeled goat anti-human IgG and Cy3 labeled goat anti-human IgA. NAPPA arrays were scanned on a Tecan PowerScanner and raw fluorescence intensities were extracted using ArrayPro Analyzer Software. The reproducibility of the protein display on arrays was assessed using an anti-GST antibody on duplicates, for which inter-array correlation coefficients were 0.95 for discovery and 0.96 for validation arrays. More than 99.7% (discovery set) and 100% (validation set) of H. pylori proteins were well expressed with raw fluorescence intensities higher than the median intensity of all negative control spots plus 3 times standard deviations. A pooled serum combining all samples in the discovery set was used as an internal positive control to assess the antibody profiling reproducibility and resulted in inter-array correlations of 0.94 for discovery and 0.96 for validation arrays, respectively (Supplementary Fig. 1B). Blinded testing was performed on all study samples. Antibody responses on both H. pylori-NAPPA arrays were normalized as Median Normalized Intensity (MNI) via dividing the raw values by the median signal intensity of all proteins within each array. Seropositivity was defined as MNI ≥ 2.0.
Anti-H. pylori antibodies in the discovery and validation sample sets
The discovery set (50 IM and 50 NAG) was profiled using both 1528-protein and 245-protein H. pylori-NAPPAs. There were high correlations (median R value, 0.89) of the MNI values between the 1528-and 245-protein H. pylori-NAPPAs for the overlapping 245 proteins tested in the discovery set. The validation set (100 IM and 100 NAG) was probed only on the 245-protein H. pylori-NAPPA. Relative seroprevalence was calculated as the percentage of IM cases or NAG controls with MNI exceeding the 90th percentile of the other group. Antibodies with relative seroprevalence ≥ 20% in either IM cases or NAG controls (in the discovery set) were selected as candidate antibodies for validation of their performance in distinguishing IM cases from NAG controls in the validation set.
Statistical analyses
The differences in the seroprevalence in IM cases and NAG controls, or between positive and negative results from wcELISA, CagA-ELISA, histology, and urease tests by case-control status were assessed by the Wilcoxon rank-sum test. Kappa coefficients measured agreement between available wcELISA and overall positivity results for the top-five most immunoreactive antibodies, as well as CagA-ELISA and CagA-NAPPA. For the discovery set, we computed the sensitivity and specificity for each antibody comparing IM and NAG, and selected antibodies with ≥ 20% sensitivity at 90% specificity for either direction for further evaluation using the validation set. When assessing the performance of each selected antibody in the validation set, we computed sensitivity and specificity using the cutoff value from the discovery set and calculated the unadjusted odds ratio (OR) and p value by the Chi-square tests. Antibodies with OR < 0.5 or > 2.0 with p value < 0.05 were considered as validated antibodies.
The area under the receiver operating characteristic (ROC) curve (AUC) values were calculated for each validated antibody using logistic regression. Correlations among the validated antibodies were assessed by Pearson correlation tests. Net reclassification improvement (NRI) was used to evaluate the reclassification of the validated model, compared to an anti-CagA-only model. In addition, we also ran a case-case comparison of corpus extension and antral restricted IM in both discovery and validation sets. The comparison between past vs. current infection among IM cases and NAG controls combined was conducted following a similar analytical approach. All tests were two-sided and p values < 0.05 were considered statistically significant. Analyses were conducted with R version 3.6 (R Core Team, R Foundation for Statistical Computing, Vienna, Austria), Stata version 15 (Stata Corp, College Station, TX, USA), and GraphPad Prism 8.0.2 (GraphPad Software, Inc., CA, USA).
Results
Anti-H. pylori antibody response and prevalence in IM cases and NAG controls in the discovery sample set
Of all antibodies profiled on the 1528-protein H. pylori-NAPPA, 62 IgG and 11 IgA antibodies showed > 10% seropositivity in IM and/or NAG groups of the discovery set (Supplementary Table 1, Supplementary Fig. 2). The proteins targeted by the 11 IgA antibodies also elicited strong reactive IgG antibodies. In IM cases and NAG controls combined, anti-HP1341/TonB/TonB2 (seropositivity = 98%), anti-HP1125/PalA/Omp18 (83%), anti-HP0596/Tipα (78%), anti-HP1199/RpI2/Rpl7/RpIL (72%), anti-HP0010/GroEL/ HSPb/Hsp60/MopA (70%), and anti-HP0547/CagA (70%) showed the highest overall seropositivity. Combining the results of wcELISA, cagA-ELISA, histology, and urease, the overall positivity of H. pylori was 80% in the discovery set. Regardless of the case-control status, H. pylori-positive individuals as defined by any of the four conventional tests (i.e., wcELISA, cagA-ELISA, histology, or urease) had a higher total number of IgG anti-H. pylori antibodies relative to H. pylori-negative individuals, while the number of IgA antibodies was comparable between the two groups (Supplementary Fig. 3).
Among our top-five most reactive antibodies, seropositivity to anti-HP0010/GroEL exhibited the highest degree of agreement with wcELISA (Kappa coefficient = 0.4), while there was low to moderate agreement between overall positivity and the other four antibodies (Kappa coefficient ranged from 0.05 to 0.3; Supplementary Table 2). The degree of agreement between cagA-ELISA and seropositivity to HP0547/CagA on H. pylori-NAPPA was moderate (coefficient = 0.6).
Discovery and validation of discriminatory anti-H. pylori antibodies between IM cases and NAG controls
Among the 73 antibodies (62 IgG and 11 IgA) identified on the 1528-protein H. pylori-NAPPA and re-tested on the 254-protein NAPPA, 12 IgG and 6 IgA antibodies showed higher relative seroprevalence in IM cases and 10 IgG antibodies showed higher relative seroprevalence in NAG controls in the validation set (Table 2, Supplementary Fig. 2). Five IgG antibodies (anti-HP0547/CagA, anti-HP1125/PalA/Omp18, anti-HP0596/Tipα, anti-HP1177 /HopQ/Omp27, and anti-HP0103/TipB) and two IgA antibodies (HP0596/Tipα and anti-HP1125/PalA/Omp18) were validated with higher sensitivity (i.e., significantly higher responses in IM cases) (Table 2). Six IgG antibodies (anti-HP0709/HpaA, anti-HP0900/HypB, anti-HP0371/FabE/AccB, anti-HP0243/NapA, anti-HP0153/RecA, and anti-HP0385) were validated with higher specificity (i.e., lower responses in IM) (Table 2). The five IgG antibodies with higher reactivity in IM cases (than in NAG controls) had AUC values ranging from 0.65 to 0.77, while the five IgG antibodies’ lower reactivity in IM had AUC values ranging from 0.50 to 0.65. The validated two IgA antibodies higher in IM had AUC values of 0.57 and 0.60.
