Abstract
Benzene is a human carcinogen whose exposure to concentrations below 1 ppm (3.19 mg·m−3) is associated with myelotoxic effects. The determination of biomarkers such as trans-trans muconic acid (AttM) and S-phenylmercapturic acid (SPMA) show exposure without reflecting the toxic effects of benzene. For this reason, in this study, the urinary metabolome of individuals exposed to low concentrations of benzene was investigated, with the aim of understanding the biological response to exposure to this xenobiotic and identifying metabolites correlated with the toxic effects induced by it. Ultra-efficient liquid chromatography coupled to a quadrupole-time-of-flight mass spectrometer (UHPLC-ESI-Q-ToF-MS) was used to identify metabolites in the urine of environmentally (n = 28) and occupationally exposed (n = 32) to benzene (mean of 22.1 μg·m−3 and 31.8 μg·m−3, respectively). Non-targeted metabolomics analysis by PLS-DA revealed nine urinary metabolites discriminating between groups and statistically correlated with oxidative damage (MDA, thiol) and genetic material (chromosomal aberrations) induced by the hydrocarbon. The analysis of metabolic pathways revealed important alterations in lipid metabolism. These results point to the involvement of alterations in lipid metabolism in the mechanisms of cytotoxic and genotoxic action of benzene. Furthermore, this study proves the potential of metabolomics to provide relevant information to understand the biological response to exposure to xenobiotics and identify early effect biomarkers.
Keywords: benzene, metabolomic untargeted, UHPLC-ESI-Q-TOF-MS, environmental toxicology, occupational toxicology
1. Introduction
Benzene is a ubiquitous pollutant, and human exposure to it occurs through occupational or environmental contamination. Benzene is used in the steel and oil industries, in addition to being released into the environment by emissions from motor vehicles (concentrations can reach up to 349 µg m−3 in high-traffic urban centers). In addition, cigarettes are another important anthropogenic source of environmental exposure [1]. It is a xenobiotic classified as a human carcinogen by the International Agency for Research on Cancer—IARC [2]. Exposure to benzene is associated with myelotoxic effects, which can cause qualitative and quantitative disturbances of blood cells such as leukopenia, thrombocytopenia, pancytopenia, aplastic anemia, and leukemia [3]. Studies have shown that benzene toxicity is greater at exposures associated with airborne concentrations below 0.1 ppm [4,5]. However, the mechanisms involved in its toxic action are still not fully elucidated.
Considering the potential risk to human health, biological monitoring is a useful exposure assessment tool to prevent adverse effects. Usually the biomarkers trans, trans muconic acid (AttM) and S-phenylmercapturic acid (S-PMA) in urine are used for this purpose [6,7,8]. However, this strategy does not reflect the adverse health effects resulting from exposure to low concentrations of this toxic agent. Therefore, an exploratory and comprehensive evaluation of the organism-benzene interaction is necessary; and metabolomics present itself as a useful alternative capable of achieving this goal.
Biomonitoring is used to indicate the occurrence of an internal dose of the chemical and its distribution in the body; which can be useful in measuring health risks. Currently, the assessment of risks arising from chronic exposure to low concentrations of toxic substances or complex mixtures represents a major challenge for modern toxicology. In many cases, the quantitative determination of exposure and effect biomarkers has been shown to be insufficient to fully characterize the harmful effects resulting from this exposure. Added to this, the inability of biomarkers to early indicate the harmful effects induced by the toxic agent [9].
In this sense, the application of “omics” sciences for toxicological evaluation has been proposed and in this context, metabolomics has stood out among the most promising [10,11]. Metabolomics consists of the comprehensive assessment of the metabolic profile of biological compartments and represents an important step toward the discovery of new biomarkers associated with environmental or occupational exposure. However, the limited number of available metabolomic biomarkers does not reflect the extent of research in this field. Among the challenges of metabolomics, we can mention the analytical process, which is critical, in addition to the difficulties in reproducing the data necessary for the replication of studies, or even its continuity after the discovery of biomarkers [12].
Disturbances in metabolic profiles are detected earlier when compared to alterations in genes or proteins [13,14]. Thus, in the field of toxicology, metabolomics represent a holistic understanding of the interaction between organism and xenobiotic, and allows the integration of information about the internal dose of the chemical and the biological response triggered, essential data to assess potential risks [15,16]. Therefore, the metabolomics approach is a powerful tool to deepen our understanding of the biochemical interactions involved in disease development from chemical exposures to the discovery of new biomarkers.
The comprehensive coverage of the chemical diversity of substances and their different concentrations in the human body is a challenge, and for this the concomitant use of different analytical techniques is necessary. Nuclear magnetic resonance—NMR [17,18] and MS-mass spectrometry [19] account for most applications. However, MS offers greater sensitivity and specificity when combined with separation techniques such as liquid or gas chromatography. Liquid chromatography is recognized as the most used technique currently in non-target metabolomics [20,21].
Untargeted metabolomics has been successfully applied in studies of environmental and occupational toxicology, providing relevant information on the mechanisms of toxic action and even identification of biomarkers [22,23,24,25,26,27,28]. However, there are few studies on the elucidation of the metabolic profile in individuals exposed to benzene.
In this study, an exploratory experimental analysis was performed in order to identify whether there are differences in the urinary metabolic profile of individuals occupationally and environmentally exposed to low concentrations of benzene. For the study of the metabolome, ultra-performance liquid chromatography coupled to a quadrupole-time-of-flight electrospray ionization mass spectrometer (UHPLC-ESI-Q-TOF-MS) was used. Multivariate statistical analysis was used to analyze the results and understand the biological response to exposure and, thus, encourage future studies in the search for new biomarkers; more specific and capable of predicting the adverse effects caused by exposure to this chemical contaminant. These new biomarkers can be used in the context of environmental and occupational health, with a preventive purpose, in order to prevent exposed individuals from becoming ill.
2. Materials and Methods
2.1. Population
The study population consisted of 60 workers of both sexes, aged between 18 and 65 years, with a history of exposure to low concentrations of benzene. These workers were part of the population described in the study by Costa-Amaral et al., (2019) [29]. The workers were divided into two groups, according to their exposure to benzene—environmental exposure and occupational exposure. The environmental group was composed of 28 workers who worked as security guards (environmental group) of a research institution located in the north of the city of Rio de Janeiro-Brazil. The occupational group included 32 gas station workers located in the west of the same city.
2.2. Ethical Aspects of Research
Urine samples from the 60 subjects were provided, in accordance with the “Ethical Principles for Medical Research Involving Human Subjects” of the World Medical Association, by the Center for Studies in Occupational Health and Human Ecology of the Oswaldo Cruz Foundation (CESTEH/FIOCRUZ), in Rio de Janeiro. of January Brazil. These samples were part of the FIOCRUZ study on the assessment of occupational exposure to benzene. The study was submitted and approved by the ethics and research committees of the Federal University of Minas Gerais—UFMG (CAAE: 57396116.1.0000.5149, opinion number: 1.691.348) and of FIOCRUZ-RJ (CAAE: 17438013.5.0000.5240, opinion number: 2.974.839).
2.3. Biochemical Biomarkers, Exposure, Oxidative Stress and Genotoxicity
Blood samples were collected to determine hematological parameters (complete blood count + platelets) and biochemical parameters (AST, ALT, total bilirubin and fractions, creatinine, GGT); which were analyzed by the Ambulatory of the National School of Public Health—ENSP/Fiocruz-RJ. The biomarkers of oxidative stress effect (enzyme activity: catalase—CAT; glutathione S transferase—GST; superoxide dismutase—SOD; thiol and malondialdehyde -MDA Group) and genotoxicity (micronucleus, chromosomal aberrations) were performed at the Toxicology Laboratory from the Center for the Study of Workers’ Health and Human Ecology—CESTEH/FIOCRUZ (COSTA-AMARAL, 2019) [29]. The comet assay (EC), another biomarker of genotoxicity; was carried out in the laboratory of Professor Dr. Andrew Richard Collins linked to the Department of Nutrition at the University of Oslo-Norway (COSTA-AMARAL, 2019) [29]. Urine samples were collected for quantification of benzene exposure biomarkers SPMA and AttM, and global metabolomics. AttM was also quantified in the Toxicology Laboratory of CESTEH/FIOCRUZ; while the SPMA and metabolomics analyzes were performed at the Laboratory of Toxicological Analysis (LATO) of the Faculty of Pharmacy at UFMG.
2.4. Standards and Reagents
The chromatographic grade solvents acetonitrile, methanol, formic acid, 2-propanol, and sodium formate were obtained from Sigma-Aldrich (San Luis, MO, USA)®.
2.5. Sample Preparation
Urine samples were frozen at −80 °C to promote metabolic extinction. To perform the analyses, the urine samples were thawed and vortexed for 30 s. Then, 100 µL were diluted in 125 µL of methanol in Eppendorf microtubes and again vortexed for 30 s. The samples were then subjected to centrifugation at 2817× g at 4 °C for 5 min for protein precipitation. Soon after, 200 µL of the supernatant were transferred to properly labeled flasks and diluted in 300 µL of an aqueous solution containing 0.1% methanoic acid. Quality control samples (QCs) were prepared by mixing 20 µL of each urine sample. Sample blanks were also prepared containing all reagents used in the preparation, with the exception of urine.
2.6. Analytical System—UHPLC-ESI-QTOF/MS
For data acquisition, after preparation, urine samples were analyzed by ultra-performance chromatography (NCS-3500RS-Thermo Scientific® model) coupled to a mass spectrometer (Bruker® Q-ToF model) and Bruker® HMDB library Metabolite 2.0. A C18 column (100 mm × 2.1 mm 1.8 µm particle) was employed in the analysis, the temperature was maintained at 40 °C with gradient elution and low flow, as shown in Table 1. The mobile phase consisted of mixing the solutions (A): water with 0.1% formic acid and (B): acetonitrile with 0.1% formic acid. The injection volume was 15 µL. Quality control (QC) samples were interleaved every five urine samples to assess instrument stability.
Table 1.
Gradient elution and rate flow used in the chromatographic separation.
| Time (min) | Rate (mL/min) | % A | % B |
|---|---|---|---|
| 0 | 0.200 | 96.0 | 4.0 |
| 2 | 0.200 | 96.0 | 4.0 |
| 7 | 0.200 | 81.7 | 18.3 |
| 12 | 0.223 | 50.0 | 50 |
| 14 | 0.400 | 0.1 | 99.9 |
| 16 | 0.480 | 0.1 | 99.9 |
| 19 | 0.480 | 96.0 | 4.0 |
| 19.10 | 0.200 | 96.0 | 4.0 |
| 20 | 0.200 | 96.0 | 4.0 |
The mass spectrometer was operated in positive mode, with electrospray ionization (IE), using collision energies of 20 and 50 eV (each 50% of the time). The ionization source was adjusted to 4500 V in positive mode with a potential plate end of −500 V. The source conditions was adjusted to: nebulizer gas (N2) at 5.0 bar; dry gas (N2) at 9.0 L/min; dry temperature at 200 °C; capillary voltage at 4.5 KV. Before analysis, sodium formate solution was used for calibration. Lock mass calibration was performed with 1 mg/mL solution of hexakis (2,2-difluoroethoxy) phosphazene in 2-propanol, generating reference ions for positive ion mode ([M + H]+ = m/z 622).
Data were obtained in a range of m/z 50 to 1200 with an acquisition rate of 4 Hz. Five most intense ions were selected for automatic fragmentation (AutoMS/MS). Data were acquired by the Hystar Application® software version 3.2 and OtofControl® (Bruker Daltonics Corporation, Bremen, Germany) and processed using DataAnalysis® 4.4 and Metaboscape® 5.0 softwares (Bruker Daltonics Corporation, Bremen, Germany).
All samples were analyzed randomly in order to avoid uncertainties and artifacts related to the injection order and to prevent the effect of gradual changes in instrument sensitivity over entire batches.
2.7. Detection and Identification of Non-Target Metabolites
The results files obtained from anlytical system were converted to the mzXML format using the Proteowizard® software (http://proteowizard.sourceforge.net/download.html (accessed on 27 January 2020)). The R® statistical software version 3.6.2 and the XCMS Bioconductor® package (Mahieu, Genenbacher, & Patti, 2016; version 3.10 http://bioconductor.org/packages/release/bioc/html/xcms.html (accessed on 27 January 2020)) [30,31] were used for the pre-treatment of the data and creating the data matrix. The results were analyzed individually using the algorithm centWave in XCMS®. The data were submitted to logarithmic transformation and Pareto scheduling. The peak intensities of each sample were normalized by the corresponding creatinine concentration; using MetaboAnalyst® version 5.0 (https://www.metaboanalyst.ca/ (accessed on 27 January 2020)) [32].
Multivariate statistical analysis using the principal component analysis (PCA) and discriminant analysis by partial least squares (PLS-DA) tests was used to identify the metabolic differences in the urine of both groups. Cross-validation were used to validate the PLS-DA model. All of these analyzes, as well as the identification of the metabolites as potential biomarkers, were also performed in MetaboAnalyst® 5.0, using the following criteria: variable importance plot (VIP) scores of the variables > 2; p-values between analyzed groups < 0.05, FDR < 0.01 and metabolites significantly correlated with oxidative stress and genotoxicity biomarkers (p < 0.05).
Compounds identification was performed with a narrow tolerance of 3.0 mDa for mass accuracy, mSigma of 40 for isotope standard and MS/MS score of 700, using Bruker Metabobase Personal Library 3.0® and Bruker HMDB Metabolite Library 2.0® (MSI level 2 [33]). For unidentified metabolites, molecular formulas were generated using Compound Crawler (Metaboscape® 5.0) with close tolerance of 3 mDa and mSigma of 30; considering CHNOPS as elements of composition. For these, the putative identification in public databases such as—Human Metabolome Database (HMDB®), Metlin®, and LIPIDMAPS® was performed by searching the m/zs, based on the accurate mass and mass spectrometric fragmentation patterns, and maximum mass error of 5 ppm.
