Abstract
Scientists have discovered many ways to treat bacteria, viruses, and parasites in aquaculture; however, there is still an impossibility in finding a permanent solution for all types of diseases. In that case, the CRISPR-Cas genome-editing technique can be the potential solution to preventing diseases for aquaculture sustainability. CRISPR-Cas is cheaper, easier, and more precise than the other existing genome-editing technologies and can be used as a new disease treatment tool to solve the far-reaching challenges in aquaculture. This technique may now be employed in novel ways, such as modifying a single nucleotide base or tagging a location in the DNA with a fluorescent protein. This review paper provides an informative discussion on adopting CRISPR technology in aquaculture disease management. Starting with the basic knowledge of CRISPR technology and phages, this study highlights the development of RNA-guided immunity to combat the Chilodonella protozoan group and nervous necrosis virus (NNV) in marine finfish. Additionally, we highlight the immunological application of CRISPR-Cas against bacterial diseases in channel catfish and the white spot syndrome virus (WSSV) in shrimp. In addition, the review summarizes a synthesis of bioinformatics tools used for CRISPR-Cas sgRNA design, and acceptable solutions are discussed, considering the limitations.
Keywords: CRISPR-Cas, fish, pathogens, phages, RNA
1. Introduction
Fish diseases are a serious barrier in the aquaculture sector, affecting more than a billion dollars yearly. Climate change and developing fish farming may influence the balance or imbalance of pathogen, host, and environmental interaction, with new infections being detected or identified annually and more known diseases arising in various global regions and species [1]. Pathogen evolution is thought to be accelerated in intensive farming systems due to the high density of vulnerable hosts, which promotes pathogen transmission and virulence [2]. Because of higher population densities and host–pathogen interactions, this aspect of the farming environment is expected to extend to biological interactions between pathogenic bacteria and their phages, viruses, and parasites. However, many of these diseases or infections have no proven or approved recommended treatments, vaccinations, or control strategies and remain a substantial barrier to the economic sustainability of aquaculture in specific regions and species [1].
Aquaculture enterprises may need new scientific procedures to increase fish production while maintaining trait quality. Several initiatives have been conducted over the last two decades to manage and treat disease in aquaculture species, with varying degrees of success [3]. Many proven aquaculture species, such as tilapia, carp, salmonids, and some marine species (sea bass, sea bream, and grouper), have commercial vaccines for a limited number of diseases and authorized treatments for specific pathogens. However, there is significant variation from country to country and even within a geographic region [1]. Several diseases that have a substantial economic impact in aquaculture are viral infections with no therapies and vaccines, which, if produced, only provide limited protection. Numerous examples of bacterial, parasitic, and fungal diseases can pose significant international economic and welfare concerns to aquaculture. The management and control of parasitic infections are critical not only for the viability of the aquaculture sector but also for preventing horizontal parasite spread to wild fish [4,5]. Chemotherapy has had some promising results and has been proposed as a potential method of treating fish parasites [6]. Such initiatives, however, are incompatible with the United Nations’ Sustainable Development Goals, as well as generally agreed fish welfare and environmental standards. As a result, additional long-term preventative approaches for parasite management must be developed. On the other hand, in aquaculture, increasing production and the frequency of diseases in aquatic animals are driving up antimicrobial use and antimicrobial resistance [7,8] across various farmed aquatic species. Antimicrobial residues in the aquatic environment affect the environmental microbiome, affecting the ecosystem’s capacity for regulation, provisioning, and sustenance [9,10]. As a result, the enrichment of naturally existing pathogens in aquaculture habitats and the usage of antimicrobial treatments provide an appealing opportunity to investigate newer, more robust solutions for aquaculture sustainability.
Currently, biotechnology research can address several issues, not just related to aquaculture farming but also environmental issues [3]. Several genome-editing tools have recently been developed, including zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and more recently clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated nucleases 9 (Cas9), which have made it possible to edit genes or knock out unwanted parts of them in various animal models [11]. The CRISPR-Cas9 system has been developed as a new elite genome engineering tool, even for organisms where genome editing would be challenging. With this promising new technology, it is possible to overcome several challenges that the aquaculture industry faces. Genomic editing using CRISPR can quickly introduce significant genome changes, making it useful for genetic improvements, disease resistance, and disease control in aquaculture [11,12]. For example, a novel mechanism for RNA-guided immunity against RNA viruses in vertebrates is provided by CRISPR-CasRx for engineering interference against RNA viruses in fish [13]. In addition, about half of all bacteria and almost all archaea possess a CRISPR-Cas system that protects them from foreign genetic elements, such as viruses and plasmids [14]. Thus, the role of the CRISPR-Cas mechanism in the aquaculture field would be crucial for future global food demand. There are several reviews on using CRISPR-based genome editing in aquaculture [15,16]; however, less effort has been made to apply CRISPR-Cas to control diseases in aquaculture. This review focuses on the pathogenic aspects of CRISPR-Cas9 genome editing relevant to aquaculture applications. Furthermore, a workflow for genomic interactions between CRISPR-Cas and phage is presented, primary techniques related to anti-RNA parasite experiments and outcome prediction of disease management in a CRISPR-Cas9 system are described, bioinformatics tools for CRISPR mechanisms are demonstrated, and the future of CRISPR-Cas9 for aquaculture disease management is briefly discussed in this study. We also mention aspects that need to be examined or improved for genome editing to effectively manage microbial diseases in fish farming. Finally, this study is intended to offer an overview of CRISPR genome-editing studies for fish disease management and treatment to encourage more genome editing research and uses in aquaculture.
2. Relationship between CRISPR-Cas and Phages
Bacteria-infecting viruses, generally known as bacteriophages, require a bacterial host to survive. Their numbers in the biosphere make them the most abundant [17]. Bacteriophages are a significant threat to bacteria due to their capacity to infect their bacterial host. When the host’s environment becomes unfavorable, phages may switch to a pseudolysogenic method. Pseudolysogeny is the development of bacteriophages in a host cell without multiplication or replication and occurs with zero degradation of the viral genome.
In such situations, prokaryotes, such as bacteria, adopt various defense mechanisms to protect themselves. CRISPR functions as a natural defense mechanism or adaptive immune system of prokaryotes against viral DNA, bacteriophages, and plasmids, which was first reported in the E. coli genome [18]. Adaptive immunity refers to the immunity that an organism acquires after exposure to an antigen, either from a pathogen or vaccine. It may be found in most lysogenic bacteria [19], including two aquaculture-related bacterial species, Flavobacterium psychrophilum [20] and Vibrio anguillarum [21]. The relationship between the phages’ life cycle and CRISPR-Cas is still poorly known [22].