Table 2.
Candidate and validated anti-H. pylori antibodies differential between intestinal metaplasia (IM) cases and non-atrophic gastritis (NAG) controls in the 245-gene NAPPA
| Discovery sample set | Validation sample set | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ig isotype | Protein | Common names | Full name | Profile | IM cases (n = 50)a, % | NAG controls (n = 50)b, % | IM cases (n = 100)a, % | NAG controls (n = 100)b, % | OR | p value | AUC |
| IgG | HP1177 | Omp27 | Outer membrane protein | High in IM | 22 | 2 | 20 | 3 | 8.08 | < 0.001 | 0.73 |
| HP0547 | CagA | Cytotoxicity-associated immunodominant antigen | 48 | 10 | 45 | 15 | 4.64 | < 0.001 | 0.77 | ||
| HP0596 | Tipα | Tumor necrosis factor alpha-inducing protein | 30 | 10 | 23 | 7 | 3.97 | 0.002 | 0.66 | ||
| HP0103 | TlpB | Methyl-accepting chemotaxis protein | 22 | 10 | 25 | 8 | 3.83 | 0.001 | 0.68 | ||
| HP1125 | PalA/OMP18 | Peptidoglycan associated lipoprotein | 32 | 10 | 37 | 16 | 3.08 | 0.001 | 0.65 | ||
| HP0908 | FlgE | Flagellar hook protein | 20 | 10 | 17 | 8 | 2.36 | 0.054 | 0.57 | ||
| HP0701 | GyrA | DNA gyrase subunit A | 20 | 10 | 16 | 10 | 1.71 | 0.207 | 0.51 | ||
| HP1172 | GlnH | Glutamine ABC transporter substrate-binding protein | 26 | 10 | 26 | 18 | 1.60 | 0.172 | 0.52 | ||
| HP1453 | HomD | Membrane protein | 20 | 10 | 16 | 12 | 1.40 | 0.415 | 0.55 | ||
| HP0373 | Uncharacterized protein | 38 | 10 | 22 | 17 | 1.38 | 0.372 | 0.53 | |||
| HP1564 | PlpA | ABC transporter substrate-binding protein | 26 | 10 | 13 | 10 | 1.34 | 0.506 | 0.56 | ||
| HP0606 | MtrC | Membrane fusion protein | 28 | 10 | 22 | 19 | 1.20 | 0.599 | 0.56 | ||
| HP0709 | Uncharacterized protein | Low in IM | 6 | 20 | 9 | 25 | 0.30 | 0.003 | 0.61 | ||
| HP0900 | HypB | Hydrogenase/urease maturation factor | 4 | 20 | 5 | 13 | 0.35 | 0.048 | 0.50 | ||
| HP0371 | AccB, FabE | Biotin carboxyl carrier protein | 10 | 20 | 9 | 21 | 0.37 | 0.017 | 0.65 | ||
| HP0243 | NapA | DNA protection during starvation protein | 4 | 20 | 11 | 24 | 0.39 | 0.016 | 0.63 | ||
| HP0385 | Hypothetical protein | 10 | 20 | 18 | 35 | 0.41 | 0.006 | 0.57 | |||
| HP0153 | RecA | Recombinase | 10 | 22 | 17 | 30 | 0.48 | 0.030 | 0.55 | ||
| HP1100 | Edd | Phosphogluconate dehydratase | 10 | 22 | 5 | 11 | 0.43 | 0.118 | 0.55 | ||
| HP1118 | Ggt | Gamma-glutamyltranspeptidase | 10 | 34 | 25 | 33 | 0.68 | 0.213 | 0.58 | ||
| HP1110 | PorA | Pyruvate flavodoxin oxidoreductase subunit alpha | 10 | 30 | 24 | 16 | 1.66 | 0.157 | 0.54 | ||
| HP0322 | ChePep | Chemotaxis regulatory protein | 10 | 24 | 19 | 11 | 1.90 | 0.113 | 0.42 | ||
| IgA | HP0596 | Tipα | Tumor necrosis factor alpha-inducing protein | High in IM | 26 | 10 | 20 | 9 | 2.53 | 0.027 | 0.57 |
| HP1125 | PalA/OMP18 | Peptidoglycan associated lipoprotein | 20 | 10 | 17 | 7 | 2.72 | 0.030 | 0.60 | ||
| HP0923 | HopK, Omp22 | Membrane protein | 30 | 6 | 21 | 12 | 1.95 | 0.086 | 0.58 | ||
| HP0547 | CagA | Cytotoxicity-associated immunodominant antigen | 24 | 8 | 14 | 23 | 0.54 | 0.101 | 0.52 | ||
| HP0477 | HopJ, Omp12 | Membrane protein | 28 | 6 | 20 | 14 | 1.54 | 0.259 | 0.52 | ||
| HP1453 | HomD | Membrane protein | 20 | 10 | 15 | 16 | 0.93 | 0.845 | 0.50 | ||
Bolded numbers are for proteins that were validated, i.e., significant in the validation set
AUC area under the curve, OR odds ratio
aCutoff generated from the 90th percentile of NAG controls
bCutoff generated from the 90th percentile of IM cases
Bold numbers are for proteins that were validated, i.e., significant in the validation set
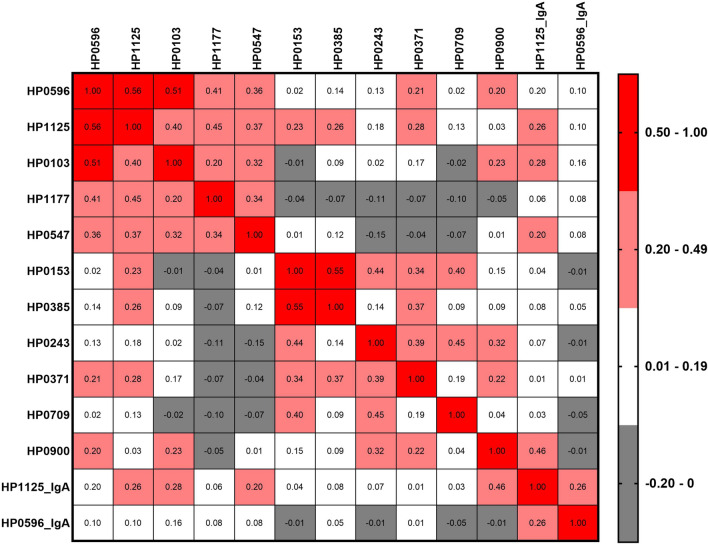
All pairwise correlations among the 13 IgG and IgA antibodies in the validation set are shown in Fig. 1. Among the five antibodies with higher seroprevalence in IM cases than in NAG controls, correlation coefficients ranged from 0.20 (anti-HP1177 vs. anti-HP0103) to 0.56 (anti-HP1125 vs. anti-HP0596). Amongst antibodies with lower seroprevalences in IM cases than in NAG controls, correlation coefficients ranged from 0.04 (anti-HP0709 vs. anti-HP0900) to 0.55 (anti-HP0153 vs. anti-HP0385). The correlations between IgG and IgA antibodies against the same antigens (anti-HP1125 and anti-HP0596) were weak (correlation coefficient < 0.3). AUC for an antibody panel with all 13 validated IgG and IgA antibodies was 0.82 (95% CI, 0.76–0.87) distinguishing IM from NAG. A model including the 11 validated IgG antibodies (AUC = 0.81; 95% CI, 0.75–0.87) had a better classification performance compared to an anti-CagA only (AUC = 0.77; 95% CI, 0.70–0.84) model (NRI = 0.44; p value = 0.001) (Supplementary Fig. 4).