2.8. Statistical Analysis
The Shapiro–Wilk test was used to assess whether the quantitative variables were normally distributed. To compare the normally distributed variables, the T test was used; for the others, the Mann–Whitney test was used. To compare the qualitative variables, the Chi-Square and Fisher’s Exact tests were used. Differences in urinary metabolite concentration between groups were investigated using the Mann–Whitney test. Multiple linear regression was used to explain the variability of effect biomarkers as a function of urinary metabolites. The linear regression model used was:
where
-
-
is the value of the oxidative stress biomarker of the ith individual;
-
-
is the kth metabolite of the ith individual;
-
-
is the coefficient of the kth metabolite;
-
-
is a random error, which follows normal distribution with mean 0 and standard deviation σ.
Through the regression study, we sought to identify the most important urinary metabolites to explain the variance of the oxidative stress biomarker. The stepwise backward elimination [34] method, using the Akaike Information Criterion [35], or AIC, as a criterion was used to identify the combination of metabolites that minimized the AIC and led to a more adjusted model, capable of explaining the variance of the effect biomarkers.
Finally, the correlation between metabolites and exposure biomarkers (SPMA and AttM, oxidative stress markers and chromosomal aberration rates) was verified using Spearman’s Correlation and illustrated by scatter diagrams. The software used in the analyzes was R® (version 4.0.2) and all statistical tests were performed assuming a significance level of 5%.
3. Results
3.1. Sociodemographic Characteristics and Results of Biological Monitoring
The descriptive analysis of sociodemographic characteristics and the results of the comparison tests between the groups of workers are shown in Table S1. The occupationally exposed population consisted of 72.5% gas station attendants; 13.7% employees with executive assignments; 5.9% general service assistants; 5.9% lubrication technician; and 2.0% convenience store clerk. The mean concentration of benzene in the atmospheric air of the evaluated stations was 31.8 μg·m−3. On the other hand, the population environmentally exposed to benzene was composed of security guards (96.6%) and doormen (3.4%), who were exposed to an average concentration of 22.1 μg·m−3 of benzene in the air atmosphere. All participants from both groups had been in this occupation for more than 3 months.
Working time in the current occupation varied greatly between groups, as shown in Table S2. In the environmental group, workers’ time in their current occupation ranged from 10 to 348 months, with a mean of 152.7 and a standard deviation of 99.2. In the occupational group, the working time ranged from 12 to 408, with a mean of 117.8 and a standard deviation of 122.8.
The complete description of the results of the descriptive analysis of the sociodemographic characteristics, hematological, biochemical parameters, oxidative stress, and genotoxicity of the workers participating in this study were previously described by Costa-Amaral et al., (2019) [29], and a summary is presented in Table S3. Oxidative stress biomarkers THIOL, MDA, and chromosomal abnormalities differed significantly between groups. Thus, oxidative damage and chromosomal aberrations were the main toxic effects induced by exposure to benzene in the population studied. For this reason, the correlation of potential metabolomic biomarkers in urine with such toxic effects was investigated in the present study.
3.2. Identification of the Urinary Metabolic Profile of Individuals Occupationally and Environmentally Exposed to Benzene
A total of 2361 molecular characteristics were detected, of which 1522 showed significant differences (p < 0.05) between occupational and environmentally exposed groups. Of these, 1267 compounds were identified; which belong to different chemical categories, as shown in Figure 1. There was a predominance of amino acids and derivatives (44%) and lipids (29%).
Figure 1.
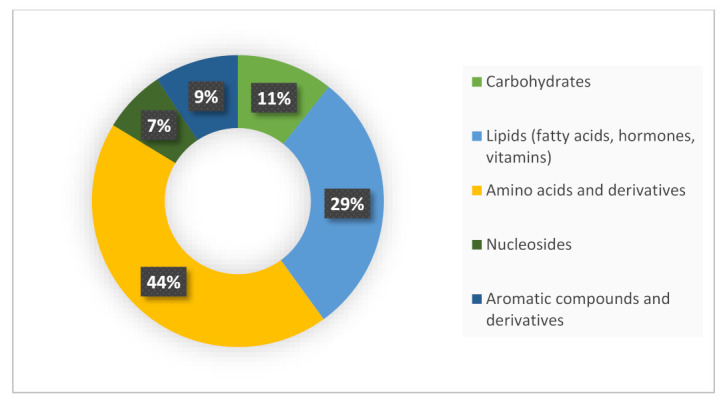
Chemical categories of metabolites identified in the urine of occupational and environmental workers exposed to benzene, investigated in this study.
The analytical results were analyzed by unsupervised pattern recognition methods—PCA (principal component analysis) and supervised—PLS-DA (partial least squares discriminative analysis). PCA analysis was used to assess the quality of data acquisition, and the strong clustering of QC samples, observed in the center of the graph (Figure 2), indicated analytical reproducibility and reliability of the results. The PCA model constructed also revealed a tenuous separation between the groups of workers with low environmental and occupational exposure to benzene.
Figure 2.
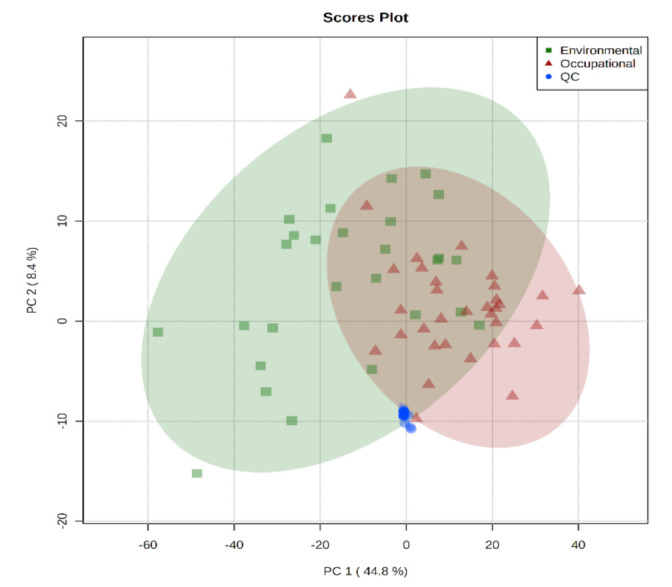
Principal component analysis-PCA (PC 1 × PC 2) of urine samples from workers exposed and occupationally to benzene, with provision for environmental quality control samples (QCs).
The supervised pattern recognition methods—discriminant partial least squares analysis (PLS-DA) and discriminant partial least squares analysis (OPLS-DA); were used to identify the metabolites capable of distinguishing the groups of workers investigated in this study. The PLS-DA model constructed is shown in Figure 3. The PLS-DA revealed a tendency to cluster samples from the same group; especially in the occupational group. This indicates that there are differences in the urinary metabolic profile between the groups of workers; and that the metabolic disorder associated with exposure differs between them. The constructed PLS-DA and OPLS-DA models were validated through the cross-validation test and the permutation test, respectively; and both presented satisfactory coefficients.
Figure 3.
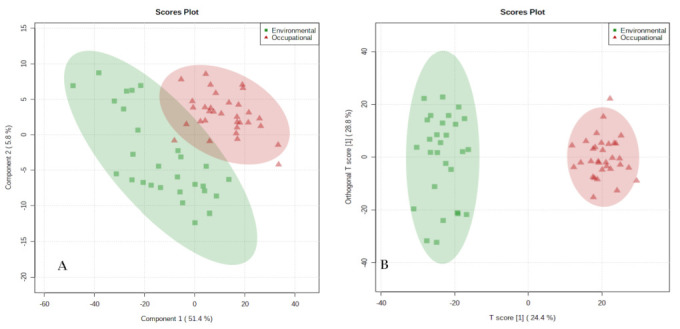
Partial least squares discriminant analysis (PLS−DA) and partial orthogonal discriminant analysis (OPLS−DA) of urine samples from workers exposed to environmental and occupational benzene, analyzed by UHPLC−ESI−Q−ToF−MS. Note: Red triangles represent samples from the occupational group and the green squares represent samples from the environmental group. (A): PLS−DA model, validation parameters: R2 = 0.98, Q2 = 0.87; (B): OPLS−DA model.
Based on the criteria for identifying the discriminating metabolites described in item 2.5, thirty-eight molecular characteristics that contributed most to the discrimination between groups were selected. Table 2 shows the identity of the metabolites, as well as their m/z ratio and chemical classification. The intensities of metabolites between groups were compared by the Mann–Whitney test, and all showed a statistically significant difference (p < 0.05). The VIP value, obtained in the PLS-DA model, indicates the importance of each metabolite in the discrimination between groups.
Table 2.
Identification of urinary metabolic biomarkers related to exposure to low concentrations of benzene.
| Metabolites | M | VIP a Score | p b Values | Chemical Category |
|---|---|---|---|---|
| Phenylalanylhydroxyproline | 279.13 | 2.42 | 0.0000 | Peptide |
| Tetrahydropteroyltri-L glutamic acid | 703.25 | 2.29 | 0.0001 | Tetrahydrofolic acid |
| Trp Gln Asp Cys Glu | 679.22 | 2.22 | 0.0000 | Peptide |
| Testosterone glucuronide | 465.24 | 2.19 | 0.0000 | Steroid glucuronide |
| 7alpha-Hydroxy-3-oxo-5beta-cholan-24-oic acid | 390.27 | 2.18 | 0.0000 | Bile acid |
| Phosphatidylcholine(44:6) | 889.65 | 2.15 | 0.0000 | Phosphatidylcholine |
| Asp Asp Phe Hys | 532.19 | 2.14 | 0.0000 | Peptide |
| Phosphatidylcholine(42:6) | 861.62 | 2.11 | 00000 | Glycerophospholipid |
| Phosphatidylcholine(42:2) | 869.68 | 2.10 | 0.0000 | Glycerophospholipid |
| Phosphatidylcholine(22:2) | 825.66 | 2.10 | 0.0000 | Glycerophospholipid |
| Folic acid | 441.13 | 2.10 | 0.0000 | Pterins |
| Phosphatidylethanolamine PGE2/22:2(13Z,16Z) | 867.56 | 2.10 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(40:3) | 891.64 | 2.10 | 0.0000 | Phosphatidylcholine |
| Cys Hys Ser Trp | 531.19 | 2.10 | 0.0000 | Peptide |
| 1,21-Henicosanediol | 328.33 | 2.09 | 0.0000 | Long chain fatty alcohol |
| Heptadecanoic carnitine | 413.35 | 2.08 | 0.0000 | Acyl carnitine |
| Phosphatidylglycerol (36:1) | 776.55 | 2.06 | 0.0000 | Phosphatidylglycerol |
| Phophatidylethanolamine(44:9) | 841.56 | 2.06 | 0.0000 | Phosphatidylethanolamine |
| 1-(9Z-heptadecenoyl)-2-(7Z, 10Z, 13Z, 16Z-docosatetraenoyl) -glycero-3-phosphoserine |
823.53 | 2.06 | 0.0000 | Glycerophospholipids |
| 1-Methylinosine | 283.28 | 2.05 | 0.0001 | Purine nucleoside |
| Cys Met Thr Tyr | 517.85 | 2.05 | 0.0000 | Peptide |
| Tetradecenoylcarnitine | 369.28 | 2.05 | 0.0000 | Acyl carnitine |
| Phosphatidylethanolamine(22:5) | 778.09 | 2.05 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(36:0) | 775.64 | 2.05 | 0.0000 | Glycerophospholipids |
| Coprocholic acid | 450.33 | 2.04 | 0.0000 | Bile acid |
| Asp Leu | 494.24 | 2.04 | 0.0000 | Peptide |
| Phosphatidylserine(38:1) | 817.58 | 2.04 | 0.0000 | Phosphatidylserine |
| Phosphatidic acid(40:1) | 758.58 | 2.03 | 0.0000 | 1,2-diacylglycerol-3-phosphate |
| Phosphatidic acid PA(18:1(12Z)-2OH(9,10)/i-15:0) | 692.46 | 2.03 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(32:1) | 731.54 | 2.02 | 0.0000 | Glycerophospholipids |
| Phosphatidylcholine(34:1) | 759.57 | 2.02 | 0.0000 | Glycerophospholipids |
| Phosphatidylethanolamine(34:2) | 715.51 | 2.02 | 0.0000 | Phosphatidylethanolamine |
| Cys Arg Trp Trp | 649.27 | 2.01 | 0.0000 | Peptide |
| Sphingomyelin (D18: 0/14: 1 (9Z) (OH)) |
688.51 | 2.01 | 0.0000 | Sphingolipid |
| Sphingomyelin (d18:1/16:0) | 702.56 | 2.01 | 0.0000 | Sphingolipid |
| Phosphatidylcholine(38:4) | 795.61 | 2.01 | 0.0000 | Glycerophosphocholine |
| Phosphatidylethanolamine(35:0) | 733.56 | 2.01 | 0.0000 | Glycerophospholipids |
| Phosphatidylethanolamine(32:3) | 671.48 | 2.00 | 0.0000 | Glycerophosphoethanolamine |
Note: M: average molecular weight. a: VIP score was obtained from the PLS-DA model. b: p-values were calculated from nonparametric Mann—Whitney test between occupational and environmental exposure groups. Thirty-eight significantly changed (VIP > 2, p value < 0.05 and FDR < 0.01) endogenous metabolites related to benzene exposure are listed in this table.
All metabolites showed higher concentration (fold change) in the urine of workers with environmental exposure to benzene, with the exception of the compound phenylalanylhydroxyproline, which showed higher concentration in the urine of gas station workers.
The correlation between potential metabolomic biomarkers and S-phenilmercapturic Acid (SPMA) and trans-trans-muconic acid (t,t-MA) exposure indicators was investigated by Spearman’s Correlation. No significant correlation was observed (p-value > 0.05) between metabolites and these biomarkers (Table S4).
3.3. Urinary Metabolites Associated with Oxidative Stress
The oxidative stress biomarkers SOD, CAT, and GST showed no significant difference between groups. To investigate the relationship of these discriminating metabolites derived from metabolomic analysis with the early toxic effects associated with low exposure to benzene identified in this study—thiol and MDA—linear regression was used. Thus, linear regression models were developed, using the stepwise backward elimination method and the Akaike information criterion (AIC), to explain thiol and MDA.
In Figure 4 it is possible to see the metabolites of the best linear regression model to explain MDA and thiol, and their respective coefficients. The metabolites that were significant are represented in blue, and those that were not significant in red. The developed models presented satisfactory performance (MDA: R2 adjusted from 0.5053 and estimated standard error was 0.1114; thiol: R2 adjusted from 0.5053, and estimated standard error of 0.1114).
Figure 4.