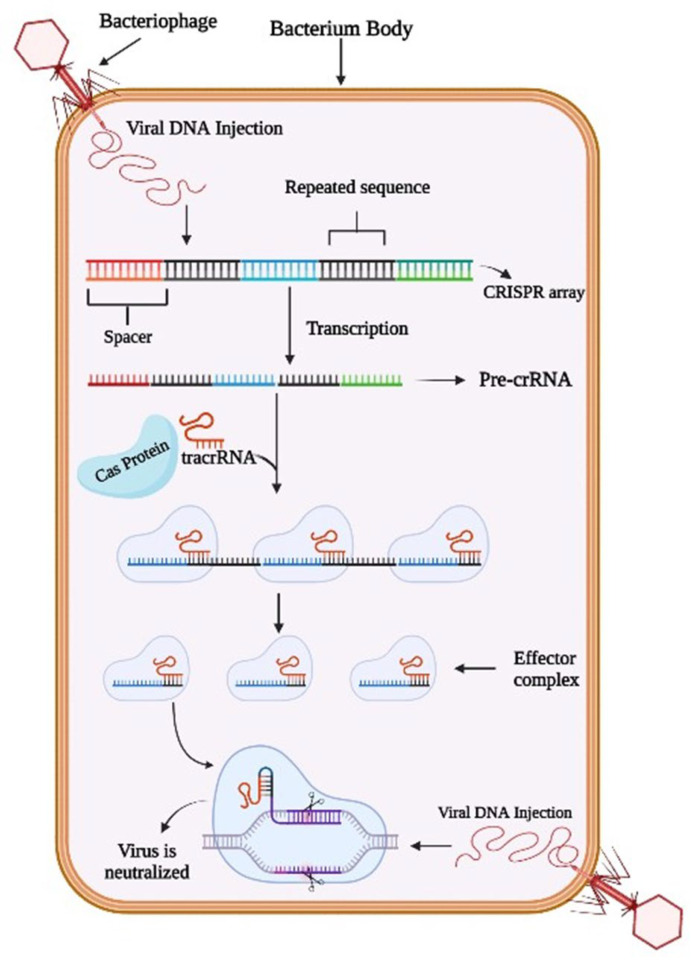
Regarding CRISPR, the repeated sequences of prokaryote DNA nucleotides are described as palindromic repeats because they are the same whether reading forwards or backward. The unique sequences nestled between the palindromic repeats are called spacers. Spacers are the DNA bits originating from the foreign mobile genetic elements (MGEs) that have previously infected the prokaryote and do not belong to the bacterium. Different spacers, potentially originating from different viruses, are sandwiched between the repeated sequences and produce a CRISPR array. In this way, bacteria retain a memory of a past infection [18]. The CRISPR array can undergo transcription to form CRISPR RNA (crRNA) called pre-crRNA. In the next step, the Cas protein becomes involved, which refers to the CRISPR-associate nuclease protein capable of cleaving DNA at specific nucleotide linkages. The presence of tracrRNA with Cas protein was also recorded. Each spacer and palindromic repeat end up with an effector complex consisting of a segment of pre-crRNA, a tracrRNA, and a Cas protein. By cleaving the strand between these complexes, the ribonuclease-3 enzyme helps the cell defend against the invader whose genome produced that crRNA. The whole process neutralizes the virus by preventing viral transcription [18]. The relation of the mechanism of CRISPR-Cas against bacteriophage interference is shown in Figure 1.
Figure 1.
Mechanism of CRISPR-Cas against bacteriophage interference.
3. CRISPR-Cas for Anti-Parasitic Action
Invertebrate parasites may either be free-living or obligatory parasites that depend on their hosts for survival and reproduction. Both obligatory and opportunistic parasites may be found in fish, but obligatory parasites are mainly responsible for causing many parasitic infections in fish. Most fish that seem to be healthy often have small numbers of different parasites on or in their bodies, by which the fish usually suffer little to no danger. However, changes in water temperature or salinity reduce fish immunity causing a significant increase in the number of parasites per fish, and parasitic disease outbreaks frequently happen. In addition, there are connections between parasitic diseases and other infections. It has been noted that cultured fish in captivity are host to a wide range of parasites. Some of these parasites have led to severe disease outbreaks or persistent subclinical effects in farmed fish, costing fish farmers a lot of money. The fish are most vulnerable in the beginning phases of the culture cycle, especially when the fish are tiny and in the hatchery and nursery stages. The three major groups of parasitic organisms that infect farmed fish are protozoa, platyhelminthes, and crustaceans. Many of these parasites can potentially spread disease and result in significant financial losses.
Several therapies and preventative measures may be used to deal with parasite assaults, including environmental disinfection, seedling disinfection, health management, nutritional supplements, copper sulfate, potassium permanganate, formalin, zinc sulfate, and ivermectin. However, the use of medications and antibiotics harms the environment and the safety of food. From this perspective, CRISPR-Cas may be a more efficient way to create a species-specific insecticide for defeating parasite infestations by a genetic operation that targets a particular place of the gene for cutting and repair. The most often researched fish parasite species of the Chilodonella protozoan group, Chilodonella piscicola, Cryptocaryon irritans, and Chilodonella uncinata, have all been subjected to CRISPR-Cas anti-parasitic activities [23,24,25]. These hypothermic protozoan parasite species oversee, causing both gill and skin diseases in freshwater fish, which restrict the development of both juvenile and adult fish, especially in spring and fall [25].
Yige Li et al. reduced the survival capacity of C. piscicola by destroying the parasite DNA at a particular location using a combination of Cas9 messenger RNA (mRNA) and single guide RNAs (sgRNA) [25]. In that experiment, the CRISPR-Cas9 system was used to design sgRNA in conjunction with the known sequences of the 18S ribosomal RNA (rRNA), internal transcribed spacer 1 (ITS-1), and 5.8S ribosomal RNA (rRNA) of C. piscicola to destroy the genetic barcode of C. piscicola. As evidence of the efficiency of the chosen sgRNA of 18S rRNA sequence and ITS-1 sequence, the survival rate of the experimental C. piscicola was decreased to 40% compared to the blank control group with zero significant difference. It demonstrated that both sites had the potential to eradicate C. piscicola successfully. According to real-time quantitative PCR (RT-qPCR), Cas9 may either act with a single sgRNA or a combination of two sgRNAs to damage the parasite DNA in a particular area [25].
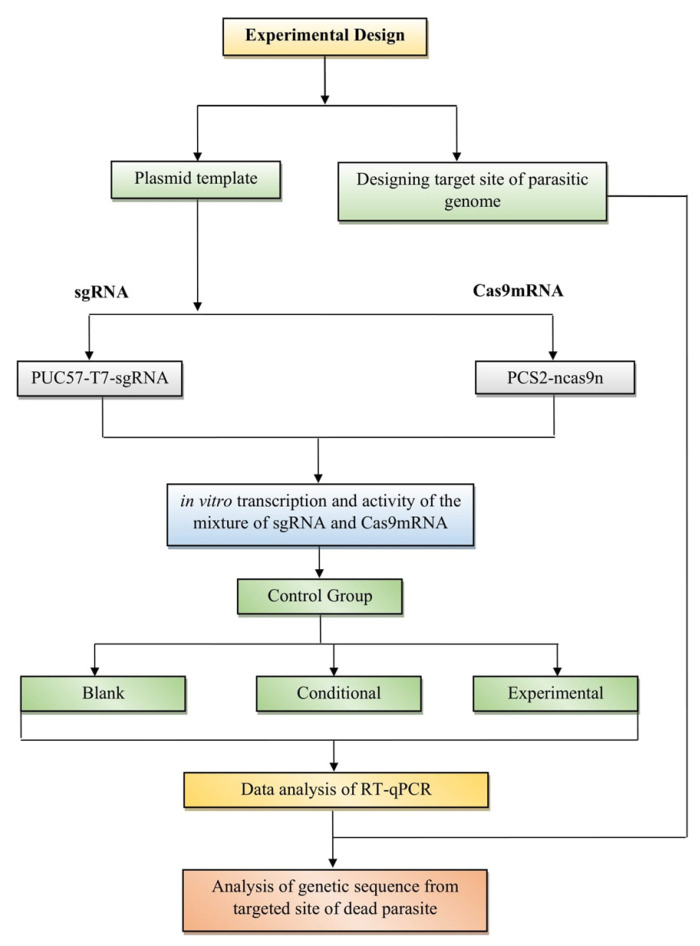
Majeed et al. also conducted a similar study to control the Aphanomyces invadans pathogen, a causative agent for epizootic ulcerative syndrome (EUS) [26]. In that experiment, scientists applied the CRISPR-Cas9 system for editing the A. invadans genome by targeting the serine protease gene in in vitro and also observed the effect on the virulence and pathogenicity of the A. invadans in vivo. They designed three single guide-RNAs (sgRNA) combined with the Cas9 to form a ribonucleoprotein (RNP) complex and transfected in A. invadans protoplasts and zoospores. Three groups of dwarf gourami (Trichogaster lalius) were taken as test species and experimentally inoculated with (i) non-treated zoospores; (ii) RNP-treated zoospores; and (iii) autoclaved pond water as a negative control to investigate the effect on the virulence in vivo. According to the in vivo results of the study, the CRISPR-Cas9-treated A. invadans zoospores did not express any signs of EUS in the fish [26]. The basic experimental design of developing an RNA anti-parasite using the CRISPR-Cas method is shown in Figure 2.