Fig. 1.
Pairwise Pearson correlations (ranged from − 0.20 to 0.56) among the 11 validated anti-H. pylori IgG and 2 IgA antibodies differential between gastric intestinal metaplasia cases and non-atrophic gastritis controls in the validation sample set. All pairwise comparisons with correlations higher than 0.2 were statistically significant with p values < 0.01
Anti-H. pylori antibodies by anatomical extension of IM
H. pylori seropositivity by wcELISA was similar (59 vs. 58%) between the case-case analysis of corpus extension (n = 73) and antral (n = 77) restricted IM subsite. Out of total proteins profiled on the 254-protein H. pylori-NAPPA, two IgG antibodies (anti-HP0516/HslU and anti-HP0385) and one IgA antibody (anti-HP1453/HomD) showed significantly higher seropositivity in antral restricted as compared to corpus extension IM (p values < 0.05) (Supplementary Table 3).
Discovery and validation of discriminatory anti-H. pylori antibodies between current vs. past H. pylori infection
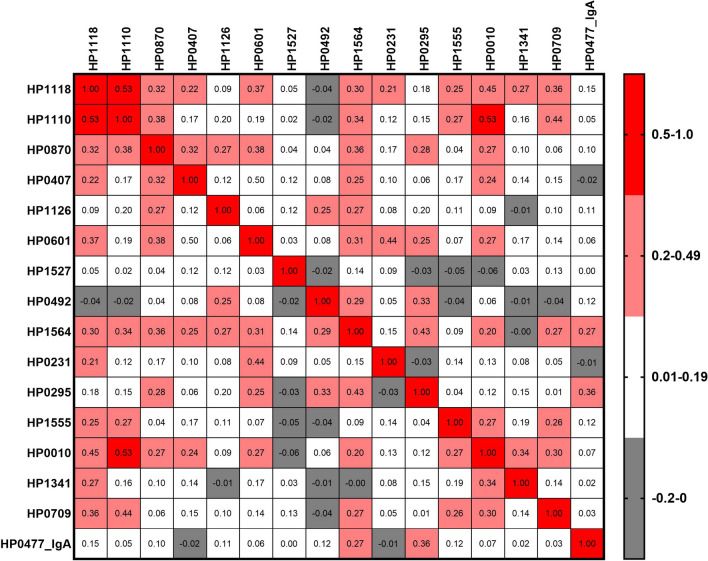
Prevalences of current and past infections were similar in both discovery and validation sets (Table 1). The agreement between histology (based on five biopsies) and urease (based on one antral biopsy) on H. pylori diagnosis was substantial with a Kappa coefficient of 0.75. Approximately 13% of individuals were classified as discordant for either test. Out of the 254 antibodies profiled, 15 IgG antibodies (anti-HP1118/Ggt, anti-HP1110/PorA, anti-HP0870/ FlgE, anti-HP0407/BisC, anti-HP0601/FlaA, anti-HP1126/TolB, anti-HP1527/ComH, anti-HP0492, anti-HP1564/ PlpA, anti-HP0231/dsbK/dsbG, anti-HP0295/Fla, anti-HP1555/Tfs, anti-HP0010/GroEL, anti-HP0709, and anti-HP1341/TonB/TonB2) and one IgA (anti-HP0477/HopJ) were strongly significantly associated with current H. pylori infection with ORs ranging from 2.19 to 7.46, regardless of case–control status (Table 3, Supplementary Fig. 5). All pairwise correlations among the 16 candidate IgG and IgA antibodies in the validation set are shown in Fig. 2. Correlation coefficients ranged from -0.06 (anti-HP0010 vs. anti-HP0492) to 0.53 (anti-HP1118 vs. anti-HP1110 and anti-HP0010 vs. anti-HP1110). AUC for a panel combining the validated 16 antibodies was 0.85 (95% CI, 0.79–0.90) distinguishing current from past H. pylori infection.
Table 3.