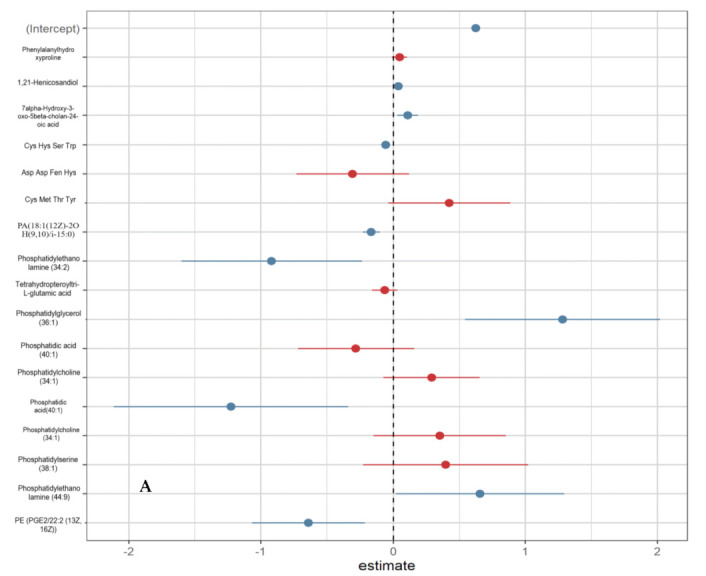
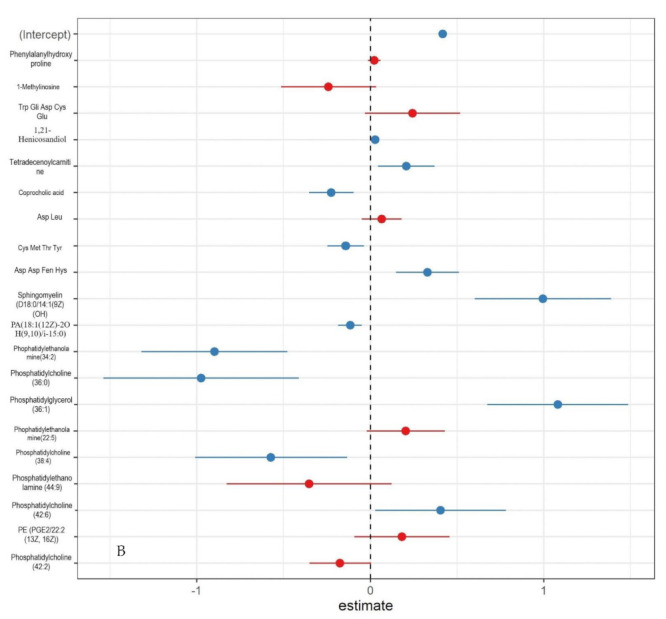
Logistic linear regression results with estimates of e of of discriminating metabolites for MDA and thiol biomarkers. Note: (A): MDA; (B): THIOL. Note: Coefficients calculated using linear regression, with a 95% confidence interval (CI). In blue, significant urinary metabolites; in red, non-significant urinary metabolites.
Multiple regression models were used to assess the influence of demographic variables and metabolites on oxidative stress biomarkers (Figure 5). The results showed that there was a significant and negative influence (Beta < 0.00; p < 0.00) of the metabolites coprocholic acid, Cys Met Tre Tyr, phosphatidylethanolamine (34:2), phosphatidylcholine (32:1), and phosphatidylcholine (38:4) on thiol levels. Thus, with the increase in the concentration of these metabolites, a decrease in serum thiol concentration is expected. In contrast, the metabolites Cys Hys Ser Trp, phosphatidylethanolamine (32:3) and phosphatidylglycerol (36:1) had a significant and positive influence (Beta > 0.00) on the same biomarker. Thus, the increase in the concentration of these metabolites in the urine is accompanied by an increase in the serum thiol concentration. Phosphatidylcholine (44:6) and PE (PGE2/22:2 (13Z, 16Z)) had a significant negative effect (Beta < 0.00; p < 0.00) on the MDA biomarker. Phosphatidylcholine (38:4) had a positive (Beta > 0.00; p < 0.00) and significant influence on serum MDA levels.
Figure 5.
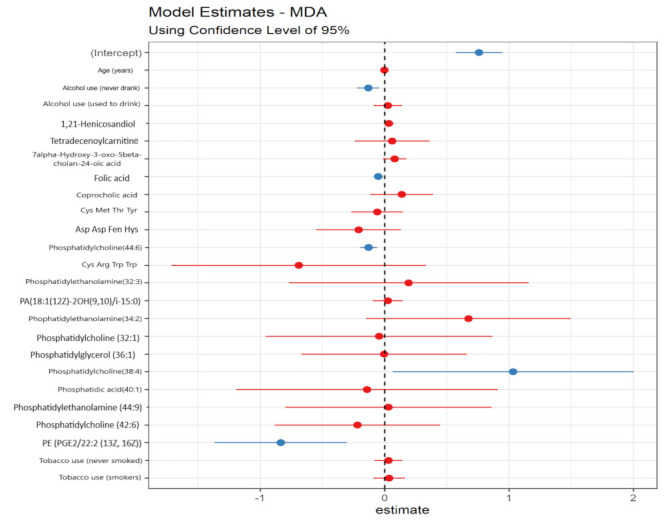
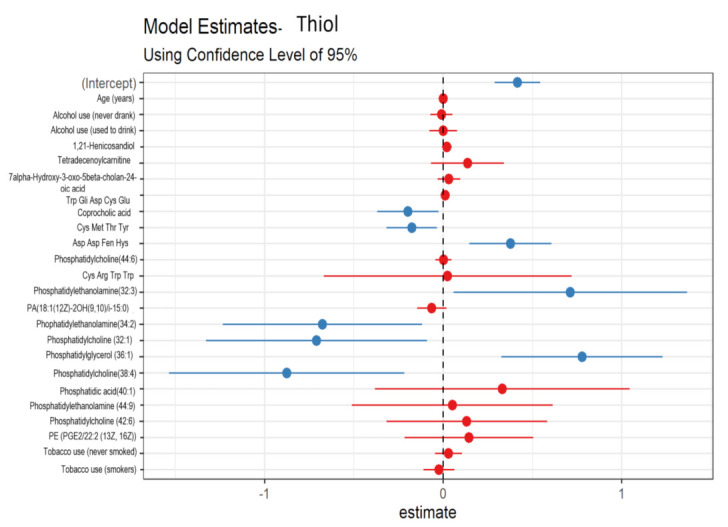
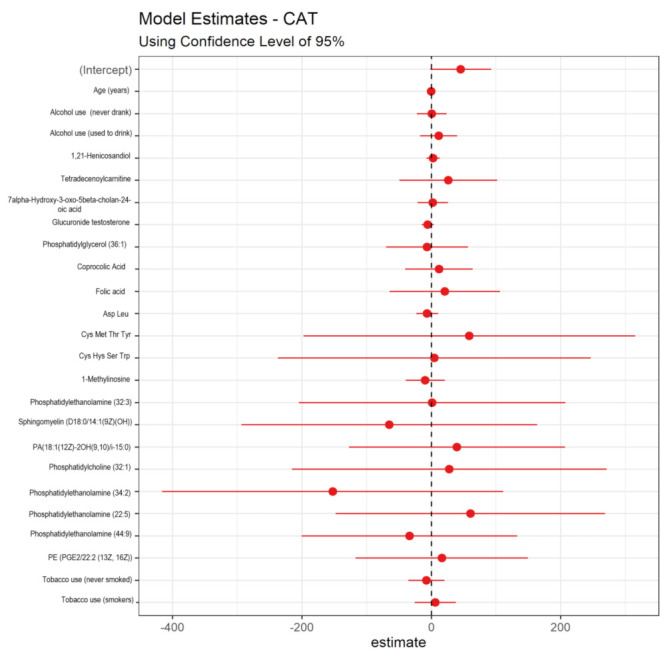
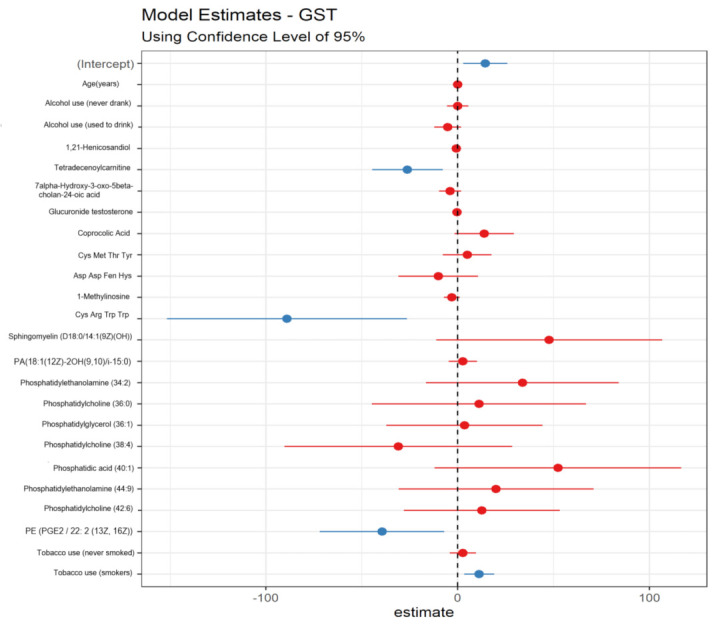
Multiple regression results and estimates of 𝛽0 and 𝛽κ for the main metabolites and sociodemographic variables for the biomarkers MDA, thiol, CAT, and GST. Note: Coefficients calculated using linear regression, with a 95% confidence interval (CI). In blue, significant urinary metabolites and in red, non-significant urinary metabolites.
No candidate metabolite proved to be significant to explain the variations of the CAT marker. On the other hand, the GST enzyme was significantly and negatively (Beta < 0.00; p < 0.00) influenced by tetradecenoylcarnitine, Cys Arg Trp Trp and PE (PGE2/22:2 (13Z, 16Z)). In addition, the smoking habit also influenced the levels of this enzyme, smokers with increased GST activity compared to former smokers. SOD enzyme activity was significantly and negatively influenced (Beta < 0.00 and p < 0.00) by phosphatidylcholine levels (44:6); and positively by 7alpha-hydroxy-3-oxo-5beta-cholan-24-oic acid (Beta > 0.00; p < 0.00).
Spearman’s correlation was used to verify the relationship between urinary metabolites and oxidative stress markers. The correlation coefficients and their significance are shown in Table 3. Many significant correlations were found (p-value < 0.05); however, the correlation coefficients showed low values, that is, close to zero.
Table 3.
Spearman correlation between urinary metabolites and oxidative stress markers.