Figure 2.
Experimental design of developing an RNA anti-parasite.
4. CRISPR-Cas for Developing RNA-Guided Immunity against RNA Viruses in Fish
One of the deadliest viruses that may infect fish is the RNA virus, which is unpredictable and challenging to prevent. They are highly dynamic pathogens because of their short generation times, enormous population numbers, and high mutation rates, among other distinctive traits. Iridoviridae, Adenoviridae, and Herpesvirdae are home to fish viruses with DNA genomes. In contrast, those with RNA genomes are found in the families Picornaviridae, Birnaviridae, Reoviridae, Rhabdoviridae, Orthomyxoviridae, Paramyxoviridae, Caliciviridae, Togaviridae, Nodaviridae, and Retroviridae.
RNA-guided immunity against RNA viruses could be developed in the fish body using the antiviral CRISPR-Cas, which had previously been successfully targeted by CRISPR-Cas13 [27,28,29]. On the other hand, CasRx, a small type VI-D effector (Cas13d), can also effectively knock down RNA in RNA viruses [13]. The RNA-targeting CRISPR-CasRx is a programmable system, and Cas13d is a CRISPR effector known as small type VI.
The two primary categories of CRISPR-Cas systems are class I, which mediates the interference via multi-effector complexes, and class II, which utilizes a unified, multi-domain effector [30]. In addition, these classes are further divided into six types and thirty-three subtypes based on the genomic architecture of the CRISPR array and its distinct interference effectors. Types II, V, and VI of the CRISPR-Cas systems are within class II. Endonucleases of types II and V are used for the DNA, while those of type VI are only used for RNA [30,31]. To provide prokaryotes with protection against RNA, class II type VI CRISPR-Cas systems use an RNA-guided and RNA-targeting mechanism [32]. The Cas13 effector protein is the same for all type VI CRISPR-Cas systems. Multiple studies have shown further variations of Cas13 proteins belonging to various Cas13 families, which have been categorized into four type VI subtypes (subtypes A–D) [28,30,33]. A novel Cas13 subtype known as CRISPR-Cas13d (CasRx) has also recently been discovered by researchers; it has minimal sequence similarity with earlier Cas13 effectors [33,34]. Compared to other Cas13 effectors, CasRx is more efficient and more robustly activated in cells when RNA-guided RNA cleavage occurs [33,34]. It provides the most effective targeting and has the smallest size, making it perfect for in vivo therapeutic applications [34].
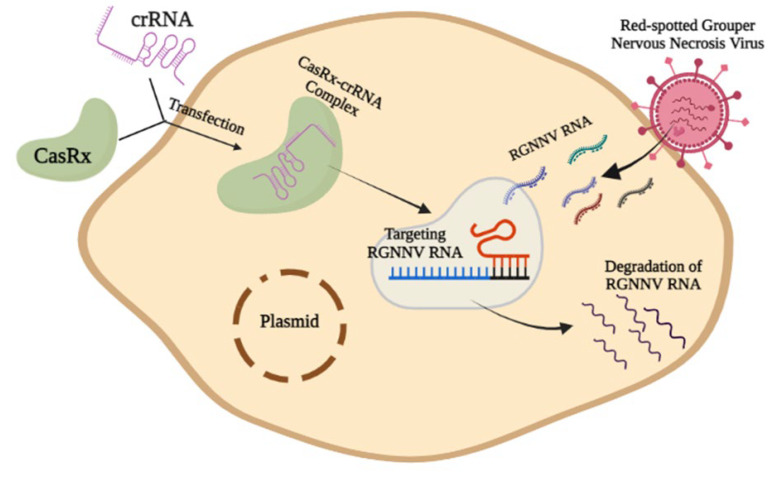
Qing Wang et al. designed synthetic mRNA coding for CasRx and used CRISPR RNAs to guide it to target the nervous necrosis virus (NNV), which is an RNA virus of fish [35,36], and applied the coding both in vitro and in vivo to observe the significance of the dominating RNA virus [13]. Scientists selected the red-spotted grouper (Epinephelus coioides) as a model species for the experiment. The red-spotted grouper NNV (RGNNV) is one of the four classified NNVs; striped jack NNV, tiger puffer NNV, and barfin flounder NNV are the remainder of them [36,37]. NNV is also found in orange-spotted grouper, Sevenband grouper, brown-marbled grouper, turbot, etc. Nervous necrosis viruses (NNV) are icosahedral non-enveloped single-stranded positive-sense RNA viruses (ssRNA+ viruses) classified in the family Nodaviridae which is the pathogen of viral nervous necrosis disease (VNN) that destroys the central nervous system of infected fish [38,39]. The in vitro RGNNV targeting via the CasRx system is shown in Figure 3.
Figure 3.
The in vitro RGNNV targeting via the CasRx system.
Scientists designed the experiment in three basic steps. Firstly, to increase the protein content of CasRx, researchers improved the codons that express it. Secondly, to enhance crRNA expression, the zebrafish U6 promoter was applied. Thirdly, they designed CasRx to eliminate the differences in cellular location.
RGNNV is composed of CP and RdRp (Clamping RNA-dependent RNA polymerase). They investigated the possibility that CasRx may target the RGNNV CP and RdRp to induce effective and reliable RNA virus interference [13]. In this research, scientists developed three crRNAs for targeting the sequences of code of CP mRNA and two crRNAs for targeting the sequences of code of RdRp mRNA. To investigate the effect of the CasRx-crRNA complex on the cellular level of RGNNV, grouper spleen (GS) cells were treated with plasmids carrying CasRx and crRNA. The first CasRx expression was observed after 6 h of transfection, and it became more expressive after 12 and 24 h, but it did not show much expression between 24 and 48 h. For each of the five crRNAs, RGNNV infection was carried out after transfecting GS cells with plasmids carrying CasRx-dNLS or CasRx-NLS. All ten combinations reduced the number of viral RNA copies. Furthermore, the virus titer data showed that transfection of cells with CasRx-dNLS or CasRx-NLS and crRNA significantly reduced viral titers and RGNNV pathogenicity. Extensive CPEs were shown when GS cells were treated with RGNNV or CasRx plus nonspecific (ns)-crRNA and RGNNV. Few CPEs were seen when cells were transfected with either CasRx-dNLS or CasRx-NLS and each crRNA. Additionally, the findings of the immunofluorescence experiment showed that GS cells subjected to only RGNNV or CasRx plus ns-crRNA and RGNNV displayed strong fluorescence signals of the RGNNV CP protein. Positive fluorescence signals were markedly reduced when cells were transfected with either CasRx-dNLS or CasRx-NLS and each crRNA. These findings demonstrated that the CasRx system effectively prevented RGNNV infections in vitro [13].
The effectiveness of CRISPR-CasRx has been proven to combat an RNA virus in vertebrates. The degradation of several viral genomic areas led to decreased CP and RdRp mRNA levels, in vitro cell vacuolation, and cumulative mortality in vivo. These results demonstrated the efficacy of using CRISPR-CasRx, a vertebrate RNA virus that may be targeted and interfered with by reducing RGNNV replication and dissemination.
5. Application of CRISPR-Cas in Fish Disease
Aquaculture industries worldwide face serious problems such as infectious and parasitic diseases, reduced viability, decreased fertility, poor development, environmental contamination by escapee fish, coastal conflicts, and disagreements over the patenting of research products [15,40]. Among these, disease outbreaks in aquaculture are a major issue that are one of the main reasons for the reduction in fish production. Many reputable shellfish farms report shellfish dying overnight because of viral assaults. In fish aquaculture, reproduction, and development [41,42], growth [43], pigment [44,45], disease resistance [46], trans-GFP usage in research [47], and omega-3 metabolism [48,49] are the qualities that are most often targeted for genetic engineering [50]. However, using molecular biological techniques to resolve diseases has become a core technology. Genomic editing (GE) has created several controls for aquatic diseases, and it will continue to do so in the future in a variety of different ways. Among these GE methods, CRISPR-Cas has been applied to modify several genes for targeting species-specific pathogens as modern technology.