Candidate and validated anti-H. pylori antibodies differential between current and past H. pylori infection in the 245-gene NAPPA
| Discovery sample seta | Validation sample seta | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ig isotype | Protein | Common names | Full name | Current infection (n = 46)b, % | Past infection (n = 34)c, % | Current infection (n = 110)b, % | Past infection (n = 60)c, % | OR | p value | AUC |
| IgG | HP1118 | Ggt | Gamma-glutamyltranspeptidase | 39 | 12 | 43 | 9 | 7.46 | 0.000 | 0.72 |
| HP1110 | PorA | Pyruvate flavodoxin oxidoreductase subunit alpha | 30 | 12 | 28 | 6 | 6.08 | 0.000 | 0.67 | |
| HP0870 | FlgE | Flagellar hook protein | 37 | 12 | 35 | 9 | 5.49 | 0.000 | 0.70 | |
| HP0407 | BisC | Biotin sulfoxide reductase | 26 | 12 | 25 | 6 | 5.04 | 0.002 | 0.64 | |
| HP0601 | FlaA | Flagellin A | 48 | 12 | 51 | 18 | 4.67 | 0.000 | 0.71 | |
| HP1126 | TolB | Tol-Pal system protein | 22 | 9 | 13 | 3 | 4.67 | 0.030 | 0.69 | |
| HP1527 | ComH | Hypothetical protein | 26 | 12 | 31 | 9 | 4.47 | 0.001 | 0.71 | |
| HP0492 | Neuraminyllactose-binding hemagglutinin | 33 | 12 | 22 | 6 | 4.33 | 0.006 | 0.64 | ||
| HP1564 | PlpA | ABC transporter substrate-binding protein | 22 | 12 | 16 | 5 | 4.11 | 0.019 | 0.76 | |
| HP0231 | dsbK, dsbG | Protein-disulfide isomerase | 39 | 9 | 32 | 11 | 3.93 | 0.001 | 0.69 | |
| HP0295 | Fla | Flagellin B homolog | 35 | 12 | 26 | 12 | 2.60 | 0.025 | 0.65 | |
| HP1555 | Tfs | Elongation factor Ts | 30 | 12 | 26 | 12 | 2.60 | 0.025 | 0.58 | |
| HP0010 | GroEL, HSPb, Hsp60, MopA | Molecular chaperone | 33 | 12 | 23 | 11 | 2.48 | 0.044 | 0.61 | |
| HP0709 | Uncharacterized protein | 22 | 6 | 23 | 11 | 2.48 | 0.044 | 0.58 | ||
| HP1341 | TonB2, TonB | Energy transducer | 28 | 12 | 33 | 18 | 2.19 | 0.036 | 0.58 | |
| HP0304 | Alginate_lyase domain-containing protein | 37 | 12 | 22 | 11 | 2.35 | 0.059 | 0.61 | ||
| HP0485 | Catalase-related peroxidase | 22 | 6 | 8 | 2 | 5.79 | 0.064 | 0.60 | ||
| HP0953 | Uncharacterized protein | 30 | 12 | 17 | 8 | 2.55 | 0.070 | 0.62 | ||
| HP0373 | Uncharacterized protein | 33 | 12 | 21 | 11 | 2.23 | 0.078 | 0.61 | ||
| HP1453 | HomD | Membrane protein | 22 | 12 | 16 | 8 | 2.39 | 0.094 | 0.60 | |
| HP0908 | FlgE | Flagellar hook protein | 24 | 12 | 17 | 9 | 2.09 | 0.132 | 0.68 | |
| HP0596 | Tipα | Tumor necrosis factor alpha-inducing protein | 30 | 12 | 19 | 11 | 1.99 | 0.136 | 0.63 | |
| HP0512 | GlnA | Glutamine synthetase | 24 | 9 | 25 | 15 | 1.82 | 0.139 | 0.57 | |
| HP0371 | AccB, FabE | Biotin carboxyl carrier protein | 39 | 12 | 38 | 27 | 1.65 | 0.139 | 0.57 | |
| HP0547 | CagA | Cytotoxicity-associated immunodominant antigen | 22 | 12 | 20 | 12 | 1.81 | 0.178 | 0.50 | |
| HP0322 | ChePep | Chemotaxis regulatory protein | 33 | 6 | 20 | 12 | 1.81 | 0.178 | 0.52 | |
| HP0175 | PpiC | Peptidylprolyl isomerase | 22 | 9 | 17 | 11 | 1.76 | 0.228 | 0.56 | |
| HP1100 | Edd | Phosphogluconate dehydratase | 24 | 12 | 9 | 5 | 2.10 | 0.264 | 0.51 | |
| HP0875 | KatA | Catalase | 22 | 12 | 25 | 18 | 1.46 | 0.325 | 0.53 | |
| HP0606 | MtrC | Membrane fusion protein | 22 | 12 | 16 | 15 | 1.10 | 0.831 | 0.56 | |
| IgA | HP0477 | HopJ, Omp12 | Membrane protein | 24 | 12 | 21 | 6 | 4.10 | 0.008 | 0.62 |
| HP1453 | HomD | Membrane protein | 39 | 6 | 27 | 17 | 1.88 | 0.107 | 0.68 | |
| HP0547 | CagA | Cytotoxicity-associated immunodominant antigen | 22 | 12 | 19 | 18 | 1.06 | 0.881 | 0.48 | |
Bolded numbers are for proteins that were validated, i.e., significant in the validation set
AUC area under the curve, OR odds ratio
aIntestinal metaplasia cases and non-atrophic gastritis controls combined
bCutoff generated from the 90th percentile of past infection
cCutoff generated from the 90th percentile of current infection
Bold numbers are for proteins that were validated, i.e., significant in the validation set
Fig. 2.
Pairwise Pearson correlations (ranged from − 0.06 to 0.53) among the 15 validated anti-H. pylori IgG and 1 IgA antibodies differential between current and past H. pylori infection in the validation sample set. All pairwise comparisons with correlations higher than 0.2 were statistically significant with p values < 0.01
Discussion
Our two-stage study represents the most comprehensive proteome-level analysis to identify anti-H. pylori antibodies to distinguish IM from NAG. We found moderate-to-strong associations with IM for 11 IgG and 2 IgA antibodies against several outer membrane proteins (OMPs) and proteins essential for bacterial survival. A combined model of the 11 IgG antibodies moderately discriminated IM from NAG. Another unique feature of our study was the comparison between current and past H. pylori infections. We identified 15 IgG and 1 IgA antibodies associated with current infection, including responses to several proteins related to colonization and persistence.
H. pylori coevolved with humans and developed efficient mechanisms to avoid the surveillance of the host immune system. As a consequence, H. pylori infection typically does not elicit a strong humoral response [8], which is consistent with the limited number of immunogenic proteins observed in our study (4%; 62/1528). Also, expanding our previous findings in gastric cancer using the same NAPPA array [5], we confirmed that IgG antibodies were more informative in determining gastric pathology compared with IgA antibodies.