| CAT | GST | THIOL | MDA | SOD | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| P¹ | p Value | P¹ | p Value | P¹ | p Value | P¹ | p Value | P¹ | p Value | |
| Phenylalanylhydroxyproline | −0.068 | 0.605 | 0.205 | 0.116 | 0.083 | 0.531 | −0.342 | 0.007 | −0.012 | 0.928 |
| Sphingomyelin (d18:1/16:0) | −0.019 | 0.887 | 0.162 | 0.218 | 0.149 | 0.256 | −0.356 | 0.005 | −0.002 | 0.987 |
| 1,21-Henicosanediol | 0.008 | 0.949 | −0.017 | 0.897 | 0.047 | 0.721 | 0.007 | 0.957 | 0.007 | 0.960 |
| Tetradecenoylcarnitine | 0.114 | 0.386 | 0.264 | 0.041 | 0.314 | 0.014 | −0.272 | 0.036 | −0.215 | 0.099 |
| 7alpha-Hydroxy-3-oxo-5beta-cholan-24-oic acid | 0.091 | 0.490 | 0.275 | 0.033 | 0.333 | 0.009 | −0.216 | 0.098 | −0.183 | 0.161 |
| Testosterone glucuronide | 0.034 | 0.796 | 0.268 | 0.039 | 0.273 | 0.035 | −0.306 | 0.017 | −0.267 | 0.039 |
| Phosphatidylglycerol (36:1) | −0.146 | 0.267 | −0.079 | 0.548 | 0.153 | 0.242 | −0.083 | 0.528 | 0.274 | 0.034 |
| Coprocholic acid | 0.135 | 0.303 | 0.302 | 0.019 | 0.238 | 0.067 | −0.305 | 0.018 | −0.280 | 0.030 |
| Folic acid | 0.064 | 0.627 | 0.281 | 0.030 | 0.331 | 0.010 | −0.276 | 0.033 | −0.251 | 0.053 |
| Asp Leu | 0.097 | 0.459 | 0.246 | 0.058 | 0.236 | 0.069 | −0.310 | 0.016 | −0.223 | 0.087 |
| Cys Met Thr Tyr | 0.141 | 0.282 | 0.266 | 0.040 | 0.214 | 0.100 | −0.303 | 0.019 | −0.236 | 0.070 |
| 1-(9Z-heptadecenoyl)-2-(7Z,10Z, 13Z, 16Z-docosatetraenoyl)-glycero-3-phosphoserine | 0.086 | 0.514 | 0.295 | 0.022 | 0.281 | 0.030 | −0.275 | 0.034 | −0.260 | 0.045 |
| Cys Hys Ser Trp | 0.030 | 0.817 | 0.275 | 0.033 | 0.254 | 0.051 | −0.248 | 0.056 | −0.261 | 0.044 |
| Asp Asp Fen Hys | 0.054 | 0.680 | 0.287 | 0.026 | 0.283 | 0.028 | −0.246 | 0.059 | −0.275 | 0.034 |
| Cys Arg Trp Trp | 0.113 | 0.391 | 0.297 | 0.021 | 0.291 | 0.024 | −0.265 | 0.041 | −0.262 | 0.043 |
| Phosphatidylethanolamine(32:3) | 0.103 | 0.433 | 0.308 | 0.017 | 0.303 | 0.019 | −0.265 | 0.041 | −0.264 | 0.042 |
| Sphingomyelin (D18: 0/14: 1 (9Z) (OH)) |
0.087 | 0.511 | 0.312 | 0.015 | 0.284 | 0.028 | −0.256 | 0.049 | −0.270 | 0.037 |
| Trp Gln Asp Cys Glu | 0.039 | 0.770 | 0.294 | 0.022 | 0.321 | 0.012 | −0.330 | 0.010 | −0.213 | 0.103 |
| Tetrahydropteroyltri-L-glutamic acid | −0.005 | 0.969 | 0.300 | 0.020 | 0.240 | 0.065 | −0.348 | 0.006 | −0.278 | 0.031 |
| Phosphatidic acid PA(18:1(12Z)-2OH(9,10)/i-15:0) | 0.095 | 0.472 | 0.304 | 0.018 | 0.268 | 0.038 | −0.253 | 0.051 | −0.256 | 0.048 |
| Phosphatidylcholine(32:1) | 0.096 | 0.464 | 0.306 | 0.017 | 0.286 | 0.027 | −0.258 | 0.047 | −0.259 | 0.045 |
| Phosphatidylethanolamine(34:2) | 0.107 | 0.414 | 0.295 | 0.022 | 0.277 | 0.032 | −0.256 | 0.049 | −0.261 | 0.044 |
| 1-Methylinosine | 0.044 | 0.739 | 0.292 | 0.024 | 0.247 | 0.057 | −0.196 | 0.132 | −0.401 | 0.002 |
| Phosphatidylethanolamine(35:0) | 0.121 | 0.358 | 0.291 | 0.024 | 0.274 | 0.034 | −0.253 | 0.051 | −0.251 | 0.053 |
| Heptadecanoic carnitine | 0.064 | 0.625 | 0.301 | 0.019 | 0.290 | 0.025 | −0.266 | 0.040 | −0.252 | 0.052 |
| Phosphatidylcholine(36:0) | 0.081 | 0.539 | 0.293 | 0.023 | 0.291 | 0.024 | −0.269 | 0.038 | −0.262 | 0.043 |
| Phophatidylethanolamine(22:5) | 0.096 | 0.468 | 0.297 | 0.021 | 0.268 | 0.038 | −0.255 | 0.049 | −0.279 | 0.031 |
| Phosphatidylcholine(38:4) | 0.091 | 0.489 | 0.308 | 0.017 | 0.266 | 0.040 | −0.256 | 0.048 | −0.251 | 0.053 |
| Phosphatidic acid(40:1) | 0.090 | 0.493 | 0.299 | 0.021 | 0.287 | 0.026 | −0.273 | 0.035 | −0.253 | 0.051 |
| Phosphatidylcholine(34:1) | 0.093 | 0.480 | 0.295 | 0.022 | 0.275 | 0.034 | −0.261 | 0.044 | −0.262 | 0.043 |
| Phosphatidylserine(38:1) | 0.088 | 0.503 | 0.296 | 0.022 | 0.277 | 0.032 | −0.259 | 0.045 | −0.252 | 0.052 |
| Phophatidylethanolamine(44:9) | 0.089 | 0.501 | 0.304 | 0.018 | 0.294 | 0.023 | −0.262 | 0.043 | −0.262 | 0.043 |
| Phosphatidylcholine(22:2) | 0.100 | 0.448 | 0.299 | 0.020 | 0.289 | 0.025 | −0.256 | 0.048 | −0.242 | 0.063 |
| Phosphatidylcholine(42:6) | 0.082 | 0.536 | 0.288 | 0.026 | 0.288 | 0.026 | −0.261 | 0.044 | −0.257 | 0.048 |
| Phosphatidylcholine(40:3) | 0.113 | 0.391 | 0.297 | 0.021 | 0.291 | 0.024 | −0.265 | 0.041 | −0.262 | 0.043 |
| Phosphatidylethanolamine PGE2/22:2(13Z, 16Z) | 0.095 | 0.473 | 0.289 | 0.025 | 0.263 | 0.043 | −0.281 | 0.029 | −0.238 | 0.067 |
| Phosphatidylcholine(42:2) | 0.079 | 0.547 | 0.311 | 0.015 | 0.285 | 0.027 | −0.274 | 0.034 | −0.279 | 0.031 |
| Phosphatidylcholine(44:6) | 0.075 | 0.569 | 0.310 | 0.016 | 0.293 | 0.023 | −0.270 | 0.037 | −0.252 | 0.052 |
P¹: Spearman’s correlation coefficient; p-values were calculated from nonparametric Mann—Whitney test.
The CAT biomarker showed low correlation with all metabolites. GST showed positive correlations with sphingomyelin (D18:0/14:1(9Z)(OH)), phosphatidylcholine (42:2), and phosphatidylcholine (44:6). On the other hand, 7alpha-hydroxy-3-oxo-5beta-cholan-24-oic acid, folic acid, and Trp Gly Asp Cys Glu were the most positively correlated with thiol. The biomarker MDA was negatively correlated (P1 < 0) with all urinary metabolites, except 1,21-henicosanediol. The highest correlations for this biomarker were observed with phenylalanylhydroxyproline, sphingomyelin (d18:1/16:0), Asp Leu, Trp Gly Asp Cys Glu, and tetrahydropteroyltri-L-glutamic acid. On the other hand, the SOD enzyme also showed negative correlations (P1 < 0) with all urinary metabolites, with the exception of 1,21-henicosanediol and was more negatively correlated with 1-methylinosine.
From these results, scatter plots were used to verify the behavior of the relationship between urinary metabolites and oxidative stress markers—thiol, MDA, CAT, SOD, and GST. The results showed that there is no linear correlation between them, therefore, the increase or decrease in the levels of oxidative stress markers does not imply a proportional change in urinary metabolites. Some correlations are shown in Figure S1.
3.4. Urinary Metabolites Associated with Chromosomal Aberrations
The relationship of discriminating urinary metabolites between groups of workers with environmental and occupational exposure to benzene, and chromosomal aberrations was also investigated using Spearman’s correlation. The following aberrations were considered: break rate, fragment rate, and metaphase rate with premature separation. The correlation coefficients and their significance are shown in Table 4. It is possible to verify that there were many significant correlations (p-value <0.05), but the correlation coefficients present low values, close to zero.
Table 4.
Spearman correlation between urinary metabolites and chromosomal aberrations.
| Break Rate | Fragments Rate | Metaphase Rate with Premature Separation | ||||
|---|---|---|---|---|---|---|
| P¹ | p Value | P¹ | p Value | P¹ | p Value | |
| Phenylalanylhydroxyproline | −0.315 | 0.014 | −0.316 | 0.014 | 0.196 | 0.136 |
| Sphingomyelin (d18:1/16:0) | −0.271 | 0.036 | −0.263 | 0.042 | 0.150 | 0.256 |
| 1,21-Henicosanediol | −0.334 | 0.009 | −0.331 | 0.010 | 0.154 | 0.243 |
| Tetradecenoylcarnitine | −0.291 | 0.024 | −0.290 | 0.024 | 0.320 | 0.013 |
| 7alpha-Hydroxy-3-oxo-5beta-cholan-24-oic acid | −0.244 | 0.060 | −0.249 | 0.055 | 0.353 | 0.006 |
| Testosterone glucuronide | −0.272 | 0.035 | −0.272 | 0.035 | 0.300 | 0.021 |
| Phosphatidylglycerol (36:1) | 0083 | 0.528 | 0.089 | 0.498 | 0.130 | 0.327 |
| Coprocholic acid | −0.287 | 0.026 | −0.298 | 0.021 | 0.275 | 0.035 |
| Folic acid | −0.241 | 0.063 | −0.243 | 0.061 | 0.349 | 0.007 |
| Asp Leu | −0.303 | 0.019 | −0.308 | 0.017 | 0.319 | 0.014 |
| Cys Met Thr Tyr | −0.315 | 0.014 | −0.313 | 0.015 | 0.269 | 0.039 |
| 1-(9Z-heptadecenoyl)-2-(7Z,10Z, 13Z, 16Z-docosatetraenoyl)-glycero-3-phosphoserine | −0.282 | 0.029 | −0.290 | 0.025 | 0.304 | 0.019 |
| Cys Hys Ser Trp | −0.258 | 0.047 | −0.250 | 0.054 | 0.281 | 0.031 |
| Asp Asp Fen Hys | −0.279 | 0.031 | −0.277 | 0.032 | 0.299 | 0.021 |
| Cys Arg Trp Trp | −0.276 | 0.033 | −0.298 | 0.021 | 0.360 | 0.005 |
| Phosphatidylethanolamine(32:3) | −0.250 | 0.054 | −0.249 | 0.055 | 0.273 | 0.037 |
| Sphingomyelin (D18: 0/14: 1 (9Z)(OH)) | −0.247 | 0.057 | −0.248 | 0.056 | 0.280 | 0.032 |
| Trp Gln Asp Cys Glu | −0.256 | 0.049 | −0.256 | 0.048 | 0.286 | 0.028 |
| Tetrahydropteroyltri-L-glutamic acid | −0.264 | 0.042 | −0.258 | 0.046 | 0.291 | 0.025 |
| Phosphatidic acid PA(18:1(12Z)-2OH(9,10)/i-15:0) | −0.277 | 0.032 | −0.283 | 0.028 | 0.357 | 0.006 |
| Phosphatidylcholine(32:1) | −0.266 | 0040 | −0.274 | 0.034 | 0.282 | 0.031 |
| Phosphatidylethanolamine(34:2) | −0.259 | 0.045 | −0.261 | 0.044 | 0.284 | 0.029 |
| 1-Methylinosine | −0.259 | 0.046 | −0.260 | 0.045 | 0.285 | 0.029 |
| Phosphatidylethanolamine(35:0) | −0.259 | 0.046 | −0.260 | 0.045 | 0.278 | 0.033 |
| Heptadecanoic carnitine | −0.257 | 0.047 | −0.260 | 0.045 | 0.294 | 0.024 |
| Phosphatidylcholine(36:0) | −0.264 | 0.042 | −0.268 | 0.039 | 0.303 | 0.020 |
| Phophatidylethanolamine(22:5) | −0.292 | 0.024 | −0.295 | 0.022 | 0.296 | 0.023 |
| Phosphatidylcholine(38:4) | −0.270 | 0.037 | −0.276 | 0.033 | 0.301 | 0.021 |
| Phosphatidic acid(40:1) | −0.259 | 0.046 | −0.262 | 0.043 | 0.299 | 0.021 |
| Phosphatidylcholine(34:1) | −0.253 | 0.051 | −0.258 | 0.047 | 0.312 | 0.016 |
| Phosphatidylserine(38:1) | −0.259 | 0.045 | −0.265 | 0.041 | 0.301 | 0.021 |
| Phophatidylethanolamine(44:9) | −0.272 | 0.035 | −0.279 | 0.031 | 0.282 | 0.030 |
| Phosphatidylcholine(22:2) | −0.258 | 0.047 | −0.259 | 0.046 | 0.280 | 0.032 |
| Phosphatidylcholine(42:6) | −0.250 | 0.054 | −0.255 | 0.049 | 0.298 | 0.022 |
| Phosphatidylcholine(40:3) | −0.255 | 0.049 | −0.261 | 0.044 | 0.306 | 0.018 |
| Phosphatidylethanolamine PGE2/22:2(13Z,16Z) | −0.258 | 0.046 | −0.263 | 0.042 | 0.276 | 0.034 |
| Phosphatidylcholine(42:2) | −0.276 | 0.033 | −0.283 | 0.029 | 0.287 | 0.027 |
| Phosphatidylcholine(44:6) | −0.257 | 0.047 | −0.267 | 0.039 | 0.321 | 0.013 |
P¹: Spearman’s correlation coefficient; p-values were calculated from nonparametric Mann—Whitney test.
Breakage rate and fragment rate were negatively correlated with all urinary metabolites (P1 < 0). Phenylalanylhydroxyproline had the highest correlation with the breakage rate. Phenylalanylhydroxyproline, 1,21-henicosanediol, and Asp Leu showed higher correlations with the rate of fragments. Tetradecenoylcarnitine, 7alpha-hydroxy-3-oxo-5beta-cholan-24-oic acid, folic acid, Asp Leu, 1-methylinosine, and phosphatidylcholine (44:6) were the most positively correlated (P1 < 0) with the rate of premature metaphases.
To verify the behavior of the relationship between them, it was verified using scatter diagrams (Figure S2). The results obtained were similar to those observed in relation to oxidative stress markers. Therefore, the correlation between metabolites and chromosome breaks, fragment and metaphases presenting premature chromatid separation is not linear.
3.5. Potential Metabolomic Biomarkers Associated with Oxidative Damage and Benzene-Induced Chromosomal Aberrations
From the regression analyses, ten metabolites were significantly related to thiol, MDA e chromosomal aberrations. Thus, these were recognized as potential metabolomic biomarkers associated with oxidative effects induced by exposure to low concentrations of benzene. Their identity, as well as p values, fold change, and biochemical pathways in which they are biologically related is shown in Table 5.
Table 5.
Urinary metabolomic biomarkers related to oxidative stress and benzene-induced chromosomal aberrations.
| Metabolite | p Value b | Fold Change c | Pathway |
|---|---|---|---|
| 1,21-Henicosanediol | <0.0001 | 2.30 | Lipid transport and lipid metabolism Fatty acid metabolism Lipid peroxidation and cell signaling. |
| Tetradecenoylcarnitine | <0.0001 | 5.05 | Lipid transport and lipid metabolism Fatty acid metabolism Lipid peroxidation and cell signaling. |
| Coprocholic acid | <0.0001 | 5.33 | Lipid transport and metabolism Fatty acid metabolism |
| Cys Met Thr Tyr | <0.0001 | 4.76 | Product of incomplete decomposition of proteins or protein catabolism |
| Asp Leu | <0.0001 | 4.72 | Product of incomplete decomposition of proteins or protein catabolism |
| Phenylalanylhydroxyproline | <0.0001 | 2.27 | Product of incomplete decomposition of proteins or protein catabolism |
| Cys Hys Ser Trp | <0.0001 | 2.31 | Product of incomplete decomposition of proteins or protein catabolism. |
| Sphingomyelin (D18: 0/14: 1 (9Z) (OH)) |
<0.0001 | 5.05 | Lipid metabolism and signaling cell. |
| PE (PGE2/22:2 (13Z, 16Z)) | <0.0001 | 4.73 | Cell signaling |
| Phophatidylethanolamine(44:9) | <0.0001 | 5.20 | Components of the lipid bilayer of cells Lipid metabolism Cell signaling |
b: p-values were calculated from nonparametric Mann—Whitney test between occupational and environmental exposure groups. c: fold change was calculated by the average concentration of the metabolite in the group with environmental exposure in relation to the group with occupational exposure.