CRISPR-Cas has been applied in immunological studies in channel catfish (Ictalurus punctatus) according to several types of research [51,52]. It enhanced the resistance of channel catfish to many diseases by injecting the alligator cathelicidin gene into the fish [53]. Additionally, this technology enhances the fish body’s natural immunity, which works against bacterial diseases or other infectious diseases such as Edwardsiella ictalurid and Flavobacterium columnare [54]. The editing of disease-resistance genes in channel catfish is an additional application of CRISPR-Cas of commercial relevance [51,52,55].
In shrimp and prawns, the eyestalk neuroendocrine complex contains suppressing/inhibiting substances that always prevent breeding and spawning under captivity. These limiting elements also hinder the process of growth. These aquatic organisms’ immune systems have reportedly been weak, making viral and bacterial diseases highly likely to strike them. Certain marine shrimps have already had their gonad-inhibiting hormone (GIH) and molt-inhibiting hormone (MIH) genes evaluated [56,57,58]. Using CRISPR-Cas technology has been able to eliminate the harmful effects of hormones on growth and reproduction, which may open the way to developing a powerful substitute for eyestalk ablation that has a comparable effect. Some researchers have tried to delete the gene using this RNA interference method [59,60]. When working on Penaeus monodon (giant tiger prawn), Treerattrakool et al. used the method of RNA interference to induce maturity in both wild and captive shrimp and reported that shrimps injected with anti-GIH double-stranded (ds) RNA showed enhanced maturation [59]. According to Das et al., RNA interference was used to silence the gonad-inhibiting hormone gene in the eyestalk neuroendocrine complex of the P. monodon (tiger shrimp) [60]. They discovered a three–five times increase in the transcript of the androgenic gland hormone (AGH) in males but no alteration in the expression of vitellogenin in females. Additionally, CRISPR-Cas technology can be utilized to manage bacterial and viral infections, particularly in shrimp and prawns. The CRISPR-Cas process in shrimp and prawns may also function similarly to that of bacteria when viral DNA attacks them. For example, CRISPR-Cas can replicate and insert portions of the white spot syndrome virus (WSSV) DNA into shrimp genomes as “spacers” between the short DNA repeats in CRISPR when WSSV invades them. By providing a template for RNA molecules to rapidly recognize and target the same DNA sequence in the case of future viral infections, these spacers improve the immune response of shrimp. The RNA molecules redirect the CRISPR complex to an incoming sequence of foreign DNA if they recognize it. There, the Plasmid Cas proteins of the shrimp cut the invading gene and render it inactive. The shrimp may be shielded against contagious infections because of this [60].
Culturing commercial species in the aquatic environment, every year, significant losses are attributed to mass mortality, rejection of aquaculture species’ shipments due to a lack of quality standards, the impact of biotic and abiotic stresses on aquaculture species, and the absence of standardized disease control and pollution-impact methods or protocols [3]. However, at this point, we require highly potent technologies to solve some of the significant problems in the aquaculture sector. With the development of CRISPR-Cas technology, it may be possible to solve any biological problems relating to genetic diseases or other problems without significantly changing the genetic makeup of aquaculture species and preventing viral and bacterial infections; CRISPR-Cas technology can prove to be a potent tool [3].
6. Advances in Bioinformatics in CRISPR-Cas
Bioinformatics is a scientific field that generates methodologies and software tools for analyzing biological data. It has been applied in various applications such as in silico studies of biological questions utilizing computational and statistical tools. It is frequently used to find potential genes and single nucleotide polymorphisms (SNPs). Furthermore, a field of study known as proteomics in bioinformatics seeks to comprehend the organizing concepts found in nucleic acid and protein sequences [61]. The main effects of bioinformatics have been the automation of microbial genome sequencing, the creation of integrated databases accessible through the internet, and genome analysis to comprehend gene and genome function. Bioinformatics is now used for a wide variety of other significant tasks in addition to the analysis of gene variation and expression, the analysis and prediction of gene and protein structure, as well as the prediction and detection of gene regulatory networks. It can analyze data more quickly to enhance the accuracy of the findings and explain the causes and phenomena of diseases at the gene/pathway level.
The first thing we must understand before relating the CRISPR-Cas system to bioinformatics is that selecting the appropriate CRISPR target gene is an essential step in successfully targeting gene editing. Bioinformatics can be used to locate and insert CRISPR-Cas into the targeted genome [3]. The choice of the target site is constrained by the possibility of off-target editing and variations in editing effectiveness. Numerous computational techniques have been created in recent years to assist researchers in choosing target sites for CRISPR knock-in/out experiments. In developing single-guide RNA (sgRNA) for CRISPR applications, these methods are likely to be helpful in both target site selection and sgRNA creation. The sgRNA design tools are specifically suitable for genetic screening and CRISPR-mediated gene regulation research has also been developed, resulting from the expansion of CRISPR applications. Computational tools have been created to analyze CRISPR genome-edited data produced by Next Generation Sequencing (NGS) systems and aid in sgRNA creation [62]. The CRISPR-Cas9 genome-editing technologies use programmable nucleases to accurately and frequently modify a particular section of the genome which may use RNA-guided nucleases [63,64,65].
CRISPR-Cas has successfully modified specific genomes in significant model species, such as zebrafish [66]. It modifies two RNAs—a transactivating CRISPR RNA (tracrRNA) that base pairs with the crRNA and a CRISPR RNA (crRNA) complementary to the targeted DNA sequence—that recruit Cas9 to the target site. The target sequence should be followed by a protospacer adjacent motif (PAM) sequence for recognition (nGG, where n can be any nucleotide). The crRNA and tracrRNA may be combined to form a single synthetic guide RNA (sgRNA) [66] that works efficiently with Cas9 to cause cleavage of the target site (~20 bp), which must come after the PAM sequence in the genome. Using in vitro transcription promoters such as T7, T3, or SP6 to create sgRNAs restricts the target sequence. Here, the CRISPR-Cas system was used to modify the Xenopus tropicalis (western clawed frog) genome, providing another tool for quick and effective targeted mutagenesis.
Cas 9 is a CRISPR-related protein adapted from a naturally occurring genome-editing system and used here as a bacterial immune defense. Most genomic restriction nucleases require substantial and complex PAM sequences that would restrict them due to reduced genome size. Distinct PAMs in the SpCas9 system are used for genome manipulation, including target gene disruption and single base-pair mutations in various organisms and cells. Developing the SpCas9 to identify more PAMs would be an alternative approach to increasing PAM specificity. Although SpCas9 is the most well-known nuclease, Cas9 can also be obtained from many bacterial species. The fundamental difference between them is the PAM sequence required for the cleavage of Cas9 nucleases from different bacteria [67].
The CRISPR-mediated genome editing tools are shown in Table 1, which are used for fish.
Table 1.
CRISPR-Cas-related genome editing resources that may be appropriate for use in aquaculture.
| Name | Function | URL | Reference |
|---|---|---|---|
| CRISPRScan |
|
https://www.crisprscan.org accessed on 1 September 2022 |
[68] |
| CHOPCHOP |
|
http://chopchop.cbu.uib.no accessed on 1 September 2022 |
[69] |
| ccTop |
|
https://crispr.cos.uni-heidelberg.de accessed on 1 September 2022 |
[70] |
| Cas-Designer |
|
http://www.rgenome.net accessed on 1 September 2022 |
[71] |
| MENTHU |
|
http://genesculpt.org/menthu accessed on 1 September 2022 |
[72] |
| CRISPR-ERA |
|
http://crispr-era.stanford.edu accessed on 1 September 2022 |
[73] |
| CRISPResso 2 |
|
http://crispresso.pinellolab.partners.org accessed on 1 September 2022 |
[74] |
| Cas-Analyzer |
|
http://www.rgenome.net/Cas-analyzer accessed on 2 September 2022 |
[75] |
| CRISPR-GA |
|
http://crispr-ga.net accessed on 2 September 2022 |
[76] |
| CRISPRz |
|
https://research.nhgri.nih.gov/CRISPRz accessed on 2 September 2022 |
[77] |
| inDelphi |
|
https://indelphi.giffordlab.mit.edu accessed on 2 September 2022 |
[78] |
| FORECasT |
|
https://partslab.sanger.ac.uk/FORECasT accessed on 2 September 2022 |
[79] |
On the other hand, many genetic disorders and undesirable features are carried on by base-pair changes in the genomic DNA. Base editing, the most recent development of CRISPR-Cas-based technologies, may directly introduce point mutations into cellular DNA without leading to a double-strand DNA break (DSB). The CRISPR-base-edit tools have recently increased by prime editing (PE), which now includes all twelve potential transition and transversion mutations in addition to minor insertion or deletion changes [77]. The base editing resources are shown in Table 2.