Anti-CagA antibodies are sustained for a long time even after successful H. pylori eradication [9, 10]. A potential mechanism for the persistent antigenic stimuli may be due to the remnants of CagA-positive strains in the deepest gastric glands surviving long years of infection. Our finding that a higher CagA seropositivity was moderately discriminative between IM cases and NAG controls aligns with two previous cross-sectional analyses [11, 12] within the Linxian Nutrition Intervention Trial cohort. Pan et al. compared antibodies against HP0547/CagA, HP0887/VacA, GroEL, UreA, HcpC, and HP1118/Ggt in patients with precancerous lesions and those with superficial gastritis, and reported a positive IM association with anti-HP0547/CagA and an inverse IM association with anti-HP1118/gGT [11]. Using a different platform, a 15-plex Luminex fluorescent bead-based immunoassay, Epplein et al. reported positive associations of IM with antibodies against HP1564/Omp, HP0547/CagA, HP0887/VacA, HcpC, HP0305, GroEL, NapA, HyuA, Cad, and HpaA [12]. These studies confirmed the important role of CagA in gastric carcinogenesis.
H. pylori OMPs serve as key factors for nutrient scavenging and evasion of host defense mechanisms [13]. In our study, two antibodies to OMPs were positively associated with IM. HP1177/Omp27/HopQ binds to carcinoembryonic antigen-related cell-adhesion molecules with high specificity [14]. Type I HopQ may be associated with carcinogenesis by transferring CagA or triggering type IV secretion system to initiate and maintain chronic inflammation mainly by activating toll-like receptor 9 via the canonical NF-KB pathway [15, 16]. Taxauer et al. showed that the HopQ-CEACAM interaction is also important for the activation of the non-canonical NF-KB pathway [17]. HP1125/PalA/Omp18 is expressed by all known H. pylori strains and is involved in persistent colonization [18]. In particular, Omp18 may alter interferon-γ levels and optimize virulence phenotype (i.e., CagA), survive oxidative stress, and anti-phagocytosis. pH taxis is another important mechanism for the long-term persistence of H. pylori [19]. HP0103/TlpB is required in chemorepulsive responses to acid, as well as the quorum-sensing molecule autoinducer-2 [20]. Interestingly, binding of polymeric G-repeats regulator to the upstream of tlpB is sufficient to regulate TlpB both at the transcript and protein level [21]. HP0596/Tipα is another tumor promoter secreted as dimers and enters the gastric cells, a process mediated through NF-KB activation [22], that also has DNA-binding activity [23]. Similar to CagA, antibodies to HP0596/Tipα may persist after bacterial eradication [24]. Notably, our analyses showed anti-HP0596/Tipα to be moderately positively correlated with other antibodies (HP1125/PalA/Omp18 and HP0103/TlpB). Although our best discriminatory model suggests independent effects for anti-HP1177/Omp27/HopQ, HP0547/CagA, HP0103/TlpB, and HP1125/PalA/Omp18, given the complex networks of these antigens, their orchestrated synergistic contribution cannot be dismissed.
In the current study, we report several inverse antibody associations with IM. These findings may indicate loss of antigenic stimuli potentially related to atrophy progression, use of antibiotics, or immune protection against carcinogenesis. In our previous NAPPA gastric cancer study, we found several inverse associations including antibodies against HP0371/FabE/AccB, HP0243/NapA, and HP0153/RecA that were also identified in this IM study. HP0371/FabE/AccB is a biotin carboxyl carrier protein of acetyl-CoA carboxylase, which is involved in fatty acid biosynthesis [25]. NapA plays dual roles, recruiting host neutrophils/monocytes and stimulating the production of reactive oxygen intermediates, but on the other hand, sequestering iron and stress-resistant to oxidative stress [26]. NapA may also promote the formation of H2O2-induced biofilm and contribute to multidrug resistance [27]. RecA is a protein that is necessary for repairing DNA damage or facilitating recombination. In addition, mutants of recA are hypersensitive to DNA-damaging agents such as metronidazole, ultraviolet, or ionizing radiation [28].
We also found some novel antibodies that were inversely associated with IM. HypB is a GTPase with a key role in nickel homeostasis [29]. H. pylori hypB-deficient mutant exhibits significantly decreased urease activity as there is a physical interaction between urease and HP0900/HypB [30]. HpaA is a lipoprotein in the flagellar sheath and outer membrane and plays an essential role in the adhesion and colonization of the gastric mucosa [31]. Although poorly characterized, HP0709 encodes an enzyme that is involved in either DNA methylation or synthesis of some branched amino acids. HP0385 is a hypothetical protein with unknown functions. The findings for HpaA or RecA are inconsistent with a previous study showing a positive association of these two proteins with IM [12]. Considering the dual-faceted roles of these antigens, along with their various parts in coordination with other proteins and the external environment, further studies should investigate host-antigen interactions of these antigens. It is also possible that these proteins are important for colonization and establishment of successful infection but less relevant when the microenvironment changes due to hypochlorhydria and other tissue transformations related to atrophy.
Extensive IM confers a higher risk of gastric cancer. Our exploratory case-case analysis identified three antibody associations (anti-HP0516/HslU, anti-HP0385, and anti-HP1453/homD) with antral restricted IM (vs. corpus extension). The corresponding bacterial antigens have not been well studied. In particular, homD is a conserved gene and little is known about its encoding OMP (HP1453/homD) [32]. Further studies are warranted to address the potential differences in the humoral responses to H. pylori by IM anatomical subsite.