The relationship between identified potential biomarkers and exposure time was investigated using Spearman’s Correlation. No significant correlation was observed (p > 0.05) (Table S5).
The ROC (receiver operating characteristic) curve was used to assess the performance of these potential early biomarkers in the assessment of toxicity induced by low concentrations of benzene. The ROC curve was used to evaluate the performance of these potential early biomarkers in the evaluation of toxicity induced by low concentrations of benzene. The ROC curve is widely used in the medical field, and consists of a graphical representation of the relationship between sensitivity and specificity of a diagnostic test. The area under the ROC curve (AUC) represents the global measure of a test’s ability to discriminate whether a specific condition is present or not. An AUC of 0.5 indicates that a test has no discriminating ability, while an AUC of 1.0 represents a test with perfect discrimination [36]. AUC > 0.85 is considered acceptable for clinical applications [37]. Based on the results of the ROC curve, all biomarker candidates performed satisfactorily, except for phenylalanylhydroxyproline and phosphatidylethanolamine (44:9) which had AUC <0.85. The ROC curve results of some potential biomarkers are shown in Figure 6.
Figure 6.
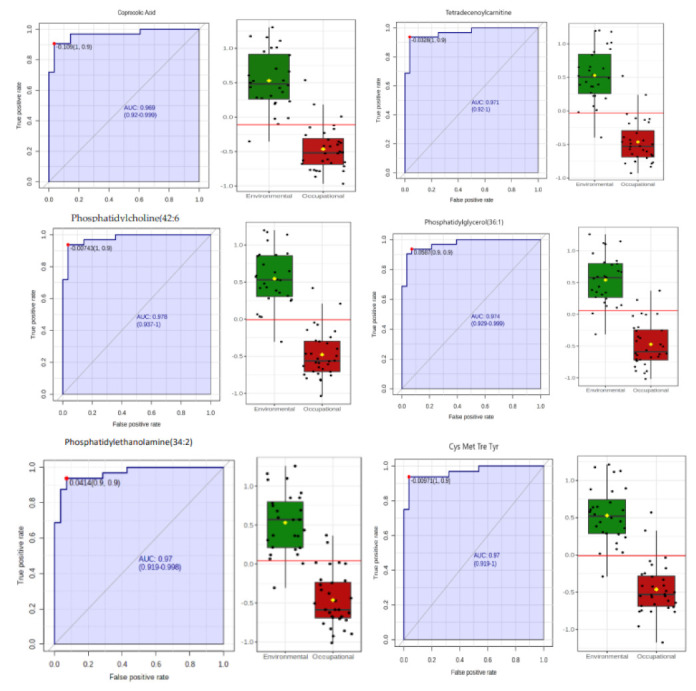
ROC curves of the discriminating urinary metabolites associated with oxidative stress and chromosomal aberrations induced by exposure to low concentrations of benzene. Note: On the left, the graphical representation of the calculated ROC curve with a 95% confidence interval for each biomarker candidate. AUC = area under the ROC curve. On the right, boxplot of metabolite concentrations in the environmental (green) and occupationally (red) exposed groups. A red horizontal line indicates the optimal cutoff point.
3.6. Metabolic Pathway Analysis
From the identification of discriminant urinary metabolites identified in the PLS-DA model, we sought to identify the metabolic pathways involved in the biological response elicited by exposure to benzene. For this, the analysis of enrichment of the metabolic pathway was performed in Metaboanalyst® 5.0 33. The results are presented in the form of bar graphs and Network View (Figure 7). It is observed that the metabolic alterations of greater impact (indicated in red) involve the bile acid synthesis pathways, lipid metabolism, amino acids, folate, mitochondrial beta-oxidation of short chain saturated fatty acids, and steroid hormone metabolism.
Figure 7.
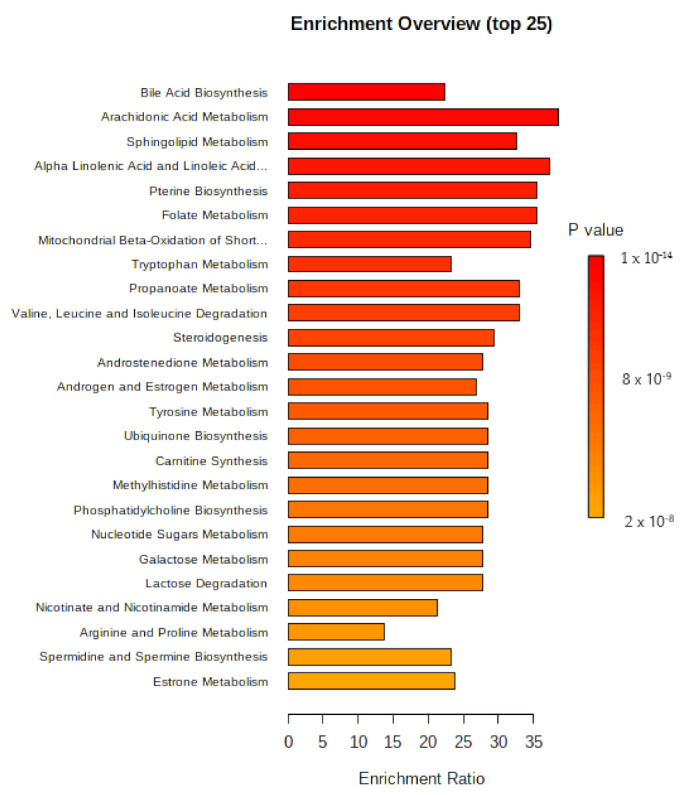
Bar graph with an overview of the analysis of enrichment of the metabolic 472 response against exposure to low concentrations of benzene.
However, it is worth noting that the other biological pathways identified as related to benzene exposure are also relevant, given that the p values are quite low. This fact indicates that exposure to benzene, even at low concentrations, is capable of disturbing numerous biological pathways that are crucial for homeostasis, and their understanding is extremely important to understand the multiplicity of toxic effects induced by benzene.
3.7. Discussion
In this study, urine was chosen as a biofluid to investigate the metabolite profile of workers exposed both environmentally and occupationally to low concentrations of benzene in the air. It is a matrix traditionally used in biological monitoring studies because it has advantages such as: simple and non-invasive collection, allowing large volumes to be obtained and presenting a lower concentration of proteins compared to blood samples [38]. The urine samples were normalized by the concentration of urinary creatinine considering that the variation in the volume of urine can cause large variations in the concentration of the excreted metabolite leading to discrepancies on results [39].
The mean concentration of benzene in the atmospheric air of the fuel stations and gates was respectively 31.8 μg·m3 and 22.1 μg·m3. These results are below the technological reference value (TRV) of 3.19 mg·m−3, established by Brazilian legislation [40]. However, it is worth noting that exposure to low concentrations of benzene does not eliminate the risk; considering that there is no safe exposure limit to carcinogenic substances such as benzene [41]. This is corroborated by the results of this study, which proved that, even at low concentrations, benzene is capable of evoking metabolic disturbances, which were statistically associated with early toxic effects—oxidative stress and chromosomal alterations.
However, despite environmental monitoring having indicated similarity of exposure to hydrocarbons for workers in the gas station and ordinances, the analysis of the benzene concentration in air by percentile showed higher concentrations of xenobiotic in the 75th and 95th percentiles for the gas stations. This represents an oscillation in the chronic exposure to benzene during the working day, and indicates that there is a difference in the intensity of exposure during the performance of certain tasks, such as the aspiration of vapors when filling tanks.
In this study, the oxidative stress biomarkers—thiol and MDA were significantly different between groups, thus representing the toxic effects induced by chronic, low-intensity exposure to benzene. Oxidative stress is recognized as one of the toxic action mechanisms of benzene [42]. Oxidative stress represents a state of imbalance between the generation of reactive species and the body’s antioxidant capacity. These reactive oxygen species (ROS), such as superoxide anion, hydrogen peroxide, hydroxyl radical, are unstable and highly reactive. They are produced through the redox cleavage of reactive products originated during the biotransformation of benzene, especially hydroquinone and benzoquinone [43]. Scientific literature indicates that such reactive species can damage biomolecules such as lipids, proteins, and genetic material [44,45,46]. Many chronic diseases such as type 2 diabetes mellitus, cardiovascular disease, Alzheimer’s disease, various types of cancer (leukemia, lung, liver, etc.,) and chronic inflammation are associated with oxidative stress. It is known that continuous oxidative stress can elicit inflammatory mechanisms, with synthesis and secretion of pro-inflammatory cytokines [47].
Inflammation is a physiological process of body protection, triggered in situations that can cause damage, such as infections by microorganisms, exposure to allergens, ionizing radiation and toxic substances. Inflammation has two stages—acute and chronic. Acute inflammation is mediated by innate immunity, has a short duration and usually results in a beneficial effect on the body. On the other hand, chronic inflammation, which lasts for a long time, is mediated by leukocytes (mast cells and monocytes) that migrate to the site of damage, where they promote an increase in oxygen uptake (respiratory explosion) with a consequent increase in the local production of ROS; production of arachidonic acid derivatives and also secretion of cytokines and chemokines; which recruit more inflammatory cells to the site and once again increase the production of ROS [48]. It should be noted that chronic inflammation is associated with a higher risk of various types of cancer [49]. Thus, there is an important relationship between oxidative stress and inflammation.
Workers occupationally exposed to benzene analyzed in this study had higher counts of monocytes and basophils in peripheral blood (leukocytes with an important role in inflammation) [29], in addition to prostaglandin PE (PGE2/22:2 (13Z, 16Z)), leukotrinene D5 and oxidized lipids were identified among the most discriminating urinary metabolites among the analyzed groups. These results indicate that oxidative stress and inflammatory state are the most relevant mechanisms of toxic action in exposure to low concentrations of benzene.
This subtle difference in exposure was not detected, for example, through exposure biomarkers—AttM and SPMA; which did not show significant difference between groups. In this context, metabolomics presents itself as a promising, sensitive, and appropriate tool to assess the risks arising from exposure to benzene. Analysis supervised by PLS-DA and OPLS-DA was able to recognize this subtle difference, thus indicating that even at low doses, the biological response induced by exposure to this hydrocarbon was different between groups.
Metabolomic analysis can offer simultaneous analysis of a large number of metabolites without any prior knowledge of these compounds [50]. However, this comprehensive analysis is not an easy task. As the metabolome is made up of an immense amount of metabolites belonging to different chemical classes such as amino acids, carbohydrates, nucleotides, and lipids that have a wide range of physicochemical properties and are present in a wide range of concentration [51,52], the analysis of the metabolic profile represents a great challenge in the field of analytical chemistry. In the field of environmental and occupational toxicology, it represents a promising tool for a holistic understanding of the interaction between chemicals and the human organism.
The validation of the developed PLS-DA and OPLS-DA models is important to confirm the quality of the developed models and attest to their predictive capacity. The obtained R2 explains the model’s variance and gives information about the goodness of fit, while the Q2 predicts the variance and provides information about the model’s predictability. A robust PLS-DA model should have high values of R2 and Q2 and not differ by more than 0.2–0.3 from each other [53,54]. Model validation is also useful in identifying the optimal number of latent variables needed to describe the entire data variance; and avoid under- or over-adjustment issues. According to Godzien et al., (2013) [55], the usual values of these coefficients for biological experiments are Q2 > 0.4 and R2 > 0.7.
Chemical exposure leads to alterations in metabolism and/or gene expression (deletion or overexpression) and such alteration may represent a specific pattern, called metabolic signature [56]. This metabolic signature may reflect this exposure, which has supported the use of metabolomics in the assessment of exposure to environmental and occupational chemical agents. According to Wishart (2016) [57], metabolites are more representative of the phenotype, as they change more quickly than genetic material and, therefore, indicate current biological events. It is noteworthy that the metabolome is compartmental, dynamic, and related to the phenotype. Thus, its assessment represents a holistic approach to the individual’s physiological and/or pathological state, since it integrates genetic and environmental factors [15,16,58]. Thus, the wealth of information acquired in undirected metabolomics is an excellent tool for a better understanding of the organism’s biochemical events in environmental exposure to chemical agents.
Therefore, the results observed in this study demonstrated that the exposure of individuals to benzene, even at low doses, was capable of causing metabolic changes in urine, which were identified through non-directed metabolomics. Due to the mildness of the alterations, including the differences between the two groups, they were only identified due to the high sensitivity of the UHPLC-ESI-Q-TOF-MS, demonstrating its robustness and capacity for wide coverage of the urinary metabolome. This justifies its choice for application in environmental and occupational metabolomics studies of chemical contaminants in low concentrations.
The discriminating metabolites significantly correlated with oxidative stress and benzene-induced chromosomal aberrations act predominantly in biological pathways related to lipid and amino acid synthesis, metabolism, and peroxidation (Table 5).
Studies on urine, plasma, and bone marrow metabolomics in C3H/He-exposed rats were conducted by Sun et al., (2012) [59] and Sun et al., (2014) [60], respectively. In urine, the metabolic disturbances reported involved the metabolism pathways of purines, spermidines, fatty acids, tryptophan, and peptides [59]. In plasma, L-acetylcarnitine, p-coumaric acid, L-tyrosine, L-phenylalanine and lysine were significantly altered; while 5-hydroxyindoleacetic acid, histamine, L-histidine, N-methylhistamine, L-acetylcarnitine, pyrrolidone carboxylic acid, and palmitoylcarnitine in bone marrow [60]. In another investigation also conducted in an animal model, Yu et al. (2021) [61] investigated the lipidome of bone marrow cells from male C57BL/6 rats, exposed to 150 mg/kg of benzene using LC-MS/MS technology, in order to understand the mechanisms associated with benzene hematotoxicity. The results showed significant changes in the levels of glycerophospholipids, sphingolipids, linoleic acid metabolism, amino acids, and unsaturated fatty acid biosynthesis. Based on this, the authors hypothesized that disturbances in the glycerophospholipid pathway affect autophagy, while the sphingolipid pathway seems to influence proliferation and apoptosis; and both processes play an important role in benzene-induced hematopoietic toxicity [61].