Table 2.
Base editing resources.
| Resources | Function | URL | References |
|---|---|---|---|
| BE-Analyzer | Used as a rapid evaluation tool for CRISPR-base edited cells of NGS data. |
http://www.rgenome.net/be-analyzer accessed on 2 September 2022 |
[80] |
| BE-Designer | Used for CRISPR base editing, a designer of guide RNA. |
http://www.rgenome.net/be-designer accessed on 2 September 2022 |
[80] |
| BEEP | Used for analysis of Sanger sequencing ab1 files for CRISPR-mediated base editing effectiveness. |
https://github.com/mitmedialab/BEEP accessed on 3 September 2022 |
[81] |
| CRISPR-SKIP | Used to select the exons that can be skipped by modifying the flanking G nucleotide. |
https://knoweng-0.igb.illinois.edu/crispr-skip accessed on 3 September 2022 |
[82] |
| CRISPResso 2 | Used as a tool for next-generation sequencing data. |
http://crispresso.pinellolab.partners.org accessed on 3 September 2022 |
[74] |
| EditR | A single Sanger sequencing run can be used to predict possible editing in a guide RNA region. |
http://baseeditr.com accessed on 3 September 2022 |
[83] |
| iSTOP | A database of sgRNAs for CRISPR-dependent base editing of STOP codons (sgSTOPs). |
https://www.ciccialab-database.com accessed on 3 September 2022 |
[84] |
| Beditor | Designing Guide RNA Libraries for CRISPR-Mediated Base Editing |
https://github.com/rraadd88/beditor accessed on 3 September 2022 |
[85] |
In addition, to targeting single-point mutations, CRISPR has provided a variety of methods for editing the genome precisely, such as functional gene knockouts and epigenome modifications. Recent research has focused on improving Cas9 selectivity and expanding target coverage; guided evolution has resulted in discovering many Cas9 variants that will significantly expand targeting coverage. Base-editing techniques have also made significant advancements in the investigation of pathogenic variations in animal models; they will speed up the functional verification of potential disease genes in model organisms and the creation of therapeutic tools for the treatment of several disorders [83]. The design of efficient sgRNAs is becoming increasingly difficult since the CRISPR-Cas9 system has swiftly become a ubiquitous gene-editing tool in biological research. To address this critical issue, various bioinformatics techniques have been created. In conclusion, by enhancing experimental planning, data integrity, and computational modeling, researchers created a novel sgRNA design tool that consistently outperformed in various experimental conditions [84].
7. Limitations of CRISPR-Cas for Aquatic Disease Perspective
The field of molecular biology is being revolutionized by the quick advancement of genome-editing technology such as CRISPR-Cas, which allows DNA modification in a broad range of species. It is being considered for several applications, from agriculture to clinical therapeutics [85]. CRISPR-Cas technology has made tremendous strides in recent years and demonstrated significant promise in several areas of life sciences’ study.
Despite the impressive CRISPR advancements, a few issues still need to be resolved to develop Cas systems to their full potential. CRISPR technologies have some primary limitations as application measures and genetic perspectives. These technical difficulties can be resolved in the present attempts to address all of those worries.
The CRISPR-Cas method is a relatively new gene-editing technology; generally, the methods associated with gene editing are quite expensive. Such an expensive technology in the molecular biological sector, such as CRISPR-Cas, is challenging to adapt in a developing or underdeveloped agriculturally dominant country. As a result, it is tough to quickly implement this technology in aquaculture in these countries, even if it is effective enough to control fish diseases. In addition to being expensive, this technology is also quite complicated. These complexities make it difficult to implement CRISPR-Cas as a commercial method. As a result, institutional education on this method is primarily essential. On the other hand, not all laboratories have sufficient equipment to run this technology except specialized and facilitated laboratories. Due to the problem of inadequate equipment, it is impossible to conduct this technology in all institutional laboratories.
Moreover, from a genetic perspective, the insufficient aquatic genomic resource is the major limitation of CRISPR technology. Although scientists have vast genetic information about some worldwide, commercially important model species (e.g., Nile Tilapia, Atlantic Salmon), they are insufficient compared to the total number of aquaculture species, which is over 600 according to the FAO [86]. Furthermore, the identification of trait-related genes is required for the genetic functioning of aquatic species to locate which gene should be targeted [86]. On the other hand, the problem of genomic duplication in fish is more than in other aquatic organisms [87]. In addition, another significant issue with using CRISPR-Cas in treating fish disease is off-target mutations at unwanted sites other than the desired on-target sites [88].
There may have some possible solutions to overcome these limitations. First, future aquatic genome-sequencing will be aided by the reduced cost of sequencing, allowing for establishing the essential genetic base. Second, the steps to start introductory institutional courses on CRISPR-Cas can be helpful in the skill development and enhancement of lab facilities. For the solutions of genetic perspectives, increasing refinements in QTL methods will result in more trait-related genes being identified [89]. On the other hand, potential candidates should be targeted for genes that impart advantageous features across species and lines. A well-designed annealing sgRNA may either avoid or identify off-target mutations by comparing it to current genome assemblies [50].
8. Future Perspective and Approaches
Although CRISPR-Cas technology is relatively new, it can be undoubtedly said that scientists will rely heavily on it in the future to diagnose and prevent fish diseases. With time, the versatile application of this technology is turning it into a multi-dimensional technology. It is possible to paint an imaginary picture of the future perspective and approaches of CRISPR-Cas. In this step, an attempt can be made to assess this technology’s future status and needs, based on several aspects of fish disease diagnosis and health management.
Antibiotics’ massive selection pressures brought on by antibiotic exposure cause commensal and pathogenic microorganisms to develop and propagate antibiotic resistance. This strategy is paradoxical for preventing the fast evolution of new antibiotic-resistant organisms because of the lengthy process of discovering new antibiotics. To deal with diseases brought on by resistant superbugs, alternative strategies including creating nucleic acid-based anti-bacterial therapeutics, anti-bacterial peptides, bacteriocins, anti-virulence chemicals, and bacteriophage therapies should be used. To address antibiotic resistance in this situation, scientists have already begun to use the recently popular CRISPR-Cas system [90]. Antibiotic-resistant superbugs are one of the major concerns today, but CRISPR technology is expected to protect from this problem if used properly;
Antibiotics target cellular processes or activities, such as nucleic acid synthesis and cell membrane formation, to impact specialized bacterial mechanisms. These processes cannot destroy specific pathogens in the diverse microbial community—antibiotics damage both the members of the beneficial microbiota and the bacteria that cause infections. There is currently no antibiotic method that targets exclusively pathogenic bacteria. The use of antibiotics nowadays is not species-specific. The CRISPR-Cas9 gene-editing technique and its applications against bacteria will be a crucial strategy to stop the clonal proliferation of dangerous bacteria, offering a novel remedy to the world-wide issue [91];
Among all of the pathogens that cause disease in the fish body, viral diseases can be considered the most dangerous. In particular, fish diseases by RNA viruses cause the most suffering to scientists and fish farmers. From that point of view, since RNA viruses show mutations or Single Nucleotide Polymorphisms (SNP) so frequently, any preventive measures designed to target a particular virus may no longer work after the mutation or SNP. The CRISPR-Cas method can play a vital role in solving this problem in the future. CRISPR-Cas technology has already experimented with the RNA virus targeting method for red-spotted grouper nervous necrosis virus (RGNNV) in fish [13]. Scientists found success in this experiment by using the CasRx-crRNA complex;
Apart from these, CRISPR has also been applied for anti-parasitic action. Scientists are also succeeding in this area [25]. Moreover, using this technology, genetically improved or modified species can be created that will be born with high immunity from the beginning of life.