Although H. pylori infection can be detected by many methods, their sensitivities and specificities are variable. Taken together, the results of four different H. pylori tests assessing current and past infections, the high-risk study population seems to have had an almost universal bacterial exposure. There are no bacterial proteins that are recognized in all H. pylori-infected individuals. Differential humoral responses to specific H. pylori antigens could be related to variable protein expression influenced by the stomach microenvironment. Loh et al. [33], documented increased expression and translocation of CagA in response to high salt conditions in rodent models. Noto et al. [34], reported a similar increased expression of CagA in strains growing under iron-limiting conditions. Our NAPPA anti-GroEL fairly agreed with a commercial wcELISA. Non-invasive identification of H. pylori-specific antibodies accurately classifying individuals with current infection could assist with clinical management (e.g., H. pylori eradication). We identified 16 antibodies as markers of current H. pylori infection, including some novel candidates. Our results align with previous studies assessing a limited number of antibodies. Comparing results of a 13-plex Luminex fluorescent bead-based immunoassay and urea breath test, Butt et al. found that seropositivity to VacA, HP0010/GroEL, HcpC, and HP1564/ PlpA indicate current infection [35]. Using combined results of wcELISA, urease, and culture to define current infection, Shafaie et al. found that seropositivity to HP0547/CagA, HP0596/Tip-α, and HP0175/ PpiC was associated with current infection [36]. In currently infected individuals, flagellar antigens exhibit seropositivity as high as 90% [37]. Our NAPPA array included a total of 28 flagellar antigens and found positive associations with HP0870/FlgE, HP0601/FlaA, HP0295/Fla that showed overall prevalences ranging from 40 to 58% in all individuals, and from 50 to 71% in currently infected individuals. Based on in vitro studies, HP1118/gGT seems to be a pathogenic factor associated with H. pylori-induced peptic ulcer disease [38]. As mentioned above, Pan et al. reported an inverse IM association with anti-HP1118-gGT [11]. Similarly, we found a suggestive association in this study and a significant inverse association in our previous NAPPA gastric cancer study [5]. In our current study, HP1341/TonB/TonB2 has shown higher seroresponse in individuals with current infection compared to those with past infection. In our previous report on gastric cancer, we only had H. pylori serology results, thus we could not determine whether the subjects had a current or past infection [5]. We speculate the slightly lower seroprevalence of HP1341/TonB/TonB2 in gastric cancer is likely due to the loss of H. pylori colonization. To our knowledge, no data are available on the functions of anti-HP1110. Future research may explore the role of these antigens in vaccine development.
Up to date, there are two bacterial Genome-Wide Association Studies (GWAS) of gastric cancer. Berthenet et al. identified 32 significant loci (HP0068, HP0102, HP0269, HP0290, HP0468, HP0524, HP0527, HP0528, HP0531, HP0532, HP0540, HP0541, HP0544, HP0555, HP0569, HP0615, HP0709, HP0747, HP0797, HP0906, HP0936, HP1004, HP1046, HP1055, HP1149, HP1177, HP1184, HP1243, HP1331, HP1421, HP1460, HP1572) by comparing hpEurope genomes from patients with gastric cancer (n = 49) with genomes from patients with gastritis (non-atrophic and atrophic with and without IM; n = 124) [39]. On the other hand, Tuan and Yahara et al. [40] identified 12 significant loci (HP0082, HP0130, HP0231, HP0463, HP0490, HP0776, HP0807, HP0915, HP1250, HP1440, HP1467, HP1523; 11 single nucleotide polymorphisms and three DNA motifs) by comparing hspEAsia genomes from patients with gastric cancer (n = 125) and genomes from patients with duodenal ulcer (n = 115). Likely due to differences in the control group (gastritis vs. duodenal ulcer) and the underlying genetic structures of hpEurope and hspEAsia populations, there are no common hits between the GWAS. Notably, two of our candidate antibodies, anti-HP1177/Omp27/HopQ (positive association with IM) and anti-HP0709 (inverse association with IM and positive association with current H. pylori infection) represent hits in the hpEurope GWAS. HP1177/Omp27/HopQ was identified under the comparison including IM in the case group, while HP0709 was identified under the comparison including IM in the control group. One of our additional candidates, anti-HP0231/dsbK/dsbG (positive association with current infection), represents a gastric cancer hit in the hspEAsia GWAS.
The strengths of our IM study include the use of a well-validated and highly reproducible state-of-the-art microarray technology. Secondly, we employed a two-stage approach with discovery and independent validation using blinded testing to ensure the rigor of our findings. Importantly, we tested well-characterized samples with a high prevalence of H. pylori infection and data from several complementary tests documenting current and past H. pylori infections. Despite these strengths, our study has some limitations. First, although comprehensive, our H. pylori-NAPPA did not include a universal set of proteins, as the panproteome of H. pylori is undetermined. Second, our findings may not be generalizable to all populations as there is a difference in the immunogenic profile among races/ethnicities [41–43]. Third, although we combined four tests, a potential misclassification in our definitions of current and past H. pylori infections cannot be averted. The sensitivity and specificity of just urease and histology combined (both > 95%) are comparable to the urea breath test [44]. Moreover, individuals with H. pylori-negative gastritis could have been included in the group with no evidence of H. pylori exposure. In addition, information on past treatment of H. pylori infection was not available. Antibiotics may alter humoral responses, thus altering the association between the antibodies and IM. We addressed antibody associations with advanced IM based on the anatomical subsite. Additional heterogeneity should be evaluated for different histological types of IM (complete vs. incomplete).
Antibodies to several specific H. pylori proteins are associated with modest gastric cancer risk in prospective cohort studies [41, 45]. To date, none of the proposed antibodies have enough discriminatory power to distinguish gastric cancer patients from cancer-free individuals for screening. Our data suggest only moderate gain in discriminating IM and NAG with a combined IgG antibody model compared to an anti-CagA-only model. Additional candidate anti-OMP antibodies could be tested in prediagnostic samples.
Besides the etiological importance of our identified H. pylori antibodies in carcinogenesis, a non-invasive blood test for IM based on the antibodies may have a potential translation to noninvasive detection of patients with this premalignant lesion. After additional validation, the candidate antibodies in combination with other biomarkers (e.g., pepsinogens), could have a direct clinical application for triaging high-risk individuals for further diagnostic procedures, particularly in places where mass gastric cancer screening resources are limited.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank the production team and NAPPA core facility at the ASU Biodesign Center for Personalized Diagnostic for providing support on H. pylori NAPPA production. We would also like to thank Peter Khan, Al Burner and Peter Wicktor at Engineering Arts, LLC for NAPPA printing. The following reagent was obtained through BEI Resources, NIAID and NIH as part of the Human Microbiome Project: H. pylori Gateway® Clone Set, Recombinant in Escherichia coli, Plates 1–20 (BEI Resources NR-19477 through NR-19496), catalog No. NR-19275. We acknowledge the original contributor “Pathogen Functional Genomics Resource Center at the J. Craig Venter Institute” and BEI resources for providing the H. pylori clones set. We also appreciate the provision of cagA clones by Laurent Terradot at Institut de Biologie et Chimie des Protéines, France.
Abbreviations
- AUC
Area under the curve
- CI
Confidence interval
- NAPPA
Nucleic acid programmable protein array
- OR
Odds ratio
- ROC
Receiver operating characteristic
- NRI
Net reclassification improvement
- wcELISA
Whole-cell enzyme-linked immunosorbent assay
Author contributions
LS and MS contributed equally through study concept and design, data analysis, data interpretation and manuscript writing. YC, SW, MS contributed through data analysis, data interpretation, and manuscript writing. CSR, JT, AHC, RG, EB and JL contributed through data collection, data interpretation, and manuscript writing. JQ and MCC are equally responsible for the entirety of the project, including obtaining funding, study concept and design, data interpretation, and manuscript writing.