In this study, the metabolism of several classes of lipids was associated with exposure to benzene, including the biosynthesis of bile acids. Coprolic acid, taurocholic acid, 3-chenodeoxycholic sulfate, 3-oxocholic acid, 7b-hydroxy-3-oxo-5b-cholanoic acid, nutriacolic acid, and hodeoxycholic acid were excreted in smaller amounts in the urine of gas station workers. Bile acids are produced in the liver from cholesterol and later converted to bile salts by conjugation with glycine or taurine. They are then stored in the gallbladder and subsequently excreted in the bile, along with its other components, in the duodenum, where they are deconjugated and reduced to urobilinogens by the action of the microbiota. These can be excreted in the feces, partially reabsorbed and about 5% excreted in the urine. Biologically, they play an important role in the intestinal absorption of lipophilic substances, participate in the regulation of cholesterol and in the metabolism of lipids and carbohydrates, stimulate bile flow, act as cellular signals, in the excretion of toxic substances, in the maintenance of a healthy organism, gut microbiota, and innate immunity [62,63]. Sun et al., (2020) [64] reported changes in the metabolic profile of the cecal content of mice exposed to benzene for 30 days. Changes were mainly observed in bile acids, fatty acids, carboxylic acids and derivatives, glycerolipids, glycerophospholipids, prenole lipids, steroids, and steroid derivatives. These results indicate that benzene is capable of causing intestinal dysbiosis with significant changes in several species, such as Proteobacteria, Bacteroidetes, and Actinobacteria, which are involved in the biotransformation of bile acids [65].
It is believed that the disturbance in bile acid excretion may also be due to the liver toxicity of benzene. Reactive metabolites formed in the liver during its biotransformation can influence the regulatory pathways of bile acid biosynthesis. Occupationally exposed individuals had higher values for liver function markers—aspartate aminotransferase (AST) and direct bilirubin. AST is an enzyme that catalyzes the interconversion of amino acids to 2-oxo-acids through the transfer of amino groups, found mainly in the heart and liver, cytoplasm, and mitochondria. Direct bilirubin, on the other hand, is a biotransformation product of the heme group, which is conjugated to glucuronic acid in the liver. Direct hyperbilirubinemia is associated with hepatocyte damage or biliary obstruction [66]. Other studies corroborate these results. In the investigation conducted by Moro et al., (2017) [67], albumin and TGO were significantly higher in gas station attendants compared to the control group. Andrea and Reddy, (2014) [68] detected higher values of liver enzyme alkaline phosphatase (ALP), aspartate amino transferase (AST), and alanine amino transferase in children accidentally exposed to benzene after a burning incident at the British Oil Refinery (BP) in the city of Texas. Additionally, the intestinal reabsorption of bile acids can be compromised by benzene-induced intestinal dysbiosis.
Koelmel et al. (2020) [14] state that lipids are ubiquitous metabolites that play different physiological roles, and are especially involved in biological processes such as oxidative stress, inflammation, obesity and endocrine disruption. As many environmental stressors exert their toxic effects through disturbances in these biological processes, the investigation of changes in lipid metabolism represents an important strategy to elucidate mechanisms of toxic action, as well as to identify new biomarkers of exposure to such stressors. Disturbances in lipid metabolism have been related to several pathologies [69,70,71,72,73].
Arachidonic acid metabolism was also implicated in the biological response of workers exposed to benzene investigated in this study. Previous studies have shown that the metabolism of this group of lipids is involved in benzene myelotoxicity [74,75]. Prostaglandins are prostanoids synthesized from arachidonic acid by cyclooxygenase enzymes (COX-1 and COX-2), and play physiological and pro-inflammatory functions. Prostanoids are often implicated in the initiation of carcinogenesis, representing the link between inflammation and cancer [76]. The increase in PGE2 was related to arachidonic acid peroxidation, increased phagocytic activity of macrophages, and decreased cellularity in the bone marrow [74,75]. In this study, a reduction in PGE1 and an increase in PGF2 in the urine of occupationally exposed workers were observed. PGE1 is a prostanoid that plays important physiological roles such as vasodilation, inhibitor of platelet aggregation, mediator of inflammation, and even hepatoprotective action. Animal studies have demonstrated hepatic cytoprotection of PGE1 through inhibition of T-cell mediated cytotoxicity, increased DNA synthesis, cyclic AMP, and liver tissue ATP levels [77,78,79]. The PGF2 is one of the most common protaglandins, is produced in many organs, and has a wide range of effects. Pabst et al. (2017) [73] reported an increase in this prostaglandin in plasma and bone marrow of patients with acute myeloid leukemia (AML).
Another metabolic pathway associated with early toxic effects induced by benzene was the metabolism of sphingolipids. These are physiologically implicated in the regulation of cell growth, differentiation, and apoptosis [80]. Among the types of sphingolipids, ceramides, structural membrane components, and secondary messengers in cell signaling have been implicated in the pathophysiology of diseases such as diabetes, cancer, Alzheimer’s disease, multiple sclerosis, and others [70]. In our study, sphingomyelin (d18:0/14:1(9Z)(OH)) and sphingomyelin (d18:1/16:0) decreased in the urine of occupationally exposed workers and was significantly associated with early toxic effects induced by benzene. Decline in plasma sphingolipids has also been described in carriers and AML [73,81]. A similar result was found by Robinson et al., (2021) [81], the authors investigated the relationship between reactive oxygen species (ROS) and the metabolome in AML cells. Sphingolipids were significantly decreased. Based on these findings, the authors postulated that ROS are important in regulating the synthesis and/or degradation of sphingolipids.
Polyunsaturated fatty acids have also been implicated in benzene-induced toxicity. In our study, in addition to fatty acids, fatty acid amides (oleamide, linoleamide and palmitoleamide) and oxylipin 14,15-DiHETrE were also detected in the urine of workers exposed to benzene, and were significantly reduced.
These lipids are considered essential and play an important role in membrane maintenance, signal transduction, and anti-inflammatory properties. Musharraf et al., (2016) [82] investigated the serum metabolome of individuals with AML, acute lymphocytic leukemia, and aplastic anemia using the CG-MS technique. The authors reported that palmitic acid was significantly reduced in patients with ALL and AML. Stearic and oleic acids, on the other hand, had higher concentrations in patients with these diseases [82]. It has been shown that AML patients have an overproduction of reactive oxygen species (ROS), and that these produce changes in carbohydrate metabolism, sphingolipids, fatty acid oxidation, purine metabolism, and amino acid homeostasis [81]. The α-linoleic acid (ALA), a polyunsaturated fatty acid, has an anti-inflammatory action due to its bioconversion into long-chain polyunsaturated fatty acids and, later, into oxylipins, bioactive lipid mediators [73]. Pabst et al., (2017) [73] reported an increase in fatty amides (oleamide and palmitoleoyl ethanolamide) in AML cells. A oleamide is an endogenous bioactive signaling molecule, similar to endocannabinoids, which participates in sleep regulation via the CB1 receptor and has anti-inflammatory action [83,84]. It is believed that the reduction of these bioactive metabolites in the urine of occupationally exposed workers is a consequence of their increased consumption, an adaptive response triggered in order to contain the toxic effects induced by benzene.
It should be noted that folate, also identified as an important pathway related to benzene exposure, is necessary for cell division and DNA synthesis; and according to Morgan and Smith (2002) [85], folate metabolism is associated with the maintenance and integrity of DNA. Folate insufficiency can lead to anemia, neutropenia, and pancytopenia [85].
Metabolomics also identified tryptophan metabolites—indoleacetic acid, 5-hydroxyindoleacetic acid and 5-hydroxy-6-methoxyindole, all of which had VIP >1. Urine indoleacetic acid was identified as a potential biomarker of benzene-induced toxicity in mice in the study by Sun et al., (2012) [59]. Evidence indicates that indoleacetic acid activates the aryl hydrocarbon receptor (AhR) and positively regulates cytochrome P-450 enzymes, implicated in tumorigenesis and cancer proliferation [86].
The propanoate metabolism and amino acid degradation of valine, leucine, and isoleucine were also listed among the most important pathways in the biological response to benzene exposure. These pathways are believed to be consequences of protein oxidation. Propanate is a short-chain fatty acid formed during the catabolism of branched-chain amino acids and methionine, and its concentration in plasma increases in amino acid oxidation states [87]. The reactive species produced during benzene bioactivation are able to form adducts with albumin, hemoglobin, and bone marrow proteins. Spatari et al., (2012) [88] found higher levels of advanced protein oxidation products in the serum of workers at an oil refinery located in Sicily, exposed to low concentrations of benzene (70.14 ± 69.71 μg/m3). Avery, (2011) [89] states that the susceptibility of proteins to oxidation depends on the relative content of oxidation-sensitive amino acid residues, the presence of metal-binding sites, location of proteins in the cell, molecular conformation, and rate of degradation. Methionine is one of the amino acid residues most prone to oxidation, and almost all organisms express methionine sulfoxide reductase enzymes to reverse this change. Oxidized cysteine and tryptophan residues are other useful markers of oxidation-modified proteins [89].
Based on the developed PLS-DA and OPLS-DA models, 38 metabolites in the urine responsible for discrimination between groups of workers exposed to benzene were identified. It is observed that they are metabolites belonging to the classes of peptides, carnitines, and predominantly lipids (Table 2).
Changes in lipid metabolism and increased oxidative stress resulting from exposure to benzene have been reported in recent studies. Rothman et al. (2021) [90] investigated the urinary metabolome of Chinese workers exposed to high concentrations of benzene (weighted average exposure of 8 h; 20 ppm), using liquid chromatography and high-resolution Fourier transform mass spectrometry (HRMS). The results showed that exposure to benzene leads to alterations in carnitine transport, fatty acid metabolism, sulfur amino acid metabolism, glycolysis, gluconeogenesis, and branched-chain amino acid metabolism. Mono- and polyunsaturated and short-chain fatty acids are also significantly associated with S-phenylmercapturic acid and leukocytes, and reported as key mediators of benzene-induced hematotoxicity in the study by Guo et al. (2022) [91].
The β-oxidation of fatty acids is a crucial pathway for hematopoietic stem cells (HSCs) and leukemic cells. Sun et al., (2014) [60] demonstrated that exposure to benzene induced disturbances in the levels of metabolites of the fatty acid β-oxidation pathway in the bone marrow of C3H/He mice. In another later study, Sun et al., (2016) [92] identified that exposure to benzene resulted in abnormal transport of fatty acids and β-oxidation of these lipids in bone marrow cells from C3H/He mice. It is worth noting that carnitine transport and fatty acid metabolism are involved in mitochondrial function.
L-carnitine (LC) is responsible for the transport of fatty acids across the mitochondrial membrane, and its levels were significantly reduced in animals treated with benzene. Mitochondrial dysfunction has been reported with a significant reduction in ATP levels and membrane potential. Furthermore, an increase in the generation of ROS and H2O2 was also observed, which led to oxidative stress and lipid peroxidation [92]. The disturbance in the β-oxidation of fatty acids decreased the production of NADPH, an important mitochondrial antioxidant, as well as the levels of ATP. These findings indicated that benzene induces mitochondrial dysfunction and oxidative stress, which may be one of the causative mechanisms of hematotoxicity [67,90]. In a recent study, Sun, Man, et al., (2020) [93] demonstrated that co-treatment with L-carnitine in K562 erythroleukemia cells treated with 1,4-benzoquinone, reduces the rate of apoptosis, DNA damage, reduces oxidative stress and promotes the β-oxidation of fatty acids.
Ten metabolites showed significant, low and non-linear correlations with biomarkers of oxidative stress and chromosomal aberrations (Table 3 and Table 4). These have been recognized as potential early-effect metabolomic biomarkers, induced by exposure to low concentrations of benzene. All of them are involved in metabolic pathways associated with the synthesis and metabolism of lipids and fatty acids. Based on these results, we speculate that oxidative damage is especially related to disturbances in lipid metabolism and transport. These results represent relevant information to guide future investigations into the mechanism underlying benzene-induced oxidative damage.
3.8. Limitations of the Study
Among the limitations of this study, the small sample size (n = 60) stands out, which may have been a source of random variation in the results of the two groups analyzed. The cross-sectional approach of the study also represents a limitation, considering that in this type of study, exposure and effect are evaluated concomitantly. Furthermore, the intensities of exposure to benzene in both groups of workers (environmental and occupational) were similar. Finally, the identification of metabolites was putatively, according to level 2, recognized by the Metabolomics Standards Initiative (MSI).
4. Conclusions
The study of metabolomic using the UHPLC-ESI-Q-TOF-MS technique revealed metabolic disturbances in workers exposed to low concentrations of benzene. Despite the history of low exposure to hydrocarbons for both groups, the intensity of exposure among workers at gas stations is greater during operations that involve direct contact with fuel.
The disturbances identified involved several metabolic pathways, with emphasis on the metabolism and transport of lipids and fatty acids. Ten urinary metabolites showed significant correlations, low and non-linear, with biomarkers of oxidative stress and chromosomal aberrations—toxic effects induced by benzene in the participants of this study, previously identified. Thus, the determination of the ten urinary metabolites can be used for a more comprehensive and accurate assessment of benzene-induced cytotoxicity and genotoxicity.
It is noteworthy that this study has advantages and limitations. Among the advantages is the unprecedented association established between chronic exposure to low doses of benzene and alterations in the urinary metabolome. On the other hand, the main limitation refers to the small size of the studied population. Furthermore, the unambiguous identification of urinary metabolites by comparing spectra with analytical standards is also necessary. Therefore, the ability of potential biomarkers to predict health outcomes needs to be confirmed through future and similar studies in a larger population.
Additionally, this study proves the power of metabolomics to provide relevant information to understand the biological response to exposure to low concentrations of xenobiotics and identification of early effect biomarkers.
Acknowledgments
The Mass Spectrometry Laboratory of the Food Research Center (FoRC) of the University of São Paulo (USP) for making the equipment and its physical structure available. Bruker Corporation is also acknowledged.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/metabo12100978/s1. Table S1: Descriptive analysis and comparison between groups for sociodemographic variables, Table S2: Time of exercise in the current occupation for workers in the environmental and occupational groups exposed to benzene, Table S3—Descriptive analysis and comparison between groups for biomarkers of exposure, effect, and biochemical and hematological parameters, Table S4— Correlation between candidate metabolites for exposure biomarkers and working time in the current occupation; Figure S1. Scatter diagrams between some metabolites and thiol and MDA, Figure S2. Scatter diagrams between some metabolites and chromosome breaks, fragment and metaphases presenting premature chromatid separation.