9. Conclusions
Aquaculture offers microbial populations semi-natural and generally ideal environments. For this reason, pathogenic attacks in aquaculture are always a significant issue that have a direct and adverse effect on production, as the fish are considered an easy host for causative agents. Among all preventive measures, we have focused on the CRISPR-Cas method in this review paper because of the popularity and dependable image created by the vast research interests and practice of CRISPR-Cas in disease management sectors. The CRISPR-Cas system has improved genome-editing technology and shown significant promise in controlling aquaculture diseases. We have reviewed the recent research and projected the uses of CRISPR-Cas in the aquaculture industry. We have discussed the case studies that have previously been conducted on the use of CRISPR-Cas for creating anti-parasitic RNA, as well as the advancement of the CasRx-crRNA complex against fish RNA-virus. Although the use of CRISPR-Cas in aquaculture disease research is still in its early stages compared to its usage in biomedical research, we have reviewed its limits and applications as a new method and attempted to relate it with bioinformatics.
Acknowledgments
All the authors of the manuscript are greatly thankful to Taif University, Saudi Arabia for the financial support.
Author Contributions
Conceptualization, M.A.F., S.I.I. and M.A. (Mazen Almehmadiand); methodology, M.A. (Mazen Almehmadiand); software, M.A.F. and N.H.; validation, S.I.I., N.H. and M.A. (Mazen Almehmadiand); formal analysis, M.A. (Mamdouh Allahyani); investigation, A.S.; resources, A.A.A.; data curation, S.I.I.; writing—original draft preparation, S.I.I., M.A.F. and N.H.; writing—review and editing, M.A. (Mamdouh Allahyani); visualization, S.I.I.; supervision, S.I.I.; project administration, M.A. (Mazen Almehmadiand); funding acquisition, M.A. (Mazen Almehmadiand). All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rodger H.D. Fish Disease Causing Economic Impact in Global Aquaculture. In: Adams A., editor. Fish Vaccines. Springer Basel; Basel, Switzerland: 2016. pp. 1–34. [Google Scholar]
- 2.Peeler E., Feist S. Human intervention in freshwater ecosystems drives disease emergence. Freshw. Biol. 2011;56:705–716. doi: 10.1111/j.1365-2427.2011.02572.x. [DOI] [Google Scholar]
- 3.Diwan A.D., Ninawe A.S., Harke S.N. Gene editing (CRISPR-Cas) technology and fisheries sector. Can. J. Microbiol. 2017;1:65–72. doi: 10.24870/cjb.2017-000108. [DOI] [Google Scholar]
- 4.Paladini G., Longshaw M., Gustinelli A., Shinn A.P. Parasitic Diseases in Aquaculture: Their Biology, Diagnosis, and Control. John Wiley & Sons Ltd.; Hoboken, NJ, USA: 2017. pp. 37–107. [Google Scholar]
- 5.Frantz A., Perga M.E., Guillard J. Parasitic versus nutritional regulation of natural fish populations. Ecol. Evol. 2018;8:8713–8725. doi: 10.1002/ece3.4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orobets V., Lisovets E., Zabashta S., Ermakov A. Control of fish parasites in aquaculture. IOP Conf. Ser. Earth Environ. Sci. 2019;403:012065. doi: 10.1088/1755-1315/403/1/012065. [DOI] [Google Scholar]
- 7.Cabello F.C. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and the environment. Environ. Microbiol. 2006;8:1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x. [DOI] [PubMed] [Google Scholar]
- 8.Cabello F.C., Godfrey H.P., Tomova A., Ivanova L., Dölz H., Millanao A., Buschmann A.H. Antimicrobial use in aquaculture re-examined: Its relevance to antimicrobial resistance and animal and human health. Environ. Microbiol. 2013;15:1917–1942. doi: 10.1111/1462-2920.12134. [DOI] [PubMed] [Google Scholar]
- 9.Sarmah A.K., Meyer M.T., Boxall A.B.A. A global perspective on the use, sales, exposure pathways, occurrence, fate, and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65:725–759. doi: 10.1016/j.chemosphere.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 10.Larsson D.J., Andremont A., Bengtsson-Palme J., Brandt K.K., de Roda Husman A.M., Fagerstedt P., Fick J., Flach C.F., Gaze W.H., Kuroda M., et al. Critical knowledge gaps and research need related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018;117:132–138. doi: 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 11.Seruggia D., Montoliu L. The new CRISPR-Cas system: RNA-guided genome engineering to efficiently produce any desired genetic alteration in animals. Transgenic Res. 2014;23:707–716. doi: 10.1007/s11248-014-9823-y. [DOI] [PubMed] [Google Scholar]
- 12.Ansai S., Mochida K., Fujimoto S., Mokodongan D.F., Sumarto B.K., Masengi K.W., Hadiaty R.K., Nagano A.J., Toyoda A., Naruse K., et al. Genome editing reveals fitness effects of a gene for sexual dichromatism in Sulawesian fishes. Nat. Commun. 2021;12:1–13. doi: 10.1038/s41467-021-21697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Liu Y., Han C., Yang M., Huang F., Duan X., Wang S., Yu Y., Liu J., Yang H., et al. Efficient RNA Virus Targeting via CRISPR/CasRx in Fish. J. Virol. 2021;95:e0046121. doi: 10.1128/JVI.00461-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian Y., Liu T., Liu C., Xu Q., Liu Q. Pathogen detection strategy based on CRISPR. Microchem. J. 2022;174:107036. doi: 10.1016/j.microc.2021.107036. [DOI] [Google Scholar]
- 15.Gratacap R.L., Wargelius A., Edvardsen R.B., Houston R.D. Potential of Genome Editing to Improve Aquaculture Breeding and Production. Trends Genet. 2019;35:672–684. doi: 10.1016/j.tig.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Wargelius A. Application of genome editing in aquatic farm animals: Atlantic salmon. Transgenic Res. 2019;28:101–105. doi: 10.1007/s11248-019-00163-0. [DOI] [PubMed] [Google Scholar]
- 17.Henry M., Debarbieux L. Tools from viruses: Bacteriophage successes and beyond. Virology. 2012;434:151–161. doi: 10.1016/j.virol.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 19.Touchon M., Bernheim A., Rocha E.P. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–2754. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castillo D., Espejo R., Middelboe M. Genomic structure of bacteriophage 6H and its distribution as prophage in Flavobacterium psychrophilum strains. FEMS Microbiol. Lett. 2014;351:51–58. doi: 10.1111/1574-6968.12342. [DOI] [PubMed] [Google Scholar]
- 21.Kalatzis P.G., Rørbo N., Castillo D., Mauritzen J.J., Jørgensen J., Kokkari C., Middelboe M. Stumbling across the same phage: Comparative genomics of widespread temperate phages infecting the fish pathogen Vibrio anguillarum. Viruses. 2017;9:122. doi: 10.3390/v9050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Almeida G.M., Laanto E., Ashrafi R., Sundberg L.R. Bacteriophage adherence to mucus mediates preventive protection against pathogenic bacteria. MBio. 2019;10:e01984-19. doi: 10.1128/mBio.01984-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan Y., Sun Y., Wang Y., Zhang Z. Barcode sequence could be a good target for developing a species-specific anti-parasite agent based on CRISPR-Cas9. FASEB J. 2020;34:9393–9404. doi: 10.1096/fj.202000118RR. [DOI] [PubMed] [Google Scholar]
- 24.Maurer-Alcalá X.X., Knight R., Katz L.A. Exploration of the germline genome of the ciliate Chilodonella uncinata through single-cell omics (transcriptomics and genomics) MBio. 2018;9:e01836-17. doi: 10.1128/mBio.01836-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y., Lai J., Zhang X., Wang Y., Zhang Z. Development of an RNA anti-parasite based on CRISPR-Cas9 against Chilodonella piscicola. Aquaculture. 2022;552:738025. doi: 10.1016/j.aquaculture.2022.738025. [DOI] [Google Scholar]