Funding
Division of Cancer Prevention, National Cancer Institute, R01 CA199948, Joshua LaBaer; Pontificia Universidad Católica de Chile, CONICYT-FONDAP 1513001, Alejandro Corvalan; Fondecyt 1191928, Alejandro Corvalan.
Data availability
The datasets that support the findings of this study are available from the corresponding authors upon reasonable request.
Declarations
Conflict of interest
The authors have no relevant financial or nonfinancial interests to disclose.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Lusheng Song and Minkyo Song shared co-first authors.
Ji Qiu and M. Constanza Camargo shared co-last authors.
References
- 1.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—first American cancer society award lecture on cancer epidemiology and prevention. Can Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 2.Kokkola A, Kosunen TU, Puolakkainen P, et al. Spontaneous disappearance of Helicobacter pylori antibodies in patients with advanced atrophic corpus gastritis. APMIS. 2003;111:619–624. doi: 10.1034/j.1600-0463.2003.1110604.x. [DOI] [PubMed] [Google Scholar]
- 3.Akbari M, Tabrizi R, Kardeh S, et al. Gastric cancer in patients with gastric atrophy and intestinal metaplasia: A systematic review and meta-analysis. PLoS One. 2019;14:e0219865. doi: 10.1371/journal.pone.0219865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dinis-Ribeiro M, Areia M, de Vries AC, et al. Management of precancerous conditions and lesions in the stomach (MAPS): guideline from the European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter Study Group (EHSG), European Society of Pathology (ESP), and the Sociedade Portuguesa de Endoscopia Digestiva (SPED) Endoscopy. 2012;44:74–94. doi: 10.1055/s-0031-1291491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song L, Song M, Rabkin CS, et al. Helicobacter pylori immunoproteomic profiles in gastric cancer. J Proteome Res. 2021;20:409–419. doi: 10.1021/acs.jproteome.0c00466. [DOI] [PubMed] [Google Scholar]
- 6.Koelblen T, Berge C, Cherrier MV, et al. Molecular dissection of protein-protein interactions between integrin alpha5beta1 and the Helicobacter pylori cag type IV secretion system. FEBS J. 2017;284:4143–4157. doi: 10.1111/febs.14299. [DOI] [PubMed] [Google Scholar]
- 7.Song LS, Wallstrom G, Yu XB, et al. Identification of antibody targets for tuberculosis serology using high-density nucleic acid programmable protein arrays. Mol Cell Proteomics. 2017;16:S277–S289. doi: 10.1074/mcp.M116.065953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejias-Luque R, Gerhard M. Immune evasion strategies and persistence of Helicobacter pylori. Curr Top Microbiol Immunol. 2017;400:53–71. doi: 10.1007/978-3-319-50520-6_3. [DOI] [PubMed] [Google Scholar]
- 9.Veijola L, Oksanen A, Sipponen P, et al. Evaluation of a commercial immunoblot, helicoblot 2.1, for diagnosis of Helicobacter pylori infection. Clin Vaccine Immunol. 2008;15:1705–1710. doi: 10.1128/CVI.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murata-Kamiya N. Pathophysiological functions of the CagA oncoprotein during infection by Helicobacter pylori. Microbes infect. 2011;13:799–807. doi: 10.1016/j.micinf.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Pan KF, Formichella L, Zhang L, et al. Helicobacter pylori antibody responses and evolution of precancerous gastric lesions in a Chinese population. Int J Cancer. 2014;134:2118–2125. doi: 10.1002/ijc.28560. [DOI] [PubMed] [Google Scholar]
- 12.Epplein M, Butt J, Zhang Y, et al. Validation of a blood biomarker for identification of individuals at high risk for gastric cancer. Cancer Epidem Biomar. 2018;27:1472–1479. doi: 10.1158/1055-9965.EPI-18-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu C, Soyfoo DM, Wu Y, et al. Virulence of Helicobacter pylori outer membrane proteins: an updated review. Eur J Clin Microbiol Infect Dis. 2020;39:1821–1830. doi: 10.1007/s10096-020-03948-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javaheri A, Kruse T, Moonens K, et al. Helicobacter pylori adhesin HopQ engages in a virulence-enhancing interaction with human CEACAMs. Nat Microbiol. 2016;2:16189. doi: 10.1038/nmicrobiol.2016.189. [DOI] [PubMed] [Google Scholar]
- 15.Xia R, Zhang B, Wang X, et al. Pathogenic interactions between Helicobacter pylori adhesion protein HopQ and human cell surface adhesion molecules CEACAMs in gastric epithelial cells. Iran J Basic Med Sci. 2019;22:710–715. doi: 10.22038/ijbms.2019.34237.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dooyema SDR, Krishna US, Loh JT, et al. Helicobacter pylori -Induced TLR9 activation and injury are associated with the virulence-associated adhesin HopQ. J Infect Dis. 2021;224:360–365. doi: 10.1093/infdis/jiaa730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taxauer K, Hamway Y, Ralser A, et al. Engagement of CEACAM1 by Helicobacter pylori HopQ is important for the activation of non-canonical NF-kappaB in gastric epithelial cells. Microorganisms. 2021;9:1748. doi: 10.3390/microorganisms9081748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan Y, Lu X, Han Y, et al. Helicobacter pylori outer membrane protein 18 (Hp1125) is involved in persistent colonization by evading interferon-gamma signaling. Biomed Res Int. 2015;2015:571280. doi: 10.1155/2015/571280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang JY, Sweeney EG, Sigal M, et al. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe. 2015;18:147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goers Sweeney E, Henderson JN, Goers J, et al. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure. 2012;20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernitzsch SR, Alzheimer M, Bremer BU, et al. Small RNA mediated gradual control of lipopolysaccharide biosynthesis affects antibiotic resistance in Helicobacter pylori. Nat Commun. 2021;12:4433. doi: 10.1038/s41467-021-24689-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suganuma M, Kurusu M, Suzuki K, et al. New tumor necrosis factor-alpha-inducing protein released from Helicobacter pylori for gastric cancer progression. J Cancer Res Clin Oncol. 2005;131:305–313. doi: 10.1007/s00432-004-0652-x. [DOI] [PubMed] [Google Scholar]