Author Contributions
Conceptualization: L.C.A. and A.L.L. Methodology: M.P.R.M., I.C.C.-A., L.V.B.C. and V.O.F. Data analyses: M.P.R.M., I.C.C.-A., L.V.B.C. and E.S.G. Writing—original draft preparation: M.P.R.M. and M.J.N.P. Writing—review and editing: L.C.A. and A.L.L. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted in accordance with the World Medical Association Code of Ethics (Declaration of Helsinki) for human testing. Informed consent was obtained from all individual participants included in the study. The study was submitted and approved by the ethics and research committees of the Fed-eral University of Minas Gerais—UFMG (CAAE: 57396116.1.0000.5149, opinion number: 1.691.348) and of FIOCRUZ-RJ (CAAE: 17438013.5.0000.5240, opinion number: 2.974.839).All procedures performed in this study that involved the participants were conducted in accordance with the ethical standards of institutional research committees and with the 1964 Declaration of Helsinki and its amendments.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The datasets generated and analyzed in this current study are not publicly available for reasons of confidentiality. However, they are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study received funding from Inova-ENSP (Innovation in Public Health Program 2013–2015 from ENSP/Fiocruz), Faperj Nº 32/2013—Support Program for Education and Research Institutions Headquartered in the State of Rio de Janeiro (Process E-26/111.732/2013) and PAPES VII (Program for Support to Strategic Health Research)/Fiocruz/CNPq (Process 401865/2015-0). Santos M.V.C. and I.C.C.-A. were supported by FAPERJ—Nota 10 fellowships (Faperj Nº E_01/2017—Scholarship Note 10—Masters—Process E-26/201.144/2017; and Faperj Nº E_04/2017 Scholarship Note 10—Pos-doctoral—Process E-26/202.437/2017). The manuscript received funding support for publication fee from Coordination of Superior Level Staff Improvement—Brazil (CAPES)—Finance Code 001.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wallace L. Environmental Exposure to Benzene: An Update. Environ. Health Perspect. 1996;104((Suppl. 6)):1129–1136. doi: 10.1289/ehp.961041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.IARC—International Agency for Research on Cancer . Benzene/IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Volume 120 IARC; Lyon, France: 2018. [Google Scholar]
- 3.Kuang S., Liang W. Clinical Analysis of 43 Cases of Chronic Benzene Poisoning. Chem. Biol. Interact. 2005;153:129–135. doi: 10.1016/j.cbi.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 4.Kim S., Vermeulen R., Waidyanatha S., Johnson B.A., Lan Q., Smith M.T., Zhang L., Li G., Shen M., Yin S., et al. Modeling Human Metabolism of Benzene Following Occupational and Environmental Exposures. Cancer Epidemiol. Biomarkers Prev. 2006;15:2246–2252. doi: 10.1158/1055-9965.EPI-06-0262. [DOI] [PubMed] [Google Scholar]
- 5.Rappaport S.M., Kim S., Thomas R., Johnson B.A., Bois F.Y., Kupper L.L. Low-Dose Metabolism of Benzene in Humans: Science and Obfuscation. Carcinogenesis. 2013;34:2–9. doi: 10.1093/carcin/bgs382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold S.M., Angerer J., Boogaard P.J., Hughes M.F., O’Lone R.B., Robison S.H., Robert Schnatter A. The Use of Biomoni toring Data in Exposure and Human Health Risk Assessment: Benzene Case Study. Crit. Rev. Toxicol. 2013;43:119–153. doi: 10.3109/10408444.2012.756455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carrieri M., Spatari G., Tranfo G., Sapienza D., Scape M., Battista G., Manno M. Biological Monitoring of Low Level Exposure to Benzene in an Oil Re Fi Nery: E Ff Ect of Modulating Factors. Toxicol. Lett. 2018;298:70–75. doi: 10.1016/j.toxlet.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 8.Lovreglio P., Palma G.D., Barbieri A., Andreoli R., Drago I., Greco L., Gallo E., Diomede L., Ricossa M.C., Fostinelli J., et al. Biological Monitoring of Exposure to Low Concentrations of Benzene in Workers at a Metallurgical Coke Production Plant: New Insights into S-Phenylmercapturic Acid and Urinary Benzene. Biomarkers. 2017;23:70–77. doi: 10.1080/1354750X.2017.1387935. [DOI] [PubMed] [Google Scholar]
- 9.Vineis P., Fecht D. Environment, Cancer and Inequalities d the Urgent Need for Prevention. Eur. J. Cancer. 2018;103:317–326. doi: 10.1016/j.ejca.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 10.Buesen R., Chorley B.N., Lima S., Daston G., Deferme L., Ebbels T., Gant T.W., Goetz A., Greally J., Hubesch B., et al. Applying ’ Omics Technologies in Chemicals Risk Assessment: Report of an ECETOC Workshop. Regul. Toxicol. Pharmacol. 2017;91:3–13. doi: 10.1016/j.yrtph.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinowska J.M., Viant M.R. ScienceDirect Toxicology Confidence in Metabolite Identification Dictates the Applicability of Metabolomics to Regulatory Toxicology. Curr. Opin. Toxicol. 2020;16:32–38. doi: 10.1016/j.cotox.2019.03.006. [DOI] [Google Scholar]
- 12.de Castro M.D.L., Capote F.P. The Analytical Process to Search for Metabolomics Biomarkers. J. Pharm. Biomed. Anal. 2017;147:341–349. doi: 10.1016/j.jpba.2017.06.073. [DOI] [PubMed] [Google Scholar]
- 13.Zhou J., Yin Y. Strategies for Large-Scale Targeted Metabolomics Quantification by Liquid Chromatography-Mass Spectrometry. Analyst. 2016;141:6362–6373. doi: 10.1039/C6AN01753C. [DOI] [PubMed] [Google Scholar]
- 14.Koelmel J.P., Napolitano M.P., Ulmer C.Z., Vasiliou V., Garrett T.J., Yost R.A., Prasad M.N.V., Godri Pollitt K.J., Bowden J.A. Environmental Lipidomics: Understanding the Response of Organisms and Ecosystems to a Changing World. Metabolomics. 2020;16:56. doi: 10.1007/s11306-020-01665-3. [DOI] [PubMed] [Google Scholar]
- 15.Niedzwiecki M.M., Walker D.I., Vermeulen R., Chadeau-hyam M., Jones D.P., Miller G.W. The Exposome: Molecules to Populations. Annu. Rev. Pharmacol. Toxicol. 2019;59:107–127. doi: 10.1146/annurev-pharmtox-010818-021315. [DOI] [PubMed] [Google Scholar]
- 16.Tzoulaki I., Ebbels T.M.D., Valdes A., Elliott P., Ioannidis J.P.A. Practice of Epidemiology Design and Analysis of Metabolomics Studies in Epidemiologic Research: A Primer on -Omic Technologies. Am. J. Epidemiol. 2014;180:129–139. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 17.Boyles M.S.P., Ranninger C., Reischl R., Rurik M., Tessadri R., Kohlbacher O., Duschl A., Huber C.G. Copper Oxide Nanoparticle Toxicity Profiling Using Untargeted Metabolomics. Part. Fibre Toxicol. 2016;13:49. doi: 10.1186/s12989-016-0160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen S., Zhang M., Bo L., Li S., Hu L., Zhao X. Metabolomic Analysis of the Toxic Effect of Chronic Exposure of Cadmium on Rat Urine. Environ. Sci. Pollut. Res. 2018;25:3765–3774. doi: 10.1007/s11356-017-0774-8. [DOI] [PubMed] [Google Scholar]
- 19.Fabian E., Bordag N., Herold M., Kamp H., Krennrich G., Looser R., Ma-hock L., Mellert W., Montoya G., Peter E., et al. Metabolite pro Fi Les of Rats in Repeated Dose Toxicological Studies after Oral and Inhalative Exposure. Toxicol. Lett. 2016;255:11–23. doi: 10.1016/j.toxlet.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 20.García-Sevillano M.A., García-Barrera T., Navarro F., Abril N., Pueyo C. Combination of Direct Infusion Mass Spectrometry and Gas Chromatography Mass Spectrometry for Toxicometabolomic Study of Red Blood Cells and Serum of Mice Mus Musculus after Mercury Exposure. J. Chromatogr. B. 2015;985:75–84. doi: 10.1016/j.jchromb.2015.01.029. [DOI] [PubMed] [Google Scholar]
- 21.Hou Y., Cao C., Bao W., Yang S., Shi H., Hao D. The Plasma Metabolic Profiling of Chronic Acephate Exposure in Rats via an Ultra-Performance Liquid Chromatography-Mass Spectrometry Based. Mol. Biosyst. 2015;11:506–515. doi: 10.1039/C4MB00523F. [DOI] [PubMed] [Google Scholar]
- 22.Wang F., Zhang H., Geng N., Ren X., Zhang B., Gong Y., Chen J. A Metabolomics Strategy to Assess the Combined Toxicity of Polycyclic Aromatic Hydrocarbons ( PAHs ) and Short-Chain Chlorinated Paraf Fi Ns. Environ. Pollut. 2018;234:572–580. doi: 10.1016/j.envpol.2017.11.073. [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Blair I.A., Mesaros C. Analytical Methods for Mass Spectrometry-Based Metabolomics Studies. Adv. Exp. Med. Biol. 2019;1140:635–647. doi: 10.1007/978-3-030-15950-4_38. [DOI] [PubMed] [Google Scholar]
- 24.Emwas A.-H., Roy R., McKay R.T., Tenori L., Saccenti E., Gowda G.A.N., Raftery D., Alahmari F., Jaremko L., Jaremko M., et al. NMR Spectroscopy for Metabolomics Research. Metabolites. 2019;9:123. doi: 10.3390/metabo9070123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Larive C.K., Barding G.A., Dinges M.M. NMR Spectroscopy for Metabolomics and Metabolic Profiling. Anal. Chem. 2015;87:133–146. doi: 10.1021/ac504075g. [DOI] [PubMed] [Google Scholar]
- 26.Ren J., Zhang A., Wang X. Advances in Mass Spectrometry-Based Metabolomics for Investigation of Metabolites. RSC Adv. 2018;8:22335–22350. doi: 10.1039/C8RA01574K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gika H., Virgiliou C., Theodoridis G., Plumb R.S., Wilson I.D. Untargeted LC/MS-Based Metabolic Phenotyping (Metabonomics/Metabolomics): The State of the Art. J. Chromatogr. B. 2019;1117:136–147. doi: 10.1016/j.jchromb.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 28.Gorrochategui E., Jaumot J., Lacorte S., Tauler R. Data Analysis Strategies for Targeted and Untargeted LC-MS Metabolomic Studies: Overview and Workflow. Trends Anal. Chem. 2016;82:425–442. doi: 10.1016/j.trac.2016.07.004. [DOI] [Google Scholar]
- 29.Costa-Amaral I.C., Carvalho L.V.B., Santos M.V.C., Teixeira L.R., Collins A.R., de Cássia O.C., Mattos O.C., Sarcinelli P.N. Environmental Assessment and Evaluation of Oxidative Stress and Genotoxicity Biomarkers Related to Chronic Occupational Exposure to Benzene. Int. J. Environ. Res. Public Health. 2019;16:2240. doi: 10.3390/ijerph16122240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahieu N.G., Genenbacher J.L., Patti G.J. A Roadmap for the XCMS Family of Software Solutions in Metabolomics. Curr. Opin. Chem. Biol. 2016;30:87–93. doi: 10.1016/j.cbpa.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tautenhahn R., Patti G.J., Rinehart D., Siuzdak G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang Z., Chong J., Zhou G., Anderson de Lima Morais D., Chang L., Barrette M., Gauthier C., Jacques P.-É., Li S., Xia J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A., Fan T.W., Fiehn O., Goodacre R., Griffin J.L., et al. Proposed Minimum Reporting Standards for Chemical Analysis. Metabolomics. 2007;3:211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venable W.B., Ripley B. Modern Applied Statistics with S. 4th ed. Springer; Berlin/Heidelberg, Germany: 2002. [Google Scholar]
- 35.Akaike H. Information Theory and an Extension of the Maximum Likelihood Principle. In: Akaike H., editor. Proceedings of the 2nd International Symposium on Information Theory. Akademiai Kiado; Budapest, Hungary: 1973. pp. 267–281. [Google Scholar]
- 36.Hoo Z.H., Candlish J., Teare D. What Is an ROC Curve? Emerg. Med. J. 2017;34:357–359. doi: 10.1136/emermed-2017-206735. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J., Shen H., Xu W., Xia Y., Barr D.B., Mu X., Wang X., Liu L., Huang Q., Tian M. Urinary Metabolomics Revealed Arsenic Internal Dose-Related Metabolic Alterations: A Proof-of-Concept Study in a Chinese Male Cohort. Environ. Sci. Technol. 2014;48:12265–12274. doi: 10.1021/es503659w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Moraes Moreau R.L., Bastos de Diqueira M.E.P. Toxicologia Analítica. 2nd ed. Guanabara Koogan; Rio de Janeiro, Brazil: 2016. [Google Scholar]
- 39.Barr D.B., Wilder L.C., Caudill S.P., Gonzalez A.J., Needham L.L., Pirkle J.L. Urinary Creatinine Concentrations in the U.S. Population: Implications for Urinary Biologic Monitoring Measurements. Environ. Health Perspect. 2005;113:192–200. doi: 10.1289/ehp.7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brazil Ministério do Trabalho . Norma Regulamentadora (NR) 15—Atividades e Operações Insalubres. Ministério do Trabalho; Rio do Janeiro, Brazil: 1995. Anexo No. 13-A. [Google Scholar]
- 41.Mendes M., Mesquita J., Machado H., Durand A., Costa-Amaral I.C., Valente D., Sydneia A., Arcuri A., Trevisan E.A., Sarcinelli P.D.N., et al. Normas Ocupacionais Do Benzeno: Uma Abordagem Sobre o Risco e Exposição Nos Postos de Revenda de Combustíveis. Rev. Bras. Saúde Ocup. 2017;6369:1–19. doi: 10.1590/2317-6369000127515. [DOI] [Google Scholar]