- 26.Majeed M., Soliman H., Kumar G., El-Matbouli M., Saleh M. Editing the genome of Aphanomyces invadans using CRISPR/Cas9. Parasites Vectors. 2018;11:1–10. doi: 10.1186/s13071-018-3134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abudayyeh O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B., Kellner M.J., Regev A., et al. RNA targeting with CRISPR-Cas13a. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox D.B., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freije C.A., Myhrvold C., Boehm C.K., Lin A.E., Welch N.L., Carter A., Metsky H.C., Luo C.Y., Abudayyeh O.O., Gootenberg J.S., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76:826–837.e11. doi: 10.1016/j.molcel.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makarova K.S., Haft D.H., Barrangou R., Brouns S.J., Charpentier E., Horvath P., Moineau S., Mojica F.J., Wolf Y.I., Yakunin A.F., et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horigome C., Oma Y., Konishi T., Schmid R., Marcomini I., Hauer M.H., Dion V., Harata M., Gasser S.M. SWR1. chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell. 2014;55:626–639. doi: 10.1016/j.molcel.2014.06.027. [DOI] [PubMed] [Google Scholar]
- 34.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173:665–676.e14. doi: 10.1016/j.cell.2018.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bandín I., Souto S. Betanodavirus and VER Disease: A 30-year. Pathogens. 2020;9:106. doi: 10.3390/pathogens9020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishizawa T., Furuhashi M., Nagai T., Nakai T., Muroga K. Genomic classification of fish nodaviruses by molecular phylogenetic analysis of the coat protein gene. Appl. Environ. Microbiol. 1997;63:1633–1636. doi: 10.1128/aem.63.4.1633-1636.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahul Hameed A.S., Ninawe A.S., Nakai T., Chi S.C., Johnson K.L. ICTV Report Consortium. ICTV Virus Taxonomy Profile: Nodaviridae. J. Gen. Virol. 2019;100:3–4. doi: 10.1099/jgv.0.001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chi S., Lo B., Lin S. Characterization of grouper nervous necrosis virus (GNNV) J. Fish Dis. 2001;24:3–13. doi: 10.1046/j.1365-2761.2001.00256.x. [DOI] [Google Scholar]
- 39.Zhou L., Wang S., Yu Q., Wei S., Liu M., Wei J., Qin Q. Characterization of novel aptamers specifically directed to red-spotted grouper nervous necrosis virus (RGNNV)-infected cells for mediating targeted siRNA delivery. Front. Microbiol. 2020;11:660. doi: 10.3389/fmicb.2020.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahmed N., Thompson S., Glaser M. Global aquaculture productivity, environmental sustainability, and climate change adaptability. Environ. Manag. 2019;63:159–172. doi: 10.1007/s00267-018-1117-3. [DOI] [PubMed] [Google Scholar]
- 41.Jin Y.H., Liao B., Migaud H., Davie A. Physiological impact and comparison of mutant screening methods in piwil2 KO founder Nile tilapia produced by CRISPR/Cas9 system. Sci. Rep. 2020;10:12600. doi: 10.1038/s41598-020-69421-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Straume A.H., Kjærner-Semb E., Skaftnesmo K.O., Güralp H., Lillico S., Wargelius A., Edvardsen R.B. A refinement to gene editing in Atlantic salmon using asymmetrical oligonucleotide donors. bioRxiv. 2021 doi: 10.1101/2021.02.08.430296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun Y., Zheng G.D., Nissa M., Chen J., Zou S.M. Disruption of mstna and mstnb gene through CRISPR/Cas9 leads to elevated muscle mass in blunt snout bream (Megalobrama amblycephala) Aquaculture. 2020;528:735597. doi: 10.1016/j.aquaculture.2020.735597. [DOI] [Google Scholar]
- 44.Xu X., Cao X., Gao J. Production of a mutant of large-scale loach Paramisgurnus dabryanus with skin pigmentation loss by genome editing with CRISPR/Cas9 system. Transgenic Res. 2019;28:341–356. doi: 10.1007/s11248-019-00125-6. [DOI] [PubMed] [Google Scholar]
- 45.Chen H., Wang J., Du J., Si Z., Yang H., Xu X., Wang C. ASIP disruption via CRISPR/Cas9 system induces black patches dispersion in Oujiang color common carp. Aquaculture. 2019;498:230–235. doi: 10.1016/j.aquaculture.2018.08.057. [DOI] [Google Scholar]
- 46.Kim J., Cho J.Y., Kim J.W., Kim D.G., Nam B.H., Kim B.S., Kim W.J., Kim Y.O., Cheong J., Kong H.J. Molecular characterization of paralichthys olivaceus MAF1 and its potential role as an anti-viral hemorrhagic septicemia virus factor in hirame natural embryo cells. Int. J. Mol. Sci. 2021;22:1353. doi: 10.3390/ijms22031353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gratacap R.L., Regan T., Dehler C.E., Martin S.A., Boudinot P., Collet B., Houston R.D. Efficient CRISPR/Cas9 genome editing in a salmonid fish cell line using a lentivirus delivery system. BMC Biotechnol. 2020;20:35. doi: 10.1186/s12896-020-00626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Datsomor A.K., Olsen R.E., Zic N., Madaro A., Bones A.M., Edvardsen R.B., Wargelius A., Winge P. CRISPR/Cas9-mediated editing of Δ5 and Δ6 desaturases impairs Δ8-desaturation and docosahexaenoic acid synthesis in Atlantic salmon (Salmo salar L.) Sci. Rep. 2019;9:16888. doi: 10.1038/s41598-019-53316-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Datsomor A.K., Zic N., Li K., Olsen R.E., Jin Y., Vik J.O., Winge P. CRISPR/Cas9-mediated ablation of elovl2 in Atlantic salmon (Salmo salar L.) inhibits elongation of polyunsaturated fatty acids and induces Srebp-1 and target genes. Sci. Rep. 2019;9:7533. doi: 10.1038/s41598-019-43862-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blix T.B., Dalmo R.A., Wargelius A., Myhr A.I. Genome editing on finfish: Current status and implications for sustainability. Rev. Aquac. 2021;13:2344–2363. doi: 10.1111/raq.12571. [DOI] [Google Scholar]
- 51.Elaswad A., Khalil K., Cline D., Page-McCaw P., Chen W., Michel M., Dunham R. Microinjection of CRISPR/Cas9 protein into channel catfish, Ictalurus punctatus, embryos for gene editing. JoVE (J. Vis. Exp.) 2018;131:e56275. doi: 10.3791/56275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elaswad A., Khalil K., Ye Z., Liu Z., Liu S., Peatman E., Odin R., Vo K., Drescher D., Gosh K., et al. Effects of CRISPR/Cas9 dosage on TICAM1 and RBL gene mutation rate, embryonic development, hatchability and fry survival in channel catfish. Sci. Rep. 2018;8:16499. doi: 10.1038/s41598-018-34738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simora R.M.C., Xing D., Bangs M.R., Wang W., Ma X., Su B., Dunham R.A. CRISPR/Cas9-mediated knock-in of alligator cathelicidin gene in a non-coding region of channel catfish genome. Sci. Rep. 2020;10:22271. doi: 10.1038/s41598-020-79409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunham R.A., Warr G.W., Nichols A., Duncan P.L., Argue B., Middleton D., Kucuktas H. Enhanced bacterial disease resistance of transgenic channel catfish Ictalurus punctatus possessing cecropin genes. Mar. Biotechnol. 2002;4:338–344. doi: 10.1007/s10126-002-0024-y. [DOI] [PubMed] [Google Scholar]
- 55.Elaswad A., Dunham R. Disease reduction in aquaculture with genetic and genomic technology: Current and future approaches. Rev. Aquac. 2018;10:876–898. doi: 10.1111/raq.12205. [DOI] [Google Scholar]
- 56.Katayama H., Nagata K., Ohira T., Yumoto F., Tanokura M., Nagasawa H. The solution structure of molt-inhibiting hormone from the Kuruma prawn Marsupenaeus japonicus. J. Biol. Chem. 2003;278:9620–9623. doi: 10.1074/jbc.M212962200. [DOI] [PubMed] [Google Scholar]