- 23.Jang JY, Yoon HJ, Yoon JY, et al. Crystal structure of the TNF-alpha-inducing protein (Tipalpha) from Helicobacter pylori: insights into its DNA-binding activity. J Mol Biol. 2009;392:191–197. doi: 10.1016/j.jmb.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 24.Voland P, Weeks DL, Vaira D, et al. Specific identification of three low molecular weight membrane-associated antigens of Helicobacter pylori. Aliment Pharmacol Ther. 2002;16:533–544. doi: 10.1046/j.1365-2036.2002.01221.x. [DOI] [PubMed] [Google Scholar]
- 25.Jung J, Lee CJ, Jeon YH, et al. Solution structure and backbone dynamics of the biotinylation domain of Helicobacter pylori biotin-carboxyl carrier protein. B Korean Chem Soc. 2008;29:347–351. doi: 10.5012/bkcs.2008.29.2.347. [DOI] [Google Scholar]
- 26.Wang G, Hong Y, Olczak A, et al. Dual roles of Helicobacter pylori NapA in inducing and combating oxidative stress. Infect Immun. 2006;74:6839–6846. doi: 10.1128/IAI.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Cai Y, Chen Z, et al. SpoT-mediated NapA upregulation promotes oxidative stress-induced Helicobacter pylori biofilm formation and confers multidrug resistance. Antimicrob Agents Chemother. 2021 doi: 10.1128/AAC.00152-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Orillard E, Radicella JP, Marsin S. Biochemical and cellular characterization of Helicobacter pylori RecA, a protein with high-level constitutive expression. J Bacteriol. 2011;193:6490–6497. doi: 10.1128/JB.05646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maroney MJ, Ciurli S. Nickel as a virulence factor in the class I bacterial carcinogen, Helicobacter pylori. Semin Cancer Biol. 2021;76:143–155. doi: 10.1016/j.semcancer.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 30.Olson JW, Mehta NS, Maier RJ. Requirement of nickel metabolism proteins HypA and HypB for full activity of both hydrogenase and urease in Helicobacter pylori. Mol Microbiol. 2001;39:176–182. doi: 10.1046/j.1365-2958.2001.02244.x. [DOI] [PubMed] [Google Scholar]
- 31.Banga Ndzouboukou JL, Lei Q, Ullah N, et al. Helicobacter pylori adhesins: HpaA a potential antigen in experimental vaccines for H. pylori. Helicobacter. 2021;26:e12758. doi: 10.1111/hel.12758. [DOI] [PubMed] [Google Scholar]
- 32.Servetas SL, Kim A, Su H, et al. Comparative analysis of the Hom family of outer membrane proteins in isolates from two geographically distinct regions: the United States and South Korea. Helicobacter. 2018;23:e12461. doi: 10.1111/hel.12461. [DOI] [PubMed] [Google Scholar]
- 33.Loh JT, Torres VJ, Cover TL. Regulation of Helicobacter pylori cagA expression in response to salt. Can Res. 2007;67:4709–4715. doi: 10.1158/0008-5472.CAN-06-4746. [DOI] [PubMed] [Google Scholar]
- 34.Noto JM, Gaddy JA, Lee JY, et al. Iron deficiency accelerates Helicobacter pylori -induced carcinogenesis in rodents and humans. J Clin Invest. 2013;123:479–492. doi: 10.1172/JCI64373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Butt J, Blot WJ, Shrubsole MJ, et al. Performance of multiplex serology in discriminating active vs past Helicobacter pylori infection in a primarily African American population in the Southeastern United States. Helicobacter. 2020;25:e12671. doi: 10.1111/hel.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafaie E, Saberi S, Esmaeili M, et al. Multiplex serology of Helicobacter pylori antigens in detection of current infection and atrophic gastritis-a simple and cost-efficient method. Microb Pathog. 2018;119:137–144. doi: 10.1016/j.micpath.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 37.Khalifeh Gholi M, Kalali B, Formichella L, et al. Helicobacter pylori FliD protein is a highly sensitive and specific marker for serologic diagnosis of H. pylori infection. Int J Med Microbiol. 2013;303:618–623. doi: 10.1016/j.ijmm.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Zhang X, Zhang J, Yang F, et al. Immunization with heat shock protein A and gamma-glutamyl transpeptidase induces reduction on the Helicobacter pylori colonization in mice. PLoS ONE. 2015;10:e0130391. doi: 10.1371/journal.pone.0130391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berthenet E, Yahara K, Thorell K, et al. A GWAS on Helicobacter pylori strains points to genetic variants associated with gastric cancer risk. BMC Biol. 2018;16:84. doi: 10.1186/s12915-018-0550-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tuan VP, Yahara K, Dung HDQ, et al. Genome-wide association study of gastric cancer-and duodenal ulcer-derived Helicobacter pylori strains reveals discriminatory genetic variations and novel oncoprotein candidates. Microb Genom. 2021 doi: 10.1099/mgen.0.000680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai H, Ye F, Michel A, et al. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J Epidemiol. 2016;45:774–781. doi: 10.1093/ije/dyw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shakeri R, Malekzadeh R, Nasrollahzadeh D, et al. Multiplex H pylori Serology and risk of gastric cardia and non cardia adenocarcinomas. Cancer Res. 2015;75:4876–4883. doi: 10.1158/0008-5472.CAN-15-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camargo MC, Beltran M, Conde-Glez CJ, et al. Serological response to Helicobacter pylori infection among Latin American populations with contrasting risks of gastric cancer. Int J Cancer. 2015;137:3000–3005. doi: 10.1002/ijc.29678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gisbert JP, Pajares JM. Review article: 13C-urea breath test in the diagnosis of Helicobacter pylori infection—a critical review. Aliment Pharmacol Ther. 2004;20:1001–1017. doi: 10.1111/j.1365-2036.2004.02203.x. [DOI] [PubMed] [Google Scholar]
- 45.Gao L, Weck MN, Michel A, et al. Association between chronic atrophic gastritis and serum antibodies to 15 Helicobacter pylori proteins measured by multiplex serology. Cancer Res. 2009;69:2973–2980. doi: 10.1158/0008-5472.CAN-08-3477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets that support the findings of this study are available from the corresponding authors upon reasonable request.