- 42.Mchale C.M., Zhang L., Smith M.T. Current Understanding of the Mechanism of Benzene-Induced Leukemia in Humans: Implications for Risk Assessment. Carcinogenesis. 2012;33:240–252. doi: 10.1093/carcin/bgr297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bolton J.L., Trush M.A., Penning T.M., Dryhurst G., Monks T.J. Role of Quinones in Toxicology. Chem. Res. Toxicol. 2000;13:135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 44.Lodovici M., Bigagli E. Oxidative Stress and Air Pollution Exposure. J. Toxicol. 2011;2011:487074. doi: 10.1155/2011/487074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moro A.M., Charão M.F., Brucker N., Durgante J., Baierle M., Bubols G., Goethel G., Fracasso R., Nascimento S., Bulcão R., et al. Genotoxicity and Oxidative Stress in Gasoline Station Attendants. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2013;754:63–70. doi: 10.1016/j.mrgentox.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 46.Uzma N., Kumar S.S., Hazari M.A.H. Exposure to Benzene Induces Oxidative Stress, Alters the Immune Response and Expression of P53 in Gasoline Filling Workers. Am. J. Ind. Med. 2010;53:1264–1270. doi: 10.1002/ajim.20901. [DOI] [PubMed] [Google Scholar]
- 47.Hussain T., Tan B., Yin Y., Blachier F., Tossou M.C.B., Rahu N. Oxidative Stress and Inflammation: What Polyphenols Can Do for Us? Oxid. Med. Cell. Longev. 2016;2016:7432797. doi: 10.1155/2016/7432797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reuter S., Gupta S.C., Chaturvedi M.M., Aggarwal B.B. Oxidative Stress, Inflammation, and Cancer: How Are They Linked? Free Radic. Biol. Med. 2010;49:1603–1616. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greten F.R., Grivennikov S.I. Inflammation and Cancer: Triggers, Mechanisms and Consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholson J.K., Lindon J.C., Holmes E. “Metabonomics”: Understanding the Metabolic Responses of Living Systems to Pathophysiological Stimuli via Multivariate Statistical Analysis of Biological NMR Spectroscopic Data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 51.Klassen A., Faccio A.T., Canuto G.A.B., Cruz P.L.R., Ribeiro H.C., Tavares M.F.M., Sussulini A. Metabolomics: Definitions and Significance in Systems Biology. Volume 965. Springer; Berlin/Heidelberg, Germany: 2017. Metabolomics: From Fundamentals to Clinical Applications; pp. 3–17. [DOI] [PubMed] [Google Scholar]
- 52.Canuto G.A.B., Costa J.L., Cruz P.L.R., Rosa S.A., Policy E.D., Fluid O., Seeds O.P. Metabolômica: Definições, Estado-da-Arte e Aplicações Representativas. Quim. Nova. 2018;41:75–91. doi: 10.21577/0100-4042.20170134. [DOI] [Google Scholar]
- 53.Santana F.B. Ph.D. Thesis. Universidade Federal de Uberlândia; Uberlândia, Brazil: 2015. Uso de Espectroscopia No Infravermelho Médio e Análise Discriminante Por Quadrados Mínimos Parciais Na Determinação de Adulterações Em Óleos de Andiroba, Prímula e Rosa Mosqueta. [Google Scholar]
- 54.Chong J., Wishart D.S., Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinforma. 2019;68:1–128. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- 55.Godzien J., Ciborowski M., Angulo S., Barbas C. From Numbers to a Biological Sense: How the Strategy Chosen for Metabolomics Data Treatment May Affect Final Results. A Practical Example Based on Urine Fingerprints Obtained by LC-MS. Electrophoresis. 2013;34:1–30. doi: 10.1002/elps.201300053. [DOI] [PubMed] [Google Scholar]
- 56.Smith M.T. Advances in Understanding Benzene Health Effects and Susceptibility. Annu. Rev. Public Health. 2010;31:133–148. doi: 10.1146/annurev.publhealth.012809.103646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wishart D.S. Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine. Nat. Rev. Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 58.Wild C.P. Future Research Perspectives on Environment and Health: The Requirement for a More Expansive Concept of Translational Cancer Research. Environ. Health. 2011;10((Suppl. 1)):1–4. doi: 10.1186/1476-069X-10-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun R., Zhang J., Xiong M., Chen Y., Yin L., Pu Y. Metabonomics Biomarkers for Subacute Toxicity Screening for Benzene Exposure in Mice. J. Toxicol. Environ. Health. 2012;75:1163–1173. doi: 10.1080/15287394.2012.699858. [DOI] [PubMed] [Google Scholar]
- 60.Sun R., Zhang J., Yin L., Pu Y. Investigation into Variation of Endogenous Metabolites in Bone Marrow Cells and Plasma in C3H / He Mice Exposed to Benzene. Int. J. Mol. Sci. 2014;15:4994–5010. doi: 10.3390/ijms15034994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu L., Sun R., Xu K., Pu Y., Huang J., Liu M., Chen M., Zhang J., Yin L. Lipidomic Analysis Reveals Disturbances in Glycerophospholipid and Sphingolipid Metabolic Pathways in Benzene-Exposed Mice. Toxicol. Res. 2021;10:706–718. doi: 10.1093/toxres/tfab053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vaz F.M., Ferdinandusse S. Bile Acid Analysis in Human Disorders of Bile Acid Biosynthesis. Mol. Aspects Med. 2017;56:10–24. doi: 10.1016/j.mam.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 63.Jia W., Xie G., Jia W. Bile Acid–Microbiota Cross-Talk in Gastrointestinal Inflammation and Carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018;15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun R., Xu K., Ji S., Pu Y., Man Z., Ji J., Chen M., Yin L., Zhang J., Pu Y. Benzene Exposure Induces Gut Microbiota Dysbiosis and Metabolic Disorder in Mice. Sci. Total Environ. 2020;705:135879. doi: 10.1016/j.scitotenv.2019.135879. [DOI] [PubMed] [Google Scholar]
- 65.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-Gut Microbiota Metabolic Interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 66.Burtis C.A., Bruns D.E., Sawyer B.G. Tietz Fundamentos de Química Clínica e Diagnóstico Molecular. 7th ed. Elsevier; Rio de Janeiro, Brazil: 2016. [Google Scholar]
- 67.Moro A.M., Brucker N., Charão M.F., Baierle M., Sauer E., Goethel G., Barth A., Nascimento S.N., Gauer B., Durgante J., et al. Biomonitoring of Gasoline Station Attendants Exposed to Benzene: Effect of Gender. Mutat. Res.—Genet. Toxicol. Environ. Mutagen. 2017;813:1–9. doi: 10.1016/j.mrgentox.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 68.Andrea M.A.D., Reddy G.K. Hematological and Hepatic Alterations in Nonsmoking Residents Exposed to Benzene Following a Flaring Incident at the British Petroleum Plant in Texas City. Environ. Health. 2014;13:1–8. doi: 10.1186/1476-069X-13-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wong M.W., Braidy N., Poljak A., Pickford R., Thambisetty M., Sachdev P.S. Dysregulation of Lipids in Alzheimer’s Disease and Their Role as Potential Biomarkers. Alzheimer’s Dement. 2017;13:810–827. doi: 10.1016/j.jalz.2017.01.008. [DOI] [PubMed] [Google Scholar]
- 70.Kurz J., Parnham M.J., Geisslinger G., Schiffmann S. Ceramides as Novel Disease Biomarkers. Trends Mol. Med. 2019;2019:20–32. doi: 10.1016/j.molmed.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 71.Ogretmen B. Sphingolipid Metabolism in Cancer Signalling and Therapy. Nat. Rev. Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mato J.M., Alonso C., Noureddin M., Lu S.C. Biomarkers and Subtypes of Deranged Lipid Metabolism in Nonalcoholic Fatty Liver Disease. World J. Gastroenterol. 2019;25:3009–3020. doi: 10.3748/wjg.v25.i24.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pabst T., Kortz L., Fiedler G.M., Ceglarek U., Idle J.R., Beyoğlu D. The Plasma Lipidome in Acute Myeloid Leukemia at Diagnosis in Relation to Clinical Disease Features. BBA Clin. 2017;7:105–114. doi: 10.1016/j.bbacli.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Subrahmanyam V.V., Ross D., Eastmond D.A., Smith M.T. Potential Role of Free Radicals in Benzene-Induced Myelotoxicity and Leukemia. Free Radic. Biol. Med. 1991;11:495–515. doi: 10.1016/0891-5849(91)90063-9. [DOI] [PubMed] [Google Scholar]
- 75.Pirozzi S.J., Schlosser M.J., Kalf G.F. Prevention of Benzene-Induced Myelotoxicity and Prostaglandin Synthesis in Bone Marrow of Mice by Inhibitors of Prostaglandin H Synthase. Immunopharmacology. 1989;18:39–55. doi: 10.1016/0162-3109(89)90029-5. [DOI] [PubMed] [Google Scholar]
- 76.Sulciner M.L., Gartung A., Gilligan M.M., Serhan C.N., Panigrahy D. Targeting Lipid Mediators in Cancer Biology. Cancer Metastasis Rev. 2018;37:557–572. doi: 10.1007/s10555-018-9754-9. [DOI] [PubMed] [Google Scholar]
- 77.Fabbri A., Magalotti D., Marchesini G., Brizi M., Bianchi G., Zoli M., Interna M., Orsola P.S. Effects of Systemic Prostaglandin E 1 on Splanchnic and Peripheral Haemodynamics in Control Subjects and in Patients with Cirrhosis. Prostaglandins Other Lipid Mediat. 1998;55:209–218. doi: 10.1016/S0090-6980(98)00020-3. [DOI] [PubMed] [Google Scholar]
- 78.Gao P., Hou L., Denslow N.D., Xiang P., Ma L.Q. Human Exposure to Polycyclic Aromatic Hydrocarbons: Metabolomics Perspective. Environ. Int. 2018;119:466–477. doi: 10.1016/j.envint.2018.07.017. [DOI] [PubMed] [Google Scholar]
- 79.Liu X.L., Fan D.M. Protective Effects of Prostaglandin E1 on Hepatocytes. World J. Gastroenterol. 2000;6:326–329. doi: 10.3748/wjg.v6.i3.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spiegel S., Merrill A.H. Sphingolipid Metabolism and Cell Growth Regulation. FASEB J. 1996;10:1388–1397. doi: 10.1096/fasebj.10.12.8903509. [DOI] [PubMed] [Google Scholar]
- 81.Robinson A.J., Davies S., Darley R.L., Tonks A. Reactive Oxygen Species Rewires Metabolic Activity in Acute Myeloid Leukemia. Front. Oncol. 2021;11:1–10. doi: 10.3389/fonc.2021.632623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Musharraf S.G., Siddiqui A.J., Shamsi T., Naz A. SERUM Metabolomics of Acute Lymphoblastic Leukaemia and Acute Myeloid Leukaemia for Probing Biomarker Molecules. Hematol. Oncol. 2016;35:769–777. doi: 10.1002/hon.2313. [DOI] [PubMed] [Google Scholar]
- 83.Moon S.M., Lee S.A., Hong J.H., Kim J.S., Kim D.K., Kim C.S. Oleamide Suppresses Inflammatory Responses in LPS-Induced RAW264.7 Murine Macrophages and Alleviates Paw Edema in a Carrageenan-Induced Inflammatory Rat Model. Int. Immunopharmacol. 2018;56:179–185. doi: 10.1016/j.intimp.2018.01.032. [DOI] [PubMed] [Google Scholar]
- 84.Naumoska K., Jug U., Metličar V., Vovk I. Oleamide, a Bioactive Compound, Unwittingly Introduced into the Human Body through Some Plastic Food/Beverages and Medicine Containers. Foods. 2020;9:549. doi: 10.3390/foods9050549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Morgan G.J., Smith M.T. Metabolic Enzyme Polymorphisms and Susceptibility to Acute Leukemia in Adults. Am. J. Pharmacogenomics. 2002;2:79–92. doi: 10.2165/00129785-200202020-00002. [DOI] [PubMed] [Google Scholar]
- 86.Bittinger M.A., Nguyen L.P., Bradfield C.A. Aspartate Aminotransferase Generates Proagonists of the Aryl Hydrocarbon Receptor. Mol. Pharmacol. 2003;64:550–556. doi: 10.1124/mol.64.3.550. [DOI] [PubMed] [Google Scholar]
- 87.Cummings J.R., Sakata T. Physiological and Clinical Aspects of Short-Chain Fatty Acids. Cambridge University Press; Cambridge, UK: 1995. [Google Scholar]
- 88.Spatari G., Saitta S., Cimino F., Sapienza D., Quattrocchi P., Carrieri M., Barbaro M., Saija A., Gangemi S. Increased Serum Levels of Advanced Oxidation Protein Products and Glycation End Products in Subjects Exposed to Low-Dose Benzene. Int. J. Hyg. Environ. Health. 2012;215:389–392. doi: 10.1016/j.ijheh.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 89.Avery S.V. Molecular Targets of Oxidative Stress. Biochem. J. 2011;434:201–210. doi: 10.1042/BJ20101695. [DOI] [PubMed] [Google Scholar]
- 90.Rothman N., Vermeulen R., Zhang L., Hu W., Yin S., Rappaport S.M., Smith M.T., Jones D.P., Rahman M., Lan Q., et al. Metabolome-Wide Association Study of Occupational Exposure to Benzene. Carcinogenesis. 2021;42:1326–1336. doi: 10.1093/carcin/bgab089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Guo X., Zhang L., Wang J., Zhang W., Ren J., Chen Y., Zhang Y., Gao A. Plasma Metabolomics Study Reveals the Critical Metabolic Signatures for Benzene-Induced Hematotoxicity. JCI Insight. 2022;7:1–14. doi: 10.1172/jci.insight.154999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun R., Cao M., Zhang J., Yang W., Wei H., Meng X., Yin L. Benzene Exposure Alters Expression of Enzymes Involved in Fatty Acid β -Oxidation in Male C3H/He Mice. Int. J. Environ. Res. Public Health. 2016;13:1068. doi: 10.3390/ijerph13111068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun R., Man Z., Ji J., Ji S., Xu K., Pu Y., Yu L., Zhang J., Yin L., Pu Y. l-Carnitine protects against 1,4-benzoquinone-induced apoptosisand DNA damage by suppressing oxidative stress and promoting fatty acid oxidation in K562 cells. Environ. Toxicol. 2020;35:1033–1042. doi: 10.1002/tox.22939. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed in this current study are not publicly available for reasons of confidentiality. However, they are available from the corresponding author upon reasonable request.