- 57.Katayama H., Ohira T., Nagata S., Nagasawa H. Structure-activity relationship of crustacean molt-inhibiting hormone from the Kuruma prawn Marsupenaeus japonicus. Biochemistry. 2004;43:9629–9635. doi: 10.1021/bi049433v. [DOI] [PubMed] [Google Scholar]
- 58.Nakatsuji T., Lee C.-Y., Watson R.D. Crustacean molt-inhibiting hormone: Structure, function, and cellular mode of action. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2009;152:139–148. doi: 10.1016/j.cbpa.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 59.Treerattrakool S., Panyim S., Udomkit A. Induction of ovarian maturation and spawning in Penaeus monodon broodstock by double-stranded RNA. Mar. Biotechnol. 2011;13:163–169. doi: 10.1007/s10126-010-9276-0. [DOI] [PubMed] [Google Scholar]
- 60.Das R., Krishna G., Priyadarshi H., Gireesh-Babu P., Pavan-Kumar A., Rajendran K.V., Chaudhari A. Captive maturation studies in Penaeus monodon by GIH silencing using constitutively expressed long hairpin RNA. Aquaculture. 2015;448:512–520. doi: 10.1016/j.aquaculture.2015.06.036. [DOI] [Google Scholar]
- 61.Lesk A. Introduction to Bioinformatics. Oxford University Press; Oxford, UK: 2019. [Google Scholar]
- 62.Choudhary S., Ubale A., Padiya J., Mikkilineni V. Application of Bioinformatics Tools in CRISPR/Cas, in CRISPR/Cas Genome Editing. Springer; Berlin/Heidelberg, Germany: 2020. pp. 31–52. [Google Scholar]
- 63.Belhaj K., Chaparro-Garcia A., Kamoun S., Nekrasov V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods. 2013;9:39. doi: 10.1186/1746-4811-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bortesi L., Fischer R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015;33:41–52. doi: 10.1016/j.biotechadv.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 65.Jung C., Capistrano-Gossmann G., Braatz J., Sashidhar N., Melzer S. Recent developments in genome editing and applications in plant breeding. Plant Breed. 2018;137:1–9. doi: 10.1111/pbr.12526. [DOI] [Google Scholar]
- 66.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Joung J.K. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park J., Bae S., Kim J.-S. Cas-Designer: A web-based tool for the choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31:4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- 69.Ata H., Ekstrom T.L., Martínez-Gálvez G., Mann C.M., Dvornikov A.V., Schaefbauer K.J., Ma A.C., Dobbs D., Clark K.J., Ekker S.C. Robust activation of microhomology-mediated end joining for precision gene editing applications. PLoS Genet. 2018;14:e1007652. doi: 10.1371/journal.pgen.1007652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu H., Wei Z., Dominguez A., Li Y., Wang X., Qi L.S. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression, and activation. Bioinformatics. 2015;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clement K., Rees H., Canver M.C., Gehrke J.M., Farouni R., Hsu J.Y., Cole M., Liu D.R., Joung J.K., Bauer D.E., et al. Analysis and comparison of genome editing using CRISPResso2. bioRxiv. 2018:392217. doi: 10.1101/392217. [DOI] [Google Scholar]
- 72.Park J., Lim K., Kim J.S., Bae S. Cas-analyzer: An online tool for assessing genome editing results using NGS data. Bioinformatics. 2017;33:286–288. doi: 10.1093/bioinformatics/btw561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Güell M., Yang L., Church G.M. Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA) Bioinformatics. 2014;30:2968–2970. doi: 10.1093/bioinformatics/btu427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Varshney G.K., Zhang S., Pei W., Adomako-Ankomah A., Fohtung J., Schaffer K., Carrington B., Maskeri A., Slevin C., Wolfsberg T., et al. CRISPRz: A database of zebrafish validated sgRNAs. Nucleic Acids Res. 2016;44:D822–D826. doi: 10.1093/nar/gkv998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018;563:646–651. doi: 10.1038/s41586-018-0686-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Páleníková P., Khodak A., Kiselev V., Kosicki M., et al. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2019;37:64–72. doi: 10.1038/nbt.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kantor A., McClements M.E., MacLaren R.E. CRISPR-Cas9 DNA base-editing and prime-editing. Int. J. Mol. Sci. 2020;21:6240. doi: 10.3390/ijms21176240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hwang G.H., Park J., Lim K., Kim S., Yu J., Yu E., Kim S.T., Eils R., Kim J.S., Bae S. Web-based design and analysis tools for CRISPR base editing. BMC Bioinform. 2018;19:542. doi: 10.1186/s12859-018-2585-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chatterjee P., Jakimo N., Jacobson J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018;4:eaau0766. doi: 10.1126/sciadv.aau0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsu P.D., Scott D.A., Weinstein J.A., Ran F., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kluesner M.G., Nedveck D.A., Lahr W.S., Garbe J.R., Abrahante J.E., Webber B.R., Moriarity B.S. EditR: A method to quantify base editing from Sanger sequencing. CRISPR J. 2018;1:239–250. doi: 10.1089/crispr.2018.0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Billon P., Bryant E.E., Joseph S.A., Nambiar T.S., Hayward S.B., Rothstein R., Ciccia A. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol. Cell. 2017;67:1068–1079.e4. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dandage R., Després P.C., Yachie N., Landry C.R. beditor: A computational workflow for designing libraries of guide RNAs for CRISPR-mediated base editing. Genetics. 2019;212:377–385. doi: 10.1534/genetics.119.302089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu K., Petree C., Requena T., Varshney P., Varshney G.K. Expanding the CRISPR toolbox in zebrafish for studying development and disease. Front. Cell Dev. Biol. 2019;7:13. doi: 10.3389/fcell.2019.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carroll D., Charo R. The societal opportunities and challenges of genome editing. Genome Biol. 2015;16:242. doi: 10.1186/s13059-015-0812-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Okoli A.S., Blix T., Myhr A.I., Xu W., Xu X. Sustainable use of CRISPR/Cas in fish aquaculture: The biosafety perspective. Transgenic Res. 2021;31:1–21. doi: 10.1007/s11248-021-00274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Glasauer S.M., Neuhauss S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014;289:1045–1060. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X.H., Tee L.Y., Wang X.G., Huang Q.S., Yang S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther.-Nucleic Acids. 2015;4:e264. doi: 10.1038/mtna.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Houston R.D., Bean T.P., Macqueen D.J., Gundappa M.K., Jin Y.H., Jenkins T.L., Selly S.L., Martin S.A., Stevens J.R., Santos E.M., et al. Harnessing genomics to fast-track genetic improvement in aquaculture. Nat. Rev. Genet. 2020;21:389–409. doi: 10.1038/s41576-020-0227-y. [DOI] [PubMed] [Google Scholar]
- 90.Aslam B., Rasool M., Idris A., Muzammil S., Alvi R.F., Khurshid M., Rasool M.H., Zhang D., Ma Z., Baloch Z. CRISPR-Cas system: A potential alternative tool to cope antibiotic resistance. Antimicrob. Resist. Infect. Control. 2020;9:131. doi: 10.1186/s13756-020-00795-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ates A., Tastan C., Ermertcan S. Gene Editing. Gene Ed. J. Young Open Free. 2020;1:30–35. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.